The pathways of DNA-protein crosslink repair in plants are characterized by the protease WSS1A, the DNA endonuclease MUS81, and tyrosyl-DNA phosphodiesterase 1.
Abstract
DNA–protein crosslinks (DPCs) represent a severe threat to the genome integrity; however, the main mechanisms of DPC repair were only recently elucidated in humans and yeast. Here we define the pathways for DPC repair in plants. Using CRISPR/Cas9, we could show that only one of two homologs of the universal repair proteases SPARTAN/ weak suppressor of smt3 (Wss1), WSS1A, is essential for DPC repair in Arabidopsis (Arabidopsis thaliana). WSS1A defective lines exhibit developmental defects and are hypersensitive to camptothecin (CPT) and cis-platin. Interestingly, the CRISPR/Cas9 mutants of TYROSYL-DNA PHOSPHODIESTERASE 1 (TDP1) are insensitive to CPT, and only the wss1A tdp1 double mutant reveals a higher sensitivity than the wss1A single mutant. This indicates that TDP1 defines a minor backup pathway in the repair of DPCs. Moreover, we found that knock out of the endonuclease METHYL METHANESULFONATE AND UV SENSITIVE PROTEIN 81 (MUS81) results in a strong sensitivity to DPC-inducing agents. The fact that wss1A mus81 and tdp1 mus81 double mutants exhibit growth defects and an increase in dead cells in root meristems after CPT treatment demonstrates that there are three independent pathways for DPC repair in Arabidopsis. These pathways are defined by their different biochemical specificities, as main actors, the DNA endonuclease MUS81 and the protease WSS1A, and the phosphodiesterase TDP1 as backup.
INTRODUCTION
DNA is constantly exposed to a wide range of damaging factors, challenging the integrity of the genome. To prevent cells from mutations, distortion by bulky adducts, or even strand breaks, a great variety of specific DNA repair mechanisms have evolved. Persisting DNA–protein crosslinks (DPC) represent a class of highly toxic DNA lesions, as they result in stalled replication forks and therefore inhibit replication. Thus, cell division is blocked, ultimately leading to cell death.
DPCs can be subdivided into enzymatic and nonenzymatic DPCs. Both types arise by exogenous and endogenous causes. Trapping of enzymatic reaction intermediates can occur either spontaneously or by enzyme poisons such as camptothecin (CPT). CPT stabilizes topoisomerase 1 (TOP1) at the DNA, thereby preventing the DNA backbone from re-ligating after the nicking of the double-stranded DNA in order to relax the DNA from torsional tension (Pommier et al., 2006). These intermediates are referred to as stabilized TOP1 cleavage complexes (TOP1cc). However, nonenzymatic DPCs can be endogenously produced by reactive aldehydes, like acetaldehyde during metabolism or formaldehyde via histone demethylation (Swenberg et al., 2011). Non-enzymatic DPCs can be induced exogenously by ionizing radiation, UV radiation, or chemical crosslinkers such as cis-platin (Zwelling et al., 1979; Olinski et al., 1987; Cadet et al., 1992; Chválová et al., 2007; Stingele et al., 2015).
Despite the major threat of DPCs by blocking DNA replication, it was only recently that the predominant mechanism of DPC repair was discovered. In 2014, the yeast metalloprotease Wss1 (weak suppressor of smt3), that was initially related to the small ubiquitin-like modifier pathway (Biggins et al., 2001; Mullen et al., 2010), was identified as a key player in DPC repair. Cells lacking Wss1 are hypersensitive in response to formaldehyde treatment, which induces unspecific DPCs. Furthermore, Wss1 possesses a crucial role in the repair of Top1ccs, as ∆wss1 ∆tdp1 (tyrosyl-DNA phosphodiesterase 1) strains exhibit severe growth defects when exposed to CPT (Stingele et al., 2014).
Based on the similarity of the zinc metalloprotease domain, the mammalian Spartan (SPRTN, also known as DVC1) was proposed to be related to the same family of repair proteases as Wss1 (Stingele et al., 2015). Humans carrying mutations in the SPRTN gene suffer from Ruijs-Aalfs syndrome. This genetic disease is associated with genomic instability featuring premature ageing and a high susceptibility for early onset of hepatocellular carcinoma (Ruijs et al., 2003; Lessel et al., 2014). SPRTN knock out murine embryonic fibroblast (MEF) cells revealed sensitivities to formaldehyde, CPT, and etoposide (inducing topoisomerase 2 cleavage complexes). Indeed, SPRTN has been identified as the mammalian protease for DPC removal, whose activity is dependent on the presence of DNA (Lopez-Mosqueda et al., 2016). SPRTN is essential for viability in mammalian cell lines. In the nematode Caenorhabditis elegans, the SPRTN ortholog Dvc-1 was found to be dispensable for viability (Mosbech et al., 2012), with dvc-1 larvae being hypersensitive to formaldehyde and cis-platin. Small interfering RNA knockdown of SPRTN in human cell lines also led to formaldehyde sensitivity, indicating SPRTN is of high importance for DPC repair. Although the proteolytic activity of SPRTN is dependent on DNA, the chromatin accessibility is controlled via ubiquitin binding (Stingele et al., 2016).
Although specialized repair proteases of the Wss1/SPRTN type are able to proteolytically degrade the protein part of the crosslink, making the lesion accessible for further processing and leaving only a small peptide remnant, other kinds of enzyme activities can also be used for the repair of specific DNA–protein adducts. In the case of a tyrosyl-DNA phosphodiester bond, direct enzymatic hydrolysis can be conducted via the TDP1. This enzyme is able to specifically hydrolyze the 3′ phosphate of the DNA and the active tyrosyl residue of class I topoisomerases (Yang et al., 1996; Pouliot et al., 1999), and belongs to the phospholipase D superfamily (Interthal et al., 2001). As yeast ∆wss1 ∆tdp1 strains display synergistic sensitivity effects after exposure to CPT, Wss1 and Tdp1 were integrated into two independent pathways of DPC repair (Stingele et al., 2014). TDP1 is present in all eukaryotes and is required for making cells resistant to the TOP1 poison CPT (Pommier et al., 2014). In plants, a single Attdp1 transfer DNA (T-DNA) insertion line was described as sensitive to CPT and exhibiting dwarfism (Lee et al., 2010).
Besides proteolytic action of repair proteases and direct hydrolysis of the crosslink, DPCs can be processed by DNA endonuclease action. The endonuclease METHYL METHANESULFONATE AND UV SENSITIVE PROTEIN 81 (MUS81) has been shown to be an important factor in DNA repair. In humans and yeast cells, MUS81 deficient lines are hypersensitive to the Top1cc-inducing agent CPT (Liu et al., 2002; Regairaz et al., 2011). This implies its involvement in the repair of DPCs. Furthermore, mutants of mammalian MUS81 and its complex partner ESSENTIAL MEIOTIC ENDONUCLEASE 1 exhibit sensitivity to the nonenzymatic DPC-inducing agent cis-platin. MUS81 has been shown to be able to cleave branched DNA structures, especially the ones arising at stalled replication forks (Abraham et al., 2003). Moreover, MUS81 acts as endonuclease in the resolution of recombination intermediates like double Holliday junctions. The importance of MUS81 in the repair of cis-platin-induced DNA damage has also been shown in the plant model Arabidopsis (Arabidopsis thaliana) by mus81 mutants being strongly sensitive to cis-platin (Hartung et al., 2006; Mannuss et al., 2010).
Phylogenetic analysis revealed that plants possess two different kinds of orthologs of the newly characterized repair proteases, belonging to the so-called ubiquitin-like–Wss1 branch, which is characterized by an N-terminal ubiquitin-like domain (Stingele et al., 2015). In this study we not only characterize the role of the two orthologs WSS1A and WSS1B in DPC repair in Arabidopsis, but we also define their role in relation to TDP1 and MUS81, indicating that plants have indeed three different pathways of DPC repair.
RESULTS
Wss1A Mutants Exhibit Serious Growth Defects
In plants, two paralogues of the repair protease SPRTN/WSS1 have been identified, WSS1A and WSS1B (Stingele et al., 2015). To characterize the functional role of these genes, Cas9-mediated mutagenesis in Arabidopsis was performed. For WSS1A, we chose two target sequences in exon 1 (5′-CTGTGAAATTGTGATGAGTT-3′, 5′-AACCTAGAGAAGATGAAGCG-3′) and one in exon 2 (5′- CAAGTGAAATTGAGGCTTAG- 3′) using Streptococcus pyogenes Cas9 (Fauser et al., 2014) and Staphylococcus aureus Cas9 (Steinert et al., 2015), respectively. This way, three mutant lines were generated, named wss1A-1 to wss1A-3. In the case of WSS1B, one target region was chosen starting 4 nucleotides after the start codon (5′-TCTCATCGCATGGAAGATTC-3′), and another one at the end of exon 1, in the region coding for the first protein domain (5′- TTCTGATGAACACTCGAGCT- 3′). The three resulting lines were named wss1B-1 to wss1B-3 (Figure 1A). In all cases frameshift mutations, leading to a premature stop codon in the respective open reading frame, were obtained, as confirmed by DNA sequencing. The induced mutations were also confirmed on mRNA level by Sanger sequencing of the complementary DNA (cDNA; Supplemental Figure 1A and B).
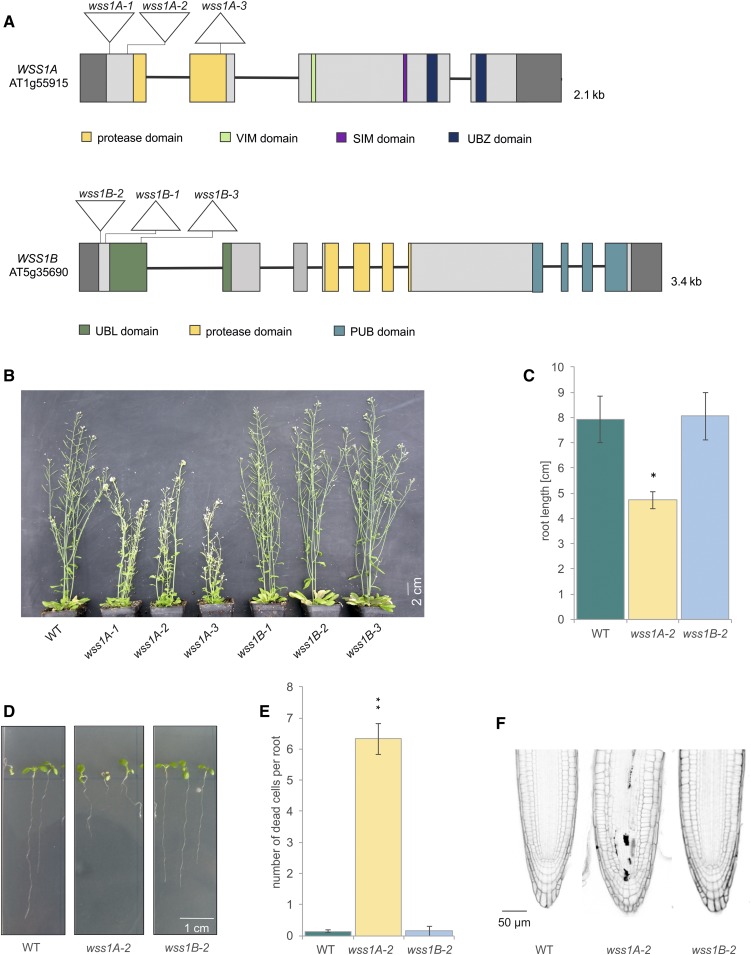
Figure 1.
Analysis of wss1A and wss1B Mutant Lines.
(A) Genomic structure and protein domains of WSS1A and WSS1B. AtWSS1A is 2.1 kb in length, consisting of 4 exons and 3 introns. The mutations introduced via CRISPR/Cas9 are located in exon 1, harboring 1 bp insertions for both wss1A-1 and wss1A-2. The 52 bp deletion of wss1A-3 is located in exon 2. AtWSS1B is 3.4 kb in length, with 10 exons and 9 introns. The 1 bp insertion, 41 bp deletion and the discontinuous 26 bp deletion of wss1B-1 to wss1B-3 are located in exon 1. Untranslated regions are colored in dark gray. UBL, ubiquitin-like. VIM, VCP (valosin containing protein) interacting motif. SIM, SUMO (small ubiquitin like modifier) interaction motif. UBZ, ubiquitin binding zinc finger. PUB, PNGase (peptide N-glycosidase)/UBA (ubiquitin associated domain).
(B) After 7 weeks of cultivation on soil, wss1A mutants displayed a fasciated growth phenotype, whereas wss1B lines were indistinguishable from wild-type (WT) plants.
(C) and (D) Mean values of root length of ten roots per line (n = 3). After 9 d of cultivation, roots of wss1A-2 seedlings reached only a length of 4.7 cm, which is significantly reduced compared with the length of wild-type roots. Wss1B-2 seedlings developed a root length comparable with wild type.
(E) and (F) Mean values of dead cells per root of ten roots per line (n = 3). Propidium iodide (PI)-stained root tips of wss1A-2 revealed a significantly elevated number of dead cells in the root meristem compared with wild type and wss1B-2. Wild-type and wss1B-2 displayed less than 1 dead cell per root.
Columns in (C) and (E) correspond to mean values, and error bars represent ±sd. Statistical differences were calculated using a two-tailed t test with unequal variances: *P < 0.05, **P < 0.01.
All three wss1A mutant lines exhibit serious growth defects, characterized by fasciation of shoots, flower buds, and leaves. The wss1B lines were indistinguishable from wild-type plants (Figure 1B). Because all mutant lines of the same genotype showed identical phenotypes, we conducted all further experiments with the wss1A-2 and wss1B-2 alleles as representative lines for each genotype. For the examination of root length, we measured the length of the roots of 9-d-old seedlings using the ImageJ add-on SmartRoot. For cell death analysis in root meristems, the roots of 5-d-old seedlings were stained with propidium iodide, which can only permeate dead cells. Besides fasciation, a significantly reduced root length (4.7 cm), as well as a significantly elevated number of dead cells in roots meristems (6.3), could be detected in the wss1A-2 line, compared with wild-type plants (7.9 cm root length and 0.1 dead cells). Wss1B-2 showed a root length (7.8 cm) and number of dead cells in root meristems (0.1) comparable with that of wild-type plants (Figures 1C to 1F).
Wss1A Mutants Are Sensitive to DPC-Inducing Agents
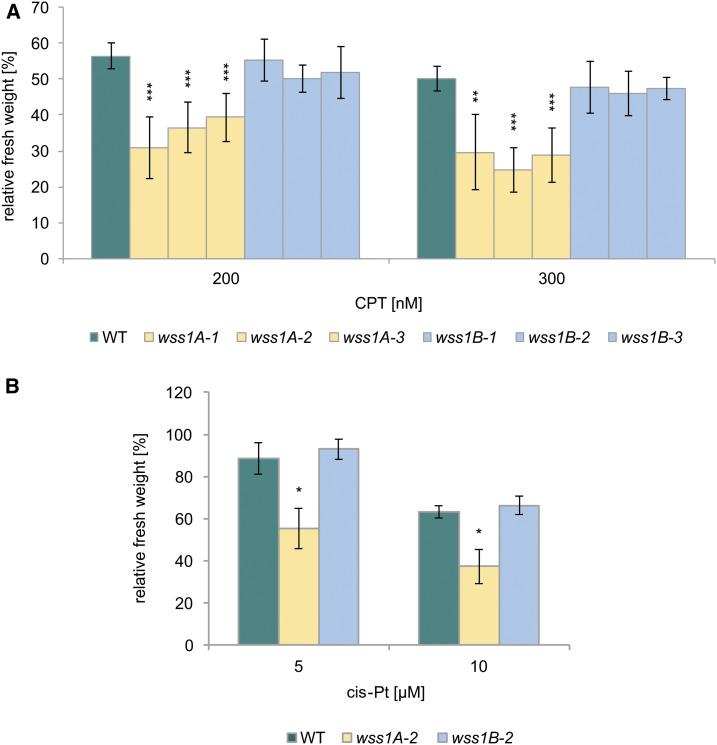
Because an involvement of the SPRTN/Wss1 proteases in the repair of stabilized topoisomerase 1 cleavage complexes (Top1cc) was reported with other organisms, we aimed to test the newly obtained Arabidopsis mutants for CPT sensitivity. One-week-old plantlets were transferred to six-well plates containing liquid media, and CPT was added to the media the following day. After two weeks of cultivation, the fresh weight of the plantlets was determined and referred to the untreated control of the respective genotype. As the CPT-sensitivity represented a key result, we conducted the analysis with all mutant alleles, and used one representative line for each for analysis of sensitivity to cis-platin. Although wss1A lines showed a severe sensitivity to CPT, wss1B mutants exhibited a relative fresh weight comparable with wild-type plants (Figure 2A). We also tested cis-platin, an agent that induces DPCs in addition to DNA crosslinks (Zwelling et al., 1979; Olinski et al., 1987; Woźniak and Walter, 2000). Although wss1B-2 was insensitive to cis-platin treatment, the wss1A-2 mutant showed a significantly higher sensitivity compared with wild-type plants (Figure 2B).
Figure 2.
Sensitivity of WSS1A and WSS1B Deficient Lines Against CPT and Cis-Platin.
(A) Mean values of fresh weights of plantlets relative to untreated controls after treatment with 200 and 300 nM CPT (n = 6). wss1A lines exhibited a statistically significant hypersensitivity in comparison with the wild type (WT) after CPT-treatment in both concentrations used. wss1B alleles were comparable with wild type.
(B) Mean values of fresh weights of plantlets relative to untreated controls after treatment with 5 and 10 µM cis-platin (cis-Pt; n = 3). All cis-platin concentrations tested led to a hypersensitivity of wss1A-2 compared with the wild type on a significant level, whereas a hypersensitivity of wss1B-2 was not detectable.
Columns in (A) and (B) correspond to mean values, and error bars represent ±sd. Statistical differences were calculated using a two-tailed t test with unequal variances: *P < 0.05, **P < 0.01, ***P < 0.001.
WSS1B Does not Play a Detectable Role in DPC Repair
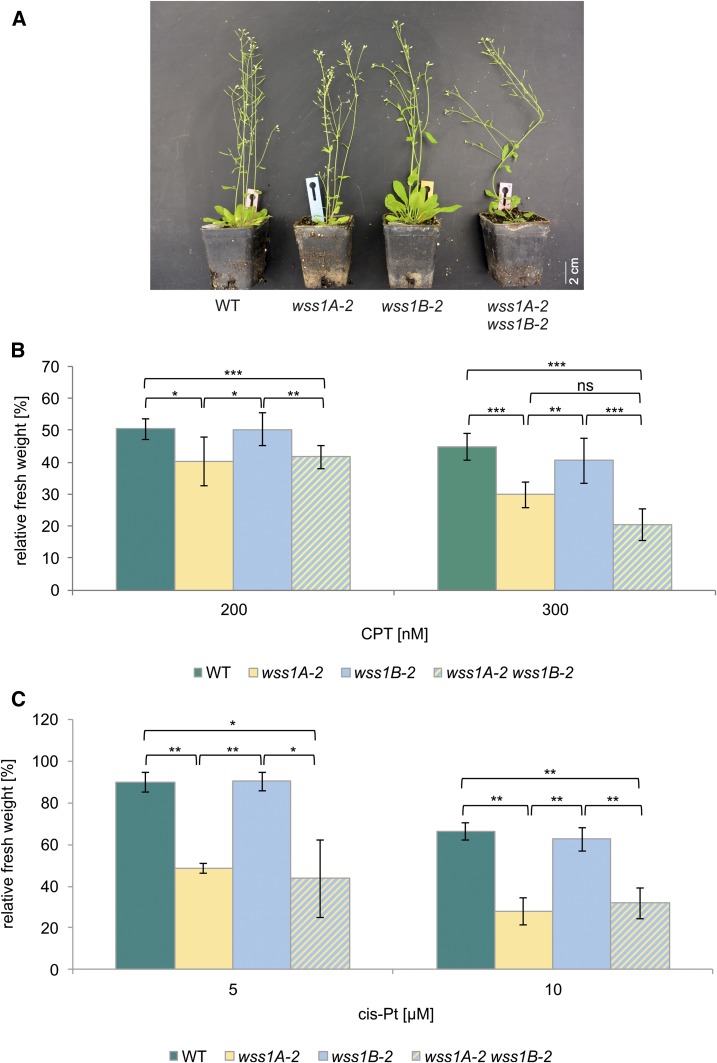
To test whether WSS1B can substitute for WSS1A functions, a wss1A-2 wss1B-2 double mutant was established via crossbreeding of the respective single mutants. The wss1A-2 wss1B-2 lines exhibit a fasciated phenotype and are indistinguishable from wss1A single mutant lines. The double mutant neither displays more severe defects nor is it further reduced in plant size (Figure 3A). Performing sensitivity assays using CPT and cis-platin, the wss1A-2 wss1B-2 double mutant exhibited sensitivities comparable with the wss1A-2 single mutant line (Figures 3B and 3C).
Figure 3.
Analysis of a wss1A-2 wss1B-2 Double Mutant.
(A) After 7 weeks of cultivation on soil, wss1A-2 wss1B-2 double mutants displayed a fasciated growth phenotype corresponding to wss1A-2. A more severe growth phenotype of the wss1A-2 wss1B-2 double mutant was not detected.
(B) Mean fresh weights of plantlets of the wss1A-2 wss1B-2 double mutant, the corresponding single mutant lines and the wild type (WT), relative to untreated controls after treatment with 200 and 300 nM CPT are shown (n = 6). Treatment of wss1A-2 wss1B-2 with CPT resulted in a decreased relative fresh weight, compared with wss1B-2 and WT. The hypersensitivity of wss1A-2 wss1B-2 did not statistically differ from wss1A-2. ns, Not significant.
(C) Mean fresh weights of plantlets of wss1A-2 wss1B-2, the corresponding single mutant lines and the wild type, relative to untreated controls after treatment with 5 and 10 µM cis-platin (cis-Pt; n = 3). Cis-platin treatment of wss1A-2 wss1B-2 was consistent with the results obtained with CPT-treatment. wss1A-2 wss1B-2 was sensitive only on wss1A-2 level and did not show an increased hypersensitive effect.
Columns in (B) and (C) correspond to mean values, and error bars represent ±sd. Statistical differences were calculated using a two-tailed t test with unequal variances: *P < 0.05, **P < 0.01, ***P < 0.001.
WSS1A Is not Required for Homologous Recombination
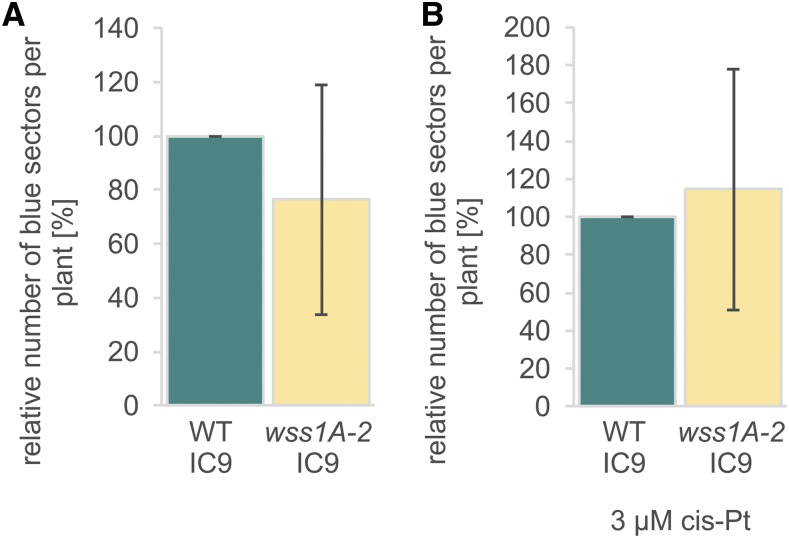
Interestingly, ∆wss1 yeast strains show increased spontaneous and formaldehyde-induced homologous recombination (HR) rates (Stingele et al., 2014), whereas for SPRTN-deficient DT40 cells HR rates were indistinguishable from wild-type cells (Nakazato et al., 2018). To investigate whether loss of WSS1A influences HR efficiencies in somatic plant cells, recombination frequencies were determined using the IC9 reporter construct (Puchta and Hohn, 2012). The interrupted gene for the β-glucuronidase with overlapping homologies can be restored by interchromosomal recombination, and HR events were subsequently detected by histochemical staining of plantlets. Both relative spontaneous and cis-platin–induced HR frequencies of wss1A-2 did not differ significantly from wild type (Figure 4).
Figure 4.
Homologous Recombination Frequencies.
(A) Mean spontaneous recombination frequency of 40 wss1A-2 plantlets relative to wild type (WT; n = 4).
(B) Mean recombination frequency of 40 plantlets of wss1A-2 relative to wild type after treatment with cis-platin (cis-Pt; n = 4).
Columns in (A) and (B) correspond to mean values, and error bars represent ±sd. Statistical differences were calculated using a two-tailed t test with unequal variances: *P < 0.05. No statistically significant differences were detected.
TDP1 Defective Lines Are not Hypersensitive to CPT
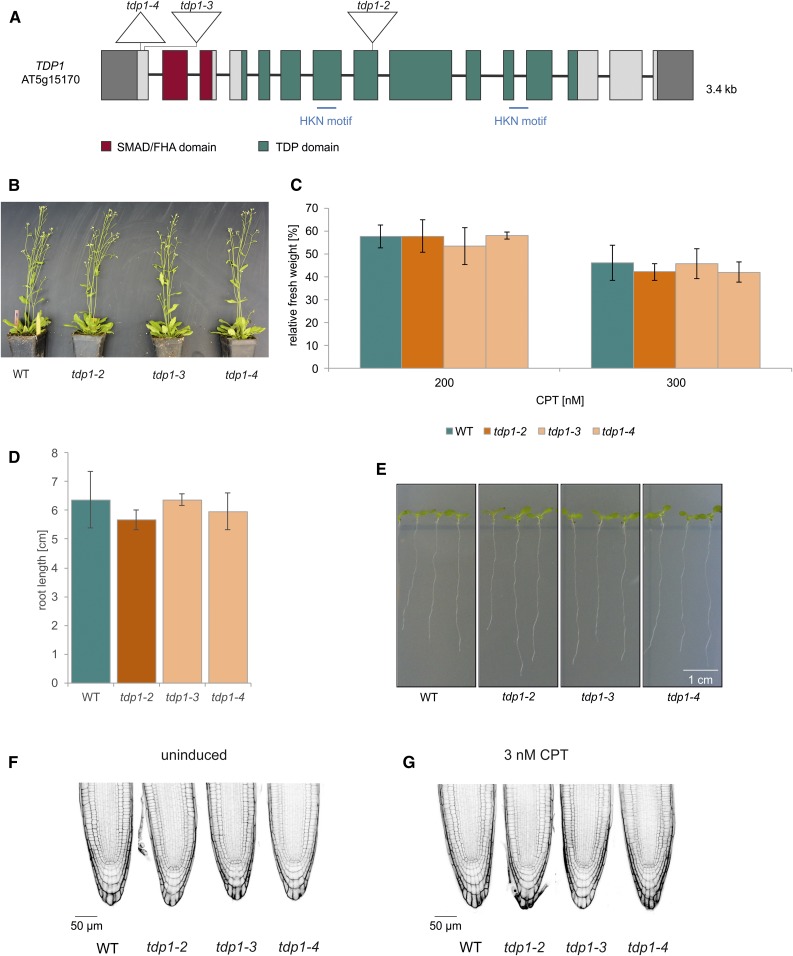
TDP1 is able to specifically hydrolyze the phosphodiester bond of the 3′ phosphate of the DNA backbone and the active tyrosyl residue of the class 1 topoisomerases (Yang et al., 1996; Pouliot et al., 1999). TDP1 has been shown to be involved in DPC repair in yeast (Pouliot et al., 2001; ; Liu et al., 2002; Vance and Wilson, 2002; Stingele et al., 2014). To check whether TDP1 does contribute to DPC repair in Arabidopsis, we analyzed three different tdp1 lines (Figure 5A). The first line we examined was the T-DNA line tdp1-2 (SALK119060) that was not characterized previously and harbors the T-DNA insertion in exon 8. The insertion was verified by sequencing (Supplemental Figure 2A), and the relative gene expression was determined both 5′ and 3′, and spanning the T-DNA insertion, via RT-quantitative PCR (RT-qPCR), and compared with that of wild type (Supplemental Figure 2B; Supplemental Methods). In tdp1-2, the expression 5′ of the insertion was reduced to 23%, whereas no expression was detected spanning the insertion. The relative expression 3' of the insertion was ∼83% of wild-type expression. In addition to the T-DNA mutant line, two CRISPR/Cas9 mutant lines, tdp1-3 and tdp1-4, were established using Cas9 from S. pyogenes (Fauser et al., 2014), targeting the first exon of the gene (5′-TTCCTTAATGGCTCACTCTC-3′). The resulting mutant lines both carry a truncated TDP1 open reading frame. The induced mutations were also confirmed by cDNA sequencing (Supplemental Figure 2C). With three available independent mutant lines, we found that the phenotype of all three mutant alleles was indistinguishable from wild-type plants (Figure 5B). Furthermore, sensitivity to CPT, as well as root length and cell death in root meristems, showed no difference to wild-type plants in all cases (Figure 5C to 5G). This was surprising because in an article published in 2010 (Lee et al., 2010), a single T-DNA insertion tdp1 mutant, tdp1-1, was described as being hypersensitive to CPT treatment and having a dwarfed phenotype. Because we could not confirm this phenotype with the three new independent mutant alleles, we regard it as highly likely that TDP1 has at most a minor role in DPC repair in plants.
Figure 5.
Analysis of tdp1 Mutant Lines.
(A) Genomic structure and protein domains of TDP1. AtTDP1 is 3.4 kb, with 15 exons and 14 introns. The mutations introduced via CRISPR/Cas9 in tdp1-3 and tdp1-4 are located in exon 1, harboring a 1-bp insertion and 14-bp deletion, respectively. The deletion in tdp1-4 includes the start codon. The T-DNA mutant line tdp1-2 harbors the T-DNA insertion in exon 8. Untranslated regions are colored in dark gray. SMAD/FHA, an acronym from the fusion of Caenorhabditis elegans Sma genes and the Drosophila Mad (Mothers against decapentaplegic)/forkhead associated. HKN, histidine, lysine and asparagine.
(B) After 7 weeks of cultivation in soil, the growth phenotype of tdp1-2 to tdp1-4 was indistinguishable from wild type (WT) plants.
(C) Mean values of plantlet fresh weights relative to untreated controls after treatment with 200 and 300 nM CPT are shown. None of the three tdp1 mutant alleles displayed any sensitivity after CPT treatment (n = 3). The relative fresh weights of the mutant lines were comparable with the fresh weight of wild type.
(D) and (E) Mean values of root length (of ten roots) measured from 9-d-old seedlings of the tdp1 mutant alleles were on wild-type level (n = 3).
(F) PI-stained root tips of all three tdp1 mutant lines did not reveal a single dead cell out of 30 roots analyzed per genotype. This was also observed with wild-type roots.
(G) PI-stained root tips of tdp1-2 to tdp1-4 and wild type were analyzed after induction with 3 nM CPT (30 roots per line). TDP1 deficient lines were indistinguishable from wild type, exhibiting less than 1 dead cell per root.
Columns in (C) and (D) correspond to mean values, and error bars represent ±sd. Statistical differences were calculated using a two-tailed t test with unequal variances: *P < 0.05.
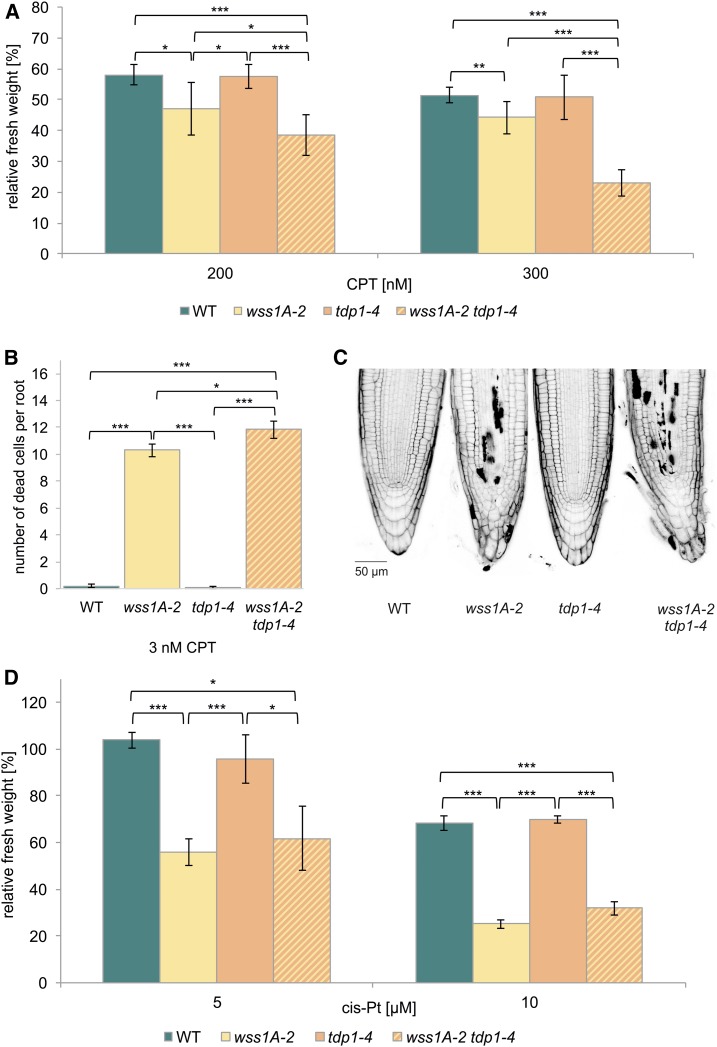
The wss1A-2 tdp1-4 Double Mutant Displays a More Severe Sensitivity to CPT, Compared With wss1A-2
As it has been previously reported that Wss1 and Tdp1 are involved in parallel pathways in the repair of DPC in budding yeast (Stingele et al., 2014), it was of special interest to investigate a wss1A tdp1 double mutant in Arabidopsis. We established the respective double mutant via crossbreeding. The obtained double mutant clearly showed a significant reduction of relative fresh weight in comparison with wss1A-2 after treatment with 200 and 300 nM CPT (Figure 6A). To support the obtained data, we checked for cell death in root meristems after induction with 3 nM CPT. Although wild type and tdp1-4 showed ∼0.1 dead cells per root, wss1A-2 already exhibited ∼10 dead cells per root. With ∼12 dead cells per root, the double mutant showed a statistically significant elevated number of dead cells, compared with wss1A-2 (Figure 6B and 6C). This result again supports the data obtained in the sensitivity assay, classifying WSS1A and TDP1 into independent pathways in the repair of CPT-induced damage. Testing of wss1B-2 tdp1-4 did not show any hypersensitivity in response to CPT (Supplemental Figure 3). Besides the TOP1cc-inducing agent CPT, we further tested wss1A-2 tdp1-4 for cis-platin sensitivity, as this agent induces different kinds of DPCs. Although the wss1A-2 mutant, in contrast with tdp1-4, displayed in both concentrations tested (5 and 10 µM) a strong hypersensitivity, the double mutant was only sensitive on wss1A-2 level. A synergistic effect was not detectable (Figure 6D). This indicates that in contrast with AtWSS1A, the action of AtTDP1 is indeed restricted to hydrolysation of phosphodiester bonds linking the DNA backbone to proteins via a tyrosine residue, but it is not involved in resolving other kinds of DNA–protein crosslinks.
Figure 6.
Analysis of a wss1A-2 tdp1-4 Double Mutant.
(A) Mean values of plantlet fresh weights of the wss1A-2 tdp1-4 double mutant, the corresponding single mutant lines and the wild type (WT), relative to untreated controls after treatment with 200 and 300 nM CPT (n = 6). The wss1A-2 tdp1-4 double mutant exhibited synergistic sensitivity effects with both CPT concentrations used, compared with both single mutants. Although the wss1A-2 allele was hypersensitive, tdp1-4 revealed relative fresh weights comparable with wild type.
(B) and (C) Mean values of ten PI-stained root tips per line of the wss1A-2 tdp1-4 double mutant, the corresponding single mutant lines, and the wild type after induction with 3 nM CPT (n = 3) confirmed the synergistic sensitivity effect. Whereas in both wild-type and tdp1-4 lines less than one dead cell per root was detectable, 10 and 12 dead cells per root were shown for wss1A-2 and the double mutant, respectively, a significantly elevated cell death level compared with both single mutants.
(D) Mean values of plantlet fresh weights of the wss1A-2 tdp1-4 double mutant, the corresponding single mutant lines, and the wild type, relative to untreated controls after treatment with 5 and 10 µM cis-platin (cis-Pt; n = 3). The wss1A-2 tdp1-4 double mutant exhibited a hypersensitivity on wss1A single mutant level with both concentrations applied. Tdp1-4 did not show a hypersensitive effect and revealed a relative fresh weight comparable with wild type.
Columns in (A), (B), and (D) correspond to mean values, and error bars represent ±sd. Statistical differences were calculated using a two-tailed t test with unequal variances: *P < 0.05, **P < 0.01, ***P < 0.001.
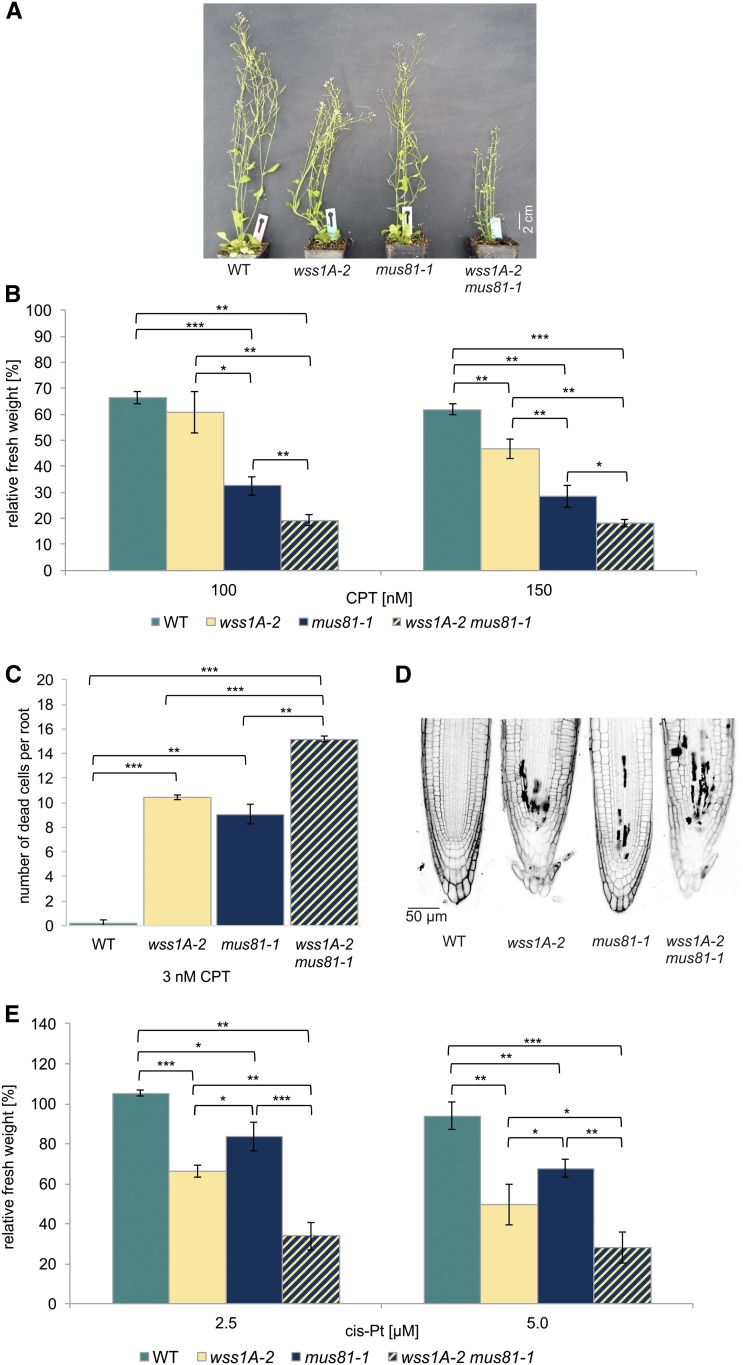
The Protease WSS1A and the Endonuclease MUS81 Define Parallel Pathways in DPC Repair
Because the endonuclease MUS81 has been identified to contribute to the repair of a wide range of DNA damage, and as being particularly active in the repair of DNA-DNA crosslinks, we regarded MUS81 as being a very interesting factor for further epistasis analysis (Interthal and Heyer, 2000; Boddy et al., 2001; Hartung et al., 2006; Ciccia et al., 2008; Mannuss et al., 2010). As MUS81-deficient human cell lines and yeast strains have already been shown to be hypersensitive to CPT treatment (Liu et al., 2002; Regairaz et al., 2011), we were strongly interested to see if there would be a similar hypersensitivity of mus81 mutants in Arabidopsis and even enhanced effects detectable in a wss1A mus81 double mutant. First, we checked for the growth phenotype of the plants. Although mus81-1 plants look perfectly like wild type, and wss1A-2 exhibits the fasciated phenotype described before, the wss1-2 mus81-1 double mutant clearly shows more severe growth defects compared with wss1A-2 (Figure 7A). Testing wild type, wss1A-2, mus81-1, and wss1A-2 mus81-1 on CPT revealed that mus81 is far more sensitive to CPT than the wss1A mutant. As the double mutant explicitly shows a significantly lower relative fresh weight, compared with both single mutants (Figure 7B), this additive effect can be reproduced by determining the number of dead cells in root meristems after induction with 3 nM CPT. Wild type shows less than one dead cell per root, with wss1A-2 and mus81-1 showing a number of about ten, whereas the double mutant exhibits an additive effect possessing ∼15 dead cells per root (Figures 7C and 7D). An additive effect was also seen for the nonspecific crosslinking agent cis-platin (Figure 7E). A wss1B-2 mus81-1 double mutant line, tested on CPT, was sensitive only on mus81-1 level, implying that WSS1B is dispensable for DPC repair (Supplemental Figure 4). Our data clearly demonstrate that in plants MUS81 is crucial for DPC repair in a pathway parallel to WSS1A.
Figure 7.
Analysis of a wss1A-2 mus81-1 Double Mutant.
(A) After 7 weeks of cultivation on soil, wss1A mutants displayed a fasciated growth phenotype. The wss1A-2 mus81-1 double mutant line exhibited a more severe growth phenotype, which was shown by an even more reduced plant size compared with wss1A-2. Mus81-1 lines were indistinguishable from wild-type (WT) plants.
(B) Mean values of plantlet fresh weights of the wss1A-2 mus81-1 double mutant, the corresponding single mutant lines, and the wild type, relative to untreated controls after treatment with 100 and 150 nM CPT (n = 6). The wss1A-2 mus81-1 double mutant exhibited a further reduced fresh weight compared with wss1A-2 and mus81-1, implying an additive hypersensitivity effect.
(C) and (D) Mean values of PI-stained root tips (10 per line) of the wss1A-2 tdp1-4 double mutant, the corresponding single mutant lines, and the wild type after induction with 3 nM CPT (n = 3). Although wild type showed less than one dead cell per root, wss1A-2 and mus81-1 exhibited 10 and 9 dead cells per root, respectively, with the double mutant displaying 16 dead cells per root.
(E) Wild type, wss1A-2, mus81-1, and the corresponding double mutant were treated with 2.5 and 5 µM cis-platin, and the fresh weight relative to an untreated control was determined. Mean values of plantlet fresh weights are given (n = 3). Although the single mutant lines exhibited a hypersensitivity in respect to cis-platin treatment compared with the wild type, the double mutant was severely more affected than both single mutants, implying an additive hypersensitive effect.
Columns in (B), (C), and (E) correspond to mean values, and error bars represent ±sd. Statistical differences were calculated using a two-tailed t test with unequal variances: *P < 0.05, **P < 0.01, ***P < 0.001.
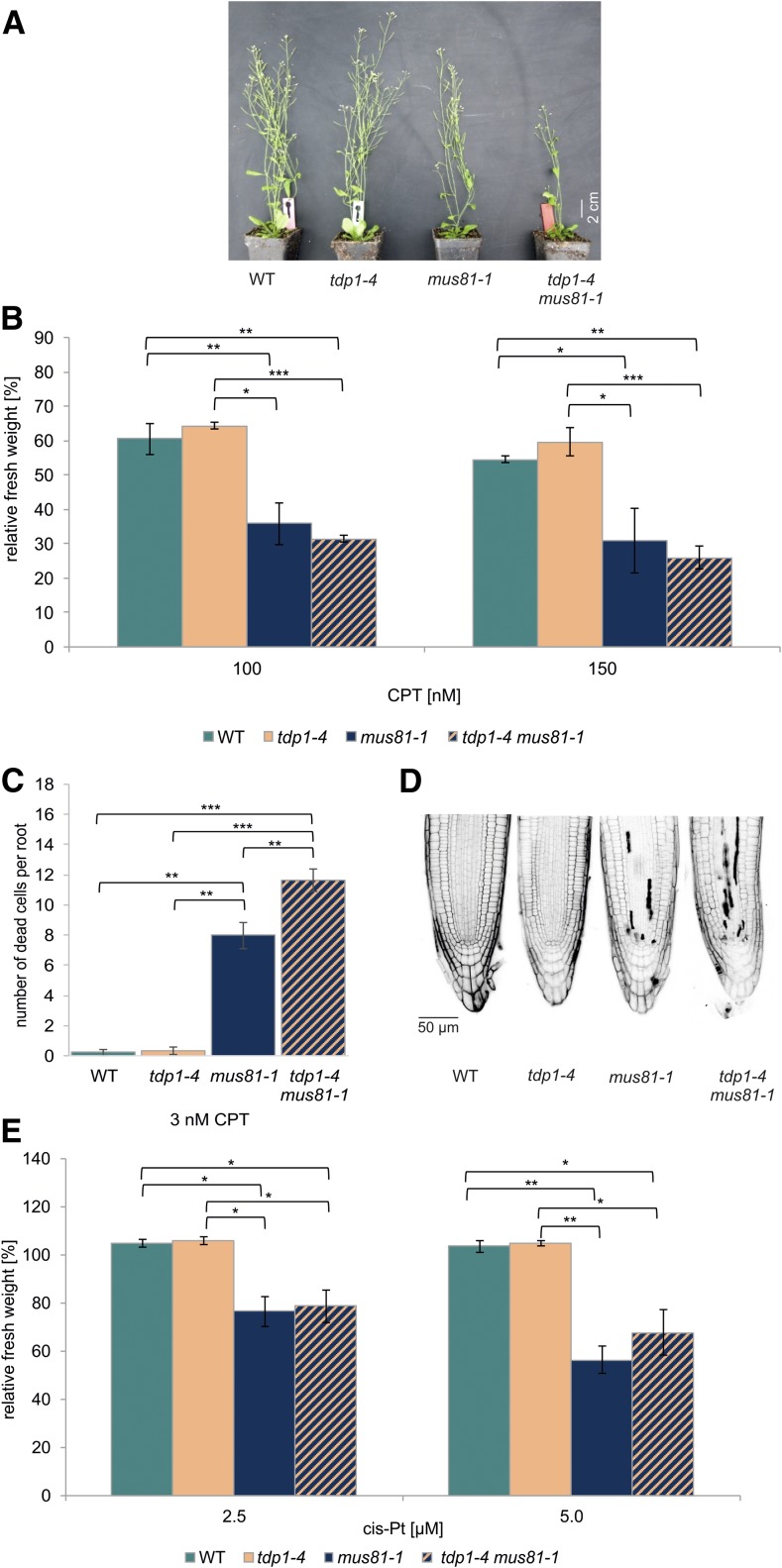
TDP1 and MUS81 Act Independently on CPT-Induced Damage
Because our results showed that both MUS81 and TDP1 work in DPC repair in Arabidopsis independently from WSS1A, it was important to elucidate whether they work in the same or different pathways. To address this question, we established a tdp1-4 mus81-1 double mutant via crossbreeding. First, we checked for the growth phenotype and, surprisingly, detected a reduction in plant size of the double mutant, wheres the respective single mutant lines have a wild-type–like phenotype (Figure 8A). Sensitivity assays in response to CPT showed that tdp1-4 did not have a sensitive effect in the tested concentrations of 100 and 150 nM CPT. The measured relative fresh weight of mus81-1 was significantly reduced, whereas the fresh weight of tdp1-4 mus81-1 was on mus81-1 level (Figure 8B). Furthermore, sensitivity to cis-platin was tested whereby we could also see that only mus81-1 and the double mutant were sensitive, both to the same extent (Figure 8E). Even though no synergistic effect of MUS81 and TDP1 could be resolved in the sensitivity assay system using whole plantlets, we could detect a more severe effect in the highly sensitive root assays. The number of dead cells per root after induction with 3 nM CPT in wild type and tdp1-4 is ∼0.2 dead cells per root. In the case of mus81-1, ∼8 dead cells were detectable, whereas the double mutant displayed ∼12 dead cells per root (Figure 8C and 8D). These results, as well as the phenotypic effect, argue that MUS81 and TDP1 are at least mainly acting in parallel pathways in the repair of DPC damage induced by CPT.
Figure 8.
Analysis of a tdp1-4 mus81-1 Double Mutant.
(A) Although tdp1-4 and mus81-1 lines were indistinguishable from wild-type (WT) plants, the tdp1-4 mus81-1 double mutant line exhibited a growth defect that was characterized by a reduced plant size compared with both single mutants. The picture was taken after 7 weeks of cultivation on soil.
(B) Mean values of plantlet fresh weights of the tdp1-4 mus81-1 double mutant, the corresponding single mutant lines, and the wild type, relative to untreated controls after treatment with 100 and 150 nM CPT (n = 6). Although tdp1-4 did not show a hypersensitivity, mus81-1 lines exhibited a decreased relative fresh weight compared with wild type. The respective double mutant was only hypersensitive on mus81-1 level.
(C) and (D) Mean values of PI-stained root tips (10 per line) of the tdp1-4 mus81-1 double mutant, the corresponding single mutant lines, and the wild type after induction with 3 nM CPT (n = 3). Although tdp1-4 was comparable with wild type, exhibiting less than one dead cell per root, mus81-1 showed 8 dead cells per root. The rate of cell death in root meristems was significantly elevated in the respective double mutant, with a number of 12 dead cells per root.
(E) Wild type, tdp1-4, mus81-1, and the corresponding double mutant were treated with 2.5 and 5 µM cis-platin (cis-Pt) and the fresh weight relative to an untreated control was determined. Mean values of plantlet fresh weights are given (n = 3). The double mutant line exhibited a hypersensitivity on the level of the mus81-1 single mutant line. Tdp1-4 did not display a sensitivity and its relative fresh weight was comparable with wild type.
Columns in (B), (C), and (E) correspond to mean values, and error bars represent ±sd. Statistical differences were calculated using a two-tailed t test with unequal variances: *P < 0.05, **P < 0.01, ***P < 0.001.
DISCUSSION
Over the last thirty years, the main pathways of DNA repair have been studied in great detail in different kinds of organisms from yeast to humans, including plants. Therefore, the existence of a novel DNA repair pathway, which had been overlooked for all of those years, came as a big surprise. Whereas the repair of single or double strand breaks, base adducts, DNA crosslinks, and base mismatches were the center of interest, the repair of DPCs was neglected for a long time. Nevertheless, DPCs are highly toxic DNA adducts and represent a severe threat to the integrity of the genome by blocking replication. In pioneering work performed in mammals and yeast, a novel repair pathway depending on the specialized repair proteases SPRTN/Wss1 was discovered only recently (Stingele et al., 2014; Lopez-Mosqueda et al., 2016; Stingele et al., 2016). Although two Arabidopsis orthologs of the repair protease, WSS1A and WSS1B, were identified via bioinformatics analysis, the mechanisms involved in repair DPCs in plants have remained unclear. Here we analyze DPC repair in Arabidopsis and characterize several subpathways that are defined by the enzymatic activity of the respective proteins involved. We demonstrate that WSS1A is indeed of crucial importance for DPC repair in Arabidopsis. By performing epistasis analysis, we could further reveal that plants possess at least three different pathways for DPC repair. Two major ones defined by the protease WSS1A and the DNA endonuclease MUS81, and a minor one defined by the TDP1.
WSS1A, but not WSS1B, Is Crucial for DPC Repair in Arabidopsis
To elucidate whether the plant homologs of SPRTN/WSS1 have a conserved function in DPC repair, we generated, using CRISPR/Cas, and characterized three knock out mutant lines for each of WSS1A and WSS1B in Arabidopsis. Although WSS1A defective lines exhibited a fasciated phenotype, a drastically reduced root length, and a significantly elevated number of dead cells in root meristems, this was not the case for WSS1B defective lines (Figure 1). Because the root meristem is a rapidly dividing tissue, deficiencies in DNA replication leading to growth retardation are detectable early in this organ. The fact that WSS1A deficient lines displayed about six dead cells per root, even without genotoxic stress, highlights how important the protease is for removing endogenously produced DPCs. Only wss1A, but not wss1B mutants, were hypersensitive to the DPC-inducing agents CPT and cis-platin, implying that WSS1A is involved as a key protease in the repair of TOP1 enzymatic and cis-platin–induced nonenzymatic DPCs (Figure 2). Interestingly, ∆wss1 yeast strains did not display CPT-sensitivity, indicating that AtWSS1A might be more important for the repair of CPT-induced lesions in plants compared with its yeast ortholog (Stingele et al., 2014). On the other side, the CPT-hypersensitivity of SPRTN-knock-out MEFs and the cis-platin sensitivity of C. elegans dvc-1 lines highly coincide with our results (Lopez-Mosqueda et al., 2016; Stingele et al., 2016).
The presence of two WSS1 orthologs in plants tempted us to speculate that WSS1A and WSS1B might be functionally redundant. However, subsequent analysis of a wss1A wss1B double mutant, with respect to CPT and cis-platin sensitivity, clearly showed that WSS1B is not able to backup WSS1A function at a detectable level and thus the biological role of WSS1B remains elusive (Figure 3). We can only speculate on functions of WSS1B, as being involved in the removal of noncovalently bound proteins from DNA during replication. Nevertheless, our analysis demonstrates that the recently discovered protease-dependent pathway for DPC repair is conserved in plants.
A unique type of DPC is caused by CPT, which induces so called stabilized TOP1ccs. Here an enzymatic reaction intermediate is trapped during topoisomerase 1 action, covalently linking TOP1 to the DNA backbone. A completely different type of DPC is induced by cis-platin by binding to N7-guanine positions of the DNA and Cys, Arg, and Lys side chains of proteins (Chválová et al., 2007; Tretyakova et al., 2015). Mechanistically, WSS1A proteolytically degrades the protein part of the DPC, enabling downstream canonical repair pathways, like translesion synthesis, to further process the remaining peptide remnant (Stingele et al., 2015). The sensitivity of the respective wss1A mutants against both, CPT and cis-platin, demonstrates that the proteolytic activity of WSS1A is not restricted to a specific type of protein adducts.
Although we could show the involvement of WSS1A in DPC repair, the loss of the respective protein did not have any impact on HR (Figure 4). In contrast with yeast, DSBs are repaired predominantly by NHEJ in plants and animals (Puchta, 2005). Although the lack of ScWss1 channels DSBs, resulting from collapsed replication forks, into repair via HR, this would presumably lead to NHEJ-mediated DSB repair in plants. This hypothesis is also in line with recent data obtained in SPRTN-deficient but HR-proficient animal cells (Nakazato et al., 2018).
TDP1 Is Involved in a Backup Pathway to WSS1A in the Repair of Protein Adducts Linked to the DNA by Phosphodiester Bonds
The TDP1 specifically mediates the hydrolysis of the phosphodiester bond of the 3′ phosphate of the DNA backbone and the class 1 topoisomerases active tyrosyl residue (Yang et al., 1996; Pouliot et al., 1999). Thus, TDP1 mechanistically provides another strategy to resolve DPCs, by directly targeting the phosphodiester bond between DNA and protein moieties. As an Attdp1 T-DNA insertion line was published as being sensitive to CPT and exhibiting dwarfism, we became interested in this factor (Lee et al., 2010). Unfortunately, we were not able to reproduce these phenotypes using a different T-DNA insertion mutant line of the SALK collection (tdp1-2), speculating whether this line exhibits a possible hypomorphic phenotype. To settle this issue, we used the power of CRISPR/Cas technology (Puchta, 2017), and obtained two further TDP1 knock out alleles, tdp1-3 and tdp1-4. Consistently, all three mutant alleles characterized in our laboratory were indistinguishable from wild-type plants in respect to growth phenotype, CPT-sensitivity, root length, and the number of dead cells in root meristems (Figure 5). In contrast with prior publication, we were only able to prove that TDP1 is involved in DPC repair at all through the analysis of double mutants. The more severe sensitivity of the wss1A-2 tdp1-4 double mutant in response to CPT clearly indicated that TDP1 is indeed able to repair certain kinds of DPCs that persist in the absence of WSS1A (Figure 6A). These results go along with reports that in yeast ∆tdp1 strains merely exhibited hypersensitive features to CPT in combination with further depletion of other DNA repair genes (Pouliot et al., 2001; Liu et al., 2002; Vance and Wilson, 2002; Stingele et al., 2014). In contrast, in humans, TDP1 seems to have a more important role: TDP1 deficiency sensitizes cells to CPT, and mutations in the TDP1 gene are responsible for the hereditary disease Spinocerebellar Ataxia with Axonal Neuropathy (Takashima et al., 2002; Alagoz et al., 2014).
Thus, WSS1A defines a major pathway of DPC repair in plants, whereas the pathway mediated by TDP1 seems merely a backup for a specific class of damage. This result could also be confirmed by analysis of cell death in root meristems after induction with CPT (Figure 6B to 6C). Interestingly, a similar effect could not be detected treating the wss1A-2 tdp1-4 double mutant with cis-platin, which induces various kinds of ternary DNA-Pt-protein complexes (Figure 6D). Therefore, the action of TDP1 seems to be restricted to protein adducts linked to the DNA by phosphodiester bonds, as we would expect from its enzymatic properties.
WSS1A, MUS81, and TDP1 Define Different Pathways of DPC Repair in Arabidopsis
The previous analysis demonstrated that the protease and the tyrosyl-phosphodiesterase based pathways of DPC repair are conserved between plants, animals, and yeast. Nevertheless, there seems to be quantitative differences when we take the sensitivity data into account: in yeast the Δwss1 phenotype seems to be less drastic than in multicellular eukaryotes, whereas TDP1 has a more important function in mammals than in plants. Therefore, we were wondering as to whether a further DPC repair pathway exists that might be of special importance for plants. The endonuclease MUS81 has been identified as a key player in DNA repair. MUS81, as part of a highly conserved nuclease complex, is able to resolve versatile DNA intermediates, reaching from 3′ flaps, replication fork structures, to displacement loops and Holliday junctions (Boddy et al., 2001; Chen et al., 2001; Doe et al., 2002; Abraham et al., 2003). In Arabidopsis, biochemical analysis showed that MUS81 is endonucleolytically active on 3′ flaps and at Holliday junctions (Geuting et al., 2009). Furthermore, sensitivity studies revealed that MUS81 is strongly involved in the repair of DNA crosslinks and alkylations induced by the genotoxic agents mitomycin C, cis-platin, methyl methanesulfonate, and ionizing radiation (Hartung et al., 2006; Berchowitz et al., 2007; Mannuss et al., 2010). Epistasis analysis indicated that the endonuclease MUS81 acts independently of the ATPase RAD5A and the DNA helicases HRQ1 and RTEL1, as well as the Translesion Polymerase zeta in the repair of cis-platin–induced damage (Mannuss et al., 2010; Recker et al., 2014; Kobbe et al., 2015; Röhrig et al., 2018). Detection of synthetically lethal phenotypes of mus81-1 lines, additionally lacking one of the helicases RECQ4A or Fanconi anaemia complementation group M protein, explicitly demonstrated that MUS81 is crucially needed for the repair of replication-associated DNA damage (Hartung et al., 2006; Mannuss et al., 2010; Dangel et al., 2014; Dorn et al., 2018). Additionally, an involvement of MUS81 in a parallel pathway to the Fanconi Anemia–associated nuclease was shown (Herrmann et al., 2015). These findings supported the concept of MUS81 being able to process aberrant DNA structures, as a backup mechanism for the removal of recombination intermediates. Besides the importance of MUS81 in somatic cells, it was found that this endonuclease is also involved in the formation of class II crossovers in meiosis (Berchowitz et al., 2007).
Based on the importance of MUS81 in DNA repair in plants and on reports on CPT-sensitivity of MUS81 deficient lines in humans and yeast (Liu et al., 2002; Regairaz et al., 2011), we decided to test whether MUS81 is also involved in DPC repair in plants. Indeed, we found a surprisingly strong hypersensitivity of mus81-1 with respect to CPT treatment, indicating an extraordinary important role of MUS81 in the repair of CPT-induced damage in Arabidopsis. As cis-platin is inducing DNA-DNA crosslinks as well as DNA–protein crosslinks, it was interesting to test whether MUS81 is also involved in the repair of the latter damage class. Therefore, we tested the wss1A-2 mus81-1 double mutant in comparison with the single mutants in response to CPT and cis-platin treatment. Indeed, for both kinds of agents, clear additive sensitivity effects could be detected. We were additionally able to confirm this effect in cell death assays in root meristems after CPT induction (Figure 7). This clearly indicates that the repair protease WSS1A and the endonuclease MUS81 act at least in most cases independently from each other in DPC repair. Nevertheless, if one pathway is absent, the other pathway can process some of the unresolved DPCs. As MUS81 deficient lines exhibit a stronger sensitivity to CPT than wss1A-2, we assume that in plants the endonuclease-mediated pathway plays an even more important role in DPC repair than the protease-dependent pathway.
Because we found out that TDP1 is involved in a backup pathway of DPC repair for CPT-induced damage, we were interested in the direct relationship of TDP1 and MUS81. Phenotypic growth defects of the respective double mutant, and an increase of dead cells in root meristems after CPT induction, indicate that TDP1 is able to process certain kinds of DPCs that are not resolved in the absence of MUS81 (Figures 8A, 8C and 8D). As tdp1 single mutants do not show any defects without additional depletion of MUS81, this points to a backup function of TDP1 also in respect to this endonuclease. The fact that TDP1 plays a minor role is indicated by sensitivity studies in response to CPT, whereby a further increase in sensitivity could not be resolved (Figure 8B). This may be due to the strong sensitivity of mus81-1. In respect to cis-platin sensitivity, as expected only an involvement of MUS81 in the repair is detectable (Figure 8E). The relationship of the two enzymes seems to be evolutionary conserved as sensitivity studies in yeast also classified Mus81 and Tdp1 in independent pathways in DPC repair (Liu et al., 2002).
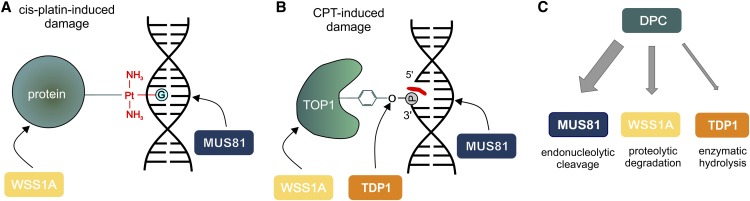
In conclusion, we showed that DPC repair in Arabidopsis is defined by at least three different pathways. Although WSS1A proteolytically degrades the protein part of the DPC, the endonuclease MUS81 targets the DNA moiety by endonucleolytic cleavage (Figure 9A). In the case of DPCs linked by a phosphodiester bond, the complex can also be hydrolyzed by TDP1 enzymatically, a minor pathway that is mainly used in absence of one or the other enzyme (Figure 9B). As the loss of MUS81 results in the strongest CPT sensitivity, the elimination of DPCs via DNA degradation seems to be the most prominent pathway of DPC repair in plants (Figure 9C). This finding is in accordance with our previous results that MUS81 is a key factor in DNA damage response in plants.
Figure 9.
Model of DPC Repair in Plants.
(A) Cis-platin induces DPCs via binding of the N7-guanine position of the DNA and Cys, Arg, and Lys side chains of proteins. This type of DPC can be repaired via either the protease WSS1A or the endonuclease MUS81.
(B) CPT-induced protein adducts are linked to the DNA via a phosphodiester bond, resulting from the attack of the tyrosyl residue of TOP1 at the 3′ phosphate of the DNA backbone. This type of DPC can be resolved via proteolytic action of WSS1A at the protein part, hydrolysis of the phosphodiester bond via TDP1 by endonucleolytic cleavage of the DNA by MUS81. CPT is depicted in red.
(C) In plants, at least three different pathways for DPC repair exist. The main pathway is defined by the endonuclease MUS81, whereas a second important pathway is dependent on the action of the newly identified repair protease WSS1A. TDP1 defines a third, minor, and mainly backup pathway, by hydrolyzing enzymatically phosphodiester bonds of a certain type of DPCs.
In the future, it will be interesting to define further factors that are involved in one or the other pathways of DPC repair in Arabidopsis and to define putative differences in plants in comparison with yeast and mammals, in more detail.
METHODS
Mutant Lines and Growth Conditions
Throughout the study, Arabidopsis (Arabidopsis thaliana) plants were used in the Columbia (Col-0) background. To characterize the candidate genes WSS1A, WSS1B, and TDP1, Cas9-mediated mutagenesis was performed. The wss1A-1, wss1A-2, wss1B-1, and wss1B-2, as well as the tdp1-3 and tdp1-4 mutant lines, were generated using Cas9 from Streptococcus pyogenes, as previously described (Fauser et al., 2014). The wss1A-3 and wss1B-3 mutant lines were established using the Cas9 from Staphylococcus aureus (Steinert et al., 2015). The T-DNA line mus81-1 (GABI_113F11) has been previously described (Hartung et al., 2006). The T-DNA insertion site of the tdp1-2 (SALK119060) mutant was characterized in frame of this study (Supplemental Methods). For the generation of double mutants, homozygous mutants were used and double mutants were identified in the F2 generation by PCR-based genotyping. Oligonucleotides used throughout the study are listed in Supplemental Table. Plants were cultivated in the greenhouse on soil (1:1 mixture of Floraton [Floragard] and vermiculite [2 to 3 mm, Deutsche Vermiculite Dämmstoff]) with 16-h light (Phillips, Master, TL-D 36W/840) and 8-h darkness at 22°C. For assays, plants were surface sterilized using 4% sodium hypochlorite, stratified overnight at 4°C and sown on germination media (GM; 4.9 g/L Murashige and Skoog medium [Duchefa], 10 g/L Suc, and 7.6 g/L; pH 5.7 with potassium hydroxide). Plates were cultivated in a CLU-36L4 plant culture chamber (Percival Scientific) with stable conditions of 16-h light at 22°C and 8-h dark at 20°C.
PCR-Based Genotyping
For PCR-based genotyping, two different primer combinations were used for each mutant line. For T-DNA lines, a T-DNA specific primer combination was used composed of a primer binding in the T-DNA region and a gene-specific primer. A second wild-type–specific primer pair consists of gene-specific primers 5′ and 3′ of the T-DNA. For CRISPR mutants, the genotyping was performed similarly, using a wild-type–specific primer pair and a mutation-specific primer combination. For wss1A-1, wss1A-2, wss1B-3, and tdp1-4 lines, which were not suitable for PCR-based genotyping because of only one nucleotide insertion mutation, a DNA fragment compromising the potentially mutated site was amplified, purified, and subsequently analyzed by Sanger sequencing. The genotyping of mus81-1 has been previously described (Hartung et al., 2006).
Sensitivity Assays
Sensitivity assays were performed as previously described (Hartung et al., 2007).Ten 7-d-old plantlets were transferred from solid GM to six-wells plates containing 5 mL liquid GM (untreated control) or 4 mL liquid GM (genotoxin-induced) per well under sterile conditions. The following day, 1 mL of genotoxin solution was added to obtain the desired concentrations. After 13 d of incubation, the fresh weight of the plants was quantified. The relative fresh weight was determined by normalizing the weight of the treated samples to the untreated control of the corresponding genotype. Means of at least three biological replicated are displayed.
Root Length Assays
Surface sterilized seeds were sown on square culture plates containing solid GM (1% agar) and cultivated in an upright position. After 9 d of cultivation, photos of the plates were taken, and the root length was determined using the SmartRoot Add-on of ImageJ (Lobet et al., 2011). Means of three biological replicates of 10 roots per genotype are depicted.
Cell Death Analysis in Roots
To determine the number of dead cells in root meristems, seeds were treated and cultured according to root length assays. Plants were transferred after 4 d of cultivation to six-well plates containing 5 mL of liquid GM supplemented with CPT (3 nM). After incubation for 24 h, the plantlets were washed several times before being placed in propidium iodide solution (5 µg/ml) on a microscope slide. The number of dead cells was determined using a confocal microscope (LSM 700 laser scanning microscope, Carl Zeiss Microscopy).
HR Assays
Recombination frequency was determined by HR assays as described using the IC9 reporter construct (Molinier et al., 2004; Hartung et al., 2007). Seeds were surface sterilized as described before and cultivated under anexic conditions on solid GM medium for 1 week. Afterward, 40 plantlets were transferred into halved Petri dishes, containing 10 mL liquid GM for genotoxin-free approaches and 9 mL for genotoxin treated approaches. For genotoxin approaches, 1 mL of liquid GM containing cis-platin (3 µM) was added. After 15 d of cultivation, histochemical β-glucuronidase staining was performed as described. Blue sectors on each plantlet were quantified using a binocular microscope. Relative HR rate of the mutant lines was determined by comparing them to wild-type HR rate. Means of four biological replicates are depicted.
Statistical Methods
For the determination of statistically significant effects, a two-sided, two-sample t test with no equal variance was performed. P-values: P* ≤ 0.05: **P < 0.01; ***P < 0.001.
Accession Numbers
Sequence data of the genes used in this article can be found at The Arabidopsis Information Resource with the following accession numbers: Arabidopsis WSS1A (At1g55915), Arabidopsis WSS1B (At5g35690), Arabidopsis MUS81 (At4g30870), and Arabidopsis TDP1 (At5g15170).
Supplemental Data
Supplemental Figure 1. cDNA analysis of wss1A and wss1B mutant lines.
Supplemental Figure 2. Validation of tdp1-2 and cDNA analysis of tdp1-3 and tdp1-4.
Supplemental Figure 3. Sensitivity analysis of a tdp1-4 wss1B-2 double mutant against CPT.
Supplemental Figure 4. Sensitivity analysis of a double wss1B-2 mus81-1 mutant against CPT.
Supplemental Methods. Expression analysis of tdp1-2 by RT-qPCR.
Supplemental Table. Oligonucleotides used for genotyping and amplification for sequencing.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
The authors thank Seline Zschach, Julia Kremer, and Waltraud Wehrle for excellent technical assistance and Amy Whitbread for critically reading the article. This work was funded by the European Research Council (ERC) (https://erc.europa.eu; grants. ERC-2010-AdG_20100317 COMREC and ERC-2016-AdG_741306 CRISBREED), as well as by Deutsche Forschungsgemeinschaft (DFG) (http://www.dfg.de; grant ERA-CAPS13.058_DeCOP Pu137/16-1 to H.P.).
AUTHOR CONTRIBUTIONS
A.D., O.T., and H.P. designed the research; J.E. and N.B. performed the research; J.E., A.D., N.B., and H.P. analyzed the data; J.E., A.D., and H.P. wrote the article.
References
- Abraham J., et al. (2003). Eme1 is involved in DNA damage processing and maintenance of genomic stability in mammalian cells. EMBO J. 22: 6137–6147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alagoz M., Wells O.S., El-Khamisy S.F. (2014). TDP1 deficiency sensitizes human cells to base damage via distinct topoisomerase I and PARP mechanisms with potential applications for cancer therapy. Nucleic Acids Res. 42: 3089–3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berchowitz L.E., Francis K.E., Bey A.L., Copenhaver G.P. (2007). The role of AtMUS81 in interference-insensitive crossovers in A. thaliana. PLoS Genet. 3: e132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggins S., Bhalla N., Chang A., Smith D.L., Murray A.W. (2001). Genes involved in sister chromatid separation and segregation in the budding yeast Saccharomyces cerevisiae. Genetics 159: 453–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddy M.N., Gaillard P.H., McDonald W.H., Shanahan P., Yates J.R. III, Russell P. (2001). Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell 107: 537–548. [DOI] [PubMed] [Google Scholar]
- Cadet J., Anselmino C., Douki T., Voituriez L. (1992). Photochemistry of nucleic acids in cells. J. Photochem. Photobiol. B 15: 277–298. [DOI] [PubMed] [Google Scholar]
- Chen X.B., Melchionna R., Denis C.M., Gaillard P.H., Blasina A., Van de Weyer I., Boddy M.N., Russell P., Vialard J., McGowan C.H. (2001). Human Mus81-associated endonuclease cleaves Holliday junctions in vitro. Mol. Cell 8: 1117–1127. [DOI] [PubMed] [Google Scholar]
- Chválová K., Brabec V., Kaspárková J. (2007). Mechanism of the formation of DNA-protein cross-links by antitumor cisplatin. Nucleic Acids Res. 35: 1812–1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A., McDonald N., West S.C. (2008). Structural and functional relationships of the XPF/MUS81 family of proteins. Annu. Rev. Biochem. 77: 259–287. [DOI] [PubMed] [Google Scholar]
- Dangel N.J., Knoll A., Puchta H. (2014). MHF1 plays Fanconi anaemia complementation group M protein (FANCM)-dependent and FANCM-independent roles in DNA repair and homologous recombination in plants. Plant J 78: 822–833. [DOI] [PubMed] [Google Scholar]
- Doe C.L., Ahn J.S., Dixon J., Whitby M.C. (2002). Mus81-Eme1 and Rqh1 involvement in processing stalled and collapsed replication forks. J. Biol. Chem. 277: 32753–32759. [DOI] [PubMed] [Google Scholar]
- Dorn A., Röhrig S., Papp K., Schröpfer S., Hartung F., Knoll A., Puchta H. (2018). The topoisomerase 3α zinc-finger domain T1 of Arabidopsis thaliana is required for targeting the enzyme activity to Holliday junction-like DNA repair intermediates. PLoS Genet. 14: e1007674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauser F., Schiml S., Puchta H. (2014). Both CRISPR/Cas-based nucleases and nickases can be used efficiently for genome engineering in Arabidopsis thaliana. Plant J 79: 348–359. [DOI] [PubMed] [Google Scholar]
- Geuting V., Kobbe D., Hartung F., Dürr J., Focke M., Puchta H. (2009). Two distinct MUS81-EME1 complexes from Arabidopsis process Holliday junctions. Plant Physiol. 150: 1062–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung F., Suer S., Bergmann T., Puchta H. (2006). The role of AtMUS81 in DNA repair and its genetic interaction with the helicase AtRecQ4A. Nucleic Acids Res. 34: 4438–4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung F., Suer S., Puchta H. (2007). Two closely related RecQ helicases have antagonistic roles in homologous recombination and DNA repair in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 104: 18836–18841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann N.J., Knoll A., Puchta H. (2015). The nuclease FAN1 is involved in DNA crosslink repair in Arabidopsis thaliana independently of the nuclease MUS81. Nucleic Acids Res. 43: 3653–3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Interthal H., Heyer W.D. (2000). MUS81 encodes a novel helix-hairpin-helix protein involved in the response to UV- and methylation-induced DNA damage in Saccharomyces cerevisiae. Mol Gen Genet 263: 812–827. [DOI] [PubMed] [Google Scholar]
- Interthal H., Pouliot J.J., Champoux J.J. (2001). The tyrosyl-DNA phosphodiesterase Tdp1 is a member of the phospholipase D superfamily. Proc. Natl. Acad. Sci. USA 98: 12009–12014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobbe S., Trapp O., Knoll A., Manuss A., Puchta H. (2015). The Translesion Polymerase ζ has roles dependent on and independent of the Nuclease MUS81 and the Helicase RECQ4A in DNA damage repair in Arabidopsis. Plant Physiol. 169: 2718–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.-Y., Kim H., Hwang H.-J., Jeong Y.-M., Na S.H., Woo J.-C., Kim S.-G. (2010). Identification of tyrosyl-DNA phosphodiesterase as a novel DNA damage repair enzyme in Arabidopsis. Plant Physiol. 154: 1460–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessel D., et al. (2014). Mutations in SPRTN cause early onset hepatocellular carcinoma, genomic instability and progeroid features. Nat. Genet. 46: 1239–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Pouliot J.J., Nash H.A. (2002). Repair of topoisomerase I covalent complexes in the absence of the tyrosyl-DNA phosphodiesterase Tdp1. Proc. Natl. Acad. Sci. USA 99: 14970–14975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobet G., Pagès L., Draye X. (2011). A novel image-analysis toolbox enabling quantitative analysis of root system architecture. Plant Physiol. 157: 29–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Mosqueda J., Maddi K., Prgomet S., Kalayil S., Marinovic-Terzic I., Terzic J., Dikic I. (2016). SPRTN is a mammalian DNA-binding metalloprotease that resolves DNA-protein crosslinks. eLife 5: e21491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannuss A., Dukowic-Schulze S., Suer S., Hartung F., Pacher M., Puchta H. (2010). RAD5A, RECQ4A, and MUS81 have specific functions in homologous recombination and define different pathways of DNA repair in Arabidopsis thaliana. Plant Cell 22: 3318–3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinier J., Ries G., Bonhoeffer S., Hohn B. (2004). Interchromatid and interhomolog recombination in Arabidopsis thaliana. Plant Cell 16: 342–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosbech A., et al. (2012). DVC1 (C1orf124) is a DNA damage-targeting p97 adaptor that promotes ubiquitin-dependent responses to replication blocks. Nat. Struct. Mol. Biol. 19: 1084–1092. [DOI] [PubMed] [Google Scholar]
- Mullen J.R., Chen C.-F., Brill S.J. (2010). Wss1 is a SUMO-dependent isopeptidase that interacts genetically with the Slx5-Slx8 SUMO-targeted ubiquitin ligase. Mol. Cell. Biol. 30: 3737–3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazato A., Kajita K., Ooka M., Akagawa R., Abe T., Takeda S., Branzei D., Hirota K. (2018). SPARTAN promotes genetic diversification of the immunoglobulin-variable gene locus in avian DT40 cells. DNA Repair (Amst.) 68: 50–57. [DOI] [PubMed] [Google Scholar]
- Olinski R., Wedrychowski A., Schmidt W.N., Briggs R.C., Hnilica L.S. (1987). In vivo DNA-protein cross-linking by cis- and trans-diamminedichloroplatinum(II). Cancer Res. 47: 201–205. [PubMed] [Google Scholar]
- Pommier Y., et al. (2006). Repair of topoisomerase I-mediated DNA damage. Prog. Nucleic Acid Res. Mol. Biol. 81: 179–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommier Y., Huang S.Y., Gao R., Das B.B., Murai J., Marchand C. (2014). Tyrosyl-DNA-phosphodiesterases (TDP1 and TDP2). DNA Repair (Amst.) 19: 114–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouliot J.J., Yao K.C., Robertson C.A., Nash H.A. (1999). Yeast gene for a Tyr-DNA phosphodiesterase that repairs topoisomerase I complexes. Science 286: 552–555. [DOI] [PubMed] [Google Scholar]
- Pouliot J.J., Robertson C.A., Nash H.A. (2001). Pathways for repair of topoisomerase I covalent complexes in Saccharomyces cerevisiae. Genes Cell 6: 677–687. [DOI] [PubMed] [Google Scholar]
- Puchta H. (2005). The repair of double-strand breaks in plants: Mechanisms and consequences for genome evolution. J. Exp. Bot. 56: 1–14. [DOI] [PubMed] [Google Scholar]
- Puchta H. (2017). Applying CRISPR/Cas for genome engineering in plants: The best is yet to come. Curr. Opin. Plant Biol. 36: 1–8. [DOI] [PubMed] [Google Scholar]
- Puchta H., Hohn B. (2012). In planta somatic homologous recombination assay revisited: A successful and versatile, but delicate tool. Plant Cell 24: 4324–4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recker J., Knoll A., Puchta H. (2014). The Arabidopsis thaliana homolog of the helicase RTEL1 plays multiple roles in preserving genome stability. Plant Cell 26: 4889–4902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regairaz M., Zhang Y.-W., Fu H., Agama K.K., Tata N., Agrawal S., Aladjem M.I., Pommier Y. (2011). Mus81-mediated DNA cleavage resolves replication forks stalled by topoisomerase I-DNA complexes. J. Cell Biol. 195: 739–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röhrig S., Dorn A., Enderle J., Schindele A., Herrmann N.J., Knoll A., Puchta H. (2018). The RecQ-like helicase HRQ1 is involved in DNA crosslink repair in Arabidopsis in a common pathway with the Fanconi anemia-associated nuclease FAN1 and the postreplicative repair ATPase RAD5A. New Phytol. 218: 1478–1490. [DOI] [PubMed] [Google Scholar]
- Ruijs M.W.G., van Andel R.N.J., Oshima J., Madan K., Nieuwint A.W.M., Aalfs C.M. (2003). Atypical progeroid syndrome: An unknown helicase gene defect? Am. J. Med. Genet. A. 116A: 295–299. [DOI] [PubMed] [Google Scholar]
- Steinert J., Schiml S., Fauser F. Puchta H. (2015). Highly efficient heritable plant genome engineering using Cas9 orthologues from Streptococcus thermophilus and Staphylococcus aureus. The Plant journal for cell and molecular biology 84: 1295–1305. [DOI] [PubMed] [Google Scholar]
- Stingele J., et al. (2016). Mechanism and regulation of DNA-protein crosslink repair by the DNA-dependent metalloprotease SPRTN. Mol. Cell 64: 688–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stingele J., Schwarz M.S., Bloemeke N., Wolf P.G., Jentsch S. (2014). A DNA-dependent protease involved in DNA-protein crosslink repair. Cell 158: 327–338. [DOI] [PubMed] [Google Scholar]
- Stingele J., Habermann B., Jentsch S. (2015). DNA-protein crosslink repair: Proteases as DNA repair enzymes. Trends Biochem. Sci. 40: 67–71. [DOI] [PubMed] [Google Scholar]
- Swenberg J.A., Lu K., Moeller B.C., Gao L., Upton P.B., Nakamura J., Starr T.B. (2011). Endogenous versus exogenous DNA adducts: Their role in carcinogenesis, epidemiology, and risk assessment. Toxicol Sci 120: S130–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takashima H., Boerkoel C.F., John J., Saifi G.M., Salih M.A.M., Armstrong D., Mao Y., Quiocho F.A., Roa B.B., Nakagawa M., Stockton D.W., Lupski J.R. (2002). Mutation of TDP1, encoding a topoisomerase I-dependent DNA damage repair enzyme, in spinocerebellar ataxia with axonal neuropathy. Nat. Genet. 32: 267–272. [DOI] [PubMed] [Google Scholar]
- Tretyakova N.Y., Groehler A. IV, Ji S. (2015). DNA-protein cross-links: formation, structural identities, and biological outcomes. Acc. Chem. Res. 48: 1631–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance J.R., Wilson T.E. (2002). Yeast Tdp1 and Rad1-Rad10 function as redundant pathways for repairing Top1 replicative damage. Proc. Natl. Acad. Sci. USA 99: 13669–13674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woźniak K., Walter Z. (2000). Induction of DNA-protein cross-links by platinum compounds. Z Naturforsch C 55: 731–736. [DOI] [PubMed] [Google Scholar]
- Yang S.W., Burgin A.B. Jr., Huizenga B.N., Robertson C.A., Yao K.C., Nash H.A. (1996). A eukaryotic enzyme that can disjoin dead-end covalent complexes between DNA and type I topoisomerases. Proc. Natl. Acad. Sci. USA 93: 11534–11539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwelling L.A., Anderson T., Kohn K.W. (1979). DNA-protein and DNA interstrand cross-linking by cis- and trans-platinum(II) diamminedichloride in L1210 mouse leukemia cells and relation to cytotoxicity. Cancer Res. 39: 365–369. [PubMed] [Google Scholar]