Cyanobacterial state transitions, which balance photosystem activities during photosynthesis, depend on the plastoquinone pool redox state but not on cytochrome b6f or phosphorylation reactions.
Abstract
Photosynthetic organisms must sense and respond to fluctuating environmental conditions in order to perform efficient photosynthesis and to avoid the formation of dangerous reactive oxygen species. The excitation energy arriving at each photosystem permanently changes due to variations in the intensity and spectral properties of the absorbed light. Cyanobacteria, like plants and algae, have developed a mechanism, named “state transitions,” that balances photosystem activities. Here, we characterize the role of the cytochrome b6f complex and phosphorylation reactions in cyanobacterial state transitions using Synechococcus elongatus PCC 7942 and Synechocystis PCC 6803 as model organisms. First, large photosystem II (PSII) fluorescence quenching was observed in State II, a result that does not appear to be related to energy transfer from PSII to PSI (spillover). This membrane-associated process was inhibited by betaine, Suc, and high concentrations of phosphate. Then, using different chemicals affecting the plastoquinone pool redox state and cytochrome b6f activity, we demonstrate that this complex is not involved in state transitions in S. elongatus or Synechocystis PCC6803. Finally, by constructing and characterizing 21 protein kinase and phosphatase mutants and using chemical inhibitors, we demonstrate that phosphorylation reactions are not essential for cyanobacterial state transitions. Thus, signal transduction is completely different in cyanobacterial and plant (green alga) state transitions.
INTRODUCTION
Photosynthetic organisms must cope with changes in the quality and quantity of incoming light. In order to survive and to optimize the use of light, they must adapt to changing environmental conditions by regulating the energy arriving at their reaction centers. Specific illumination of photosystem II (PSII) or photosystem I (PSI) creates an energy imbalance that leads to the over-reduction or over-oxidation of the intersystem electron transport chain. Murata (1969) and Bonaventura and Myers (1969) were the first to propose a mechanism, called “state transitions,” which rebalances the activity of reaction centers I and II. Two states were defined: State I, induced by light preferentially absorbed by PSI and characterized by a high PSII to PSI fluorescence ratio; State II, induced by light preferentially absorbed by PSII and characterized by a low PSII to PSI fluorescence ratio. The transition from one state to the other is triggered by changes in the redox state of the plastoquinone (PQ) pool (Allen et al., 1981; Mullineaux and Allen, 1990): oxidation of the PQ pool induces the transition to State I and its reduction induces the transition to State II.
In plants and green algae, reduction of the PQ pool induces the activation of a specific kinase that phosphorylates the membrane-bound light-harvesting complex II (LHCII). The phosphorylated LHCII detaches from PSII and attaches to PSI during the transition from State I to State II. Oxidation of the PQ pool deactivates the kinase and a phosphatase dephosphorylates LHCII, which again migrates to PSII. The migration of LHCII from one photosystem to the other allows for a readjustment in the distribution of excitation energy arriving at PSI and PSII (see review in Minagawa, 2011).
In red algae and cyanobacteria, the principal PSII antenna is the phycobilisome (PBS), a large extramembrane complex constituted by phycobiliproteins organized in a core from which rods radiate (reviews can be found in Glazer, 1984; MacColl, 1998; and Adir, 2008). As a consequence, the processes involved in state transitions in these organisms differ. In red algae, the large fluorescence quenching induced by the illumination of dark-adapted cells is related to two different mechanisms: a PSII non-photochemical-quenching mechanism induced by a low luminal pH (Delphin et al., 1995, 1996; Kowalczyk et al., 2013; Krupnik et al., 2013), in which the fluorescence quenching occurs at the level of the reaction centers (Krupnik et al., 2013); and state transitions induced by changes in the redox state of the PQ pool, which involve changes in energy transfer from PSII to PSI (spillover; Ley and Butler, 1980; Kowalczyk et al., 2013). The relative importance of each mechanism varies among strains (Delphin et al., 1996; Kowalczyk et al., 2013). In cyanobacteria, the molecular mechanism of the PQ-pool dependent state transitions remains largely obscure. This process, which involves fluorescence changes occurring upon illumination of dark-adapted cells or under illumination with light absorbed more specifically by PSII or PSI, indeed remains an open question, despite the many studies resulting in the proposal of several hypotheses and models.
In the mobile-PBS model, the movement of PBSs induces changes in direct energy transfer from PBS to PSII and PSI (Allen et al., 1985; Mullineaux and Allen, 1990; Mullineaux et al., 1997). This model attributes the low PSII fluorescence yield in State II to a lower amount of energy transfer from PBSs to PSII, together with larger energy transfer to PSI. The observations that PBSs are able to rapidly move on the thylakoid surface (Mullineaux et al., 1997) and that chemicals inhibiting PBS diffusion also inhibit state transitions support this model (Joshua and Mullineaux, 2004; Li et al., 2004, 2006). In the spillover model, the energy transfer from PBS to PSII remains equal in both states, but the excess energy absorbed by PSII is transferred to PSI (spillover) in State II via a process involving the movement of photosystems (Ley and Butler, 1980; Bruce and Biggins, 1985; Olive et al., 1986; Biggins and Bruce, 1989; Biggins et al., 1989; Vernotte et al., 1992; El Bissati et al., 2000; Federman et al., 2000). The hypothesis that changes at the level of photosystems are responsible for state transitions is supported by various observations: state transitions occur in mutants lacking PBSs (Bruce et al., 1989; Olive et al., 1997; El Bissati et al., 2000); state transitions are accompanied by structural changes in membranes (Vernotte et al., 1992; Folea et al., 2008) and PSI monomerization/trimerization (Kruip et al., 1994; Schluchter et al., 1996; Aspinwall et al., 2004); and membrane fluidity influences state transitions (El Bissati et al., 2000). Nevertheless, there has been no clear demonstration that the spillover is larger in State II than in State I, although some studies have suggested this (see Mullineaux et al., 1991; Bruce and Salehian, 1992). It was also proposed that these two mechanisms coexist and are responsible for the fluorescence changes observed in state transitions: movement of PBS (changes in direct energy transfer from PBSs to photosystems) and movement of photosystems (changes in spillover; Scott et al., 2006). However, more recent studies have questioned the definition of cyanobacterial state transitions as a rebalance of excitation energy arriving to one or another photosystem. The increase in fluorescence in State I has principally been associated with the functional detachment of PBS from the photosystems (Kaňa et al., 2009; Kaňa, 2013; Chukhutsina et al., 2015), whereas the decrease in fluorescence in State II was mainly attributed to a specific fluorescence quenching of PSII not involving spillover (Ranjbar Choubeh et al., 2018).
Furthermore, the states of plants and cyanobacteria in darkness differ: while plants are generally in State I, cyanobacteria are in State II (Aoki and Katoh, 1982; Mullineaux and Allen, 1986). In cyanobacteria, respiration and photosynthesis occur in the thylakoid membranes, and PQ, cytochrome (cyt) b6f and plastocyanin (or cyt c6) are electron carriers common to both electron transport chains (see a review in Mullineaux, 2014). During respiration, the homologs of mitochondrial complex I (NDH-1) and complex II (succinate dehydrogenase, SDH) reduce the PQ pool, and different oxidases oxidize it (for a review, see Mullineaux, 2014). Different cyanobacterial strains present different PQ pool reduction states in darkness, giving different levels of dark PSII fluorescence (see, for example, Misumi et al., 2016). Upon illumination, PSI is activated and the PQ pool becomes more oxidized, leading to State I (Mullineaux and Allen, 1990; Campbell et al., 1998).
In plants and green algae, the redox sensor of the PQ pool is the cyt b6f complex, which interacts with a specific kinase of the major membrane chlorophyll (Chl) antenna, LHCII (Wollman and Lemaire, 1988). Phosphorylation of LHCII trimers induces their detachment from PSII and partial (or total) attachment to PSI, inducing the transition to State II (Kyle et al., 1984). Two reports suggest that cyt b6f also plays a role in cyanobacterial state transitions (Mao et al., 2002; Huang et al., 2003). However, further evidence is still needed to confirm its direct involvement. In this sense, the relationship between phosphorylation and cyanobacterial state transitions is also an open question. Allen and coworkers suggested that specific types of phosphorylation could occur during state transitions (Allen et al., 1985), but this was not confirmed in more recent works. Nevertheless, analysis of phospho-proteomes showed that phosphorylation takes place in PBSs and photosystems (Yang et al., 2013; Chen et al., 2015; Spät et al., 2015). In addition, when residues Ser22, 49, and 154 and Thr94 of phycocyanin (PC; PBS protein) were mutated to non-phosphorylatable amino acids in Synechocystis cells, the kinetic and amplitude of transition to State I induced by light illumination of dark-adapted cells appeared to be affected (Chen et al., 2015). Some functions of Ser/Thr kinases (Spk) have already been described. For example, SpkA is involved in the control of cell motility (Kamei et al., 2001; Panichkin et al., 2006); SpkB participates in the oxidative stress response by phosphorylating glycyl-tRNA-synthetase β-subunit (Mata-Cabana et al., 2012); SpkE might be involved in the regulation of nitrogen metabolism (Galkin et al., 2003); SpkD might be involved in adjusting the pool of tricarboxylic acid cycle metabolites (Laurent et al., 2008); SpkG plays an essential role in high-salt resistance (Liang et al., 2011); SpkC, SpkF, and SpkK are involved in the phosphorylation of the GroES chaperone protein (Zorina et al., 2014); and SpkG is involved in the phosphorylation of ferredoxin 5 protein (Angeleri et al., 2018).
As a whole, the molecular mechanism behind cyanobacterial state transitions is still a matter of discussion, although many hypotheses have been proposed. Therefore, we decided to further study this mechanism by specifically addressing the role of the cyt b6f complex in this process.
In the past decades, state transitions have mainly been studied in the cyanobacteria Synechocystis PCC 6803 (hereafter Synechocystis; for example, Vernotte et al., 1992; Emlyn-Jones et al., 1999; McConnell et al., 2002; Kondo et al., 2009; Chukhutsina et al., 2015), Synechococcus PCC7002 (McConnell et al., 2002; Dong and Zhao, 2008; Dong et al., 2009) and Spirulina platensis (Li et al., 2004, 2006). In these strains, the changes in fluorescence related to state transitions (dark versus blue [or far-red] illumination) are rather small, which makes mechanistic studies difficult. The differences in PSII fluorescence in darkness (State II) and under blue light illumination (State I) are significantly larger in Synechococcus elongatus strain than in Synechocystis. Therefore, in our study, we characterized state transitions in S. elongatus and compared them to those in Synechocystis, which allowed us to obtain clearer conclusions. Our results confirm data demonstrating that a large amplitude of PSII fluorescence quenching is induced in State II in S. elongatus (Ranjbar Choubeh et al., 2018). This PSII quenching appears to be unrelated to spillover. In addition, not only do we show that the results and arguments used to link the cyt b6f complex with state transitions were not conclusive, but we also demonstrate that cyt b6f and protein phosphorylation reactions do not participate in this process in cyanobacteria. Thus, different signaling pathways are involved in state transitions in cyanobacteria compared with plants and green algae.
RESULTS
State Transitions in S. elongatus and Synechocystis
77 K Fluorescence Spectra in State II and State I
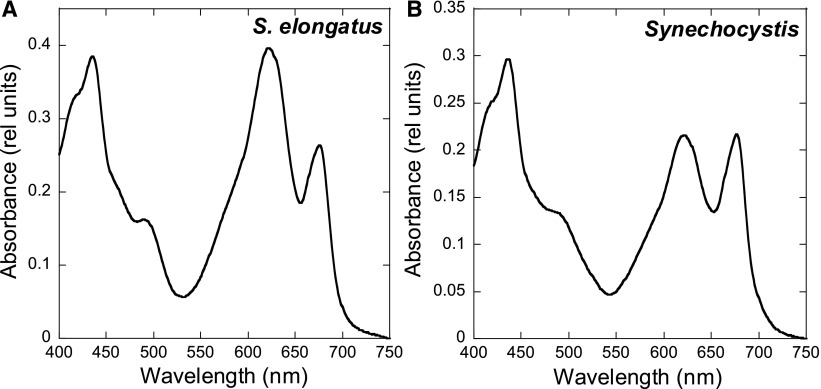
The absorbance spectra indicate that, under our growth conditions, S. elongatus presents a higher PC (absorbance at 620 nanometers [nm]) to Chl (absorbance at 680 nm) ratio than Synechocystis (Figure 1). In addition, the PSI to PSII ratio [measured by (FA, FB)− and Tyrosine-D+ (TyrD+) electron paramagnetic resonance (EPR) signals] was around 2:3 in S. elongatus and 4:5 in Synechocystis cells (for details, see Methods).
Figure 1.
Absorption Spectra of S. elongatus (A) and Synechocystis (B) Cells.
The spectra were recorded at a Chl concentration of 2.5 μg/mL. The curves shown are the average of three independent biological replicates (as described in Methods).
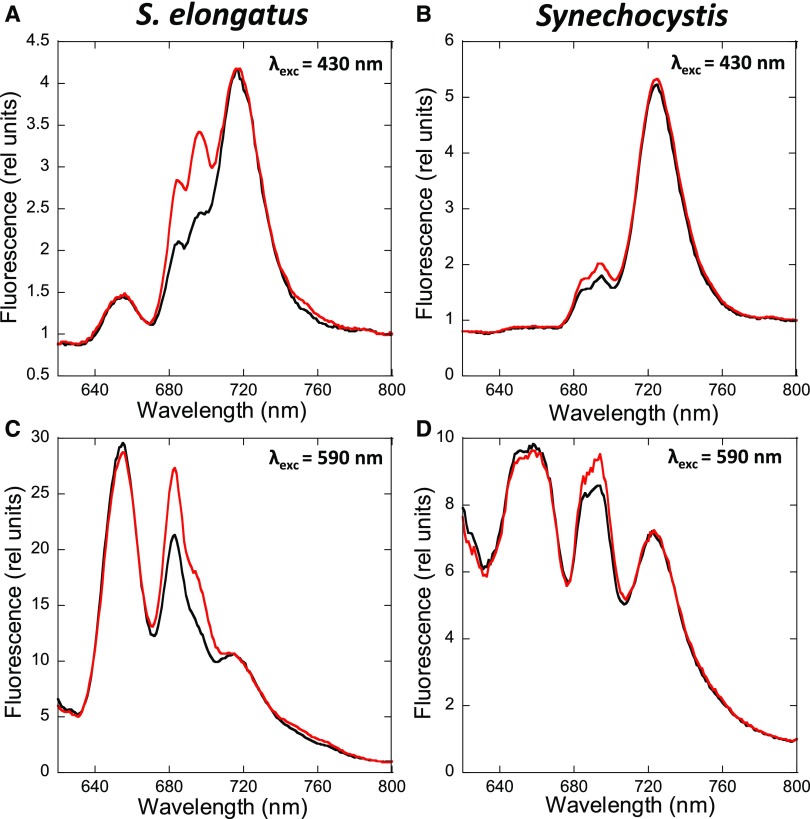
Figure 2 compares the low temperature (−196.15°C) 77 K fluorescence emission spectra of dark- and blue light-adapted S. elongatus (A, C) and Synechocystis (B, D) cells. At this temperature, the fluorescence of both photosystems is visible; while at room temperature, only PSII-related fluorescence is observed.
Figure 2.
State Transitions in S. elongatus and Synechocystis.
Shown are 77 K fluorescence emission spectra of dark- (black, State II) and blue light- (red, 85 µM photons m−2 s−1, State I) adapted S. elongatus (A) and (C) and Synechocystis (B) and (D) cells. The excitation was done at 430 nm (A) and (B) and 590 nm (C) and (D). Normalization was done at 800 nm, and the spectra are the average of at least three independent biological replicates.
Supplemental Figures 1 and 2 show the Gaussian decomposition of these spectra, providing a visualization of different components of the spectra. When the PBSs were preferentially excited (excitation at 590 nm), we observed a large peak at 650 to 660 nm related to PC and allophycocyanin fluorescence; a peak at 683 nm related to the Chl binding protein CP43 and the last emitters of PBS; a peak (or shoulder in S. elongatus) at 695 nm corresponding to reaction center II and CP47; and a peak at 718 nm (S. elongatus) or 722 nm (Synechocystis) related to PSI fluorescence (Van Dorssen et al., 1987; Siefermann-Harms, 1988). The PSII-related peaks at 683 and 695 nm were higher in blue light-adapted cells (State I) than in dark-adapted cells (State II). The differences between the peaks in State I and II were larger in S. elongatus than in Synechocystis. Regarding PSI, the fluorescence was similar in dark- and light-adapted cells of both strains.
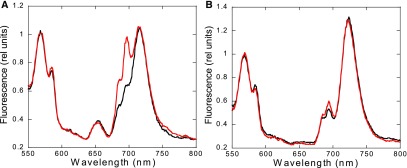
When the Chl was preferentially excited (excitation at 430 nm), the PSI emission peak (at 718 nm in S. elongatus and 722 nm in Synechocystis) was the highest in both strains. In addition, the PSII-related peaks at 685 and 695 nm were much higher in S. elongatus than in Synechocystis, corresponding to a lower PSI to PSII ratio (2:3 in S. elongatus versus 4:5 in Synechocystis). The PSI-related peaks were similar in darkness and blue light illumination. The absence of changes in PSI-related fluorescence during state transitions was confirmed by normalizing the S. elongatus spectra with an external dye (Rhodamine B; Figure 3). The PSII-related peaks increased upon blue light illumination in both strains. Nevertheless, the emission at 695 nm increased more than the one at 683 nm (Supplemental Figures 1 and 2).
Figure 3.
77 K Fluorescence Emission Spectra of the Wild-Type S. elongatus (A) and Synechocystis (B) Cells Using Rhodamine B as an Internal Standard.
Cells were dark adapted for 15 min before the measurements (black lines) or incubated under blue light illumination for 5 min (red lines) before the addition of Rhodamine B (0.4 µM). The excitation was done at 430 nm. Normalization was done at the peak of Rhodamine B at 568 nm, and the spectra are the average of at least three independent biological replicates.
Effect of Hyperosmotic Buffers on State Transitions
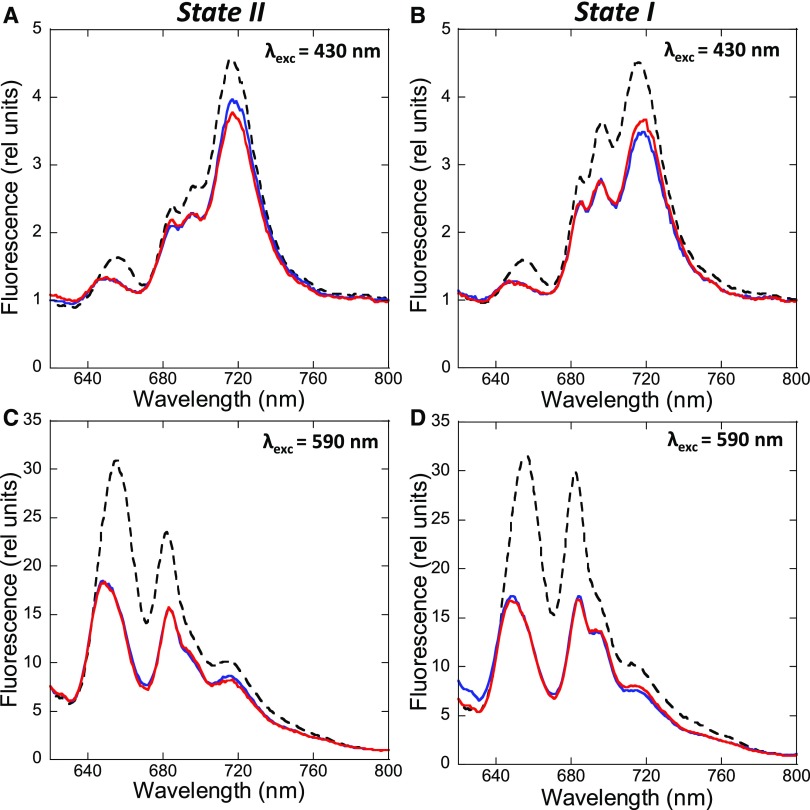
We investigated the effects of the hyperosmotic buffers betaine (1 M, pH 7.0), Suc (1 M), and phosphate (0.5 M, pH 7.5) on state transitions in S. elongatus cells (Figure 4; Supplemental Figure 3). Control cells were incubated in the absence of chemicals in the dark (cells-II) or under blue light illumination (cells-I). Aliquots of these cells were rapidly frozen. Cells-II and cells-I were then incubated with chemicals for 5 min under identical conditions (dark for cells-II, blue light illumination for cells-I). Cells-II were then transferred to blue light for 5 min before freezing, whereas cells-I were transferred to dark for 5 min before freezing. We measured 77 K fluorescence spectra with excitation at 430 nm and at 590 nm. The initial effect observed upon addition of betaine and Suc was a large, general quenching of fluorescence, suggesting that these chemicals have an effect not only on PBSs but also on membranes (Figure 4; Supplemental Figure 3). The quenching effect of hyperosmotic media was previously reported in Synechocystis by (Papageorgiou et al., 1999), who attributed it to alterations in membrane fluidity. Nevertheless, the PBS quenching seemed to be larger than that of Chl, because the 683/695 and 660/695 ratios decreased after the addition of betaine and Suc with excitation at 590 nm. By contrast, phosphate addition had a more specific quenching effect on PBS fluorescence, as seen by the relatively large decrease in fluorescence at 660 nm (Supplemental Figure 3).
Figure 4.
77 K Fluorescence Emission Spectra of S. elongatus Cells Treated with Betaine.
Cells were dark adapted for 15 min (A) and (C) or incubated under blue light illumination (85 photons m−2 s−1) for 5 min (B) and (D). 77 K spectra were measured (dashed lines). Then, samples were taken after 5 min incubation with betaine (1 M) and measured (blue lines). Finally, State I (A) and (C) or State II (B) and (D) was tentatively induced by blue light illumination (85 µM photons m−2 s−1) or darkness, respectively, for 5 min more (red lines). The excitation was done at 430 nm (A) and (B) or 590 nm (C) and (D). Normalization was done at 800 nm, and the spectra are the average of at least three independent biological replicates.
As previously observed (Joshua and Mullineaux, 2004; Li et al., 2004, 2006), no fluorescence changes were detected in 77 K emission spectra obtained by excitation at 590 nm when state transitions were tentatively induced in the presence of betaine, Suc, or phosphate (Figure 4; Supplemental Figure 3). In addition, the chemicals also inhibited the fluorescence changes observed in the 77 K emission spectra obtained with 430 nm excitation (Figure 4; Supplemental Figure 3). These results demonstrate that betaine, Suc, and phosphate also block the changes produced in the membranes. In conclusion, the effect of these hyperosmotic buffers on state transitions could be due to the inhibition of PBS movement and/or processes occurring in the membrane.
State Transitions Kinetics and the Redox State of the PQ Pool in Darkness
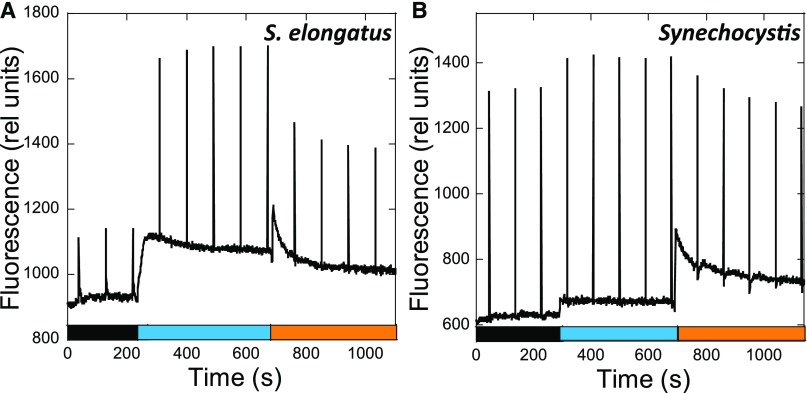
Figure 5 shows typical traces of room-temperature fluorescence kinetics measured with a pulse-amplitude-modulation (PAM) fluorometer in dark-adapted S. elongatus (A) and Synechocystis (B) cells successively illuminated by low intensities of blue and orange light. Dark-adapted cells presented a low dark maximal fluorescence (Fmd), indicating that the cells were in State II. Upon illumination with blue light, which preferentially excites Chl, a large and rapid increase in Fm’ was observed (arriving at a maximal Fmb′ level), indicating the transition to State I. The ratio Fvb to Fvd [variable fluorescence (Fv) = Fm − F0] was ∼4.0 in S. elongatus but only 1.2 in Synechocystis cells (Figure 5). The dark Fvd/F0 was much smaller in S. elongatus than in Synechocystis (0.22 versus 1.09). By contrast, small differences were observed in the Fvb/F0 ratio (0.83 versus 1.26). These data suggest that dark-adapted S. elongatus cells were in a “stronger” State II than Synechocystis cells.
Figure 5.
State Transitions in S. elongatus (A) and Synechocystis (B).
The fluorescence changes were followed with a PAM fluorometer. Dark-adapted cells (Chl concentration 2.5 μg/mL) were successively illuminated with blue light (85 µM photons m−2 s−1) and orange light (20 µM photons m−2 s−1). Saturating pulses (400 ms × 1200 µM photons m−2 s−1) were applied every 90 s.
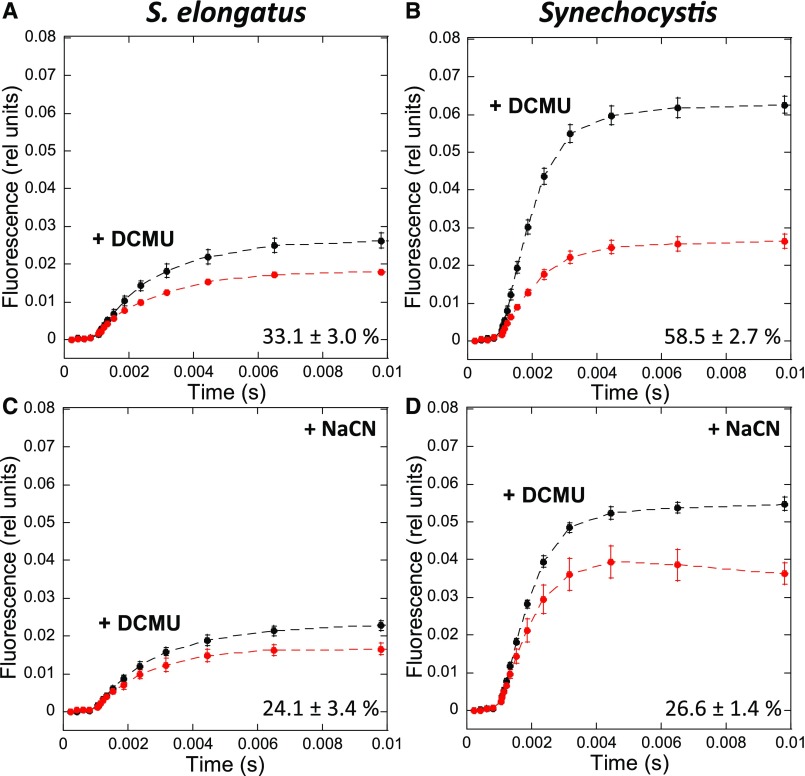
In order to elucidate whether this could be explained by a more reduced PQ pool in dark-adapted S. elongatus, we estimated the redox state of the PQ pool in each strain by measuring fluorescence induction curves in the absence and presence of DCMU, which inhibits electron transfer between the primary (QA) and secondary (QB) quinones in PSII (Figure 6). When dark-adapted cells are illuminated, the PQ pool becomes more reduced and the photochemical centers become partially closed. As a consequence, a concomitant fluorescence increase is observed until a steady state level is reached. The increased kinetics depends on both the initial redox state of the PQ pool and its rate of photochemical reduction under illumination. However, when DCMU is present, a maximum level of fluorescence is reached in which all the centers are closed and the rate of fluorescence increase depends only on the antenna size and is independent of the dark redox state of the PQ pool. Figures 7A and 7B show the fluorescence induction curves in the presence and absence of DCMU for S. elongatus and Synechocystis dark-adapted cells. The area between the curves is much larger for dark-adapted Synechocystis than for S. elongatus cells (59% versus 33% of the DCMU area, respectively).
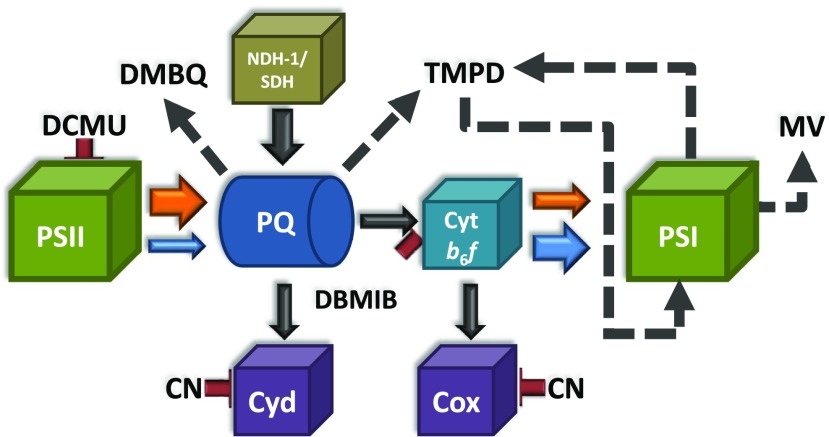
Figure 6.
Schematic Model of the Effects of Different Chemicals on the Intersystem Electron Transport Chain in Cyanobacteria.
The arrows indicate the direction of electron transport between different protein complexes/chemical compounds. The size of each arrow is proportional to the quantity of electron transfer under different conditions. Orange and blue arrows indicate electron transfer under orange or blue light illumination, respectively. Black solid arrows indicate electron transfer occurring under dark and light conditions. Dashed arrows indicate electrons taken/given by the added chemicals. Red lines indicate the inhibition of the different complexes by the given chemicals. Protein complexes: PSII, photosystem II; PSI, photosystem I; PQ, plastoquinone pool; Cyt b6f, cytochrome b6f; NDH-1, NAD(P)H dehydrogenase type 1; SDH, succinate dehydrogenase; Cyd, cytochrome bd-type quinol oxidase; Cox, cytochrome c oxidase. Chemical compounds: DCMU, 3-(3,4-dichlorophenyl)-1,1-dimethylurea; DMBQ, 2,6-dimethoxy-1,4-benzoquinone; DBMIB, 2,5-dibromo-3-methyl-6-isopropyl-p-benzoquinone; TMPD, N, N, N′, N′-tetramethyl-p-phenylenediamine; MV, methyl viologen; CN, cyanide.
Figure 7.
Fluorescence Induction in the Absence and Presence of NaCN in S. elongatus and Synechocystis Cells.
Whole cells (Chl concentration 2.5 μg/mL) were dark adapted for 15 min at 31°C before illumination with orange light (180 µM photons m−2 s−1).
(A) and (B) Fluorescence induction in the absence of NaCN.
(C) and (D) Fluorescence induction after 3-min incubation in the presence of NaCN (80 µM).
Black lines show fluorescence induction in the presence of DCMU (10 µM). Red lines show fluorescence induction in the absence of DCMU. The detection wavelength was ≥695 nm (far-red). The curves are the average of at least three independent biological replicates. The percentages shown are the area between the curves relative to the area under the curve with DCMU. The errors bars correspond to the sd of the data shown.
Because the area between the curves is at least partially proportional to the amount of dark-oxidized PQ (Bennoun, 1982; Srivastava et al., 1995), these results suggest that the PQ pool is more reduced in dark-adapted S. elongatus cells than in Synechocystis. However, under illumination, the rates of PQ reduction by PSII and of PQH2 (reduced PQ) reoxidation by PSI also affect the kinetics of fluorescence induction. These rates could be different between the strains, as the PSI to PSII and the phycobiliprotein to Chl ratios are different in Synechocystis and S. elongatus. Therefore, we performed a control experiment in which the PQ dark reduction level in each strain was modified without changing the size of the antenna or PSI and PSII activities. The dark PQ redox state depends on the relative activities of NDH-1/SDH and of cyanide-sensitive terminal oxidases (Cyd and Cox; Figure 6); thus, the addition of sodium cyanide (NaCN) should lead to a large PQ-reduction level. A large effect of NaCN was observed in Synechocystis cells, in which the area between the curves (±DCMU) decreased from 59 in the absence of NaCN to 27% in its presence (Figure 7). By contrast, the effect was much smaller in S. elongatus cells, with only a decrease from 33 to 24% (Figure 7). These results strongly suggest that the PQ pool was more strongly reduced in S. elongatus than in Synechocystis.
The illumination of blue light-adapted cells with orange light (which preferentially excites phycobiliproteins) induced a decrease in Fm’ in both cyanobacterial strains (Figure 5). In Synechocystis cells, the steady state Fmo was similar to Fmd, whereas in S. elongatus, the Fmo was higher. As already mentioned, the “strength” of State II depends on the concentration of reduced PQ. Thus, our results indicate that the PQ pool was more strongly reduced in dark-adapted S. elongatus than in orange light-illuminated cells. This was not the case in Synechocystis cells, where the redox state of the PQ pool appeared to be similar under both conditions.
Is Cyt b6f Involved in State Transitions in S. elongatus?
Effect of DMBQ and DBMIB on State Transitions
Mao et al. (Mao et al., 2002) and Huang et al. (Huang et al., 2003) proposed that cyt b6f is involved in the signaling pathway of cyanobacterial state transitions, as observed in green algae and plants. They based their proposal on the results obtained by chemically inducing state transitions in Synechocystis and Synechococcus PCC 7002 using 2,6-dimethoxy-1,4-benzoquinone (DMBQ), p-benzoquinone (PBQ), and 2,5-dibromo-3-methyl-6-isopropyl-p-benzoquinone (DBMIB). DMBQ and PBQ accept electrons from the PQ pool (Preston and Critchley, 1988), while DBMIB inhibits cyt b6f activity by attaching to the Qo site (the PQH2 binding site), preventing reoxidation of the PQ pool (Roberts and Kramer, 2001; Figure 6). These authors found that the addition of DMBQ (or PBQ) to dark-adapted cells induced an increase in Fmd, which they attributed to a partial transition to State I triggered by oxidation of the PQ pool. The simultaneous addition of DMBQ and DBMIB inhibited this increase. Both authors hypothesized that under these conditions, the PQ pool remained oxidized, which led them to conclude that the binding of DBMIB to the cyt b6f Qo site was primarily involved in the transition to State II. However, none of these studies demonstrated that the PQ pool remained oxidized under these conditions.
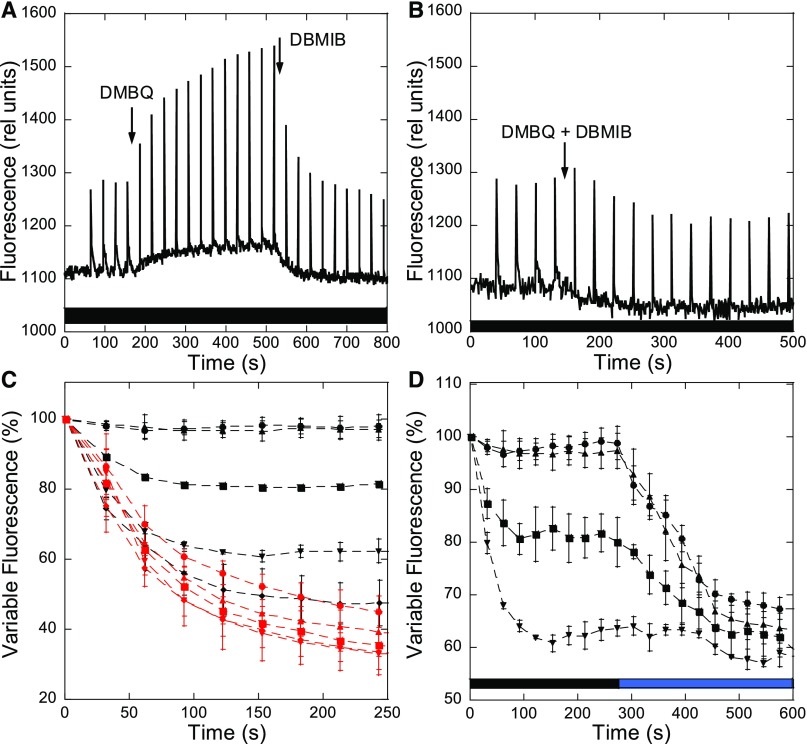
Figure 8 shows the effect of DMBQ and DBMIB on Fmd in dark-adapted S. elongatus cells. The addition of DMBQ (250 µM) induced a slow increase in Fmd (and F0) related to a partial transition to State I. Higher concentrations of DMBQ cannot be used because they induce fluorescence quenching. When DBMIB (20 µM) was subsequently added, a rapid decrease of Fmd (and F0) was observed (Figure 8A). When both chemicals were simultaneously added, the DMBQ-induced increase in fluorescence was inhibited (Figure 8B). These effects of chemicals were therefore similar to those previously observed in Synechocystis and Synechococcus PCC 7002 cells (Huang et al., 2003; Mao et al., 2003).
Figure 8.
Fluorescence Changes Induced by Chemicals.
S. elongatus cells were dark adapted (15 min) before the successive addition of DMBQ (250 µM) and DBMIB (20 µM) (A) or the simultaneous addition of DMBQ (250 µM) and DBMIB (20 µM) (B). The measurements were done using a PAM fluorometer (measuring light 650 nm). Saturating pulses (400 ms × 1200 µM photons m−2 s−1) were applied every 30 s. Typical experiments are shown.
(C) Variable fluorescence traces of different concentrations of DBMIB, added at time 0, when State I was induced by blue light (red lines, 85 µM photons m−2 s−1) or by DMBQ (250 µM) in darkness (black lines). The values were normalized to the variable fluorescence of State I (induced by light or DMBQ, respectively).
(D) Variable fluorescence traces of different concentrations of DBMIB when State I was induced by DMBQ (250 µM) in darkness, and the cells were then illuminated with blue light (85 µM photons m−2 s−1). The values were normalized to the variable fluorescence of State I induced by DMBQ.
The averages of at least three independent biological replicates are shown. The errors bars correspond to the sd of the data shown. (●) 2.5 µM DBMIB; (▴) 5 µM DBMIB; (■) 10 µM DBMIB; (▾) 15 µM DBMIB; (♦) 20 µM DBMIB.
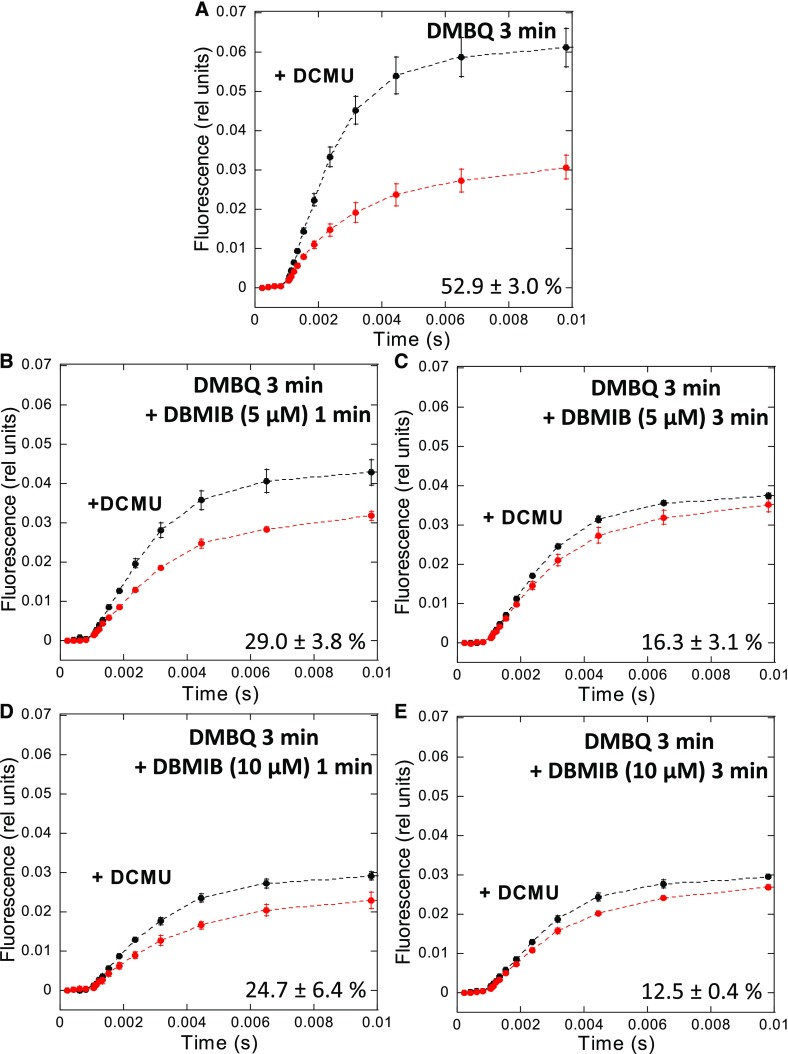
We then explored the dark redox state of the PQ pool in S. elongatus cells by analyzing the kinetics of fluorescence induction in the presence and absence of DCMU (as described in the previous section). The measurements were performed in dark-adapted cells (15 min) in the presence of DMBQ alone (250 µM) or DMBQ and DBMIB (5 and 10 µM). The measurements were done after 3 min of incubation with DMBQ and at 1 and 3 min after the addition of DBMIB (Figure 9). As previously mentioned, the PQ pool is reduced in dark-adapted S. elongatus cells: the area between the curves (±DCMU) is small and Fmd is low (Figure 7A). The addition of DMBQ increased the level of Fmd and the area between the two curves (±DCMU), suggesting that the PQ pool had become oxidized (Figure 9A). Because DMBQ also accepts electrons from the PQ pool during the light measurement, the larger area between the curves is also partially related to its activity during the measurement.
Figure 9.
Estimation of the PQ Pool Redox State.
Fluorescence induction in the presence (black lines) or absence (red lines) of DCMU (10 µM) in dark-adapted S. elongatus cells. Whole cells (Chl concentration 2.5 μg/mL) were dark adapted for 15 min (at 31°C) before illumination with orange light (180 µM photons m−2 s−1).
(A) Cells were incubated for 3 min with DMBQ (250 µM).
(B) and (C) Cells were incubated with DMBQ (250 µM) for 3 min and then with DBMIB (5 µM) for 1 min or 3 min, respectively.
(D) and (E) Cells were incubated with DMBQ (250 µM) for 3 min and then with DBMIB (10 µM) for 1 min or 3 min, respectively.
Black lines show fluorescence induction in the presence of DCMU. Red lines show fluorescence induction in the absence of DCMU. The detection wavelength was ≥695 nm (far-red). The curves are the average of at least three independent biological replicates. The percentages shown are the area between the curves relative to the area under the curve with DCMU. The errors bars correspond to the sd of the data shown.
The addition of 5 µM DBMIB induced a rapid decrease of Fmd and of the area between the curves. The smaller area indicated that the PQ pool was more strongly reduced in darkness and during the light measurement in the presence of DBMIB (Figure 9B). Longer periods of incubation with DBMIB induced a larger decrease in area, indicating a larger reduction of the PQ pool even in the presence of DMBQ (Figure 9C). In line with these results, the addition of 10 µM DBMIB had a faster and larger effect (Figures 9D and 9E). Thus, the DMBQ-induced transition to State I is inhibited by DBMIB, likely because DBMIB treatment leads to the reduction of the PQ pool, even in the presence of DMBQ, at least in S. elongatus.
We further studied the effects of DBMIB by adding different concentrations of this chemical (2.5, 5, 10, 15, and 20 µM) to S. elongatus cells adapted to State 1 by illumination with blue-green light or by adding DMBQ in darkness (Figure 8C). Under illumination, all concentrations of DBMIB efficiently induced a large transition to State 2 with rather similar kinetics (red curves). However, concentrations higher than 5 µM were necessary to induce the transition to State 2 in darkness and in the presence of DMBQ. In this case, the rate and amplitude of the transition depended on DBMIB concentration. Thus, the same concentration of DBMIB had different effects under illumination and in darkness, in the presence of DBMQ. For example, 5 µM DBIMB induced almost maximum fluorescence quenching under illumination but was unable to induce any quenching in darkness in the presence of DMBQ. Upon illumination of dark-adapted cells in the presence of DMBQ and different concentrations of DBMIB (2.5, 5, 10, and 15 µM), larger fluorescence quenching was induced (Figure 8D). These results strongly suggest that the transition to State II is induced by the reduction of the PQ pool and not by the binding of DBMIB to the Qo site in cyt b6f.
Effect of the TMPD on State Transitions
We then looked for another chemical compound that does not interact with cyt b6f and can oxidize the PQ pool in the presence of DBMIB to further confirm that this complex does not play a role in state transitions. N, N′, N′-tetramethyl-p-phenylenediamine (TMPD) represents such a compound, because it restores oxygen evolution by reversing the inhibitory effect of DBMIB on the photosynthetic electron transport chain (Figure 6; Draber et al., 1970; Nanba and Katoh, 1985). Nanba and Katoh demonstrated that TMPD accepts electrons from the PQ pool and directly donates them to P700+, bypassing the DBMIB-poisoned cyt b6f complex. However, TMPD is also a good electron acceptor from PSI and can generate cyclic electron transfer around PSI (Figure 6; Hiyama and Ke, 1972). This chemical has opposite effects on state transitions, depending on the preferential electron donor to TMPD (PQ pool versus PSI).
It is expected that, under blue light in the absence of DBMIB (State I), the addition of TMPD should have no effect on fluorescence if TMPD is efficient at oxidizing reduced PQ, as the PQ pool should remain oxidized. However, exactly the opposite effect was observed: a large quenching of fluorescence was induced upon TMPD addition. The amplitude of quenching depended on the TMPD concentration (Supplemental Figures 4A to 4D). This effect can be explained by assuming that, under these conditions, TMPD is primarily involved in cyclic electron transport and functions poorly as an oxidizer of reduced PQ. Cyclic electron transport also limits linear electron flow through cyt b6f and consequently contributes to PQ reduction. In accordance with the poor efficiency of TMPD in oxidizing reduced PQ under these conditions, the addition of TMPD to DBMIB-poisoned cells led to no change in fluorescence, indicating that State II was maintained (Supplemental Figure 4E). In addition, TMPD did not hinder the effect of DBMIB on state transitions: large fluorescence quenching was observed even in the presence of TMPD (Supplemental Figure 4F).
In an attempt to modify the behavior of TMPD, we added methyl viologen (MV) to the reaction. MV is a good electron acceptor from PSI that could compete with TMPD at this level. This approach was found to be successful: the addition of DBMIB following that of TMPD led to only a small decrease in fluorescence, indicating that under these conditions, TMPD was efficient at performing electron uptake from the PQ pool (Supplemental Figure 4G). Notably, the presence of MV did not affect state transitions in the absence of TMPD (Supplemental Figures 4F and 4G).
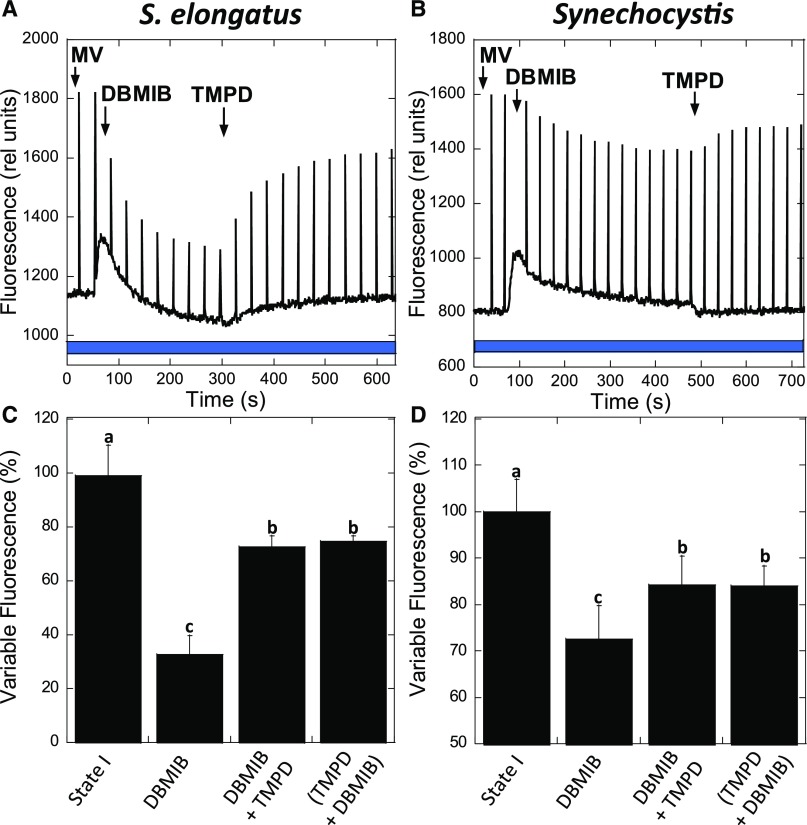
More importantly, when TMPD was added in the presence of MV to largely quenched DBMIB-poisoned cells, it induced a large increase in fluorescence related to the transition to State 1 (Figure 10). The final Fv was 70% that of cells under blue light illumination in the absence of chemicals. The transition to State 1 induced by TMPD under blue light illumination was larger than that induced by DMBQ in darkness (55%). Thus, TMPD is able to reverse the effect of DBMIB by taking electrons from PQ and giving them to PSI, bypassing the inhibited cyt b6f complex. Similar results were obtained with Synechocystis cells using 3 mM MV and 7.5 µM TMPD (Figure 10). In conclusion, the transition to State I can be induced even when cyt b6f is inhibited by DBMIB by partially oxidizing the PQ pool.
Figure 10.
Fluorescence Changes Induced by TMPD in the Presence of MV and DBMIB.
S. elongatus (A) and Synechocystis (B) cells were dark adapted (15 min) before the transition to State I by blue light illumination (85 µM photons m−2 s−1) in the presence of MV (2 mM or 3 mM, respectively). DBMIB (10 µM for S. elongatus or 7.5 µM for Synechocystis) was then added. After reaching a steady State II, TMPD (10 µM) was added. Saturating pulses were applied every 30 s.
(C) and (D) Graphs show the percentage of variable fluorescence for S. elongatus and Synechocystis, respectively, after blue light illumination (State I); after the addition of DBMIB; DBMIB and then TMPD or when both chemicals were added at the same time.
The mean and sd of four independent biological replicates are shown. Columns without common letters differ significantly (Tukey test, P < 0.05).
Are Protein Phosphorylation Reactions Required for Cyanobacterial State Transitions?
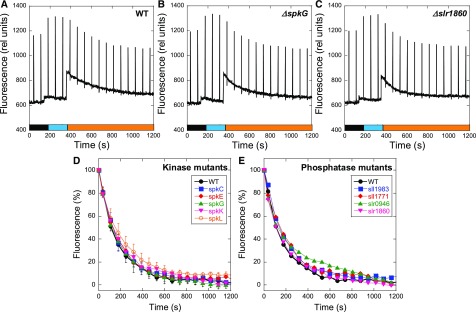
In plants and green algae, conformational changes induced in the cyt b6f complex (especially in the Rieske protein) by the occupancy of the Qo site by a PQH2 molecule (Zhang et al., 1998; Zito et al., 1999; Breyton, 2000; Finazzi et al., 2001) activates a specific kinase (STN7/Stt7; Depège et al., 2003; Bellafiore et al., 2005), which phosphorylates the mobile trimers of LHCII, inducing their detachment from PSII. As previously mentioned, at least two studies suggested that protein phosphorylation by one specific Ser/Thr kinase could also trigger cyanobacterial state transitions (Allen et al., 1985; Chen et al., 2015). To test this hypothesis, we created Synechocystis protein kinase and phosphatase mutants. Synechocystis has 12 genes encoding putative Ser/Thr kinases (SPTKs). Seven of these genes encode proteins belonging to the Ser/Thr-protein kinase N2 subfamily (spkA to spkG) and five belonging to the ATP-binding cassette transporter1 subfamily (spkH to spkL; Zorina, 2013). Each gene was individually deleted by replacing it with a kanamycin resistance cassette (see Methods for details). We also individually deleted genes encoding nine phosphatases: slr0328 (Protein tyrosine phosphatase family), sll1771, slr1860, sll1033, sll0602, slr0114, slr1983 (protein phosphatase Mg2+- or Mn2+-dependent family), sll1387 (phosphoprotein phosphatase family) and slr0946. Thus, we created 12 single Synechocystis kinase mutants and 9 single Synechocystis phosphatase mutants. To determine if the mutants were affected in state transitions, we illuminated dark-adapted mutant cells with blue light to induce the transition to State I, followed by orange light to induce the transition to State II (Figure 11). All of the dark-adapted single mutants went to State I upon blue light illumination and then to State II during orange light illumination (Figure 11; Supplemental Figure 5). The rates of decrease in fluorescence during the State I to State II transition were similar in the wild-type and mutant cells (Figure 11; Supplemental Figure 5). These experiments indicate that no specific Ser/Thr kinase or phosphatase is involved in cyanobacterial state transitions, as it is the case in green algae and plants.
Figure 11.
State Transitions in Synechocystis Kinase and Phosphatase Mutants.
The fluorescence changes were followed with a PAM fluorometer. Dark-adapted cells (Chl concentration 2.5 μg/mL) were successively illuminated with blue light (85 µM photons m−2 s−1) and orange light (40 µM photons m−2 s−1). Typical experiments are shown for the Synechocystis wild-type (WT) (A), ΔspkG (B), and Δslr1860 (C) cells. Saturating pulses (400 ms × 1200 µM photons m−2 s−1) were applied every 70 s.
(D) and (E) Variable fluorescence (%) decrease induced by orange illumination of blue light-adapted cells (State I to State II transition) of kinase and phosphatase mutants, respectively. The values were normalized, with the variable fluorescence of State I equal to 100% and the variable fluorescence of State II equal to 0%.
(D) The values for the wild type (WT) and ΔspkC, ΔspkE, ΔspkG, ΔspkK, and ΔspkL mutants are shown.
(F) The values for the wild type (WT) and Δsll1983, Δsll1771, Δslr0946, and Δslr1860 mutants are shown.
The averages of at least three independent biological replicates are shown. The errors bars correspond to the sd of the data shown.
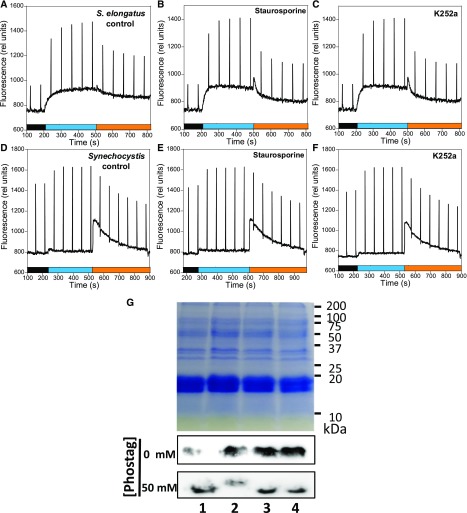
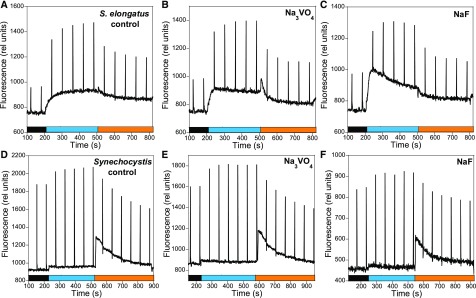
To confirm that phosphorylation reactions are not essential for cyanobacterial state transitions, we tested the effects of kinase and phosphatase inhibitors on Synechocystis and S. elongatus. Staurosporine and K252a (a derivative of staurosporine) are potent inhibitors of Ser/Thr and Tyr kinases that interact with their ATP binding sites (Fernandez et al., 2006; Nakano and Omura, 2009). NaF inhibits Ser/Thr and acid phosphatases and Na3VO4 inhibits Tyr and alkaline phosphatases (Delphin et al., 1995; McCartney et al., 1997; and references inside). Staurosporine and NaF were shown to inhibit state transitions in the green alga Chlamydomonas reinhardtii (Delphin et al., 1995 and references inside). Figure 12 shows that the presence of staurosporine or K252a, which were added in excess (21 µM and 1 µM, with the half maximal inhibitory concentration of these compounds 0.6 µM and 96 nM, respectively (Nakano and Omura, 2009) did not inhibit state transitions in Synechocystis or S. elongatus.
Figure 12.
Effects of Kinase Inhibitors on State Transitions and PII Phosphorylation.
S. elongatus (A) to (C) and Synechocystis (D) to (F) cells were incubated in the absence and presence of kinase inhibitors for 90 min in darkness. Then the cells (Chl concentration 2.5 μg/mL) were successively illuminated with blue light (85 µM photons m−2 s−1) and orange light (40 µM photons m−2 s−1), and the fluorescence changes were followed using a PAM fluorometer.
(A) and (D) Control cells containing DMSO (0.1% v/v).
(B) and (E) Cells treated with stausporine (21 µM).
(C) and (F) Cells treated with K252a (1.07 µM). Saturating pulses (400 ms × 1200 µM photons m−2 s−1) were applied every 60 s.
(G) Gel electrophoresis containing Phos-tag to detect phosphorylated proteins and immunoblot using an anti-PII protein. (1) BG11 medium; (2) nitrogen-depleted BG11 medium; (3) nitrogen-depleted BG11 medium supplemented with K252a (1.07 µM); (4) nitrogen-depleted BG11 medium supplemented with staurosporine (21 µm).
To confirm that these kinase inhibitors are able to enter cyanobacterial cells and inhibit phosphorylation reactions, we tested their effects on the phosphorylation of the PII protein in S. elongatus. The PII protein (glnB gene product) is involved in the tight coordination of carbon and nitrogen assimilation. Its activity involves the phosphorylation and dephosphorylation of a Ser residue (Forchhammer and Tandeau de Marsac, 1995a, 1995b). In ammonium-grown cells, PII is completely dephosphorylated. The transfer of cells to medium lacking combined nitrogen induces phosphorylation of PII (Forchhammer and Tandeau de Marsac, 1995a, 1995b). The phosphorylation state of PII can be analyzed by gel electrophoresis in a phos-tag gel SDS-PAGE system and by immunoblot detection. In this system, phosphorylated proteins migrate more slowly than non-phosphorylated ones (Kinoshita and Kinoshita-Kikuta, 2011). Figure 12G shows that the presence of staurosporine and K252a completely inhibited the phosphorylation of the PII protein under nitrogen starvation conditions. This result indicates that both kinase inhibitors entered into S. elongatus cells and were able to inhibit protein phosphorylation.
Finally, phosphatase inhibitors (NaF and Na3VO4) also did not affect state transitions (Figure 13). Nevertheless, NaF induced the partial inhibition of the oxygen-evolving activity of PSII (Supplemental Figure 6) and a general decrease in fluorescence (Figure 13) in Synechocystis, indicating that this chemical entered the cells. In conclusion, our experiments show that phosphorylation reactions are not involved in cyanobacterial state transitions.
Figure 13.
Effect of Phosphatase Inhibitors on State Transitions.
S. elongatus (A) to (C) and Synechocystis (D) to (F) cells were incubated in the absence and presence of phosphatase inhibitors for 90 min in darkness. Then the cells (Chl concentration 2.5 μg/mL) were successively illuminated with blue light (85 µM photons m−2 s−1) and orange light (40 µM photons m−2 s−1) and the fluorescence changes were followed using a PAM fluorometer.
(A) and (D) Control cells containing NaCl (100 mM).
(B) and (E) Cells treated with Na3VO4 (1 mM).
(C) and (F) Cells treated with NaF (50 mM). Saturating pulses (400 ms × 1200 µM photons m−2 s−1) were applied every 60 s.
DISCUSSION
While the mechanism of state transitions in plants and green algae has been largely elucidated, it remains to be characterized in cyanobacteria. Contradictory hypotheses have been proposed about cyanobacterial state transitions based on studies addressing different aspects of the mechanism: the movement of PBS, spillover, reorganization of membrane complexes, involvement of cyt b6f, and/or phosphorylation reactions in signal transduction. However, none of these hypotheses has been definitively supported. Our results help to elucidate open questions about the mechanism behind the large fluorescence quenching observed in State II and the alleged role of the cyt b6f complex in the signaling pathway involved in cyanobacterial state transitions.
The Contributions of PBS Versus the Membrane to State Transitions
It was previously shown that PBS can easily move and that high concentrations of betaine, Suc, and phosphate inhibit the diffusion of PBS and state transitions (Joshua and Mullineaux, 2004; Li et al., 2004, 2006). Li et al. (2004) also observed that, in Spirulina platensis, betaine inhibits changes in 77 K emission spectra with 430 nm excitation. However, the authors did not discuss this last result. Based on these works, it was concluded that the movement of PBS from one photosystem to the other was the main reason for the observed changes in fluorescence. By contrast, we demonstrated that these chemicals, in addition to hindering PBS movement, inhibit fluorescence changes that depend on membrane processes. No fluorescence change in 77 K emission spectra was detected in the presence of betaine, Suc, or phosphate upon cells’ illumination, not only when PBSs were excited but also when Chl was excited. Thus, based on these experiments, it cannot be concluded that the movement of PBS is the main contributor to state transitions in cyanobacteria.
Our experiments did not allow us to distinguish which changes, if any, occur at the level of PBS during state transitions: detachment of PBS from one or both photosystems, or changes in energy transfer from PBS to one or the other photosystem. Nevertheless, we expect the contribution of these changes to be small in both S. elongatus and Synechocystis, because the main increase in fluorescence emission from State II to State I was related to PSII (G3, 695 nm emission peak; Supplemental Figures 1 and 2).
PSII Quenching Is Involved in State II
Ranjbar Choubeh et al. (2018) recently showed that state transitions involve a reversible quenching of PSII fluorescence independently of spillover changes in S. elongatus cells. They measured the fluorescence decay kinetics of cells in State II and State I with a streak camera using 430 nm or 577 nm excitation at 77 K. By performing global analysis of the data, the authors obtained decay-associated spectra. When 430 nm excitation was used, PSII emission decreased, but the decay-associated spectra showed that PSI emission was similar in State I and State II. This argues against a change in spill-over during state transitions. This was also observed in Synechocystis cells (Ranjbar Choubeh et al., 2018).
Our results confirm these observations. We showed that both the 683 nm and 695 nm peaks decrease in State II (under Chl and PBS excitation), whereas the 718 nm peak did not change (see Supplemental Figures 1 and 2, including peak deconvolution to visualize different components of the spectra). The absence of changes in PSI-related fluorescence during state transitions was confirmed by normalizing the spectra with an external dye (Rhodamine B; Figure 3). These results indicate that PSII emission is largely quenched in State II and that spillover from PSII to PSI does not contribute to this quenching. The PSII-related quenching mechanism remains to be elucidated.
The Role of Cyt b6f in Cyanobacterial State Transitions
One of the big questions that remain to be answered about the mechanism of cyanobacterial state transitions is how the signal is transmitted from the PQ pool to the PBS and/or photosystems to induce their movements or fluorescence quenching. In plants and green algae, the binding (and subsequent release) of PQH2 in the Qo site of the cyt b6f complex plays a critical role in the activation of a specific Ser/Thr kinase that phosphorylates LHCII (Vener et al., 1995, 1997; Zito et al., 1999). The phosphorylated LCHII detaches from PSII and totally (or partially) associates with PSI (Vener et al., 1997; Wollman, 2001). The role of the Qo site in state transitions was first suggested based on the effect of DBMIB in Chlamydomonas, where DBMIB inhibits State I to State II transition, although in its presence, the PQ pool is largely reduced (Finazzi et al., 2001). By contrast, in cyanobacteria, DBMIB induces the State I to State II transition. This occurs even in the presence of DCMU (which inhibits photoreduction of the PQ pool by PSII) or DMBQ (which oxidizes the PQ pool by taking electrons from PQH2, as shown in present results and in Mao et al. [2002] and Huang et al. [2003]). These authors proposed that the action of DBMIB is related to its binding to the Qo site and not to the reduction of the PQ pool. The authors assumed that the PQ pool remains oxidized in the presence of both DBMIB and DMBQ (or PBQ).
Here, we demonstrated that this is not true, because the addition of DBMIB induced reduction of the PQ pool even in the presence of DMBQ. Moreover, we demonstrated that the same concentration of DBMIB has different effects on state transitions depending on the experimental conditions. For instance, DBMIB at 5 µM was unable to induce the transition to State II in dark-adapted cells in the presence of DMBQ, whereas it induced large quenching in blue-green-light-adapted cells in the absence or presence of DMBQ. The amplitude of fluorescence quenching induced by 10 µM DBMIB was also larger under illumination than in darkness in the presence of DMBQ. Altogether, these experiments strongly suggested that the cyt b6f complex is not involved in cyanobacterial state transitions. The finding that the addition of TMPD to DBMIB-poisoned cells induced a large increase in fluorescence related to PQ oxidation and transition to State I indicates that DBMIB binding to the Qo site of cyt b6f is not involved in the transition to State II. Under these conditions, DBMIB remained attached to the Qo site, and the cyt b6f complex was inactive. As an alternative, it was recently proposed that the single Chl a molecule present in cyt b6f could act as a redox sensor and signal transmitter during state transitions (Vladkova, 2016). This Chl a molecule is evolutionarily conserved and is present in all oxygen-evolving photosynthetic species (Vladkova, 2016). However, changes in this Chl a were induced by binding of DBMIB or PQH2 to the Qo site. Thus, based on our results, the involvement of this Chl a molecule in cyanobacterial state transitions is not likely.
In addition, the characterization of 12 kinase and 9 phosphatase single mutants demonstrated that no specific protein kinase and/or phosphatase is necessary for cyanobacterial state transitions. More generally, the use of kinase and phosphatase inhibitors demonstrated that phosphorylation reactions are not essential for state transitions in Synechocystis and S. elongatus. Thus, signal transduction from the PQ pool to the antenna and the photosystems is completely different in cyanobacteria versus green algae and plants.
While DCMU and DBMIB have opposite effects on cyanobacterial state transitions, they have the same effect on the transcription of photosynthetic genes (Alfonso et al., 1999, 2000; El Bissati and Kirilovsky, 2001). Both DCMU and DBMIB induced an increase in psbA transcription and a decrease in psaE transcription when added into Synechocystis cells under white and orange illumination. These findings strongly suggest that cyt b6f is involved in the redox transcriptional regulation of photosynthetic genes in Synechocystis (Alfonso et al., 1999, 2000; El Bissati and Kirilovsky, 2001). In conclusion, cyt b6f is not involved in cyanobacterial state transitions, but it appears to be involved in redox transcriptional regulation.
Because it seems that in cyanobacteria, the principal effect of PQ reduction is an increase in PSII quenching and that cyt b6f is not involved in the signaling pathway, it is tempting to hypothesize that PSII itself senses the redox state of the PQ pool. Sensing cannot be linked to QA− accumulation because both the presence of DCMU and the reduction of the PQ pool increase QA− concentration; but while DCMU induces the transition to State I, the reduction of the PQ pool induces the transition to State II. DCMU binding and over-reduction of the PQ pool were previously found to accelerate photoinhibition but through different mechanisms (Kirilovsky et al., 1994; Fufezan et al., 2005; Fischer et al., 2006). In line with this observation, the QB site could be modified differently in the presence of DCMU or PQH2, leading to different effects on QA redox potential, recombination reactions, and the generation of PSII quenching related to State II.
The PQ pool redox state could be sensed not only at the level of the QB site but also by the QC hydrophobic tunnel. The existence of this QC tunnel formed by cyt b559 and psbJ was suggested by the x-ray crystallographic structural model of PSII of Thermosynechococcus elongatus at 2.9 Å resolution (Guskov et al., 2009). The function of this tunnel as a quinone binding site remains to be confirmed, because later structures at higher resolution did not contain a quinone in this hydrophobic pocket (Umena et al., 2011). However, Synechocystis mutants containing mutations around the proposed QC site show altered state transitions, making this site another interesting target of study (Huang et al., 2016).
In addition of these QB/C sites, there are other ways by which the redox state of the PQ pool could be sensed; the participation of other known proteins or novel factors in the signaling pathway cannot be ruled out. Overall, while the PSII-quenching mechanism and the redox sensor of the PQ pool remain to be elucidated, our results rule out the involvement of cyt b6f in this process.
METHODS
Culture Conditions and Replicates
The cyanobacteria Synechocystis PCC 6803 and Synechococcus elongatus PCC 7942 strains were grown photo-autotrophically in blue-green (BG11) medium (Herdman et al., 1973). The cells were incubated in a rotary shaker (120 rpm) at 31°C illuminated by fluorescence white lamps (50 µM photons m−2 s−1) under a CO2-enriched atmosphere. The cells were maintained in their logarithmic phase of growth for all experiments. The kinase and phosphatase mutants were grown in the presence of kanamycin (40 µg/mL).
A biological replicate is a batch of cells on a particular day. The measurements were performed several times using the same batch of cells (technical replicates). The mean of each batch was calculated. The biologically independent experiments were performed on different days separated by at least a week. Thus, completely different cells were tested.
Construction of Kinase and Phosphatase Mutants
To obtain the kinase mutants (∆spkB, ∆spkD, ∆spkE, ∆spkF, ∆spkG, ∆spkI, ∆spkJ, ∆spkK, and ∆spkL) a 500 bp fragment in the upstream region of each kinase gene was cloned into the pMD T-18 vector (Takara, Japan) and digested with XbaI. The PRL446 plasmid containing the kanamycin cassette was also digested with XbaI. Both linear fragments were ligated to generate the plasmid use to transform the Synechocystis wild-type cells in order to obtain the knockout kinase mutants. The strategy to obtain the ∆spkA and the ∆spkC mutants was similar with only two minor modifications: 1) the plasmid containing the 500 bp upstream fragment was digested with SmaI instead of XbaI. 2) To obtain the final construction, the linearized plasmid was ligated to the kanamycin cassette with blunt ends, and the pPM-kinase-upper-kanamycin was obtained.
In parallel, a 500 bp fragment in the downstream region of each kinase gene was cloned into the pMD T-18 vector (Takara) and digested with SalI to generate the downstream fragment. Blunt ends were generated in the downstream fragment and in the pPM-kinase-upper-kanamycin linearized using the SacI/SphI enzyme. The resulting blunt-end DNAs were ligated together. After testing the direction of the inserted downstream fragment by PCR, the pPM-kinase-upper-kanamycin-down was used to transform the wild-type Synechocystis cells.
To obtain the phosphatase mutants (∆slr0328, ∆sll1771, ∆slr1860, ∆sll1033, ∆sll0602, ∆slr0114, ∆slr1983, ∆sll1387, and ∆slr0946), a 500 bp fragment upstream of each phosphatase gene, the kanamycin cassette, and a 500 bp fragment downstream of each phosphatase gene were spliced together using the PCR overlap extension method to obtain the upper-kanamycin-down DNA fragment for each phosphatase gene. The resulting DNA fragments were supplemented with a thymine at both termini and inserted into the T-cloning vector pMD T-18 (Takara) to generate the final pPM-phosphatase-upper-kanamycin-down plasmids.
The Synechocystis wild-type cells were transformed with these plasmids to obtain the knockout kinase and phosphatase mutants. The presence of the kanamycin cassette replacing the kinase and phosphatase genes and the complete segregation of each mutant were tested by PCR amplification and sequencing.
The oligonucleotides used in these constructions are described in Supplemental Table.
The ΔspkH Synechocystis mutant construction is described in (Zorina et al., 2011).
Fluorescence Measurements
PAM Fluorometer
State transitions were monitored using a pulse amplitude modulated fluorometer (101/102/103-PAM; Walz) in a 1 × 1-cm square stirred cuvette. All experiments were performed at 31°C on dark-adapted (15 min) whole cells at a Chl concentration of 2.5 μg/mL. State I was induced by treatment with 85 µM photons m−2 s−1 of blue-green light (halogen white light filtered by a Corion cut-off 550-nm filter, 400 to 500 nm). State II was induced by treatment with 25 (or 40) µM photons m−2 s−1 of orange light (halogen white light filtered by a Melles Griot 03 FIV 046 filter, 600 to 640 nm) or by dark incubation. Saturating flashes (400 ms × 1200 µM photons m−2 s−1) were given to probe the maximum fluorescence level. The fluorescence parameters used in the analysis are the following: F0, basal fluorescence; Fmd, maximum fluorescence in darkness; Fm’, maximum fluorescence under illumination; Fmb’, maximum fluorescence under blue light illumination; Fmo’, maximum fluorescence under orange light illumination; variable fluorescence (Fv) = Fm − F0; Fvd, variable fluorescence in darkness; Fvb, variable fluorescence under blue light illumination.
State transitions in the ΔspkH mutant was measured in Turku (Finland) with a dual-PAM and compared with its own wild type. State I was induced by treatment with 50 µM photons m−2 s−1 of blue light (460 nm) and then State II by treatment with 50 µM photons m−2 s−1 of orange light (635 nm). The measuring light was at 620 nm.
When mentioned, 2,6-dimethoxy-1,4-benzoquinone (DMBQ, 250 µM) and/or 2,5-dibromo-3-methyl-6-isopropyl-p-benzoquinone (DBMIB, 2.5 to 20 µM), methyl viologen (2 mM or 3 mM) or N, N, N′, N′-tetramethyl-p-phenylenediamine (TMPD, 7.5 or 10 µM) were added to the stirred cuvette.
When mentioned, staurosporine (21 µM), K252a (1.07 µM), NaF (50 or 100 mM) or Na3VO4 (1 mM) were added to dark-adapted cells. The kinase inhibitors were incubated for 90 min and the phosphatase inhibitors for 1 hour. Longer incubations gave the same results. State transitions were then measured.
PQ Pool Redox State Estimations
The PQ pool redox state was estimated by measuring fluorescence induction curves in the presence and absence of DCMU in a PSI fluorometer (PSI Instruments). Whole cells (Chl concentration 2.5 μg/mL) were dark-adapted for 15 min at 31°C, and illuminated (orange light 180 µM photons m−2 s−1, λ = 630 nm) in the 1-ms to 1-s time range with or without 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU, 10 μM). The measuring light for these experiments was blue (λ = 460 nm), and detection was in the far-red region (≥695 nm). The fluorescence induction curves were followed in each case, and the area between them was considered to be proportional to the oxidation state of the PQ pool. In addition, measurements in the presence of DMBQ (250 µM) and/or DBMIB (5 or 10 µM), with or without DCMU (10 μM) were performed.
Fluorescence Emission Spectra
We monitored 77 K fluorescence emission spectra in a CARY Eclipse spectrophotometer (Varian, Santa Clara, CA, USA). In all cases, whole cells (Chl concentration 5.0 μg/mL) were dark-adapted for 15 min before the measurements. Then, spectra were recorded corresponding to State II. For State I spectra, cells were illuminated for 5 min with 85 µM photons m−2 s−1 of blue-green light (halogen white light filtered by a Corion cut-off 550-nm filter; 400 to 500 nm). Samples were collected in nulear magnetic resonance tubes and frozen by immersion in liquid nitrogen. Excitation was made at 430 nm or 590 nm, and emission was scanned from 620 nm to 800 nm. All the spectra were normalized by the signal intensity at 800 nm. When Rhodamine B (0.4 µM) was added as an internal standard, excitation was made at 430 nm and emission was scanned from 550 nm to 800 nm. In these cases, the spectra were normalized to the Rhodamine B peak at 568 nm.
To address the effects of high osmotic buffers in state transitions, betaine (1 M), Suc (1 M), or K2HPO4/KH2PO4 buffer (0.5 M, pH 7.5) was added to cells pre-adapted to State I or State II. The cells were incubated with the different buffers for 5 min, before State II or I was induced by darkness or blue light illumination (5 min), respectively. Samples were taken before and after 5-min incubation with the chemicals and at the end of the light/dark treatment. Excitation was at 430 nm or 590 nm, and emission was scanned from 620 nm to 800 nm. All of the spectra were normalized by the signal intensity at 800 nm.
EPR Measurements
To assess the PSI/PSII ratio in the cyanobacterial strains, reduced FA/FB Fe-S centers and TyrD+ were measured as an estimation of PSI and PSII levels, respectively. First, 300 mL of cells (OD800 = 1.0) were harvested and washed with 50 mL of washing buffer [50 mM HEPES) pH 8.0, 5 mM MgCl2]. The cells were then centrifuged at 6000 rpm for 10 min at 20°C and washed again with 25 mL of washing buffer. This step was repeated, and the cells were washed with 2 mL of buffer. Finally, the cells were centrifuged at 3500 rpm for 10 min at 20°C and resuspended in 500 μL of buffer.
Calibrated EPR tubes were prepared with 150 μL of concentrated cells and 20 μL of 200 mM potassium ferricyanide. The tubes were illuminated for 30 s and incubated in darkness for 5 s before freezing. This treatment elicited full oxidation of TyrD+, with no or very little P700+ (a few percent). In case some P700+ was present, its contribution was subtracted in order to obtain a pure TyrD+ line shape before spin quantitation. The spectra were recorded at 20 K with an ESR300D X-band spectrometer (Bruker, Rheinstetten, Germany), using a TE102 resonator equipped with a front grid for sample illumination within the cavity. Illumination was performed using a halogen lamp (250 W). The temperature was controlled with a helium cryostat (Oxford Instruments, UK). Samples were measured in darkness for TyrD+ spectra and FA/FB baseline, and then, after 2-min illumination at 20 K, for the singly reduced FA/FB spectra. The FA/FB difference light-induced spectrum was used for quantitation after suppressing the P700+ signal. Isolated PSI was used to check that charge separation was 100% efficient at 20 K by comparing the dithionite-reduced (FA−/FB−) spectrum (two spins per P700) to the light-induced, singly reduced FA/FB difference spectrum (one spin per PSI). The following EPR parameters were used: for TyrD+, modulation amplitude: 2 G; microwave power: 2 µW; number of scans: eight. For FA/FB spectra, modulation amplitude: 10 G; microwave power: 0.8 mW; number of scans: two. These microwave powers were found to be nonsaturating at 20 K. The singly reduced (FA/FB) and TyrD+ relative spin amounts were calculated by double integration of the EPR signals, with correction for differences in microwave power and modulation amplitude.
Determining the Effects of Kinase Inhibitors by Phos-tagTM Gel SDS-PAGE
S. elongatus cells were pre-grown in fresh BG11 medium. When the cells were at 0.8 OD800, they were harvested and washed with nitrogen-free BG11 medium (BG11−N). The pellet was resuspended in BG11 medium containing 5 mM NH4Cl (BG11NH4) as the nitrogen source and incubated for 2 h. The culture was then separated into four samples. One sample was kept in the same BG11NH4 medium to obtain a completely dephosphorylated PII protein.
A second sample was washed and transferred to BG11-N medium for two hours to obtain a fully phosphorylated PII. The other two samples (after the 2-h incubation in the presence of NH4Cl) were supplemented with a final concentration of 21 µM staurosporine and 1.07 µM K252a, respectively, and incubated for an additional hour. Finally, both samples were washed and transferred into fresh BG11−N medium in the presence of inhibitors and incubated for two more hours. The cells of all the samples were washed quickly with pre-chilled 50 mM Tris-HCl (pH 6.8) and harvested by centrifugation at 4°C.
The cell-free extracts were prepared as follows. The cells were resuspended in 500 μL of 50 mM Tris-HCl (pH 6.8) with protease (1 mM caproic acid, 1 mM phenylmethylsulfonyl fluoride, 1 mM benzamidine) and phosphatase inhibitors, and broken via five cycles of vortexing (1 min) in the presence of glass beads. Unbroken cells were removed by centrifugation and the supernatant was recovered. The phosphorylation state of PII was tested by loading the samples onto a normal 15% SDS-PAGE (polyacrylamide gel electrophoresis) gel with or without 50 µM Zn-Phos-tag (Kinoshita and Kinoshita-Kikuta, 2011). The gels were washed with 10 mM EDTA for 3 × 15 min, followed by a 10-min washing with Phos-tag gel running buffer. The gels were then blotted onto polyvinylidene difluoride membranes using a Trans-blot Turbo System (Bio-Rad Laboratories, Hercules, CA, USA). The PII band was revealed by immunoblotting with an anti-PII antibody (dilution 1:2500, kindly provided by Professor Karl Forchhammer) using a chemoluminescent detection system (Pierce).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under the following accession numbers: spkA (sll1575), AB046597; spkB (slr1697), AB046598; spkC (slr0599), AB046599; spkD (sll0776), AB046600; spkE (slr1443), AB046602; spkF (slr1225), AB046601; spkG (slr0152), BAA18552; spkH (sll0005), BAA10206; spkI (sll1770), BAA17672; spkJ (slr0889), BAA17617; spkK (slr1919), BAA17147; spkL (sll0095), BAA10646; slr0328, BAA10030; sll1771, BAA17671; slr1860, X75568; sll1033, BAA16771; sll0602, BAA10367; slr0114, BAA10651; slr1983, BAA18225; sll1387, BAA18237; slr0946, ALJ68053.
Supplemental Data
Supplemental Figure 1. Gaussian decomposition of the 77 K fluorescence emission spectra of S. elongatus, the wild-type strains.
Supplemental Figure 2. Gaussian decomposition of the 77 K fluorescence emission spectra of Synechocystis strains.
Supplemental Figure 3. 77 K fluorescence emission spectra of the wild-type S. elongatus cells treated with Suc or phosphate buffer.
Supplemental Figure 4. Fluorescence changes induced by TMPD in the presence or absence of MV.
Supplemental Figure 5. State transitions in Synechocystis kinase and phosphatase mutants.
Supplemental Figure 6. Oxygen evolving activity in the presence of NaF.
Supplemental Table. Oligonucleotides used for the construction of the kinase and phosphatase mutants.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
The authors thank Dr. A. Zorina for the gift of the ΔspkH mutant to Turku’s laboratory and Professor Karl Forchhammer for the gift of the anti-PII antibody. This work was supported by the Agence Nationale de la Recherche’s RECYFUEL project (grant ANR-16-CE05-0026), and from the European Union’s Horizon 2020 research and innovation program (grant 675006, SE2B). P.C.’s salary is financed by the Agence Nationale de la Recherche’s RECYFUEL project. The research was also supported by the Centre National de la Recherche Scientifique (CNRS), the Commissariat à l’Energie Atomique (CEA) and the National Natural Science Foundation of China (grants 31700107 and 31770128). The French Infrastructure for Integrated Structural Biology (FRISBI) (grant ANR-10-INBS-05) also partially supported this research.
AUTHOR CONTRIBUTIONS
P.C. performed all the experiments related to the characterization of state transitions and the role of cyt b6f, and analyzed data; J.Z. constructed all the kinase and phosphatase mutants, and realized almost all the experiments related with the role of phosphorylations and analyzed the data; P.S. designed, performed, analyzed, and supervised experiments; C.L, N.B., and Q.W. designed and supervised some experiments; D.K. conceived the project, designed and supervised almost all the experiments, and analyzed data; the article was written by D.K., P.S., and P.C.
Footnotes
Articles can be viewed without a subscription.
References
- Adir N. (2008). Structure of the phycobilisome antennae in cyanobacteria and red algae. In Photosynthetic protein complexes: A structural approach, Fromme P., ed (Hoboken: Wiley-VCH Verlag GmbH & Co. KGaA; ), pp. 243–274. [Google Scholar]
- Alfonso M., Perewoska I., Constant S., Kirilovsky D. (1999). Redox control of psbA expression in cyanobacteria Synechocystis strains. J. Photochem. Photobiol. Biol 48: 104–113. [Google Scholar]
- Alfonso M., Perewoska I., Kirilovsky D. (2000). Redox control of psbA gene expression in the cyanobacterium Synechocystis PCC 6803. Involvement of the cytochrome b6f complex. Plant Physiol. 122: 505–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen J.F., Bennett J., Steinback K.E., Arntzen C.J. (1981). Chloroplast protein phosphorylation couples plastoquinone redox state to distribution of excitation energy between photosystems. Nature 291: 25–29. [Google Scholar]
- Allen J.F., Sanders C.E., Holmes N.G. (1985). Correlation of membrane protein phosphorylation with excitation energy distribution in the cyanobacterium Synechococcus 6301. FEBS Lett. 193: 271–275. [Google Scholar]
- Angeleri M., Zorina A., Aro E.M., Battchikova N. (2018). Interplay of SpkG kinase and the Slr0151 protein in the phosphorylation of ferredoxin 5 in Synechocystis sp. strain PCC 6803. FEBS Lett. 592: 411–421. [DOI] [PubMed] [Google Scholar]
- Aoki M., Katoh S. (1982). Oxidation and reduction of plastoquinone by photosynthetic and respiratory electron transport in a cyanobacterium Synechococcus sp. Biochim. Biophys. Acta 682: 307–314. [Google Scholar]
- Aspinwall C.L., Sarcina M., Mullineaux C.W. (2004). Phycobilisome mobility in the cyanobacterium Synechococcus sp. PCC7942 is influenced by the trimerisation of Photosystem I. Photosynth. Res. 79: 179–187. [DOI] [PubMed] [Google Scholar]
- Bellafiore S., Barneche F., Peltier G., Rochaix J.D. (2005). State transitions and light adaptation require chloroplast thylakoid protein kinase STN7. Nature 433: 892–895. [DOI] [PubMed] [Google Scholar]
- Bennoun P. (1982). Evidence for a respiratory chain in the chloroplast. Proc. Natl. Acad. Sci. USA 79: 4352–4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggins J., Bruce D. (1989). Regulation of excitation energy transfer in organisms containing phycobilins. Photosynth. Res. 20: 1–34. [DOI] [PubMed] [Google Scholar]
- Biggins J., Tanguay N.A., Frank H.A. (1989). Electron transfer reactions in photosystem I following vitamin K1 depletion by ultraviolet irradiation. FEBS Lett. 250: 271–274. [DOI] [PubMed] [Google Scholar]
- Bonaventura C., Myers J. (1969). Fluorescence and oxygen evolution from Chlorella pyrenoidosa. Biochim. Biophys. Acta 189: 366–383. [DOI] [PubMed] [Google Scholar]
- Breyton C. (2000). Conformational changes in the cytochrome b6f complex induced by inhibitor binding. J. Biol. Chem. 275: 13195–13201. [DOI] [PubMed] [Google Scholar]
- Bruce D., Biggins J. (1985). Mechanism of the light state transition in photosynthesis V. 77K linear dichroism of Anacystis nidulans in state 1 and state 2. Biochim. Biophys. Acta 810: 295–301. [Google Scholar]
- Bruce D., Salehian O. (1992). Laser-induced optoacustic calorimetry of cyanobacteria. The efficiency of primary photosynthetic processes in state 1 and state 2. Biochim. Biophys. Acta 1100: 242–250. [Google Scholar]
- Bruce D., Brimble S., Bryant D.A. (1989). State transitions in a phycobilisome-less mutant of the cyanobacterium Synechococcus sp. PCC 7002. Biochim. Biophys. Acta 974: 66–73. [DOI] [PubMed] [Google Scholar]
- Campbell D., Hurry V., Clarke A.K., Gustafsson P., Öquist G. (1998). Chlorophyll fluorescence analysis of cyanobacterial photosynthesis and acclimation. Microbiol. Mol. Biol. Rev. 62: 667–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Zhan J., Chen Y., Yang M., He C., Ge F., Wang Q. (2015). Effects of phosphorylation of β subunits of phycocyanins on state transition in the model cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 56: 1997–2013. [DOI] [PubMed] [Google Scholar]
- Chukhutsina V., Bersanini L., Aro E.M., van Amerongen H. (2015). Cyanobacterial light-harvesting phycobilisomes uncouple from photosystem I during dark-to-light transitions. Sci. Rep. 5: 14193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delphin E., Duval J.C., Kirilovsky D. (1995). Comparison of state 1 state 2 transitions in the green alga Chlamydomonas reinhardtii and in the red alga Rhodella violacea: Effect of kinase and phosphatase inhibitors. Biochimica et Biophysica Acta—Bioenergetics 1232: 91–95. [Google Scholar]
- Delphin E., Duval J.C., Etienne A.L., Kirilovsky D. (1996). State transitions or delta pH-dependent quenching of photosystem II fluorescence in red algae. Biochemistry 35: 9435–9445. [DOI] [PubMed] [Google Scholar]
- Depège N., Bellafiore S., Rochaix J.D. (2003). Role of chloroplast protein kinase Stt7 in LHCII phosphorylation and state transition in Chlamydomonas. Science 299: 1572–1575. [DOI] [PubMed] [Google Scholar]
- Dong C., Zhao J. (2008). ApcD is required for state transition but not involved in blue-light induced quenching in the cyanobacterium Anabaena sp. PCC7120. Chin. Sci. Bull. 53: 3422–3424. [Google Scholar]
- Dong C., Tang A., Zhao J., Mullineaux C.W., Shen G., Bryant D.A. (2009). ApcD is necessary for efficient energy transfer from phycobilisomes to photosystem I and helps to prevent photoinhibition in the cyanobacterium Synechococcus sp. PCC 7002. Biochim. Biophys. Acta 1787: 1122–1128. [DOI] [PubMed] [Google Scholar]
- Draber W., Trebst A., Harth E. (1970). On a new inhibitor of photosynthetic electron-transport in isolated chloroplasts. Z. Naturforsch. B 25: 1157–1159. [DOI] [PubMed] [Google Scholar]
- El Bissati K., Kirilovsky D. (2001). Regulation of psbA and psaE expression by light quality in Synechocystis species PCC 6803. A redox control mechanism. Plant Physiol. 125: 1988–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Bissati K., Delphin E., Murata N., Etienne A., Kirilovsky D. (2000). Photosystem II fluorescence quenching in the cyanobacterium Synechocystis PCC 6803: Involvement of two different mechanisms. Biochim. Biophys. Acta 1457: 229–242. [DOI] [PubMed] [Google Scholar]
- Emlyn-Jones D., Ashby M.K., Mullineaux C.W. (1999). A gene required for the regulation of photosynthetic light harvesting in the cyanobacterium Synechocystis 6803. Mol. Microbiol. 33: 1050–1058. [DOI] [PubMed] [Google Scholar]
- Federman S., Malkin S., Scherz A. (2000). Excitation energy transfer in aggregates of Photosystem I and Photosystem II of the cyanobacterium Synechocystis sp. PCC 6803: Can assembly of the pigment-protein complexes control the extent of spillover? Photosynth. Res. 64: 199–207. [DOI] [PubMed] [Google Scholar]
- Fernandez P., Saint-Joanis B., Barilone N., Jackson M., Gicquel B., Cole S.T., Alzari P.M. (2006). The Ser/Thr protein kinase PknB is essential for sustaining mycobacterial growth. J. Bacteriol. 188: 7778–7784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finazzi G., Zito F., Barbagallo R.P., Wollman F.A. (2001). Contrasted effects of inhibitors of cytochrome b6f complex on state transitions in Chlamydomonas reinhardtii: The role of Qo site occupancy in LHCII kinase activation. J. Biol. Chem. 276: 9770–9774. [DOI] [PubMed] [Google Scholar]
- Fischer B.B., Eggen R.I., Trebst A., Krieger-Liszkay A. (2006). The glutathione peroxidase homologous gene Gpxh in Chlamydomonas reinhardtii is upregulated by singlet oxygen produced in photosystem II. Planta 223: 583–590. [DOI] [PubMed] [Google Scholar]
- Folea I.M., Zhang P., Aro E.M., Boekema E.J. (2008). Domain organization of photosystem II in membranes of the cyanobacterium Synechocystis PCC6803 investigated by electron microscopy. FEBS Lett. 582: 1749–1754. [DOI] [PubMed] [Google Scholar]
- Forchhammer K., Tandeau de Marsac N. (1995a). Functional analysis of the phosphoprotein PII (glnB gene product) in the cyanobacterium Synechococcus sp. strain PCC 7942. J. Bacteriol. 177: 2033–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forchhammer K., Tandeau de Marsac N. (1995b). Phosphorylation of the PII protein (glnB gene product) in the cyanobacterium Synechococcus sp. strain PCC 7942: Analysis of in vitro kinase activity. J. Bacteriol. 177: 5812–5817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fufezan C., Drepper F., Juhnke H.D., Lancaster C.R.D., Un S., Rutherford A.W., Krieger-Liszkay A. (2005). Herbicide-induced changes in charge recombination and redox potential of QA in the T4 mutant of Blastochloris viridis. Biochemistry 44: 5931–5939. [DOI] [PubMed] [Google Scholar]
- Galkin A.N., Mikheeva L.E., Shestakov S.V. (2003). Insertsionnaia inaktivatsiaa genov, kodiruiushchikh serin/treoninovye proteinkinazy éukarioticheskogo tipa u tsianobakterii Synechocystis sp. PCC 6803 [Insertional inactivation of genes encoding eukaryotic type serine/threonine protein kinases in cyanobacterium Synechocystis sp. PCC 6803]. Mikrobiologiia 72: 64–69. [PubMed] [Google Scholar]
- Glazer A.N. (1984). Phycobilisome, a macromolecular complex optimized for light energy transfer. Biochim. Biophys. Acta 768: 29–51. [Google Scholar]
- Guskov A., Kern J., Gabdulkhakov A., Broser M., Zouni A., Saenger W. (2009). Cyanobacterial photosystem II at 2.9-A resolution and the role of quinones, lipids, channels and chloride. Nat. Struct. Mol. Biol. 16: 334–342. [DOI] [PubMed] [Google Scholar]
- Herdman M, Delaney SF, Carr NG (1973). A new medium for the isolation and growth of auxotrophic mutants of the blue-green alga Anacystis nidulans. J. Gen. Microbiol. 79: 233–237. [Google Scholar]
- Hiyama T., Ke B. (1972). Difference spectra and extinction coefficients of P 700. Biochim. Biophys. Acta 267: 160–171. [DOI] [PubMed] [Google Scholar]
- Huang C., Yuan X., Zhao J., Bryant D.A. (2003). Kinetic analyses of state transitions of the cyanobacterium Synechococcus sp. PCC 7002 and its mutant strains impaired in electron transport. Biochim. Biophys. Acta 1607: 121–130. [DOI] [PubMed] [Google Scholar]
- Huang J.Y., Chiu Y.F., Ortega J.M., Wang H.T., Tseng T.S., Ke S.C., Roncel M., Chu H.A. (2016). Mutations of cytochrome b559 and PsbJ on and near the QC site in Photosystem II influence the regulation of short-term light response and photosynthetic growth of the cyanobacterium Synechocystis sp. PCC 6803. Biochemistry 55: 2214–2226. [DOI] [PubMed] [Google Scholar]
- Joshua S., Mullineaux C.W. (2004). Phycobilisome diffusion is required for light-state transitions in cyanobacteria. Plant Physiol. 135: 2112–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamei A., Yuasa T., Orikawa K., Geng X.X., Ikeuchi M. (2001). A eukaryotic-type protein kinase, SpkA, is required for normal motility of the unicellular cyanobacterium Synechocystis sp. strain PCC 6803. J. Bacteriol. 183: 1505–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaňa R. (2013). Mobility of photosynthetic proteins. Photosynth. Res. 116: 465–479. [DOI] [PubMed] [Google Scholar]
- Kaňa R., Prášil O., Komárek O., Papageorgiou G.C., Govindjee R. (2009). Spectral characteristic of fluorescence induction in a model cyanobacterium, Synechococcus sp. (PCC 7942). Biochim. Biophys. Acta 1787: 1170–1178. [DOI] [PubMed] [Google Scholar]
- Kinoshita E., Kinoshita-Kikuta E. (2011). Improved Phos-tag SDS-PAGE under neutral pH conditions for advanced protein phosphorylation profiling. Proteomics 11: 319–323. [DOI] [PubMed] [Google Scholar]
- Kirilovsky D., Rutherford A.W., Etienne A.L. (1994). Influence of DCMU and ferricyanide on photodamage in photosystem II. Biochemistry 33: 3087–3095. [DOI] [PubMed] [Google Scholar]
- Kondo K., Mullineaux C.W., Ikeuchi M. (2009). Distinct roles of CpcG1-phycobilisome and CpcG2-phycobilisome in state transitions in a cyanobacterium Synechocystis sp. PCC 6803. Photosynth. Res. 99: 217–225. [DOI] [PubMed] [Google Scholar]
- Kowalczyk N., Rappaport F., Boyen C., Wollman F.A., Collén J., Joliot P. (2013). Photosynthesis in Chondrus crispus: The contribution of energy spill-over in the regulation of excitonic flux. Biochim. Biophys. Acta 1827: 834–842. [DOI] [PubMed] [Google Scholar]
- Kruip J., Bald D., Boekema E., Rögner M. (1994). Evidence for the existence of trimeric and monomeric Photosystem I complexes in thylakoid membranes from cyanobacteria. Photosynth. Res. 40: 279–286. [DOI] [PubMed] [Google Scholar]
- Krupnik T., Kotabová E., van Bezouwen L.S., Mazur R., Garstka M., Nixon P.J., Barber J., Kaňa R., Boekema E.J., Kargul J. (2013). A reaction center-dependent photoprotection mechanism in a highly robust photosystem II from an extremophilic red alga, Cyanidioschyzon merolae. J. Biol. Chem. 288: 23529–23542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyle D.J., Ohad I., Arntzen C.J. (1984). Membrane protein damage and repair: Selective loss of a quinone-protein function in chloroplast membranes. Proc. Natl. Acad. Sci. USA 81: 4070–4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent S., Jang J., Janicki A., Zhang C.C., Bédu S. (2008). Inactivation of spkD, encoding a Ser/Thr kinase, affects the pool of the TCA cycle metabolites in Synechocystis sp. strain PCC 6803. Microbiology 154: 2161–2167. [DOI] [PubMed] [Google Scholar]
- Ley A.C., Butler W.L. (1980). Energy distribution in the photochemical apparatus of Porphyridium cruentum in state I and state II. Biochim. Biophys. Acta 592: 349–363. [DOI] [PubMed] [Google Scholar]
- Li D., Xie J., Zhao J., Xia A., Li D., Gong Y. (2004). Light-induced excitation energy redistribution in Spirulina platensis cells: “Spillover” or “mobile PBSs”? Biochim. Biophys. Acta 1608: 114–121. [DOI] [PubMed] [Google Scholar]
- Li H., Li D., Yang S., Xie J., Zhao J. (2006). The state transition mechanism—Simply depending on light-on and -off in Spirulina platensis. Biochim. Biophys. Acta 1757: 1512–1519. [DOI] [PubMed] [Google Scholar]
- Liang C., Zhang X., Chi X., Guan X., Li Y., Qin S., Shao H.B. (2011). Serine/threonine protein kinase SpkG is a candidate for high salt resistance in the unicellular cyanobacterium Synechocystis sp. PCC 6803. PLoS One 6: e18718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacColl R. (1998). Cyanobacterial phycobilisomes. J. Struct. Biol. 124: 311–334. [DOI] [PubMed] [Google Scholar]
- Mao H.B., Li G.F., Ruan X., Wu Q.Y., Gong Y.D., Zhang X.F., Zhao N.M. (2002). The redox state of plastoquinone pool regulates state transitions via cytochrome b6f complex in Synechocystis sp. PCC 6803. FEBS Lett. 519: 82–86. [DOI] [PubMed] [Google Scholar]
- Mao L., Wang Y., Hu X. (2003). Π-Π stacking interactions in the peridinin-chlorophyll-protein of Amphidinium carterae. J. Phys. Chem. B 107: 3963–3971. [Google Scholar]
- Mata-Cabana A., García-Domínguez M., Florencio F.J., Lindahl M. (2012). Thiol-based redox modulation of a cyanobacterial eukaryotic-type serine/threonine kinase required for oxidative stress tolerance. Antioxid. Redox Signal. 17: 521–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCartney B., Howell L.D., Kennelly P.J., Potts M. (1997). Protein tyrosine phosphorylation in the cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 179: 2314–2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell M.D., Koop R., Vasil’ev S., Bruce D. (2002). Regulation of the distribution of chlorophyll and phycobilin-absorbed excitation energy in cyanobacteria. A structure-based model for the light state transition. Plant Physiol. 130: 1201–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minagawa J. (2011). State transitions—The molecular remodeling of photosynthetic supercomplexes that controls energy flow in the chloroplast. Biochim. Biophys. Acta 1807: 897–905. [DOI] [PubMed] [Google Scholar]
- Misumi M., Katoh H., Tomo T., Sonoike K. (2016). Relationship between photochemical quenching and non-photochemical quenching in six species of cyanobacteria reveals species difference in redox state and species commonality in energy dissipation. Plant Cell Physiol. 57: 1510–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullineaux C.W. (2014). Co-existence of photosynthetic and respiratory activities in cyanobacterial thylakoid membranes. Biochim. Biophys. Acta 1837: 503–511. [DOI] [PubMed] [Google Scholar]
- Mullineaux C.W., Allen J.F. (1986). The state 2 transition in the cyanobacterium Synechococcus 6301 can be driven by respiratory electron flow into the plastoquinone pool. FEBS Lett. 205: 155–160. [Google Scholar]
- Mullineaux C.W., Allen J.F. (1990). State 1 State 2 transitions in the cyanobacterium Synechococcus 6301 are controlled by the redox state of electron carriers between Photosystems I and II. Photosynth. Res. 23: 297–311. [DOI] [PubMed] [Google Scholar]
- Mullineaux C.W., Griebenow S., Braslavsky S.E. (1991). Photosynthetic energy storage in cyanobacterial cells adapted to light-states 1 and 2. A laser-induced optoacustic study. Biochim. Biophys. Acta 1060: 315–318. [Google Scholar]
- Mullineaux C.W., Tobin M.J., Jones G.R. (1997). Mobility of photosynthetic complexes in thylakoid membranes. Nature 390: 421–424. [Google Scholar]
- Murata N. (1969). Control of excitation transfer in photosynthesis. I. Light-induced change of chlorophyll a fluorescence in Porphyridium cruentum. Biochim. Biophys. Acta 172: 242–251. [DOI] [PubMed] [Google Scholar]
- Nakano H., Omura S. (2009). Chemical biology of natural indolocarbazole products: 30 years since the discovery of staurosporine. J. Antibiot. (Tokyo) 62: 17–26. [DOI] [PubMed] [Google Scholar]
- Nanba M., Katoh S. (1985). Restoration by tetramethyl-p-phenylenediamine of photosynthesis in dibromothymoquinone-inhibited cells of the cyanobacterium Synechococcus sp. Biochim. Biophys. Acta 809: 74–80. [Google Scholar]
- Olive J., M’Bina I., Vernotte C., Astier C., Wollman F.A. (1986). Randomization of the Ef particles in thylakoid membranes of Synechocystis 6714 upon transition from state-I to state-II. FEBS Lett. 208: 308–312. [Google Scholar]
- Olive J., Ajlani G., Astier C., Recouvreur M., Vernotte C. (1997). Ultrastructure and light adaptation of phycobilisome mutants of Synechocystis PCC 6803. Biochim. Biophys. Acta 1319: 275–282. [Google Scholar]
- Panichkin V.B., Arakawa-Kobayashi S., Kanaseki T., Suzuki I., Los D.A., Shestakov S.V., Murata N. (2006). Serine/threonine protein kinase SpkA in Synechocystis sp. strain PCC 6803 is a regulator of expression of three putative pilA operons, formation of thick pili, and cell motility. J. Bacteriol. 188: 7696–7699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papageorgiou G.C., Govindjee R., Mimuro M., Stamatakis K., Alygizaki-Zorba A., Murata N. (1999). Light-induced and osmotically-induced changes in chlorophyll a fluorescence in two Synechocystis sp. PCC 6803 strains that differ in membrane lipid unsaturation. Photosynth. Res. 59: 125–136. [Google Scholar]
- Preston C., Critchley C. (1988). Interaction of electron acceptors with thylakoids from halophytic and non-halophytic species. Photosynth. Res. 16: 187–202. [DOI] [PubMed] [Google Scholar]
- Ranjbar Choubeh R., Wientjes E., Struik P.C., Kirilovsky D., van Amerongen H. (2018). State transitions in the cyanobacterium Synechococcus elongatus 7942 involve reversible quenching of the photosystem II core. Biochim Biophys Acta Bioenerg 1859: 1059–1066. [DOI] [PubMed] [Google Scholar]
- Roberts A.G., Kramer D.M. (2001). Inhibitor “double occupancy” in the Qo pocket of the chloroplast cytochrome b6f complex. Biochemistry 40: 13407–13412. [DOI] [PubMed] [Google Scholar]
- Schluchter W.M., Shen G., Zhao J., Bryant D.A. (1996). Characterization of psaI and psaL mutants of Synechococcus sp. strain PCC 7002: A new model for state transitions in cyanobacteria. Photochem. Photobiol. 64: 53–66. [DOI] [PubMed] [Google Scholar]
- Scott M., McCollum C., Vasil’ev S., Crozier C., Espie G.S., Krol M., Huner N.P., Bruce D. (2006). Mechanism of the down regulation of photosynthesis by blue light in the Cyanobacterium Synechocystis sp. PCC 6803. Biochemistry 45: 8952–8958. [DOI] [PubMed] [Google Scholar]
- Siefermann-Harms D. (1988). Application of chlorophyll fluorescence. In Fluorescence properties of isolated chlorophyll-protein complexes., Lichtenthaler HK, ed (Dordrecht: Kluwer Academic Publisher; ), pp. 45–54. [Google Scholar]
- Spät P., Maček B., Forchhammer K. (2015). Phosphoproteome of the cyanobacterium Synechocystis sp. PCC 6803 and its dynamics during nitrogen starvation. Front. Microbiol. 6: 248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava A., Strasser R.J., Govindjee R. (1995). Polyphasic rise of chlorophyll a fluorescence in herbicide-resistant D1 mutants of Chlamydomonas reinardtii. Photosynth. Res. 43: 131–141. [DOI] [PubMed] [Google Scholar]
- Umena Y., Kawakami K., Shen J.R., Kamiya N. (2011). Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 Å. Nature 473: 55–60. [DOI] [PubMed] [Google Scholar]
- Van Dorssen RJ, Breton J, Plijter JJ, Satoh K, Van Gorkom HJ, Amesz J (1987). Spectroscopic properties of the reaction center and of the 47 kDa chlorophyll protein of photosystem II. Biochim Biophys Acta 893: 267–274. [Google Scholar]
- Vener A.V., Van Kan P.J., Gal A., Andersson B., Ohad I. (1995). Activation/deactivation cycle of redox-controlled thylakoid protein phosphorylation. Role of plastoquinol bound to the reduced cytochrome bf complex. J. Biol. Chem. 270: 25225–25232. [DOI] [PubMed] [Google Scholar]
- Vener A.V., van Kan P.J., Rich P.R., Ohad I., Andersson B. (1997). Plastoquinol at the quinol oxidation site of reduced cytochrome bf mediates signal transduction between light and protein phosphorylation: Thylakoid protein kinase deactivation by a single-turnover flash. Proc. Natl. Acad. Sci. USA 94: 1585–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernotte C., Picaud M., Kirilovsky D., Olive J., Ajlani G., Astier C. (1992). Changes in the photosynthetic apparatus in the cyanobacterium Synechocystis sp. PCC 6714 following light-to-dark and dark-to-light transitions. Photosynth. Res. 32: 45–57. [DOI] [PubMed] [Google Scholar]
- Vladkova R. (2016). Chlorophyll a is the crucial redox sensor and transmembrane signal transmitter in the cytochrome b6f complex. Components and mechanisms of state transitions from the hydrophobic mismatch viewpoint. J. Biomol. Struct. Dyn. 34: 824–854. [DOI] [PubMed] [Google Scholar]
- Wollman F.A. (2001). State transitions reveal the dynamics and flexibility of the photosynthetic apparatus. EMBO J. 20: 3623–3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollman F.A., Lemaire C. (1988). Studies on kinase-controlled state transitions in Photosystem II and b6f mutants from Chlamydomonas reinhardtii which lack quinone-binding proteins. Biochim. Biophys. Acta 933: 85–94. [Google Scholar]
- Yang M.K., Qiao Z.X., Zhang W.Y., Xiong Q., Zhang J., Li T., Ge F., Zhao J.D. (2013). Global phosphoproteomic analysis reveals diverse functions of serine/threonine/tyrosine phosphorylation in the model cyanobacterium Synechococcus sp. strain PCC 7002. J. Proteome Res. 12: 1909–1923. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Huang L., Shulmeister V.M., Chi Y.I., Kim K.K., Hung L.W., Crofts A.R., Berry E.A., Kim S.H. (1998). Electron transfer by domain movement in cytochrome bc1. Nature 392: 677–684. [DOI] [PubMed] [Google Scholar]
- Zito F., Finazzi G., Delosme R., Nitschke W., Picot D., Wollman F.A. (1999). The Qo site of cytochrome b6f complexes controls the activation of the LHCII kinase. EMBO J. 18: 2961–2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorina A.A. (2013). Eukaryotic protein kinases in cyanobacteria. Russ. J. Plant Physiol. 60: 589–596. [Google Scholar]
- Zorina A., Stepanchenko N., Novikova G.V., Sinetova M., Panichkin V.B., Moshkov I.E., Zinchenko V.V., Shestakov S.V., Suzuki I., Murata N., Los D.A. (2011). Eukaryotic-like Ser/Thr protein kinases SpkC/F/K are involved in phosphorylation of GroES in the cyanobacterium synechocystis. DNA Res. 18: 137–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorina A.A., Bedbenov V.S., Novikova G.V., Panichkin V.B., Los D.A. (2014). [Involvement of serine/threonine protein kinases in cold stress response in the cyanobacterium Synechocystis sp. PCC 6803: Functional characterization of a protein kinase SpkE]. Mol. Biol. (Mosk.) 48: 452–462. [PubMed] [Google Scholar]