Abstract
BACKGROUND:
Hepatitis E virus (HEV) can inapparently infect blood donors. To assess transfusion transmission of HEV in the United States, which has not been documented, a donor-recipient repository was evaluated.
STUDY DESIGN AND METHODS:
To identify donations that contained HEV RNA and were linked to patient-recipients with antibody evidence of HEV exposure, we assayed samples from the Retrovirus Epidemiology Donor Study (REDS) Allogeneic Donor and Recipient repository that represents 13,201 linked donations and 3384 transfused patients. Post-transfusion samples, determined to contain IgG anti-HEV by ELISA, were re-assayed along with corresponding pre-transfusion samples for seroconversion (incident exposure) or ≥4-fold IgG anti-HEV increase (re-exposure). HEV-exposed patients were linked to donations in which HEV RNA was then detected by RT-qPCR, confirmed by Transcription Mediated Amplification (TMA), and phylogenetically analyzed as sub-genomic cDNA sequences.
RESULTS:
Among all patients, 19 of 1036 (1.8%) who had IgG anti-HEV before transfusion were re-exposed; 40 of 2348 (1.7%) without pre-transfusion IgG anti-HEV seroconverted. These 59 patients were linked to 257 donations, 1 of which was positive by RT-qPCR and TMA. Plasma from this donation contained 5.5 log10 IU/mL of HEV RNA that grouped with HEV genotype 3, clade 3abchij. The patient-recipient of pRBC from this donation had a >8-fold IgG increase; however, clinical data are unavailable.
CONCLUSIONS:
This is the first report of probable HEV transmission via transfusion in the US, although it has been frequently observed in Europe and Japan. Additional data on the magnitude of the risk in the US are needed.
Keywords: hepatitis E virus, transfusion-transmitted virus, hepatitis, IgG anti-viral antibodies, viral RNA
INTRODUCTION
Hepatitis E virus (HEV) is a global pathogen that, among humans, is represented by a single serotype with four genotypes 1,2. The virus is commonly acquired by enteric transmission and, in developing countries, genotypes 1 and 2 can cause large waterborne epidemics associated with monsoon rain or in humanitarian emergencies with contaminated supplies of drinking water. More recently, autochthonous HEV infections have been frequently reported among populations in industrialized countries. Such infections are associated with genotypes 3 or 4, and usually occur as isolated cases or in small clusters. They commonly include asymptomatic infections of adults who acquire HEV from contaminated food, especially solid-organ meats from swine, wild boar, deer, or raw shellfish.
HEV transmission by transfusion has been reported since 2004 from Europe and Japan 3–12. A study of 225,000 southeastern UK donors identified 79 (0.035%) with detectable HEV RNA 8. Among 43 patients who were transfused with these donors’ HEV RNA-containing products, 18 (42%) became infected. Chinese, European, and American investigators have detected HEV RNA in blood products, including pooled plasma, from otherwise acceptable donors 6,13–17. In Japan, a total of 20 patients were reported to have acquired HEV by transfusion of blood products 12. Consequently, blood centers in Hokkaido prefecture of northern Japan have routinely screened donors for HEV RNA during the past ten years to prevent transmission by transfusion 11,12.
Despite these international reports, limited data have been reported from blood centers in the United States. A study of 1939 donors at the National Institutes of Health Clinical Center (NIH CC, Bethesda, MD), who were sampled in 2006 and 2012, found 18.8% with IgG anti-HEV and 0.4% with IgM anti-HEV but none had detectable HEV RNA 18. A study of 18,829 American Red Cross (ARC) donation samples, collected during 2013, identified 2 (0.01%) with HEV RNA, 7.7% with IgG anti-HEV, and 0.58% with IgM anti-HEV 16. Another study of ARC donors, 5040 who were sampled in 2015, detected IgG anti-HEV among 11.4%; 0.18% had IgM anti-HEV detected by each of three assays among which there was, however, only 22% agreement 19. HEV transmission in these US studies could not be assessed, however, because donations were not linked to blood-product recipients.
To evaluate the risk of HEV transmission by transfusion in a US population, we tested samples from the Retrovirus Epidemiology Donor Study (REDS) Allogeneic Donor and Recipient (RADAR) repository 20. This collection was organized between 2000 and 2003 by seven US blood centers. It links 13,201 donations, from 12,408 donors, with 3575 patients in eight California, Florida, Maryland, Michigan, Oklahoma, and Pennsylvania hospitals; these patients had cardiac, vascular or orthopedic operations. The RADAR repository contains plasma samples from donors, and paired plasma samples that were collected from patients before or immediately after transfusion, and 6 to 12 months later.
MATERIALS AND METHODS
Patient-recipient and donation samples
The RADAR repository 20 is maintained by BioLINCC (Biologic Specimen and Data Repository Information Coordinating Center, NHLBI, NIH; c/o Information Management Services, Calverton, MD; see Web Resources). We initially obtained all 3384 post-transfusion samples that were available from the 3575 patient-recipients, and subsequently obtained selected pre-transfusion and donation samples according to the testing algorithm below.
Reference materials
We conducted limited assessments of assay performance with two World Health Organization (WHO) reference materials and a characterized research-specimen. These WHO materials were: WHO Reference Reagent for HEV Antibody, reconstituted with water to 100 U/mL (NIBSC code 95/584; National Institute for Biological Standards and Control, Potters Bar, Hertfordshire, UK); and 1st WHO International Standard for Hepatitis E Virus RNA Nucleic Acid Amplification Techniques-Based Assays, reconstituted with water to 5.7 log10 International Units (IU) per mL (PEI code 6329/10; Paul Ehrlich Institut, Langen, Germany). The latter contains genotype 3 strain HRC-HE104, complete genomic sequence of which has GenBank accession AB630970 21. The characterized research-specimen was bile, containing ≈ 9.8 log10 IU/mL of HEV subtype 2a, from a cynomolgus monkey that was experimentally infected with strain Mexico-14 in human feces; GenBank M74506 and KX578717 correspond to HEV in the fecal specimen 22–24.
Testing algorithm for repository specimens
Our approach was intended to identify patients who had antibody evidence of HEV exposure during the pre- to post-transfusion sampling interval, and then HEV RNA-containing donations that were likely sources of such exposure (Figure 1). We have used “exposure”, rather than “infection”, because the latter term might imply degrees of HEV replication and HEV-associated disease that could not be determined.
FIGURE 1. Testing algorithm for patient-recipient and donation samples from the RADAR repository.
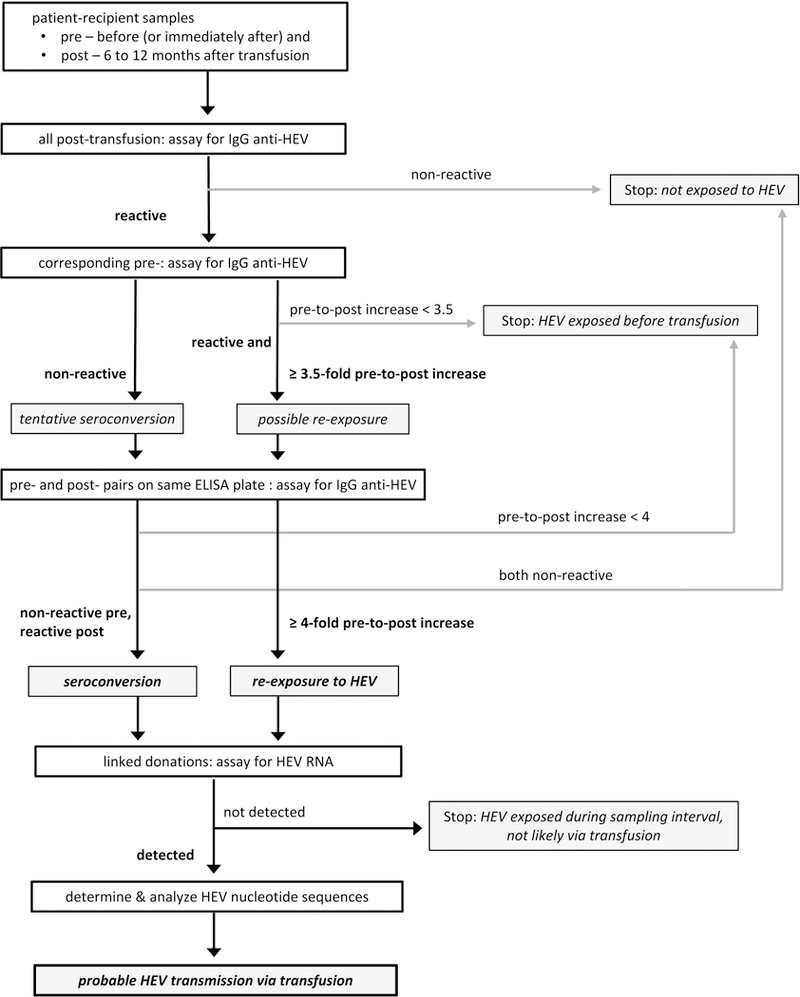
We assayed post-transfusion specimens for IgG anti-HEV and, for those with reactive results, tested corresponding pre-transfusion specimens in subsequent assay-runs. Each specimen-pair that yielded preliminary evidence of HEV exposure during the sampling interval, as manifested by seroconversion or by ≥ 3.5-fold increase in IgG anti-HEV S/CO value, was re-assayed on a single ELISA plate. We then assayed for HEV RNA in donations that were linked to patients who had single-plate confirmed seroconversion or ≥ 4-fold increase of IgG anti-HEV concentration. Finally, we determined and analyzed partial nucleotide sequences of any detected HEV RNA in donation samples.
Detection and semi-quantitation of IgG antibodies to HEV
We assayed patients’ plasma specimens by using a commercially available enzyme-linked immunosorbent assay (ELISA), Wantai HEV-IgG ELISA (WE-7296, Beijing Wantai Biological Pharmacy Enterprise, Beijing, PRC), generally following manufacturer’s instructions. Minor modifications included re-assaying selected patients’ specimens, which had yielded post-transfusion IgG anti-HEV sample-to-cutoff values (S/COpost) > 6.0, diluted 1:4 or 1:8 in phosphate-buffered saline, pH 7.0, with 1% wt/vol bovine serum albumin (PBS/BSA). By assaying such 2-fold dilutions of the WHO Reference Reagent for HEV Antibody, we determined analytical sensitivity of the IgG anti-HEV ELISA to be 1 U/mL according to pre-assay concentration, or 0.091 U/mL in-ELISA (Supplementary Table 1).
When a post-transfusion specimen was non-reactive for IgG anti-HEV, the corresponding pre-transfusion sample was not tested, and the patient interpreted as not HEV exposed before or during the sampling interval. When a post-transfusion specimen was reactive, we tested the corresponding pre-transfusion sample and compared results with those obtained earlier for post-transfusion specimens. We defined patients as having tentatively seroconverted when pre-transfusion results were non-reactive or equivocal (S/COpre < 1.1), and possibly increased IgG anti-HEV concentration when S/COpost was at least 3.5-fold higher than S/COpre ≥ 1.
We then re-assayed, on the same ELISA plate, each specimen-pair that yielded such preliminary evidence of sampling-interval exposure. Same-plate IgG anti-HEV results were used to identify (a) incident exposure, or seroconversion, defined as pre-transfusion non-reactive (S/COpre < 1) and post-transfusion reactive; (b) re-exposure, as both specimens reactive and post-transfusion concentration at least 4-fold higher than before transfusion (S/COpost after 4-fold dilution ≥ S/COpre); and (c) past exposure, as both specimens reactive and S/COpost after 4-fold dilution ≤ S/COpre or, without dilution, S/COpost < 6.0 and < 4 × S/COpre. Our re-exposure criterion was based on a linear and approximately 1:1 correlation between WHO U/mL and IgG anti-HEV S/CO ranging from 0.25 to 6.0 (Supplementary Table 1).
Detection and quantitation of HEV RNA
We tested donation samples, identified as linked to patients who had serologic evidence of HEV exposure during the sampling interval, by using assays that are based on polymerase chain reaction (PCR) or transcription-mediated amplification (TMA).
To generate templates for reverse-transcription quantitative polymerase chain reaction (RT-qPCR) and for sequence analysis (below), we added ≈ 4.7 log10 pfu of coliphage MS2 25 and 8 μg of yeast tRNA to 200 μL of plasma or reference material. We then purified RNA and DNA by using MagNA Pure LC Total Nucleic Acid Isolation - High Performance kits with the MagNA Pure LC 2.0 instrument (Roche Diagnostics, Indianapolis, IN), eluting into 100 μL of proprietary (Roche) buffer.
We detected and quantified HEV RNA with a RT-qPCR assay that our Johns Hopkins Bloomberg School of Public Health (JHBSPH) laboratory implemented for environmental and plasma samples 26,27 and then adapted to increase sensitivity and throughput. We mixed 5 μL of purified nucleic acids into a 20-μL reaction with VeriQuest Probe One-Step qRT-PCR Master Mix (Affymetrix/USB, Santa Clara, CA) and oligonucleotide sets for amplifying a highly conserved segment of the HEV genome 28 (primers, 500 nM; probe, 250 nM) and for MS2 25 (primers, 250 nM; probe, 125 nM); for sequences, see Supplementary Table 2. RNAs were reverse transcribed and then amplified in an Applied Biosystems StepOnePlus Real-Time PCR system (Thermo Fisher Scientific, Foster City, CA) by incubating at 50° C for 15 min and 95° C for 10 min; and then 45 cycles of 95° C for 15 sec, 55° C for 20 sec, and 60° C for 20 sec. The quantitation standard was cloned HEV complementary DNA (cDNA), assayed as a 10-fold dilution series of concentrations between 0.5 log10 and 5.5 log10 copies/reaction. Samples that yielded an HEV threshold-cycle (CT) value ≤ 38.0 were considered to be positive. Analytical sensitivity was 2.5 log10 IU/mL of plasma, or 0.5 log10 IU/reaction; 0.5 log10 IU corresponded to 1.5 log10 copies of cloned HEV cDNA.
To confirm selected RT-qPCR results, donation specimens were tested with the TMA-based Procleix HEV assay (Hologic, San Diego, CA; and Grifols Diagnostic Solutions, Emeryville, CA) that has a 95% detection probability of 0.90 log10 IU/mL 16. This assay requires 0.7 mL of specimen for singulate testing; because the volume of many RADAR samples is extremely limited, selected samples were diluted as much as 8-fold (i.e., 0.1 mL of sample with 0.7 mL of proprietary buffer).
Determination and analysis of HEV cDNA nucleotide sequences
We synthesized and then amplified HEV cDNA via nested PCR with primers that represent segments of HEV open reading frame (ORF) 1 and ORF2 29–31 (Supplementary Table 2; JR Ticehurst and MS Forman, unpublished data). Sanger sequence-reads were generated from nested PCR products by using an Applied Biosystems 3500 Genetic Analyzer and then base-called, trimmed to amplicon-length without primers, and assembled by Aligner v8.0.1 (CodonCode, Centerville, MA) and BioEdit v 7.2.5 (for availability, see Web Resources) to yield sequences for phylogenetic analysis.
We constructed maximum-likelihood phylogenetic trees via PhyML 32 at the website of Le Laboratoire d’Informatique, de Robotique et de Microélectronique de Montpellier (Université Montpellier, Montpellier, France; see Web Resources) with this laboratory’s default parameters; we did not choose optional Gblocks curation 33. Before submitting for analysis, we used BioEdit v 7.2.5 to trim reference-sequences to RADAR HEV cDNA length, and to align all sequences via ClustalW 34 (included with BioEdit). We initially constructed a partial-ORF1 tree by using 158 unique HEV reference-sequences 35 and the Approximate Likelihood-Ratio Test for branch-assessment 33. For clarity in this presentation, we re-made the partial-ORF1 tree with 32 taxa, 27 of which represent consensus reference strains for the four human HEV genotypes and a genotype 5 representative as outgroup 36, plus 3 other well characterized human strains 35. For this tree, we statistically assessed branches by bootstrapping with 100 re-samplings, outgroup-rendered the tree with TreeView v.1.6.6 (for availability, see Web Resources), and annotated it by using PowerPoint 2016 (Microsoft, Redmond, WA). We similarly generated trees from partial ORF2 sequences (not shown). We also compared nucleotide sequences representing our JHBSPH laboratory’s HEV strains to determine if those representing the RADAR-donation are unique; i.e., not the result of contamination.
RESULTS
IgG anti-HEV in single and paired specimens from RADAR patient-recipients
Among all 3384 patients, 1036 (30.6%) had detectable IgG anti-HEV before transfusion (Table 1). Based on changes in IgG anti-HEV reactivity at 6 to 12 months after transfusion, 59 patients (1.7%) were determined to have seroconverted or been re-exposed after the pre-transfusion specimen was collected. Incident exposures occurred in 40 among 2348 patients (1.7% of 2308 + 40; Table 1) who had not been HEV-exposed before transfusion. The 19 re-exposures represent 1.8% of the 1036 previously exposed patients.
TABLE 1.
Evidence of HEV exposure among 3384 RADAR patients, based on comparison of pre- and post-transfusion results for IgG anti-HEV
| After transfusion |
Before transfusion |
Pre- to
post-transfusion |
||||
|---|---|---|---|---|---|---|
| IgG anti-HEV result | No. | IgG anti-HEV result | No. | Change | No. | Interpretation |
| Non-reactive | 40 | Seroconversion | 40 | Incident exposure | ||
| Reactive | 1076 | Reactive | 1036 | ≥ 4-fold increase* | 19 | Re-exposure |
| < 4-fold increase | 1017 | Past exposure (without re-exposure) | ||||
| Non-reactive | 2308 | Not tested (presumed non-reactive) | 2308 | None determined | 2308 | No exposure |
| Total | 3384 | |||||
≥ 4-fold increase, S/CO value of 1:4 diluted post-transfusion sample greater than or equal to S/CO value of undiluted pre-transfusion sample (see Materials and Methods and Supplementary Table 1).
Detection and analysis of HEV RNA
The 59 RADAR patients who had evidence of HEV exposure were linked to 257 donations from 257 donors, all of which were assayed for HEV RNA by RT-qPCR. Fifteen (5.8% of 257) were positive: one yielded a CT of 26.9, the fourteen other CT values ranged between 33.9 and 37.5. Seventeen of these 257 donations, including 14 RT-qPCR positives and 1 that yielded an invalid (MS2 internal control not detected) result, were also assayed by using the Procleix HEV assay. One (0.008% of 13,201 linked donations) RT-qPCR-positive was confirmed, that for which CT = 26.9. We also RT-qPCR assayed remaining plasma (50 μL pre-transfusion, 200 μL post-transfusion) from the recipient of the HEV RNA-containing donation; neither had detectable HEV RNA.
By RT-qPCR, the HEV RNA-confirmed specimen contained 5.5 log10 IU per mL of plasma. This RNA phylogenetically represents HEV genotype 3, clade 3abchij 37, based on HEV ORF1 (Figure 2) and ORF2 (data not shown) nucleotide sequences that are also distinct from those of all other strains in our JHBSPH laboratory. Analogous subgenomic sequences of co-amplified cDNAs that represent the 1st WHO International Standard for HEV RNA (genotype 3, grouping with clade 3abchij 37), are identical to those in GenBank and are represented in Figure 2. Sequences representing HEV subtype 2a strain Mexico-14 in monkey bile, determined from separately amplified cDNA, are 99.7-100% identical to those in GenBank. (See Web Resources for new accession numbers.)
FIGURE 2. Phylogenetic tree of a 530-nucleotide segment of HEV ORF1 from 31 reference-taxa and a RADAR donation.
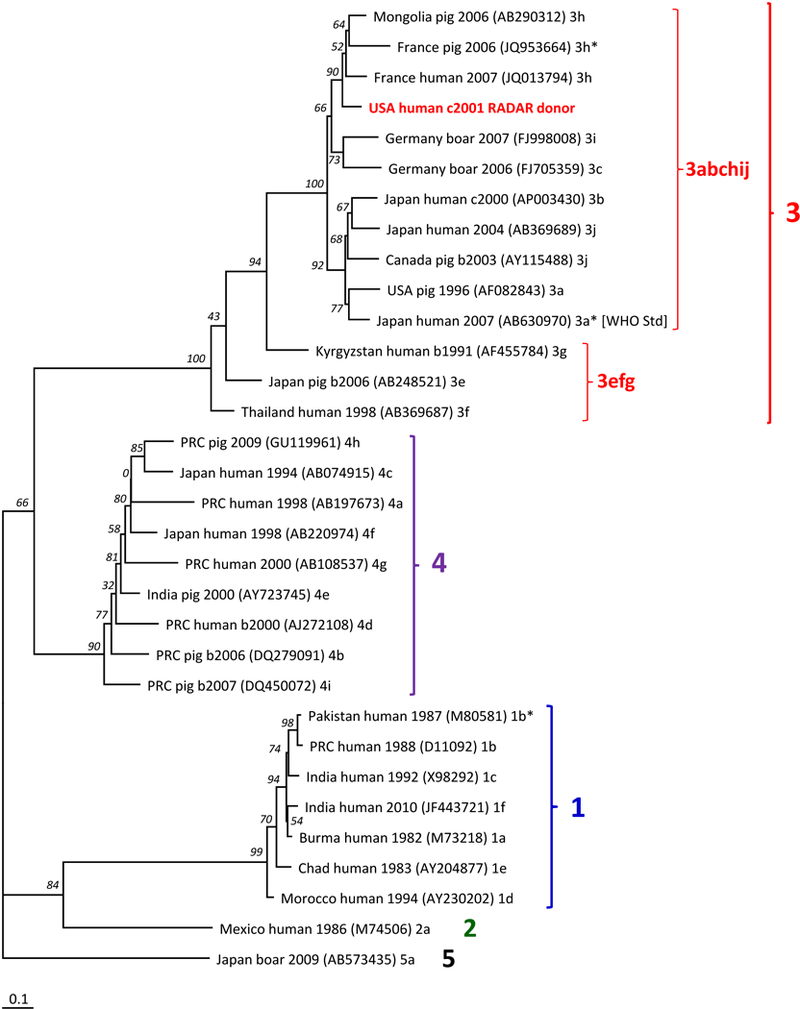
This tree is a rectangular phylogram with an HEV genotype 5 outgroup. Bootstrap values, as per cent of 100 re-samplings, are indicated by italicized numerals near branch-points. Reference-sequences 35,36 are designated by country; host; collection-year (b, before the earlier of GenBank deposition or publication; c, circa, the midpoint in a range of possible years); GenBank accession number, in parentheses; [WHO Std], 1st WHO International Standard for Hepatitis E Virus RNA Nucleic Acid Amplification Techniques-Based Assays; and clade assignment by Smith et al. 36,37 or, with asterisk, Vina-Rodriguez et al. 35. All recognized human subtypes of genotypes 1, 2, 3, and 4 are represented except 3d, for which only ORF2 sequences have been reported 36, and 3ra that primarily represents rabbits and for which there is one reported human-strain sequence that includes the pertinent ORF1 segment 57,58. “USA human c2001 RADAR donor” designates sequence from this study. Largest numerals and brackets indicate genotypes; numeral 3 followed by letters indicate proposed monophyletic groups 37. Bar indicates genetic distance.
Characteristics of the donation with detectable HEV RNA and selected patient-recipients
The HEV RNA–containing donation was from an individual who made a single donation that was documented in the RADAR archive. This donation was transfused as packed red blood cells (pRBC) to one patient who received three other pRBC units, each from one donor. This patient’s IgG anti-HEV concentration increased more than 8-fold after transfusion; i.e., S/CO of 1:8 diluted post-transfusion sample was greater than S/CO of neat pre-transfusion sample (Table 2). As noted above, HEV RNA was not detected in either of this patient’s specimens.
TABLE 2.
IgG anti-HEV S/CO values for recipient of HEV RNA-containing packed red blood cells, demonstrating increased concentration of IgG anti-HEV after transfusion *
| pre-transfusion |
post-transfusion |
|||||
|---|---|---|---|---|---|---|
| ELISA plates | neat | 1:4 | 1:8 | neat | 1:4 | 1:8 |
| separate (initial-testing runs) | 8.91 | 17.11 | ||||
| same (re-assay run) | 10.16 | 3.63 | 1.29 | 16.15 | 14.93 | 14.12 |
neat, assayed without dilution; 1:4 and 1:8, assayed after respectively diluting 4- and 8-fold in PBS/BSA.
Nine of the other 58 IgG anti-HEV seroconversions and re-exposures were linked to a HEV RNA-negative donation that was also linked to a second recipient. In two such instances, both recipients seroconverted. Otherwise, the second recipient did not have evidence of exposure during the sampling interval: both specimens were reactive for IgG anti-HEV without a pre-to-post-transfusion increase, or both were non-reactive (data not shown).
It is not known if any donor or patient developed symptoms or signs of HEV-associated disease, or if patients had foodborne or other types of exposure to HEV, because repository data do not contain such information about RADAR subjects. Certain demographic characteristics of RADAR donors and patient-recipients are available; BioLINCC and NHLBI do not allow such characteristics in publications, however, because of privacy concerns.
DISCUSSION
Our report provides the first documentation of probable HEV transmission via transfusion in the US, from an HEV RNA-containing donation to a patient who had antibody evidence of HEV exposure. Our data are suggestive of re-exposure because the patient had IgG anti-HEV that increased in concentration after transfusion; IgG anti-HEV evidence of HEV re-exposure has been reported 38. Because the RADAR database does not include subjects’ clinical data, we cannot determine if this patient-recipient developed any HEV-associated illness. Pathogenic association with either clade 3abchij, with which the RADAR-donation HEV RNA phylogenetically grouped (Figure 2), or clade 3efg, was not identified by an analysis of genotype 3 infections in the UK and western Europe during 2003 to 2015 37.
To detect HEV transmission that was temporally associated with transfusion, our strategy was to evaluate all possible incident and secondary exposures by assaying linked donations for HEV RNA (Figure 1). By testing paired recipient-specimens for IgG anti-HEV, we identified patients who had antibody evidence of exposure during the pre-to-post-transfusion interval, thereby reducing the number of donations to assay for HEV RNA. The 59 identified HEV exposures are based on same-plate ELISA testing that reproduced earlier results from separate runs in which identity of specimen pairs was blinded.
One cannot unambiguously conclude that a blood product is the source of HEV transmission unless the donor is determined to circulate infectious HEV, which most likely would require inoculation of a susceptible primate. While more definitive evidence of transfusion transmission would include a donation and linked recipient with identical or nearly identical HEV RNAs, it is extremely unlikely that RADAR post-transfusion samples, like others collected from immunocompetent patients at least 6 months after exposure 12,39–41, would contain HEV RNA. We also cannot rule out a temporal association, without transmission, between the HEV RNA-containing donation and linked patient-recipient: better evidence would include detectable or increased anti-HEV in post-transfusion specimens collected sooner than those in the RADAR repository.
Transfusion transmission accounts for a minority of all HEV infections except possibly those among highly transfusion-dependent patients. Based on an estimated 0.2% annual HEV incidence in the UK, investigators there estimated that the ratio of foodborne to transfusion-acquired HEV was approximately 13:1 42. This ratio may be higher in the US because, among the 59 incident exposures and re-exposures that we identified, only one could be associated with a HEV RNA-containing donation.
We may have underestimated transfusion-transmission risk, however, because RADAR patients also received 11,141 blood-components from donors who were not enrolled in the study and therefore could not be linked to HEV-exposed patient-recipients and screened for HEV RNA 20. In addition, RADAR patients who died less than 6 months after transfusion were not studied because post-transfusion specimens could not be collected. Patients who contributed paired specimens were generally immunocompetent but others who might have been at higher risk of HEV infection (e.g., organ-transplant recipients) were not included 20. We also may have under-detected HEV RNA-containing donations because the confirmatory assay for HEV RNA was considerably more sensitive than that we used for initial HEV RNA detection.
We encountered other limitations that are worth noting. First, our RT-qPCR assay yielded positive results that failed confirmation via the more sensitive TMA-based assay, and we generated HEV sequences only from the TMA-confirmed donation and two HEV RNA reference materials. While it is very unlikely that these false-positives resulted from cross-contamination, several pertinent samples yielded human DNA after nested PCR with primers for HEV ORF1 or ORF2 (data not shown). Computer-assisted searches did not reveal high identity between GenBank human sequences and our RT-qPCR oligonucleotides for HEV ORF3 and coliphage MS2. Other groups have noted failure to reproduce initial HEV RNA detection 16 or have successfully co-amplified HEV and MS2 cDNAs 43. Second, we attempted to generate IgM anti-HEV data with a commercial μ-capture ELISA and an analogous approach to that for detecting IgG anti-HEV. The overall frequency of IgM anti-HEV reactivity was higher than in other studies, and most reactive results were implausible or uninterpretable; some such reactivity may have been caused by non-specific binding between captured IgM or other specimen-material and reagent HEV ORF2 protein (data not shown). Furthermore, sample-collection timing made it impossible to determine if any patient developed IgM anti-HEV soon after transfusion and then “seroreverted” to undetectable when the post-transfusion specimen was collected. While post-transfusion IgM and IgG anti-HEV might be considered as more definitive evidence for exposure, assays for IgM anti-viral antibodies are typically configured to yield predominantly non-reactive results by six months into convalescence. Third, although inter-run repeatability was a requirement for incident- or re-exposure categorization, we cannot exclude the possibility of false-positive IgG anti-HEV results because we did not independently verify reactivity (e.g., via Western blot immunoassay) and our 31% frequency of IgG anti-HEV reactivity is high. Noting that RADAR patient-recipients were older (91% and 74% at least 50 and 60 years, respectively) and predominantly male (54%) 20, our IgG anti-HEV frequency is consistent with several of those reported for older US sub-populations 18,19,44,45; see discussion below about prevalence.
The only other HEV study of US recipients and linked donors, by Xu et al. 18, investigated 362 patients in the NIH CC, Suburban Hospital (both Bethesda, MD), and Children’s National Medical Center (Washington, DC), starting in 2001. Two patient-recipients, including one who received an HEV RNA-containing and a “high titer” anti-HEV product shortly before death, became reactive for IgG anti-HEV but the authors concluded that neither patient had a transfusion-associated exposure to HEV. Another publication reported the results of retrospectively assaying cryopreserved specimens that were collected during the 1960s, prior to routine donor screening for viral markers, from 66 NIH CC cardiac-surgery patients who developed post-transfusion hepatitis: 4 (6%), 20 (31%), and 1 (2%) were respectively infected with hepatitis B virus (HBV), hepatitis C virus (HCV), and HEV 46. It is very likely that many transfusion-transmissions of HEV have gone unrecognized in the US. Linking donations to American recipients has been difficult because most blood-product collection, processing, and distribution are centralized, and the products are often transfused after a substantial interval. Also, foodborne transmission of HEV genotype 3 is likely to be more common than infection from a transfusion in the US, a likelihood with which our data are consistent. In addition, the lack of FDA-licensed assays for detecting serologic or virologic evidence of HEV infection, as well as US clinicians’ unfamiliarity with hepatitis E and extrahepatic manifestations of HEV infection 47, are important barriers to diagnosis.
However, large population-based surveys have documented high anti-HEV prevalence in the general US population. A study by Kuniholm et al. 44, of 18,695 individuals from the Third National Health and Nutrition Examination Survey (NHANES) that represents the 1988–94 US population, found an IgG anti-HEV prevalence of 21% by using an assay that was developed at NIH 48. Another study, using an ELISA that has been reportedly 49 less analytically sensitive than the Wantai HEV-IgG ELISA (that we used) and the NIH-developed assay, determined a decline in IgG anti-HEV prevalence from that in the 1998–94 NHANES population (10%, weighted; 17%, unweighted) to that in the 2005–06 NHANES population (6%, weighted or unweighted) 50. Regardless of diminishing IgG anti-HEV prevalence, which others have recognized 18,51,52, these reports have provided persuasive evidence that HEV infections, which often are subclinical, are common in the US.
Population-based studies have consistently detected increasing IgG anti-HEV seroprevalence with age, and other studies have reported high anti-HEV frequency among older Americans, especially men, who were sampled at about the same time as RADAR patient-recipients. The above-cited study that demonstrated declining NHANES anti-HEV prevalence 50 detected, among subjects who were US-born and at least 50 years old, 25% unweighted IgG anti-HEV reactivity in the 1988–94 NHANES sub-population and 11% in the 2005–06 sub-population. Among 1988–94 NHANES US-born males studied by Kuniholm et al. 44, ≈ 31% of those who were 50 to 59 and ≈ 39% of those ≥ 60 years old had IgG anti-HEV. A 2002 publication 45 reported using the same NIH-developed ELISA as Kuniholm et al. 44, and detecting IgG anti-HEV among 27% of 120 blood donors who were ≥ 50 years old. Among 574 blood donors > 45 years old who were sampled during 2006 at the NIH CC, 30% had IgG anti-HEV detected by a Wantai ELISA 18 that was likely to be similar to the ELISA we used. In a study of more recently collected (during 2015) ARC samples, which included ≈1600 from donors ≥ 50 years old and testing with a Wantai IgG anti-HEV ELISA, reactive frequencies ranged from ≈ 16% for 50-to-55-year-old donors to ≈ 44% for those between 80 and 93 years 19. Incident infections are also likely to be frequent in older men 53; although the frequency of incident HEV exposures among RADAR patient-recipients was higher than that reported for general populations 1,53, our data may reflect higher incidence in an older and predominantly male RADAR population 20.
During recent years, there has been increasing recognition of the risk of transmitting HEV by transfusion outside of the US. While infections with HEV genotype 3 are common among adults and frequently asymptomatic in the US and Europe 1,2, a substantial portion of patients in industrialized countries who need transfusions are immunocompromised. Several European countries and Japan’s Hokkaido Prefecture have considered or adopted selective screening of blood products for transfusion into high risk patients, or routine screening of all donors 11,12,54. The UK has elected to screen all donors for HEV RNA because a high proportion of transfusion recipients, those who are immunocompromised, may be at increased risk of more severe HEV 54; chronic progressive hepatitis E has been reported among immunocompromised patients, especially those with solid organ transplants 12.
A recent publication from the Netherlands concluded that screening of blood donors for HEV could have a reasonable cost-benefit ratio 55. Among US donations, reported HEV RNA detection-frequencies (76, 102, and 23 per million respectively during 2000–03 [this study], 2013 16, and 2015 17) are similar to those for HBV DNA, HCV RNA, and human immunodeficiency virus type 1 RNA (respectively 76, 200, and 28 per million during 2011–12 56), for which testing is currently performed; the latter frequencies are 41 to 240 times higher than corresponding infection-frequencies (1, 0.83, and 0.67 per million 16). It is not known if US infection- and illness-frequencies for HEV are comparable, for example, to those reported for the southeastern UK, where 18 of 43 recipients of HEV RNA-containing blood products became infected, among whom 5 had elevated serum concentrations of alanine aminotransferase, including 1 with clinically apparent hepatitis, and 10 developed prolonged or persistent infection 8. Health-economic analysis, similar to that performed for the Netherlands 55, could be important for the US; however, the data on HEV transmission in the US are too scarce to do such an analysis at present.
In conclusion, we detected one case of likely transfusion transmission of HEV among a population of 3384 transfused patients in the US. These recipients were exposed to approximately 25,000 blood-components among which 13,800 were from linked donations 20. We were able to identify this case even though the RADAR population was much smaller than the UK linked study-population 8. Our study’s source donor who likely transmitted HEV had an HEV RNA plasma-concentration of 5.5 log10 IU/mL. This level of HEV RNA was consistently associated with HEV transmission from donors in the large UK study 8, and is much greater than those of the two HEV RNA positive donors in the ARC study 16 or the three in a recent study of US plasma-donors 17. Our report documents for the first time that the risk of transfusion-transmitted HEV probably exists in the US. Further quantifying this risk, and potentially developing a strategy to prevent HEV transfusion-transmission to US patients at high risk of complicated infections, should be priorities.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Robin Cory, Tim Shin, Anh Hoang, and Graham Anderson for their technical assistance. We also acknowledge BioLINCC and NHLBI staff for considering and fulfilling our requests.
Support: This work was supported by grant R21 HL121740 from NHLBI, NIH. CDH was supported by EW “Al” Thrasher Award 10287 from the Thrasher Research Fund; grant 1316318 from NSF as part of the joint NSF-NIH-USDA Ecology and Evolution of Infectious Diseases program; grant K01OH010193 from NIOSH, CDC; and grant R01ES026973 NIEHS, NIH.
Abbreviations
- ARC
American Red Cross
- anti-HEV
antibodies to HEV
- CDC
Centers for Disease Control and Prevention, US Department of Health and Human Services
- cDNA
complementary DNA
- CT
threshold cycle (of a RT-qPCR run)
- DNA
deoxyribonucleic acid
- ELISA
enzyme-linked immunosorbent assay
- FDA
Food and Drug Administration, US Department of Health and Human Services
- HBV
hepatitis B virus
- HCV
hepatitis C virus
- HEV
hepatitis E virus
- IU
International Units (of HEV RNA)
- JHBSPH
Johns Hopkins Bloomberg School of Public Health
- NHANES
National Health and Nutritional Evaluation Survey (National Center for Health Statistics, CDC)
- NIEHS
National Institute of Environmental Health Sciences, NIH
- NHLBI
National Heart, Lung, and Blood Institute, NIH
- NIH
National Institutes of Health, US Department of Health and Human Services
- NIH CC
NIH Clinical Center
- NIOSH
National Institute for Occupational Safety and Health, CDC
- NSF
National Science Foundation, US
- ORF
open reading frame
- PBS/BSA
phosphate-buffered saline, pH 7.0, with 1% wt/vol bovine serum albumin
- PCR
polymerase chain reaction
- pRBC
packed red blood cells
- RADAR
REDS Allogeneic Donor and Recipient
- REDS
Retrovirus Epidemiology Donor Study
- RNA
ribonucleic acid
- RT-qPCR
reverse-transcription quantitative PCR
- S/CO
sample to cutoff value
- S/COpost
post-transfusion S/CO
- S/COpre
pre-transfusion S/CO
- TMA
transcription-mediated amplification
- U
Units (of anti-HEV)
- US
United States of America
- USDA
US Department of Agriculture
- WHO
World Health Organization
Conflict of interest: JRT provides professional services, as a part-time contractor, to CSL Plasma Inc. EO and JML were Hologic Inc employees when the reported findings were generated. The other authors do not have any conflicts of interest to disclose.
WEB RESOURCES
We obtained RADAR samples from BioLINCC (biolincc.nhlbi.nih.gov/studies/radar). BioEdit software is available from author Tom Hall at www.mbio.ncsu.edu/bioedit/page2.html. We generated phylogenetic trees at the Phylogeny Analysis page (phylogeny.lirmm.fr/phylo_cgi/phylogeny.cgi) of Le Laboratoire d’Informatique, de Robotique et de Microélectronique de Montpellier, Université Montpellier, Montpellier, France 33. Each of these sites was accessed 16 September 2018. TreeView was obtained from author Rod Page at taxonomy.zoology.gla.ac.uk/rod/treeview.html, a site that was no longer accessible on 17 September 2018, when it was available at treeview.software.informer.com/download.
Nucleotide sequences, described above and representing segments of ORF1 and ORF2 of HEV RNA in RADAR-donation plasma, have been deposited to GenBank with accession numbers MK385653 and MK385654. Nucleotide sequences that represent segments of ORF1, the region in which ORFs 1–3 overlap, and ORF2 of HEV in cynomolgus monkey bile have accession numbers MK385655–MK385657.
Reprints: will not be available from the authors
REFERENCES
- 1.Hoofnagle JH, Nelson KE, Purcell RH. Hepatitis E. N.Engl.J.Med. 2012. September 26;367(13):1237–44. [DOI] [PubMed] [Google Scholar]
- 2.Nelson KE, Heaney CD, Kmush BL. The epidemiology and prevention of hepatitis E virus infection. Curr.Epidemiol.Rep. 2017;1–13. [Google Scholar]
- 3.Ankcorn MJ, Tedder RS. Hepatitis E: the current state of play. Transfus.Med. 2017. April 1;27(2):84–95. [DOI] [PubMed] [Google Scholar]
- 4.Boxall E, Herborn A, Kochethu G, Pratt G, Adams D, Ijaz S, Teo CG. Transfusion-transmitted hepatitis E in a ‘nonhyperendemic’ country. Transfus.Med. 2006;16(2):79–83. [DOI] [PubMed] [Google Scholar]
- 5.Colson P, Coze C, Gallian P, Henry M, De Micco P, Tamalet C. Transfusion-associated hepatitis E, France (letter). Emerg.Infect.Dis. 2007. April 1;13(4):648–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haïm-Boukobza S, Ferey MP, Vétillard AL, Jeblaoui A, Pélissier E, Pelletier G, Teillet L, Roque-Afonso AM. Transfusion-transmitted hepatitis E in a misleading context of autoimmunity and drug-induced toxicity. J.Hepatol. 2012. August 9;57(6):1374–8. [DOI] [PubMed] [Google Scholar]
- 7.Huzly D, Umhau M, Bettinger D, Cathomen T, Emmerich F, Hasselblatt P, Hengel H, Herzog R, Kappert O, Maassen S, et al. Transfusion-transmitted hepatitis E in Germany, 2013. Euro Surveill. 2014. May 29;19(21):pii=20812. [DOI] [PubMed] [Google Scholar]
- 8.Hewitt PE, Ijaz S, Brailsford SR, Brett R, Dicks S, Haywood B, Kennedy ITR, Kitchen A, Patel P, Poh J, et al. Hepatitis E virus in blood components: a prevalence and transmission study in southeast England. Lancet 2014. July 28;384(9956):1766–73. [DOI] [PubMed] [Google Scholar]
- 9.Mitsui T, Tsukamoto Y, Yamazaki C, Masuko K, Tsuda F, Takahasi M, Nishizawa T, Okamoto H. Prevalence of hepatitis E virus infection among hemodialysis patients in Japan: Evidence for infection with genotype 3 HEV by blood transfusion. J.Med.Virol. 2004;74:563–72. [DOI] [PubMed] [Google Scholar]
- 10.Matsubayashi K, Nagaoka Y, Sakata H, Sato S, Fukai K, Kato T, Takahashi K, Mishiro S, Imai M, Takeda N, et al. Transfusion-transmitted hepatitis E caused by apparently indigenous hepatitis E virus strain in Hokkaido, Japan. Transfusion 2004. June 1;44(6):934–40. [DOI] [PubMed] [Google Scholar]
- 11.Matsubayashi K, Kang JH, Sakata H, Takahashi K, Shindo M, Kato M, Sato S, Kato T, Nishimori H, Tsuji K, et al. A case of transfusion-transmitted hepatitis E caused by blood from a donor infected with hepatitis E virus via zoonotic food-borne route. Transfusion 2008. July 1;48(7):1368–75. [DOI] [PubMed] [Google Scholar]
- 12.Satake M, Matsubayashi K, Hoshi Y, Taira R, Furui Y, Kokudo N, Akamatsu N, Yoshizumi T, Ohkohchi N, Okamoto H, et al. Unique clinical courses of transfusion-transmitted hepatitis E in patients with immunosuppression. Transfusion 2017. February 1;57(2):280–8. [DOI] [PubMed] [Google Scholar]
- 13.Wang M, He M, Wu B, Ke L, Han T, Wang J, Shan H, Ness P, Guo N, Liu Y, et al. The association of elevated alanine aminotransferase levels with hepatitis E virus infections among blood donors in China. Transfusion 2017. February 1;57(2):273–9. [DOI] [PubMed] [Google Scholar]
- 14.Slot E, Hogema BM, Riezebos-Brilman A, Kok TM, Molier M, Zaaijer HL. Silent hepatitis E virus infection in Dutch blood donors, 2011 to 2012. Euro Surveill. 2013;18(31):pii=20550. [DOI] [PubMed] [Google Scholar]
- 15.Baylis SA, Corman VM, Ong E, Linnen JM, Nübling CM, Blümel J. Hepatitis E viral loads in plasma pools for fractionation. Transfusion 2016. October 1;56(10):2532–7. [DOI] [PubMed] [Google Scholar]
- 16.Stramer SL, Moritz ED, Foster GA, Ong E, Linnen JM, Hogema BM, Mak M, Chia CP, Dodd RY. Hepatitis E virus: seroprevalence and frequency of viral RNA detection among US blood donors. Transfusion 2016. February 1;56(2):481–8. [DOI] [PubMed] [Google Scholar]
- 17.Roth NJ, Schäfer W, Alexander R, Elliott K, Elliott-Browne W, Knowles J, Wenzel JJ, Simon TL. Low hepatitis E virus RNA prevalence in a large-scale survey of United States source plasma donors. Transfusion 2017. December 1;57(12):2958–64. [DOI] [PubMed] [Google Scholar]
- 18.Xu C, Wang RY, Schechterly CA, Ge S, Shih JW, Xia NS, Luban NLC, Alter HJ. An assessment of hepatitis E virus (HEV) in US blood donors and recipients: no detectable HEV RNA in 1939 donors tested and no evidence for HEV transmission to 362 prospectively followed recipients. Transfusion 2013. October 1;53(10 part 2):2505–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zafrullah M, Zhang X, Tran C, Nguyen M, Kamili S, Purdy MA, Stramer SL. Disparities in detection of antibodies against hepatitis E virus in US blood donor samples using commercial assays. Transfusion 2018. March 9;58(5):1254–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kleinman SH, Glynn SA, Higgins MJ, Triulzi DJ, Smith JW, Nass CC, Garratty G, Murphy EL, LeParc GF, Schreiber GB, et al. The RADAR repository: a resource for studies of infectious agents and their transmissibility by transfusion. Transfusion 2005. July 1;45(7):1073–83. [DOI] [PubMed] [Google Scholar]
- 21.Baylis SA, Hanschmann KM, Blümel J, Nübling CM. Standardization of hepatitis E virus (HEV) nucleic acid amplification technique-based assays: an initial study to evaluate a panel of HEV strains and investigate laboratory performance. J.Clin.Microbiol. 2011. April 1;49(4):1234–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ticehurst J, Rhodes LL Jr., Krawczynski K, Asher LVS, Engler WF, Mensing TL, Caudill JD, Sjogren MH, Hoke CH Jr., LeDuc JW, et al. Infection of owl monkeys (Aotus trivirgatus) and cynomolgus monkeys (Macaca fascicularis) with hepatitis E virus from Mexico. J.Infect.Dis. 1992. May;165:835–45. [DOI] [PubMed] [Google Scholar]
- 23.Huang C-C, Nguyen D, Fernandez J, Yun KY, Fry KE, Bradley DW, Tam AW, Reyes GR. Molecular cloning and sequencing of the Mexico isolate of hepatitis E virus (HEV). Virology 1992;191:550–8. [DOI] [PubMed] [Google Scholar]
- 24.Kaiser M, Kamili S, Hayden T, Blümel J, Baylis SA. Genome sequence of a genotype 2 hepatitis E virus World Health Organization reference strain. Genome Announc. 2017. February 16;5(7):e01664–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rolfe KJ, Parmar S, Mururi D, Wreghitt TG, Jalal H, Zhang H, Curran MD. An internally controlled, one-step, real-time RT-PCR assay for norovirus detection and genogrouping. J.Clin.Virol. 2007. August;39(4):318–21. [DOI] [PubMed] [Google Scholar]
- 26.Gentry-Shields J, Myers K, Pisanic N, Heaney C, Stewart J. Hepatitis E virus and coliphages in waters proximal to swine concentrated animal feeding operations. Sci.Total Environ. 2015. February 1;505:487–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sue PK, Pisanic N, Heaney CD, Forman M, Valsamakis A, Jackson AM, Ticehurst J, Montgomery RA, Schwarz KB, Nelson K, et al. Hepatitis E virus infection among solid organ transplant recipients at a North American transplant center. Open Forum Infect.Dis. 2016. January 17;3(1):ofw006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jothikumar N, Cromeans TL, Robertson BH, Meng XJ, Hill VR. A broadly reactive one-step real-time RT-PCR assay for rapid and sensitive detection of hepatitis E virus. J.Virol.Methods 2006. January 1;131(1):65–71. [DOI] [PubMed] [Google Scholar]
- 29.Dong C, Meng J, Dai X, Liang JH, Feagins AR, Meng XJ, Belfiore NM, Bradford C, Corn JL, Cray C, et al. Restricted enzooticity of hepatitis E virus genotypes 1 to 4 in the United States. J.Clin.Microbiol. 2011. December 16;49(12):4164–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Y, Ling R, Erker JC, Zhang H, Li H, Desai S, Mushahwar IK, Harrison TJ. A divergent genotype of hepatitis E virus in Chinese patients with acute hepatitis. J.Gen.Virol. 1999. January 1;80(1):169–77. [DOI] [PubMed] [Google Scholar]
- 31.Zhai L, Dai X, Meng J. Hepatitis E virus genotyping based on full-length genome and partial genomic regions. Virus Res. 2006. September;120(1–2):57–69. [DOI] [PubMed] [Google Scholar]
- 32.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst.Biol. 2010. May 1;59(3):307–21. [DOI] [PubMed] [Google Scholar]
- 33.Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008. July 1;36(suppl_2):W465–W469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vina-Rodriguez A, Schlosser J, Becher D, Kaden V, Groschup HM, Eiden M. Hepatitis E virus genotype 3 diversity: phylogenetic analysis and presence of subtype 3b in wild boar in Europe. Viruses 2015;7(5):2704–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith DB, Simmonds P, Izopet J, Oliveira-Filho EF, Ulrich RG, Johne R, Koenig M, Jameel S, Harrison TJ, Meng XJ, et al. Proposed reference sequences for hepatitis E virus subtypes. J.Gen.Virol. 2016. March;97(3):537–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith DB, Ijaz S, Tedder RS, Hogema B, Zaaijer HL, Izopet J, Bradley-Stewart A, Gunson R, Harvala H, Kokki I, et al. Variability and pathogenicity of hepatitis E virus genotype 3 variants. J.Gen.Virol. 2015. November 30;96(Pt 11):3255–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baylis SA, Crossan C, Corman VM, Blümel J, Scobie L, Dalton HR. Unusual serological response to hepatitis E virus in plasma donors consistent with re-infection. Vox Sang. 2015. November 1;109(4):406–9. [DOI] [PubMed] [Google Scholar]
- 39.Clayson ET, Myint KSA, Snitbhan R, Vaughn DW, Innis BL, Chan L, Cheung P, Shrestha MP. Viremia, fecal shedding, and IgM and IgG responses in patients with hepatitis E. J.Infect.Dis. 1995;172(October):927–33. [DOI] [PubMed] [Google Scholar]
- 40.Aggarwal R, Kini D, Sofat S, Naik SR, Krawczynski K. Duration of viraemia and faecal viral excretion in acute hepatitis E. Lancet 2000. September 23;356(9235):1081–2. [DOI] [PubMed] [Google Scholar]
- 41.Chandra NS, Sharma A, Malhotra B, Rai RR. Dynamics of HEV viremia, fecal shedding and its relationship with transaminases and antibody response in patients with sporadic acute hepatitis E. Virol.J. 2010;7(1):213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tedder RS, Ijaz S, Kitchen A, Ushiro-Lumb I, Tettmar KI, Hewitt P, Andrews N. Hepatitis E risks: pigs or blood − that is the question. Transfusion 2017. February 1;57(2):267–72. [DOI] [PubMed] [Google Scholar]
- 43.Germer JJ, Ankoudinova I, Belousov YS, Mahoney W, Dong C, Meng J, Mandrekar JN, Yao JD. Hepatitis E virus (HEV) detection and quantification by a real-time reverse transcription-PCR assay calibrated to the World Health Organization standard for HEV RNA. J.Clin.Microbiol. 2017. May 1;55(5):1478–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuniholm MH, Purcell RH, McQuillan GM, Engle RE, Wasley A, Nelson KE. Epidemiology of hepatitis E virus in the United States: results from NHANES III, 1988–1994. J.Infect.Dis. 2009. July 1;200(1):48–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meng XJ, Wiseman B, Elvinger F, Guenette DK, Toth TE, Engle RE, Emerson SU, Purcell RH. Prevalence of antibodies to hepatitis E virus in veterinarians working with swine and in normal blood donors in the United States and other countries. J.Clin.Microbiol. 2002. January 1;40(1):117–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Engle RE, Bukh J, Alter HJ, Emerson SU, Trenbeath JL, Nguyen HT, Brockington A, Mitra T, Purcell RH. Transfusion-associated hepatitis before the screening of blood for hepatitis risk factors. Transfusion 2014. June 1;54(11):2833–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kamar N, Marion O, Abravanel F, Izopet J, Dalton HR. Extrahepatic manifestations of hepatitis E virus. Liver Int 2016. April 1;36(4):467–72. [DOI] [PubMed] [Google Scholar]
- 48.Tsarev SA, Tsareva TS, Emerson SU, Kapikian AZ, Ticehurst J, London W, Purcell RH. ELISA for antibody to hepatitis E virus (HEV) based on complete open-reading frame-2 protein expressed in insect cells: identification of HEV infection in primates. J.Infect.Dis. 1993;168(2):369–78. [DOI] [PubMed] [Google Scholar]
- 49.Kodani M, Kamili NA, Tejada-Strop A, Poe A, Denniston MM, Drobeniuc J, Kamili S. Variability in the performance characteristics of IgG anti-HEV assays and its impact on reliability of seroprevalence rates of hepatitis E. J.Med.Virol. 2017;89(6):1055–61. [DOI] [PubMed] [Google Scholar]
- 50.Teshale EH, Denniston MM, Drobeniuc J, Kamili S, Teo CG, Holmberg SD. Decline in hepatitis E virus antibody prevalence in the United States from 1988−1994 to 2009–2010. J.Infect.Dis. 2015. February 1;211(3):366–73. [DOI] [PubMed] [Google Scholar]
- 51.Kuniholm MH, Engle RE, Purcell RH, Nelson KE. Hepatitis E virus seroprevalence in the United States: No easy answers (letter). Hepatology 2014. July 4;61(4):1441–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Petrik J, Lozano M, Seed CR, Faddy HM, Keller AJ, Prado Scuracchio PS, Wendel S, Andonov A, Fearon M, Delage G, et al. Hepatitis E. Vox Sang. 2015. July 21;110(1):93–103. [DOI] [PubMed] [Google Scholar]
- 53.Rein DB, Stevens GA, Theaker J, Wittenborn JS, Wiersma ST. The global burden of hepatitis E virus genotypes 1 and 2 in 2005. Hepatology 2012. April 1;55(4):988–97. [DOI] [PubMed] [Google Scholar]
- 54.Domanovic D, Tedder R, Blümel J, Zaaijer H, Gallian P, Niederhauser C, Sauleda Oliveras S, O’Riordan J, Boland F, Harritshøj L, et al. Hepatitis E and blood donation safety in selected European countries: a shift to screening? Euro Surveill. 2017. April 20;22(16):pii=30514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.de Vos AS, Janssen MP, Zaaijer HL, Hogema BM. Cost-effectiveness of the screening of blood donations for hepatitis E virus in the Netherlands. Transfusion 2017. February 1;57(2):258–66. [DOI] [PubMed] [Google Scholar]
- 56.Dodd RY, Notari EP, Nelson D, Foster GA, Krysztof DE, Kaidarova Z, Milan-Benson L, Kessler DA, Shaz BH, Vahidnia F, et al. Development of a multisystem surveillance database for transfusion-transmitted infections among blood donors in the United States. Transfusion 2016. August 25;56(11):2781–9. [DOI] [PubMed] [Google Scholar]
- 57.Izopet J, Dubois M, Bertagnoli S, Lhomme S, Marchandeau S, Boucher S, Kamar N, Abravanel F, Guérin JL. Hepatitis E virus strains in rabbits and evidence of a closely related strain in humans, France. Emerg.Infect.Dis. 2012. August;18(8):1274–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Abravanel F, Lhomme S, El Costa H, Schvartz B, Peron JM, Kamar N, Izopet J. Rabbit hepatitis E virus infections in humans, France. Emerg.Infect.Dis. 2017. July;23(7):1191–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


