Recruitment of the chloroplast Min-system protein ARC3 to the organelle division site by PARC6 enables ARC3 to interact with and facilitate remodeling of the FtsZ ring during chloroplast division.
Abstract
Chloroplast division is initiated by assembly of the stromal Z ring, composed of cytoskeletal Filamenting temperature-sensitive Z (FtsZ) proteins. Midplastid Z-ring positioning is governed by the chloroplast Min (Minicell) system, which inhibits Z-ring assembly everywhere except the division site. The central Min-system player is the FtsZ-assembly inhibitor ACCUMULATION AND REPLICATION OF CHLOROPLASTS3 (ARC3). Here, we report Arabidopsis (Arabidopsis thaliana) chloroplasts contain two pools of ARC3: one distributed throughout the stroma, which presumably fully inhibits Z-ring assembly at nondivision sites, and the other localized to a midplastid ring-like structure. We show that ARC3 is recruited to the middle of the plastid by the inner envelope membrane protein PARALOG OF ARC6 (PARC6). ARC3 bears a C-terminal Membrane Occupation and Recognition Nexus (MORN) domain; previous yeast two-hybrid experiments with full-length and MORN-truncated ARC3 showed the MORN domain mediates ARC3-PARC6 interaction but prevents ARC3-FtsZ interaction. Using yeast three-hybrid experiments, we demonstrate that the MORN-dependent ARC3-PARC6 interaction enables full-length ARC3 to bind FtsZ. The resulting PARC6/ARC3/FtsZ complex enhances the dynamics of Z rings reconstituted in a heterologous system. Our findings lead to a model whereby activation of midplastid-localized ARC3 by PARC6 facilitates Z-ring remodeling during chloroplast division by promoting Z-ring dynamics and reveal a novel function for MORN domains in regulating protein–protein interactions.
INTRODUCTION
Chloroplasts originated through endosymbiosis, when a eukaryotic cell engulfed a free-living cyanobacterium roughly one billion years ago (Keeling, 2010; Zimorski et al., 2014). Analogous to their prokaryotic ancestor, chloroplasts in plant cells multiply by binary fission of preexisting organelles, which ensures their faithful inheritance during cell division and increases their populations during leaf cell expansion and plant growth. Chloroplast division is powered by a dynamic macromolecular machine positioned at the middle of the organelle. Ring-like contractile structures assembled on both sides of the surrounding membranes simultaneously pull from the inside and squeeze from the outside to generate the two new daughter chloroplasts. Proteins in the inner and outer envelope membranes coordinate the formation and constriction of these ring structures across the two membranes (reviewed in Miyagishima, 2011; Miyagishima et al., 2011; Osteryoung and Pyke, 2014; Chen et al., 2018a).
The foundation of the chloroplast division machinery and first contractile structure to assemble is the Filamenting temperature-sensitive Z (FtsZ) ring (Z ring), which forms on the stromal surface of the inner envelope membrane (IEM; Miyagishima et al., 2001). FtsZ is a tubulin-like, self-assembling cytoskeletal GTPase that functions in the division of bacteria (Bi and Lutkenhaus, 1991; Löwe and Amos, 1998; Erickson et al., 2010; Busiek and Margolin, 2015), chloroplasts (Osteryoung and Vierling, 1995; Osteryoung et al., 1998; Strepp et al., 1998; TerBush et al., 2013), and some mitochondria (Beech et al., 2000; Gilson et al., 2003; Miyagishima et al., 2004; Leger et al., 2015). In land plants and green algae, which comprise the green lineage, the chloroplast Z ring is composed of two types of FtsZ called FtsZ1 and FtsZ2 (Osteryoung et al., 1998; McAndrew et al., 2001; Vitha et al., 2001; Miyagishima et al., 2004; TerBush et al., 2013). These FtsZs, and all other organelle-localized chloroplast division proteins in land plants, are encoded in the nuclear genome and targeted to the chloroplast by N-terminal transit peptides that are cleaved upon import and thus are not part of the functional gene products. FtsZ1 and FtsZ2 are both highly conserved with their bacterial homologs, but only FtsZ2 retains a C-terminal peptide (CTP) conserved among bacterial FtsZs that interacts with membrane proteins to tether the Z ring to the membrane (Ma and Margolin, 1999; Osteryoung and McAndrew, 2001; TerBush et al., 2013). The FtsZ2 CTP binds to the transmembrane IEM protein ACCUMULATION AND REPLICATION OF CHLOROPLASTS6 (ARC6; Vitha et al., 2003; Maple et al., 2005; Schmitz et al., 2009), and this interaction is thought to be the primary basis for chloroplast Z-ring tethering (Johnson et al., 2013). Although FtsZ2 and FtsZ1 copolymerize as heteropolymers that presumably comprise the Z ring in vivo (McAndrew et al., 2001; Vitha et al., 2001; Olson et al., 2010; Yoshida et al., 2016), they have nonredundant functions in chloroplast division (Schmitz et al., 2009). FtsZ2 seems to impart structural stability to the Z ring, whereas FtsZ1 promotes the dynamic exchange of subunits in and out of the ring (turnover) (Yoder et al., 2007; TerBush and Osteryoung, 2012; Yoshida et al., 2016; TerBush et al., 2018). Subunit turnover is likely essential for full Z-ring and chloroplast constriction based on studies of bacterial Z-ring dynamics (Erickson et al., 2010; Osawa and Erickson, 2011; Busiek and Margolin, 2015).
In bacteria, FtsZ self-assembly and Z-ring positioning are controlled by the Min (Minicell) system, a negative regulatory system that inhibits Z-ring assembly everywhere but at the midcell division site (Rothfield et al., 2005; Lutkenhaus, 2007; Rowlett and Margolin, 2013; Monahan et al., 2014). The direct inhibitor of bacterial Z-ring assembly is MinC (Hu et al., 1999; Lutkenhaus, 2007; Arumugam et al., 2014; Park et al., 2018). In Escherichia coli and cyanobacteria, MinC exhibits a pole-to-pole oscillation driven by the regulators MinD and MinE (Hu and Lutkenhaus, 1999; Raskin and de Boer, 1999a, 1999b; Rothfield et al., 2005; MacCready et al., 2017). Oscillation results in a time-averaged concentration of MinC that is highest at the poles and lowest at the midcell, permitting Z-ring assembly only at the division site. Although homologs of MinD and MinE are retained throughout the green lineage, wherein they likewise function in regulating chloroplast Z-ring placement (Colletti et al., 2000; Itoh et al., 2001; Fujiwara et al., 2004, 2008; Aldridge and Møller, 2005), MinC has been lost in many green-lineage organisms. Instead, a host-derived stromal protein called ACCUMULATION AND REPLICATION OF CHLOROPLASTS3 (ARC3) has replaced MinC in the chloroplast Min system as the direct inhibitor of FtsZ assembly at nondivision sites (Shimada et al., 2004; Maple et al., 2007; Yang et al., 2008; TerBush and Osteryoung, 2012; Zhang et al., 2013; Osteryoung and Pyke, 2014; Shaik et al., 2018). However, the localization of ARC3 in the chloroplast has not yet been well established, including whether it oscillates like MinC, although previous data suggest it localizes partly to the midplastid where the Z ring forms (Shimada et al., 2004; Maple et al., 2007). The significance of this has been unclear.
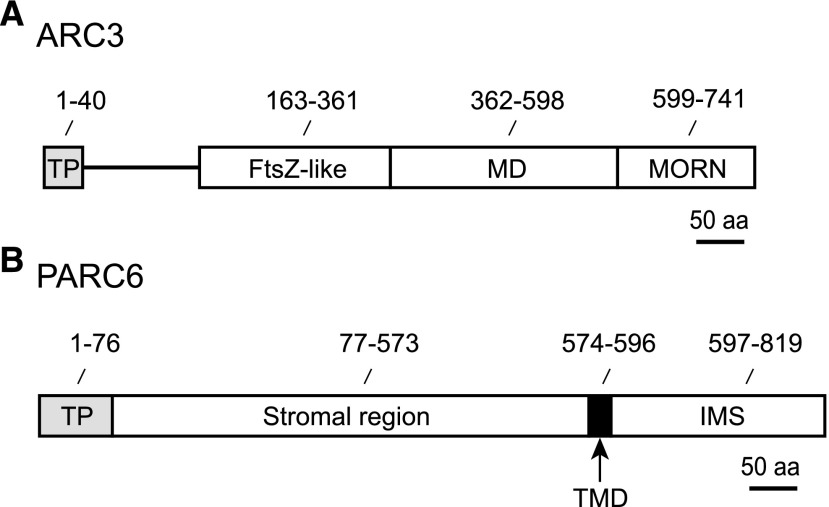
Arabidopsis (Arabidopsis thaliana) ARC3 harbors three distinct domains: an FtsZ-like domain, a middle domain, and a Membrane Occupation and Recognition Nexus (MORN) domain (Figure 1A; Shimada et al., 2004; Maple et al., 2007). The FtsZ-like and possibly middle domains mediate interactions with FtsZ1 and FtsZ2, but yeast two-hybrid (Y2H) experiments with full-length and MORN-truncated forms of ARC3 have shown that the presence of the MORN domain prevents FtsZ interaction with ARC3 (Maple et al., 2007; Zhang et al., 2013). Consistent with these interaction data, only the truncated version of ARC3 lacking the MORN domain inhibited assembly of FtsZ2 filaments in vitro; full-length ARC3 did not (Shaik et al., 2018). However, overexpression of either MORN-truncated or full-length ARC3 inhibited Z-ring assembly in Arabidopsis (Zhang et al., 2013), suggesting the existence of a mechanism that enables FtsZ to bind to full-length ARC3 in vivo despite the presence of the MORN domain.
Figure 1.
Structural Features of ARC3 and PARC6.
(A) and (B) The sequences depicted are from Arabidopsis: ARC3 (A) and PARC6 (B). aa, amino acids; MD, middle domain; TMD, transmembrane domain; TP, predicted chloroplast transit peptide. In plants, the TP is cleaved upon protein import to the chloroplast and is not part of the mature protein.
While the MORN domain prevents FtsZ-ARC3 interaction in Y2H and other assays, it is required for interaction of ARC3 with another chloroplast division protein, PARALOG OF ARC6 (PARC6, also called Chloroplast Division Site Positioning1 and Homolog of ARC6; Glynn et al., 2009; Zhang et al., 2009; Ottesen et al., 2010). Like ARC6, PARC6 is an IEM protein with a single transmembrane helix (Figure 1B) that localizes at least partly to the division site (Glynn et al., 2009). As in ARC6, the stromal region of PARC6 interacts specifically with FtsZ2 via the FtsZ2 CTP (Zhang et al., 2016). However, whereas ARC6 is a positive regulator of FtsZ assembly (Vitha et al., 2003; Sung et al., 2018), mutant and overexpression studies in Arabidopsis demonstrated that PARC6 is a negative regulator in vivo (Glynn et al., 2009). But PARC6 did not inhibit assembly of FtsZ2 filaments in a heterologous system despite direct FtsZ2-PARC6 interaction, leading us to hypothesize that the negative regulation of FtsZ assembly by PARC6 in vivo may be a consequence of its interaction with ARC3 (Zhang et al., 2016). We further postulated that MORN domain–dependent ARC3-PARC6 interaction in vivo might sequester the MORN domain and thereby promote ARC3-FtsZ interaction, allowing ARC3 to exert an inhibitory effect on Z-ring assembly in chloroplasts (Glynn et al., 2009; Zhang et al., 2016).
Here, we investigate the subcellular localization of ARC3 in Arabidopsis, assess the role of PARC6 in the regulation of ARC3 localization, and test the hypothesis that PARC6 activates the inhibitory activity of ARC3 on FtsZ assembly through binding to the MORN domain. Our findings demonstrate the presence of two pools of ARC3 within chloroplasts, one disbursed in the stroma and one concentrated at the midplastid, and we show that ARC3 is recruited to the midplastid by PARC6. Our results also provide evidence that ARC3 activation by PARC6 prevents ectopic Z-ring assembly in the vicinity of the division site and enhances the subunit exchange from Z rings reconstituted in a heterologous system. Our findings lead to a model in which ARC3-PARC6 interaction at the midplastid division site, rather than fully inhibiting Z-ring assembly, facilitates Z-ring remodeling during chloroplast division by promoting Z-ring dynamics. Our results also reveal a novel function for a MORN domain in the regulation of protein–protein interactions.
RESULTS
Localization of ARC3
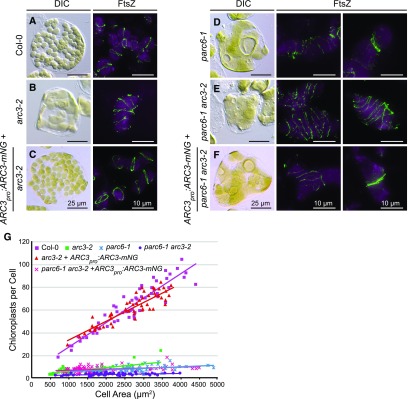
To investigate the localization of ARC3 in Arabidopsis, we expressed an ARC3 fluorescent fusion protein in the Arabidopsis arc3-2 mutant. This mutant has a heterogeneous population of enlarged, irregularly shaped chloroplasts with multiple misplaced Z rings and constrictions (Figure 2B; Shimada et al., 2004; Maple et al., 2007; Zhang et al., 2013). The fluorescent protein mNeonGreen (mNG) was fused to the C terminus of ARC3, generating ARC3-mNG, whose expression was driven by the native ARC3 promoter (ARC3pro:ARC3-mNG). mNG was chosen to increase the brightness and photostability of the fusion protein when expressed in vivo (Shaner et al., 2013). The functionality of ARC3-mNG was validated by its ability to complement both the chloroplast morphology defect and the formation of multiple Z rings in arc3-2 (Figures 2A to 2C and 2G).
Figure 2.
ARC3pro:ARC3-mNG Fusion Construct Is Functional In Vivo.
(A) to (G) Test of the functionality of the ARC3pro:ARC3-mNG fusion construct. (A) to (F) Chloroplast morphology (left panels) and FtsZ localization (right panels) were visualized using differential interference contrast microscopy and immunofluorescence staining of FtsZ2-1 (FtsZ), respectively, in mesophyll cells of (A) the wild-type Col-0, (B) arc3-2, (C) arc3-2 expressing ARC3pro:ARC3-mNG, (D) parc6-1, (E) parc6-1 arc3-2, and (F) parc6-1 arc3-2 expressing ARC3pro:ARC3-mNG. The middle and right panels show merged images of immunostained FtsZ2-1 (green) and chlorophyll autofluorescence (magenta). Bars = 25 μm for differential interference contrast images and 10 μm for immunofluorescence images. Samples analyzed were from young emerging leaves (∼3 to 5 mm long) of T1 or T2 transgenic seedlings. (G) Quantitative analysis of chloroplast number versus cell size in the nontransgenic and transgenic lines shown in (A) to (F). Slopes of the best-fit lines are as follows: Col-0, 0.021 (R2 = 0.88); arc3-2, 0.003 (R2 = 0.48); arc3-2 + ARC3pro:ARC3-mNG, 0.017 (R2 = 0.82); parc6-1, 0.001 (R2 = 0.11); parc6-1 arc3-2, 0.002 (R2 = 0.13); parc6-1 arc3-2 + ARC3pro:ARC3-mNG, 0.001 (R2 = 0.17). DIC, differential interference contrast.
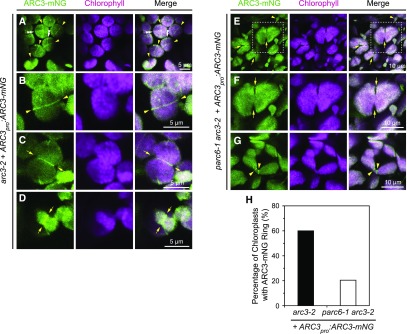
To observe the spatial distribution of ARC3 in vivo, confocal laser scanning microscopy was used to acquire images from young leaves of transgenic seedlings. Expression of the ARC3-mNG fusion protein in transgenic plants was confirmed by detection of the mNG fluorescence signal in nongreen plastids in roots and petals, where potential interference from chlorophyll autofluorescence is excluded (Supplemental Figures 1C, 1I, and 1J). In mesophyll cells, the ARC3-mNG signals were associated exclusively with chloroplasts and exhibited two distinct localization patterns: one distributed diffusely across the chloroplast stroma, and the other concentrated in a ring at the middle of the chloroplast (Figures 3A to 3C; Supplemental Figures 1E to 1H). At the midplastid, ARC3-mNG was associated with both unconstricted chloroplasts (Figures 3A and 3B, arrowheads; Supplemental Figures 1F and 1G, arrowheads) and constricted chloroplasts (Figures 3C and 3D, arrows; Supplemental Figure 1H, arrows). Occasionally, short fragments and patches of ARC3-mNG and additional ARC3-mNG strands not associated with the midplastid were also observed (Figures 3A to 3C; Supplemental Figures 1E and 1G). We also detected ARC3-mNG in tissues other than leaf, including root tips and flower buds (sepals, petals, stamens, and pistils; Supplemental Figures 1C, 1I, and 1J). We note that a fraction of the mNG fluorescence signal detected in leaf samples of transgenic plants arose from bleed-through of chlorophyll autofluorescence into the mNG channel, as indicated by detection of fluorescence in the mNG channel in chloroplasts of nontransformed plants (Supplemental Figure 1A). However, the diffuse signal was much brighter in plants expressing the ARC3pro:ARC3-mNG transgene, and no fluorescence in rings or strands was detected in nontransformed plants. ARC3-mNG was also present in the smaller chloroplasts of pavement cells (Figure 3D), as well as in nongreen plastids in roots and petals, although ARC3-mNG ring structures were less obvious in the latter two organs (Supplemental Figures 1C, 1I, and 1J).
Figure 3.
ARC3-mNG Localization in Chloroplasts.
(A) to (D) The mNG fluorescence (ARC3-mNG, green) and chlorophyll autofluorescence (chlorophyll, magenta) signals were detected by confocal laser scanning microscopy. Merged images are shown. Bars are as indicated. Localization of ARC3-mNG in T2 transgenic arc3-2 expressing ARC3pro:ARC3-mNG (arc3-2 + ARC3pro:ARC3-mNG). Midplastid-localized ARC3-mNG ring structures are associated with both (A) and (B) unconstricted chloroplasts (yellow arrowheads) and (C) and (D) constricted chloroplasts (yellow arrows). White double-arrowheads in (A) denote additional ARC3-mNG strands that are not associated with the division site.
(E) to (G) Localization of ARC3-mNG in T1 transgenic parc6-1 arc3-2 expressing ARC3pro:ARC3-mNG (parc6-1 arc3-2 + ARC3pro:ARC3-mNG). Arrows in (E) and (F) point out constriction sites. Arrowheads in (E) and (G) denote ARC3-mNG ring structures. The white boxed regions in (E) are magnified in (F) for better visualization.
(H) Quantification of ARC3-mNG ring structures in chloroplasts of transgenic arc3-2 and parc6-1 arc3-2 plants expressing ARC3pro:ARC3-mNG. The percentage of chloroplasts with ARC3-mNG rings was 60.3% (n = 156) in arc3-2 and 21.5% (n = 148) in parc6-1 arc3-2. n is the total number of chloroplasts observed in 8 images (arc3-2) or 13 images (parc6-1 arc3-2) obtained in each case from two independent transgenic lines.
In E. coli and the cyanobacterium Synechococcus elongatus, MinC and MinD oscillate from pole to pole with periodicities of ∼30 and 60 s, respectively (Hu and Lutkenhaus, 1999; Raskin and de Boer, 1999a, 1999b; MacCready et al., 2017), but oscillation does not occur in Bacillus subtilis (Lutkenhaus, 2007; Gregory et al., 2008; Rowlett and Margolin, 2013), indicating multiple mechanisms for the control of Z-ring placement in bacteria. In an effort to assess whether the chloroplast Min system oscillates, we monitored ARC3-mNG distribution in young leaves of transgenic arc3-2 mutants complemented by ARC3pro:ARC3-mNG using time-lapse imaging every 40 s over 7 to 8 min (Supplemental Figure 2). No clear change in distribution of the fluorescence signal was observed that would suggest oscillatory behavior. Thus, to date there is no evidence for Min-system oscillation in chloroplasts.
PARC6 Recruits ARC3 to the Midplastid Division Site
Previous studies demonstrated that ARC3 interacts with PARC6, which localizes partly to the midplastid division site (Glynn et al., 2009; Zhang et al., 2016). Therefore, we hypothesized that PARC6 plays a role in recruiting the midplastid pool of ARC3. Toward testing this, we first generated a parc6-1 arc3-2 double mutant. As reported previously (Glynn et al., 2009; Zhang et al., 2009), chloroplasts in the parc6-1 single mutant were enlarged and variable in size, resembling those in arc3-2, and parc6-1 exhibited multiple Z rings and spirals (Figure 2D). In addition, Z rings in parc6‐1 were often clustered near chloroplast constrictions (Figure 2D, right panel; Supplemental Figure 3, right panels), indicting ectopic Z-ring assembly near the division site in the absence of PARC6. In the parc6-1 arc3-2 double mutant, the division defect was more pronounced, as indicated by the presence of fewer and larger chloroplasts and visibly more FtsZ filaments and rings, and obvious constrictions were rarely observed (Figures 2E and 2G). Furthermore, clustered Z rings were never observed in the double mutant (Figure 2E, middle and right panels).
To gain insight into the localization of ARC3 in the absence of PARC6, we transformed ARC3pro:ARC3-mNG into parc6-1 arc3-2. Transgenic plants showed both heterogeneous chloroplast morphologies and multiple clustered Z rings, as well as dispersed rings (Figure 2F), similar to those in parc6-1, indicating that the ARC3pro:ARC3-mNG transgene effectively compensated for the loss of ARC3 function in the double mutant background. Expression of the ARC3-mNG fusion protein was verified by detection of the mNG fluorescence signal in roots of T1 seedlings (Supplemental Figure 1D). As in complemented arc3-2 mutants, ARC3-mNG localized diffusely throughout the chloroplasts in transformed parc6-1 arc3-2 mutants (Figures 3E to 3G). However, unlike in arc3-2 wherein ARC3-mNG was also frequently localized to ring-like structures (60.3%, n = 156; Figure 3H), ARC3-mNG rings were far less common in the double mutant (21.5%, n = 148; Figures 3G, arrowheads; and 3H), even though chloroplast constrictions were still evident (Figure 3E and 3F, arrows). These results provide evidence that PARC6 plays a key role in recruiting ARC3 to the division site. However, they also hint at the possible involvement of an additional unidentified factor in the midplastid localization of ARC3. Given that ARC3 is an inhibitor of FtsZ assembly, the genetic data also suggest that the presence of ectopic Z rings near constrictions in parc6-1 may not be due directly to the loss of PARC6 but rather to the reduction or absence of ARC3 at these sites in the parc6-1 mutant.
Conserved Residues within the MORN Domain of ARC3 Are Required for ARC3-PARC6 Interaction but Interfere with ARC3-FtsZ Interaction in Y2H Assays
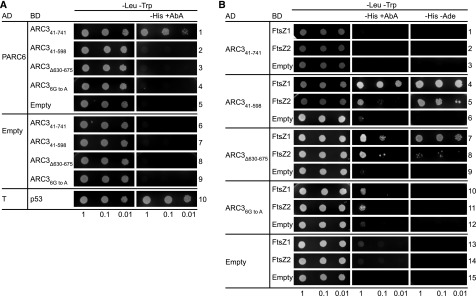
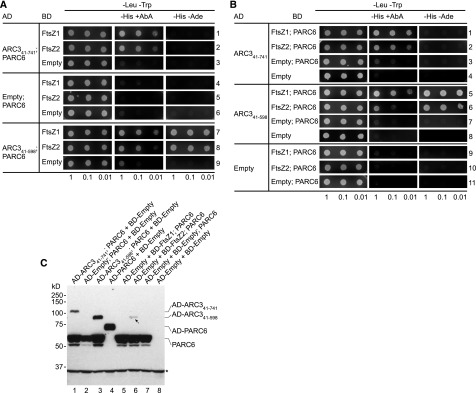
The ARC3 MORN domain bears three MORN motifs: MORN1 (amino acids 612 to 628), MORN2 (amino acids 630 to 652), and MORN3 (amino acids 653 to 675; Supplemental Figure 4). MORN motifs are short repeats found in a wide range of proteins, but their functions are poorly understood (Takeshima et al., 2000; Mueller-Roeber and Pical, 2002; Ma et al., 2006; Habicht et al., 2015). Toward further defining the requirements for ARC3‐PARC6 interaction, we used multiple sequence alignments to identify conserved amino acids within the ARC3 MORN motifs and found six Gly residues within MORN2 and MORN3 whose distribution pattern is conserved in MORN motifs in plants and green algae (G632, G641, G643, G655, G664, and G666; Supplemental Figure 4, asterisks) as well as in other eukaryotes (Takeshima et al., 2000; Mueller-Roeber and Pical, 2002; Ma et al., 2006; Habicht et al., 2015). We generated ARC3 derivatives in which these two MORN motifs were deleted (ARC3Δ630-675) or in which the six conserved Gly residues were mutated to Ala (ARC36G to A) and used Y2H assays to test their ability to interact with the stromal region of PARC6, PARC677-573 (Figure 1B). In control assays and as previously reported (Zhang et al., 2016), PARC6 interacted with the full-length form of ARC3, ARC341-741 (lacking the predicted chloroplast transit peptide; Figure 1A), but did not interact with a truncated form, ARC341-598, that completely lacks the MORN domain (Figure 4A, rows 1 and 2; Supplemental Figure 5, rows 6 and 7). Similar to the latter results, neither ARC3Δ630-675 nor ARC36G to A could interact with PARC6 (Figure 4A, rows 3 and 4), suggesting the two MORN motifs and the conserved Gly residues within them are required for this interaction.
Figure 4.
Conserved Gly Residues Within the MORN Domain of ARC3 Are Required for Its Interaction With PARC6.
(A) and (B) Y2H assays of (A) PARC677-573 (PARC6) with ARC3 derivatives and (B) ARC3 derivatives with FtsZ proteins. Constructs were expressed in Y2HGold cells from the pGAD-T7 vector (AD) or pGBK-T7 vector (BD), and transformants were selected on medium lacking Leu and Trp (−Leu −Trp). Interactions were assayed based on activation of the HIS3 and AUR1-C reporter genes, as indicated by growth on medium lacking His (−His) supplemented with the toxic drug AbA (+AbA) at (A) 20 ng mL−1 or (B) 40 ng mL−1. Activation of the AUR1-C reporter by a positive interaction confers resistance of the Y2HGold strain to AbA (Zhang et al., 2016). In (B), an additional assay was used based on activation of the ADE2 reporter gene by growth in the absence of Ade (−Ade). Representative images in (A) show transformed yeast cells grown on selective medium for 3 d after streaking out the diluted cultures. Representative images in (B) show cells grown for 4 d on −Leu −Trp and −Leu −Trp −His +AbA medium, and 5 d on −Leu −Trp −His −Ade medium, except cells in rows 7 to 12 on −Leu −Trp −His +AbA medium, which were grown for 5 d due to weak interactions in these strains. Empty vectors (Empty) were used as negative controls. Interaction between AD-Simian Virus 40 large T-antigen (T) and BD-p53 (p53) (row 10 in [A]) was used as a positive control as suggested by the manufacturer. Dilutions from the same starting culture are indicated at the bottom. Assays were replicated three times with similar results.
While the MORN domain is required for ARC3-PARC6 interaction, it inhibits interaction of ARC3 with the FtsZ proteins in Y2H assays, as shown by experiments demonstrating that ARC341-598, but not ARC341-741, interacts with FtsZ1 and FtsZ2 (Figure 4B, rows 1 to 6; Maple et al., 2007; Zhang et al., 2013). Both ARC3Δ630-675 and ARC36G to A were also able to interact with FtsZ1 and FtsZ2 (Figure 4B, rows 7 to 12; Supplemental Figure 6). However, the two mutants may bind FtsZ with lower affinity than ARC341‐598, as suggested by reduced growth of transformed yeast cells on selective medium (Figure 4B, rows 7 to 12). This was particularly evident for ARC36G to A; strains with this mutant grew less well on medium containing the toxin aureobasidin A (AbA; see “Methods”; Figure 4, legend; Figure 4B, rows 10 to 12, middle) than strains with ARC341-598 or ARC3Δ630-675 (Figure 4B, rows 4 to 9). However, the ability of ARC36G to A to interact with the FtsZs was clearly demonstrated by growth of transformed cells on less stringent medium lacking AbA (Supplemental Figure 6). Thus, mutation of conserved residues in MORN2 and MORN3 or deletion of these MORN motifs not only abolishes the ability of ARC3 to interact with PARC6 but also enables ARC3 to interact with FtsZ.
Binding of PARC6 to the MORN Domain Enables Full-Length ARC3 to Interact with FtsZ Proteins in Y3H Assays
We hypothesized that PARC6 may promote interaction of full-length ARC3 with FtsZ by binding to the MORN domain. Toward testing this hypothesis, we implemented a yeast three-hybrid (Y3H) strategy in Schizosaccharomyces cerevisiae wherein a gene product fused to either the GAL4 activation domain (AD) or binding domain (BD) was expressed from the same vector as the stromal region of PARC6 that was not fused to either the AD or BD (Supplemental Figure 7). Activation of the reporter genes permitting growth of yeast on selective medium should result only from direct interaction between the AD and BD fusion proteins expressed from the cotransformed vectors, and not from direct interaction between PARC6 and any AD or BD fusion protein. This was verified in a set of control assays. For example, when the vector encoding both AD-ARC341-598 and PARC6 (pAD-ARC341-598; PARC6; Supplemental Figure 7E) was cotransformed into yeast with a vector encoding BD-FtsZ2 (pBD-FtsZ2), growth was observed on selective medium (Figure 5A, row 8), consistent with previous Y2H and other data showing interaction between ARC341-598 and FtsZ2 (Figure 4B, row 5; Zhang et al., 2013; Shaik et al., 2018). Although AD-PARC6 (Supplemental Figure 7B) also interacts with BD-FtsZ2 in standard Y2H assays (Supplemental Figure 5, row 9; Zhang et al., 2016), in the Y3H system growth of the strain expressing AD-ARC341-598, PARC6, and BD-FtsZ2 (Figure 5A, row 8) was not due to PARC6 interaction with BD-FtsZ2 (although this interactions presumably occurs in these cells) because growth was not observed in a control strain cotransformed with a vector expressing only PARC6 (pAD-Empty; PARC6; Supplemental Figure 7C) and pBD-FtsZ2 (Figure 5A, row 5). As all PARC6 and ARC3 constructs bore hemagglutinin (HA) tags (Supplemental Figure 7), expression of PARC6 and AD-ARC341-598 from the pAD-Empty; PARC6 and pAD-ARC341-598; PARC6 vectors was verified by immunoblotting with an anti–HA antibody (Figure 5C, lanes 2 and 3).
Figure 5.
PARC6 Promotes Interaction Between Full-Length ARC3 and FtsZ.
(A) and (B) Y3H assays to test for interactions between the indicated ARC3 derivatives and the FtsZ proteins in the presence and absence of PARC677-573 (PARC6). Vectors and assays are as described in Figure 4 except that in vectors expressing PARC677-573, PARC677-573 was not fused to the GAL4 activation domain or binding domain. The PARC677-573 expression cassette was inserted into (A) the pGAD-T7 vectors (AD) expressing the AD-ARC341-741, AD-Empty, and AD-ARC341-598 constructs or (B) the pGBK-T7 vectors (BD) expressing the BD-FtsZ1, BD-FtsZ2, and BD-Empty constructs (Supplemental Figure 7). In medium containing AbA (+AbA), AbA was used at 40 ng mL−1 in rows 5 to 8 in (B) and at 20 ng mL−1 in all other assays. Cells on −Leu −Trp and −Leu −Trp −His −Ade medium were grown for 3 and 5 d, respectively. Cells on −Leu −Trp −His +AbA medium in rows 7 to 9 in (A) and in rows 5 to 8 in (B) were grown for 2 d due to leakiness of the HIS3 reporter; all other cells on these media were grown for 3 d in (A) and for 4 d in (B). Dilutions from the same starting culture are indicated at the bottom. Assays were replicated three times with similar results.
(C) Immunoblot assays of yeast cells transformed with the indicated constructs. Total proteins were separated on 10% SDS-PAGE gels, and membranes were probed with an anti–HA antibody, as all PARC6 and ARC3 constructs bore HA tags. The predicted masses of the proteins detected were as follows: AD-ARC341-741, 96 kD; PARC677-573, 60.4 kD; AD-ARC341-598, 80.8 kD; and AD-PARC677-573, 72.7 kD. AD-ARC341-741 ran slightly larger than predicted. Arrow indicates a potential complex formed by PARC677-573 and FtsZ2. Asterisk denotes a nonspecific band recognized by the anti–HA antibody in all samples. Bands just beneath PARC6 may represent a degradation product of PARC677-573 when it is not fused to the GAL4 AD. Lanes 1 to 3 correspond to rows 3, 6, and 9 in (A); lane 4 corresponds to row 10 in Supplemental Figure 5; lanes 5 to 7 correspond to rows 9 to 11 in (B); and lane 8 corresponds to row 15 in Figure 4B.
We used our Y3H system to test whether PARC6 could promote interaction between full-length ARC3 bearing the MORN domain (ARC341-741) and FtsZ. In control Y2H assays, growth was not observed in cells cotransformed with pAD-ARC341-741 and either pBD-FtsZ1 or pBD-FtsZ2 (Figure 4B, rows 1 and 2), as expected since the MORN domain inhibits ARC3-FtsZ interaction (Maple et al., 2007; Zhang et al., 2013; Shaik et al., 2018). By contrast, growth was observed in cells cotransformed with pAD-ARC341-741; PARC6 and either pBD-FtsZ1 or pBD-FtsZ2 (Figure 5A, rows 1 and 2, middle). PARC6 coexpressed with BD-FtsZ2 did not activate the reporter genes (Figure 5A, row 5) since PARC6 was not fused to the AD, whereas AD-PARC6 did (Supplemental Figure 5, row 9; Zhang et al., 2016). Immunoblotting confirmed expression of PARC6, AD-PARC6, and AD-ARC341-741 from the relevant vectors (Figure 5C, lanes 1 and 4). Together, these results demonstrate that interaction between AD-ARC341-741 and BD-FtsZ2 to activate the reporter genes only occurs in the presence of PARC6, suggesting the formation of an AD-ARC341-741/PARC6/BD-FtsZ2 complex that promotes interaction of full-length ARC3 with FtsZ2. Because the MORN domain also prevents ARC3-FtsZ1 interaction (Figure 4B, rows 1 and 4; Maple et al., 2007; Zhang et al., 2013), reporter-gene activation in cells expressing AD-ARC341-741, PARC6, and BD-FtsZ1 (Figure 5A, row 1, middle panel) suggests formation of a similar AD-ARC341-741/PARC6/BD-FtsZ1 complex.
To further verify the Y3H results, we also generated constructs in which PARC6 was instead coexpressed from the same vector with either BD-FtsZ1 (pBD-FtsZ1; PARC6) or BD-FtsZ2 (pBD-FtsZ2; PARC6; Figure 5C, lanes 5 and 6; Supplemental Figures 7G and 7H) and cotransformed each vector with pAD-ARC341-598 as a positive interaction control (Figure 5B, rows 5 and 6) or with pAD-ARC341-741. Similar to the Y3H results above, interaction between AD-ARC341-741 and BD-FtsZ1 or BD-FtsZ2 only occurred in the presence of PARC6 (Figure 4B rows 1 and 2; Figure 5B, rows 1 and 2), further suggesting the formation of ARC341-741/PARC6/FtsZ complexes in yeast. As a negative control, AD-ARC341-741 coexpressed with PARC6 did not activate the reporter genes (Figure 5B, row 3) since PARC6 was not fused to the BD.
Together, the Y3H findings support our hypothesis that PARC6-ARC3 interaction sequesters the MORN domain and/or alters the conformation of ARC3, enabling full-length ARC3 to interact with FtsZ. They also provide evidence that ARC3 is capable of binding PARC6 and FtsZ proteins simultaneously in a complex.
Activation of ARC3 by PARC6 Enables Full-Length ARC3 to Inhibit the Assembly of FtsZ Filaments in a Heterologous Yeast System
The Y3H results prompted us to ask whether ARC3-PARC6 interaction has any effect on ARC3-mediated inhibition of FtsZ assembly. Toward addressing this question, we used a heterologous system, the fission yeast (Schizosaccharomyces pombe). Because yeasts lack endogenous FtsZs and their many assembly regulators, functional analysis of FtsZ assembly and dynamics, as well as the effects of individual assembly regulators, can be performed in isolation from other division components in this heterologous system, yet in a cellular milieu (Srinivasan et al., 2007, 2008; TerBush and Osteryoung, 2012; Zhang et al., 2013; TerBush et al., 2016, 2018). Fluorescently tagged FtsZ1 and FtsZ2 each assemble filaments in S. pombe, and ARC3 lacking the MORN domain inhibits their assembly in a dose-dependent manner in this system (TerBush and Osteryoung, 2012; Zhang et al., 2013). Because PARC6 interacts directly only with FtsZ2 and colocalizes with FtsZ2 filaments in S. pombe (Zhang et al., 2016), in the current experiments we used FtsZ1, which does not colocalize with or influence the assembly of FtsZ1 filaments when the two proteins are expressed together in this system (Zhang et al., 2016).
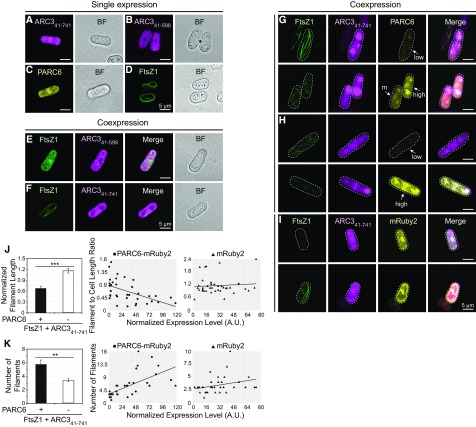
FtsZ1, ARC3 with or without the MORN domain, and the stromal region of PARC6 were fused at their C termini to mVenus, mCerulean, or mRuby2, respectively, creating the constructs FtsZ1-mVenus, ARC341-741-mCerulean, ARC341-598-mCerulean, and PARC6-mRuby2. When expressed alone in S. pombe, the ARC3 and PARC6 constructs were all distributed throughout the transformed yeast cells (Figures 6A to 6C). In the case of ARC341-741-mCerulean, intense bright spots were observed in some cells (Supplemental Figure 8A, arrows), suggesting this phenomenon may be due to the presence of the MORN domain in full-length ARC3. As shown previously (TerBush et al., 2016, 2018), in control experiments FtsZ1-mVenus assembled filaments when expressed alone (Figure 6D), whereas filament formation was reduced when FtsZ1-mVenus was coexpressed with ARC341-598-mCerulean (Figure 6E), as indicated by the absence of long FtsZ filaments and presence of much shorter filaments and diffuse FtsZ1-mVenus distribution. By contrast, ARC341-741-mCerulean did not inhibit FtsZ1 filament formation (Figure 6F), consistent with the Y2H data showing that only ARC341-598 interacts with FtsZ (Maple et al., 2007; Zhang et al., 2013) (Figure 4B, rows 1 to 6).
Figure 6.
Activation of Full-Length ARC3 by PARC6 Inhibits Assembly of FtsZ1 Filaments in S. pombe.
Epifluorescence and bright-field micrographs of transformed S. pombe cells expressing the indicated proteins are shown. In epifluorescence images, mVenus, mCerulean (mCer), and mRuby2 signals are falsely colored green, magenta, and yellow, respectively. Bars = 5 μm. BF, bright-field micrograph.
(A) to (D) Single-expression strains: (A) ARC341-741-mCerulean, (B) ARC341-598-mCerulean, (C) PARC677-573-mRuby2, and (D) FtsZ1-mVenus.
(E) and (F) Effect of (E) ARC341-598-mCerulean and (F) ARC341-741-mCerulean on assembly of FtsZ1-mVenus filaments in coexpression strains.
(G) to (I) Effect of ARC341-741-mCerulean on assembly of FtsZ1-mVenus filaments in strains also coexpressing (G) and (H) PARC677-573-mRuby2 or (I) mRuby2. White arrows in (G) and (H) designate cells expressing PARC677-573-mRuby2 at high, moderate, or low levels as indicated by the mRuby2 signal intensity. Outlines of the imaged yeast cells are indicated by dashed lines. Additional details are described in the text. M, moderate.
(J) and (K) Quantitative analysis of the effect of ARC341-741-mCerulean on the assembly of FtsZ1-mVenus filaments in the presence of PARC677-573-mRuby2 (n = 36 cells) or mRuby2 (n = 37 cells). The effect on assembly was evaluated in individual cells based on (J) the length of FtsZ1-mVenus filaments and (K) the number of FtsZ1-mVenus filaments. In (J), filament length was normalized to cell length because cell length constrains filament length (TerBush and Osteryoung, 2012). Left panels show means and error bars are sem. ***P < 0.0001; **P < 0.001 as determined by the t test. In the middle and right panels, the x axis indicates the level of PARC677-573-mRuby2 or mRuby2 expression as indicated by the total fluorescence intensity normalized to the cell area. Slopes of the best-fit lines are as follows: (J) PARC677-573-mRuby2, −0.006 (R2 = 0.275); mRuby2, 0.002 (R2 = 0.006); (K) PARC677-573-mRuby2, 0.081 (R2 = 0.406); mRuby2, 0.026 (R2 = 0.089). A.U., arbitrary units.
To test whether full-length ARC3 could inhibit FtsZ1 assembly in the presence of PARC6, we generated a construct in which PARC6-mRuby2 and ARC341-741-mCerulean were expressed from the same vector and cotransformed it into S. pombe with the vector expressing FtsZ1-mVenus (Figures 6G and 6H). As described previously, expression of fusion proteins in S. pombe is variable within a single culture as indicated by differences in fluorescence intensity in different cells, facilitating analysis of the dose dependency of coexpressed proteins on FtsZ assembly (TerBush and Osteryoung, 2012; TerBush et al., 2016). In cells with readily detectable levels of FtsZ1-mVenus and ARC341-741-mCerulean but low levels of PARC6-mRuby2, long FtsZ1 filaments were observed that often extended the length of the cell or wrapped around the cell (Figures 6G and 6H, top), similar to filaments in cells expressing only FtsZ1-mVenus and ARC341-741-mCerulean (Figure 6F). By contrast, shorter filaments were observed in cells with higher levels of PARC6-mRuby2 (Figures 6G and 6H, bottom; Supplemental Figures 8G to 8I). The observed effect was attributable to PARC6 because mRuby2 alone did not have a similar effect (Figure 6I). Quantitative analysis further showed that filament length was significantly reduced and the number of filaments significantly increased in the presence of PARC6-mRuby2 compared with mRuby2 (Figures 6J and 6K, left). Moreover, filament length was negatively correlated, and filament number positively correlated, with the level of PARC6-mRuby2 expression, but not with mRuby2 expression (Figures 6J and 6K, middle and right panels), indicating a dose-dependent effect of PARC6 on ARC341-741-mediated inhibition of FtsZ1 assembly. Inhibition of FtsZ assembly by ARC341-741-mCerulean and PARC6-mRuby2 (Figures 6G and 6H, bottom; Supplemental Figures 8G to 8I) was not caused by reduced levels of FtsZ1-mVenus in these cells because long filaments were also observed in cells expressing FtsZ1-mVenus at very low levels (Figure 6I, top left; Supplemental Figure 8E; TerBush and Osteryoung, 2012). However, in cells expressing very high levels of FtsZ1-mVenus, many long filaments were observed even in the presence of PARC6-mRuby2 and ARC341-741-mCerulean (Supplemental Figure 8J), suggesting that the ratios between FtsZ, ARC3, and PARC6 are important in determining the extent to which FtsZ assembly is inhibited by ARC3 and PARC6.
Together, the Y3H results and FtsZ1 assembly experiments in S. pombe provide evidence that PARC6 not only promotes FtsZ interaction with ARC3 by binding to the MORN domain but also activates the inhibitory activity of ARC3 on FtsZ assembly.
ARC3-PARC6 Interaction Enhances the Dynamics of Z Rings Reconstituted in the Heterologous Yeast Pichia pastoris
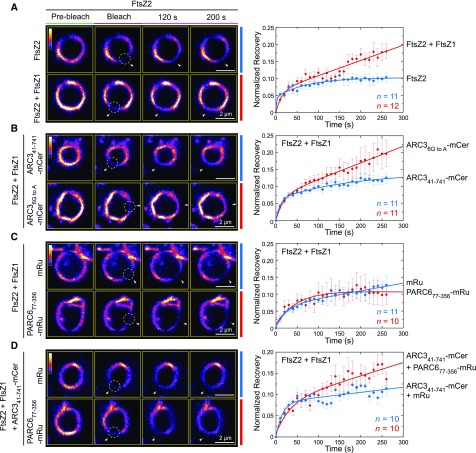
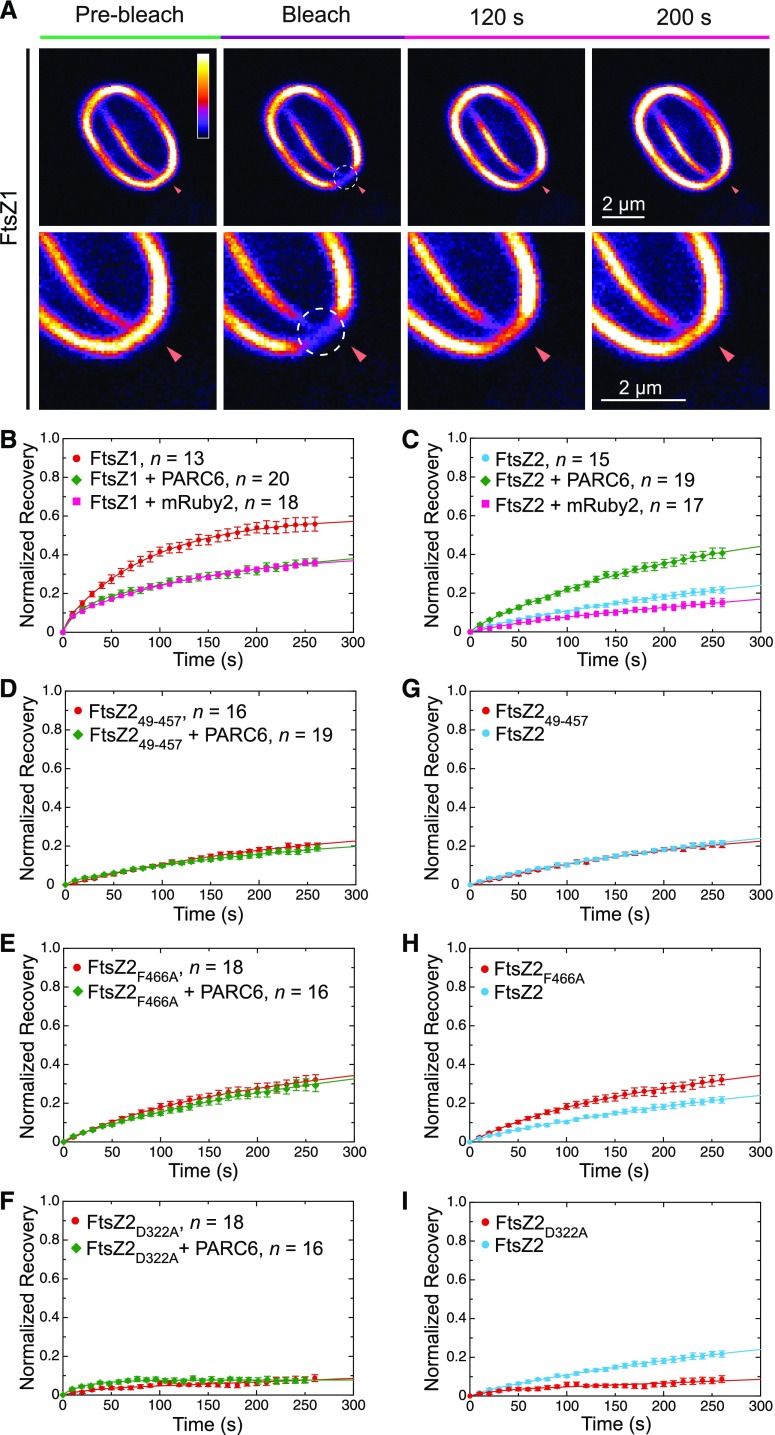
A previous study reported that the chloroplast Z ring is less dynamic in Arabidopsis arc3-2 mutants than in the wild-type plants (Johnson et al., 2015b), suggesting a role for ARC3 in promoting Z-ring remodeling at the division site. However, studies of Z-ring dynamics using fluorescently tagged FtsZ fusion proteins in transgenic plants are challenging because chloroplast division and Z-ring morphology are disrupted by relatively small changes in FtsZ protein levels (Stokes et al., 2000; Schmitz et al., 2009). Therefore, to investigate the effect of ARC3 and PARC6 on Z-ring dynamics, we used another heterologous yeast system, P. pastoris, in which we previously reconstituted Z rings composed of fluorescently tagged FtsZ2 only, or of both FtsZ1 and FtsZ2, by fusing a membrane tethering sequence (MTS) to the C terminus of FtsZ2 (Yoshida et al., 2016). The MTS binds directly to the plasma membrane of the yeast cells, mimicking the role of the FtsZ2 C terminus in tethering the Z ring to the inner chloroplast envelope membrane through interaction with ARC6 (Maple et al., 2005; Schmitz et al., 2009; Yoshida et al., 2016). In a control experiment, FtsZ2-eYFP-MTS (hereafter FtsZ2-eYFP [enhanced yellow fluorescent protein]; Yoshida et al., 2016) was expressed alone or with FtsZ1 (with no fluorescent tag or MTS). Consistent with previous results showing that FtsZ1 promotes turnover of FtsZ subunits from filaments and rings when the two proteins are coassembled (TerBush and Osteryoung, 2012; TerBush et al., 2016, 2018; Yoshida et al., 2016), fluorescence recovery after photobleaching (FRAP) demonstrated that Z rings in Pichia cells coexpressing FtsZ1 and FtsZ2-eYFP were more dynamic than those in cells expressing FtsZ2-eYFP alone (Figure 7A; Supplemental Figure 9C), confirming that reconstituted rings in the FtsZ1 + FtsZ2-eYFP coexpression strain contained both proteins. Strains expressing FtsZ1 and FtsZ2-eYFP were used in all additional experiments in Pichia.
Figure 7.
ARC3-PARC6 Interaction Increases the Dynamics of Z Rings Reconstituted in Pichia.
Pichia cells were transformed with vectors expressing the indicated combinations of FtsZ2-eYFP (FtsZ2), FtsZ1 (unfused), ARC341-741-mCerulean (ARC341-741-mCer), ARC36G to A-mCerulean (ARC36G to A-mCer), PARC677-356-mRuby2 (PARC677-356-mRu), and mRuby2 (mRu). In strains expressing more than one protein, all constructs were expressed from a single vector.
(A) to (D) FRAP analysis of the FtsZ2-eYFP signal in Pichia cells expressing the indicated proteins. Recovery of eYFP fluorescence was recorded for 270 s following photobleaching. Fluorescence images in the left panels show Z rings prior to photobleaching (Pre-bleach), at the time of photobleaching (Bleach), and at 120 and 200 s after photobleaching. Circles and arrowheads indicate the photobleached regions. The color scale bar indicates the FtsZ2-eYFP fluorescence intensity from white (highest) to black (lowest). Bars = 2 μm. Panels on the right show recovery curves. A two-binding-state equation was used to conduct curve fitting using averaged recovery data (Sprague et al., 2004; Yoshida et al., 2016). The resultant parameters are summarized in Supplemental Table 2. n is the number of cells used to obtain the averaged FRAP data for each experiment. Error bars represent sem at the indicated time point.
Constructs encoding variants of ARC3 fused to mCerulean and/or PARC6 fused to mRuby2 were coexpressed in different combinations in Pichia along with FtsZ1 and FtsZ2-eYFP, and the effects on Z-ring dynamics were assessed by FRAP analysis on FtsZ2-eYFP. In these experiments, all proteins in a given Pichia strain were expressed from the same vector. Expression of the ARC3 and PARC6 fusion proteins in the transformed Pichia cells was verified by detection of the mCerulean and mRuby2 fluorescence signals, respectively (Supplemental Figures 9A and 9B). In control FRAP assays, we found that the dynamic turnover of FtsZ2-eYFP from Z rings in cells coexpressing either unfused mCerulean or unfused mRuby2 was reduced compared with FtsZ2-eYPF turnover from rings in cells expressing only the FtsZ proteins (Figures 7A and 7C; Supplemental Figure 9C). Therefore, in all FRAP experiments investigating the effects of ARC3 and PARC6 on Z-ring dynamics, FRAP recovery curves were compared with those in the control strains expressing unfused mCerulean and mRuby2, respectively.
We first compared the ability of ARC341-741-mCerulean and the ARC3 MORN mutant ARC36G to A-mCerulean to influence the dynamics of the reconstituted Z rings. We used the latter mutant because our Y2H data suggested it interacts more weakly with the FtsZ proteins than does ARC341-598 (Figure 4B, rows 4, 5, 10, and 11; Supplemental Figure 6), similar to the interaction of the ARC341-741-PARC6 complex with FtsZ proteins in the Y3H assays (Figures 5A, rows 1 and 2; and 5B, rows 1 and 2). Consistent with the ability of ARC36G to A, but not ARC341-741, to interact with FtsZ (Figure 4B, rows 1 to 3 and 10 to 12; Supplemental Figure 6), ARC36G to A-mCerulean enhanced fluorescence recovery into Z rings in comparison to the control strain expressing unfused mCerulean, whereas ARC341-741-mCerulean did not (Figure 7B; Supplemental Figure 9C). We then tested the effect of PARC6. To exclude the potential effect of the full stromal region of PARC6 on Z-ring dynamics due to its interaction with FtsZ2 (Supplemental Figure 5, row 9; Zhang et al., 2016), we used PARC677-356, which is able to interact with full-length ARC3, but not with the FtsZ proteins (Supplemental Figure 5, rows 1, 3, and 4; Glynn et al., 2009; Zhang et al., 2016). As expected, PARC677-356-mRuby2 did not affect the turnover of reconstituted Z rings in comparison to the unfused mRuby2 control (Figure 7C). By contrast, recovery was enhanced in cells expressing both PARC677-356-mRuby2 and ARC341-741-mCerulean (Figure 7D). Together with the Y3H data, these findings provide evidence that ARC3-PARC6 interaction via the MORN domain enhances chloroplast Z-ring dynamics in Pichia, suggesting it may do so in vivo.
PARC6 Is Not Required for Activation of ARC3 at Nondivision Sites
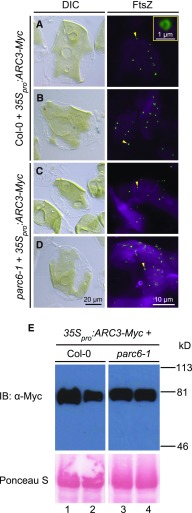
Previous work has shown that overexpression of ARC3 inhibits Z-ring assembly and disrupts chloroplast division in Arabidopsis (Maple et al., 2007; Zhang et al., 2013). Our current finding that interaction of ARC3 with PARC6 enables full-length ARC3 to inhibit FtsZ1 assembly in S. pombe led us to test whether PARC6 is required for the inhibitory effect of ARC3 overexpression on Z-ring assembly in vivo. An ARC3-Myc fusion construct, which complements the arc3-2 mutant when driven by the native ARC3 promoter (Zhang et al., 2013), was introduced into the wild-type Columbia (Col-0) and parc6-1 mutant plants under control of the 35S promoter, and immunofluorescence staining was performed to investigate FtsZ localization in T2 transgenics. As reported by Zhang et al., 2013), chloroplasts were drastically enlarged and assembly of normal Z rings was disrupted in wild-type plants overexpressing ARC3 (Figures 8A and 8B). FtsZ mini-rings were observed in some chloroplasts (Figure 8, right panels, arrowheads and inset), similar to those described previously by Zhang et al. (2013) and Johnson et al. (2015a); the significance of these structures remains unclear. ARC3 overexpression in parc6-1 similarly inhibited Z-ring assembly and chloroplast division (Figures 8C and 8D). Immunoblotting showed comparable levels of ARC3-Myc in Col-0 and parc6-1 transgenics (Figure 8E). These results imply that PARC6 is not required for activating the assembly-inhibitory activity of ARC3 away from the division site in vivo and, combined with the role of PARC6 in recruiting ARC3 to the midplastid (Figures 3E and 3F), suggest it may only be required for activating midplastid-localized ARC3. Because full-length ARC3 can only interact with FtsZs in Y3H assays and inhibit FtsZ assembly in S. pombe when PARC6 is bound to the MORN domain (Figures 5 and 6), the ability of full-length ARC3 to inhibit Z-ring assembly in the absence of PARC6 in vivo suggests the possibility that a different factor activates the assembly-inhibitory activity of ARC3 at nondivision sites in chloroplasts, perhaps through interaction with the MORN domain.
Figure 8.
Overexpression of ARC3 Inhibits Z-Ring Assembly in the parc6-1 Mutant.
(A) to (D) Chloroplast morphology (left panels) and FtsZ localization (right panels) in mesophyll cells of T2 transgenic plants expressing 35Spro:ARC3-Myc in (A) and (B) Col-0 and (C) and (D) parc6-1. Untransformed Col-0 and parc6-1 controls are shown in Figures 2A and 2D, respectively. Chloroplast morphology and FtsZ localization were visualized using differential interference contrast microscopy and immunofluorescence staining of FtsZ2-1 (FtsZ), respectively. For immunofluorescence, merged images of FtsZ2-1 (green) and chlorophyll autofluorescence (magenta) are shown. The inset in the right panel in (A) is a magnified image of the mini-ring pointed to the arrowhead. Bars = 20 μm for differential interference contrast microscopy (DIC) images and 10 μm for all immunofluorescence images except the inset, in which it is 1 μm. Arrowheads indicate mini-rings.
(E) Immunoblot analysis of the ARC3-Myc fusion protein in T2 transgenic Col-0 (left) and parc6-1 (right) plants expressing 35pro:ARC3-Myc. Total proteins extracted from 4- to 5-week-old plants were separated on a 10% SDS-PAGE gel, and membranes were probed with an anti–Myc antibody. Ponceau S–stained large subunit of Rubisco (bottom panels) served as a loading control. Molecular mass markers are shown on the right. Lanes 1 to 4 are samples prepared from the same plants imaged in (A) to (D), respectively.
PARC6 Has an ARC3-Independent Effect on FtsZ Dynamics
In a previous study, PARC6 did not obviously affect the assembly or morphology of FtsZ filaments in S. pombe (Zhang et al., 2016). However, genetic analysis showed that both the chloroplast division defect and misplacement of Z rings are more severe in the parc6-1 arc3-2 double mutant than in either single mutant (Figures 2B, 2D, 2E, and 2G), hinting that PARC6 might contribute to the negative regulation of Z-ring formation independently of ARC3. To test whether PARC6 influences FtsZ dynamics, we coexpressed PARC677‐573-mRuby2 with either FtsZ1-mVenus or FtsZ2-mCerulean in S. pombe and conducted FRAP analysis on the assembled FtsZ filaments in the transformed yeast cells (Figure 9A). Similar to the observations in Pichia, in control S. pombe strains unfused mRuby2 reduced the recovery of fluorescence into photobleached FtsZ1-mVenus and FtsZ2-mCerulean filaments (Figures 9B and 9C). Consistent with previous data showing that PARC677‐573 interacts only with FtsZ2 in Y2H assays and colocalizes only with FtsZ2 in S. pombe (Supplemental Figures 5, rows 8 and 9; and Supplemental Figures 10F and 10G; Zhang et al., 2016), PARC677‐573-mRuby2 did not alter fluorescence recovery of FtsZ1 filaments but did enhance recovery of FtsZ2 filaments compared with recoveries in control cells expressing unfused mRuby2 (Figures 9B and 9C; Supplemental Figures 10A, 10B, 10F, 10G, 10K, and 10L). PARC677‐573-mRuby2 did not enhance recovery of filaments formed by FtsZ249-457-mCerulean or FtsZ2F466A-mCerulean (Figures 9D and 9E; Supplemental Figures 10H and 10I), mutants of FtsZ2 that retain filament-turnover dynamics (Figures 9G and 9H; Supplemental Figures 10C and 10D) but are much less efficient at binding PARC6 (Supplemental Figures 10H and 10I; Zhang et al., 2016). These findings suggest that PARC6 may promote Z-ring remodeling through direct interaction with FtsZ2 as well as indirectly by activating ARC3. However, PARC677‐573-mRuby2 did not enhance turnover of filaments formed by FtsZ2D322A-mCerulean (Figure 9F; Supplemental Figures 10E and 10J), an FtsZ2 mutant that is competent for assembly (Olson et al., 2010; TerBush and Osteryoung, 2012) and colocalizes with FtsZ2 in S. pombe (Supplemental Figure 10J) but is deficient in GTPase activity (Olson et al., 2010), making FtsZ2D322A filaments static compared with the wild-type FtsZ2 filaments (Figure 9I; Supplemental Figures 10B and 10E; TerBush and Osteryoung, 2012; TerBush et al., 2018). This result suggests that enhancement of FtsZ2 dynamics by PARC6 (Figure 9C) may require GTPase-dependent filament turnover.
Figure 9.
PARC6 Increases the Dynamic Turnover of FtsZ2 Filaments in S. pombe.
(A) to (I) S. pombe cells were transformed with vectors expressing the indicated combinations of FtsZ1-mVenus (FtsZ1), FtsZ2-mCerulean (FtsZ2), FtsZ249-457-mCerulean (FtsZ249-457), FtsZ2F466A-mCerulean (FtsZ2F466A), FtsZ2D322A-mCerulean (FtsZ2D322A), PARC677-573-mRuby2 (PARC6), and mRuby2. FRAP assays were performed based on recovery of (A) and (B) the mVenus signal in FtsZ1-mVenus filaments or (C) to (F) the mCerulean signal in FtsZ2-mCerulean, FtsZ249-457-mCerulean, FtsZ2F466A-mCerulean, or FtsZ2D322A-mCerulean filaments. Recovery of fluorescence into photobleached regions was recorded for 270 s following photobleaching. A two-binding-state equation was used to conduct curve fitting using averaged recovery data (Sprague et al., 2004; Yoshida et al., 2016; TerBush et al., 2018). The resulting parameters are summarized in Supplemental Table 3. n is the number of cells used to obtain the averaged FRAP data for each experiment. Error bars represent sem at the indicated time points.
(A) Example of images from a FRAP analysis performed on FtsZ1-mVenus filaments in an S. pombe cell. Images are shown prior to photobleaching (Pre-bleach), at the time of photobleaching (Bleach), and at 120 and 200 s after photobleaching. Circles and arrowheads indicate the photobleached regions. The color scale bar indicates the FtsZ1-mVenus fluorescence intensity from white (highest) to black (lowest). The lower panels are ×2 magnifications of the same images shown in the top panels. Bars = 2 μm. Sample images of other cells analyzed by FRAP in panels (B) to (F) are shown in Supplemental Figure 10.
(B) FRAP assays comparing the dynamics of filaments formed by FtsZ1-mVenus in the presence and absence of PARC677-573-mRuby2 or mRuby2.
(C) FRAP assays comparing the dynamics of filaments formed by FtsZ2-mCerulean in the presence and absence of PARC677-573-mRuby2 or mRuby2.
(D) to (F) FRAP assays comparing the dynamics of filaments formed by (D) FtsZ249-457-mCerulean, (E) FtsZ2F466A-mCerulean, and (F) FtsZ2D322A-mCerulean in the presence and absence of PARC677-573-mRuby2.
(G) to (I) FRAP assays comparing the dynamics of filaments formed by FtsZ2-mCerulean with those formed by (G) FtsZ249-457-mCerulean, (H) FtsZ2F466A-mCerulean, and (I) FtsZ2D322A-mCerulean. The red recovery curves in (G) to (I) are the same as those shown in (D) to (F), respectively.
DISCUSSION
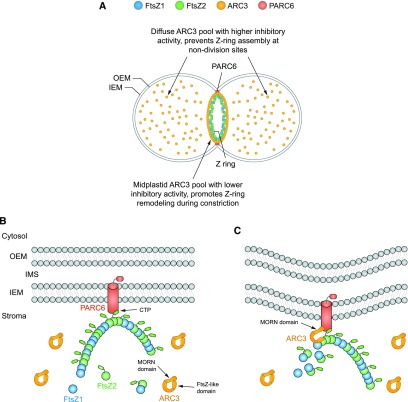
In this study, we have established that the Arabidopsis FtsZ-assembly inhibitor ARC3 exhibits both diffuse and midplastid localization patterns in the chloroplast and that ARC3 is recruited to the midplastid by the IEM protein PARC6 (Figure 10A). We investigated the functional significance of midplastid ARC3 localization using a combination of Y3H assays and experiments in heterologous yeast systems. Our results revealed that PARC6 interaction with the ARC3 C-terminal MORN domain (Figure 1) promotes FtsZ binding to ARC3. PARC6/ARC3/FtsZ interaction in turn activates the inhibitory activity of ARC3 on FtsZ assembly and increases the dynamic turnover of Z rings reconstituted in yeast. Collectively, our findings lead to a working model in which the recruitment and activation of ARC3 by PARC6 at the chloroplast division site enhances Z-ring remodeling to facilitate constriction (Figures 10B and 10C).
Figure 10.
Working Model of the Roles of ARC3 and PARC6 in the Regulation of Z-Ring Assembly and Dynamics during Chloroplast Division.
(A) ARC3 exhibits two distinct localization patterns within chloroplasts. The diffuse pool of ARC3 has higher anti–FtsZ assembly activity and functions in preventing Z-ring assembly at nondivision sites (Zhang et al., 2013). The midplastid ARC3 pool has lower anti–FtsZ assembly activity and promotes Z-ring remodeling at the division site to facilitate chloroplast constriction.
(B) The Z ring, composed of FtsZ1 and FtsZ2 heteropolymers (Olson et al., 2010; Yoshida et al., 2016), interacts with PARC6 via the FtsZ2 CTP (Zhang et al., 2016). ARC3 in its unbound state is inhibited from interacting with FtsZ by the MORN domain (Maple et al., 2007; Zhang et al., 2013), possibly because of the close proximity of the MORN domain to the FtsZ-like region of ARC3 (Shaik et al., 2018).
(C) PARC6 recruits ARC3 to the midplastid through direct interaction with the ARC3 MORN domain. MORN-domain binding to PARC6 alters the conformation of ARC3, allowing ARC3 to interact with FtsZ proteins in the Z ring. The latter interactions enhance the exchange of subunits from the Z ring during constriction, possibly by increasing FtsZ2 GTPase activity and/or reducing lateral interactions between multiple protofilaments that may comprise the Z ring (shown in [A]) (Shaik et al., 2018). FtsZ1 (TerBush and Osteryoung, 2012; Yoshida et al., 2016) and possibly PARC6 also promote dynamic remodeling of the Z ring independently of ARC3. OEM, outer envelope membrane.
Previous data on the subcellular localization of Arabidopsis ARC3 based on immunostaining of fixed leaf samples (Shimada et al., 2004) and transient expression experiments in tobacco (Nicotiana tabacum cv Samsun; Maple et al., 2007) suggested that ARC3 localizes partly to the stroma and partly to a midplastid ring, but were not entirely clear. Our analysis of stable transgenic Arabidopsis lines expressing an ARC3-mNG fusion protein that fully complements the chloroplast division phenotypes in the arc3-2 mutant (Figures 2B, 2C, and 2G) indicates that ARC3 localizes to two distinct pools within chloroplasts: one distributed diffusely across the stroma where FtsZ assembly must be fully inhibited, and the other concentrated at the division site where the Z ring forms (Figures 3A to 3D and Figure 10A). We presume that the diffuse pool, in cooperation with other components of the chloroplast Min system (Zhang et al., 2013; Chen et al., 2018b), is primarily responsible for preventing Z-ring formation at nondivision sites. This is suggested in part by the fact that arc3 mutants exhibit multiple Z rings across their chloroplasts (Glynn et al., 2007; Zhang et al., 2013) as well as by studies demonstrating that ARC3 is the direct inhibitor of FtsZ assembly in the chloroplast Min system (TerBush and Osteryoung, 2012; Zhang et al., 2013; Irieda and Shiomi, 2017; Shaik et al., 2018). A recent in vitro study using purified FtsZ2 and MORN-truncated ARC3 (Shaik et al., 2018) suggests that ARC3 may not act by inhibiting FtsZ polymerization per se, but rather by promoting debundling of assembled FtsZ filaments, somewhat similar to a mechanism proposed for bacterial MinC (Park et al., 2018), and by slightly stimulating FtsZ2 GTPase activity, which could facilitate filament disassembly by destabilizing interactions between FtsZ subunits (Erickson et al., 2010; Shaik et al., 2018). However, in contrast with MinC, which oscillates from pole to pole in E. coli and cyanobacteria (Hu and Lutkenhaus, 1999; Raskin and de Boer, 1999b; MacCready et al., 2017), we did not observe oscillation of the diffuse pool of ARC3-mNG in chloroplasts (Supplemental Figure 2). While we cannot completely rule out an oscillatory mechanism for Z-ring positioning in chloroplasts, we note that these organelles are considerably larger than most bacterial cells and that the mature form of ARC3 (∼78 kD) is much larger than MinC (∼25 to 29 kD), which may be incompatible with oscillation. Additionally, many mechanisms for regulating Z-ring placement in bacteria do not involve Min-system oscillation (Monahan et al., 2014).
A role for ARC3 in promoting Z-ring dynamics was initially suggested by FRAP experiments showing that turnover of fluorescently tagged FtsZ1 from filaments and Z rings in transgenic Arabidopsis was considerably slower in arc3 mutants than in the wild-type plants (Johnson et al., 2015b). Our finding that full-length ARC3 increases the dynamic turnover of Z rings reconstituted in Pichia cells in the presence of PARC6 (Figure 7D) provides additional compelling evidence for this. Moreover, our data indicate that PARC6 plays a key role both in recruiting ARC3 to the division site and in activating ARC3 to enhance Z-ring dynamics, and suggest that both functions involve PARC6 interaction with the ARC3 MORN domain. The requirement of the MORN domain for midplastid ARC3 localization is suggested by data showing that only ARC3 bearing the MORN domain, as well as the MORN domain alone, interacts with PARC6 (Figure 4A, rows 1 and 2; Glynn et al., 2009; Zhang et al., 2016). Furthermore, while full-length ARC3 cannot interact with FtsZ1 or FtsZ2 on its own in Y2H assays (Figure 4B, rows 1 and 2; Maple et al., 2007; Zhang et al., 2013), our Y3H data demonstrate that it can interact with the FtsZ proteins when PARC6 is present (Figures 5A, rows 1 and 2; and 5B, rows 1 and 2). MORN-dependent ARC3-PARC6 interaction also activates ARC3, as suggested by the reduced assembly of FtsZ1 filaments in S. pombe and increased turnover of reconstituted Z rings in Pichia only in cells in which full-length ARC3 is coexpressed with PARC6 (Figures 6G, 6H, 6J, and 6K and 7D). These data strongly suggest that ARC3 is recruited to the midplastid and activated by PARC6 to enhance Z-ring remodeling at the chloroplast division site (Figure 10C). ARC3 binds FtsZ at least partly via its FtsZ-like region (Figure 1A; Maple et al., 2007), which in unbound full-length ARC3 may be inaccessible to FtsZ due to its close proximity to the MORN domain, as suggested by single-particle analysis (Figure 10B; Shaik et al., 2018). Therefore, binding of PARC6 to the MORN domain may activate ARC3 by altering its conformation in a manner that enables it to interact with FtsZ subunits within the Z ring, which may in turn destabilize the Z ring to facilitate Z-ring turnover and remodeling (Figure 10C). However, while ARC3 is present in many land plants and some green algae (but not all), PARC6 has only been identified in vascular plants (Glynn et al., 2009; Osteryoung and Pyke, 2014). It would be interesting to learn whether ARC3 localizes to the chloroplast division site in organisms that lack PARC6.
An additional function of midplastid-localized ARC3 appears to be to prevent ectopic Z-ring formation in the vicinity of the division site. Our observation that multiple clustered Z rings are observed in parc6‐1 single mutants, but not in parc6‐1 arc3-2 double mutants (Figures 2D and 2E; Supplemental Figure 3, right), indicates that this phenotype is not caused directly by loss of PARC6 in parc6‐1 because otherwise it should also be observed in parc6‐1 arc3-2. Because ARC3 is the only protein known to directly inhibit chloroplast FtsZ assembly (Zhang et al., 2013; Shaik et al., 2018) and is recruited to the midplastid by PARC6 (Figures 3E and 3F), we hypothesize that the clustered Z rings in parc6‐1 mutants result from the substantial reduction of ARC3 at the division site. We suggest that such Z-ring clustering is not observed in arc3-2 because the lack of ARC3 functionality in the diffuse pool prevents confinement of Z-ring assembly to the middle of the plastid, enabling multiple Z rings to form across the chloroplast in arc3 mutants (Glynn et al., 2007; Zhang et al., 2013).
We predict that the inhibitory activity of ARC3 on FtsZ assembly is regulated differently for the two pools of ARC3. The diffuse pool should possess more robust inhibitory activity in order to fully prevent the formation of nascent FtsZ filaments and Z rings at nondivision sites. Our data showing that PARC6 is not required for activation of ARC3 at nondivision sites (Figure 8) suggest that the diffuse pool of ARC3 might be activated by a different protein that confers such robust inhibitory activity. By contrast, we suggest that the assembly-inhibitory activity of ARC3 at the midplastid should be more constrained to allow for Z-ring establishment and maintenance at the division site (Figure 10A). Although the mechanisms underlying the regulation of ARC3 activity at the division site are not yet known either, several observations hint that interaction with PARC6 may indeed constrain the activity of ARC3. Y2H and Y3H assays suggest that MORN-truncated ARC341-598, which does not interact with PARC6 (Figure 4A, row 2; Glynn et al., 2009; Zhang et al., 2016), binds more strongly to FtsZ1 and FtsZ2 than does the ARC341-741-PARC6 complex, as demonstrated by the relative growth of yeast strains on the most stringent quadruple dropout medium (Figures 4B, right, rows 4 and 5; and 5A, right, rows 1 and 2; and 5B, right l, rows 1 and 2). Consistent with these observations, formation of FtsZ1 filaments in S. pombe cells appears more inhibited by ARC341-598 than by the ARC341-741-PARC6 complex (Figures 6E, 6G, and 6H; Supplemental Figures 8G to 8J TerBush and Osteryoung, 2012) , suggesting the latter has lower inhibitory activity. Furthermore, in other studies the assembly of FtsZ2 filaments, which are less dynamic than FtsZ1 filaments (TerBush and Osteryoung, 2012; Yoshida et al., 2016) and may therefore be more resistant to ARC3, was inhibited by ARC341-598 in S. pombe (Zhang et al., 2013), but was not visibly inhibited in S. cerevisiae cells expressing both ARC341-741 and PARC6 (Shaik et al., 2018). These data lead us to speculate that the FtsZ binding region of ARC3, which remains to be fully defined (Maple et al., 2007), may be less accessible to, or have a lower affinity for, the FtsZ proteins when ARC3 is bound to PARC6 than when the MORN domain is completely removed. Thus, ARC3-PARC6 interaction could be part of a mechanism that restrains the inhibitory activity of ARC3 at the chloroplast division site enough to support the assembly of Z rings, while still preserving sufficient activity to enhance Z-ring dynamics. However, previous work has shown that ARC3 activity on FtsZ assembly is dose dependent (TerBush and Osteryoung, 2012; Zhang et al., 2013; Shaik et al., 2018), and we noticed that inhibition of FtsZ1 assembly by the ARC341-741-PARC6 complex in S. pombe depended on the levels of all three proteins (Figures 6G, 6H, 6J, and 6K; Supplemental Figures 8F to 8J). Additionally, Z rings in chloroplasts are almost certainly composed of coassembled FtsZ2, which forms the structural framework of the ring, and FtsZ1 (Vitha et al., 2001; Olson et al., 2010; TerBush and Osteryoung, 2012), which also enhances turnover of coassembled filaments and Z rings (TerBush and Osteryoung, 2012; Yoshida et al., 2016). Moreover, increased dynamic turnover of Z rings reconstituted in Pichia was associated with their constriction (Yoshida et al., 2016). Therefore, dynamic Z-ring remodeling and constriction in vivo are likely to be influenced by multiple factors, including the relative stoichiometries of ARC3, FtsZ2, and FtsZ1 associated with the Z ring, and possibly by PARC6 itself since PARC6 independently promotes the turnover of FtsZ2 filaments in S. pombe (Figures 9C and 10C).
MORN domains are protein regions containing variable numbers of tandemly repeated MORN motifs, which are 14 to 23 amino acids in length (Mueller-Roeber and Pical, 2002; Habicht et al., 2015). MORN domains are found in a wide array of proteins in animals, plants, and protists that function in diverse cellular processes, including cell division (Gubbels et al., 2006; Lorestani et al., 2010), apicoplast division (van Dooren et al., 2009; Lorestani et al., 2010), flagellum biogenesis (Ju and Huang, 2004; Matsuoka et al., 2005; Satouh et al., 2005; Morriswood and Schmidt, 2015), axonal degeneration (Bhattacharya et al., 2012), and plant phosphatidylinositol signaling (Mueller-Roeber and Pical, 2002; Ma et al., 2006). They also occur in bacterial proteins (Habicht et al., 2015), but their functions have not been explored in prokaryotes. MORN motifs were first described in mammalian junctophilin1, in which they were proposed to mediate the formation of endoplasmic reticulum-plasma membrane contact sites in muscle cells by binding to phospholipids in the plasma membrane (Takeshima et al., 2000), and in several other proteins they have also been reported to interact with lipids and to regulate association with membranes (Ma et al., 2006; Im et al., 2007; Habicht et al., 2015). However, the functions of these regions are largely unstudied. In plants, MORN domains are found in a few other types of proteins in addition to ARC3, including EMBRYO DEFECTIVE1211/TRANSLOCON AT THE INNER ENVELOPE MEMBRANE OF CHLOROPLASTS 100, a component of the chloroplast protein import complex (Liang et al., 2010; Kikuchi et al., 2013) whose MORN domain has not been functionally explored, and BrMORN from Brassica rapa and its Arabidopsis homolog AtRGP (Reduction in Growth and Productivity), which promote vegetative growth, seed production, and freezing tolerance (Lee et al., 2010, 2014). The BrMORN MORN domain was reported to target BrMORN to the chloroplast (Lee et al., 2010), but was not further investigated. The best-studied MORN-domain proteins in plants are the phosphatidylinositol 4-phosphate 5-kinases (PIP5Ks), which catalyze the phosphorylation of phosphatidylinositol 4-phosphate to form phosphatidylinositol-(4,5)-bisphosphate [PI(4,5)P2] during phosphoinositide signaling (reviewed in Boss and Im, 2012; Gerth et al., 2017). Nine PIP5Ks in Arabidopsis and many in other plants bear seven to nine MORN repeats at their N termini, a feature unique to plant PIP5Ks (Mueller-Roeber and Pical, 2002; Ma et al., 2006; Mikami et al., 2010). In some PIP5Ks, the MORN domains have been shown to be required for (Kusano et al., 2008), or capable of directing (Ma et al., 2006), localization to the plasma membrane, but for others the MORN domain was not required for plasma membrane localization (Mikami et al., 2010; Stenzel et al., 2012). However, in two cases, interaction of Arabidopsis PIP5K MORN domains with other molecules has been shown to regulate their catalytic activities. Binding of the AtPIPK1 MORN domain to phosphatidic acid or its product PI(4,5)P2 stimulated its kinase activity in vitro, as did deletion of the MORN domain. The authors therefore hypothesized that MORN-domain binding to phosphatidic acid or PI(4,5)P2 induced a conformational change that made the AtPIPK1 active site more accessible to its phosphatidylinositol 4-phosphate substrate (Im et al., 2007). The MORN domain of AtPIP5K2 was shown to interact with all five members of the Rab-E clade of Rab GTPases in Arabidopsis, and this interaction both stimulated AtPIP5K2 kinase activity in vitro and caused relocalization of a Rab-E GTPase from the Golgi to the plasma membrane in vivo (Camacho et al., 2009). The ability of PIP5K MORN domains to stimulate catalytic activity upon interaction with other molecules is somewhat similar to the role of the MORN domain in ARC3. However, unlike plant PIP5Ks, ARC3 is unlikely to have any catalytic activity, has only three MORN motifs positioned near its C terminus (Supplemental Figure 4), and is localized in the chloroplast (Shimada et al., 2004; Maple et al., 2007). The ability of the ARC3 MORN domain to modulate ARC3 interaction with FtsZ1 and FtsZ2 based on its PARC6 binding status represents an additional function for a eukaryotic MORN domain in regulating multiple protein–protein interactions within a macromolecular complex.
While the stromal region of PARC6 interacts with ARC3 and FtsZ2, its intermembrane space (IMS) region (Figure 1B) interacts with the outer envelope membrane protein PLASTID DIVISION1 (PDV1), and these interactions play a role in coordinating the inner and outer division machineries during chloroplast division (Glynn et al., 2009; Zhang et al., 2016). PARC6-PDV1 interaction is similar to the interaction between ARC6 and PDV2 (Glynn et al., 2008). PDV2 induces dimerization of the IMS regions of two ARC6 molecules, as reported by a recent study (Wang et al., 2017), which is critical for normal ARC6 localization at the division site and thus for chloroplast division, suggesting that components of the outer division machinery could affect the behavior and/or interactions of inner components. PDV1-PARC6 interaction in the IMS could likewise influence the division machinery on the stromal side of the IEM (Chen et al., 2018a). For example, because clustered Z rings are present in the vicinity of the division site in pdv1 mutants (Miyagishima et al., 2006), reminiscent of those found in parc6-1 mutants (Figure 2D, right; Supplemental Figure 3, right; Glynn et al., 2009), it is possible that the recruitment and/or activation of ARC3 by PARC6 at the midplastid might be regulated partly by PDV1. However, in addition to clustered Z rings around the division site, parc6-1 mutants exhibit multiple Z rings dispersed across chloroplasts (Figure 2D, middle; Supplemental Figure 3, left; Glynn et al., 2009), suggesting a classic Min-system function for PARC6, although the underlying basis for this phenotype is unclear. A recent study reported a possible interaction between PARC6 and MinD1 (the Arabidopsis homolog of bacterial MinD) (Itoh et al., 2018). Therefore, PARC6 may help control Z-ring placement through interaction with other components of the chloroplast Min system in addition to ARC3.
Like ARC3, other chloroplast Min-system proteins, including MinD1, MinE1 (the Arabidopsis homolog of bacterial MinE), and the plant-specific MULTIPLE CHLOROPLAST DIVISION SITE1, have been reported to localize partly to the midplastid in Arabidopsis (Nakanishi et al., 2009; Miyagishima et al., 2011; Chen et al., 2018b), but their functions there remain unknown. Analysis of Z-ring morphology in a series of double mutants generated between arc3 and other Min mutants suggests that ARC3’s activity is regulated by these proteins in vivo (Zhang et al., 2013; Chen et al., 2018b). Both MinD1 and MinE1 (but not MULTIPLE CHLOROPLAST DIVISION SITE1) interact directly with ARC3 in Y2H assays (Maple et al., 2007; Chen et al., 2018b), but the MORN domain prevents such interactions (Maple et al., 2007). Binding of the MORN domain by PARC6 may therefore allow ARC3 to interact with MinD1 and MinE1 as well as the FtsZs at the division site, which may impose additional regulation on ARC3 activity. Future studies are necessary for further understanding of the mechanistic significance of Min-system function at the chloroplast division site.
METHODS
Plant Materials, Growth Conditions, and Generation of Double Mutant
The T-DNA insertion mutants arc3-2 (SALK_057144) and parc6-1 (SALK_100009) in the Arabidopsis (Arabidopsis thaliana) Col-0 background were described previously (Shimada et al., 2004; Maple et al., 2007; Glynn et al., 2009; Zhang et al., 2009, 2013). The parc6-1 arc3-2 double mutant was obtained by crossing parc6-1 with arc3-2, and the double homozygote was validated by PCR with MZ1123/MZ1124 (for the wild-type) and LBb1.3/MZ1124 (for T-DNA insertion) primer sets for arc3-2 (SALK_057144) and MZ1127/MZ1128 (for the wild-type) and LBb1.3/MZ1128 (for T-DNA insertion) primer sets for parc6-1 (SALK_100009). All seeds used in this study were surface sterilized and sown on half-strength Murashige-Skoog medium plates supplemented with 0.7% (w/v) phytagar (Gibco BRL). Plates were kept in the dark for 2 d at 4°C before being moved into environmentally controlled chambers. The germinated seedlings were transferred to soil after 1 week of growth. The growth chamber conditions were set to a 16-h-light/8-h-dark photoperiod with white fluorescent light (bulb type: PHILIPS, F17T8/TL841 ALTO, 17 W, with two tubes; SYLVANIA FO96/841/ECO, 59 W, with five tubes; 110 μol m−2 s−1) and 70% humidity at 21°C.
Plasmid Design and Construction
All PCR amplifications were conducted with Phusion High-Fidelity DNA Polymerase (Thermo Fisher Scientific) in a PTC-225 thermocycler (MJ Research). All restriction enzymes were purchased from New England Biolabs. Recombinant DNA constructs used in this study were generated by Gibson assembly (Gibson et al., 2009), except where otherwise indicated. All chloroplast division genes (without the predicted transit peptide sequences) were PCR amplified from the cDNA sequences except where specifically noted. Primers are listed in Supplemental Table 1 and the finished constructs were verified by sequencing prior to use in experiments.
To obtain ARC3pro:ARC3-mNG, primers CC-47/CC-48 and CC-41/CC-44 (Supplemental Table 1) were used to amplify an ARC3 genomic sequence, including 1463 nucleotides upstream of the start codon as annotated in The Arabidopsis Information Resource from the bacterial artificial chromosome clone F25A4 purchased from the Arabidopsis Biological Resource Center. The mNG fragment was PCR amplified with the CC-45/CC-46 primer set. The purified PCR fragments were directly inserted into the backbone vector pCAMBIA-1300-20 (Zhang et al., 2013) digested with PstI and SacI. Construction of 35Spro:ARC3-Myc was described in a previous report (Zhang et al., 2013).
The pGADT7 (AD) and pGBKT7 (BD) vectors (Clontech) were used as backbones to generate all the constructs used in the Y2H and Y3H assays. The CC-122/CC-123 and CC-124/CC-125 primer sets and CC-122/CC-126 and CC-127/CC-125 primer sets were used to amplify ARC3Δ630-675 and ARC36G to A, respectively, from pGADT7-ARC341-741 (Zhang et al., 2013), and the resulting PCR fragments were inserted into the BD vector digested with NdeI and BamHI, yielding pBD-ARC3Δ630-675 and pBD-ARC36G to A. Subsequently, the primer set CC-130/CC-131 was used to amplify fragments from pBD-ARC3Δ630-675 and pBD-ARC36G to A, and the purified fragments were inserted into the NdeI/BamHI-digested AD vector, generating pAD-ARC3Δ630-675 and pAD-ARC36G to A. The remaining constructs used in the Y2H assays were from prior studies (Glynn et al., 2009; Zhang et al., 2013, 2016).
Plasmids constructed for Y3H assays are diagrammed in Supplemental Figure 7. To make AD-ARC341-741; PARC6, CC-168/CC-169 and CC-170/CC-171 primer sets were used to amplify the PARC677-573 expression cassette (without the GAL4 AD domain) from pGADT7-PARC677-573 (Zhang et al., 2016). The PCR fragments were inserted into pGADT7-ARC341-741 vector digested with NotI. Similarly, these fragments were inserted into NotI-digested pGADT7-ARC341-598 (Zhang et al., 2013) and pGADT7 empty vectors, respectively, in order to generate AD-ARC341-598; PARC6 and AD-Empty; PARC6 constructs. The primer sets CC-172/CC-169 and CC-173/CC-171 were used when the PARC677-573 expression cassette was cloned into the BD vectors. The resulting PCR products were inserted into AvrII-digested pGBKT7-FtsZ1, pGBKT7-FtsZ2 (Glynn et al., 2009), and pGBKT7 empty vectors, respectively, yielding the BD-FtsZ1; PARC6, BD-FtsZ2; PARC6, and BD-Empty; PARC6 constructs.
The pREP41X and pREP42X vectors (http://www-bcf.usc.edu/∼forsburg/vectortable.html), containing the nmt1* promoter, were used as backbones for generating constructs for expressing chloroplast division proteins in fission yeast (Schizosaccharomyces pombe). The ARC341-741 and ARC341-598 fragments were amplified with the primer sets CC-17/CC-15 and CC-17/CC-19, respectively. The mCerulean fragment was PCR amplified using the CC-6/CC-86 primers. These fragments were then cloned into pREP41X digested with XhoI and BamHI to generate ARC341-741-mCerulean and ARC341-598-mCerulean constructs, respectively. To generate PARC6-mRuby2, CC-4/CC-8 and CC-85/CC-86 primer sets were used to amplify PARC677-573 and mRuby2 fragments, respectively, followed by subsequent insertion of the fragments into pREP41X digested with XhoI and BamHI. The FtsZ1, FtsZ2, and mVenus fragments were obtained using the AT262/AT298, AT264/AT299, and AT7/AT297 primer sets, respectively. The resulting FtsZ1 and mVenus as well as FtsZ2 and mVenus fragments were cloned into BamHI-digested pREP42X, yielding FtsZ1-mVenus and FtsZ2-mVenus constructs, respectively. To generate the construct where ARC341-741 and PARC6 fusion proteins were expressed from the same vector, primers AT109/AT110 were used to amplify the PARC677-573-mRuby2 expression cassette (including the nmt1* promoter and terminator sequences) from pREP41X-PARC6-mRuby2, and the resulting PCR product was inserted into AatII-digested pREP41X-ARC341-741-mCerulean, yielding ARC341-741-mCerulean; nmt1*pro:PARC6-mRuby2 construct. Likewise, the AT109/AT110 primer set was used to amplify the mRuby2 expression cassette (including the nmt1* promoter and terminator sequences) from pREP42-mRuby2, followed by insertion into pREP41X-ARC341-741-mCerulean digested with AatII, generating ARC341-741-mCerulean; nmt1*pro:mRuby2 construct as a control. To generate FtsZ2D322A-mCerulean, AT264/AT299 and AT7/AT297 primer sets were used to amplify FtsZ2D322A and mCerulean fragments, respectively, followed by insertion into BamHI-digested pREP42X. The CC-98/CC-86 primers were used to amplify the mRuby2 fragment, and the resulting product was inserted into BamHI-digested pREP41X or pREP42X, yielding pREP41X-mRuby2 and pREP42X-mRuby2 vectors. Other vectors used in S. pombe, including pREP41X-mVenus, pREP42X-mCerulean, pREP42X-FtsZ2-mCerulean, pREP42X-FtsZ249-457-mCerulean as well as pREP42X-FtsZ2F466A-mCerulean, were described in previous reports (TerBush et al., 2016, 2018; Zhang et al., 2016).
The pPICZA vector (Invitrogen), bearing the alcohol oxidase1 promoter designed for integration of exogenous genes into the Pichia genome and inducible expression of the inserted genes, was used as backbone to express chloroplast division genes in Pichia pastoris (strain X-33). First, the relevant chloroplast division gene with or without a fluorescent tag, or the fluorescent tag itself, was cloned into pPICZA digested with NotI. The resulting constructs served as PCR templates for amplification of expression cassettes used to generate constructs for coexpression of two or more fusion proteins from the same vector. In brief, CC-219/CC-220 and CC-221/CC-220 primer sets were used to amplify mCerulean and ARC341-741-mCerulean fragments, respectively, to generate pPICZA-mCerulean and pPICZA-ARC341-741-mCerulean vectors. To generate pPICZA-ARC36G to A-mCerulean, the CC-221/CC-220 primer set was used to amplify the ARC36G to A-mCerulean fragment. The primer sets CC-222/CC-223 and CC-225/CC-226 were used to obtain the mRuby2 and FtsZ1 PCR products, respectively, followed by insertion into the NotI-digested pPICZA to generate pPICZA-mRuby2 and pPICZA-FtsZ1 vectors. To generate the pPICZA-PARC677-356-mRuby2, CC-224/CC-5 and CC-85/CC-223 primer sets were used to amplify PARC677-356 and mRuby2 fragments, respectively, followed by insertion of these two fragments together into NotI-digested pPICZA vector. To generate the construct coexpressing FtsZ1 and FtsZ2-eYFP, CC-227/CC-228, CC-229/CC-230, and CC-231/CC-232 primer sets were used to amplify fragments from the pPICZA, pPICZA-FtsZ1, and pPICZA-FtsZ2-eYFP-MTS (Yoshida et al., 2016) vectors, respectively, and the resulting PCR products were assembled by Gibson reaction to yield the pPICZA-FtsZ1; FtsZ2-eYFP-MTS vector. An NheI site was introduced into the pPICZA-FtsZ1; FtsZ2-eYFP-MTS vector for subsequent linearization to increase the efficiency of transformation into Pichia cells according to the manufacturer’s protocol and a prior study (Yoshida et al., 2016). pPICZA-FtsZ1; FtsZ2-eYFP-MTS was then used as the backbone vector to make the following five vectors for coexpression of three distinct proteins from the same vector. The CC-233/CC-234 primer set was used to amplify distinct expression cassettes from pPICZA-PARC677-356-mRuby2, pPICZA-mRuby2, pPICZA-ARC341-741-mCerulean, and pPICZA-mCerulean. The resulting PCR fragments were then cloned separately into BamHI-digested pPICZA-FtsZ1; FtsZ2-eYFP-MTS, generating pPICZA-FtsZ1; FtsZ2-eYFP-MTS; PARC677-356-mRuby2, pPICZA-FtsZ1; FtsZ2-eYFP-MTS; mRuby2, pPICZA-FtsZ1; FtsZ2-eYFP-MTS; ARC341-741-mCerulean, pPICZA-FtsZ1; FtsZ2-eYFP-MTS; ARC36G to A-mCerulean, and pPICZA-FtsZ1; FtsZ2-eYFP-MTS; mCerulean vectors. To further make the constructs for coexpressing four distinct proteins from the same vector, the expression cassettes PARC677-356-mRuby2 and mRuby2 were amplified from pPICZA-PARC677-356-mRuby2 and pPICZA-mRuby2, respectively, using the CC-235/CC-236 primer set. The PCR fragments were inserted separately into NdeI-digested pPICZA-FtsZ1; FtsZ2-eYFP-MTS; ARC341-741-mCerulean to create pPICZA-FtsZ1; FtsZ2-eYFP-MTS; ARC341-741-mCerulean; PARC677-356-mRuby2 and pPICZA-FtsZ1; FtsZ2-eYFP-MTS; ARC341-741-mCerulean; mRuby2 vectors.
Generation of Transgenic Plants
The ARC3pro:ARC3-mNG and 35Spro:ARC3-Myc constructs were transformed into Agrobacterium tumefaciens GV3101 strain by electroporation according to the manufacturer’s instructions (Bio-Rad). The transformed Agrobacterium strains were then introduced into plants through floral dipping (Zhang et al., 2006). Selection of transgenic plants was conducted by sowing the T1 seeds on half-strength Murashige-Skoog plates supplemented with 20 μg mL−1 hygromycin (Calbiochem). After 1 week of growth on the selective plates, positive T1 seedlings were transferred to soil and grown in the chamber as described above. T1 plants were used for transgenic parc6-1 arc3-2 expressing ARC3pro:ARC3-mNG, while both T1 (Supplemental Figures 1E to 1J and 2) and T2 (Figures 3A to 3D; Supplemental Figure 1C) plants were used for transgenic arc3-2 expressing ARC3pro:ARC3-mNG. T2 plants were used for the transgenic wild-type Col-0 and parc6-1 expressing 35Spro:ARC3-Myc.
Chloroplast Phenotype Analysis
Quantitative analysis of chloroplast number in the wild-type Col-0, mutants, and transgenic plants was performed as previously described (Pyke and Leech, 1991), with minor modifications. In brief, leaf samples were harvested from expanded rosette leaves of 3- to 4-week-old plants. After fixation in 3.5% (v/v) glutaraldehyde for 1 h, leaf samples were transferred into 0.1 M EDTA, disodium salt dihydrate, pH 9.0, and heated at 50°C for 2 h before observation. The samples were imaged using an inverted microscope (DMI3000B, Leica) equipped with a digital camera (DFC320, Leica) as described previously (Pyke and Leech, 1991; Schmitz et al., 2009). The cell area was measured using Fiji (ImageJ) software (http://fiji.sc/Fiji), and the number of chloroplasts was manually counted in individual cells. The chloroplast number per mesophyll cell was plotted against the cell size. As reported previously (Pyke and Leech, 1991), there is a tight correlation (R2 > 0.8) between the number of chloroplasts per mesophyll cell and the size of the mesophyll cell in the wild-type Arabidopsis plants, but this correlation breaks down in chloroplast division mutants because of the compensatory relationship between chloroplast number and chloroplast size (Pyke and Leech, 1992).
Immunofluorescence Staining of FtsZ
FtsZ assembly and Z-ring positioning were analyzed by immunofluorescence staining with an anti–FtsZ2-1 antibody as described previously (Vitha et al., 2001; Yoder et al., 2007; Vitha and Osteryoung, 2011). Briefly, leaf tips were usually cut from 3- to 4-week-old plants. The wax-embedded tissues were sectioned with an 820 Spencer rotatory microtome (American Optical) with the thickness set at 5 μm. Subsequently, the sections were incubated with anti–FtsZ2-1 antibody (1:3500 dilution) (Yoder et al., 2007) followed by the addition of Alexa Fluor 488-conjugated goat anti-rabbit secondary antibody (1:500 dilution) (lot no. 514957, Invitrogen). Samples were examined under a DMRA2 microscope (Leica) as described in “Confocal and Epifluorescence Microscopy.”
Y2H and Y3H Assays
Yeast strain Y2HGold (Clontech) was used for all Y2H and Y3H assays conducted in this study. Growth and transformation of Y2HGold cells were performed according to the manufacturer’s protocol. Positive transformants were selected based on growth on dropout medium lacking Leu and Trp (−Leu −Trp). The reporter genes HIS3, ADE2, and AUR1-C in the Y2HGold cells were used to detect positive interactions as indicated by growth of the transformed cells in the absence of His (−His) and either the presence of AbA (Clontech) (+AbA) or absence of Ade (−Ade) compared with growth of negative control cells. Activation of the AUR1-C reporter gene by a positive interaction confers resistance of the Y2HGold strain to AbA. The stringency of selective media used to detect positive interactions in this study was from highest to lowest: −Leu −Trp −His −Ade, −Leu −Trp −His +AbA and −Leu −Trp −His.
Five hundred nanograms of each plasmid was used for yeast cell transformation. After growth on the synthetic dropout (SD/−Leu −Trp) agar plate for 3 d at 30°C, positive colonies were washed with liquid SD/−Leu −Trp medium and the obtained mixture was streaked onto a new SD/−Leu −Trp plate and the plate was then kept at 30°C for another 3 d. Subsequently, the dropout assay was conducted with the activated transformants according to the manufacturer’s protocol. Ten microliters of the original (OD600 = 1.0) and diluted cultures (OD600 = 0.1, 0.01) were streaked onto SD/−Leu −Trp, SD/−Leu −Trp −His supplemented with either 20 or 40 ng mL−1 AbA, and SD/−Leu −Trp −His −Ade agar plates as indicated. Plates were kept at 30°C for 2 to 5 d as indicated in the figure legends. Images were acquired with a C600 instrument (Azure Biosystems) using the auto exposure mode. Interactions were validated in at least three independent assays, and representative images are shown in the figures. Positive and negative controls for the Y2H assays were included as indicated.
Protein Extraction and Immunoblot Assay
Total protein was extracted from Y2HGold yeast cells according to a previous report (Kushnirov, 2000), with modifications. Briefly, 3 mL of yeast cell culture (OD600 ∼ 4.0) was harvested and pelleted down by centrifugation at 16,000g for 1 min. The supernatant was removed, and the cells were resuspended in 100 μL of water. Then, 100 μL 0.2 M sodium hydroxide was added and the mixture was kept at room temperature for 10 min before centrifugation at 16,000g for 10 min. After removing the supernatant, 100 μL of 2× SDS-PAGE loading buffer (20% [v/v] glycerol, 120 mM Tris-HCl, pH 6.8, 4% [w/v] SDS, 0.02% [w/v] bromophenol blue, and 2.5% [v/v] β-mercaptoethanol) was added to the pellet and heated at 95°C for 3 min. Finally, the samples were centrifuged at 16,000g for 10 min, and 25 μL of the supernatant containing total proteins was loaded.
To prepare total protein samples from transgenic plants expressing 35Spro:ARC3-Myc, rosette leaves from 4- to 5-week-old plants were collected and frozen instantly in liquid nitrogen. The leaf tissue was then thoroughly homogenized using a FastPrep-24 tissue and cell homogenizer (MP Biomedicals). The samples were boiled in 4× SDS-PAGE loading buffer (40% [v/v] glycerol, 240 mM Tris-HCl, pH 6.8, 8% [w/v] SDS, 0.04% [w/v] bromophenol blue, and 5% [v/v] β-mercaptoethanol) for 5 min. After centrifugation at 16,000g for 2 min, 10 μL of the supernatant (equal to 5 mg of fresh weight of the leaf sample) was loaded.
Proteins were separated on 10% SDS-PAGE gels and then transferred onto nitrocellulose membranes. After blocking in Tris-buffered saline, 0.2% (v/v) Tween 20 buffer containing 2% (v/v) fat-free milk, the membrane was either probed with anti–HA antibody (1:1000 dilution; MBL) for yeast samples or anti–c-Myc antibody (1:2000 dilution; Thermo Fisher Scientific) for plant samples, following by incubation with a horseradish peroxidase–conjugated goat anti–mouse secondary antibody (1:8000 or 1:10000 dilution; Thermo Fisher Scientific) for 1 h at room temperature. Immunoreactive bands were revealed using SuperSignal West Pico Chemiluminescent Substrate (Thermo Fisher Scientific) according to the manufacturer’s protocol.
Expression of Chloroplast Division Proteins in S. pombe
S. pombe strain MBY192 (h− leu1-32 ura4-D18) was used in this study. Transformation of S. pombe cells with various constructs encoding chloroplast division fusion proteins was performed using the classic lithium acetate method as described previously (TerBush and Osteryoung, 2012). The wild-type S. pombe cells were inoculated and grown in 50 mL of Yeast Extract with Supplements (5 g L−1 yeast extract; 30 g L−1 Glc; 225 mg L−1 adenine, histidine, Leu, uracil, and lysine hydrochloride) liquid medium at 32°C with shaking at 250 rpm. Cells were harvested at 4000g at room temperature when the culture reached OD600 = 0.5 (∼107 cells mL−1), which usually took almost 2 d. The pellet was then washed with half culture volume of 1× Tris-EDTA (TE) buffer (10 mM Tris-HCl, pH 7.5, and 1 mM EDTA), resuspended, and pelleted again. Cells were finally resuspended in 1 mL of 100 mM lithium acetate, pH 7.5, in TE buffer and kept at 30°C for 30 min. Then, 200 μL of the S. pombe cell suspension was transferred to a microfuge tube and 200 μg of salmon sperm DNA (Agilent Technologies) and 1 μg of each plasmid DNA was added, followed by vigorous vortexing to mix. Subsequently, 1.2 mL of polyethylene glycol solution (40% [w/v] polyethylene glycol 11 and 100 mM lithium acetate, pH 7.5, in TE buffer) was added to the tube followed by 10 s of vortexing to mix. The mixture was then incubated at 30°C for 30 min with shaking at 200 rpm. After heating at 42°C for 15 min, cells were collected at 7000g for 30 s, and the pellet was resuspended in 150 μL of TE buffer. Finally, the culture was streaked on the selective synthetic dropout Pombe Glutamate medium (see
http://www-bcf.usc.edu/∼forsburg/media.html for details) containing 15 μM thiamine but lacking either Leu (for pREP41X) or uracil (for pREP42X) or both (for cotransformation of pREP41X and pREP42X). The plates were then kept at 30°C until isolated colonies formed. Three to four colonies for each transformed line were inoculated into the relevant synthetic dropout Pombe Glutamate liquid medium without thiamine to allow protein expression driven by the nmt1* promoter. The cultures were grown at 32°C for 36 to 40 h and expression of fusion proteins in the transformed cells was detected by epifluorescence microscopy. To quantify the effect of coexpression of ARC341-741-mCerulean and PARC6-mRuby2 or ARC341-741-mCerulean or mRuby2 on the length of FtsZ1-mVenus filaments in distinct cells, the length of the FtsZ1-mVenus filament (the longest one) within the cell was measured and normalized to the cell length. The number of the filaments was manually counted. The protein expression level of PARC6-mRuby2 and mRuby2 was directly measured by the fluorescence intensity of the mRuby2 signal within the cell area, and the obtained value was further divided by the cell area to normalize for protein expression level in distinct cells.
Expression of Chloroplast Division Proteins in Pichia
Pichia strain X-33 was used in this study. Growth and transformation of Pichia cells were conducted according to a previous study (Yoshida et al., 2016), with minor modifications. Briefly, 10 μg of linearized plasmid (pPICZA as backbone vector) was used for transformation of Pichia cells via electroporation. The exogenous DNA was integrated into the yeast genome through homologous recombination, leading to stable transformants with even expression levels of the introduced fusion proteins in distinct cells (Cregg et al., 1985, 1989; Cregg and Higgins, 1995). Selection of the transformed Pichia cells was conducted with medium supplemented with 10 μg mL−1 phleomycin (InvioGen). To prepare cells for FRAP analysis, transformed yeast cells were first cultured overnight in 2 mL of Buffered Glycerol Complex medium (1% [w/v] yeast extract, 2% [w/v] peptone, 100 mM potassium phosphate, pH 6.0, 1.34% [w/v] Yeast Nitrogen Base [Sigma Aldrich], 0.00004% [w/v] biotin, and 1% [v/v] glycerol) containing 5 μg mL−1 phleomycin and then centrifuged at 2200g for 2 min at room temperature. The pellet was resuspended in 500 μL of Buffered Complex medium (1% [w/v] yeast extract, 2% [w/v] peptone, 100 mM potassium phosphate, pH 6.0, 1.34% [w/v] Yeast Nitrogen Base, and 0.00004% [w/v] biotin) supplemented with 5 μg mL−1 phleomycin and then spun down at 2200g for 2 min at room temperature. Cells were resuspended in 400 μL of Buffered Complex medium and kept at 30°C for 36 h with continuous shaking to achieve moderate induction of chloroplast division proteins prior to FRAP analysis (Yoshida et al., 2016).
FRAP Analysis
FRAP analysis was conducted as previously described (Yoshida et al., 2016; TerBush et al., 2018), with slight modification. All FRAP data were acquired at room temperature with a spectral-based FluoView 1000 laser scanning confocal microscope (Olympus) equipped with either a PlanApo 60× oil immersion objective with a numerical aperture (NA) of 1.42 for S. pombe or a UPlanSApo 100× oil immersion objective with a NA of 1.40 for Pichia cells. FRAP images were recorded using FV1000 ASW software (Olympus).
Transformed S. pombe cells were grown at 32°C for 36 to 40 h, and 2 μL of the liquid culture was pipetted onto a poly-l-lysine–coated glass slide (Sigma-Aldrich) and then covered with a No. 1.5 cover slip. Cells with high expression levels (as observed under the confocal microscope) of the fusion proteins were chosen to perform FRAP. For proteins fused with mCerulean, photobleacheing was performed with a 458 nm laser at 40% intensity, and the fluorescence recovery was acquired at 7% intensity so as to reduce photobleaching. For proteins fused with mVenus, photobleaching was conducted with a 515 nm laser at 13% intensity, while recovery was recorded at 0.7% intensity. The mRuby2 fluorescence signal was detected with a 559 nm laser at 5% intensity. The fluorescence signals were photobleached for 20 ms using the Tornado scanning tool within the FV1000 ASW software. Recovery of the fluorescence signals was monitored at 10-s intervals for a total of 270 s. Prior to photobleaching, three images were taken at 10-s intervals to serve as prebleach control. The bleach area was set to be 20 pixels (∼1.28 μm) in diameter. In order to correct and normalize the FRAP data, fluorescence intensities were recorded during each experiment at a region of the fluorescence signal away from the photobleached spot, which accounts for photobleaching due to the continual acquisition, and at a region of the background signal, which accounts for random noise. Pichia cells were prepared as described in "Expression of Chloroplast Division Proteins in Pichia", and 2 μL of the cell culture was used. FRAP analysis was performed as for S. pombe cells with the modification that 100% intensity of the 515 nm laser was used to conduct photobleaching of the fluorescence signal of the FtsZ2-eYFP fusion protein, while 1% laser intensity was used to acquire the recovery. The number of yeast cells used for each sample is indicated in corresponding figures. Normalized FRAP data were used to perform curve fitting with pro Fit software (QuantumSoft). The data were fit to the function of a two-binding-state model (Sprague et al., 2004; Yoshida et al., 2016): f(t) = (1 − r)(1 − Ceq1 e−koff1*t − Ceq2 e−koff2*t), where t is time (s), Ceq1 and Ceq2 refer to fractions of bound molecules, koff1 and koff2 refer to dissociation rate constants, and r is an additional added parameter to take the incomplete recovery effect into account (Sprague et al., 2004). The resulting parameters are summarized in Supplemental Tables 2 and 3 for FRAP analysis in Pichia and S. pombe, respectively.
Confocal and Epifluorescence Microscopy
Localization of the ARC3-mNG fusion protein in transgenic plants was imaged using an A1 laser scanning confocal microscope (Nikon). The 100× oil immersion objective (1.49 NA) was used to capture the images. The laser intensity to detect the mNG fluorescence signal (488 nm excitation/500 to 550 nm emission) and the chlorophyll autofluorescence (561 nm excitation/662 to 737 nm emission) was 0.3 and 0.2%, respectively, unless otherwise stated. Identical settings on the confocal microscope were applied when images of the nontransformed wild-type Col-0 were acquired as controls. T1 or T2 transgenic plants were first selected on half MS plates supplemented with 20 μg mL−1 hygromycin (Calbiochem). After 5 d, root tips of the selected seedlings were examined under the confocal microscope with identical settings as described for leaf samples before transferring plants to soil. The adaxial side of leaf samples (∼1 to 4 mm2) collected from 2- to 3-week-old plants was imaged to explore the subcellular localization of ARC3 in vivo, unless otherwise indicated. To quantify the number of chloroplasts with ARC3-mNG ring structures, at least eight images from two independent transgenic lines were analyzed for each type of transgenic plant. Confocal microscope used for FRAP analysis in yeast cells was described in “FRAP Analysis.”
Fluorescent images of chloroplast division proteins expressed in transformed S. pombe and Pichia yeast cells, and of immunostained FtsZ2 in plants, were acquired with a DMRA2 microscope (Leica) equipped with a 100× oil immersion objective (HCX PL FLUOTAR; NA 1.30 to 0.60) and a charge-coupled device camera (Retiga Exi, QImaging) as previously described (Zhang et al., 2016). For yeast samples, 2 μL of liquid culture was placed onto a poly-l-lysine–coated glass slide (Sigma-Aldrich) and covered with a No. 1.5 cover slip before observation at room temperature. The fluorescence signals from mCerulean, mVenus or eYFP, and mRuby2 fusion proteins were captured using Leica filters CFP (436 nm excitation/455 nm emission), YFP (500 nm excitation/515 nm emission), and TX2 (560 nm excitation/595 nm emission), respectively. Both z stacks with a 0.5-μm interval (Figure 6D) and single snap acquisition (Figures 6A to 6C, 6E to 6I; Supplemental Figure 8) were performed with Image-Pro Plus 7.0 software (Media Cybernetics). The exposure time was 400 ms for all channels except in Figure 6H and Supplemental Figures 8H to 8J, where the exposure time was 500 ms for both FtsZ1-mVenus and PARC677-573-mRuby2 and 200 ms for ARC341-741-mCerulean. Nearest-neighbor deconvolution with 70% haze removal was conducted for all fusion proteins except ARC341-741-mCerulean due to the presence of intensive bright spots found in some transformed S. pombe cells. For immunofluorescence staining of FtsZ2, leaf sections were mounted on the poly-l-lysine–coated slide (Sigma-Aldrich) followed by immunostaining as described in "Immunofluorescence Staining of FtsZ". Final prepared samples were imaged with a DMRA2 microscope (Leica) using filters L5 (480 nm excitation/505 nm emission) and TX2 (560 nm excitation/595 nm emission) to detect Alexa Fluor 488 and chlorophyll fluorescence, respectively. z stacks with a 0.5-μm interval were taken for both channels. The exposure time was 400 ms for both channels, and deconvolution was performed. Projection of all obtained z stacks was conducted with Fiji (ImageJ) software (http://fiji.sc/Fiji) using the max intensity algorithm. False color for distinct fluorescent proteins is as indicated in figure legends.
Accession Numbers
Sequence data from this article can be found in The Arabidopsis Information Resource database (https://www.arabidopsis.org/) under the following names and accession numbers: PARC6 (AT3G19180), ARC3 (AT1G75010), FtsZ1 (AT5G55280), and FtsZ2 (AT2G36250). The single mutants used in this study are parc6-1 (SALK_100009) and arc3-2 (SALK_057144). Accession number for protein sequences harboring MORN domains can be found in the legend for Supplemental Figure 4.
SUPPLEMENTAL DATA
Supplemental Figure 1. ARC3 localization in various plant tissues.
Supplemental Figure 2. ARC3-mNG does not oscillate in chloroplasts.
Supplemental Figure 3. FtsZ localization in mesophyll cells of parc6-1.
Supplemental Figure 4. Multiple sequence alignment of MORN domains in proteins from various species.
Supplemental Figure 5. Reinvestigation of interactions between PARC6 derivatives and ARC3 or FtsZ proteins.
Supplemental Figure 6. The ARC3 MORN mutant, ARC36G to A, interacts with FtsZ proteins.
Supplemental Figure 7. Generation of constructs for yeast three-hybrid assays.
Supplemental Figure 8. Effect of full-length ARC3 on assembly of FtsZ1 filaments in the presence of PARC6 in S. pombe.
Supplemental Figure 9. Morphology and dynamics of reconstituted Z rings in the presence of full-length ARC3 and PARC6 in Pichia.
Supplemental Figure 10. Effect of PARC6 on the dynamic turnover of FtsZ filaments polymerized in S. pombe.
Supplemental Table 1. List of primers used for constructs.
Supplemental Table 2. FRAP analysis of the effect of ARC3-PARC6 complex on the dynamics of Z ring reconstituted in Pichia.
Supplemental Table 3. FRAP analysis of the effect of PARC6 on the dynamics of FtsZ filaments in S. pombe.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Daniel Ducat and Joshua MacCready for providing the anti–HA antibody and the template for mNG, Min Zhang for generating transgenic parc6-1 expressing 35Spro:ARC3-Myc, Allan TerBush for technical help with FRAP experiments and data analysis, and Yamato Yoshida and Yuko Mogi for critical comments on the manuscript. This work was supported by the U.S. Department of Energy, Office of Science, Basic Energy Sciences under Award DE-FG02-06ER15808 and by the National Science Foundation (Award 1719376 to K.W.O.).
AUTHOR CONTRIBUTIONS
C.C. and K.W.O. conceived the project and designed the experiments; C.C. performed most of the experiments and analyzed data; L.C. performed the experiments in Pichia and analyzed data; Y.Y. generated the parc6-1 arc3-2 double mutant and analyzed the phenotype; K.J.P. helped with data analysis; and C.C. and K.W.O. wrote the article with contributions from all authors.
References
- Aldridge C., Møller S.G. (2005). The plastid division protein AtMinD1 is a Ca2+-ATPase stimulated by AtMinE1. J. Biol. Chem. 280: 31673–31678. [DOI] [PubMed] [Google Scholar]
- Arumugam S., Petrašek Z., Schwille P. (2014). MinCDE exploits the dynamic nature of FtsZ filaments for its spatial regulation. Proc. Natl. Acad. Sci. USA 111: E1192–E1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beech P.L., Nheu T., Schultz T., Herbert S., Lithgow T., Gilson P.R., McFadden G.I. (2000). Mitochondrial FtsZ in a chromophyte alga. Science 287: 1276–1279. [DOI] [PubMed] [Google Scholar]
- Bhattacharya M.R.C., Gerdts J., Naylor S.A., Royse E.X., Ebstein S.Y., Sasaki Y., Milbrandt J., DiAntonio A. (2012). A model of toxic neuropathy in Drosophila reveals a role for MORN4 in promoting axonal degeneration. J. Neurosci. 32: 5054–5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi E.F., Lutkenhaus J. (1991). FtsZ ring structure associated with division in Escherichia coli. Nature 354: 161–164. [DOI] [PubMed] [Google Scholar]
- Boss W.F., Im Y.J. (2012). Phosphoinositide signaling. Annu. Rev. Plant Biol. 63: 409–429. [DOI] [PubMed] [Google Scholar]
- Busiek K.K., Margolin W. (2015). Bacterial actin and tubulin homologs in cell growth and division. Curr. Biol. 25: R243–R254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho L., Smertenko A.P., Pérez-Gómez J., Hussey P.J., Moore I. (2009). Arabidopsis Rab-E GTPases exhibit a novel interaction with a plasma-membrane phosphatidylinositol-4-phosphate 5-kinase. J. Cell Sci. 122: 4383–4392. [DOI] [PubMed] [Google Scholar]
- Chen C., MacCready J.S., Ducat D.C., Osteryoung K.W. (2018a). The molecular machinery of chloroplast division. Plant Physiol. 176: 138–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Sun B., Gao W., Zhang Q.-Y., Yuan H., Zhang M. (2018b). MCD1 associates with FtsZ filaments via the membrane-tethering protein ARC6 to guide chloroplast division. Plant Cell 30: 1807–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colletti K.S., Tattersall E.A., Pyke K.A., Froelich J.E., Stokes K.D., Osteryoung K.W. (2000). A homologue of the bacterial cell division site-determining factor MinD mediates placement of the chloroplast division apparatus. Curr. Biol. 10: 507–516. [DOI] [PubMed] [Google Scholar]
- Cregg J.M. and Higgins D.R. (1995). Production of foreign proteins in the yeast Pichia pastoris. Can. J. Bot. 73: 891– 897. [Google Scholar]
- Cregg J.M., Barringer K.J., Hessler A.Y., Madden K.R. (1985). Pichia pastoris as a host system for transformations. Mol. Cell. Biol. 5: 3376–3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cregg J.M., Madden K.R., Barringer K.J., Thill G.P., Stillman C.A. (1989). Functional characterization of the two alcohol oxidase genes from the yeast Pichia pastoris. Mol. Cell. Biol. 9: 1316–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson H.P., Anderson D.E., Osawa M. (2010). FtsZ in bacterial cytokinesis: Cytoskeleton and force generator all in one. Microbiol. Mol. Biol. Rev. 74: 504–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara M.T., Nakamura A., Itoh R., Shimada Y., Yoshida S., Møller S.G. (2004). Chloroplast division site placement requires dimerization of the ARC11/AtMinD1 protein in Arabidopsis. J. Cell Sci. 117: 2399–2410. [DOI] [PubMed] [Google Scholar]
- Fujiwara M.T., Hashimoto H., Kazama Y., Abe T., Yoshida S., Sato N., Itoh R.D. (2008). The assembly of the FtsZ ring at the mid-chloroplast division site depends on a balance between the activities of AtMinE1 and ARC11/AtMinD1. Plant Cell Physiol. 49: 345–361. [DOI] [PubMed] [Google Scholar]
- Gerth K., Lin F., Menzel W., Krishnamoorthy P., Stenzel I., Heilmann M., Heilmann I. (2017). Guilt by association: A phenotype-based view of the plant phosphoinositide network. Annu. Rev. Plant Biol. 68: 349–374. [DOI] [PubMed] [Google Scholar]
- Gibson D.G., Young L., Chuang R.-Y., Venter J.C., Hutchison C.A. III, Smith H.O. (2009). Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6: 343–345. [DOI] [PubMed] [Google Scholar]
- Gilson P.R., Yu X.-C., Hereld D., Barth C., Savage A., Kiefel B.R., Lay S., Fisher P.R., Margolin W., Beech P.L. (2003). Two Dictyostelium orthologs of the prokaryotic cell division protein FtsZ localize to mitochondria and are required for the maintenance of normal mitochondrial morphology. Eukaryot. Cell 2: 1315–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glynn J.M., Miyagishima S.-Y., Yoder D.W., Osteryoung K.W., Vitha S. (2007). Chloroplast division. Traffic 8: 451–461. [DOI] [PubMed] [Google Scholar]
- Glynn J.M., Froehlich J.E., Osteryoung K.W. (2008). Arabidopsis ARC6 coordinates the division machineries of the inner and outer chloroplast membranes through interaction with PDV2 in the intermembrane space. Plant Cell 20: 2460–2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glynn J.M., Yang Y., Vitha S., Schmitz A.J., Hemmes M., Miyagishima S.-Y., Osteryoung K.W. (2009). PARC6, a novel chloroplast division factor, influences FtsZ assembly and is required for recruitment of PDV1 during chloroplast division in Arabidopsis. Plant J. 59: 700–711. [DOI] [PubMed] [Google Scholar]
- Gregory J.A., Becker E.C., Pogliano K. (2008). Bacillus subtilis MinC destabilizes FtsZ-rings at new cell poles and contributes to the timing of cell division. Genes Dev. 22: 3475–3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubbels M.J., Vaishnava S., Boot N., Dubremetz J.F., Striepen B. (2006). A MORN-repeat protein is a dynamic component of the Toxoplasma gondii cell division apparatus. J. Cell Sci. 119: 2236–2245. [DOI] [PubMed] [Google Scholar]
- Habicht J., Woehle C., Gould S.B. (2015). Tetrahymena expresses more than a hundred proteins with lipid-binding MORN motifs that can differ in their subcellular localisations. J. Eukaryot. Microbiol. 62: 694–700. [DOI] [PubMed] [Google Scholar]
- Hu Z., Lutkenhaus J. (1999). Topological regulation of cell division in Escherichia coli involves rapid pole to pole oscillation of the division inhibitor MinC under the control of MinD and MinE. Mol. Microbiol. 34: 82–90. [DOI] [PubMed] [Google Scholar]
- Hu Z., Mukherjee A., Pichoff S., Lutkenhaus J. (1999). The MinC component of the division site selection system in Escherichia coli interacts with FtsZ to prevent polymerization. Proc. Natl. Acad. Sci. USA 96: 14819–14824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im Y.J., Davis A.J., Perera I.Y., Johannes E., Allen N.S., Boss W.F. (2007). The N-terminal membrane occupation and recognition nexus domain of Arabidopsis phosphatidylinositol phosphate kinase 1 regulates enzyme activity. J. Biol. Chem. 282: 5443–5452. [DOI] [PubMed] [Google Scholar]
- Irieda H., Shiomi D. (2017). ARC6-mediated Z ring-like structure formation of prokaryote-descended chloroplast FtsZ in Escherichia coli. Sci. Rep. 7: 3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh R., Fujiwara M., Nagata N., Yoshida S. (2001). A chloroplast protein homologous to the eubacterial topological specificity factor minE plays a role in chloroplast division. Plant Physiol. 127: 1644–1655. [PMC free article] [PubMed] [Google Scholar]
- Itoh R.D., Ishikawa H., Nakajima K.P., Moriyama S., Fujiwara M.T. (2018). Isolation and analysis of a stromule-overproducing Arabidopsis mutant suggest the role of PARC6 in plastid morphology maintenance in the leaf epidermis. Physiol. Plant. 162: 479–494. [DOI] [PubMed] [Google Scholar]
- Johnson C.B., Tang L.K., Smith A.G., Ravichandran A., Luo Z., Vitha S., Holzenburg A. (2013). Single particle tracking analysis of the chloroplast division protein FtsZ anchoring to the inner envelope membrane. Microsc. Microanal. 19: 507–512. [DOI] [PubMed] [Google Scholar]
- Johnson C.B., Long Z., Luo Z., Shaik R.S., Sung M.W., Vitha S., Holzenburg A. (2015a). In situ structure of FtsZ mini-rings in Arabidopsis chloroplasts. Adv. Struct. Chem. Imaging 1: 1–12. [Google Scholar]
- Johnson C.B., Shaik R., Abdallah R., Vitha S., Holzenburg A. (2015b). FtsZ1/FtsZ2 turnover in chloroplasts and the role of ARC3. Microsc. Microanal. 21: 313–323. [DOI] [PubMed] [Google Scholar]
- Ju T.-K., Huang F.-L. (2004). MSAP, the meichroacidin homolog of carp (Cyprinus carpio), differs from the rodent counterpart in germline expression and involves flagellar differentiation. Biol. Reprod. 71: 1419–1429. [DOI] [PubMed] [Google Scholar]
- Keeling P.J. (2010). The endosymbiotic origin, diversification and fate of plastids. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365: 729–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi S., Bédard J., Hirano M., Hirabayashi Y., Oishi M., Imai M., Takase M., Ide T., Nakai M. (2013). Uncovering the protein translocon at the chloroplast inner envelope membrane. Science 339: 571–574. [DOI] [PubMed] [Google Scholar]
- Kusano H., Testerink C., Vermeer J.E.M., Tsuge T., Shimada H., Oka A., Munnik T., Aoyama T. (2008). The Arabidopsis phosphatidylinositol phosphate 5-kinase PIP5K3 is a key regulator of root hair tip growth. Plant Cell 20: 367–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushnirov V.V. (2000). Rapid and reliable protein extraction from yeast. Yeast 16: 857–860. [DOI] [PubMed] [Google Scholar]
- Lee J., Han C.-T., Hur Y. (2010). Overexpression of BrMORN, a novel ‘membrane occupation and recognition nexus’ motif protein gene from Chinese cabbage, promotes vegetative growth and seed production in Arabidopsis. Mol. Cells 29: 113–122. [DOI] [PubMed] [Google Scholar]
- Lee J., Han C.-T., Kim H., Hur Y. (2014). A MORN-domain protein regulates growth and seed production and enhances freezing tolerance in Arabidopsis. Plant Biotechnol. Rep. 8: 229–241. [Google Scholar]
- Leger M.M., Petrů M., Žárský V., Eme L., Vlček Č., Harding T., Lang B.F., Eliáš M., Doležal P., Roger A.J. (2015). An ancestral bacterial division system is widespread in eukaryotic mitochondria. Proc. Natl. Acad. Sci. USA 112: 10239–10246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Q., Lu X., Jiang L., Wang C., Fan Y., Zhang C. (2010). EMB1211 is required for normal embryo development and influences chloroplast biogenesis in Arabidopsis. Physiol. Plant. 140: 380–394. [DOI] [PubMed] [Google Scholar]
- Lorestani A., Sheiner L., Yang K., Robertson S.D., Sahoo N., Brooks C.F., Ferguson D.J.P., Striepen B., Gubbels M.-J. (2010). A Toxoplasma MORN1 null mutant undergoes repeated divisions but is defective in basal assembly, apicoplast division and cytokinesis. PLoS One 5: e12302–e12315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löwe J., Amos L.A. (1998). Crystal structure of the bacterial cell-division protein FtsZ. Nature 391: 203–206. [DOI] [PubMed] [Google Scholar]
- Lutkenhaus J. (2007). Assembly dynamics of the bacterial MinCDE system and spatial regulation of the Z ring. Annu. Rev. Biochem. 76: 539–562. [DOI] [PubMed] [Google Scholar]
- Ma X., Margolin W. (1999). Genetic and functional analyses of the conserved C-terminal core domain of Escherichia coli FtsZ. J. Bacteriol. 181: 7531–7544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H., Lou Y., Lin W.H., Xue H.W. (2006). MORN motifs in plant PIPKs are involved in the regulation of subcellular localization and phospholipid binding. Cell Res. 16: 466–478. [DOI] [PubMed] [Google Scholar]
- MacCready J.S., Schossau J., Osteryoung K.W., Ducat D.C. (2017). Robust Min-system oscillation in the presence of internal photosynthetic membranes in cyanobacteria. Mol. Microbiol. 103: 483–503. [DOI] [PubMed] [Google Scholar]
- Maple J., Aldridge C., Møller S.G. (2005). Plastid division is mediated by combinatorial assembly of plastid division proteins. Plant J. 43: 811–823. [DOI] [PubMed] [Google Scholar]
- Maple J., Vojta L., Soll J., Møller S.G. (2007). ARC3 is a stromal Z-ring accessory protein essential for plastid division. EMBO Rep. 8: 293–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka Y., Nishimura H., Numazawa K., Tsuchida J., Miyagawa Y., Tsujimura A., Matsumiya K., Okuyama A., Nishimune Y., Tanaka H. (2005). Sperm flagella protein components: Human meichroacidin constructed by the membrane occupation and recognition nexus motif. Reprod. Med. Biol. 4: 213–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAndrew R.S., Froehlich J.E., Vitha S., Stokes K.D., Osteryoung K.W. (2001). Colocalization of plastid division proteins in the chloroplast stromal compartment establishes a new functional relationship between FtsZ1 and FtsZ2 in higher plants. Plant Physiol. 127: 1656–1666. [PMC free article] [PubMed] [Google Scholar]
- Mikami K., Saavedra L., Hiwatashi Y., Uji T., Hasebe M., Sommarin M. (2010). A dibasic amino acid pair conserved in the activation loop directs plasma membrane localization and is necessary for activity of plant type I/II phosphatidylinositol phosphate kinase. Plant Physiol. 153: 1004–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagishima S.-Y. (2011). Mechanism of plastid division: from a bacterium to an organelle. Plant Physiol. 155: 1533–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagishima Sy, Takahara M., Mori T., Kuroiwa H., Higashiyama T., Kuroiwa T. (2001). Plastid division is driven by a complex mechanism that involves differential transition of the bacterial and eukaryotic division rings. Plant Cell 13: 2257–2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagishima S.-Y., Nozaki H., Nishida K., Nishida K., Matsuzaki M., Kuroiwa T. (2004). Two types of FtsZ proteins in mitochondria and red-lineage chloroplasts: The duplication of FtsZ is implicated in endosymbiosis. J. Mol. Evol. 58: 291–303. [DOI] [PubMed] [Google Scholar]
- Miyagishima S.Y., Froehlich J.E., Osteryoung K.W. (2006). PDV1 and PDV2 mediate recruitment of the dynamin-related protein ARC5 to the plastid division site. Plant Cell 18: 2517–2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagishima S.-Y., Nakanishi H., Kabeya Y. (2011). Structure, regulation, and evolution of the plastid division machinery. Int. Rev. Cell Mol. Biol. 291: 115–153. [DOI] [PubMed] [Google Scholar]
- Monahan L.G., Liew A.T.F., Bottomley A.L., Harry E.J. (2014). Division site positioning in bacteria: one size does not fit all. Front. Microbiol. 5: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morriswood B., Schmidt K. (2015). A MORN repeat protein facilitates protein entry into the flagellar pocket of Trypanosoma brucei. Eukaryot. Cell 14: 1081–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller-Roeber B., Pical C. (2002). Inositol phospholipid metabolism in Arabidopsis. Characterized and putative isoforms of inositol phospholipid kinase and phosphoinositide-specific phospholipase C. Plant Physiol. 130: 22–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi H., Suzuki K., Kabeya Y., Miyagishima S.Y. (2009). Plant-specific protein MCD1 determines the site of chloroplast division in concert with bacteria-derived MinD. Curr. Biol. 19: 151–156. [DOI] [PubMed] [Google Scholar]
- Olson B.J.S.C., Wang Q., Osteryoung K.W. (2010). GTP-dependent heteropolymer formation and bundling of chloroplast FtsZ1 and FtsZ2. J. Biol. Chem. 285: 20634–20643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa M., Erickson H.P. (2011). Inside-out Z rings–Constriction with and without GTP hydrolysis. Mol. Microbiol. 81: 571–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osteryoung K.W., McAndrew R.S. (2001). The plastid division machine. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52: 315–333. [DOI] [PubMed] [Google Scholar]
- Osteryoung K.W., Pyke K.A. (2014). Division and dynamic morphology of plastids. Annu. Rev. Plant Biol. 65: 443–472. [DOI] [PubMed] [Google Scholar]
- Osteryoung K.W., Vierling E. (1995). Conserved cell and organelle division. Nature 376: 473–474. [DOI] [PubMed] [Google Scholar]
- Osteryoung K.W., Stokes K.D., Rutherford S.M., Percival A.L., Lee W.Y. (1998). Chloroplast division in higher plants requires members of two functionally divergent gene families with homology to bacterial ftsZ. Plant Cell 10: 1991–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottesen E., Zhong R., Lamppa G.K. (2010). Identification of a chloroplast division mutant coding for ARC6H, an ARC6 homolog that plays a nonredundant role. Plant Sci. 178: 114–122. [Google Scholar]
- Park K.-T., Dajkovic A., Wissel M., Du S., and Lutkenhaus J. (2018). MinC and FtsZ mutant analysis provides insight into MinC/MinD-mediated Z Ring disassembly. J. Biol. Chem. 293: 5834–5846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyke K.A., Leech R.M. (1991). Rapid image analysis screening procedure for identifying chloroplast number mutants in mesophyll cells of Arabidopsis thaliana (L.) Heynh. Plant Physiol. 96: 1193–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyke K.A., Leech R.M. (1992). Chloroplast division and expansion is radically altered by nuclear mutations in Arabidopsis thaliana. Plant Physiol. 99: 1005–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raskin D.M., de Boer P.A. (1999a). MinDE-dependent pole-to-pole oscillation of division inhibitor MinC in Escherichia coli. J. Bacteriol. 181: 6419–6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raskin D.M., de Boer P.A. (1999b). Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli. Proc. Natl. Acad. Sci. USA 96: 4971–4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothfield L., Taghbalout A., Shih Y.-L. (2005). Spatial control of bacterial division-site placement. Nat. Rev. Microbiol. 3: 959–968. [DOI] [PubMed] [Google Scholar]
- Rowlett V.W., Margolin W. (2013). The bacterial Min system. Curr. Biol. 23: R553–R556. [DOI] [PubMed] [Google Scholar]
- Satouh Y., Padma P., Toda T., Satoh N., Ide H., Inaba K. (2005). Molecular characterization of radial spoke subcomplex containing radial spoke protein 3 and heat shock protein 40 in sperm flagella of the ascidian Ciona intestinalis. Mol. Biol. Cell 16: 626–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz A.J., Glynn J.M., Olson B.J.S.C., Stokes K.D., Osteryoung K.W. (2009). Arabidopsis FtsZ2-1 and FtsZ2-2 are functionally redundant, but FtsZ-based plastid division is not essential for chloroplast partitioning or plant growth and development. Mol. Plant 2: 1211–1222. [DOI] [PubMed] [Google Scholar]
- Shaik R.S., Sung M.W., Vitha S., Holzenburg A. (2018). Chloroplast division protein ARC3 acts on FtsZ2 by preventing filament bundling and enhancing GTPase activity. Biochem. J. 475: 99–115. [DOI] [PubMed] [Google Scholar]
- Shaner N.C., Lambert G.G., Chammas A., Ni Y., Cranfill P.J., Baird M.A., Sell B.R., Allen J.R., Day R.N., Israelsson M., Davidson M.W., Wang J. (2013). A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum. Nat. Methods 10: 407–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada H., Koizumi M., Kuroki K., Mochizuki M., Fujimoto H., Ohta H., Masuda T., Takamiya K. (2004). ARC3, a chloroplast division factor, is a chimera of prokaryotic FtsZ and part of eukaryotic phosphatidylinositol-4-phosphate 5-kinase. Plant Cell Physiol. 45: 960–967. [DOI] [PubMed] [Google Scholar]
- Sprague B.L., Pego R.L., Stavreva D.A., McNally J.G. (2004). Analysis of binding reactions by fluorescence recovery after photobleaching. Biophys. J. 86: 3473–3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan R., Mishra M., Murata-Hori M., Balasubramanian M.K. (2007). Filament formation of the Escherichia coli actin-related protein, MreB, in fission yeast. Curr. Biol. 17: 266–272. [DOI] [PubMed] [Google Scholar]
- Srinivasan R., Mishra M., Wu L., Yin Z., Balasubramanian M.K. (2008). The bacterial cell division protein FtsZ assembles into cytoplasmic rings in fission yeast. Genes Dev. 22: 1741–1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenzel I., Ischebeck T., Quint M., Heilmann I. (2012). Variable regions of PI4P 5-kinases direct PtdIns(4,5)P(2) toward alternative regulatory functions in tobacco pollen tubes. Front. Plant Sci. 2: 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes K.D., McAndrew R.S., Figueroa R., Vitha S., Osteryoung K.W. (2000). Chloroplast division and morphology are differentially affected by overexpression of FtsZ1 and FtsZ2 genes in Arabidopsis. Plant Physiol. 124: 1668–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strepp R., Scholz S., Kruse S., Speth V., Reski R. (1998). Plant nuclear gene knockout reveals a role in plastid division for the homolog of the bacterial cell division protein FtsZ, an ancestral tubulin. Proc. Natl. Acad. Sci. USA 95: 4368–4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung M.W., Shaik R., TerBush A.D., Osteryoung K.W., Vitha S., Holzenburg A. (2018). The chloroplast division protein ARC6 acts to inhibit disassembly of GDP-bound FtsZ2. J. Biol. Chem. 293: 10692–10706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshima H., Komazaki S., Nishi M., Iino M., Kangawa K. (2000). Junctophilins: A novel family of junctional membrane complex proteins. Mol. Cell 6: 11–22. [DOI] [PubMed] [Google Scholar]
- TerBush A.D., Osteryoung K.W. (2012). Distinct functions of chloroplast FtsZ1 and FtsZ2 in Z-ring structure and remodeling. J. Cell Biol. 199: 623–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TerBush A.D., Yoshida Y., Osteryoung K.W. (2013). FtsZ in chloroplast division: structure, function and evolution. Curr. Opin. Cell Biol. 25: 461–470. [DOI] [PubMed] [Google Scholar]
- TerBush A.D., Porzondek C.A., Osteryoung K.W. (2016). Functional analysis of the chloroplast division complex using Schizosaccharomyces pombe as a heterologous expression system. Microsc. Microanal. 22: 275–289. [DOI] [PubMed] [Google Scholar]
- TerBush A.D., MacCready J.S., Chen C., Ducat D.C., Osteryoung K.W. (2018). Conserved dynamics of chloroplast cytoskeletal FtsZ proteins across photosynthetic lineages. Plant Physiol. 176: 295–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dooren G.G., Reiff S.B., Tomova C., Meissner M., Humbel B.M., Striepen B. (2009). A novel dynamin-related protein has been recruited for apicoplast fission in Toxoplasma gondii. Curr. Biol. 19: 267–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitha S., Osteryoung K.W. (2011). Immunofluorescence microscopy for localization of Arabidopsis chloroplast proteins. Methods Mol. Biol. 774: 33–58. [DOI] [PubMed] [Google Scholar]
- Vitha S., McAndrew R.S., Osteryoung K.W. (2001). FtsZ ring formation at the chloroplast division site in plants. J. Cell Biol. 153: 111–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitha S., Froehlich J.E., Koksharova O., Pyke K.A., van Erp H., Osteryoung K.W. (2003). ARC6 is a J-domain plastid division protein and an evolutionary descendant of the cyanobacterial cell division protein Ftn2. Plant Cell 15: 1918–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., et al. (2017). Structural insights into the coordination of plastid division by the ARC6-PDV2 complex. Nat. Plants 3: 17011. [DOI] [PubMed] [Google Scholar]
- Yang Y., Glynn J.M., Olson B.J.S.C., Schmitz A.J., Osteryoung K.W. (2008). Plastid division: Across time and space. Curr. Opin. Plant Biol. 11: 577–584. [DOI] [PubMed] [Google Scholar]
- Yoder D.W., Kadirjan-Kalbach D., Olson B.J.S.C., Miyagishima S.Y., Deblasio S.L., Hangarter R.P., Osteryoung K.W. (2007). Effects of mutations in Arabidopsis FtsZ1 on plastid division, FtsZ ring formation and positioning, and FtsZ filament morphology in vivo. Plant Cell Physiol. 48: 775–791. [DOI] [PubMed] [Google Scholar]
- Yoshida Y., Mogi Y., TerBush A.D., Osteryoung K.W. (2016). Chloroplast FtsZ assembles into a contractible ring via tubulin-like heteropolymerization. Nat. Plants 2: 16095. [DOI] [PubMed] [Google Scholar]
- Zhang M., Hu Y., Jia J., Li D., Zhang R., Gao H., He Y. (2009). CDP1, a novel component of chloroplast division site positioning system in Arabidopsis. Cell Res. 19: 877–886. [DOI] [PubMed] [Google Scholar]
- Zhang M., Schmitz A.J., Kadirjan-Kalbach D.K., Terbush A.D., Osteryoung K.W. (2013). Chloroplast division protein ARC3 regulates chloroplast FtsZ-ring assembly and positioning in Arabidopsis through interaction with FtsZ2. Plant Cell 25: 1787–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Chen C., Froehlich J.E., TerBush A.D., Osteryoung K.W. (2016). Roles of Arabidopsis PARC6 in coordination of the chloroplast division complex and negative regulation of FtsZ assembly. Plant Physiol. 170: 250–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Henriques R., Lin S.-S., Niu Q.-W., Chua N.-H. (2006). Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 1: 641–646. [DOI] [PubMed] [Google Scholar]
- Zimorski V., Ku C., Martin W.F., Gould S.B. (2014). Endosymbiotic theory for organelle origins. Curr. Opin. Microbiol. 22: 38–48. [DOI] [PubMed] [Google Scholar]