Brassinosteroid signaling tightly controls the degradation of the transcription factor BRASSINAZOLE RESISTANT1 specifically in roots.
Abstract
Brassinosteroid (BR) regulates a wide range of physiological responses through the activation of BRASSINAZOLE RESISTANT1 (BZR1), whose activity is tightly controlled by its phosphorylation status and degradation. Although BZR1 appears to be degraded in distinct ways in response to different hormonal or environmental cues, little is known about how BR signaling regulates its degradation. Here we show that the BR-regulated U-box protein PUB40 mediates the proteasomal degradation of BZR1 in a root-specific manner in Arabidopsis (Arabidopsis thaliana). BZR1 levels were strongly reduced by plant U-box40 (PUB40) overexpression, whereas the pub39 pub40 pub41 mutant accumulated much more BZR1 than wild type in roots. The bzr1-1D gain-of-function mutation reduced the interaction with PUB40, which suppressed PUB40-mediated BZR1 degradation in roots. The cell layer-specific expression of PUB40 in roots helps induce selective BZR1 accumulation in the epidermal layer. Both BR treatment and loss-of-function of PUB40 expanded BZR1 accumulation to most cell layers. In addition, BZR1 accumulation increased the resistance of pub39 pub40 pub41 to low inorganic phosphate availability, as observed in bzr1-1D. BRASSINOSTEROID-INSENSITIVE2-induced phosphorylation of PUB40, which mainly occurs in roots, gives rise to BZR1 degradation through enhanced binding of PUB40 to BZR1 and PUB40’s stability. Our results suggest a molecular mechanism of root-specific BZR1 degradation regulated by BR signaling.
INTRODUCTION
The ubiquitin-proteasome system mediates the selective degradation of target proteins in eukaryotic cells (Smalle and Vierstra, 2004; Vierstra, 2009). In plants, ubiquitination has been implicated in a wide range of cellular processes, including the cell cycle, circadian rhythm control, hormone signaling, senescence, disease resistance, photomorphogenesis, and flowering (Vierstra, 2009; Sadanandom et al., 2012). Ubiquitination occurs via the sequential activation of three enzymes. A ubiquitin-activating enzyme (E1) transfers ubiquitin to a ubiquitin-conjugating enzyme (E2; Schulman and Harper, 2009). E2 then forms a complex with ubiquitin ligase (E3) to transfer ubiquitin from E2 to the target protein (Spratt et al., 2012). In addition to proteasomal degradation, ubiquitination regulates protein interactions, activation, and localization (Komander and Rape, 2012).
During the ubiquitination process, E3 ligases play crucial roles in determining the substrate specificity of a target protein (Smalle and Vierstra, 2004). Plant ubiquitin E3 ligases are classified into several types based on the mechanisms for target recognition and ubiquitin tagging and the presence of specific domains such as HECT, RING, or U-box domains (Mazzucotelli et al., 2006; Vierstra, 2009). Of these, S PHASE KINASE-ASSOCIATED PROTEIN1-Cullin-F-box (SCF) complexes containing a RING ligase have been extensively studied as key regulators of plant hormone signaling (Kelley and Estelle, 2012). Interestingly, repressor proteins involved in the signaling of plant hormones such as auxin (Gray et al., 2001), gibberellin (Dill et al., 2004), jasmonic acid (Thines et al., 2007), strigolactone (Wang et al., 2013), and brassinosteroids (BRs; Zhu et al., 2017) appear to be degraded by SCF complexes (Kelley and Estelle, 2012).
Ubiquitin E3 ligases of the U-box type, with a modified RING domain, were discovered most recently (Trujillo, 2018). Whereas 2- and 21-U–box genes were identified in yeast (Saccharomyces cerevisiae) and the human genomes, respectively, plant genomes contain many more U-box genes (64 in Arabidopsis [Arabidopsis thaliana] and 77 in rice [Oryza sativa]; Yee and Goring, 2009). This finding suggests that plant-U–box (PUB) proteins might regulate cellular processes that are specific to plant growth and development. Indeed, recent studies have begun to demonstrate that PUB proteins are involved in plant-specific responses including biotic/abiotic stress responses, self-incompatibility, and hormone pathways (Yee and Goring, 2009). However, the functional roles of most Arabidopsis PUBs remain unclear.
BRs are steroidal compounds that regulate many aspects of plant growth and development (Yang et al., 2011). The physiological importance of BR as an essential plant hormone has been established by biochemical and genetic studies of BR-related mutants (Zhu et al., 2013). BR regulates cell growth, photomorphogenesis, vascular differentiation, reproductive organ development, stomatal cell development, and stress responses (Yang et al., 2011). A variety of BR functions are mediated by BR signaling and its crosstalk with other physiological and developmental pathways (Wang et al., 2013; Zhu et al., 2013).
Unlike animal steroid hormones, as a chemical signal, BR is perceived by the cell-surface receptor kinases BR INSENSITIVE1 (BRI1) and BRI1-ASSOCIATED KINASE1 (Sun et al., 2013). The binding of BR to the BRI1/BRI1-ASSOCIATED KINASE1 receptor complex ultimately activates the BR-responsive transcription factors BRASSINAZOLE RESISTANT1 (BZR1) and bri1 EMS SUPPRESSOR1 (BES1)/BZR2 through a sequential signal relay mediated by phosphorylation and dephosphorylation (Kim and Wang, 2010; Wang et al., 2012). In the absence of BR, the glycogen synthase kinase-3–like (GSK3-like) kinase BRASSINOSTEROID-INSENSITIVE2 (BIN2) constitutively inactivates BZR1 and BES1 through phosphorylation (He et al., 2002). In the presence of BR, BR Signaling Kinase1 and Constitutive Differential Growth1 phosphorylated by BRI1 activate the phosphatase BRI1 Suppressor1, which inhibits BIN2 (Tang et al., 2008; Kim et al., 2011). Meanwhile, Protein Phosphatase 2A (PP2A) dephosphorylates BZR1 and BES1, allowing for their accumulation in the nucleus and transcriptional regulation (Tang et al., 2011).
In addition to phosphorylation and dephosphorylation, protein degradation also plays a pivotal role in regulating BIN2 and BZR1/BES1. The F-box protein Kink Suppressed in bzr1-1D (KIB1) mediates BR-induced ubiquitination and proteasomal degradation of BIN2 (Zhu et al., 2017). In addition to BIN2 degradation, the binding of KIB1 to BIN2 blocks its binding to substrates. Thus, the ubiquitin E3 ligase, KIB1, acts as a positive regulator of BR signaling.
Three different types of proteins involved in the proteasomal degradation of BZR1/BES1 have been identified. The F-box protein MORE AXILLARY GROWTH LOCUS2 (MAX2), a subunit of the SCF ubiquitin E3 ligase complex that regulates strigolactone signaling, appears to mediate BES1 degradation (Wang et al., 2013). MAX2-mediated BES1 degradation increases in response to strigolactone treatment, and the bes1-D gain-of-function mutant (with increased branching) is less sensitive to strigolactone than the wild type. Two other types of E3 ligases, CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1) and Seven-IN-Absentia of Arabidopsis thaliana (SINATs), also modulate BZR1/BES1 stability (Kim et al., 2014; Yang et al., 2017). Early studies suggested that phosphorylated BZR1 and BES1 are degraded by the 26S proteasome (He et al., 2002). However, recent studies have shown that COP1 degrades phosphorylated BZR1/BES1 in the dark, whereas the RING finger E3 ligases, SINATs, degrade dephosphorylated BZR1/BES1 in the light (Kim et al., 2014; Yang et al., 2017).
In contrast to the proteasomal degradation of BIN2, the degradation of BZR1/BES1 is mediated by autophagy as well as the proteasomal pathway (Zhang et al., 2016; Nolan et al., 2017). Sugar signaling appears to enhance BZR1 accumulation via the Target of Rapamycin pathway (Zhang et al., 2016). Under starvation conditions, inactivated Target of Rapamycin causes autophagy-mediated BZR1 degradation to inhibit plant growth. A selective autophagic pathway of BES1 has also been reported (Nolan et al., 2017). Under stress conditions, DOMINANT SUPPRESSOR OF KAR2, a ubiquitin receptor protein, interacts with BES1 and SINATs, resulting in autophagy-mediated BES1 degradation through interaction with AUTOPHAGY8. Therefore, BZR1/BES1 are degraded in multiple ways under different hormonal and environmental conditions.
In this study, we identified another ubiquitin E3 ligase that degrades BZR1 in a distinct way. PUB40 interacts with BZR1 in vitro and in vivo. The gain-of-function bzr1-1D mutation greatly decreases the interaction of this protein with PUB40. In particular, PUB40 mediates BZR1 degradation in a root-specific manner. Endogenous BZR1 levels were greatly reduced by PUB40 overexpression and increased by the pub40 loss-of-function mutation. We also demonstrated a physiological role for PUB40-mediated BZR1 degradation in roots. Like bzr1-1D, a triple mutant for PUB40 and its homologs displayed root insensitivity to inorganic phosphate (Pi) deprivation. Furthermore, we found that BIN2 interacts with and phosphorylates PUB40. BIN2-induced phosphorylation of PUB40 in roots increases the interaction with BZR1 as well as the stability of PUB40. Our results provide evidence that BZR1 degradation mediated by the U-box ubiquitin E3 ligase PUB40 determines tissue-specific BZR1 accumulation in roots.
RESULTS
PUB40 Interacts with BZR1 In Vitro and In Vivo
BR signaling is often compared with the WINGLESS/INTEGRATED (Wnt) pathway of mammals because the ligand is perceived by cell surface receptors and GSK3 acts as a negative regulator. Downstream of Wnt signaling, β-catenin, the best-known mammalian Armadillo (ARM) protein, plays an essential role in regulating Wnt-responsive transcription factors (MacDonald et al., 2009; Clevers and Nusse, 2012). ARM is a sequence motif composed of ∼42 amino acids that creates diverse three-dimensional structures for protein–protein interactions (Peifer et al., 1994; Tewari et al., 2010). However, the functional role of ARM-repeat proteins in BR signaling is yet to be determined. Thus, based on the similarity of these two pathways, we hypothesized that the BR-responsive transcription factor BZR1 is also regulated by an ARM-repeat protein. Given that most proteins with ARM-repeat domains in Arabidopsis are ubiquitin E3 ligases of the U-box type (Mudgil et al., 2004), we tried to identify a PUB protein that interacts with BZR1.
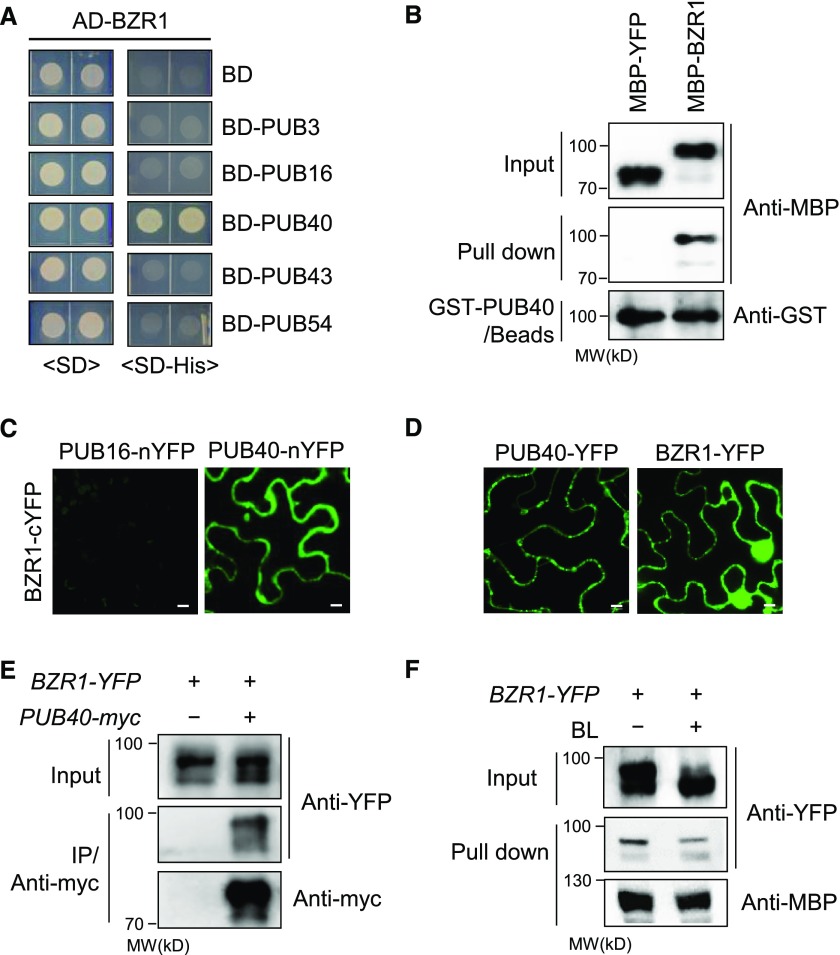
Of the six clusters of PUB groups presented by Wiborg et al. (2008), we selected five PUB genes (PUB3, PUB16, PUB40, PUB43, and PUB54) from each group, except for one group where relatively many members have been identified. In a yeast two-hybrid assay to test the interaction with BZR1, PUB40 interacted with BZR1 (Figure 1A). PUB40 is a ubiquitin E3 ligase composed of an N-terminal U-box domain and five C-terminal ARM domains (Supplemental Figure 1A). We investigated the interaction of PUB40 with BZR1. In an in vitro pull-down assay, Maltose Binding Protein (MBP)-BZR1 but not MBP-YFP (yellow fluorescent protein) was pulled down by glutathione S-transferase (GST)-PUB40–bound beads (Figure 1B). In a bimolecular fluorescence complementation (BiFC) assay, strong fluorescent signals were detected in the cytoplasm of wild tobacco (Nicotiana benthamiana) epidermal cells coexpressing PUB40 fused to the N-terminal half of YFP (nYFP) and BZR1 fused to the C-terminal half of YFP (cYFP; Figure 1C; Supplemental Figures 1B and 1C). When we compared the subcellular localization of BZR1 and PUB40 in plant cells, BZR1-YFP localized to both the nucleus and cytoplasm, whereas PUB40-YFP was detected only in the cytoplasm (Figure 1D).
Figure 1.
PUB40 Interacts with BZR1 In Vivo and In Vitro.
(A) Yeast two-hybrid assay to examine the interaction between activation domain (AD)-fused BZR1 and DNA binding domain (BD)-fused PUBs. Yeast cells expressing the indicated constructs were grown on synthetic dropout (SD) or SD-His medium.
(B) In vitro pull-down assay between BZR1 and PUB40. MBP-YFP or MBP-BZR1 was pulled down with GST-PUB40 immobilized on glutathione agarose beads. The immunoblot was probed with anti-MBP and anti-GST antibodies.
(C) BiFC assay using PUB40 fused to the N-terminal half of YFP and BZR1 fused to the C-terminal half of YFP. The indicated constructs were cotransformed into wild tobacco epidermal cells. PUB16-nYFP was used as a negative control. Scale bars = 10 μm.
(D) Subcellular localization of PUB40-YFP and BZR1-YFP in plant cells. C-terminal YFP-fused PUB40 or BZR1 was transiently expressed in wild tobacco epidermal cells and observed by confocal microscopy. Scale bars = 10 μm.
(E) Coimmunoprecipitation between PUB40 with BZR1 in vivo. Protein extracts of transgenic Arabidopsis seedlings coexpressing PUB40-myc and BZR1-YFP were immunoprecipitated with anti-myc antibody. Immunoblots were probed with anti-myc and anti-YFP antibodies.
(F) PUB40 binding affinity based on BZR1-induced phosphorylation status. MBP-PUB40 protein immobilized on amylose beads was incubated with protein extracts of untreated BZR1pro-BZR1-YFP seedlings or seedlings treated with 100 nM of BL for 1 h. The immunoblot was probed with anti-YFP and anti-MBP antibodies.
Given that phosphorylated BZR1 is retained in the cytoplasm by the interaction with 14-3-3 protein and degraded by the 26S proteasome, the cytoplasmic localization of PUB40 might be correlated with the degradation of phosphorylated BZR1 (Gampala et al., 2007). Thus, we investigated whether the binding of PUB40 to BZR1 is altered by BZR1’s phosphorylation status. In a coimmunoprecipitation assay, phosphorylated forms of BZR1-YFP strongly coimmunoprecipitated with PUB40-myc (Figure 1E). Next, we incubated immobilized MBP-PUB40 with protein extracts from BZR1-YFP plants subjected to mock treatment or treatment with brassinolide (BL, most active BR). Phosphorylated BZR1-YFP was mainly pulled down by MBP-PUB40 (Figure 1F). This finding indicates that the cytoplasmic accumulation of phosphorylated BZR1 causes PUB40 binding.
PUB40 Mediates BZR1 Degradation in a Root-Specific Manner
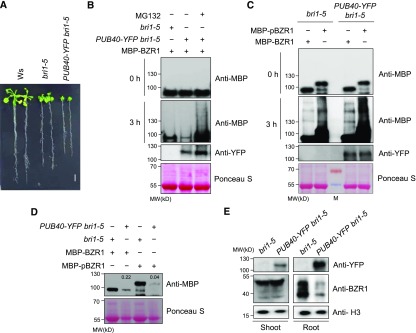
To investigate whether PUB40 regulates BR signaling, we analyzed the phenotype of the bri1-5 mutant overexpressing PUB40-YFP. PUB40-YFP bri1-5 showed a more severe dwarf phenotype than bri1-5 (Figure 2A; Supplemental Figures 2A, 2B, and 2C). In addition, in the dark, PUB40-YFP bri1-5 displayed shorter hypocotyls and roots than bri1-5 (Supplemental Figure 2D). These results suggest that PUB40 might act as a negative regulator of BR signaling.
Figure 2.
PUB40 Is Involved in BZR1 Degradation in Arabidopsis.
(A) Phenotypes of the Ws, bri1-5, and bri1-5 overexpressing PUB40-YFP. PUB40-YFP was expressed under the control of the 35S promoter. The plants were grown on MS medium for 14 d. Scale bar = 1 cm.
(B) Cell-free protein degradation assay of BZR1 in PUB40-YFP overexpression seedlings. MBP-BZR1 was incubated with protein extracts from bri1-5 or PUB40-YFP bri1-5. PUB40-YFP bri1-5 was treated with mock or 50 μM of MG132 for 3 h. Protein levels of MBP-BZR1 and PUB40-YFP were detected with anti-MBP and anti-YFP antibodies, respectively. Ponceau S staining was used as a loading control.
(C) Semi-in vivo assay to compare the modification of BZR1 and phosphorylated BZR1 by protein extracts from PUB40-YFP. MBP-BZR1 or MBP-pBZR1 was incubated with protein extracts obtained from bri1-5 or PUB40-YFP bri1-5 treated with 50 μM of MG132 for 3 h. Protein levels of MBP-BZR1 and PUB40-YFP were detected with anti-MBP and anti-YFP antibodies, respectively. Ponceau S staining was used as a loading control.
(D) BZR1 degradation upon phosphorylation by BIN2. MBP-BZR1 or MBP-BZR1 phosphorylated by BIN2 (MBP-pBZR1) was incubated with protein extracts of bri1-5 or PUB40-YFP bri1-5 for 2 h. Numbers in lanes 2 and 4 indicate relative signal intensities normalized to the controls (lanes 1 and 3). Ponceau S staining was used as a loading control.
(E) Endogenous BZR1 levels in the shoots and roots of 14-d-old bri1-5 plants overexpressing PUB40-YFP. Protein levels of BZR1 and PUB40-YFP were detected with anti-BZR1 and anti-YFP antibodies. Histone H3 (H3) detected with anti-Histone H3 antibody was used as a loading control.
To examine PUB40-mediated BZR1 degradation, we performed a cell-free degradation assay. When equal amounts of MBP-BZR1 were incubated with protein extracts from bri1-5 or PUB40-YFP bri1-5, MBP-BZR1 degradation was greatly increased by the overexpression of PUB40-YFP. In addition, MBP-BZR1 degradation in PUB40-YFP bri1-5 was inhibited by MG132 treatment (Figure 2B), suggesting that PUB40 causes 26S proteasomal degradation of BZR1. When BZR1 or phosphorylated BZR1 was incubated with protein extracts from PUB40-YFP, slowly migrating bands, most likely ubiquitinated proteins (Figure 2C), accumulated to higher levels with phosphorylated BZR1 versus BZR1. Consistently, PUB40 more effectively degraded phosphorylated versus nonphosphorylated BZR1 (Figure 2D). These results are consistent with the observation that PUB40 primarily bound to phosphorylated BZR1 (Figures 1E and 1F).
Based on the results described above, we investigated the endogenous BZR1 levels in PUB40-YFP bri1-5 and bri1-5. Interestingly, although no obviously difference in BZR1 levels was observed in the shoots of PUB40-YFP bri1-5, BZR1 levels were lower in PUB40-YFP bri1-5 roots than in bri1-5 roots (Figure 2E).
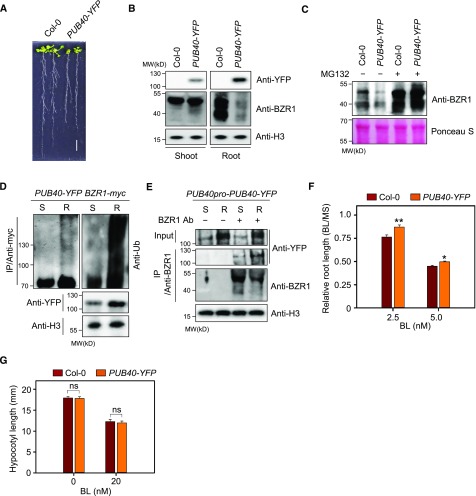
We then analyzed the phenotype of transgenic Arabidopsis overexpressing PUB40-YFP in a wild-type background. PUB40-YFP showed stunted growth (Figure 3A; Supplemental Figure 3A). Consistent with the results shown in Figure 2E, endogenous BZR1 levels in roots but not in shoots, were greatly reduced by PUB40-YFP overexpression (Figure 3B). Another independent transgenic plant overexpressing PUB40-YFP showed a similar phenotype and reduced BZR1 levels (Supplemental Figures 3, B and C). Moreover, the reduction in BZR1 levels in PUB40-YFP roots was reversed by MG132 treatment (Figure 3C). When we examined BZR1 mRNA in wild type and PUB40-YFP, BZR1 was highly expressed in the roots of PUB40-YFP compared with the wild type (Supplemental Figure 3D), indicating that the reduced BZR1 levels in PUB40-YFP were due to proteasomal degradation of BZR1. In addition, we performed immunoblot analysis to investigate the ubiquitination of BZR1 induced by PUB40. BZR1-myc immunoprecipitated from transgenic plants coexpressing PUB40-YFP and BZR1-myc was detected with anti-myc and anti-Ub antibodies. Consistent with root-specific BZR1 degradation by PUB40-YFP (Figure 3B), poly-ubiquitinated BZR1-myc was detected in the roots but not the shoots of a transgenic plant overexpressing PUB40-YFP (Figure 3D). Indeed, the interaction of PUB40 with BZR1 also appeared to be stronger in roots versus shoots under native conditions. When PUB40-YFP was expressed under the control of its own promoter, the amount of PUB40-YFP that coimmunoprecipitated with anti-BZR1 antibody was higher in the roots than the shoots (Figure 3E).
Figure 3.
Root-Specific Degradation of BZR1 in PUB40-Overexpressing Plants.
(A) Phenotypes of two wild-type Col-0 and PUB40-overexpressing plants (PUB40-YFP). PUB40-YFP was expressed under the control of the 35S promoter. Plants were grown on MS medium for 14 d. Scale bar = 1 cm.
(B) Endogenous BZR1 levels in the shoots and roots of the plants shown in (A). BZR1 was detected with anti-BZR1 antibody, and H3 was used as a loading control.
(C) MG132-induced BZR1 accumulation in the roots of 35S-PUB40-YFP. 14-d-old seedlings were treated with 10 μM of MG132 for 4 h. Total protein extracts were analyzed by immunoblotting using anti-BZR1 antibody. Ponceau S staining was used as a loading control.
(D) Comparison of PUB40-mediated BZR1 ubiquitination in shoots and roots. Transgenic plants coexpressing PUB40-YFP and BZR1-myc were treated with 50 μM of MG132 for 12 h. Protein extracts from shoots (S) or roots (R) were immunoprecipitated with anti-myc antibody for 1 h in the presence of 50 μM of MG132. Immunoblots were probed with anti-myc, anti-Ub, and anti-YFP antibodies.
(E) Coimmunoprecipitation of PUB40-YFP with BZR1 in roots and shoots. Protein A beads with or without anti-BZR1 antibody were incubated with protein extracts from transgenic plants expressing PUB40-YFP driven by the PUB40 promoter. Immunoblots were probed with anti-myc, anti-YFP, and anti-H3 antibodies. S, shoots; R, roots.
(F) BL sensitivity of PUB40-overexpressing plants in a root growth inhibition assay. Col-0 and PUB40-YFP were grown for 7 d in the light on MS medium with or without BL. Error bars = ±se (n > 40 per genotype). Asterisks indicate statistically significant differences from wild-type Col-0 (Student’s t test, *P < 0.05, **P < 0.001). The data shown are representative of three independent experiments.
(G) BL sensitivity of PUB40-overexpressing plants in a hypocotyl growth inhibition assay. Col-0 and PUB40-YFP were grown for 5 d in the dark on MS medium with or without 20 nM of BL. (Student’s t test, ns, not significant.) Error bars = ±se (n > 30 per genotype). The data shown are representative of three independent experiments.
When we examined the sensitivity of PUB40-YFP to BL in terms of root and hypocotyl growth, only the roots of PUB40-YFP were less sensitive to BL than the wild type (Figure 3F; Supplemental Figure 3E). Similarly, there was no difference in BL sensitivity in terms of etiolated hypocotyl growth of PUB40-YFP compared with the wild type (Figure 3G). Together, these results indicate that PUB40 mediates the ubiquitination of BZR1 in a root-specific manner.
A Loss-of-Function Mutant of PUB40 and Its Homologs Accumulates a Large Quantities of BZR1 in Roots
We obtained two independent transfer DNA (T-DNA) insertion mutants for PUB40 from the Arabidopsis Biological Resource Center (https://abrc.osu.edu/) and isolated homozygous mutants. However, both the pub40-1 and pub40-2 mutants showed only subtle phenotypic differences compared with the wild type (Supplemental Figures 4A and 4B), although more BZR1 accumulated in the roots of these mutants (Supplemental Figure 4C). Thus, we investigated the possibility that PUB40 might share functional redundancy with its homologs. In the Arabidopsis PUB family, a subclade including PUB40 is composed of four members: PUB38, PUB39, PUB40, and PUB41 ( Supplemental Figure 5A; Supplemental File). In an in vitro pull-down assay, three homologs of PUB40 also interacted with BZR1 (Supplemental Figure 5B), implying that PUB40’s close homologs are also involved in BZR1 degradation. Because a T-DNA insertion mutant for PUB38 was not available, we generated the triple knockout mutant for PUB39, PUB40, and PUB41 (Supplemental Figures 5C and 5D).
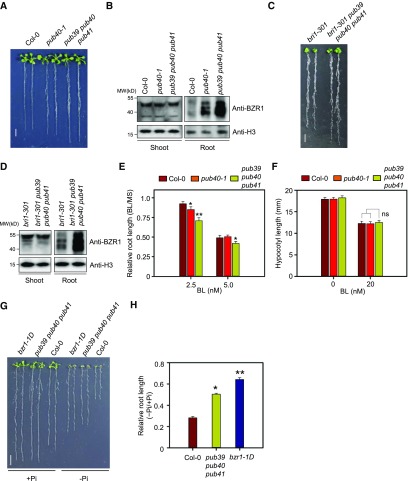
The pub39 pub40 pub41 mutant only displayed enhanced root growth in terms of length and fresh weight (Figure 4A; Supplemental Figure 5E), although the root-specific BZR1 accumulation observed in the pub40 mutant was even greater in the triple mutant (Figure 4B). When the 35S-PUB40-YFP construct was introduced into the pub39 pub40 pub41 mutant, the enhanced root growth and BZR1 accumulation were suppressed by PUB40 overexpression (Supplemental Figure 6). Given that bzr1-1D suppressed the growth defect of the roots as well as the shoots of the bri1-116 null mutant (Supplemental Figure 7), it is clear that BZR1 activity is also essential for root growth and development. Similarly, the short root phenotype and BZR1 levels of bri1-301 (Xu et al., 2008) were reversed when the bri1-301 mutant was crossed to pub39 pub40 pub41 (Figures 4C and 4D). Thus, the root phenotype of pub39 pub40 pub41 corresponds to the physiological role of BZR1 in roots.
Figure 4.
BZR1 Accumulation in the Roots of pub40 Loss-of-Function Mutants.
(A) Phenotypes of pub40 loss-of-function mutants. Wild-type Col-0, pub40, and pub39 pub40 pub 41 were grown on MS medium for 14 d. Scale bar = 1 cm.
(B) BZR1 accumulation in the roots of the pub40 and pub39 pub40 pub41 mutants shown (A). BZR1 was detected with anti-BZR1 antibody. H3 was used as a loading control.
(C) Phenotypes of the bri1-301 pub39 pub40 pub41 mutant. Wild-type Col-0, bri1-301, and bri1-301 pub39 pub40 pub41 were grown on MS medium for 14 d. Scale bar indicates 1 cm.
(D) BZR1 accumulation in the roots of the bri1-301 pub39 pub40 pub41 mutant shown (C). BZR1 was detected with anti-BZR1 antibody. H3 was used as a loading control.
(E) BL sensitivity of Col-0, pub40-1, and pub39 pub40 pub41 in a root growth inhibition assay. Plants were grown on MS medium with or without BL for 5 d. Error bars = ±se (n > 30 per genotype). Asterisks indicate statistically significant differences by Student’s t test (*P < 0.05, **P < 0.001). The data shown are representative of three independent experiments.
(F) BL sensitivity of Col-0, pub40-1, and pub39 pub40 pub41 in a hypocotyl growth inhibition assay. Plants were grown on MS medium with or without 20 nM BL for 5 d in the dark. Error bars = ±se (n > 30 per genotype). The data shown are representative of three independent experiments.
(G) Root growth of Col-0, pub39 pub40 pub41, and bzr1-1D in MS medium containing low Pi levels. Seedlings grown in normal MS medium for 3 d were transferred onto MS medium containing adequate Pi (1.25 mM, +Pi) or low Pi (1 μM, −Pi) and grown for another 7 d. Scale bar = 1 cm.
(H) Measurement of root length of the plants shown in (G). Error bars = ±se (n > 40 per genotype). Asterisks indicate statistically significant differences by Student’s t test (*P < 0.05, **P < 0.001; ns, not significant). The data shown are representative of three independent experiments.
In a root growth inhibition assay, both pub40-1 and pub39 pub40 pub41 were hypersensitive to 2.5 nM BL, whereas only pub39 pub40 pub41 showed a significantly increased response to 5 nM of BL treatment (Figure 4E). However, neither pub40-1 nor pub39 pub40 pub41 showed increased sensitivity to BL in terms of hypocotyl growth (Figure 4F; Supplemental Figure 8A).
To further clarify the physiological role of PUB40-mediated BZR1 degradation in roots, we noted a previous report stating that increased BZR1 activity confers resistance to Pi starvation (Singh et al., 2014). Low Pi availability causes severe inhibition of root growth in wild-type plants, but not in the gain-of-function mutant of BZR1, bzr1-1D (Wang et al., 2002). Thus, we investigated the sensitivity of pub39 pub40 pub41 roots to low Pi stress. Indeed, the growth of pub39 pub40 pub41 roots was less sensitive to Pi starvation compared with the wild type, like that of bzr1-1D (Figures 4G and 4H; Supplemental Figure 8B). These results indicate that PUB40-mediated degradation of BZR1 helps modulate root growth according to changes in environmental conditions.
Differentially Expressed Genes in pub39 pub40 pub41 Significantly Overlap with BZR1-Regulated Genes
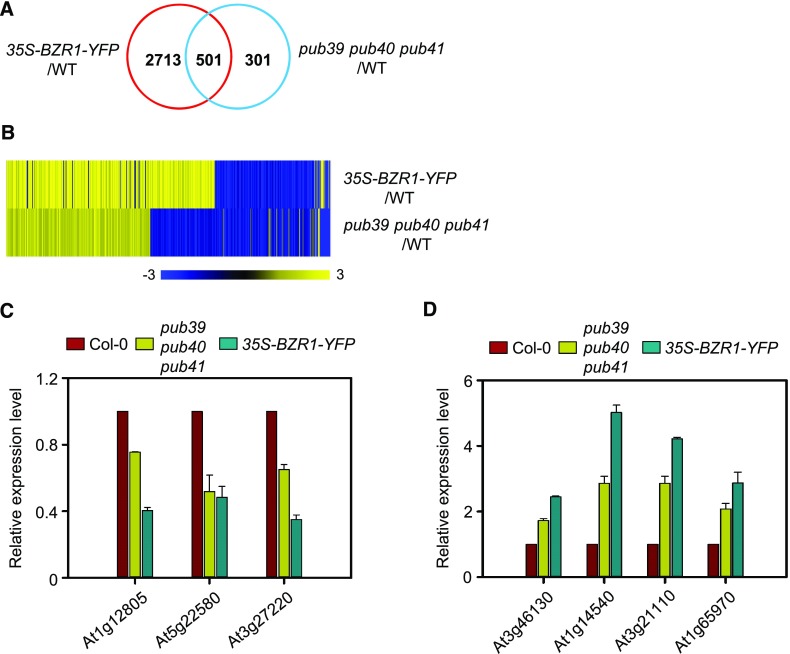
To compare the genes that are differentially expressed in pub39 pub40 pub41 and 35S-BZR1-YFP versus the wild type, we isolated total RNA from whole roots of 14-d-old seedlings and subjected them to RNA sequencing (RNA-Seq) analysis. We identified 3,214 and 812 differentially expressed genes (fold change > 1.3) in 35S-BZR1-YFP and pub39 pub40 pub41 compared with the wild type, respectively (Figure 5A; Supplemental Data Set). In addition, 501 genes of the 812 differentially expressed genes identified from pub39 pub40 pub41 were also regulated by BZR1-YFP overexpression (Figure 5B). Heatmap analysis of these genes showed that 367 genes (73.3%) were regulated in the same manner as those of 35S-BZR1-YFP (Figure 5B). Of the 258 genes upregulated by PUB and the 243 downregulated by PUB, 240 (93%) and 127 (52.3%) genes were up- and downregulated by BZR1-YFP overexpression, respectively (Supplemental Data Set). Reverse transcription-quantitative PCR (RT-qPCR) analysis further confirmed that the expression patterns of genes down- and upregulated by BR were similar in pub39 pub40 pub41 and 35S-BZR1-YFP (Figures 5C and 5D; Supplemental Figures 9A and 9B). In addition, the expression of two well-known BR marker genes, CONSTITUTIVE PHOTOMORPHOGENESIS AND DWARFISM and EXPANSIN8, in the roots but not the shoots was more sensitive to BL in pub39 pub40 pub41 compared with the wild type (Supplemental Figures 9C and 9D). These results support the notion that gene expression affected by the mutation of three PUBs corresponds to transcriptional regulation by BZR1.
Figure 5.
Comparison of Gene Expression Profiles of 35S-BZR1-YFP and pub39 pub40 pub41 Plants.
(A) Venn diagram of differentially expressed genes in 35S-BZR1-YFP and the pub39 pub40 pub41 compared with the wild type. Total RNA was extracted from whole roots of seedlings grown on MS medium for 14 d. The data were obtained from RNA-Seq analysis.
(B) Heat map of coregulated genes by BZR1 and PUBs. Scale bar = fold changes.
(C) RT-qPCR analysis of BZR1-downregulated gene expression in Col-0, pub39 pub40 pub41, and 35S-BZR1-YFP. Error bars = ±se. The data shown are representative of two independent experiments. Data represent the means from two technical replicates.
(D) RT-qPCR analysis of BZR1-upregulated gene expression in Col-0, pub39 pub40 pub41, and 35S-BZR1-YFP. Error bars = ±se. The data shown are representative of two independent experiments. Data represent the means from two technical replicates.
The bzr1-1D Mutation Reduces PUB40-Mediated BZR1 Degradation
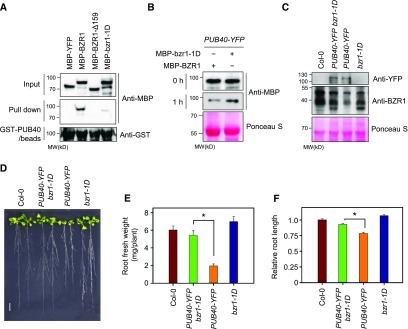
The bzr1-1D gain-of-function mutation enhances the interaction of BZR1 with PP2A and the subsequent dephosphorylation of BZR1, leading to the accumulation of active BZR1. The bzr1-1D mutation (P234L) is located within BZR1’s PEST (Pro, Glu, Ser, and Thr-rich) domain, which is thought to mediate protein degradation (Rechsteiner and Rogers, 1996). Thus, we investigated whether the binding of PUB40 to BZR1 is also affected by the gain-of-function bzr1-1D mutation. As shown in Figure 6A, an in vitro pull-down assay showed that a deletion mutation of 159 amino acids (∆149 to ∆307) including the PEST domain abolished the binding of PUB40 to BZR1. In addition, the bzr1-1D mutation greatly reduced the interaction of PUB40 with BZR1 (Figure 6A). These results indicate that the PEST domain of BZR1 is required for the interaction of PUB40 with BZR1. Consistent with the weak interaction of bzr1-1D with PUB40, the degradation rate of MBP-bzr1-1D was slower than that of MBP-BZR1 when protein extracts of PUB40-YFP were incubated with equal amounts of MBP-BZR1 or MBP-bzr1-1D (Figure 6B).
Figure 6.
The bzr1-1D Mutation Reduces PUB40-Mediated BZR1 Degradation.
(A) The binding affinity of PUB40 for bzr1-1D protein. Equal amounts of MBP-YFP, MBP-BZR1, MBP-BZR1-∆159 (∆149 to Δ307), or MBP-bzr1-1D (P234L) were pulled down with GST-PUB40–bound beads. Immunoblots were probed with anti-MBP and anti-GST antibodies.
(B) bzr1-1D degradation by PUB40. Equal amounts of MBP-YFP and MBP-bzr1-1D were incubated with protein extracts from 35S-PUB40-YFP plants. Ponceau S staining was used as a loading control.
(C) Endogenous BZR1 levels in the roots of PUB40-YFP bzr1-1D. PUB40-YFP and BZR1 were detected with anti-YFP and anti-BZR1 antibodies, respectively. Ponceau S staining was used as a loading control.
(D) Phenotypes of Col-0, PUB40-YFP, bzr1-1D, and bzr1-1D overexpressing PUB40-YFP. Plants were grown on MS medium for 14 d. Scale bar = 1 cm.
(E) Measurement of root fresh weight per seedling shown in (D). Error bars = ±se. Asterisks (*) indicate significant differences by Student’s t test (P < 0.001). Three roots were grouped and weighed at one time (n = 10). The data shown are representative of three independent experiments.
(F) Measurement of root length in the seedlings shown in (D). Error bars = ±se (n > 30 per genotype). Asterisks (*) indicate significant differences by Student’s t test (P < 0.001). The data shown are representative of two independent experiments.
To further explore whether bzr1-1D avoids PUB40-mediated degradation in plant tissues, we generated bzr1-1D plants overexpressing PUB40-YFP and examined bzr1-1D levels. PUB40-mediated BZR1 degradation in the roots was suppressed by the bzr1-1D mutation (Figure 6C). Consistently, the root growth defect caused by PUB40-YFP overexpression was restored by bzr1-1D mutation (Figures 6D, 6E, and 6F).
PUB40 Expression Pattern Determines BZR1 Accumulation in the Epidermal Layer of Roots
To further understand how BZR1 degradation is regulated by PUB40 in roots, we compared the expression patterns of PUB40 and BZR1 in root tips at the transcript and protein levels. Both PUB40 and BZR1 were expressed in the shoots and roots, but their transcript levels were higher in roots than in shoots (Supplemental Figures 3D and 10A). In addition, in transgenic plants expressing PUB40-YFP driven by its native promoter or the 35S promoter, much more PUB40-YFP accumulated in roots than in shoots (Supplemental Figures 10B and 10C). By contrast, there was no notable difference between BZR1-YFP levels in the roots versus shoots of plants harboring BZR1promoter-BZR1-YFP (Supplemental Figure 10D).
Birnbaum et al. (2003) provided a gene expression map for Arabidopsis root tissues. Based on their studies, we extracted data and compared the expression patterns of BZR1 and PUB40 in four cell layers (epidermis, cortex, endodermis, and stele) of roots. PUB40 appeared to be highly expressed in the stele, whereas BZR1 was ubiquitously expressed in all root tissues (Supplemental Figure 11A).
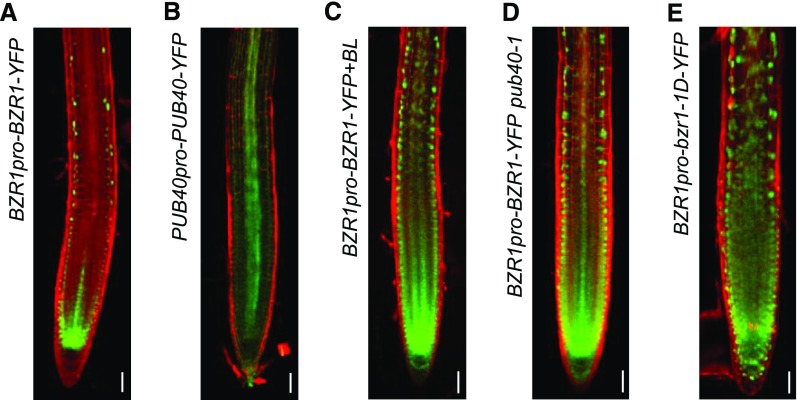
Next, we performed confocal microscopy to observe the root tips of transgenic plants expressing BZR1-YFP driven by the BZR1 promoter. Unlike the broad expression pattern of BZR1 mRNA in root tips (Supplemental Figure 11A), BZR1-YFP fluorescence was detected in the nucleus of the epidermal layer and the root apical meristem including quiescent cells (Figure 7A). A similar pattern of BZR1-YFP accumulation was observed in the root tips of 35S-BZR1-YFP plants (Supplemental Figure 11B). By contrast, PUB40-YFP expressed under the control of its native promoter mainly accumulated in the stele, which corresponded with its mRNA expression pattern (Supplemental Figure 11A; Figure 7B).
Figure 7.
PUB40-Mediated Cell-Layer–Specific Accumulation of BZR1 in Roots.
(A and B) Protein accumulation pattern of BZR1-YFP (A) and PUB40-YFP (B) in Arabidopsis roots.
(C) BZR1-YFP accumulation in response to 1 h of 100 nM of BL treatment.
(D) Protein accumulation of BZR1-YFP in BZR1pro-BZR1-YFP pub40-1.
(E) bzr1-1D-YFP accumulation in wild-type roots. Proteins were expressed under the control of their native promoters. PI-stained root tips were observed by confocal microscopy. Green = fluorescent signals of YFP-fused proteins. Scale bars = 10 μm.
Consistent with a previous report (Chaiwanon and Wang, 2015), BL treatment promoted BZR1-YFP accumulation in the cortex, endodermis, and stele, as well as the epidermis (Figure 7C). Similar to BR’s effect on BZR1 accumulation in root tips, BZR1-YFP in the pub40-1 mutant was also detected in the inner layers of the roots (Figure 7D; Supplemental Figure 11C). Remarkably, a large amount of bzr1-1D-YFP expressed under the control of the BZR1 promoter accumulated in all root tissues of the wild type (Figure 7E), suggesting that the accumulation of BZR1 in the inner layers of roots is regulated by PUB40 and that the bzr1-1D mutation suppresses PUB40-mediated BZR1 degradation in the roots.
PUB40 Is Regulated by the GSK3-Like Kinase BIN2
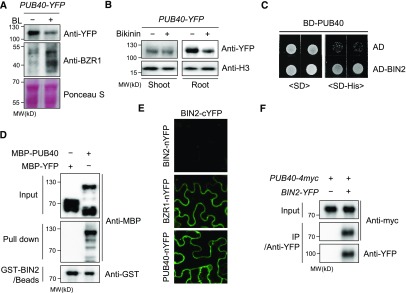
Next, we investigated whether BR signaling regulates PUB40. In 35S-PUB40-YFP plants, PUB40-YFP levels in roots were greatly reduced by BL treatment (Figure 8A). Consistently, BZR1 levels in 35S-PUB40-YFP plants increased in response to BL treatment. In addition, the plant GSK3-like kinase inhibitor Bikinin also reduced PUB40-YFP levels, which was reversed by treatment with the 26S proteasomal inhibitor MG132 (Supplemental Figure 12A). Similarly, PUB40-YFP expressed under the control of its native promoter was destabilized by both BL and Bikinin (Supplemental Figure 12B). When we compared the effects of Bikinin on PUB40 destabilization in shoots and roots, PUB40-YFP signals were specifically reduced by Bikinin treatment in roots but not shoots, suggesting that BIN2 regulates PUB40 stability in a root-specific manner (Figure 8B). These results indicate that BR signaling regulates PUB40 stability through proteasomal degradation and that PUB40 acts downstream of BIN2.
Figure 8.
BIN2 Interacts with PUB40 In Vitro and In Vivo.
(A and B) Both BL (A) and Bikinin (B) reduce PUB40 stability. 35S-PUB40-YFP was treated with Bikinin (30 μM) or BL (50 nM) for 3 h. PUB40-YFP and BZR1 levels were detected with anti-YFP and anti-BZR1 antibodies, respectively.
(C) PUB40 interacts with BIN2 in yeast cells. The BD fused to PUB40, the AD fused to BIN2, and AD constructs were cotransformed into yeast cells as indicated. Yeast cells were grown on SD or SD-His medium. BD, binding domain; AD, activation domain; SD, synthetic dropout.
(D) In vitro pull-down assay to test the interaction between PUB40 and BIN2. MBP-YFP or MBP-PUB40 was incubated with GST-BIN2–bound beads. Proteins pulled down by GST-BIN2 were analyzed using anti-MBP and anti-GST antibodies.
(E) BiFC assays to test the in vivo interaction between BIN2 and PUB40. The indicated constructs were cotransformed into wild-tobacco–leaf epidermal cells. The BZR1-nYFP and BIN2-cYFP pair was used as a positive control in the BiFC assay.
(F) Coimmunoprecipitation between PUB40 and BIN2 in vivo. BIN2-YFP protein was immunoprecipitated by anti-YFP antibody from wild tobacco cells coexpressing PUB40-myc and BIN2-YFP and detected with anti-YFP and anti-myc antibodies.
Considering that PUB40 was destabilized when GSK3-like kinase activity was inhibited by Bikinin and the ARM-repeat protein β-catenin is known to be phosphorylated by GSK3 during Wnt signaling, we investigated whether the GSK3-like kinase BIN2 regulates PUB40, which possesses Arm-repeat domains. Both a yeast two-hybrid assay and an in vitro pull-down assay indicated that BIN2 interacted with PUB40 (Figures 8C and 8D). Three homologs of PUB40 also appeared to interact with BIN2 in vitro (Supplemental Figure 13A). In addition, a BiFC assay demonstrated that BIN2-cYFP interacted with PUB40-nYFP as well as BZR1-nYFP in the cytoplasm of wild tobacco epidermal cells (Figure 8E; Supplemental Figure 13B). Furthermore, PUB40-myc was coimmunoprecipitated with BIN2-YFP by anti-YFP antibodies in wild tobacco cells coexpressing PUB40-myc and BIN2-YFP (Figure 8F).
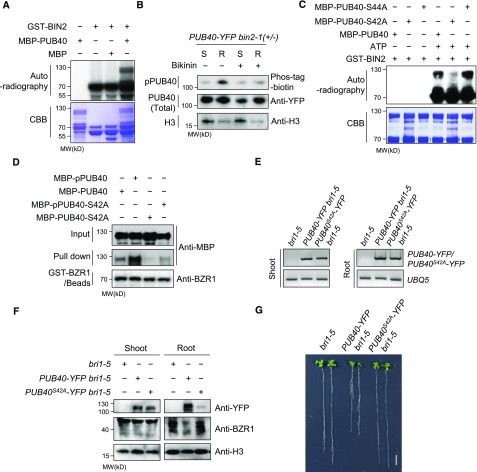
In addition to the binding of BIN2 to PUB40, an in vitro kinase assay showed that the GSK3-like kinase BIN2 phosphorylates PUB40 (Figure 9A). We investigated whether the phosphorylation of PUB40 is regulated by BIN2 in vivo. When we compared the phosphorylation status of PUB40 in roots and shoots with Phos-tag-biotin, phosphorylated PUB40 was more highly accumulated in the roots than the shoots (Figure 9B). Importantly, the amount of phosphorylated PUB40 greatly decreased in response to Bikinin treatment in roots, suggesting that BIN2 specifically phosphorylates PUB40 in roots.
Figure 9.
BIN2 Stabilizes PUB40 through the Phosphorylation of Ser-42 in Roots.
(A) BIN2 phosphorylates PUB40 in vitro. CBB, Coomassie brilliant blue.
(B) BIN2 specifically phosphorylates PUB40 in roots. PUB40-YFP was treated with mock or Bikinin (30 μM) for 3 h. The amount of input was adjusted to immunoprecipitate a comparable amount of PUB40-YFP from the shoots (S) and the root (R). The immunoblot was probed with Phos-tag-streptavidin-HRP and anti-YFP and H3 antibodies.
(C) Identification of BIN2 phosphorylation site on PUB40. MBP-PUB40, MBP-PUB40-S42A, or MBP-PUB40-S44A was incubated with GST-BIN2 and 32P-γATP. CBB, Coomassie brilliant blue.
(D) BIN2-induced phosphorylation of PUB40 increases the binding of PUB40 to BZR1. MBP-PUB40 or MBP-PUB40-S42A preincubated with GST-BIN2 and ATP was purified to remove GST-BIN2. MBP-PUB40, phosphorylated MBP-PUB40 (MBP-pPUB40), MBP-PUB40-S42A, or phosphorylated MBP-PUB40-S42A (MBP-pPUB40-S42A) was pulled down with equal amounts of GST-BZR1–bound beads.
(E) RT-PCR analysis of PUB40-YFP or PUB40S42A-YFP in bri1-5, PUB40-YFP bri1-5, and PUB40S42A-YFP bri1-5. UBQ5 was used as a loading control.
(F) Protein accumulation of PUB40-YFP or PUB40S42A-YFP in the shoots and roots of bri1-5, PUB40-YFP bri1-5, and PUB40S42A-YFP bri1-5. PUB40 and BZR1 were analyzed by immunoblotting using anti-YFP and anti-BZR1 antibodies.
(G) Phenotypes of bri1-5 overexpressing PUB40-YFP or PUB40S42A-YFP. The plants were grown for 14 d on MS medium.
Next, we searched for predicted phosphorylation sites of PUB40 in PhosPhAt 4.0 (http://phosphat.uni-hohenheim.de/). An N-terminal region of PUB40 possessing seven consecutive serines (residues 42 to 48) was predicted as a putative phosphorylation site. As predicted, a phospho-peptide (WRTSLSRSSSSSSSNNNSPTK) including residues 42 to 48 was identified by mass spectrometry analysis of PUB40-YFP immunoprecipitated from roots of PUB40-YFP (Supplemental Figure 14A). However, we were unable to perform a precise assignment of the phosphorylation site because the phospho-peptide contains many Ser and Thr residues, including seven consecutive Ser residues. The fragmentation pattern indicated that one of the six residues (Thr-37, Ser-38, Ser-40, Ser-42, Ser-43, and Ser-44) was phosphorylated. To further determine phosphorylation site of PUB40, we generated a mutant PUB40 with deletion of the seven serines (PUB40-Δ7) and performed an in vitro kinase assay. These results indicate that BIN2-induced phosphorylation of PUB40 was greatly reduced by the deletion mutation (Supplemental Figure 14B), implying that the consecutive Ser residues might contain a BIN2 phosphorylation site. Indeed, further investigation using mutant PUB40 protein harboring a point mutation confirmed that BIN2 phosphorylates PUB40 at Ser-42 (Figure 9C).
Based on these results, we investigated whether the phosphorylation of PUB40 by BIN2 regulates the interaction between PUB40 and BZR1. We incubated MBP-PUB40 with GST-BIN2 in the presence of ATP and purified phosphorylated MBP-PUB40 (MBP-pPUB40) by affinity purification. When equal amounts of MBP-PUB40 and MBP-pPUB40 were incubated with immobilized GST-BZR1, MBP-pPUB40 interacted more strongly with GST-BZR1 than did MBP-PUB40 (Figure 9D). These results suggest that the binding of PUB40 to BZR1 increases in response to BIN2-induced phosphorylation. Importantly, the binding of PUB40 to BZR1 due to BIN2-induced phosphorylation was abolished by Ser-42 substitution to Ala (Figure 9D), suggesting that BIN2 enhances the interaction of PUB40 with BZR1 through the phosphorylation of Ser-42 in PUB40. Finally, we generated transgenic bri1-5 plants overexpressing PUB40S42A-YFP and compared their phenotype with PUB40-YFP bri1-5. Although the mRNA levels of PUB40S42A–YFP were similar to those of PUB40-YFP in both shoots and roots (Figure 9E), the protein level of PUB40S42A-YFP in roots, but not in shoots, was much lower than that of PUB40-YFP (Figure 9F). Therefore, unlike wild-type PUB40, PUB40S42A overexpression in the bri1-5 background did not inhibit root growth (Figure 9G). The results strongly suggest that the phosphorylation of Ser-42 in PUB40 by BIN2 helps stabilize PUB40 in roots.
DISCUSSION
Recent studies have suggested that the degradation of BZR1 and BES1 is mediated in different ways in response to diverse environmental stimuli and developmental signals (Yang and Wang, 2017). Under stress conditions such as drought and starvation, BZR1 and BES1 undergo autophagic degradation (Zhang et al., 2016; Nolan et al., 2017). By contrast, distinct ubiquitination complexes determine the stability of BZR1 and BES1 according to light conditions and developmental programs. Light and dark conditions activate two different types of RING finger E3 ligases to regulate hypocotyl growth (Kim et al., 2014; Yang et al., 2017). SINATs cause the degradation of dephosphorylated BZR1/BES1 in the light to inhibit hypocotyl elongation (Yang et al., 2017). Conversely, in the dark, COP1 mediates the degradation of phosphorylated BZR1/BES1 to promote hypocotyl elongation (Kim et al., 2014). The F-box protein MAX2, which regulates strigolactone signaling as a subunit of an SCF complex, mediates BES1 degradation to regulate shoot branching (Wang et al., 2013).
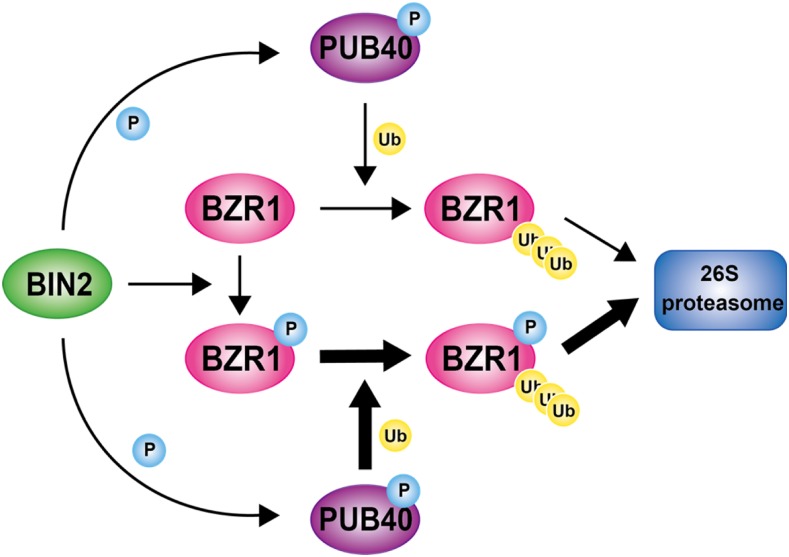
Our results further suggest that BZR1 stability is regulated in a variety of ways. We propose a regulatory model for BZR1 degradation mediated by PUB40 in the roots (Figure 10). According to this model, when BR levels are low, BIN2 is constitutively active and phosphorylates PUB40 as well as BZR1, which promotes the binding of PUB40 to BZR1. Phosphorylated BZR1 is retained in the cytoplasm and ubiquitinated by PUB40, resulting in the 26S proteasomal degradation of BZR1. When BR levels are high, BIN2 activity is inhibited and it is degraded by upstream BR signaling. Dephosphorylated BZR1 accumulates in the nucleus and regulates BR-responsive gene expression. Meanwhile, PUB40 is destabilized due to the lack of BIN2-induced phosphorylation.
Figure 10.
A Proposed Model of PUB40-Mediated Degradation of BZR1 in Arabidopsis Roots.
In the absence of BR, BIN2 constitutively phosphorylates both BZR1 and PUB40 in roots. BIN2-induced phosphorylation increases PUB40 stability as well as the binding of PUB40 to BZR1. PUB40 primarily ubiquitinates BZR1, leading to 26S proteasomal degradation of BZR1 in roots. The thick arrow indicates prominent ubiquitination. P, phosphorylation. Ub, ubiquitination.
Although PUB40 mainly degrades phosphorylated BZR1, our results indicate that dephosphorylated BZR1 also interacts with and is degraded by PUB40. Considering the differential subcellular localization of BZR1 depending on its phosphorylation status, the main target of cytoplasmic PUB40 is thought to be phosphorylated BZR1. However, we cannot rule out the possibility that PUB40 degrades dephosphorylated BZR1 in plant cells because the intracellular localization of PUB40 and dephosphorylated BZR1 might overlap. Dephosphorylated BZR1, although relatively small in amount, can be present in the cytoplasm. For example, newly translated BZR1 could transiently localize to the cytoplasm. In addition, PP2A dephosphorylates BZR1 in the cytoplasm as well as the nucleus (Tang et al., 2011).
The most interesting finding of our study is that PUB40 degrades BZR1 only in roots. Our biochemical analyses suggest that this is mainly caused by the differential regulation of PUB40 by BIN2 in roots versus shoots. Bikinin treatment greatly reduced PUB40 levels in roots but not in shoots, suggesting that BIN2-induced phosphorylation stabilizes PUB40 in roots. Indeed, in transgenic Arabidopsis harboring PUB40-YFP driven by the 35S promoter, much more PUB40-YFP accumulated in roots than in shoots. PUB40-YFP appeared to be highly phosphorylated in roots but not in shoots (Figure 8B). Consistently, phosphorylated PUB40-YFP levels in roots were reduced by Bikinin treatment. Our results indicate that BIN2-induced phosphorylation of PUB40 mainly occurs in roots, leading to the stabilization of PUB40. Moreover, we demonstrated that PUB40 more effectively binds to BZR1 when phosphorylated by BIN2 (Figure 9D). The binding of PUB40 to BZR1 due to BIN2-induced phosphorylation is abolished by the S42A mutation (Figure 9D). Taken together, our results suggest that BIN2-induced phosphorylation of PUB40 in roots gives rise to PUB40-mediated BZR1 degradation due to effective BZR1 binding as well as the stabilization of PUB40. However, further investigation is needed to understand why BIN2-induced phosphorylation of PUB40 mainly occurs in roots.
Only a small amount of PUB40 protein was detected in shoots compared with roots, but PUB40 might also function in shoots. In shoots, PUB40-mediated BZR1 degradation might occur with the help of another signaling component under a specific environmental or physiological condition. Alternatively, given that the structural dynamics of the ARM-repeat domain of PUBs might contribute to a broad range of substrate specificities, it is possible that PUB40 also has the potential to modulate the stability of certain proteins other than BZR1 in the shoots.
In the roots, PUB40 overexpression greatly reduced endogenous BZR1 levels and inhibited root growth (Figures 3A and 3B). Interestingly, the aerial parts of PUB40 overexpression plants also displayed growth retardation. However, BZR1 levels in shoots were not altered by PUB40 overexpression, and the leaf phenotype differed from that of typical BR mutants. Considering that overall plant growth depends on the appropriate growth and functioning of roots, which are essential for the uptake of nutrients and water, the growth retardation observed in the aerial portions of plants overexpressing PUB40 might be due to functional defects in roots.
The epidermal accumulation of dephosphorylated BZR1 in root tips is essential for promoting root growth (Chaiwanon and Wang, 2015). Our study clarifies how BZR1 accumulates in the epidermal layer of roots. Whereas BZR1 mRNA is transcribed in most cell layers of the root, BZR1 protein accumulates in the outermost layer (Figure 7). We show that BZR1 is degraded by PUB40 enriched in the inner layers, resulting in a distinct pattern of BZR1 distribution in roots. Upon BR treatment, BIN2-mediated PUB40 phosphorylation is inhibited, leading to the destabilization of PUB40 in the inner layers. BR appears to increase phosphorylated BZR1 (pBZR1) as well as BZR1 levels in the inner layers because PUB40 degrades both phosphorylated BZR1 and BZR1, although there is a difference in efficiency. We also showed that the bzr1-1D mutation greatly reduces the binding of PUB40 to BZR1 (Figure 6). Thus, bzr1-1D protein accumulates in the inner layers to avoid degradation by PUB40.
Epidermal BZR1 levels in the pub40-1 mutant were considerably higher than those of wild type, which is similar to the response to BL treatment (Figures 7A, 7C, and 7D). This finding suggests that PUB40 in epidermal cells can regulate BZR1-mediated root growth with relatively low levels of expression. Nevertheless, the prominent localization of PUB40 in the stele suggests that PUB40 regulates BZR1 levels mainly in the inner layers. However, it remains to be determined whether and how BZR1 that has accumulated in the inner layers mediates BR-regulated root growth. Notably, genetic studies of BR receptor family members (BRI1, BRL1, and BRL3) demonstrated that BR is also required for vasculature development (Savaldi-Goldstein et al., 2007; Kang et al., 2017). It would be worthwhile to further investigate how vascular BR signaling utilizes the relationship between BZR1 and PUB40 to exert its physiological activity in roots.
Although BZR1 levels were greatly increased in pub39 pub40 pub41 mutant roots, the triple mutant showed a subtle phenotypic difference in root growth and mild hypersensitivity to BR in a root growth inhibition assay. Not surprisingly, the overexpression of wild-type BZR1 did not significantly alter plant growth phenotypes, and it slightly alleviated BR sensitivity. In addition, unlike bzr1-1D, BZR1-overexpressing plants were not resistant to the BR biosynthesis inhibitor Brassinazole, because the phosphorylation status rather than the amount of BZR1 largely determines BZR1 activity in plant cells. Meanwhile, Singh et al. (2014) showed that the bzr1-1D mutant is resistant to root growth inhibition caused by phosphate-depletion conditions. In their study, roots rather than shoots of bzr1-1D appeared to be unaffected under depleted phosphate conditions. Consistently, we found that BZR1 accumulation in the pub39 pub40 pub41 roots led to resistance to low phosphate availability. Thus, our results further support the notion that BZR1 plays a crucial role in regulating root growth in response to environmental changes to nutrient acquisition.
Recent studies have highlighted the functional importance of PUB proteins. Most of their functions are related to physiological responses to biotic and abiotic stress, as well as phytohormones (Yee and Goring, 2009). PUB proteins regulate multiple physiological processes (Trujillo, 2018). For example, PUB12 and PUB13 mediate the degradation of not only the pattern-recognition receptor FLS2 but also the abscisic acid (ABA) coreceptor ABI1 (Lu et al., 2011; Kong et al., 2015). In addition, PUB22 and PUB23 act as negative regulators of Pattern-Triggered Immunity and ABA-mediated drought responses (Trujillo et al., 2008; Seo et al., 2012). Interestingly, the stability of PUB22 is also regulated by phosphorylation. In a negative feedback loop of immune responses, MPK3 interacts with and phosphorylates PUB22 (Furlan et al., 2017). Similar to BIN2-induced phosphorylation of PUB40, MPK3-induced phosphorylation of PUB22 occurs at N-terminal residues (Thr-62 and Thr-88) within and adjacent to the U-box domain, which increases the stability of PUB22 by inhibiting its dimerization. It would be worth investigating whether PUB40, whose domain arrangement is similar to that of PUB22, forms dimers and whether this dimerization is directly regulated by BIN2-induced phosphorylation.
METHODS
Plant Materials and Growth Conditions
PUB40 was overexpressed in the Arabidopsis (Arabidopsis thaliana) Columbia ecotype (Col-0) background or in bri1-5 plants of the Wassilewskija background. All other BR-related mutants (bri1-116, bri1-301, bzr1-1D) are in the Col-0 background. The T-DNA insertional mutants (pub40-1; SALK_01282, pub40-2; SALK_035132, pub39; SALK_136984, pub41; and SALK_099012) were obtained from the Arabidopsis Biological Resource Center. Sterilized seeds were imbibed with distilled water for 3 d at 6°C and planted on 1/2 Murashige and Skoog medium (1/2 MS; Duchefa Biochemie) containing 1% (w/v) Suc and 0.8% Phytoagar (Duchefa Biochemie). Plants were grown in a plant growth chamber at 22°C under long-day conditions (16-h light, 8-h dark). The light intensity was maintained at 104 μmol·m−2·s−1 (fluorescent lamp).
To test plant growth under low phosphate availability, MS basal salts without Pi (MSP19; Caisson Laboratories) and micropropagation agar-type I (A038; Caisson Laboratories) were used. Arabidopsis seedlings were grown on 1/2 MS (MSP19) medium containing 1.25 mM of KH2PO4, 1% (w/v) Suc, and 0.8% agar (A038) for 3 d. The seedlings were transferred into adequate (1.25 mM) or low (1 μM) phosphate medium and grown for an additional 7 d. To prepare phosphate-deficient medium, KH2PO4 was replaced with equimolar KCl with pH adjustment (pH 5.8).
Arabidopsis Transformation
PUB40 was overexpressed in Col-0 or in bri1-5 plants. Agrobacterium tumefaciens (GV1301) carrying the plasmid for PUB40-YFP or PUB40-myc was resuspended with transformation medium (1/2 MS at pH 5.7, 5% Suc, and 0.05% Silwet L-77 [PhytoTechnology Laboratories]) and transformed by the floral dip method (Clough, 2005).
Plasmid Constructs
All plasmid constructs were cloned into pENTR/SD/D-TOPO vectors (Invitrogen). Mutations were induced with a QuikChange Site-Directed Mutagenesis Kit (Stratagene). Entry clones were subcloned into Gateway-compatible vectors by LR reaction (Invitrogen). To express proteins in Escherichia coli, gcpMALc2 and pDEST15 were used. For expression in plants, pEarleyGate101 (35S-PUB40-YFP, 35S-BZR1-YFP, and 35S-BIN2-YFP), gcpCAMBIA1390-4myc-6His (35S-PUB40-myc and 35S-BZR1-myc), and BiFC vectors (Gampala et al., 2007) were used. To generate a Gateway-compatible pEG-TW1 vector carrying no promoter-Gateway cassette-YFP, the pEarleyGate301 vector was digested by NcoI and PacI and ligated with a YFP-containing fragment digested by NcoI and PacI from pEarleyGate101. A genomic fragment of PUB40 including its promoter and gene was cloned into the pEG-TW1 vector.
BiFC Assay
To produce plasmid constructs for BiFC assay, Gateway-compatible binary vectors described by Gampala et al. (2007) were used. Agrobacterium (GV3101) cells harboring the construct expressing nYFP or cYFP were mixed as indicated and infiltrated into wild tobacco leaves with infiltration medium (10 mM of MES at pH 5.6, 10 mM of MgCl2, and 150 μM of acetosyringone). At 36 h after infiltration, fluorescence signals were detected by confocal microscopy.
Yeast Two Hybrid Assay
Each entry clone was cloned into pXDGATcy86 vectors containing the DNA binding domain or the gcpGADT7 vector containing the activation domain. Two plasmids paired according to the combination of AD and BD fusion were cotransformed into yeast AH109 cells. Transformed yeast cells were spotted onto synthetic dropout (SD; -Leu/-Trp) and SD-His (-Leu/-Trp/-His) medium and incubated at 30°C for 4 d.
Purification of Recombinant Proteins
To purify the MBP- or GST-fusion proteins, E. coli BL21 cells were grown at 37°C to optical density 0.5 to 0.6 and subjected to isopropyl β-d-1-thiogalactopyranoside (0.2 mM) induction. After incubation at 37°C (MBP-fusion proteins) or 28°C (GST-fusion proteins) for 3 h, the cells were harvested and stored at −20°C until use. Harvested cells were resuspended in lysis buffer (20 mM of Tris-Cl at pH 7.4, 200 mM of NaCl, and 1 mM of EDTA) for MBP-fusion proteins or in 1× PBS for GST-fusion proteins. After sonication, cell lysates were centrifuged at 12,000g for 20 min. The supernatants were incubated with amylose resins (New England BioLabs) or glutathione resins (GenScript) for 1.5 h at 4°C, loaded onto a microchromatography column, and washed with lysis buffer or 1× PBS buffer. The MBP- or GST-fusion proteins were eluted by elution buffer (20 mM of Tris-Cl at pH 7.4 and 200 mM of NaCl) containing 10 mM of maltose or 5 mM of glutathione.
In Vitro Pull-down and Coimmunoprecipitation Assay
For the in vitro pull-down assay, GST-PUB40 bound to glutathione agarose beads was incubated with recombinant MBP-YFP or MBP-BZR1 in binding buffer (50 mM of Tris-Cl at pH 7.5, 150 of mM NaCl, 0.2% [v/v] NP-40, and 0.05 of mg/mL BSA). After incubation for 1.5 h, the beads were washed with buffer (50 mM of Tris-Cl at pH 7.5, 100 of mM NaCl, and 0.1% [v/v] NP-40). Associated proteins were then eluted with 2× SDS sample buffer (24 mM of Tris-Cl at pH 6.8, 10% glycerol, 0.8% of SDS, and 2% 2-mercaptoethanol) and analyzed by immunoblotting. The antibodies used in our study are listed in Supplemental Table 1.
To perform the coimmunoprecipitation assay, plant materials were pretreated 10 μM of MG132 for 12 h at 23°C. After grinding with liquid N, the samples were resuspended in immunoprecipitation (IP) extraction buffer (20 mM of HEPES at pH 7.5, 40 mM of KCl, 1 mM of EDTA, 1% Triton X-100, 0.2 mM of phenylmethylsulfonyl fluoride, and 1× protease inhibitor cocktail). After filtering through Miracloth (EMD Millipore), the extracts were centrifuged at 12,000 rpm for 10 min. The supernatants were incubated with Protein A beads (Thermo Fisher Scientific) bound to the indicated antibodies for 1 h. The beads were washed with IP extraction buffer containing 0.1% Triton X-100 and eluted with 2× SDS sample buffer.
For Figure 1F, 7-d-old transgenic plants expressing BZR1pro-BZR1-YFP were pretreated with mock or 100 nM of BL for 1 h. Each sample extracted by IP extraction buffer was incubated with MBP-PUB40 protein immobilized on amylose beads for 1 h. The beads were washed with IP extraction buffer containing 0.1% Triton X-100 and eluted with 2× SDS sample buffer.
Cell-Free Protein Degradation Assay
PUB40-YFP bri1-5 and bri1-5 plants were ground into a powder with liquid N and proteins were extracted in degradation assay buffer (50 mM of Tris-Cl at pH 7.5, 50 mM of NaCl, 1% Triton X-100, 0.2 of mM phenylmethylsulfonyl fluoride, 5 mM of MgCl2, 5 mM of DTT, and 5 mM of ATP). After centrifugation, an equal amount of each supernatant was incubated with 100 ng of MBP-BZR1 or MBP-bzr1-1D. To generate MBP-pBZR1, MBP-BZR1 that was preincubated with GST-BIN2 and ATP in kinase assay buffer (20 mM of Tris at pH 7.5, 1 mM of MgCl2, 100 mM of NaCl, and 1 mM of DTT) was purified by affinity purification. After incubation for the indicated times at 30°C, the reactions were stopped by adding 2× SDS sample buffer.
Confocal Microscopy
Fluorescence of YFP, BiFC (YFP), and propidium iodide (PI) were visualized under a confocal microscope (C2 Plus; Nikon). YFP signals were visualized after excitation by Kr/Ar 488-nm laser and emission through a 500- to 545-nm filter. For PI staining, 5-d-old Arabidopsis roots were stained in 10 μg/mL PI for 1 min and rinsed in water before confocal microscopy observation. PI was detected with a 570- to 670-nm filter.
RNA Analysis
Total RNA was extracted from 14-d-old wild-type Col-0, 35S-BZR1-YFP, and pub39 pub40 pub41 seedlings using a Spectrum Plant Total RNA Kit (Sigma-Aldrich). RNA-Seq analysis was performed following Illumina HiSeq 2500 (Macrogen). Reads that aligned with the Arabidopsis genome sequence in TAIR10 (NCBI) were quantified. Differentially expressed genes were defined based on a 1.3-fold difference between samples. Quantitative RT-PCR was performed using the CFX96 Real-Time PCR Detection System (Bio-Rad) using iQ SYBR Green Supermix (Bio-Rad). The cycling condition was 95°C for 3 min, followed by 45 cycles of 95°C for 10 s, 60°C for 15 s, and 75°C 15 s. Gene expression levels were normalized to PP2A. Primers used in qPCR are listed in Supplemental Table 2.
In Vitro Kinase Assay
MBP-PUB40, MBP-PUB40-S42A, and MBP-PUB40-S44A were incubated with GST-BIN2 in kinase assay buffer (100 μM of ATP, and [γ-32P] ATP, 10 μCi) for 2 h at 30°C. After the reaction, the samples were separated by SDS-PAGE) and stained with Coomassie brilliant blue. Protein phosphorylation was analyzed on a phosphor-image analyzer (Cyclone; Packard).
Detection of Phosphorylated PUB40
The in vivo phosphorylation status of PUB40 was analyzed using biotinylated Phos-tag BTL (Wako). To prepare the Phos-tag-biotin and streptavidin-conjugated Horseradish Peroxidase (HRP) complex, 0.02 mM of Phos-tag BTL solution was incubated with 4 μg of streptavidin-conjugated HRP (Pierce) in 1× Tris Buffered Saline with Tween buffer (20 mM of Tris-Cl at pH 7.5, 150 mM of NaCl, and 0.1% TWEEN 20) containing 0.2 mM of Zn (NO3)2 for 30 min at room temperature. After incubation, free Phos-tag-biotin was removed by centrifugation at 14,000g for 20 min using a 30K Amicon Ultra-0.5 mL centrifugal filter (Millipore). The remaining Phos-tag-streptavidin-HRP solution was diluted with 20 mL of 1× Tris Buffered Saline with Tween.
For Figure 9B, bin2-1 (+/-) overexpressing PUB40-YFP were treated with mock or 30 μM Bikinin for 3 h. Proteins immunoprecipitated from the shoots or roots of PUB40-YFP with anti-YFP antibody were separated by SDS-PAGE and transferred to a polyvinylidene difluoride membrane. The blot was probed with 20 mL of Phos-tag-streptavidin-HRP and anti-YFP antibody (1/5,000).
Mass Spectrometry Analysis
PUB40-YFP immunoprecipitated from the roots of 14-d-old PUB40-YFP plants were separated by SDS-PAGE. The PUB40-YFP protein band was excised and subjected to in-gel digestion with Lysyl Endopeptidase (Wako Chemicals). Peptide mixtures desalted using C18 ZipTips (EMD Millipore) were analyzed using the EasyLC1200 system (Thermo Fisher Scientific) connected to a Q Exactive HF high performance quadrupole Orbitrap mass spectrometer (Thermo Fisher Scientific). An Easy-Spray C18 column (75 μm × 250 mm, ES802; Thermo Fisher Scientific) was used at a flow rate of 300 nL/min and 120-min gradient. Peptides were eluted by a gradient from 3% to 28% (v/v) solvent B (80% [v/v] acetonitrile/0.1% [v/v] formic acid) over 100 min and from 28% to 44% (v/v) solvent B over 20 min, followed by a short wash in 90% (v/v/) solvent B.
For mass spectrometry analysis, a precursor scan was performed from mass-to-charge ratio (m/z) 375 to 1,600, and the top 20 most intense multipally charged precursors were selected for fragmentation. Peptides were fragmented with higher-energy collision dissociation with normalized collision energy of 27. Tandem mass spectrometry data were converted to a peak list using the in-house script PAVA, and data were searched using ProteinProspector against peptide sequences from TAIR10 (https://www.arabidopsis.org/), concatenated with sequence randomized versions of each protein (a total of 35,386 entries). A precursor mass tolerance of 10 ppm and a fragment mass error tolerance of 20 ppm were allowed. A tandem mass spectrometry spectrum was obtained using Xcalibur (Thermo Fisher Scientific), and the assignment of modified peptide was manually checked.
Accession Numbers
Sequence data from this article can be found in the European Molecular Biology Laboratory/GenBank data libraries under the following accession numbers: PUB38; At5g65200.1, PUB39; At3g47820.1, PUB40; At5g40140.1, PUB41; AT5G62560.1, BIN2; At4g18710.1, and BZR1; AT1G75080.1. PUB3; At3g54790.1; PUB16; At5G01830.1, PUB43; At1G76390.1, PUB54; At1G01680.1. The Gene Expression Omnibus accession number for the RNA-Seq data is GSE52966.
Supplemental Data
Supplemental Figure 1. Domain configuration of PUB40 and expression of BiFC constructs.
Supplemental Figure 2. Phenotypes of bri1-5 plants overexpressing PUB40-YFP.
Supplemental Figure 3. Growth phenotypes of PUB40 overexpression plants and BZR1 levels in shoots and roots.
Supplemental Figure 4. BZR1 accumulation in two pub40 knockout mutants.
Supplemental Figure 5. Three homologs of PUB40 and isolation of the pub39 pub40 pub41 triple mutant.
Supplemental Figure 6. Complementation of the root phenotype and BZR1 level of pub39 pub40 pub41 by PUB40 overexpression.
Supplemental Figure 7. BZR1 mediates BR-regulated root growth.
Supplemental Figure 8. BL-induced hypocotyl and root growth in response to low Pi levels in pub39 pub40 pub41.
Supplemental Figure 9. qRT-PCR analysis of BL-regulated gene expression.
Supplemental Figure 10. Expression levels of PUB40 and BZR1 in shoots and roots.
Supplemental Figure 11. Expression of BZR1 and PUB40 in roots and BZR1 accumulation in the pub40-1 mutant.
Supplemental Figure 12. BR-regulated proteasomal degradation of PUB40.
Supplemental Figure 13. Interactions of BIN2 with close homologs of PUB40 and protein expression of BiFC constructs.
Supplemental Figure 14. Identification of the in vivo phosphorylation site for PUB40.
Supplemental Table 1. The antibodies used in this study.
Supplemental Table 2. Primers used in this study.
Supplemental Data Set. Differentially expressed genes in BZR1ox and the pub39 pub40 pub41 mutant.
Supplemental File. Alignment used for phylogenetic tree shown in Supplemental Figure 5A.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
This work was supported by the Basic Science Research Program, National Research Foundation of Korea funded by the Ministry of Science, ICT, and Future Planning (NRF-2018R1D1A1B07043928 to S.K.K. and NRF-2017R1A2B4004274 to T.W.K.). We thank the Carnegie Mass Spectrometry Facility.
Author Contributions
E.J.K., Z.Y.W., S.K.K., and T.W.K. designed the experiments; E.J.K., S.H.L., C.H.P., S.H.K., C.C.H., and S.X. performed the experiments; E.J.K., S.K.K., and T.W.K. wrote the article; all authors read and approved the article.
References
- Birnbaum K., Shasha D.E., Wang J.Y., Jung J.W., Lambert G.M., Galbraith D.W., Benfey P.N. (2003). A gene expression map of the Arabidopsis root. Science 302: 1956–1960. [DOI] [PubMed] [Google Scholar]
- Chaiwanon J., Wang Z.Y. (2015). Spatiotemporal brassinosteroid signaling and antagonism with auxin pattern stem cell dynamics in Arabidopsis roots. Curr. Biol. 25: 1031–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H., Nusse R. (2012). Wnt/β-catenin signaling and disease. Cell 149: 1192–1205. [DOI] [PubMed] [Google Scholar]
- Clough S.J. (2005). Floral dip: Agrobacterium-mediated germ line transformation. Methods Mol. Biol. 286: 91–102. [DOI] [PubMed] [Google Scholar]
- Dill A., Thomas S.G., Hu J., Steber C.M., Sun T.P. (2004). The Arabidopsis F-box protein SLEEPY1 targets gibberellin signaling repressors for gibberellin-induced degradation. Plant Cell 16: 1392–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furlan G., Nakagami H., Eschen-Lippold L., Jiang X., Majovsky P., Kowarschik K., Hoehenwarter W., Lee J., Trujillo M. (2017). Changes in PUB22 ubiquitination modes triggered by MITOGEN-ACTIVATED PROTEIN KINASE3 dampen the immune response. Plant Cell 29: 726–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gampala S.S., et al. (2007). An essential role for 14-3-3 proteins in brassinosteroid signal transduction in Arabidopsis. Dev. Cell 13: 177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray W.M., Kepinski S., Rouse D., Leyser O., Estelle M. (2001). Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins. Nature 414: 271–276. [DOI] [PubMed] [Google Scholar]
- He J.X., Gendron J.M., Yang Y., Li J., Wang Z.Y. (2002). The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 99: 10185–10190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y.H., Breda A., Hardtke C.S. (2017). Brassinosteroid signaling directs formative cell divisions and protophloem differentiation in Arabidopsis root meristems. Development 144: 272–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley D.R., Estelle M. (2012). Ubiquitin-mediated control of plant hormone signaling. Plant Physiol. 160: 47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.W., Wang Z.Y. (2010). Brassinosteroid signal transduction from receptor kinases to transcription factors. Annu. Rev. Plant Biol. 61: 681–704. [DOI] [PubMed] [Google Scholar]
- Kim B., Jeong Y.J., Corvalán C., Fujioka S., Cho S., Park T., Choe S. (2014). Darkness and gulliver2/phyB mutation decrease the abundance of phosphorylated BZR1 to activate brassinosteroid signaling in Arabidopsis. Plant J. 77: 737–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.W., Guan S., Burlingame A.L., Wang Z.Y. (2011). The CDG1 kinase mediates brassinosteroid signal transduction from BRI1 receptor kinase to BSU1 phosphatase and GSK3-like kinase BIN2. Mol. Cell 43: 561–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D., Rape M. (2012). The ubiquitin code. Annu. Rev. Biochem. 81: 203–229. [DOI] [PubMed] [Google Scholar]
- Kong L., Cheng J., Zhu Y., Ding Y., Meng J., Chen Z., Xie Q., Guo Y., Li J., Yang S., Gong Z. (2015). Degradation of the ABA co-receptor ABI1 by PUB12/13 U-box E3 ligases. Nat. Commun. 6: 8630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D., Lin W., Gao X., Wu S., Cheng C., Avila J., Heese A., Devarenne T.P., He P., Shan L. (2011). Direct ubiquitination of pattern recognition receptor FLS2 attenuates plant innate immunity. Science 332: 1439–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald B.T., Tamai K., He X. (2009). Wnt/beta-catenin signaling: Components, mechanisms, and diseases. Dev. Cell 17: 9–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzucotelli E., Belloni S., Marone D., De Leonardis A., Guerra D., Di Fonzo N., Cattivelli L., Mastrangelo A. (2006). The e3 ubiquitin ligase gene family in plants: Regulation by degradation. Curr. Genomics 7: 509–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudgil Y., Shiu S.H., Stone S.L., Salt J.N., Goring D.R. (2004). A large complement of the predicted Arabidopsis ARM repeat proteins are members of the U-box E3 ubiquitin ligase family. Plant Physiol. 134: 59–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan T.M., Brennan B., Yang M., Chen J., Zhang M., Li Z., Wang X., Bassham D.C., Walley J., and Yin Y. (2017). Selective autophagy of BES1 mediated by DSK2 balances plant growth and survival. Dev Cell 41: 33–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peifer M., Berg S., Reynolds A.B. (1994). A repeating amino acid motif shared by proteins with diverse cellular roles. Cell 76: 789–791. [DOI] [PubMed] [Google Scholar]
- Rechsteiner M., Rogers S.W. (1996). PEST sequences and regulation by proteolysis. Trends Biochem. Sci. 21: 267–271. [PubMed] [Google Scholar]
- Sadanandom A., Bailey M., Ewan R., Lee J., Nelis S. (2012). The ubiquitin-proteasome system: Central modifier of plant signalling. New Phytol. 196: 13–28. [DOI] [PubMed] [Google Scholar]
- Savaldi-Goldstein S., Peto C., Chory J. (2007). The epidermis both drives and restricts plant shoot growth. Nature 446: 199–202. [DOI] [PubMed] [Google Scholar]
- Schulman B.A., Harper J.W. (2009). Ubiquitin-like protein activation by E1 enzymes: The apex for downstream signalling pathways. Nat. Rev. Mol. Cell Biol. 10: 319–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo D.H., Ryu M.Y., Jammes F., Hwang J.H., Turek M., Kang B.G., Kwak J.M., Kim W.T. (2012). Roles of four Arabidopsis U-box E3 ubiquitin ligases in negative regulation of abscisic acid-mediated drought stress responses. Plant Physiol. 160: 556–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A.P., Fridman Y., Friedlander-Shani L., Tarkowska D., Strnad M., Savaldi-Goldstein S. (2014). Activity of the brassinosteroid transcription factors BRASSINAZOLE RESISTANT1 and BRASSINOSTEROID INSENSITIVE1-ETHYL METHANESULFONATE-SUPPRESSOR1/BRASSINAZOLE RESISTANT2 blocks developmental reprogramming in response to low phosphate availability. Plant Physiol. 166: 678–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle J., Vierstra R.D. (2004). The ubiquitin 26S proteasome proteolytic pathway. Annu. Rev. Plant Biol. 55: 555–590. [DOI] [PubMed] [Google Scholar]
- Spratt D.E., Wu K., Kovacev J., Pan Z.Q., Shaw G.S. (2012). Selective recruitment of an E2∽ubiquitin complex by an E3 ubiquitin ligase. J. Biol. Chem. 287: 17374–17385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Han Z., Tang J., Hu Z., Chai C., Zhou B., Chai J. (2013). Structure reveals that BAK1 as a co-receptor recognizes the BRI1-bound brassinolide. Cell Res. 23: 1326–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W., et al. (2011). PP2A activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating BZR1. Nat. Cell Biol. 13: 124–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W., Kim T.W., Oses-Prieto J.A., Sun Y., Deng Z., Zhu S., Wang R., Burlingame A.L., Wang Z.Y. (2008). BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science 321: 557–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tewari R., Bailes E., Bunting K.A., Coates J.C. (2010). Armadillo-repeat protein functions: Questions for little creatures. Trends Cell Biol. 20: 470–481. [DOI] [PubMed] [Google Scholar]
- Thines B., Katsir L., Melotto M., Niu Y., Mandaokar A., Liu G., Nomura K., He S.Y., Howe G.A., Browse J. (2007). JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448: 661–665. [DOI] [PubMed] [Google Scholar]
- Trujillo M. (2018). News from the PUB: Plant U-box type E3 ubiquitin ligases. J. Exp. Bot. 69: 371–384. [DOI] [PubMed] [Google Scholar]
- Trujillo M., Ichimura K., Casais C., Shirasu K. (2008). Negative regulation of PAMP-triggered immunity by an E3 ubiquitin ligase triplet in Arabidopsis. Curr. Biol. 18: 1396–1401. [DOI] [PubMed] [Google Scholar]
- Vierstra R.D. (2009). The ubiquitin-26S proteasome system at the nexus of plant biology. Nat. Rev. Mol. Cell Biol. 10: 385–397. [DOI] [PubMed] [Google Scholar]
- Wang Y., Sun S., Zhu W., Jia K., Yang H., Wang X. (2013). Strigolactone/MAX2-induced degradation of brassinosteroid transcriptional effector BES1 regulates shoot branching. Dev. Cell 27: 681–688. [DOI] [PubMed] [Google Scholar]
- Wang Z.Y., Nakano T., Gendron J., He J., Chen M., Vafeados D., Yang Y., Fujioka S., Yoshida S., Asami T., Chory J. (2002). Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev. Cell 2: 505–513. [DOI] [PubMed] [Google Scholar]
- Wang Z.Y., Bai M.Y., Oh E., Zhu J.Y. (2012). Brassinosteroid signaling network and regulation of photomorphogenesis. Annu. Rev. Genet. 46: 701–724. [DOI] [PubMed] [Google Scholar]
- Wiborg J., O’Shea C., Skriver K. (2008). Biochemical function of typical and variant Arabidopsis thaliana U-box E3 ubiquitin-protein ligases. Biochem. J. 413: 447–457. [DOI] [PubMed] [Google Scholar]
- Xu W., Huang J., Li B., Li J., Wang Y. (2008). Is kinase activity essential for biological functions of BRI1? Cell Res. 18: 472–478. [DOI] [PubMed] [Google Scholar]
- Yang M., Wang X. (2017). Multiple ways of BES1/BZR1 degradation to decode distinct developmental and environmental cues in plants. Mol. Plant 10: 915–917. [DOI] [PubMed] [Google Scholar]
- Yang C.J., Zhang C., Lu Y.N., Jin J.Q., Wang X.L. (2011). The mechanisms of brassinosteroids’ action: From signal transduction to plant development. Mol. Plant 4: 588–600. [DOI] [PubMed] [Google Scholar]
- Yang M., Li C., Cai Z., Hu Y., Nolan T., Yu F., Yin Y., Xie Q., Tang G., and Wang X. (2017). SINAT E3 ligases control the light-mediated stability of the brassinosteroid-activated transcription factor BES1 in Arabidopsis. Dev Cell 41: 47–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee D., Goring D.R. (2009). The diversity of plant U-box E3 ubiquitin ligases: From upstream activators to downstream target substrates. J. Exp. Bot. 60: 1109–1121. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Zhu J.Y., Roh J., Marchive C., Kim S.K., Meyer C., Sun Y., Wang W., Wang Z.Y. (2016). TOR signaling promotes accumulation of BZR1 to balance growth with carbon availability in Arabidopsis. Curr. Biol. 26: 1854–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J.Y., Sae-Seaw J., Wang Z.Y. (2013). Brassinosteroid signalling. Development 140: 1615–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J.Y., Li Y., Cao D.M., Yang H., Oh E., Bi Y., Zhu S., and Wang Z.Y. (2017). The F-box Protein KIB1 mediates brassinosteroid-induced inactivation and degradation of GSK3-like kinases in Arabidopsis. Mol Cell 66: 648–657 [DOI] [PMC free article] [PubMed] [Google Scholar]