A putative galactose oxidase, RUBY, promotes cell-to-cell adhesion between seed coat epidermal cells and modifies galactosylated RG-I pectin in the seed coat mucilage of Arabidopsis.
Abstract
Cell-to-cell adhesion is essential for establishment of multicellularity. In plants, such adhesion is mediated through a middle lamella composed primarily of pectic polysaccharides. The molecular interactions that influence cell-to-cell adhesion are not fully understood. We have used Arabidopsis (Arabidopsis thaliana) seed coat mucilage as a model system to investigate interactions between cell wall carbohydrates. Using a forward-genetic approach, we have discovered a gene, RUBY PARTICLES IN MUCILAGE (RUBY), encoding a protein that is annotated as a member of the Auxiliary Activity 5 (AA5) family of Carbohydrate-Active Enzymes (Gal/glyoxal oxidases) and is secreted to the apoplast late in the differentiation of seed coat epidermal cells. We show that RUBY is required for the Gal oxidase activity of intact seeds; the oxidation of Gal in side-chains of rhamnogalacturonan-I (RG-I) present in mucilage-modified2 (mum2) mucilage, but not in wild-type mucilage; the retention of branched RG-I in the seed following extrusion; and the enhancement of cell-to-cell adhesion in the seed coat epidermis. These data support the hypothesis that RUBY is a Gal oxidase that strengthens pectin cohesion within the middle lamella, and possibly the mucilage of wild-type seed coat epidermal cells, through oxidation of RG-I Gal side-chains.
INTRODUCTION
The emergence of multicellularity necessitated the development of mechanisms that promote cell-to-cell adhesion. In plants, cell adhesion is mediated largely through the middle lamella, an extracellular matrix rich in pectins and structural proteins that is shared between the walls of two adjacent cells (Zamil and Geitmann, 2017). Three major types of pectic polysaccharides have been described: homogalacturonan (HG), rhamnogalacturonan I (RG-I), and RG-II. HG has the simplest structure consisting of α-(1→4)–linked galacturonic acid (GalUA) monosaccharides (McNeil et al., 1984). By contrast, RG-I has a backbone composed of alternating rhamnose (Rha) and GalUA monosaccharides linked as [→2)-α-L-Rhap-(1→4)-α-D-GalpUA-(1→] (McNeil et al., 1980). In addition, the rhamnose can act as branch points for diverse oligosaccharide side-chains composed of Ara and/or Gal (Lau et al., 1987; Lerouge et al., 1993). RG-II has a backbone that resembles HG, but its side-chains are more complex, consisting of 12 monosaccharides linked in a specific manner that is conserved among many plant species (O’Neill et al., 2004). Immunolabeling experiments indicate that middle lamellae between cells contain HG, RG-I, and Hyp-rich glycoproteins (HRGPs; Moore et al., 1986; Smallwood et al., 1994; Bush et al., 2001; Willats et al., 2001). Mutants affecting cell adhesion have defects in one of the three pectic polysaccharides. Defects in biosynthesis of HG in quasimodo1 (qua1) and quasimodo2 (qua2) mutants result in loss of cell adhesion in hypocotyls and leaves (Bouton et al., 2002; Mouille et al., 2007). Similarly, when an HG-degrading polygalacturonase is ectopically expressed in apples (Malus domestica cv Royal Gala), the result is a loss of cell adhesion (Atkinson et al., 2002). Mutants for the putative polygalacturonase gene of Arabidopsis (Arabidopsis thaliana), QUARTET3, and its functional partner, the pectin methylesterase QUARTET1, lack pollen tetrad separation, which requires HG degradation (Rhee et al., 2003; Francis et al., 2006). A mutant affecting cell adhesion of Nicotiana plumbaginifolia callus, nolac-H18, lacks a portion of the RG-II side-chain, which prevents RG-II cross-linking through borate ions (Iwai et al., 2002). A number of studies have implicated the involvement of arabinans and galactans in cell-to-cell adhesion. For example, the N. plumbaginifolia cell adhesion mutant nolac-H14 lacks arabinans (Iwai et al., 2001). The tomato (Lycopersicon esculentum) Cnr mutant has a cell adhesion phenotype and a change in arabinan distribution in the cell walls of fruit pericarp (Orfila et al., 2001). Arabidopsis plants deficient in FRIABLE1, a putative O-fucosyltransferase, exhibit cell adhesion and organ fusion phenotypes, as well as changes in Ara and Gal-containing oligosaccharides in the Golgi apparatus (Neumetzler et al., 2012). A hallmark of fruit softening in many species is the removal of arabinan and galactan side-chains. For example, the softening of nectarines (Prunus persica) is associated with the degradation of RG-I–associated galactans and solubilization of pectins rich in arabinans (Dawson et al., 1992). In apples, these side-chains were specifically assigned to RG-I (Peña and Carpita, 2004). In ripe carambola (Averrhoa carambola cv B10) fruit, β-galactosidase was found to be involved in the removal of galactans, and responsible for solubilization of pectins and tissue softening (Balasubramaniam et al., 2005). Despite this evidence, the mechanism(s) through which arabinans and galactans associated with RG-I and cell wall structural proteins influence cell-to-cell adhesion remain unclear.
Arabidopsis seed coat mucilage is a useful tool for using genetics to study interactions between cell wall components (Haughn and Western, 2012). Seed coat mucilage is produced by seed coat epidermal cells that differentiate from integument cells of the ovule following fertilization. During the first few days of differentiation, the seed coat epidermal cells increase in size ∼3-fold and change in shape from cuboid to hexagonal. Mucilage is then synthesized in large amounts and deposited into a specific domain of the apoplast to form a doughnut-shaped pocket surrounding a volcano-shaped cytoplasmic column (Beeckman et al., 2000; Western et al., 2000; Windsor et al., 2000). After mucilage deposition into the apoplast, the cells deposit a thick cellulosic secondary wall, called the columella, that completely replaces the cytoplasm of the cell by seed maturity (Western et al., 2000; Mendu et al., 2011). Upon imbibition, the mucilage expands, breaks the primary walls of the epidermal cells, and extrudes to envelop the seed (Western et al., 2000; Windsor et al., 2000).
Mucilage contains all the major components of plant primary cell walls: cellulose, hemicelluloses, proteins, and pectins. Of these, pectin, and more specifically RG-I, is the most abundant component (Western et al., 2000; Macquet et al., 2007a;). RG-I in extruded wild-type seed coat mucilage is mostly unbranched (Dean et al., 2007; Macquet et al., 2007a;). Mutants with seed mucilage RG-I that has an increased number of side-chains with Gal and/or Ara, beta-xylosidase1 (bxl1) and mucilage-modified2 (mum2), exhibit strong mucilage extrusion defects (Western et al., 2001; Dean et al., 2007; Macquet et al., 2007b; Arsovski et al., 2009). BXL1 encodes a bifunctional β-D-xylosidase/α-L-arabinofuranosidase that acts primarily as an α-L-arabinofuranosidase on (1→5)-α-L-arabinan in the mucilage and primary radial cell walls of the seed coat epidermal cells (Arsovski et al., 2009). MUM2 encodes a β-galactosidase that is believed to remove terminal Gal residues from RG-I in the mucilage. The mum2 mutants, lacking this β-galactosidase activity, produce mature seed mucilage with more highly branched RG-I that cannot expand when exposed to water, thus preventing normal extrusion (Western et al., 2001; Dean et al., 2007; Macquet et al., 2007b). The availability of viable mutants affecting RG-I properties through side-chain modifications makes mucilage a much better system for investigation of these phenomena than middle lamellae, which play essential biological roles.
To investigate the role of galactans and arabinans in pectin cohesion, we undertook a forward genetic approach to find suppressors of the mum2 phenotype. Here we demonstrate that one suppressor mutation, ruby particles in mucilage (ruby), ameliorates the ability of mum2 mucilage to expand, and, in addition, disrupts cell adhesion in the seed coat epidermis. RUBY encodes a putative Gal oxidase that appears to strengthen the middle lamellae and perhaps mucilage through branched RG-I. These data suggest a new type of reinforcement of the middle lamellae between seed coat epidermal cells, provide evidence for a biological role of plant Gal oxidases, and demonstrate the importance of arabinogalactan side-chains and oxidation in cell wall biology.
RESULTS
Mutations in RUBY can Suppress the mum2 Mucilage Extrusion Phenotype
To investigate the mechanism by which RG-I side-chains influence mucilage extrusion, we used a genetic modifier screen to find suppressor mutations of mum2. A population of mum2-1 seeds was mutagenized with ethyl methanesulfonate (EMS), and M3 seeds from individual M2 plants were screened for wild-type–like mucilage extrusion when exposed to water. From 2469 M2 lines screened, 3 lines extruded a mucilage capsule similar to wild type, but with small particles that stained dark red when treated with ruthenium red (Figures 1C, 1D, 1G, and 1H). Based on phenotypic ratios in a cross of the ruby-1 mum2-1 double mutant to mum2-1, ruby-1 segregated as a single nuclear recessive mutation (3 mum2:1 suppressor; Χ2 = 0.0045662, df = 1, P = 0.9461, n = 73). Allelism tests confirmed that all three mutants were homozygous for mutant alleles of the same gene (Figures 1I to 1K). Based on the novel phenotype (Figure 1D, arrowheads), we named this gene RUBY, and accordingly the mutant alleles ruby-1, ruby-2, and ruby-3. To determine the mucilage phenotype of the ruby single mutants, ruby single mutants were isolated from the F2 of crosses between ruby mum2-1 double mutants and wild type. The extruded mucilage of ruby-1 and ruby-3 was similar to that of mum2-1 ruby-1 and mum2-1 ruby-3, respectively (compare Figures 1C and 1D, and 1G and 1H) in all aspects of the observed phenotypes, whereas ruby-2 resembled wild type more than mum2-1 ruby-2 (compared with Figures 1E and 1F). In addition, mum2-1 ruby-1 and mum2-1 ruby-3 (Figures 1C and 1G) demonstrated stronger suppression than mum2-1 ruby-2 (Figure 1E). Further phenotypic characterization was done on ruby-1.
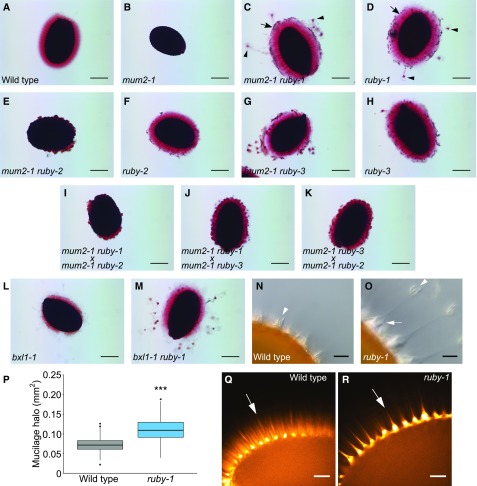
Figure 1.
ruby Exhibits Multiple Seed Coat Mucilage Phenotypes.
(A) to (M) Seeds agitated in water for 2 h and stained with ruthenium red. Bars = 200 μm.
(A) to (H) Three suppressor lines homozygous for an allele of ruby and mum2. Shown are Wild type (Col-2) (A), mum2-1 (B), mum2-1 ruby-1 (C), ruby-1 (D), mum2-1 ruby-2 (E), ruby-2 (F), mum2-1 ruby-3 (G), and ruby-3 (H). Black arrowheads indicate ruby particles in mucilage. Black arrows indicate primary cell wall being released in sheets.
(I) to (K) F2 seeds harvested from individual F1 plants resulting from crosses between different suppressor lines. Shown are mum2-1 ruby-1 × mum2-1 ruby-2 cross (I), mum2-1 ruby-1 × mum2-1 ruby-3 cross (J), and mum2-1 ruby-3 × mum2-1 ruby-2 cross (K). Note that all pairwise crosses failed to complement each other with respect to ruby suppression.
(L) and (M) ruby can suppress bxl1-1. Shown are bxl1-1 (L) and bxl1-1 ruby-1 (M).
(N) and (O) Nonstained wild-type (N) and ruby-1 (O) seeds imaged using differential interference contrast (DIC) microscopy after shaking in water. White arrow indicates columella without primary cell wall attached in the mutant. White arrowhead indicates primary cell wall attached to the top of columella in wild-type seed and detached in the mutant. Bars = 20 μm.
(P) Box-plot showing surfaces of adherent mucilage halo for wild type and ruby-1. Asterisks indicate P < 0.001 based on Welch’s t test. n = 130.
(Q) and (R) Seeds hydrated and agitated in water, followed by staining with Pontamine Fast Scarlet 4B. Shown are wild type (Q) and ruby-1 (R). Arrows indicate cellulosic rays. Bars = 50 μm.
RUBY is Required for Mucilage Integrity
Aside from the strong suppression of the mum2 extrusion phenotype and the presence of ‘ruby’ particles, the extruded mucilage of ruby seeds possessed several other phenotypes that could be identified by light microscopy. First, the ruby adherent mucilage halo was not smooth like wild type, but had rough edges giving the halo a dishevelled appearance (Figures 1C and 1D, arrows). Second, the outer primary cell walls of ruby epidermal cells frequently detached from seeds (Figure 1O, arrowhead), instead of remaining attached to the top of columellae (Figure 1N). Third, the adherent mucilage halo appeared larger than that of wild type. To confirm this observation, we measured the area occupied by the adherent mucilage of ruthenium red–stained seeds as described by Voiniciuc et al. (2015). Based on pair-wise comparison using two-tailed Welch’s t test, the ruby halo is significantly larger than that of wild type (Figure 1P). These data indicate that RUBY may have a role in making the mucilage halo more compact. Fourth, we compared the extrusion dynamics of ruby to wild type by exposing mature dry seeds to 0.02% ruthenium red and filming mucilage extrusion. Wild-type seeds readily extruded mucilage within seconds, and the nonadherent halo expanded rapidly (Supplemental Figure 1A; Supplemental Movie 1). When compared with the nonextruding mum2 (Figure 1B; Supplemental Movie 2), the ruby mutant appears to extrude at a similar rate to wild type (Supplemental Figure 1D; Supplemental Movie 3), but, unlike wild type, parts of ruptured primary cell walls are lifted from the seed surface. Fifth, when seeds are stained for cellulose with Pontamine Fast Scarlet 4B, wild-type seeds show prominent rays attached to the surface of columellae (Figure 1Q, arrow; Griffiths et al., 2014). By contrast, in ruby-1 mutant seeds we observed what seems to be a collapsed cellulosic ray above the columellae, changing the shape of rays from rod-like to conical (Figure 1R, arrow). This implicates RUBY in the organization of mucilage cellulose.
To understand whether the ability of the ruby mutation to suppress mucilage extrusion defects is specific to mum2, we crossed ruby to bxl1. The bxl1-1 mutant exhibits patchy, reduced, mucilage extrusion (Figure 1L) that occurs more slowly than in wild type (Supplemental Figure 1E), as well as an increase in terminal Ara residues on RG-I (Arsovski et al., 2009). The bxl1-1 ruby-1 double mutant extrudes mucilage more rapidly than bxl1-1 (Figure 1M; Supplemental Figure 1F), demonstrating that ruby is a suppressor of bxl1-1. These data show that the ability of ruby mutations to suppress the loss of mucilage expansion is not specific to mum2 mucilage.
RUBY is Required for Cell-to-Cell Adhesion
One of the most striking visual phenotypes is the presence of small particles, which we termed rubies, at the edge of the adherent mucilage both in ruby and mum2 ruby seeds (Figures 1C and 1D, arrowheads). Based on the shape and size of the particles, we hypothesized that they were seed coat epidermal cells that had separated from the underlying cell layer (palisade). To test this hypothesis, mature hydrated seeds were stained with the β-glucan stain calcofluor white to observe the seed surface. In wild-type seeds, epidermal cells are in close contact separated only by middle lamellae (Figures 2A and 2C), whereas in ruby seeds separation can be observed between the individual cells and in some places, large cell-sized gaps are apparent on the seed surface (Figures 2B and 2D, arrowheads) indicating that columellae had detached. The number of the observed gaps depends on the degree of shaking during hydration (Supplemental Figure 2). To test whether the cell-to-cell adhesion also requires divalent cation bridges formed between HG domains, ruby seeds were treated with a chelator, EDTA, to remove divalent cations, and CaCl2 to provide calcium. The addition of EDTA enhanced the loss of epidermal cells, whereas the addition of CaCl2 suppressed the loss of epidermal cells, suggesting that as in most cell types, Ca2+ bridges have roles in mediating cell adhesion. Therefore, RUBY activity and calcium bridges both contribute to this process. Because EDTA treatment of wild-type seeds alone does not result in loss of cell-to-cell adhesion (Western et al., 2001; Rautengarten et al., 2008; Voiniciuc et al., 2013), the separation of epidermal cells in ruby suggests that RUBY-mediated cell-to-cell adhesion seems to be more important in the seed coat.
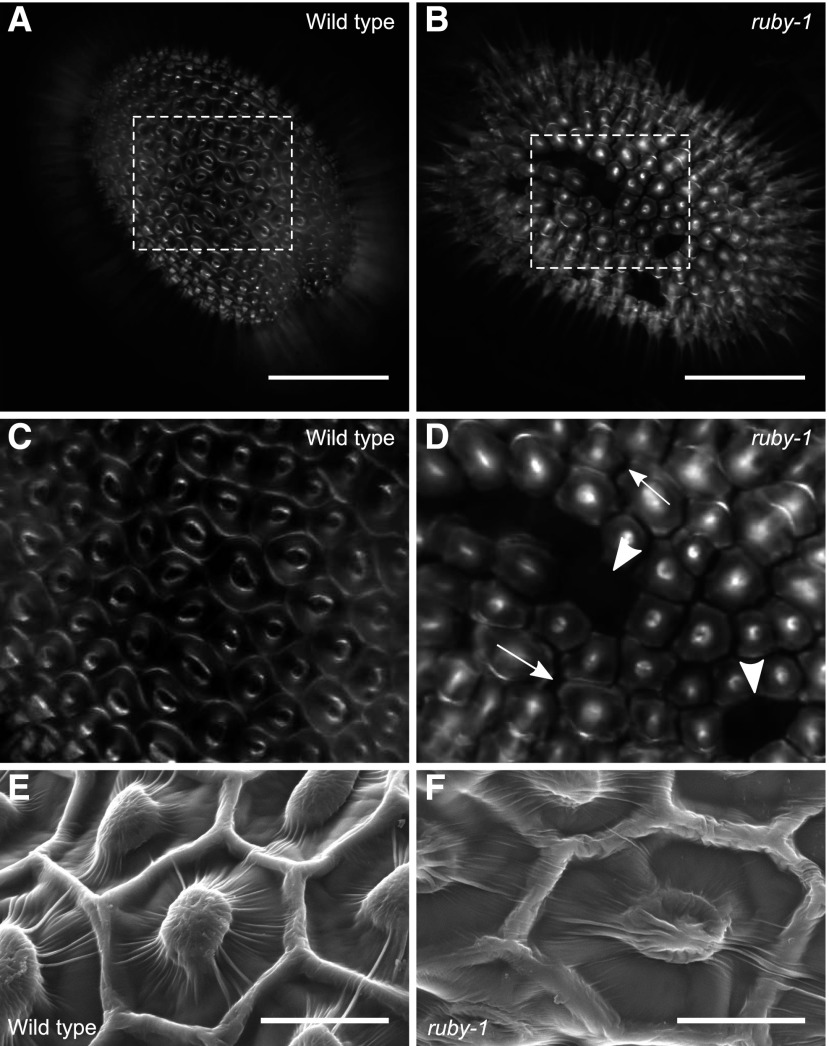
Figure 2.
RUBY is Involved in Cell-To-Cell Adhesion between Seed Coat Epidermal Cells as well as between Seed Coat Epidermal Cells and Palisade Cells.
(A) to (D) Mature seeds agitated in water for 2 h and stained with calcofluor white. Bars = 200 μm. Shown is the surface of wild-type (Col-2) (A) and ruby-1 seed (B).
(C) and (D) Magnified areas of seeds marked by dashed rectangles in (A) and (B). White arrowheads indicate two of the spaces where epidermal cells are missing. White arrows indicate two of the cracks between epidermal cells.
(E) and (F) Scanning electron micrographs of dry seeds. Bars = 20 μm. Shown are wild type (E) and ruby-1 (F).
To determine whether the cell separation occurs before mucilage extrusion, the surfaces of dry wild-type and mutant seeds were examined using scanning electron microscopy. No significant difference in the appearance of the cell edges of dry mature seeds were apparent between wild type and ruby (n = 6 seeds; Figures 2E and 2F), indicating that the cell-to-cell adhesion defects are apparent only upon hydration. Indeed, the ruby epidermal cells were observed to lift from the surface of the seed during extrusion (Figure 1O, arrow; Supplemental Movie 4), corroborating the hypothesis that the cells separate due to mechanical forces generated by the mucilage extrusion.
RUBY is Needed for Retention of an Arabinogalactan-Branched RG-I to the Seed
The visual aspects of ruby phenotypes suggest that structural changes occur in the mucilage and/or middle lamella. To test this possibility, mucilage was extracted from wild-type, ruby, mum2, and mum2 ruby seeds, and the monosaccharide composition determined. Surprisingly, large increases in the amount of Rha (1.29×), GalUA (1.3×), Ara (49.6×), and Gal (17×) relative to wild type were observed in the mucilage of ruby seeds (Figure 3A). Stoichiometrically, there were approximately two molecules of Ara and one of Gal for every new molecule of Rha and GalUA, suggesting that ruby mucilage contains an arabinogalactan-branched RG-I fraction not previously observed in extruded wild-type mucilage. This additional pectin may explain why the halo size is increased in the ruby mutant. The appearance of a novel polysaccharide observed in the mutant may have occurred in one of three ways. First, the new RG-I may be tightly linked to the epidermal cells of wild-type seeds, including the middle lamella, and not released with the mucilage. Second, the novel RG-I may be synthesized and present in the mucilage of the mutant, but not the wild type. Third, the branched RG-I may be synthesized in both wild type and ruby but processed differently, resulting in wild type having only occasional Gal branches and ruby having frequent branches containing both Gal and Ara. To distinguish between these hypotheses, a monosaccharide analysis of the whole seed alcohol-insoluble residue was completed for wild type and ruby. The only significant difference observed between wild-type and ruby seed monosaccharide content was an increase in Ara in ruby samples (Figure 3B; Supplemental Data Sets 1 and 2). The levels of Gal, Rha, and GalUA in wild-type and ruby seeds were similar, suggesting that the ruby mutation results in the release of a branched RG-I that is present but not released from wild-type seed. The increase in Ara suggests that some of the Ara residues may be absent in the side-chains of the branched RG-I present in wild type.
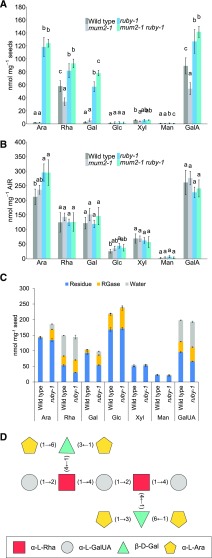
Figure 3.
Branched RG-I is Present in ruby Mucilage.
(A) Monosaccharide composition of Na2CO3-extracted mucilage as mean values of 4 biological replicates ±sd.
(B) Monosaccharide composition of the whole seed alcohol-insoluble residue as mean values of 4 biological replicates ±sd. Letters above bars represent groups determined by Tukey’s HSD test (α = 0.05), following one-way ANOVA performed for each monosaccharide independently (Supplemental Data Sets 1 and 2).
(C) Monosaccharide composition of sequentially extracted mucilage using water (nonadherent mucilage) and RGase (adherent mucilage), and residue left after the extractions. Each stack represents a mean of 3 biological replicates for a given fraction ±sd.
(D) Model of RG-I released in ruby mucilage based on monosaccharide composition, linkage analysis (Table 1), and LC-MS/MS analysis of RGase digests (Supplemental Figure 3C).
To confirm the existence of a branched RG-I that is released with mucilage in the ruby mutant but not in wild type, nonadherent mucilage was extracted with water and then adherent mucilage was extracted from the same seeds with RGase enzyme, and a monosaccharide analysis of each mucilage layer and the remaining “naked” seeds was performed (Figure 3C). Both mucilage layers were enriched in Ara, Gal, Rha, and GalUA, and reduced in the “naked” seeds in ruby samples, demonstrating that the ruby mutation indeed results in the release of a branched RG-I that is present but not released from wild-type seeds. The increase in total Ara in ruby seeds was confirmed.
The structure of this novel polysaccharide in ruby mucilage was investigated by carbohydrate permethylated alditol acetate (PMAA) linkage analysis of neutral monosaccharides in the nonadherent mucilage layer. The largest differences in mole percentages of linkages observed in ruby mucilage relative to that of wild type were increases in (t)-Araf, 3,6-Gal, and 2,4-Rha, and a decrease in 2-Rha (Table 1). These data suggest that the novel RG-I released in ruby mucilage is composed primarily of (t)-Araf, 3,6-Gal, and 2,4-Rha. The decrease in the percentage of 2-Rha in ruby mucilage reflects the fact that unbranched RG-I constitutes a lower percentage of the total. The mole percentages of monosaccharide linkages that showed increase and values for the same monosaccharides in nanomoles permilligram of seed from the monosaccharide composition were used to calculate the ratios to ascertain the structure of the polysaccharide from which they were derived. Total mole percent values for all PMAAs of a single monosaccharide were added, and the percentage of a specific linkage for that given monosaccharide was calculated and multiplied by the absolute values obtained from monosaccharide compositional analysis. For each 2,4-Rha there were 1.1 3,6-Gal, and for each 3,6-Gal there were 2.25 t-Araf, consistent with a branched RG-I where each branched backbone rhamnose has a molecule of Gal attached to it by a β-(1→4) linkage, which in turn has two molecules of t-Araf attached through C-3 and C-6 positions of the pyranose ring. This structure was confirmed by digesting wild-type and ruby adherent mucilage RG-I with RGase and analyzing products by high performance anion-exchange chromatography with pulsed amperometric detection (HPAEC-PAD) and Ion Pairing-Reverse Phase (IP-RP)–ultra-HPLC tandem mass spectrometry (UHPLC-MS/MS). The digest of wild type showed four distinct peaks with retention time lower than 15 min (Supplemental Figure 3A, a-d). These peaks were found to contain one to three rhamnoses (R) and two to three galacturonic acids (U), indicating an RG-I with a “naked” backbone with no branching. Additional peaks with higher retention time were identified in ruby, but not wild-type mucilage digests (Supplemental Figure 3B, asterisks). To reveal the structure of these carbohydrate molecules, they were analyzed using IP-RP UHPLC-MS/MS. In the first step, the digest of wild type and the digest of ruby were compared by IP-RP-UHPLC-MS (Supplemental Figure 3C). The main retention process in IP-RP is based on the number of acidic functional groups. In the area of species that contained two acidic functional groups (between retention time 8.0 and 11.0 min), a peak, specific to ruby, was identified at retention time 9.54 min. This oligosaccharide was selected for IP-RP-UHPLC-MS/MS analysis. Based on the spectrum obtained (Supplemental Figure 3D) the oligosaccharide that was present in ruby extracts but not in wild-type extracts, which contained the fewest acidic functional groups and the lowest molecular weight, contained two hexoses and four pentoses in addition to two Rha and two GalUA residues usually found in digests of RG-I backbone. By using the intense fragment at m/z 747.3, which corresponds to a Z2 and/or B2, we can postulate that each dimer (Rha-GalUA) in the characterized peak carries a lateral group consisting of 1 hexose and 2 pentoses.
Table 1. Monosaccharide Linkage Analysis of Wild-Type and ruby-1 Mucilage.
| Monosaccharide and Linkage | Wild type | ruby-1 |
|---|---|---|
| Fuc | ||
| t-Fuc | 0.20 ± 0.28 | ND |
| Rhamnose | ||
| t-Rha | 2.15 ± 0.07 | 0.70 ± 0.28 |
| 2-Rha | 64.5 ± 1.56 | 12.20 ± 4.81 |
| 2,3-Rha | 2.10 ± 0.28 | 0.50 ± 0.28 |
| 2,4-Rha | 8.35 ± 4.74 | 19.15 ± 0.49 |
| 2,3,4-Rha | 2.45 ± 0.64 | 1.70 ± 0.14 |
| Ara | ||
| t-Araf | 1.30 ± 1.27 | 24.25 ± 1.20 |
| t-Arap | 0.60 ± 0.14 | ND |
| 3-Araf | ND | 0.30 ± 0 |
| 5-Araf | 1.55 ± 0.21 | 1.80 ± 0.57 |
| 3,5-Araf | ND | 1.20 ± 1.41 |
| Xyl | ||
| 4-Xyl | 1.80 ± 0 | 0.20 ± 0 |
| 2,4-Xyl | 0.70 ± 0 | ND |
| Man | ||
| t-Man | ND | 0.10 ± 0.14 |
| 2-Man | 1.10 ± 0.28 | ND |
| 4-Man | 1.25 ± 0.35 | 0.40 ± 0.14 |
| 4,6-Man | 0.80 ± 0 | ND |
| Gal | ||
| t-Gal | 1.10 ± 0.14 | 0.70 ± 0.14 |
| 3-Gal | 0.55 ± 0.07 | 1.65 ± 0.07 |
| 4-Gal | ND | 2.90 ± 2.05 |
| 6-Gal | 0.10 ± 0.14 | 1.25 ± 0.21 |
| 3,6-Gal | ND | 28.35 ± 4.31 |
| 2,4,6-Gal | ND | 0.30 ± 0.14 |
| 3,4,6-Gal | ND | 0.50 ± 0.14 |
| Glc | ||
| t-Glc | 3.25 ± 0.35 | 0.50 ± 0.28 |
| 4-Glc | 6.05 ± 0.07 | 1.65 ± 0.49 |
The values indicate mean mol% ±sd of two biological replicates. ND, not detected. Major changes are marked in bold.
When monosaccharide composition and PMAA linkage data are considered, the hexose molecules are most likely Gal, whereas the pentose molecules are most likely Ara, suggesting an RG-I that has one Gal linked to each branched rhamnose in β-(1→4) linkage, and two Ara linked to each Gal, one in α-(1→3) and one in α-(1→6) linkage (Figure 3D).
RUBY Encodes a Putative Gal Oxidase
The ruby-1 mutation was mapped using a positional cloning approach. Thirty-two mutants were selected from an F2 population made by crossing mum2-1 ruby-1 to wild type Ler, and used to map RUBY to chromosome 1 between DNA markers on BACs F14F17 and F10B61. The genomic DNA of the 32 individuals was also pooled and sequenced, and the low heterozygosity of Col/Ler SNPs used to verify the position on chromosome 1. A mutation in this region was identified in At1g19900. The sequencing of At1g19900 in plants homozygous for two additional alleles of ruby (ruby-2 and ruby-3) also identified mutations, suggesting that At1g19900, a gene encoding a putative glyoxal oxidase-related protein, is RUBY (Figure 4A).
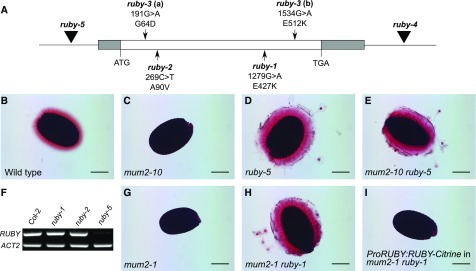
Figure 4.
RUBY Encodes a Glyoxal Oxidase-Like Protein.
(A) Schematic of the region of chromosome 1 containing RUBY (At1g19900). The gray bars represent untranslated regions, whereas the white bar represents the coding sequence. Black triangles indicate the positions of T-DNA insertions, and arrows show the positions of different EMS-induced point mutations. Numbers under EMS allele labels represent positions of mutations in the coding sequence of RUBY, and letters represent nucleotide changes. Predicted amino acid changes in the deduced protein resulting from the mutation are shown beneath the nucleotide changes.
(B) to (E) Ruthenium red–stained seeds demonstrating that the ruby-5 line with insertion upstream of At1g19900 has a similar phenotype to other ruby alleles. Shown are wild type (Col-0) (B), mum2-10 (C), ruby-5 (D), and mum2-10 ruby-5 (E).
(F) RT-PCR analysis of RUBY transcript levels in 11 DPA siliques. ACT2 (At3g18780) was used as an internal control.
(G) to (I) Ruthenium red–stained seeds showing that genomic At1g19900 can complement the ruby-1 mutation. Shown are mum2-1 (G), mum2-1 ruby-1 (H), and ProRUBY:RUBY-Citrine (I) in the mum2-1 ruby-1 background. Bars = 200 μm.
Reverse genetic analysis was used to further verify the identity of RUBY. Seeds from lines homozygous for T-DNA insertions in the At1g19900 gene were examined for seed mucilage phenotypes. One line, WiscDsLoxHs097_11H, displayed a phenotype similar to other ruby alleles (Figure 4D; Supplemental Figures 4A to 4C). The insertion was confirmed to be 162 bp upstream of the predicted transcription initiation site of At1g19900 (Figure 4A) and designated ruby-5. RT-PCR analysis of At1g19900 using whole siliques at 11 d post anthesis (DPA) suggested that At1g19900 has reduced transcript levels relative to wild type and plants homozygous for ruby alleles with point mutations (Figure 4F). The ruby-5 mutation, when introduced into a line carrying a T-DNA allele of mum2 (mum2-10), was able to suppress the mum2 phenotype (Figures 4B to 4E). An additional line, named ruby-4 (SALK_020627C), showed no aberrant phenotype (Supplemental Figures 4D and 4E) most likely because it carries an insertion downstream of the At1g19900 coding region (Figure 4A). These data support the hypothesis that At1g19900 is RUBY and that ruby mutations are able to suppress different alleles of mum2.
Additional evidence supporting the hypothesis that RUBY is At1g19900 was obtained by constructing an in-frame fusion of Citrine to the carboxyl terminus of a genomic clone of At1g19900. This genomic clone included the entire 5′ genomic region upstream of At1g19900. When transformed into mum2 ruby plants, this construct successfully complemented ruby, as demonstrated by T2 seeds of all independent transgenic lines exhibiting mum2-like phenotypes (Figures 4G, 4H, and 4I).
RUBY is annotated as a member of the Carbohydrate-Active Enzymes Database Auxiliary Activity 5 (AA5) family comprising AA5_1 subfamily (glyoxal oxidases, EC 1.1.3.15) and AA5_2 subfamily (Gal oxidases, EC 1.1.3.9). Amino acid sequence alignment was performed using sequences of AA5 enzymes with experimentally confirmed activities (Avigad et al., 1962; Kersten and Kirk, 1987; Kersten, 1990; McPherson et al., 1992; Leuthner et al., 2005; Aparecido Cordeiro et al., 2010; Paukner et al., 2014, 2015; Yin et al., 2015; Daou et al., 2016; Andberg et al., 2017). Based on amino acids involved in copper coordination and substrate oxidation in the active site, RUBY more closely resembles glyoxal oxidases than Gal oxidases (GalOx; Supplemental Figure 5). A Trp residue that was proposed to be necessary for the substrate specificity of GalOx enzymes is missing from the RUBY amino acid sequence and is replaced by Gly. However, based on research that used site-directed mutagenesis, such a substitution does not abolish GalOx activity (Rogers et al., 2007), indicating that it is possible that RUBY is a GalOx.
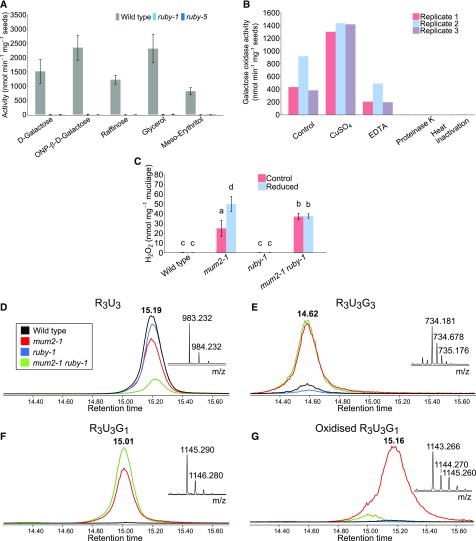
To test the enzymatic activity of RUBY, we attempted to express RUBY protein in multiple heterologous systems. Protein was expressed without its predicted signal sequence in yeasts to obtain properly folded and modified protein. We tested cytosolic and secreted expression in Saccharomyces cerevisiae, and secreted expression in Pichia pastoris, but the protein was either insoluble or degraded, and never successfully secreted. Escherichia coli expression yielded soluble protein when we expressed it at 11°C in Arctic Express cells, but the ensuing protein showed no activity against monosaccharides, glyoxal, methylglyoxal, or glyoxylic acid. We did observe that dry wild-type seeds were able to generate hydrogen peroxide (H2O2) in the presence of glycerol, whereas ruby-1 and ruby-5 seeds showed no such activity. This suggests that RUBY is required for such a reaction. Multiple monosaccharides, disaccharides, trisaccharides, polyols, alcohols, and carbonyl compounds were tested as substrates (Supplemental Table 1), and the reaction was further investigated for the compounds showing positive reactions. The only tested monosaccharide oxidized by wild-type seeds was Gal, suggesting that the enzyme performing the oxidation is a GalOx (Figure 5A). The reaction was inhibited by heating at 95°C and by Proteinase K, suggesting that the reaction is dependent on a protein (Figure 5B). Preincubation in CuSO4 substantially increased the activity, as has been previously observed for fungal GalOxs (Spadiut et al., 2010), and also reduced differences between biological replicates (Figure 5B). This suggests that the seed GalOx is activated by Cu2+, a cofactor in the active sites of GalOx enzymes. Likewise, preincubation in EDTA, a chelator of divalent cations, reduced the activity by about half (Figure 5B). Like fungal GalOx enzymes, seed GalOx uses glycerol and also meso-erythritol, as a substrate (Figure 5A). In addition, Gal-containing raffinose was oxidized, consistent with previous reports of GalOx enzyme activity (Avigad et al., 1962; Paukner et al., 2014, 2015). Oxidation of lactose was observed, but the reaction was very weak, again consistent with other GalOx enzymes (Avigad et al., 1962; Xu et al., 2000). For this reason, we were not able to further analyze the activity of the seed enzyme on lactose. Because raffinose, containing α-Gal, was oxidized, we tested whether this GalOx has a preference for the Gal α-anomer over the β-anomer using p-nitrophenyl-β-D-Gal (PNP-β-D-Gal) and o-nitrophenyl-β-D-Gal (ONP-β-D-Gal). Both were readily oxidized by wild-type seeds (Figure 5A; Supplemental Table 1), suggesting that the seed GalOx can use Gal regardless of anomerism. Gal-containing polysaccharides, i.e., galactomannan, guar gum, xyloglucan, and linear galactan, were not oxidized. Because the assays were done on seeds, it is possible that these large substrates could not access the enzyme, which is likely inside the columella, a cellulosic secondary cell wall. Oxidation of p-nitrophenyl-α-L-arabinofuranoside was observed (Supplemental Table 1), but the reaction was weak and thus not tested in a kinetic assay. These data demonstrate that RUBY is required for Gal oxidase activity of intact, newly hydrated seeds.
Figure 5.
RUBY is a Putative Gal Oxidase with mum2 Mucilage as a Substrate.
(A) Gal oxidase activity measured on whole seeds of wild type, ruby-1, and ruby-5 using HRP and TMB as a chromogenic substrate for detection of H2O2. Bars = means (n = 3 biological replicates); error bars represent standard deviations.
(B) Oxidation of D-gal by wild-type seeds assayed using HRP-TMB. The control was no treatment before activity measurement; seeds were assayed after pretreatment with CuSO4, EDTA, proteinase K, or heat (95°C). Bars = independently grown biological replicates.
(C) Oxidation of Na2CO3-extracted mucilage by commercial gal oxidase. H2O2 was measured using HRP-TMB. The mucilage samples were reduced using NaBH4 (reduced) or left untreated (control). Bars = means ±sd of 3 independently grown biological replicates. Letters above bars represent groups based on Tukey’s HSD test (α = 0.05), following two-way ANOVA. The main effect of genotype on substrate availability was significant (df = 3, F-value = 153.670, P < 0.001), as well as the effect of treatment (reduction) of samples on the substrate availability (df = 1, F-value = 13.347, P < 0.005), and interaction between genotype and treatment (df = 3, F-value = 12.966, P < 0.001).
(D) to (G) Extracted-ion chromatograms of RGase-digested mucilage obtained by IP-RP-UHPLC-MS for the four genotypes (wild type [Col-2], black trace; mum2-1, red trace; ruby-1, blue trace; mum2-1 ruby-1, green trace). Next to the chromatogram, the mass spectrum for the peak is represented. Separated oligosaccharides are labeled with regards to the number of R (Rha), U (GalUA), and G (Gal) they contain. Shown are (D) R3U3 isolated as [M-H]− at m/z 983.23; (E) R3U3G3 isolated as [M-2H]2− at m/z 734.18; (F) R3U3G1 isolated as [M-H]− at m/z 1145.29; and (G) oxidized R3U3G1 isolated as [M-H]− at m/z 1143.27. Exact masses of each compound were selected with a mass window of ± 0.1 Da.
It has been reported that unlike that of wild type, mum2 mucilage was found to have oxidized Gal residues attached to Rha (Macquet et al., 2007b). We extracted mucilage using Na2CO3 and tested it for the amounts of oxidizable Gal residues using commercial GalOx. As expected, mum2-1 and mum2-1 ruby-1 have the highest amounts of nonoxidized Gal (Figure 5C) due to an increase in t-Gal. This indicates that RUBY can promote oxidation of mum2 mucilage, but the seed GalOx may not act on wild-type mucilage, as suggested by the low levels of oxidation and lack of difference between wild type and ruby-1. To determine whether oxidation of mum2 mucilage makes it insoluble, we divided extracted mucilage samples, and then reduced putative aldehydes in one sample using NaBH4 and processed the other sample as a control without reduction. Strikingly, reduced mum2 mucilage showed twice as much available substrate than the corresponding nonreduced control (Figure 5C), suggesting that approximately half of the Gal is oxidized in extracted mum2 mucilage. This increase was not observed in reduced mum2 ruby mucilage compared with control, suggesting that the oxidation of mum2 mucilage is dependent on RUBY.
We further analyzed adherent mucilage from wild-type, ruby-1, mum2-1, and mum2-1 ruby-1 seeds. We first extracted nonadherent mucilage sequentially using mild acid and then mild alkali, and then further hydrolyzed the remaining adherent mucilage surrounding seeds with rhamnogalacturonan hydrolase, as described by Macquet et al. (2007b). Hydrolysates were analyzed by IP-RP-UHPLC-MS. Besides unbranched RG-I oligosaccharides, represented by R2U2 and R3U3, that were present in all samples (Figure 5D), several galactosylated RG-I oligosaccharides were detected in mum2-1 and mum2-1 ruby-1 (Figures 5E and 5F), with R3U3G1 being particularly abundant (Figure 5F). Interestingly, R3U3G1 is present in both mum2-1 and mum2-1 ruby-1 (Figure 5F), but oxidized forms of this oligosaccharide are present in mum2-1 only (Figure 5G). This shows that some Gal units are indeed oxidized in mum2 mucilage, and that this oxidation of mum2 mucilage is dependent on RUBY.
RUBY is Expressed in Seeds after Mucilage Secretion into the Mucilage Pocket and Localizes to the Apoplast in Columella
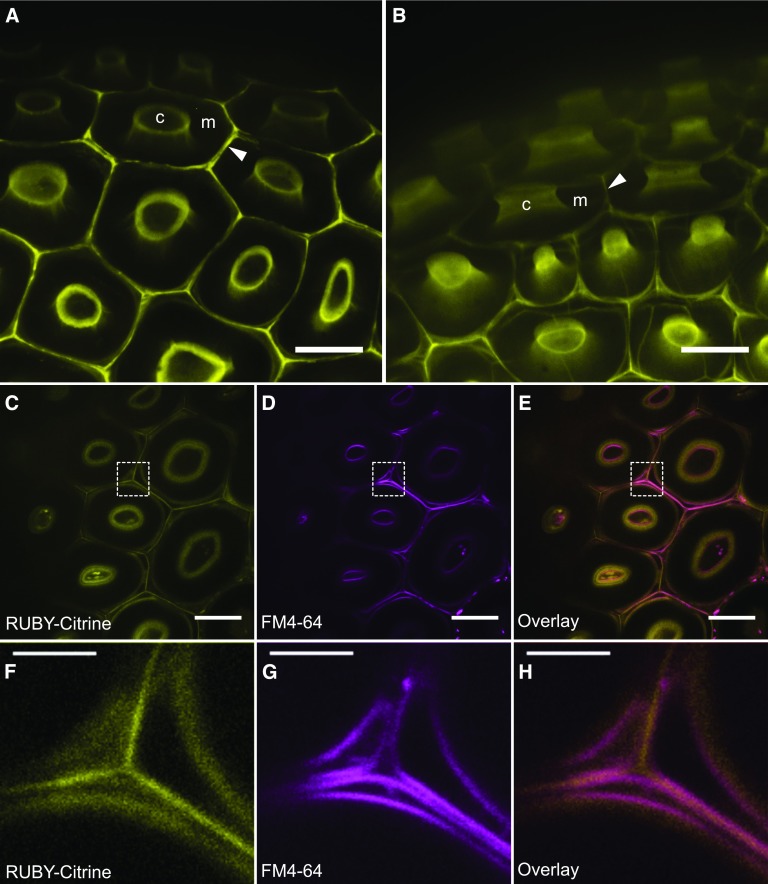
The ProRUBY:RUBY-Citrine construct that complemented ruby was used to study the temporal and spatial expression, and the subcellular localization of RUBY. Developing seeds of T2 plants were removed from siliques and imaged by spinning-disc confocal microscopy. No signal was observed before 9–10 DPA, indicating that RUBY is expressed following the completion of mucilage secretion into the mucilage pocket. RT-PCR analysis of RNA from developing siliques also demonstrated that RUBY is primarily expressed in siliques late in the development (Supplemental Figure 6D). The signal at 10 DPA was localized in the secondary wall of developing columella, and in the primary cell walls and middle lamellae around the cells (Figure 6A). At 13 DPA when the columella is fully developed (Western et al., 2000), signal continues to accumulate in the columella and primary cell walls surrounding epidermal cells (Figure 6B). The signal does not lose intensity even in fully developed dry seeds. These results demonstrate that RUBY, consistent with its roles in mum2 mucilage modification and cell-to-cell adhesion, localizes to the columella adjacent to the mucilage pocket, as well as to the primary cell walls surrounding cells. The signal is also visible around the cells in the underlying palisade cell layer (Figure 6B).
Figure 6.
RUBY-Citrine is Expressed After Mucilage Secretion Into the Mucilage Pocket and Localizes to the Apoplast.
Developing seeds carrying ProRUBY:RUBY-Citrine in the mum2-1 ruby-1 background imaged on a spinning disc confocal microscope are shown.
(A) Seed surface at 10 DPA. Bar = 20 μm.
(B) Seed surface at 13 DPA. Bar = 20 μm.
(C) to (E) Top view of the seed surface stained with FM4-64 dye. Bars = 20 μm.
(F) to (H) Magnified areas marked by dashed rectangles in (C) to (E). Bars = 5 μm.
(C) and (F) RUBY-Citrine shown in yellow.
(D) and (G) FM4-64 imaged shown in magenta.
(E) and (H) Overlay of RUBY-Citrine (yellow) and FM4-64 (magenta) demonstrating that RUBY-Citrine localizes outside of the plasma membrane. c, columella; m, mucilage pocket; arrowhead, middle lamella.
To test whether RUBY is indeed secreted, we examined localization in the apoplast by staining plasma membrane of ProRUBY:RUBY-Citrine developing seeds with the dye FM4-64. In the overlay of RUBY-Citrine (yellow; Figure 6C) and plasma membrane (magenta; Figure 6D) images, it is evident that RUBY localizes outside of the plasma membrane (Figure 6E), demonstrating extracellular localization of the protein. At earlier stages of development (9–10 DPA), it is possible to observe fluorescence inside the cells in punctate or reticulate patterns, most likely representing the protein in the secretory pathway before deposition into the apoplast.
Pectin Cohesion is Increased by Gal Oxidation
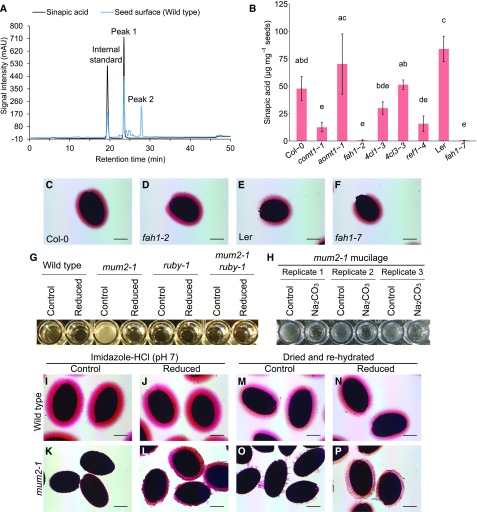
Cross-linking of RG-I via dimerization of ferulic acid (FA) attached to Ara and Gal side-chains of sugar beet RG-I, or Ara side-chains of arabinoxylan, has been previously reported (Grabber et al., 1995; Saulnier and Thibault, 1999; Fry, 2004; Ralet et al., 2005). Oxidative coupling of arabinoxylan-FA occurs in the presence of H2O2 and peroxidases (Encina and Fry, 2005; Burr and Fry, 2009). Because the RUBY reaction generates H2O2, we investigated whether RUBY functions to cross-link cell walls through dimerization of hydroxycinnamate esters.
We first treated mature seeds with 2 M NaOH to extract any ester-linked phenolic compounds that are present in the mucilage and the columella surface, and analyzed extracts by HPLC-UV. The most abundant phenolic compound detected was sinapic acid, confirmed by comparison with retention time (Supplemental Figure 7A) and UV absorbance of a standard, as well as a molecular mass [M-H]− of 223.0599 (Supplemental Figure 7C). Like FA, sinapic acid can also form dimers (Bunzel et al., 2003). If wild type, expressing functional RUBY, makes dimers of sinapic acid, it would be expected to have lower levels of sinapic acid (monomers) than ruby. However, we did not observe differences in sinapic acid between wild type and ruby (Supplemental Figure 7A), suggesting that it exists only as a monomer on the seed surface. A second compound was detected and was reduced by ∼30% in ruby compared with wild type (Supplemental Figure 7B). Based on a molecular mass [M-H] − of 447.0906 (Supplemental Figure 7C) and published results on Arabidopsis seed phenolics (Routaboul et al., 2006), this compound is most likely quercetin-3-O-rhamnoside.
To test whether lack of sinapate has an effect on the whole seed mucilage phenotype, we stained seeds of mutants known to be involved in sinapic acid biosynthesis, with ruthenium red. Based on HPLC-UV quantification, fah1-2 and fah1-7 mutants have almost complete reduction in sinapic acid (Figure 7B). However, we observed no difference in seed mucilage phenotype between these mutants and wild type (Figures 7C to 7F), indicating that hydroxycinnamates are likely not involved in RUBY-mediated cross-linking.
Figure 7.
RUBY Modifies Mucilage Properties directly through Aldehydes.
(A) HPLC-UV chromatograms representing sinapic acid standard (black) and surface phenolics of wild-type (Col-2) seeds released by 2 M NaOH (blue). Vanillin was added as an internal standard.
(B) Quantification of sinapic acid released from seed surface of mutants for genes in the sinapic acid biosynthetic pathway. Bars = mean ±sd of 3 independently grown biological replicates. Letters above bars represent groups assigned based on Tukey’s HSD test, following one-way ANOVA (df = 8, F-value = 21.939, P < 0.001).
(C) to (F) Seeds agitated in water and stained with ruthenium red. Shown are Col-0 (wild type) (C), fah1-2 mutant (Col-0 background) (D), Ler (wild type) (E), and fah1-7 mutant (Ler background) (F).
(G) Photograph of rehydrated mucilage samples in 96-well plate after Na2CO3 extraction, reduction, and purification, demonstrating solubility/insolubility of mucilage.
(H) Na2CO3 can break cross-links made by RUBY. Mucilage extracted from mum2-1 seeds with Na2CO3, purified, dried, and rehydrated. One sample was mixed with water (Control) and the other with Na2CO3.
(I) to (L) Reduction promotes mucilage extrusion from mum2 seeds. Seeds were incubated with NaBH4 at neutral pH. Shown are wild type (Col-2) without NaBH4 (I), wild type (Col-2) with NaBH4 (J), mum2-1 without NaBH4 (K), and mum2-1 with NaBH4 (L).
(M) to (P) Drying promotes insolubility of mum2 mucilage through aldehydes. Images show rehydrated seeds after base-extraction of mucilage, reduction, and air-drying. Shown are wild type (Col-2) without NaBH4 (M), wild type (Col-2) with NaBH4 (N), mum2-1 without NaBH4 (O), and mum2-1 with NaBH4 (P). Scale bars = 200 μm.
Further investigation of mechanisms by which RUBY may function was guided by the observation that purified nonreduced (control) mum2 mucilage was unable to fully hydrate in water, resulting in increased opacity of the solution (Figure 7G). The reduced mum2 sample, however, rehydrated to a higher degree, resulting in a transparent solution (Figure 7G). This result suggests that oxidation reduces the solubility of mum2 mucilage. When mixed with Na2CO3, mum2 mucilage became more soluble, as evident by a loss of opacity (Figure 7H). This result demonstrates that the cause of the poor hydration of oxidized mucilage can be disrupted by Na2CO3.
To determine the relevance of Gal oxidation for mucilage extrusion, we tested whether the reduction of carbonyls by NaBH4 can release mucilage from mum2. NaBH4 reductions are usually performed in basic solutions to prevent its decomposition, but basic solutions can also extract mucilage from mum2 seeds, which would obscure the effects of reduction on mucilage hydration properties. To avoid using basic solutions and to prevent pH shift upon addition of NaBH4, we performed reduction in imidazole-HCl buffer at pH 7, which was previously suggested to increase stability of NaBH4 (Kim and Carpita, 1992). We observed a release of mucilage from mum2-1 seeds in the presence of the reductant (Figure 7L), whereas control seeds showed patchy extrusion only sporadically (Figure 7K). Seeds that extrude even small amounts of mucilage and seeds with no mucilage after the NaBH4 treatment were counted and compared with control seeds (Supplemental Figure 8). NaBH4-treated mum2 seeds displayed extrusion 80% to 90% of the time, whereas only 10% to 20% of control seeds extruded. When observed, the extrusion in control seeds was limited and patchy. The reason for extrusion may be a pH higher than that of water, which may promote extrusion to a limited extent. The wild-type seeds displayed no clear difference with regard to the treatment (Figures 7I and 7J). This result indicates that the oxidation of Gal into an aldehyde makes mucilage insoluble in vivo.
It has been suggested that polysaccharides oxidized by GalOx can form insoluble aerogels when dried through the formation of hemiacetals (Mikkonen et al., 2014). To test whether drying can make mucilage insoluble, we reduced wild-type and mum2 seeds in a basic solution to ensure mucilage extrusion, air-dried them, and rehydrated them in water. The mucilage of mutant seeds rehydrated only after reduction with NaBH4 (Figure 7P), whereas mucilage of nonreduced control remained collapsed (Figure 7O). Similar to other experiments, wild type showed no difference between treatments (Figures 7M and N). The fact that oxidized mucilage becomes insoluble when dried is consistent with the hypothesis that hemiacetal formation could be responsible, as has been observed in studies of aerogels.
DISCUSSION
We have identified an Arabidopsis gene, RUBY, which encodes a protein annotated as a GalOx in the AA5 family and is secreted by seed coat epidermal cells late in seed development. Functional RUBY protein is required for GalOx activity of intact seeds, the retention of a branched class of RG-I in hydrated seed, the oxidation of Gal side-chains of RG-I in mum2 mucilage, and normal cell adhesion and mucilage structure in the seed coat of Arabidopsis. The gene is expressed late in the differentiation of seed coat epidermal cells, and is secreted to the apoplast, consistent with a role in modification of the middle lamellae and mucilage. Taken together, these results suggest that RUBY is a GalOx that oxidizes Gal side-chains of RG-I to promote greater cohesion in pectin of the middle lamellae and mucilage. However, because we were unable to demonstrate that purified RUBY has GalOx activity, we cannot eliminate the possibility that the requirement of RUBY for seed GalOx activity is indirect. Therefore, RUBY is most accurately described as a putative GalOx, and the seed oxidase as RUBY-associated seed oxidase activity.
The RUBY-associated GalOx activity detected in intact seed appears very similar to that of fungal GalOxs, which have been well-characterized enzymatically (Avigad et al., 1962). Gal is the only monosaccharide that is oxidized, whereas glycerol, raffinose, and meso-erythritol can also serve as substrates. In addition, Cu2+ is needed for full activity. The exact substrate for the RUBY-associated GalOx detected in wild-type seed coat epidermal cells is not known, but several lines of evidence suggest Gal in RG-I side-chains as at least one substrate. First, RUBY promotes cell adhesion, which normally occurs via the pectin of the middle lamella, and RG-I is a pectin with an abundance of Gal side-chains. Second, mutations in ruby result in the release of a branched RG-I from the seed coat epidermal cells, which is present but not released in wild type, suggesting that RUBY functions to cross-link the RG-I to the cell surface. Finally, RG-I in mum2 mucilage contains oxidized Gal whose formation is dependent on RUBY (Figures 5C, 5F, and 5G; Macquet et al., 2007b).
The branched RG-I extracted with the mucilage of ruby mutants has a distinct structure, not previously described, where each molecule of branched rhamnose is covalently bonded to one molecule of β-D-Gal, which in turn is linked, via carbons 3 and 6, to two molecules of Ara (Figure 3D). This RG-I appears to be present in wild-type epidermal cells, but is extracted from the surface of the seed with the mucilage only in the absence of functional RUBY. These data suggest that RUBY is involved in the cross-linking of branched RG-I to the seed coat epidermal cell even though the molecule has no terminal Gal substrate to be oxidized. This cross-linking could be explained in one of three ways. The terminal Ara residues on the RG-I side-chains might interact with oxidized Gal on other carbohydrates. Because some oxidation on PNP-α-L-Araf was observed (Supplemental Table 1), the second possibility is that the t-Araf of the RG-I side-chains can be used as substrates in the absence of Gal. Alternatively, we did observe a consistent increase in total Ara in ruby versus wild-type seeds (Figure 3B), suggesting that at least some of the Ara is not present in the wild-type cells but is added to the branched RG-I in the ruby mutant. If so, the branched RG-I present in the wild type that is bound to the seed surface may lack many of the terminal Ara molecules observed in ruby mucilage and instead have primarily Gal side-chains, making it a possible direct substrate for seed GalOx(s). The exact location of the branched RG-I is unknown, but it must be in the middle lamella, and/or on the surface of the columella and primary wall of the epidermal cells. At least some of the branched RG-I released from ruby seeds could be associated with the epidermal cells that separate from the seed surface with the mucilage.
Our results suggest that RUBY promotes connections between the middle lamellae of adjacent seed coat epidermal cells, and between seed coat epidermal cells and the underlying palisade cells via oxidation of Gal. The oxidation of Gal could promote carbohydrate cross-linking through at least two nonmutually exclusive mechanisms. First, it has been shown in a variety of plant species that hydroxycinnamic acids can be covalently bonded to Gal or Ara and then oxidatively cross-linked by H2O2 and peroxidases (Ralph et al., 1994; Grabber et al., 1995; Saulnier and Thibault, 1999; Bunzel et al., 2003; Fry, 2004; Encina and Fry, 2005; Ralet et al., 2005; Burr and Fry, 2009). Therefore, the availability of Gal or Ara side-chains on RG-I in the apoplast of seed coat epidermal cells could provide substrate for the covalent bonding to a hydroxycinnamic acid. Oxidation of Gal by GalOx would generate the H2O2 needed to cross-link two molecules of hydroxycinnamic acid attached to different carbohydrate chains. Whereas this hypothesis is consistent with much of our data, we were unsuccessful in our attempt to find direct evidence to support the involvement of hydroxycinnamic acids as a structural element in either middle lamellae or mucilage. Sinapic acid was the only hydroxycinnamic acid we could detect in seeds, and fah1-2 and fah1-7 mutant seeds lacking sinapic acid (Figures 7C to 7F) did not show seed coat epidermal defects in either cell-to-cell adhesion or mucilage cohesion. These data suggest that cross-linking of hydroxycinnamic acids is not a mechanism used in the apoplast of seed coat epidermal cells. Worth noting is that an additional metabolite with reduced levels in ruby mutant was detected on the seed surface (Figure 7A; Supplemental Figure 7B). Based on molecular mass (Supplemental Figure 7D) and previous studies on flavonoids in Arabidopsis seeds (Routaboul et al., 2006), this is most likely quercetin-3-O-rhamnoside. Because flavonoids are known scavengers of reactive oxygen species (Husain et al., 1987), we hypothesize that in the presence of RUBY, quercetin-3-O-rhamnoside is synthesized to quench H2O2, a by-product of Gal oxidation. In the absence of RUBY, its levels are reduced as there is no need for protection from H2O2. Like hydroxycinnamic acids, Tyr amino acids present in HRGPs, which are highly glycosylated structural proteins present in algal and plant cell walls, could be cross-linked in the presence of H2O2 and peroxidases (Waffenschmidt et al., 1993; Kjellbom et al., 1997; Fry, 2004). Thus, it is also possible that the H2O2 generated by RUBY is used to cross-link HRGPs present in the mucilage and middle lamellae. HRGPs have been identified in seed mucilage through proteomic analyses, but mutations in genes encoding such proteins exhibit no phenotypes (Tsai et al., 2017).
NaBH4, a reducing agent that can reduce Gal aldehydes, increases solubility of mum2 mucilage (Figures 7K and 7L), suggesting that the oxidation of Gal itself promotes this insolubility. Based on this observation, a second possibility for the formation of cross-links in the middle lamellae and the mucilage is the formation of hemiacetals between oxidized Gal and adjacent carbohydrates. When oxidized, Gal and Gal-containing substrates rarely appear as aldehydes, and mostly assume the hydrate form in aqueous solutions (Schmitz and Eichhorn, 1967; Andberg et al., 2017), or form hemiacetals with alcohols (Andberg et al., 2017). The formation of hemiacetals between oxidized Gal and hydroxy groups on neighboring polysaccharides has been demonstrated (Parikka et al., 2012; Merlini et al., 2015). Such direct cross-linking of polysaccharides in the plant cell walls has not been shown, but at least two studies have suggested that enzymatic oxidations of hemicellulosic polysaccharides in vitro can lead to cross-linking through hemiacetals. Both galactomannan (GM) and xyloglucan formed gels in the presence of GalOx, horseradish peroxidase (HRP), and catalase (Parikka et al., 2010), whereas fenugreek (Trigonella foenum-graecum) GM formed a gel when treated with a combination of laccase and 2,2,6,6-tetramethyl-1-piperidinyloxy radical (Rossi et al., 2016). In both cases, Gal side-chains were oxidized at the C-6 position to aldehydes, and hemiacetal formation with another hydroxy group in the proximity was proposed as a cross-linking mechanism. Two-dimensional NMR spectroscopy showed the existence of the hemiacetal bond between oxidized Gal and C-4 of Man in the backbone of fenugreek GM (Merlini et al., 2015). These data indicate that Gal oxidation could directly result in the formation of stable hemiacetal cross-links and, therefore, that RUBY-mediated oxidation may result in cross-links in the apoplast in the presence of pectin-rich environments like mucilage and the middle lamellae. Hemiacetal formation and breakdown are both catalyzed by acids and bases (Schmitz and Eichhorn, 1967). Our observation that a base, Na2CO3, can solubilize the mum2 mucilage (Figure 7H) is in favor of this hypothesis. Additionally, oxidation of monosaccharides into aldehydes in polysaccharides results in the formation of insoluble aerogels upon drying, owing to hemiacetal formation (Christensen et al., 2001; Köhnke et al., 2014; Mikkonen et al., 2014; Ghafar et al., 2015). Our data suggest that once the base is removed by washing or dialysis and samples dried, mucilage becomes less soluble again (Figures 7G, 7O, and 7P). This reduced solubility of mum2 mucilage can be prevented by the reduction of aldehydes (Figures 7O and 7P). Therefore, we propose that RUBY promotes oxidation of terminal Gal residues on RG-I to create aldehydes. In an aqueous environment of the cell wall, these aldehydes are expected to exist as hydrates, but they may be replaced by formation of hemiacetals during seed dehydration. In the cell wall, an environment rich in carbohydrates, the abundance of hydroxy groups around newly formed aldehydes may promote formation of hemiacetals between oxidized Gal and carbohydrates in its proximity. Once the pectin is extracted with basic solutions, it is necessary to dry it to bring these two functional groups together and re-form hemiacetals. Cross-linking through hemiacetals would have an advantage over hydroxycinnamate or HRGP cross-linking because it requires only a single enzyme. RUBY functions at the time when the epidermal cells are undergoing programmed cell death; thus, the generation of the H2O2 as a by-product of Gal oxidation may not be harmful to the seed, eliminating the need for peroxidases. Even though supported by the literature and some of our experiments, the presence of hemiacetals in the mucilage still needs to be confirmed by techniques like Fourier-TransformInfrared Spectroscopyor NMR spectroscopy.
The strengthening of pectin by GalOx enzymes can be beneficial in tissues where the cell wall needs additional reinforcements due to a high exposure to mechanical stress. In the seed coat epidermis, the middle lamella must resist the shear forces generated by rapid extrusion of the mucilage. Indeed, we have shown that one role of RUBY is to strengthen the middle lamellae between adjacent seed coat epidermal columellae, as well as between seed coat epidermal cells and the underlying palisade. The fact that, in ruby mutants, cell separation was not evident in mature dry seed (Figures 2E and 2F), but it was obvious only following hydration and mucilage extrusion (Figures 2A to 2D; Supplemental Movie 4) suggests that ruby cell separation requires mucilage extrusion.
In addition to the attachment of the columellae to the seed surface, RUBY appears to be required for the connection of the primary cell wall to the top of the columellae. In wild-type cells mucilage extrusion breaks the radial portion of the primary wall. The resulting cell wall fragment remains firmly attached to the top of the columellae (Figure 1N; Western et al., 2000). By contrast, these cell wall fragments typically separate from the columellae and are observed within the adherent mucilage of ruby mutants (Figure 1N), suggesting that RUBY strengthens connections between the primary wall and the columellae. This hypothesis is consistent with the appearance of RUBY in the columellae late in seed coat differentiation (Figures 6A and 6B).
In contrast with the middle lamellae, mucilage needs to expand upon hydration, so cross-links throughout mucilage would limit its ability to extrude during hydration. Our data suggest that the removal of RG-I Gal side-chains by MUM2 is necessary to allow mucilage extrusion in the presence of active RUBY. However, it is less clear whether RUBY influences wild-type mucilage pectin once MUM2 has removed the Gal side-chains. The mucilage of the ruby single mutant has a dishevelled appearance and a larger adherent mucilage halo (Figure 1P; Supplemental Figure 4C), indicating that the mucilage is not normal. It is possible that the removal of Gal side-chains from RG-I by MUM2 is not complete, and that RUBY protein in the columellae adjacent to the mucilage pocket (Figures 6A and 6B) establishes the correct cohesiveness of wild-type mucilage. However, the dishevelled mucilage could be an indirect effect of the loosened epidermal cells and/or primary cell walls detaching from the columella, whereas the larger halo may be the result of the release of additional pectin from the middle lamellae.
One curious aspect of the ruby mutant phenotype is the collapsed cellulosic rays observed in the adherent mucilage (Figures 1Q and 1R). Collapsed rays have been observed in mutants that fail to synthesize the mucilage galactoglucomannans that surround the cellulose rays of extruded mucilage (Yu et al., 2014; Voiniciuc et al., 2015). Because the Gal side-chains of galactoglucomannans are a potential substrate of RUBY, it is tempting to speculate that RUBY may play a role in the strengthening of the mucilage ray structure, but confirmation of such a role would require additional evidence.
In addition to the seed coat epidermal cells, RUBY may be expressed at low levels in the root epidermis starting at the elongation zone (Supplemental Figures 6A to 6D). To date, we have been unable to identify ruby phenotypes in these cell types. RUBY is one of seven homologous genes present in the Arabidopsis genome; therefore, it is possible that redundancy obscures some mutant phenotypes. Molecular genetic and biochemical analyses of these RUBY homologs may shed additional light on the role of Gal oxidation in pectin cross-linking.
METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) plants were grown in growth rooms or growth chambers under continuous light at 20°–22°C and low light intensity (80-120 μmol m−2 s−1; Philips F32T8/TL950 fluorescent tubes, 5000 K color temperature). Seeds were surface-sterilized with 70% ethanol and germinated on Arabidopsis thaliana (AT) minimal medium (Haughn and Somerville, 1986) with 0.7% (w/v) agar. Seedlings were transferred to soil mix (Sunshine Mix #4, Sun Gro Horticulture) once true leaves formed.
ruby-1, ruby-2, and ruby-3 were isolated from an EMS-mutagenized mum2-1 mutant population, which was generated in the Col-2 ecotype (Western et al., 2001). ruby-4 (SALK_020627C; Alonso et al., 2003), ruby-5 (WiscDsLoxHs097_11H; Woody et al., 2007), aomt1-1 (GK-007F02-014809; Kleinboelting et al., 2012), comt1-1 (SALK_002373; Alonso et al., 2003), 4cl1-3 (SAIL_350_H10; Sessions et al., 2002), 4cl3-3 (SALK_003025; Alonso et al., 2003), fah1-2, and fah1-7 were ordered from the Arabidopsis Biological Resource Center through The Arabidopsis Information Resource ; (www.arabidopsis.org). ref1-4 was a gift from Dr. Clint Chapple (University of Purdue). bxl1-1 is a T-DNA insertional mutant in the Ws-2 ecotype (Arsovski et al., 2009). ruby-4, ruby-5, aomt1-1 (Fellenberg et al., 2012), comt1-1, 4cl3-3, and mum2-10 (SALK_011436; Dean et al., 2007) are T-DNA insertional lines in the Col-0 ecotype. 4cl1-3 is a T-DNA insertional mutant in the Col-3 ecotype. ref1-4 and fah1-7 are EMS mutants generated in the Col-0 ecotype, whereas fah1-2 is an EMS mutant in the Landsberg erecta (Ler) ecotype (Chapple et al., 1992; Ruegger and Chapple, 2001).
Statistical Analyses
In the analyses, a biological replicate represents a batch of seeds harvested from 6 plants of the same genotype from the same pot, grown in the same tray with other genotypes used in the analysis. Separate biological replicates were grown under the same conditions at different times to ensure statistical independence and account for variations between separate growth trials. Experiments were repeated at least once.
Statistical tests were done in R (https://www.r-project.org/) using RStudio (https://www.rstudio.com/). With use of the “car” package, Levene’s test was used to assess whether variances across samples were equal. If the equal variance assumption was satisfied, one-way analysis of variance (ANOVA) was used on data sets with more than two groups, and two-way ANOVA was used on data sets with more than two groups and two independent variables (two treatments). Tukey’s Honestly Significant Difference (HSD) test was done after ANOVA tests, and the letters designating groups were generated using the “multcompView” package. Pair-wise comparisons were done using Welch’s t test. Data frames for plotting were arranged using the “plyr” and “reshape2” packages, and results were plotted using the “ggplot2” package.
Microscopy
For light microscopy of mucilage, dry seeds were shaken in water for 2 h, washed twice to remove nonadherent mucilage, stained with 0.02% (w/v) ruthenium red (Cat# R2751, Sigma) for 10 min, and washed twice in water. The seeds were imaged using a Leica DFC450 C camera (Leica Microsystems) attached to a Zeiss AxioSkop2 light microscope (Carl Zeiss). Imaging of the primary cell wall detachment was done with seeds shaken in water for 1 h using the same microscope with differential interference contrast configuration.
Filming of mucilage extrusion was completed on a single seed placed on a microscope slide. The seed was covered with a cover slip, which was held in place by the weight of metal forceps to prevent movement of the seed during filming. Immediately after filming started, 0.02% (w/v) ruthenium red was added under the cover slip.
Cellulose on the seed surface was stained using calcofluor white M2R (CFW; Cat# F3543, Sigma) or Pontamine Fast Scarlet 4B (Cat# S479896, Sigma) in 0.1 M NaCl. Seeds were shaken for 1 h in water, washed twice, shaken in CFW/Pontamine for 1 h protected from light, washed twice in water, and kept in the dark until imaged. Seeds were imaged using a Hamamatsu C9100-02 charge coupled device camera (Hamamatsu Photonics) attached to Leica DMI6000 inverted microscope (Leica Microsystems) combined with a PerkinElmer UltraVIEW VoX Spinning Disk Confocal system (PerkinElmer), with excitation at 405 nm and 460/50 nm emission filter for CFW, or excitation at 561 nm, emission filter 595/50 nm for Pontamine.
The seed surface was imaged with a Hitachi S4700 scanning electron microscope (Hitachi High-Technologies), after coating dry seeds with Au/Pd using EMPrep2 sputter coater (Nanotech).
For imaging of RUBY expression and localization in developing seeds, first open flowers, corresponding to 0 DPA of T2 and T3 plants expressing RUBY-Citrine, were marked using nontoxic water-soluble paint. Valves of siliques at the desired developmental stage were peeled using forceps, and seeds were mounted in water on a glass slide. Seeds were imaged using the same spinning-disc confocal system as for CFW staining, except that excitation and emission were set at 514 nm and 540/30 nm, respectively. For plasma membrane colocalization, seeds of the same plants were immersed in 10 μM FM4-64 (Cat# T-3166, Thermo Fisher Scientific), vacuum-infiltrated for 5 min, incubated in the dark for another 10 min at room temperature, and washed in water. After being mounting on slides, they were imaged at excitation 514 nm, emission filter 540/30 nm for RUBY-Citrine, and 561 nm excitation laser and 650/75 nm emission filter for FM4-64.
Scale bars were added using Fiji/ImageJ (Schindelin et al., 2012).
Mutagenesis, Genetic Analysis, and Positional Cloning of ruby
Seeds homozygous for the mum2-1 mutation were mutagenized with EMS (Cat# M0880, Sigma Aldrich). Seeds (100 mg; ∼5000) were placed into 50 mL Falcon tubes and imbibed in 30 mL sterile water. EMS stock solution (120 μL) was added, and seeds were left rotating gently overnight (16 to 17 h). Seeds were then allowed to settle, and EMS solution was removed to a flask containing 100 mM sodium thiosulfate (Na2S2O3). Seeds were washed three times with 30 mL of 100 mM Na2S2O3, rotating gently for 15 min in Na2S2O3 each time. Seeds were similarly washed with 30 mL distilled water (15 min each), and then mixed into 400 mL 0.1% (w/v) agarose (cooled to room temperature). Seeds (20–50) were plated onto solid AT medium to test germination rate. The remaining seeds were distributed 10 mL per pot into 40, 12 cm diameter, round pots, keeping seeds mixed in agarose so that they were distributed evenly. M1 seeds were harvested in bulk (each pot is one pooled M1 stock), and 70–100 M2 seeds from each M1 pool were planted for screening for suppression of the mum2 phenotype. M3 seeds were harvested individually from each M2 plant making sure to track from which M1 pool they originated. Seeds exhibiting suppression of the mum2 phenotype were individually selected from pools of mutagenized seeds and placed on AT medium. Seeds that germinated were transferred to soil and their seeds (M3) were tested to see if the suppression was a heritable trait, indicating a suppressor mutation in that line.
To confirm that the mutations were recessive and in single nuclear genes, mum2-1 suppressor plants were crossed to mum2-1. The F2 generation of the cross was harvested and F3 seeds screened to see if the phenotypic segregation ratio resembles that of a single nuclear mutation (3 mum2:1 suppressor).
Once confirmed to be recessive, suppressor lines were crossed to each other to determine whether they fell into different complementation groups. F2 seeds (harvested from F1 plants) from these crosses were stained with ruthenium red to determine whether they exhibited a suppressor phenotype. If the phenotype was suppressor, and not mum2, they were determined to be allelic.
Rough mapping of ruby was performed by first crossing mum2-1 ruby-1 to the Landsberg erecta (Ler) ecotype to generate plants that were heterozygous for Col-2 and Ler molecular markers. The segregating F2 population was genotyped for the mum2-1 mutation, and seeds of mum2-1 homozygous plants (F3 seeds) were screened for suppression. Only DNA of suppressors was used for positional cloning. Markers used for mapping are indicated in Supplemental Data Set 1. Linkage was inferred from molecular markers associated with the Col-2 ecotype that segregated with the suppressor phenotype.
After rough mapping, a Next Generation Mapping approach was used to identify the mutation in the suppressor line, as described in Austin et al. (2011). Briefly, DNA was extracted from F4 seedlings, progeny of 45 F3 lines exhibiting the suppressor phenotype, using PowerPlant Pro DNA Isolation Kit (Cat# 13400-50, MO BIO Laboratories) after ∼100 mg of tissue was homogenized using a Precellys 24 homogenizer (Bertin Technologies) and 2 mm zirconium oxide beads (Next Advance). DNA was resuspended in 10 mM Tris-HCl, pH 8.0, and quantified using a NanoDrop 8000 (Thermo Fisher Scientific), and an equal amount of DNA (275 ng) from each sample was added to a pooled sample. The solution was evaporated in a Savant DNA120 SpeedVac concentrator (Thermo Fisher Scientific) and resuspended to 100 ng μL−1 in 10 mM Tris-HCl, pH 8. Sequencing and analysis were performed at the Centre for the Analysis of Genome Evolution and Function at the University of Toronto, Toronto, ON, Canada (http://www.cagef.utoronto.ca/services/next-generation-genomics/). Candidate genes were identified using the Next Generation EMS Mutation Mapping online tool (http://bar.utoronto.ca/ngm/), and the identity of RUBY was confirmed by Sanger sequencing all 5 candidate genes in ruby-2 and ruby-3 lines. In all three mutants, mutations were present only in At1g19900. To confirm that RUBY is At1g19900, additional T-DNA insertional mutants were obtained, and transgene complementation was done by transforming ProRUBY:RUBY-Citrine into mum2-1 ruby-1.
Molecular Cloning and Transgenic Plants
RUBY (At1g19900) genomic DNA was amplified using Phusion High-Fidelity DNA polymerase (Thermo Fisher Scientific), including 4477 bp upstream of START codon, from Col-2 genomic DNA using forward primer including an EcoRI restriction site (5′- GCTTAGAATTCGCTAACCCAATCTCAATCGAACC-3′), and reverse primer with an XbaI site (5′- ATAGTCTAGACCTCCTTCTAACTTTACCC-3′). The amplicon was gel-purified using an EZ-10 Spin Column DNA Gel Extraction Kit (Bio Basic) according to manufacturer’s instructions, digested with EcoRI and XbaI, and purified using an EZ-10 Spin Column PCR Products Purification Kit (Bio Basic). The resultant insert was ligated into pCambia2300 (Cambia) plasmid digested with EcoRI and XbaI, using T4 Ligase (New England Biolabs). Plasmid was cultured in a DH5α Escherichia coli strain (New England Biolabs) in liquid Lysogeny Broth (LB) medium (Cat# 1.10285.5007, Millipore Sigma) with 50 μg mL−1 kanamycin (Kan; Cat# K-120-25, Gold Biotechnology), purified using an EZ-10 Spin Column Plasmid DNA Miniprep Kit (Bio Basic), and sequenced. For fluorescent tagging of RUBY, the Citrine-encoding sequence, together with a linker on its 5′ end and nosT on its 3′ end, was amplified from pAD vector (DeBono, 2011) using forward primer with a SalI restriction site (5′-CTAGAGTCGACCCCTGGAGGTGGAGGTGGAGC-3′), and reverse primer containing an SbfI restriction site (5′-GCATGCCTGCAGGAGTAACATAGATGACACCGCGC-3′). The insert was subsequently subcloned into pCambia2300/ProRUBY-RUBY the same way as described above. The final sequence-confirmed binary vector was transformed into Agrobacterium tumefaciens strain GV3101 (pMP90). Bacteria were selected on LB plates containing 50 μg mL−1 Kan, 25 μg mL−1 rifampicin (Cat# R-120-5, Gold Biotechnology), and 25 μg mL−1 gentamycin (Gent; Cat# G-400-10, Gold Biotechnology), at 28°C. A colony was grown in 5 mL LB broth overnight with same selection antibiotics, and subcultured in 200 mL LB broth with Kan/Gent overnight at 28°C. mum2-1 ruby-1 plants were transformed using the floral dip method (Clough and Bent, 1998). T1 plants were selected on AT plates containing 35 μg mL−1 Kan, and positive transformants were transferred to soil.
Transcript Analysis
A modified protocol based on Meisel et al. (2005) was used for total RNA extraction. Tissue (20–100 mg) was snap-frozen on dry ice, left at −70°C overnight, and ground to fine powder on dry ice using a prechilled mortar and pestle, or with 2 mm zirconium oxide beads (Next Advance) on a Precellys 24 tissue homogenizer (Bertin Technologies) at 6000 rpm for 20 s. Tissue was kept frozen until resuspended in 500 μL of cetyltrimethylammonium bromide buffer preheated to 65°C, containing 2% (w/v) cetyltrimethylammonium bromide (Cat# H5882, Sigma Aldrich); 1.4 M NaCl (Cat# S271-3, Fisher Scientific); 20 mM EDTA (Cat# S311-100, Thermo Fisher Scientific); 100 mM Tris-HCl, pH 8.0; 1% (w/v) polyvinylpyrolidone MW 40 000 (Cat# 529504, Millipore Sigma Aldrich); 0.05% (w/v) spermidine trihydrochloride (Cat# 85578, Sigma Aldrich); and 5 mM DL-DTT (Cat# D0632, Sigma Aldrich). Spermidine and DTT were added to the buffer right before use. Samples were mixed well on a vortex mixer and incubated at 65°C for 15 min. An equal volume (500 μL) of 24:1 (v/v) chloroform/isoamyl alcohol was added to samples, mixed on a vortex mixer, and centrifuged at 12 000 g for 10 min at room temperature. The upper, aqueous phase was re-extracted one more time and collected by carefully avoiding the interphase. RNA was precipitated overnight at −20°C by adding 10 M LiCl (Cat# L9650, Sigma Aldrich) to 2 M final concentration. RNA was pelleted by centrifugation for 20 min at 16 000 g at 4°C, washed carefully in 75% (v/v) ethanol in diethyl pyrocarbonate-treated Milli-Q water, and centrifuged for 2 min at 16 000 g at 4°C. The wash was repeated once, the ethanol was removed, and the pellet was air-dried at room temperature. Dry pellet was resuspended in 10-15 μL diethyl pyrocarbonate-treated water, and total RNA was quantified using a NanoDrop 8000 spectrophotometer (Thermo Fisher Scientific). The integrity of native RNA was inspected by agarose gel electrophoresis.
cDNA for RT-PCR was synthesized from 250 ng total RNA, treated with DNase I (Cat# 18068015, Thermo Fisher Scientific) according to the manufacturer’s instructions. After inactivation of DNase, cDNA was synthesized using 5x All-in-One room temperature MasterMix (Cat# G490, Applied Biological Materials) according to the manufacturer’s instructions in 10 μL final volume. For RT-PCR, 1 μL of cDNA was used in a 10 μL reaction. ACT2 (At3g18780) and GAPC1 (At3g04120) were employed as reference genes. RT-PCR primers are described in Supplemental Data Set 1.
Monosaccharide Composition Analysis using HPAEC-PAD
The procedures used to analyze seed monosaccharides are summarized in Supplemental Table 2.
For analysis of soluble mucilage, 20 mg of dry seeds hydrated in 1.4 mL of Milli-Q water or 20 mM Na2CO3, 5 μL of 5 mg mL−1 meso-erythritol (Cat# E7500, Sigma Aldrich) was added as internal standard, and samples were shaken for 2 h. External standards for calibration were prepared by serial dilution (2 mM, 1 mM, 0.5 mM, 0.25 mM, 0.125 mM, 62.5 μM) from 100 mM monosaccharide mixture of L-Fuc (Cat# F2252, Sigma Aldrich), L-rhamnose (Cat# R3875, Sigma Aldrich), L-Ara (Cat# A3256), D-Gal (Cat# G6404, Sigma Aldrich), D-Glc (Cat# G8270, Sigma Aldrich), D-Xyl (Cat# X3877, Sigma Aldrich), D-Man (Cat# M2069, Sigma Aldrich), and D-galacturonic acid (Cat# Aldrich 85,728-9, Sigma Aldrich). Standards were spiked with the same amount of meso-erythritol and processed the same way as the samples. Then 1 mL of mucilage extract was collected and dried under N2(g) at 60°C. Samples were incubated in 72% (w/v) H2SO4 on ice for 2 h to swell cellulose. Na2CO3 in samples was neutralized by adding additional H2SO4. After 2 h, H2SO4 was diluted to 4% w/v and the sample volume was brought to 0.5 mL with Milli-Q water. Samples were hydrolyzed at 121°C at 15 psi for 1 h in an autoclave. Hydrolysates were filtered through 0.45 μm Millex HVsyringe filters (Millipore) and loaded into HPLC vials. Then, 15 μL samples were injected using a Spectra AS 3500 autoinjector (Spectra-Physics) onto a Dionex DX-600 HPLC (Thermo Fisher Scientific) and separated using a Dionex CarboPac PA1 column (Thermo Fisher Scientific). Neutral monosaccharides were separated at 1 mL min−1, with isocratic elution with water for 35 min, and sugars were detected using a pulsed-amperometric detector with a gold electrode after the postcolumn addition of 200 mM NaOH at a flowrate of 0.5 mL min−1. The column was washed after each separation for 10 min with 250 mM NaOH and re-equilibrated with water. For acid sugars (GalUA), the same samples were separated at 0.4 mL min−1, using linear gradient elution from 10 mM to 400 mM sodium acetate (NaOAc) for 30 min, and NaOH was kept at a constant concentration of 100 mM. The column was washed for 10 min with 10 mM NaOAc, 300 mM NaOH, then re-equilibrated with 10 mM NaOAc, 100 mM NaOH. Peak areas were integrated in Chromeleon software (Thermo Fisher Scientific), and data were processed using Python 2.7 with NumPy and SciPy packages.
For whole seed analysis, 20 mg of seeds were frozen on dry ice, then ground on dry ice using a mortar and pestle. Alternatively, frozen seeds were ground with 2 mm zirconium oxide beads (Next Advance) on a Precellys 24 tissue homogenizer (Bertin Technologies) at 6000 rpm for 20 s. Samples were suspended in 1 mL 70% (v/v) aqueous ethanol, incubated for 10 min at 65°C, vortexed for 10 min at room temperature, and centrifuged at 16,000 g for 30 s at room temperature. The samples were then washed in 70% (v/v) ethanol, followed by a 10-min vortex, and the centrifugation was repeated once. Three more washes were completed to prepare the final alcohol-insoluble residue (AIR): 80% (v/v) methanol, 100% methanol, and acetone. Finally, acetone was evaporated under a gentle stream of N2(g), and the AIR weighed and transferred to glass tubes for hydrolysis. Meso-erythritol (20 μL of 5 mg mL−1 stock) was added to samples, dried under N2(g), and processed and analyzed the same as the mucilage samples (see paragraph above). The final volume of H2SO4 was adjusted so that, once diluted, the final concentration was 4% (w/v) H2SO4 in 2 mL.
For sequential extraction of mucilage, outer nonadherent mucilage was extracted from 100 mg of intact seeds with water (3 mL) for 3 h at room temperature. After centrifugation (8 000 g, 5 min) at room temperature, supernatants were carefully removed, filtered through a disposable glass microfiber filter (13 mm diameter, 2.7 µm pore size; Whatman), and retained for analysis. Seeds were then rinsed with 5 mL of 50 mM sodium acetate buffer, pH 4.5 (three changes), and rhamnogalacturonan hydrolase (UniProt Q00018; Novozymes) was added to the washed seeds (0.8 nkat). The inner adherent mucilage extracts were recovered as described previously (Sullivan et al., 2011). Seeds following mucilage removal were carefully rinsed with distilled water (three changes), freeze-dried, and ground with a mortar and pestle. They were further delipidated with 2 mL methanol/chloroform (1/2; v/v) overnight with head-over-tail mixing at room temperature, centrifuged (8 000 g, 10 min, room temperature), and the supernatant was removed. The treatment was repeated twice with 2 h head-over-tail mixing, after which the samples were dried overnight at 40°C. Uronic acid (as GalUA) was determined by an automated m-hydroxybiphenyl method (Thibault, 1979) either directly (mucilage extracts) or after prehydrolysis (H2SO4 72%, 30 min at room temperature) and hydrolysis (H2SO4 2 N, 6 h at 100°C) for demucilaged delipidated seeds. Individual neutral sugars were analyzed as their alditol acetate derivatives (Blakeney et al., 1983) by gas-liquid chromatography after hydrolysis with 2 M trifluoroacetic acid at 121°C for 2.5 h for mucilage extracts or prehydrolysis (H2SO4 72%, 30 min at room temperature) and hydrolysis (H2SO4 2 N, 6 h at 100°C) for demucilaged delipidated seeds.
Per-O-Methylation and Linkage Analysis of Neutral Sugars
Mucilage was prepared for PMAA glycosyl linkage analysis by shaking 20 mg of Col-2 and ruby-1 seeds in 25 mM Na2CO3 for 2 h. The seeds were left to settle, and the supernatant was removed and dried under N2(g) at 60°C. Dry mucilage was sent to the Complex Carbohydrate Research Center, University of Georgia for analysis. The analysis was done on biological duplicates, as described by York et al. (1986).
Initially, dry sample was suspended in ∼300 μL of dimethyl sulphoxide and placed on a magnetic stirrer for 1 week. The sample was permethylated according to Ciucanu and Kerek (1984). Solid NaOH was added to the sample and incubated for 15 min at room temperature, followed by the addition of methyl iodide (CH3I) and a 45-min incubation at room temperature. Additional NaOH was added, followed by 10 min incubation at room temperature, and finally more CH3I was added and samples were incubated for 40 min at room temperature. Following the derivatization, the permethylated material was hydrolyzed using 2 M trifluoroacetic acid (TFA) for 2 h in a sealed tube at 121°C, reduced with NaBD4, and acetylated using acetic anhydride/TFA. The resulting PMAAs were analyzed on a Hewlett Packard/Agilent 7890A GC gas chromatograph (Agilent Technologies Inc.) interfaced to a 5975C MSD (mass selective detector, electron impact ionization mode EI-MS; Agilent Technologies Inc.); separation was performed on a 30 m Supelco SP-2380 bonded phase fused silica capillary column (Sigma-Aldrich).
Digestion and Analysis of Mucilage RG-I
For the analysis of Gal oxidation in the mucilage, outer soluble mucilage was extracted from wild type (Col-2), ruby, mum2, and ruby mum2 seeds (100 mg) with 0.05 M HCl (5 mL) for 30 min at 85°C, and then with 5 mL of 0.3 M NaOH as previously described (Macquet et al., 2007b). Seeds were rinsed 3 times with 5 mL of water, acidified to pH 4.5 with 0.05 M HCl, and then rinsed with 5 mL of 50 mM sodium acetate buffer, pH 4.5 (three changes). Rhamnogalacturonan hydrolase (UniProt Q00018; Novozymes) was added to the washed seeds (0.8 nkat), and inner adherent mucilage extracts were recovered as described previously (Sullivan et al., 2011). The oligosaccharides were analyzed using HPAEC-PAD as described by Ralet et al. (2010) and using the IP-RP-UHPLC-MS/MS method described below.
IP-RP-UHPLC-MS/MS
Chromatographic separation was achieved on a UHPLC (Acquity H-Class, Waters), with a Ethylene Bridged Hybrid C18 column (100 mm × 1 mm, packed with 1.7 µm porosity particles; Waters). A ternary gradient was used (A: Milli-Q water, B: 100% methanol, and C: 20 mM heptylammonium formate, pH 6), from 2% to 25% of solvent B for 10 min, then up to 73% at 23.5 min and maintained at 73% for 4 min. Percentage of solvent C was kept constant at 25%. The flow rate was 0.15 mL min−1, and the column was heated to 45°C.
Mass spectrometry measurements were performed using a Synapt G2 Si HDMS (Waters) in negative ionization mode and in sensitivity time-of-flight measurements mode. The parameters used for electrospray were the following: capillary voltage: 2.5 kV; sampling cone: 50; desolvation temperature: 250°C; desolvation gas: 350 L/h. MS/MS measurements were performed in the trap cell of the triwave cell using an energy of 50.
Sequence Alignment
Multiple sequence alignment was performed with the Multiple Alignment using Fast Fourier Transform algorithm (Katoh and Standley, 2013) using amino acid sequences of characterized members of the AA5 Carbohydrate-Active Enzymes Database family of enzymes and RUBY. Based on the initial alignment of sequences, the ends of sequences were truncated to remove components outside the catalytic domain. Truncated sequences were aligned again using Multiple Alignment using Fast Fourier Transform algorithm, and the alignment was visualized using JalView (Waterhouse et al., 2009).
Protein Expression and Purification in E. coli
RUBY was cloned with and without predicted signal sequence, based on a SignalP 4.1 prediction. The gene was amplified from pCambia2300/ProRUBY:RUBY-Citrine:nosT vector together with 3′ linker sequence using a forward primer with the SacI restriction site for both full-length (5′-aattcgagctccggaggtatggcagcacgagcaacattc-3′) and sequence without the signal sequence (5′-aattcgagctccggaggtgcccgaggattatggaaatacatcg-3′). The same reverse primer was used to amplify both inserts, and it contained an XhoI site (5′-tggtgctcgagtccacctccacctccaggg-3′). After digestion and purification, the inserts were ligated into the pET-21b (+) vector (Novagen) in-frame with the C-terminal His6-tag. Plasmid was then cultured in DH5α, purified, and sequenced. For soluble protein expression, plasmids were transformed into Arctic Express E. coli (Agilent Technologies). Expression was assessed on a small scale, and full-length protein was found to be poorly expressed; thus protein without the signal peptide was used in the purification. For purification, starter culture was grown overnight at 37°C in LB broth with 100 μg mL−1 Carbenicillin (Cat# BP2648, Fisher Scientific) and 25 μg mL−1 Gent. The next day, cells were subcultured in 500 mL Terrific Broth medium (2.4% yeast extract, 1.2% tryptone, 0.4% glycerol, 100 mM KPi buffer, pH 7.5) with 50 μg mL−1 Carbenicillin. Culture was grown in a 2 liter flask at 37°C at 250 rpm until an O.D. 600 of 0.8–1.2 was achieved. Expression was induced by adding isopropyl-β-D-thiogalactopyranoside (Cat# 10724815001, Roche) to 0.1 mM. Protein was expressed at 11°C for 36 h, after which the culture was centrifuged for 20 min at 10 000 g at 4°C. The pellet was resuspended in 35 mL of extraction buffer (100 mM NaPi buffer pH 7.4, 10% [w/v] Suc, 500 mM NaCl, 0.3 mM PMSF); lysozyme (Cat# L6876, Sigma Aldrich) was added at 0.5 mg mL−1, and the suspension was incubated on ice for 1 h. Cells were lysed using a Vibra Cell™ VC505 sonicator fitted with a CV334 converter and 13-mm tip probe (Sonics and Materials, Inc.) at 40% amplitude, for 15 min (5 s ON, 5 s OFF). The lysate was brought to 50 mL with cold buffer and centrifuged for 25 min at 12,000 g and 4°C. The supernatant was loaded on equilibrated HisPur Ni-NTA agarose resin (Thermo Fisher Scientific) in a glass column, and resin with sample was incubated for 1 h at 4°C, after which the flow-through was collected, and resin washed twice with 10 mL of 100 mM NaPi buffer pH 7.4, 10% (w/v) Suc, 500 mM NaCl, 30 mM imidazole, then twice with 10 mL of 100 mM NaPi buffer pH 7.4, 10% (w/v) Suc, 500 mM NaCl, 30 mM imidazole, 5 mM ATP, 5 mM MgCl2, followed by 3 washes with 10 mL 100 mM NaPi buffer pH 7.4, 10% (w/v) Suc, 500 mM NaCl. Protein was eluted 5 times with 2 mL 100mM NaPi buffer pH 7.4, 10% (w/v) Suc, 500 mM NaCl, 250 mM imidazole. The A280 of eluted fractions was read on a NanoDrop 8000 Spectrophotometer (Thermo Fisher Scientific), and fractions with the highest reads were combined and desalted in 100 mM NaPi pH 6.0, 10% (w/v) Suc, 500 mM NaCl on an Econo-Pac 10DG column (Bio Rad) at 4°C using a minimal dilution protocol. For activation, 0.5 mM CuSO4 was added to the buffer, as described by Spadiut et al. (2010), and mixed on a head-over-tail mixer at 4°C overnight. Proteins were analyzed using SDS-PAGE and protein gel blot with a 1:1000 dilution of His-Probe (H-3) mouse monoclonal antibody (200 μg mL−1; Cat# sc-8036, Santa Cruz Biotechnology).
For activity assays, cells carrying pET-21b empty vector were grown under the same conditions alongside cultures with pET-21b/RUBY-His6 and used as a negative control in activity assays. With use of buffers described in the section "Enzyme Activity Assays with Seeds", and Gal and glycerol as substrates, activity was assayed on crude lysates, supernatants, and all the fractions derived from the purification.
Enzyme Activity Assays with Seeds
For substrate screening using dry seeds, ∼50 seeds (Col-2 and ruby-1) were incubated in 100 μL of 50 mM NaPi buffer pH 6.0, 2 U mL−1 horseradish peroxidase, Type II (Cat# P8250, Sigma Aldrich), 100 μM 3,3′,5,5′-tetramethylbenzidine (TMB; Cat# 229280010, Acros Organics), and 10 mM substrate. Samples were incubated overnight at room temperature protected from light. The compound was considered a substrate if a blue color developed in Col-2, but not ruby-1 samples.
The effects of activators and inactivators were assayed on 3 mg of seeds (exact mass recorded) of 3 biological replicates of Col-2 seeds. Seeds were preincubated for 1 h in 1 mL Milli-Q water at room temperature, washed twice, and incubated for 1 h in Milli-Q water at room temperature (control), 1 mg mL−1 Proteinase K, 1 mM CuSO4, 1 mM EDTA, or Milli-Q water at 95°C (heat inactivation). After 1 h preincubation, the samples were washed twice in 2 mL 200 mM NaPi buffer, pH 6.0, and Proteinase K was inactivated at 65°C for 10 min. Activity was assayed in 2 mL of 50 mM NaPi buffer pH 6.0, 2 U mL−1 HRP, 100 μM TMB, and 200 mM D-Gal at room temperature (22°C). Seeds were mixed by inverting, and 150 μL were sampled at multiple time points once the color started to develop. Sampling was done within the linear range (blue color). Immediately before reading samples at A450 on a Synergy HT microplate reader (BioTek), 10 μL of 1 M H2SO4 was added to each sample to stop the reactions. H2O2 in the solutions was calculated based on calibration curves of standards with known concentrations of H2O2 within a linear range of detection. Standard solutions were loaded on the same microtiter plates in the same volumes as the samples, and values were calculated based on the Beer-Lambert law. For calculation of specific activity (nmol min−1 mg−1 seeds), only time points that showed linearity were used.
Specific activity was assayed on 3 mg of seeds (exact mass recorded) of 3 biological replicates, using the Col-2, ruby-1, and ruby-5 genotypes. Seeds were preincubated in Milli-Q water for 2 h at room temperature, washed twice, and left in 1 mL 2× reaction buffer (100 mM NaPi buffer pH 6.0, 4 U mL−1 HRP, 200 μM TMB). Reactions were initiated by addition of 1 mL of 2× substrates in Milli-Q water to give final substrate concentrations of 200 mM (D-galactose, raffinose, meso-erythritol, and glycerol) or 40 mM (ONP-β-D-Gal). Data were collected and processed the same way as for activation and inactivation assays.
Detection of Gal Oxidase Substrates in the Mucilage
Col-2, mum2-1, ruby-1, and mum2-1 ruby-1 seeds (200 mg; 3 biological replicates) were shaken in 5 mL of 20 mM Na2CO3 for 2 h at room temperature. After the seeds settled at the bottom, the supernatant was collected, and seeds were washed in 5 mL of Milli-Q water. Both supernatants were pooled, and then separated into two samples of equal volumes. NaBH4 was added to 0.1% (w/v) from a 1% (w/v) stock in 0.2 M NaOH, and the other sample was left untreated (control). After 15 min, the samples were neutralized by adding glacial acetic acid, precipitated by adding 4 volumes of 95% (v/v) ethanol, and pelleted by 10 min centrifugation at 7000 g at 4°C. Supernatants were discarded, and pellets resuspended in Milli-Q water and dialyzed in cellulose acetate tubing (12-14 kD molecular weight cut-off) against Milli-Q water. Samples were then frozen on dry ice and lyophilized using a FreeZone 4.5 liter freeze dry system (Labconco). Dry mucilage was weighed and re-suspended to 2 mg mL−1 in Milli-Q water. Samples were mixed on a vortex mixer for 2 h at room temperature to enhance resuspension and stored at 4°C until the analysis.
Extracted mucilage was imaged for opacity/transparency by loading 150 μL of 2 mg mL−1 mucilage samples into a 96-well microtiter plate and taking images with a Nikon Coolpix 8400 digital camera. The effect of Na2CO3 on mum2 mucilage solubility was examined by splitting 2 mg mL−1 mum2-1 mucilage samples into two 140 μL samples. The samples were loaded into a 96-well plate, 10 μL of water was added to one (control) and 10 μL of 0.5 M Na2CO3 to the other (Na2CO3). The plate was incubated at room temperature for 10 min and imaged the same as above. Three biological replicates were used.
Availability of Gal as a substrate was measured by adding 50 μg of mucilage in buffer containing 50 mM NaPi pH 6.0, 100 μM TMB, 1 U mL−1 HRP, and 1 U mL−1 Gal oxidase (GalOx, Cat# G7400, Sigma) to a final volume of 150 μL in a 96-well polystyrene microtiter plate. The plate was left in the dark at room temperature for 1 h, reactions were stopped by adding 10 μL of 1 M H2SO4, and absorbance at 450 nm was measured within 10 min using a Synergy HT microplate reader (BioTek). The concentration of H2O2 was measured using standard solutions with the same volumes as the samples.
Mucilage Insolubility Experiments
For reduction at neutral pH, 50–100 seeds were mixed with 900 μL of 1 M imidazole-HCl, pH 7.0, and left for 30 min to prehydrate, followed by addition of 100 μL of fresh 1% (w/v) NaBH4 in 10 mM NaOH (or only 10 mM NaOH for the control samples). Seeds were mixed on a head-over-tail mixer for 2 h at room temperature, after which the pH was measured using pH indicator strips to ensure that it remained stable. Seeds were washed twice with H2O, once in 1% (w/v) aqueous glacial acetic acid, twice in H2O, stained with 0.02% (w/v) ruthenium red, and imaged.
Dehydration–rehydration experiments were completed as follows: mucilage was released by mixing 50–100 seeds in 1425 μL of 20 mM Na2CO3 for 2 h at room temperature. Then, 75 μL of 1% (w/v) NaBH4 in 0.2 M NaOH (0.2 M NaOH only to control samples) was added for reduction, and samples were mixed for another hour. Seeds were washed twice with water, once in 1% (w/v) aqueous glacial acetic acid, twice in water, vacuum-filtered, and dried with filter papers (Whatman) on silica beads overnight. The next day, seeds were collected into plastic tubes, hydrated in water, mixed for 2 h at room temperature, stained with 0.02% (w/v) ruthenium red, and imaged.
Extraction and Analysis of Seed Surface Phenolics
To extract ester-linked hydroxycinnamic acids from the mucilage and cell walls of the seed surface, 20 mg of wild-type and mutant seeds (exact mass recorded) were hydrated in 1 mL of degassed 2 M NaOH. Then, 20 μL of 5 mg mL−1 vanillin (Cat# V2375, Sigma Aldrich) was added as an internal standard, and samples were flushed with N2(g) and incubated at 30°C for 2 h. Liquid was transferred to glass tubes, acidified with 150 μL 72% (w/v) H2SO4, and extracted three times with 1 mL of ethyl acetate (EtOAc). All three EtOAc extracts were combined and evaporated under N2(g). Samples were resuspended in 0.5 mL of 50% (v/v) aqueous acetonitrile (MeCN), 0.1% (v/v) TFA, filtered through 0.45μm HV filters, and loaded into plastic HPLC vials. Sinapic acid (Cat# D13,460-0, Sigma Aldrich) standards were made by serial dilution from 50 mM stock in methanol, spiked with the same amount of vanillin as the samples, evaporated under N2(g), and resuspended in 50% (v/v) MeCN, 0.1% (v/v) TFA. Then, 15 μL of sample was injected by an ASI-100 autoinjector (Dionex) and separated on a Symmetry C18 column (5 μm, 4.6 mm × 250 mm; Waters) at 35°C on a Summit HPLC system (Dionex) fitted with a PDA-100 Photodiode Array Detector (Dionex). Water with 0.1% (v/v) TFA (solvent A) and MeCN/MeOH 3:1 with 0.1% (v/v) TFA (solvent B) were used for separation at a flowrate of 0.7 mL min−1 using a linear gradient from 5% to 45% solvent B, 0–40 min. Analytes were detected by measuring absorbance at 255 nm and 320 nm. The column was washed for 10 min with 75% solvent B, and re-equilibrated for 20 min with 5% solvent B.
For LC-Electrospray Ionization (ESI)-MS identification of seed surface phenolics, Col-2 and ruby-1 samples were processed as described above and delivered dry under N2(g) to the Mass Spectrometry Services, Department of Chemistry, Simon Fraser University (https://www.sfu.ca/chemistry/research/facilities/massspec.html). Samples were analyzed on Agilent 1200 SL LC system (Agilent Technologies). They were separated on a Zorbax XDB C18 column (1.8 μm, 50 mm × 4.6 mm; Agilent Technologies), at 30°C, with a flowrate of 0.6 mL min−1. Water with 0.1% (v/v) formic acid (Solvent A) and MeCN with 0.1% (v/v) formic acid (Solvent B) were used for separation as follows: 10% B at 0 min, 10% B at 1min, 40% B at 8 min, 100% B at 9 min, 100% B at 10 min, 10% B at 10.1 min, 10% B at 12 min (stop). Analytes were detected using a maXis Impact Ultra-High Resolution tandem TOF (ultra-high resolution-Qq-TOF) mass spectrometer (Bruker) in 50–1500 D mass range (ESI in negative mode; source capillary 4.5 kV; gas temperature 200°C; gas flow 9 liter min−1).
Accession Numbers
Arabidopsis sequence data from this article can be found on The Arabidopsis Information Resource (https://www.arabidopsis.org/) under accession numbers: At1g19900 (RUBY), At5g63800 (MUM2), At5g49360 (BXL1), At3g18780 (ACT2), At3g04120 (GAPC1), At4g36220 (FAH1), At3g24503 (REF1), At5g54160 (COMT1), At1g15950 (CCR1), At1g51680 (4CL1), At1g65060 (4CL3), and At4g34050 (AOMT1). Gal oxidase and glyoxal oxidase amino acid sequence data from this article can be found in the UniProt (https://www.uniprot.org/) data library under accession numbers: P0CS93 (FgGalOx), A0A089QAB6 (FsaGaoA), V5NQ89 (FoGalOx), E7CHE7 (FsuGaoB), E7CHE8 (FvGaoB), L2FIQ2 (CglAlcOx), E3QHV8 (CgrAlcOx), E3R0R1 (CgRaOx), Q93Z02 (RUBY), Q01772 (PcGlx1), A0A191VY32 (PciGLOX1), A0A060SYB0 (PciGLOX2), Q7Z868 (UmGlo1), Q7Z866 (UmGlo3).
Supplemental Data
Supplemental Figure 1. ruby suppresses mum2 and bxl1 phenotypes to a different degree.
Supplemental Figure 2. Cell adhesion defects of ruby-1 are enhanced by chelator (EDTA) and suppressed by calcium (CaCl2).
Supplemental Figure 3. RGase releases novel branched RG-I from ruby-1 mucilage.
Supplemental Figure 4. Insertional mutant, ruby-5, resembles ruby-1 in all phenotypic aspects.
Supplemental Figure 5. RUBY contains catalytic amino acids required for galactose/glyoxal oxidase function.
Supplemental Figure 6. RUBY is expressed in root epidermis and seed coat.
Supplemental Figure 7. Most abundant phenolics on seed surface are sinapic acid and a flavonol glycoside.
Supplemental Figure 8. NaBH4 promotes mucilage extrusion in mum2 seeds.
Supplemental Table 1. List of compounds tested as substrates for oxidases on dry mature seeds using HRP-TMB assay.
Supplemental Table 2. Summary of mucilage/cell wall extraction and hydrolysis procedures.
Supplemental Movie 1. Seed coat mucilage extrusion of wild-type (Col-2) seed.
Supplemental Movie 2. Seed coat mucilage extrusion of mum2-1 seed.
Supplemental Movie 3. Seed coat mucilage extrusion of mum2-1 ruby-1 seed.
Supplemental Movie 4. Seed coat mucilage extrusion of ruby-1 seed.
Supplemental Data Set 1. ANOVA tables.
Supplemental Data Set 2. List of primers used for positional cloning and genotyping.
Supplemental Movie. Supplemental movie legends.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Faride Unda (University of British Columbia) for assistance with the HPAEC-PAD and HPLC-UV analyses; Marie-Jeanne Crépeau and Jacqueline Vigouroux (Institut National de la Recherche Agronomique, Nantes) for assistance with the sugar analyses; Hongwen Chen (Mass Spectrometry Services, Department of Chemistry, Simon Fraser University) for performing the LC-ESI-MS analysis; Radnaa Naran (Complex Carbohydrate Research Center, University of Georgia) for performing the PMAA linkage analysis; Yunchen Gong (Centre for the Analysis of Genome Evolution and Function, University of Toronto) for performing the Next Generation Sequencing and data processing; Derrick Horne (University of British Columbia) for assistance with scanning electron microscopy; Kevin Hodgson (University of British Columbia) for assistance with confocal microscopy, Yann Mathieu (University of British Columbia) for help with the Pichia pastoris expression experiments; Katharina Vollheyde (University of Goettingen) for help with the E. coli expression experiments; Robert McGee (University of British Columbia) and Jana Phan (University of Adelaide) for developing a protocol for filming mucilage extrusion; Clint Chapple (Purdue University) for sharing the ref1-4 mutant; Gillian Dean, Lacey Samuels, and Harry Brumer (University of British Columbia) for reading the article; Harry Brumer, Gillian Dean, Ljerka Kunst, and Yaseen Mottiar (University of British Columbia) for comments and discussion; and University of British Columbia Bioimaging Facility for microscopy training and support. This work was supported by the Gouvernement du Canada | Natural Sciences and Engineering Research Council of Canada (Conseil de Recherches en Sciences Naturelles et en génie du Canada) (NSERC) (Discovery Grants to G.W.H. and S.D.M.), the internal funding of the Institut National de la Recherche Agronomique (INRA, Nantes) and the Deutsche Forschungsgemeinschaft (DFG) for providing funding for “PRoTECT” (IRTG 2172) of the Goettingen Graduate School of Neurosciences, Biophysics, and Molecular Biology.
Author Contributions
K.Š., E.J.G., D.R., I.F., S.D.M., M.-C.R., and G.W.H. designed the experiments and interpreted the results; K.Š., E.J.G., D.R., L.W., and M.-C.R. performed the experiments and analyzed the data; K.Š. wrote the first draft; all authors reviewed and edited the article; I.F., S.D.M, M.-C.R., and G.W.H. provided resources and supervised the research.
References
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657. [DOI] [PubMed] [Google Scholar]
- Andberg M., Mollerup F., Parikka K., Koutaniemi S., Boer H., Juvonen M., Master E., Tenkanen M., Kruus K. (2017). A novel Colletotrichum graminicola raffinose oxidase in the AA5 family. Appl. Environ. Microbiol. 83: 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparecido Cordeiro F., Bertechini Faria C., Parra Barbosa-Tessmann I. (2010). Identification of new galactose oxidase genes in Fusarium spp. J. Basic Microbiol. 50: 527–537. [DOI] [PubMed] [Google Scholar]
- Arsovski A.A., Popma T.M., Haughn G.W., Carpita N.C., McCann M.C., Western T.L. (2009). AtBXL1 encodes a bifunctional β-d-xylosidase/α-L-arabinofuranosidase required for pectic arabinan modification in Arabidopsis mucilage secretory cells. Plant Physiol. 150: 1219–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson R.G., Schröder R., Hallett I.C., Cohen D., MacRae E.A. (2002). Overexpression of polygalacturonase in transgenic apple trees leads to a range of novel phenotypes involving changes in cell adhesion. Plant Physiol. 129: 122–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin R.S., Vidaurre D., Stamatiou G., Breit R., Provart N.J., Bonetta D., Zhang J., Fung P., Gong Y., Wang P.W., McCourt P., Guttman D.S. (2011). Next-generation mapping of Arabidopsis genes. Plant J. 67: 715–725. [DOI] [PubMed] [Google Scholar]
- Avigad G., Amaral D., Asensio C., Horecker B.L. (1962). The d-galactose oxidase of Polyporus circinatus. J. Biol. Chem. 237: 2736–2743. [PubMed] [Google Scholar]
- Balasubramaniam S., Lee H.C., Lazan H., Othman R., Ali Z.M. (2005). Purification and properties of a β-galactosidase from carambola fruit with significant activity towards cell wall polysaccharides. Phytochemistry 66: 153–163. [DOI] [PubMed] [Google Scholar]
- Beeckman T., De Rycke R., Viane R., Inzé D. (2000). Histological study of seed coat development in Arabidopsis thaliana. J. Plant Res. 113: 139–148. [Google Scholar]
- Blakeney A.B., Harris P.J., Henry R.J., Stone B.A. (1983). A simple and rapid preparation of alditol acetates for monosaccharide analysis. Carbohydr. Res. 113: 291–299. [Google Scholar]
- Bouton S., Leboeuf E., Mouille G., Leydecker M.-T., Talbotec J., Granier F., Lahaye M., Höfte H., Truong H.-N. (2002). QUASIMODO1 encodes a putative membrane-bound glycosyltransferase required for normal pectin synthesis and cell adhesion in Arabidopsis. Plant Cell 14: 2577–2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunzel M., Ralph J., Kim H., Lu F., Ralph S.A., Marita J.M., Hatfield R.D., Steinhart H. (2003). Sinapate dehydrodimers and sinapate-ferulate heterodimers in cereal dietary fiber. J. Agric. Food Chem. 51: 1427–1434. [DOI] [PubMed] [Google Scholar]
- Burr S.J., Fry S.C. (2009). Feruloylated arabinoxylans are oxidatively cross-linked by extracellular maize peroxidase but not by horseradish peroxidase. Mol. Plant 2: 883–892. [DOI] [PubMed] [Google Scholar]
- Bush M.S., Marry M., Huxham I.M., Jarvis M.C., McCann M.C. (2001). Developmental regulation of pectic epitopes during potato tuberisation. Planta 213: 869–880. [DOI] [PubMed] [Google Scholar]
- Chapple C.C.S., Vogt T., Ellis B.E., Somerville C.R., Chapple C.C.S., Vogt T., Ellis B.E., Somerville C.R. (1992). An Arabidopsis mutant defective in the general phenylpropanoid pathway. Plant Cell 4: 1413–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen B.E., Aasprong E., Stokke B.T. (2001). Gelation of periodate oxidised scleroglucan (scleraldehyde). Carbohydr. Polym. 46: 241–248. [Google Scholar]
- Ciucanu I., Kerek F. (1984). A simple and rapid method for the permethylation of carbohydrates. Carbohydr. Res. 131: 209–217. [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Daou M., Piumi F., Cullen D., Record E., Faulds C.B. (2016). Heterologous production and characterization of two glyoxal oxidases from Pycnoporus cinnabarinus. Appl. Environ. Microbiol. 82: 4867–4875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson D.M., Melton L.D., Watkins C.B. (1992). Cell wall changes in nectarines (Prunus persica): Solubilization and depolymerization of pectic and neutral polymers during ripening and in mealy fruit. Plant Physiol. 100: 1203–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean G.H., Zheng H., Tewari J., Huang J., Young D.S., Hwang Y.T., Western T.L., Carpita N.C., McCann M.C., Mansfield S.D., Haughn G.W. (2007). The Arabidopsis MUM2 gene encodes a β-galactosidase required for the production of seed coat mucilage with correct hydration properties. Plant Cell 19: 4007–4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBono A. (2011). The role and behavior of Arabidopsis thaliana lipid transfer proteins during cuticular wax deposition. PhD dissertation (Vancouver: University of British Columbia; ).. [Google Scholar]
- Encina A., Fry S.C. (2005). Oxidative coupling of a feruloyl-arabinoxylan trisaccharide (FAXX) in the walls of living maize cells requires endogenous hydrogen peroxide and is controlled by a low-Mr apoplastic inhibitor. Planta 223: 77–89. [DOI] [PubMed] [Google Scholar]
- Fellenberg C., van Ohlen M., Handrick V., Vogt T. (2012). The role of CCoAOMT1 and COMT1 in Arabidopsis anthers. Planta 236: 51–61. [DOI] [PubMed] [Google Scholar]
- Francis K.E., Lam S.Y., Copenhaver G.P. (2006). Separation of Arabidopsis pollen tetrads is regulated by QUARTET1, a pectin methylesterase gene. Plant Physiol. 142: 1004–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry S.C. (2004). Oxidative coupling of tyrosine and ferulic acid residues: Intra- and extra-protoplasmic occurrence, predominance of trimers and larger products, and possible role in inter-polymeric cross-linking. Phytochem. Rev. 3: 97–111. [Google Scholar]
- Ghafar A., Parikka K., Sontag-Strohm T., Österberg M., Tenkanen M., Mikkonen K.S. (2015). Strengthening effect of nanofibrillated cellulose is dependent on enzymatically oxidized polysaccharide gel matrices. Eur. Polym. J. 71: 171–184. [Google Scholar]
- Grabber J.H., Hatfield R.D., Ralph J., Zoń J., Amrhein N. (1995). Ferulate cross-linking in cell walls isolated from maize cell suspensions. Phytochemistry 40: 1077–1082. [Google Scholar]
- Griffiths J.S., Tsai A.Y.-L., Xue H., Voiniciuc C., Šola K., Seifert G.J., Mansfield S.D., Haughn G.W. (2014). SALT-OVERLY SENSITIVE5 Mediates Arabidopsis Seed Coat Mucilage Adherence and Organization through Pectins. Plant Physiol. 165: 991–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haughn G.W.G.W., Somerville C. (1986). Sulfonylurea-resistant mutants of Arabidopsis thaliana. Mol. Gen. Genet. 204: 430–434. [Google Scholar]
- Haughn G.W., Western T.L. (2012). Arabidopsis seed coat mucilage is a specialized cell wall that can be used as a model for genetic analysis of plant cell wall structure and function. Front. Plant Sci. 3: 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husain S.R., Cilurd J., Cillard P. (1987). Hydroxyl radical scavenging activity of flavonoids. Phytochemistry 26: 2489–2491. [Google Scholar]
- Iwai H., Ishii T., Satoh S. (2001). Absence of arabinan in the side chains of the pectic polysaccharides strongly associated with cell walls of Nicotiana plumbaginifolia non-organogenic callus with loosely attached constituent cells. Planta 213: 907–915. [DOI] [PubMed] [Google Scholar]
- Iwai H., Masaoka N., Ishii T., Satoh S. (2002). A pectin glucuronyltransferase gene is essential for intercellular attachment in the plant meristem. Proc. Natl. Acad. Sci. USA 99: 16319–16324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Standley D.M. (2013). MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersten P.J. (1990). Glyoxal oxidase of Phanerochaete chrysosporium: Its characterization and activation by lignin peroxidase. Proc. Natl. Acad. Sci. USA 87: 2936–2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersten P.J., Kirk T.K. (1987). Involvement of a new enzyme, glyoxal oxidase, in extracellular H2O2 production by Phanerochaete chrysosporium. J. Bacteriol. 169: 2195–2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.-B., Carpita N.C. (1992). Changes in esterification of the uronic acid groups of cell wall polysaccharides during elongation of maize coleoptiles. Plant Physiol. 98: 646–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjellbom P., Snogerup L., Stöhr C., Reuzeau C., McCabe P.F., Pennell R.I. (1997). Oxidative cross-linking of plasma membrane arabinogalactan proteins. Plant J. 12: 1189–1196. [DOI] [PubMed] [Google Scholar]
- Kleinboelting N., Huep G., Kloetgen A., Viehoever P., Weisshaar B. (2012). GABI-Kat SimpleSearch: New features of the Arabidopsis thaliana T-DNA mutant database. Nucleic Acids Res. 40: D1211–D1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhnke T., Elder T., Theliander H., Ragauskas A.J. (2014). Ice templated and cross-linked xylan/nanocrystalline cellulose hydrogels. Carbohydr. Polym. 100: 24–30. [DOI] [PubMed] [Google Scholar]
- Lau J.M., McNeil M., Darvill A.G., Albersheim P. (1987). Treatment of rhamnogalacturonan I with lithium in ethylenediamine. Carbohydr. Res. 168: 245–274. [Google Scholar]
- Lerouge P., O’Neill M.A., Darvill A.G., Albersheim P. (1993). Structural characterization of endo-glycanase-generated oligoglycosyl side chains of rhamnogalacturonan I. Carbohydr. Res. 243: 359–371. [DOI] [PubMed] [Google Scholar]
- Leuthner B., Aichinger C., Oehmen E., Koopmann E., Müller O., Müller P., Kahmann R., Bölker M., Schreier P.H. (2005). A H2O2-producing glyoxal oxidase is required for filamentous growth and pathogenicity in Ustilago maydis. Mol. Genet. Genomics 272: 639–650. [DOI] [PubMed] [Google Scholar]
- Macquet A., Ralet M.-C., Kronenberger J., Marion-Poll A., North H.M. (2007a). In situ, chemical and macromolecular study of the composition of Arabidopsis thaliana seed coat mucilage. Plant Cell Physiol. 48: 984–999. [DOI] [PubMed] [Google Scholar]
- Macquet A., Ralet M.-C., Loudet O., Kronenberger J., Mouille G., Marion-Poll A., North H.M. (2007b). A naturally occurring mutation in an Arabidopsis accession affects a β-d-galactosidase that increases the hydrophilic potential of rhamnogalacturonan I in seed mucilage. Plant Cell 19: 3990–4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeil M., Darvill A.G., Albersheim P. (1980). Structure of plant cell walls: HAMNOGALACTURONAN I, a structurally complex pectic polysaccharide in the walls of suspension-cultured sycamore cells. Plant Physiol. 66: 1128–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeil M., Darvill A.G., Fry S.C., Albersheim P. (1984). Structure and function of the primary cell walls of plants. Annu. Rev. Biochem. 53: 625–663. [DOI] [PubMed] [Google Scholar]
- McPherson M.J., Ogel Z.B., Stevens C., Yadav K.D., Keen J.N., Knowles P.F. (1992). Galactose oxidase of Dactylium dendroides. Gene cloning and sequence analysis. J. Biol. Chem. 267: 8146–8152. [PubMed] [Google Scholar]
- Meisel L., Fonseca B., González S., Baeza-Yates R., Cambiazo V., Campos R., Gonźalez M., Orellana A., Retamales J., Silva H. (2005). A rapid and efficient method for purifying high quality total RNA from peaches (Prunus persica) for functional genomics analyses. Biol. Res. 38: 83–88. [DOI] [PubMed] [Google Scholar]
- Mendu V., Griffiths J.S., Persson S., Stork J., Downie A.B., Voiniciuc C., Haughn G.W., DeBolt S. (2011). Subfunctionalization of cellulose synthases in seed coat epidermal cells mediates secondary radial wall synthesis and mucilage attachment. Plant Physiol. 157: 441–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlini L., Boccia A.C., Mendichi R., Galante Y.M. (2015). Enzymatic and chemical oxidation of polygalactomannans from the seeds of a few species of leguminous plants and characterization of the oxidized products. J. Biotechnol. 198: 31–43. [DOI] [PubMed] [Google Scholar]
- Mikkonen K.S., Parikka K., Suuronen J.-P., Ghafar A., Serimaa R., Tenkanen M. (2014). Enzymatic oxidation as a potential new route to produce polysaccharide aerogels. RSC Advances 4: 11884– 11892. [Google Scholar]
- Moore P.J., Darvill A.G., Albersheim P., Staehelin L.A. (1986). Immunogold localization of xyloglucan and rhamnogalacturonan I in the cell walls of suspension-cultured sycamore cells. Plant Physiol. 82: 787–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouille G., Ralet M.C., Cavelier C., Eland C., Effroy D., Hématy K., McCartney L., Truong H.N., Gaudon V., Thibault J.F., Marchant A., Höfte H. (2007). Homogalacturonan synthesis in Arabidopsis thaliana requires a Golgi-localized protein with a putative methyltransferase domain. Plant J. 50: 605–614. [DOI] [PubMed] [Google Scholar]
- Neumetzler L., Humphrey T., Lumba S., Snyder S., Yeats T.H., Usadel B., Vasilevski A., Patel J., Rose J.K.C., Persson S., Bonetta D. (2012). The FRIABLE1 gene product affects cell adhesion in Arabidopsis. PLoS One 7: e42914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill M.A., Ishii T., Albersheim P., Darvill A.G. (2004). Rhamnogalacturonan II: Structure and function of a borate cross-linked cell wall pectic polysaccharide. Annu. Rev. Plant Biol. 55: 109–139. [DOI] [PubMed] [Google Scholar]
- Orfila C., Seymour G.B., Willats W.G., Huxham I.M., Jarvis M.C., Dover C.J., Thompson A.J., Knox J.P. (2001). Altered middle lamella homogalacturonan and disrupted deposition of (1→5)-α-L-arabinan in the pericarp of Cnr, a ripening mutant of tomato. Plant Physiol. 126: 210–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikka K., Leppänen A.S., Pitkänen L., Reunanen M., Willför S., Tenkanen M. (2010). Oxidation of polysaccharides by galactose oxidase. J. Agric. Food Chem. 58: 262–271. [DOI] [PubMed] [Google Scholar]
- Parikka K., Ansari F., Hietala S., Tenkanen M. (2012). Thermally stable hydrogels from enzymatically oxidized polysaccharides. Food Hydrocoll. 26: 212–220. [Google Scholar]
- Paukner R., Staudigl P., Choosri W., Sygmund C., Halada P., Haltrich D., Leitner C. (2014). Galactose oxidase from Fusarium oxysporum--expression in E. coli and P. pastoris and biochemical characterization. PLoS One 9: e100116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paukner R., Staudigl P., Choosri W., Haltrich D., Leitner C. (2015). Expression, purification, and characterization of galactose oxidase of Fusarium sambucinum in E. coli. Protein Expr. Purif. 108: 73–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña M.J., Carpita N.C. (2004). Loss of highly branched arabinans and debranching of rhamnogalacturonan I accompany loss of firm texture and cell separation during prolonged storage of apple. Plant Physiol. 135: 1305–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralet M.-C., André-Leroux G., Quéméner B., Thibault J.F. (2005). Sugar beet (Beta vulgaris) pectins are covalently cross-linked through diferulic bridges in the cell wall. Phytochemistry 66: 2800–2814. [DOI] [PubMed] [Google Scholar]
- Ralet M.C., Tranquet O., Poulain D., Moïse A., Guillon F. (2010). Monoclonal antibodies to rhamnogalacturonan I backbone. Planta 231: 1373–1383. [DOI] [PubMed] [Google Scholar]
- Ralph J., Quideau S., Grabber J.H., Hatfieida R.D. (1994). ldentification and synthesis of new ferulic acid dehydrodimers present in grass cell walls. J. Chem. Soc., Perkin Trans. 1 23: 3485–3498. [Google Scholar]
- Rautengarten C., Usadel B., Neumetzler L., Hartmann J., Büssis D., Altmann T. (2008). A subtilisin-like serine protease essential for mucilage release from Arabidopsis seed coats. Plant J. 54: 466–480. [DOI] [PubMed] [Google Scholar]
- Rhee S.Y., Osborne E., Poindexter P.D., Somerville C.R. (2003). Microspore separation in the quartet 3 mutants of Arabidopsis is impaired by a defect in a developmentally regulated polygalacturonase required for pollen mother cell wall degradation. Plant Physiol. 133: 1170–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers M.S., et al. (2007). The stacking tryptophan of galactose oxidase: A second-coordination sphere residue that has profound effects on tyrosyl radical behavior and enzyme catalysis. Biochemistry 46: 4606–4618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi B., Campia P., Merlini L., Brasca M., Pastori N., Farris S., Melone L., Punta C., Galante Y.M. (2016). An aerogel obtained from chemo-enzymatically oxidized fenugreek galactomannans as a versatile delivery system. Carbohydr. Polym. 144: 353–361. [DOI] [PubMed] [Google Scholar]
- Routaboul J.M., Kerhoas L., Debeaujon I., Pourcel L., Caboche M., Einhorn J., Lepiniec L. (2006). Flavonoid diversity and biosynthesis in seed of Arabidopsis thaliana. Planta 224: 96–107. [DOI] [PubMed] [Google Scholar]
- Ruegger M., Chapple C. (2001). Mutations that reduce sinapoylmalate accumulation in Arabidopsis thaliana define loci with diverse roles in phenylpropanoid metabolism. Genetics 159: 1741–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saulnier L., Thibault J.F. (1999). Ferulic acid and diferulic acids as components of sugar-beet pectins and maize bran heteroxylans. J. Sci. Food Agric. 79: 396–402. [Google Scholar]
- Schindelin J., et al. (2012). Fiji: An open-source platform for biological-image analysis. Nat. Methods 9: 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz E., Eichhorn I. (1967). Acetals and hemiacetals. In Patai S, ed, The Ether Linkage, John Wiley & Sons, New York, pp 309–351. [Google Scholar]
- Sessions A., et al. (2002). A high-throughput Arabidopsis reverse genetics system. Plant Cell 14: 2985–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smallwood M., Beven A., Donovan N., Neill S.J., Peart J., Roberts K., Knox J.P. (1994). Localization of cell wall proteins in relation to the developmental anatomy of the carrot root apex. Plant J. 5: 237–246. [Google Scholar]
- Spadiut O., Olsson L., Brumer H. III (2010). A comparative summary of expression systems for the recombinant production of galactose oxidase. Microb. Cell Fact. 9: 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan S., Ralet M.-C., Berger A., Diatloff E., Bischoff V., Gonneau M., Marion-Poll A., North H.M. (2011). CESA5 is required for the synthesis of cellulose with a role in structuring the adherent mucilage of Arabidopsis seeds. Plant Physiol. 156: 1725–1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibault J. (1979). Automatisation du dosage des substances pectiques par la methode au meta-hydroxydiphenyl. Lebensm. Wiss. Technol. 12: 247–251. [Google Scholar]
- Tsai A.Y.-L., Kunieda T., Rogalski J., Foster L.J., Ellis B.E., Haughn G.W. (2017). Identification and characterization of Arabidopsis seed coat mucilage proteins. Plant Physiol. 173: 1059–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voiniciuc C., Dean G.H., Griffiths J.S., Kirchsteiger K., Hwang Y.T., Gillett A., Dow G., Western T.L., Estelle M., Haughn G.W. (2013). Flying saucer1 is a transmembrane RING E3 ubiquitin ligase that regulates the degree of pectin methylesterification in Arabidopsis seed mucilage. Plant Cell 25: 944–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voiniciuc C., Schmidt M.H.-W., Berger A., Yang B., Ebert B., Scheller H.V., North H.M., Usadel B., Günl M. (2015). MUCILAGE-RELATED10 produces galactoglucomannan that maintains pectin and cellulose architecture in Arabidopsis seed mucilage. Plant Physiol. 169: 403–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waffenschmidt S., Woessner J.P., Beer K., Goodenough U.W. (1993). Isodityrosine cross-linking mediates insolubilization of cell walls in Chlamydomonas. Plant Cell 5: 809–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse A.M., Procter J.B., Martin D.M.A., Clamp M., Barton G.J. (2009). Jalview version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics 25: 1189–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Western T.L., Skinner D.J., Haughn G.W. (2000). Differentiation of mucilage secretory cells of the Arabidopsis seed coat. Plant Physiol. 122: 345–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Western T.L., Burn J., Tan W.L., Skinner D.J., Martin-McCaffrey L., Moffatt B.A., Haughn G.W. (2001). Isolation and characterization of mutants defective in seed coat mucilage secretory cell development in Arabidopsis. Plant Physiol. 127: 998–1011. [PMC free article] [PubMed] [Google Scholar]
- Willats W.G.T., Orfila C., Limberg G., Buchholt H.C., van Alebeek G.J.W.M., Voragen A.G.J., Marcus S.E., Christensen T.M.I.E., Mikkelsen J.D., Murray B.S., Knox J.P. (2001). Modulation of the degree and pattern of methyl-esterification of pectic homogalacturonan in plant cell walls. Implications for pectin methyl esterase action, matrix properties, and cell adhesion. J. Biol. Chem. 276: 19404–19413. [DOI] [PubMed] [Google Scholar]
- Windsor J.B., Symonds V.V., Mendenhall J., Lloyd A.M. (2000). Arabidopsis seed coat development: Morphological differentiation of the outer integument. Plant J. 22: 483–493. [DOI] [PubMed] [Google Scholar]
- Woody S.T., Austin-Phillips S., Amasino R.M., Krysan P.J. (2007). The WiscDsLox T-DNA collection: An arabidopsis community resource generated by using an improved high-throughput T-DNA sequencing pipeline. J. Plant Res. 120: 157–165. [DOI] [PubMed] [Google Scholar]
- Xu F., Golightly E.J., Schneider P., Berka R.M., Brown K.M., Johnstone J.A. (2000). Expression and characterization of a recombinant Fusarium spp. galactose oxidase. Appl. Biochem. Biotechnol. 88: 23–32. [Google Scholar]
- Yin D.T., Urresti S., Lafond M., Johnston E.M., Derikvand F., Ciano L., Berrin J.G., Henrissat B., Walton P.H., Davies G.J., Brumer H. (2015). Structure-function characterization reveals new catalytic diversity in the galactose oxidase and glyoxal oxidase family. Nat. Commun. 6: 10197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- York W.S., Darvill A.G., McNeil M., Stevenson T.T., Albersheim P. (1986). Isolation and characterization of plant cell walls and cell wall components. Methods Enzymol. 118: 3–40. [Google Scholar]
- Yu L., Shi D., Li J., Kong Y., Yu Y., Chai G., Hu R., Wang J., Hahn M.G., Zhou G. (2014). CELLULOSE SYNTHASE-LIKE A2, a glucomannan synthase, is involved in maintaining adherent mucilage structure in Arabidopsis seed. Plant Physiol. 164: 1842–1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamil M.S., Geitmann A. (2017). The middle lamella - more than a glue. Physiol. Biol. 14: 015004. [DOI] [PubMed] [Google Scholar]