Abstract
We have developed a simple and efficient procedure for adding an epitope-encoding tail to one or more genes of interest in the bacterial chromosome. The procedure is a modification of the gene replacement method of Datsenko and Wanner [Datsenko, K. A. & Wanner, B. L. (2000) Proc. Natl. Acad. Sci. USA 97, 6640–6645]. A DNA module that begins with the epitope-encoding sequence and includes a selectable marker is amplified by PCR with primers that carry extensions (as short as 36 nt) homologous to the last portion of the targeted gene and to a region downstream from it. Transformation of a strain expressing bacteriophage λ red functions yields recombinants carrying the targeted gene fused to the epitope-encoding sequence. The resulting C-terminal-tagged protein can be identified by standard immuno-detection techniques. In an initial application of the method, we have added the sequences encoding the FLAG and 3xFLAG and influenza virus hemagglutinin epitopes to various genes of Salmonella enterica serovar Typhimurium, including putative and established pathogenic determinants present in prophage genomes. Epitope fusion proteins were detected in bacteria growing in vitro, tissue culture cells, and infected mouse tissues. This work identified a prophage locus specifically expressed in bacteria growing intracellularly. The procedure described here should be applicable to a wide variety of Gram-negative bacteria and is particularly suited for the study of intracellular pathogens.
The accumulation of genome sequence data from an increasing number of organisms is changing classical genetic approaches. Rather than identifying genes from mutant phenotypes, it is becoming more common to start from the gene and move to the study of its function and regulation by using recombinant DNA techniques. The growing importance of this “reverse genetics” generates a demand for simple, one-step procedures for disrupting and/or tagging genes directly in their natural chromosomal context. In the yeast Saccharomyces cerevisiae, the ability of DNA fragments to undergo recombination with the chromosome in the presence of stretches of sequence homology as short as 35 bp has led to the development of PCR-based methods for gene replacement and modification (1). In their latest refinements, these techniques have allowed the introduction of epitope tags into genes of interest (2–4). In enteric bacteria, similar approaches have been hampered by the instability of linear DNA, rapidly degraded by the nuclease activity (ExoV) of the RecBCD recombinase. Efforts to circumvent this problem have involved the use of mutants or conditions inhibiting ExoV (5–9). However, these systems are not efficient enough to promote recombination of PCR fragments with short homology extensions. Recently, the problem has been solved upon recognizing the effectiveness of bacteriophage λ recombination system (the red system) in reactions involving linear DNA (10, 11). Using Escherichia coli cells expressing red functions, three independent groups succeeded in achieving gene replacement by transformation with PCR fragments carrying short extensions (12–14). In the system developed by Datsenko and Wanner (12), presence of the red operon on a plasmid makes it suitable for use in bacteria other than E. coli. The amplifiable modules include an antibiotic resistance marker (KnR or CmR) flanked by FRT sites (FLP recombinase recognition targets). The latter feature allows the “pop-out” of the drug resistance gene by site-specific recombination once the construct is obtained (15).
We have made extensive use of the Datsenko and Wanner procedure to analyze virulence genes present on the Gifsy-1 and Gifsy-2 prophages of Salmonella enterica serovar Typhimurium (9, 16). In the course of this work, it became evident that the system could be further engineered to generate protein fusions to known antigens. To test this idea, we chose the sequences specifying the FLAG, 3xFLAG, and hemagglutinin (HA) peptides that have been widely used for protein tagging and are recognized by commercially available mAbs (17–20). By applying the scheme outlined in Results, we were able to fuse the epitope-encoding sequences at the 3′ ends of a number of prophage genes. The resulting C-terminal-tagged proteins were detected by Western blotting. This study provided valuable information on the expression patterns of some of the prophage loci in vitro and in bacteria growing intracellularly. Some chromosomal genes also were tagged, including the essential lepB gene coding for signal peptidase I. The ease of the manipulations, the sensitivity of the detection method, and the adaptability to additional applications should make the system described here a powerful tool in the study of genes and their products in bacteria.
Materials and Methods
Plasmids.
E. coli K12 strains carrying plasmids pKD46, pKD3, pKD4, and pCP20 (12) were obtained from B. Wanner (Purdue University, West Lafayette, IN) by means of S. Maloy (University of Illinois, Urbana, IL). Plasmid pKD46 is a low-copy number, temperature-sensitive replicon that carries bacteriophage λ red genes (γ, β, and exo) under the control of arabinose-inducible ParaBAD promoter (12). Plasmids pKD3 and pKD4 are π-dependent plasmids carrying chloramphenicol- and kanamicin-resistance genes (CmR and KnR), respectively, flanked by the recognition sites (FRT sites) of the yeast FLP recombinase in direct repeats (12). Plasmid pCP20 is a temperature-sensitive replicon expressing the FLP gene (15). π-Dependent plasmid pGP704 (21) and its derivatives were maintained in E. coli K12 strain CC118 λpir (22).
Bacterial Strains and Culture Conditions.
This work was carried out by using strains of S. enterica serovar Typhimurium derived from strain ATCC14028s (23). Strain MJW141 (ATCC14028s ssrB∷Cm/pMJW120), described in ref. 24, was a gift of M. J. Worley and F. Heffron (Oregon Health Sciences University, Portland, OR). Strain MA6897 (wild-type ATCC14028s/pKD46), constructed at the onset of this work, served as general recipient for the recombinational exchanges described here. For the construction of FLAG and 3xFLAG-tagged derivatives of cat and sodCIII genes, two additional ATCC14028s-derived, pKD46-harboring strains were used: MA6987, carrying a cat gene insertion in the ilvIH operon (ilvI3305∷Tn10dTac-cat/pKD46) and MA7088, a Gifsy-2-cured strain lysogenic for the Fels-1 phage (Gifsy-2 [-] Fels-1[+]/pKD46). Bacteria were grown in LB media (Difco) unless indicated otherwise. NCE medium (25) was supplemented with 0.2% glycerol. M9 medium (26) was supplemented with 0.2% glucose or 0.2% arabinose. When required, antibiotics were used at the following concentrations: chloramphenicol, 10 μg⋅ml−1; kanamycin, 50 μg⋅ml−1; and ampicillin, 50 μg⋅ml−1.
Recombinant DNA Techniques and Electrotransformation.
Restriction enzymes and T4 DNA ligase (New England Biolabs) were used according to the manufacturer's recommendations. In ligation reactions involving noncomplementary protruding 3′ ends, DNA fragments were treated with DNA polymerase (Klenow fragment; New England Biolabs) to generate blunt ends. Transformation of Salmonella with plasmid DNA was accomplished by electroporation by using a Bio-Rad Gene Pulser, under the conditions specified by the manufacturer.
Construction of Template Plasmids.
DNA from plasmid pGP704 was cleaved with EcoRV and BglII restriction endonucleases and religated. This step eliminated the BamHI site in the pGP704 polylinker. The resulting plasmid (pSU310) was digested with BamHI, and a 1.9-kb fragment carrying the R6K oriV and the βla gene was circularized to obtain plasmid pSU311. DNA fragments spanning the KnR and CmR cassettes of plasmids pKD4 and pKD3, respectively, and including the flanking FRT sites, were amplified by PCR by using primers carrying oligonucleotide extensions with EcoRI recognition sequences at their 5′ ends. The extension in the “forward” primer included a 24-nt segment corresponding to the coding sequence of the FLAG peptide (ref. 20; Fig. 1B). PCR products were digested with EcoRI nuclease and ligated to EcoRI-cleaved plasmid pSU311 DNA. The resulting recombinant plasmids, pSU312 (KnR) and pSU313 (CmR), were used as templates for the amplification DNA fragments carrying FLAG sequence fusions. Additional template plasmids pSU314 (CmR) and pSU315 (KnR), carrying the influenza virus HA epitope sequence (27) and pSUB7 (KnR) encoding the 6His epitope, were similarly constructed (Fig. 1B). The procedure that produced plasmid pSU312 was repeated by using the latter as the template and the same reverse primer as above. The forward primer was an oligonucleotide containing the complete 3xFLAG sequence (20) ending with an EcoRI nuclease recognition site. EcoRI enzyme digestion of the amplification product and ligation to EcoRI-cleaved pSU311 DNA yielded plasmid pSUB11 (Fig. 1B).
Figure 1.
(A) The recombinational tagging protocol. A DNA module that begins with the epitope-encoding sequence (filled box) and includes an antibiotic-resistance cassette (KnR or CmR) flanked by FRT sites (hatched boxes) is amplified with primers carrying extensions (36–40 nt) homologous to the region immediately preceding the translation stop signal of the target gene (shaded box) and to a region downstream from it (checkered box). The amplified fragment is introduced into a strain that expresses bacteriophage λ red operon and antibiotic resistant recombinants are selected. Recombinant bacteria synthesize the target protein with the epitope sequence (filled beads) fused to its carboxy terminus. When needed, the antibiotic resistance gene can be “popped-out” by the FLP recombinase, allowing the above procedure to be repeated with another target gene. (B) Primer-annealing sequences. The annealing segments in the forward primers (epitope-side primers) are indicated by filled bars above the sequence. The constant annealing region in the reverse primer (corresponding to priming site 2 of ref. 12) is indicated by a filled bar underneath the sequence.
Tagging.
Recombinational transfer of the FLAG sequence into chromosomal genes was achieved by following the PCR-based method of Datsenko and Wanner (12) with a few modifications. Primers carried extensions (between 36 and 40 nt) homologous to the last portion of the targeted gene (forward primer) and to a region downstream from it (reverse primer). For most of the genes studied in this work, both FLAG-tagged and 3xFLAG-tagged derivatives were made. The sequences of the primers used for the latter constructs (template plasmid pSUB11) are shown in Table 1. Taq polymerase (Promega) and Pfu (Stratagene) were used for all amplifications as described by Datsenko and Wanner (12). PCR products were purified by passage through Qiagen spin columns and used directly for electro-transformation. Bacteria (strain MA6897 or its derivatives; see above) to be made electrocompetent were grown at 30°C in LB medium supplemented with 100 μg⋅ml−1 ampicillin and 1 mM arabinose to an A600 of 0.5. Cells were collected by centrifugation, washed three times with ice-cold 10% glycerol, and concentrated 200-fold. Then 50-μl aliquots of the suspensions were mixed to 0.5–1 μg of PCR product in a chilled cuvette (0.2-cm electrode gap) and subjected to a single pulse of 12.5 kV/cm (2.5 kV, 200 Ω, 25 μF). After an 1-h recovery at 37°C in SOC medium (2% Bacto Tryptone/0.5% yeast extract/10 mM NaCl/2.5 mM KCl/10 mM MgCl2/10 mM MgSO4/20 mM glucose), bacteria were spread on LB agar plates supplemented with antibiotics for the selection of CmR or KnR recombinants. Typically, these occurred at frequencies of 20–200 colonies/μg of DNA. Phage P22 lysates were made on the transformants and used to transfer the antibiotic resistance gene and the linked epitope fusion into a fresh ATCC14028s strain background. Concomitantly, transduction of appropriate recipient strains allowed to verify the correct map position of the construct. The genetic markers used for these tests are listed in Table 2. When sought, the FRT-flanked antibiotic resistance cassette was eliminated after transformation with pCP20, as described (15).
Table 1.
Primers used with template plasmid pSUB11
| Primer | Sequence, 5′–3′* | Target gene | GenBank |
|---|---|---|---|
| pp190 (fw) | TTTACACTCCTCCTCCTGGAAAGACTGGTGTACCATTTTAGACTACAAAGACCATGACGG | gtgE | AF147699 |
| pp145 (rv) | CGACGGCTTTATTCCCCAGTTTCACCCCATAGCTTCCATATGAATATCCTCCTTAG | ||
| pp180 (fw) | TGGTGGCGGTGCACGTTTTGCCTGTGGTGTCATTGAGAAAGACTACAAAGACCATGACGG | sodCI | AF007380 |
| pp182 (rv) | TTTTCCACTGATGCTGACAGTGACGTCAAAGCTGGAACCTCATATGAATATCCTCCTTAG | ||
| pp193 (fw) | CAATACAGGCATTCTGCAGAGGTTTTCCCGGATGAAGACTACAAAGACCATGACGG | gogC | — |
| pp128 (rv) | TGGTTTTTGTCACGCGTTTTATGGATAGTTTTCTGCCATATGAATATCCTCCTTAG | ||
| pp189 (fw) | GGCGGTGGTGCACGAATCGCCTGTGGCGTTATAAAAGACTACAAAGACCATGACGG | sodCIII | AF254764 |
| pp167 (rv) | GGTAATGCCCTGCCCGTGCTGTACGGCGAAATGCTGCATATGAATATCCTCCTTAG | ||
| pp187 (fw) | AGTTGGTTGGGATAAATATAAGCCTAAAAATAGAAATCGTGACTACAAAGACCATGACGG | gogB | AF254761 |
| pp138 (rv) | TACTAACAACCACAAACCTGAGACCAATTCAGTTGCTTCACATATGAATATCCTCCTTAG | ||
| pp188 (fw) | TACCTTTAAAAAGCAAAAATATTCCTTAATAGGTAAAATGGACTACAAAGACCATGACGG | gtgB | AF254763 |
| pp140 (rv) | AAGAAAACCACCTCACCCTCATAACTCAGTAAGCGTCCCGCATATGAATATCCTCCTTAG | ||
| pp215 (fw) | TGAATTACAACAGTACTGCGATGAGTGGCAGGGCGGGGCGGACTACAAAGACCATGACGG | cat | J01841 |
| pp136 (rv) | CCATTCATCCGCTTATTATCACTTATTCAGGCGTAGCACCCATATGAATATCCTCCTTAG | ||
| pp194 (fw) | AGCGTTCCTCGCCATTCTGCATGTCGGTAAAGACAATAAAGACTACAAAGACCATGACGG | lepA | X54933 |
| pp169 (rv) | GCAAACATGTTCGCCATGCCAACTCCTTAGGGATTATTTACATATGAATATCCTCCTTAG | ||
| pp220 (fw) | ACAGGCGTACGCCTGAGTCGTATCGGCGGTATTCACGACTACAAAGACCATGACGG | lepB | X54933 |
| pp126 (rv) | GCCGTGGCGACCAAACGGCGCCAACCTAAAACTTCGCATATGAATATCCTCCTTAG |
Primers were designed to anneal to the beginning of the 3xFLAG-coding sequence (fw) and on the opposite side from the KnR cassette (rv) of template plasmid pSUB11 (in italics), and carry extensions homologous to the last 36–40 nts of the target gene coding sequence (fw) and to a region downstream from it (rv). fw, forward; rv, reverse.
Table 2.
Genetic markers used for map verification of epitope fusion constructs*
| Tagged gene, donor | Linked marker, recipient | Percentage cotransduction |
|---|---|---|
| gtgE | pyrD2266∷Tn10 | 70 |
| sodCI | sodCI∷Cm | 100 |
| gogC | gogB∷Cm | 85 |
| sodCIII | Fels-1 prophage† | 100 |
| gogB | gogB∷Cm | 100 |
| gtgB | pyrD2266∷Tn10 | 60 |
| cat | leuA3241∷MudJ | 80 |
| lepA | gogB∷Cm | 80 |
| lepB | gogB∷Cm | 70 |
Only markers used for verification of KnR constructs are indicated. Phage P22 lysates prepared on strains harboring the putative fusions (donor) were used to transduce recipient strains carrying the indicated markers. Linkage is expressed as percentage of KnR transductants acquiring the marker configuration of the donor. Not <20 transductants were analyzed in each case.
Initial verification of the sodCIII epitope fusion was based on obtaining KnR transductants with a strain lysogenic for the Fels-1 prophage but not on a strain lacking it.
Immunodetection Analysis.
FLAG and 3xFLAG fusion proteins were immunodetected by the use of anti-FLAG M2 mAbs from Sigma. The HA epitope was detected with anti-HA mAbs from Sigma (clone HA-7). The 6His epitope was recognized by the Penta-His mAbs from Qiagen. Strains carrying the epitope-tagged gene(s) were grown in 1.5-ml cultures to stationary phase and centrifuged. Bacterial pellets were resuspended in 100 μl of H2O and immediately mixed with 100 μl of Laemmli lysis buffer (28). Suspensions were incubated at 100°C for 5–10 min. The resulting lysates were quickly centrifuged to remove cell debris and used, straight or suitably diluted, for SDS/PAGE. Bacterial proteins resolved by SDS/PAGE were transferred to poly(vinylidene difluoride) membranes and probed with mAbs (1:1,000) and horseradish peroxidase-conjugated goat anti-mouse IgG [1:5,000 (Sigma)]. Detection was performed by enhanced chemioluminescence (ECL, Amersham Pharmacia).
Epithelial Cell Infections.
Human laryngeal epithelial (Hep-2) cells were maintained in RPMI 1640 as described (29). Confluent monolayers were inoculated with bacteria grown standing overnight in LB broth, at an multiplicity of infection of ≈10:1 without centrifugation. Infected monolayers were then incubated for 1 h in a tissue culture incubator, washed twice with PBS, and then overlaid with tissue culture medium supplemented with 100 μg⋅ml−1 gentamicin. After 2 h, the cell culture was washed once with RPMI medium 1640 and incubated for an additional 20 h in the same medium containing 10 μg⋅ml−1 gentamicin. Finally, monolayers were washed three times with PBS and lysed with 1% Triton X-100 in PBS to release intracellular bacteria. An aliquot of this suspension was used to determine the number of intracellular bacteria by plating serial dilutions onto LB agar plates. Released bacteria were then prepared for immunoblotting analysis as described above. In selected experiments, intracellular colony-forming units (cfus) were counted both after 3 h and 23 h from the infection to determine the replication rate of intracellular salmonellae.
Mouse Infection.
Female BALB/c mice (8 wk old) were inoculated i.p. with ≈2 × 103 cfu/mouse as described (30). Mice were killed 4 days after infection. Spleens were removed, and bacteria were recovered as described (31). The bacterial contents of each spleen were resuspended in 50 μl of H20 and mixed with 50 μl of Laemmli lysis buffer. Bacterial suspensions were lysed as described above and used for SDS/PAGE directly or suitably diluted.
Results
Rationale.
The basic strategy applied here (outlined in Fig. 1A) involves chemically synthesizing the junction between a target gene and an epitope-encoding sequence and then amplifying the resulting sequence as part of a DNA fragment that includes a selectable marker. The amplifiable segment begins on one side with the epitope sequence and comprises a drug-resistance gene flanked by FRT sites. Primer pairs (between 56 and 60 nt long) are designed to anneal to templates over constant regions of 20–21 bp and have between 36- and 40-nt extensions homologous to the last portion of the targeted gene (forward primer) and to a region downstream from it (reverse primer) (Fig. 1B). The products of the PCR, suitably purified, are introduced into Salmonella strains carrying plasmid pKD46 (12) and recombinants incorporating the amplified sequence into the chromosome are selected. Using the above strategy with template plasmids pSU312 and pSU313, we were able to introduce the FLAG epitope into a variety of prophage and chromosomal genes of S. enterica. Gene products were detected by immunoblotting with commercial anti-FLAG mAbs (see below). Some variability in the binding affinities of subsequent mAbs lots prompted us to replace the FLAG epitope with the more sensitive 3xFLAG variant on a new template plasmid (pSUB11). Additional plasmids designed for the introduction of influenza virus HA epitope and the 6His peptide were constructed in the later stages of this work.
Epitope Tagging and Detection of Salmonella Proteins.
We have been involved in the characterization of the prophage complements of virulent Salmonella strains. Two such elements, prophages Gifsy-1 and Gifsy-2, contain genes enhancing pathogenicity in mice (9, 30, 32, 33). In addition, both elements include a number of loci that, albeit apparently dispensable for murine salmonellosis, can be linked to virulence based on sequence homologies or expression patterns (9, 24, 34). A putative [Cu, Zn] superoxide dismutase gene named sodCIII is encoded within the prophage Fels-1, specifically found in Salmonella Typhimurium strain LT2 (9).
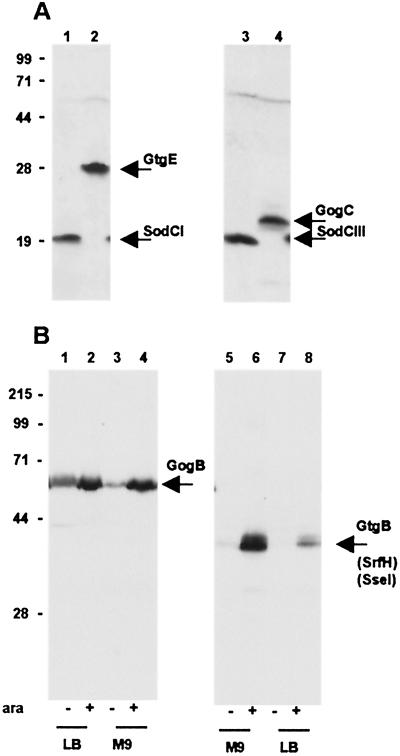
As an initial test for the tagging technique described here, we have fused the FLAG and/or 3xFLAG epitopes to known or presumptive genes present on the prophage genomes. Two Gifsy-1 loci were chosen, both are at the right end of the prophage map: the gogB locus, which specifies a putative leucine-rich protein similar to several type III-translocated proteins, and an unknown ORF named gogC (9). The Gifsy-2 genes that were tagged include the sodCI gene coding for a [Cu, Zn] superoxide dismutase implicated in virulence (30, 35, 36), an uncharacterized ORF named gtgE, and the gtgB gene (9). The latter, variably called sseI or srfH, is under the control of the SsrB activator protein and directs the synthesis of a protein translocated inside eukaryotic cells (24, 34). Finally, the sodCIII gene of the Fels-1 prophage was also tagged. Tagged recombinants were obtained as described above, and cultures from these strains grown in LB medium were processed for the detection of the FLAG epitope. As shown in Fig. 2A, the anti-FLAG M2 mAb recognizes the predicted fusion products for mature SodCI and SodCIII enzymes (lanes 1 and 3) and for GtgE and GogC proteins (lanes 2 and 4), respectively. A weak band corresponding to the tentative gogB gene product was also visible (data not shown) whereas the analysis failed to detect the GtgB protein. Transcription of the gtgB gene (srfH) was reported to require the SsrB activator protein, whose synthesis is normally repressed in LB medium (24). Therefore, the above constructs were transferred by transduction into strain MJW141, which carries a plasmid-borne ssrB gene fused to the araBAD promoter (24). Immunoblot analysis of cell extracts from bacteria grown in the presence or absence of arabinose confirmed the gtgB gene to be strongly induced by the SsrB effector, particularly in bacteria growing in minimal medium (Fig. 2B, lanes 5–8). These experiments revealed that the gogB gene is also under SsrB control. Fig. 2B shows that synthesis of the GogB protein is greatly enhanced in the presence of arabinose, albeit the control appears less tight than with gtgB and less dependent on the growth medium (lanes 1–4). No appreciable changes in the levels of SodCI and GogC proteins were observed after ssrB gene induction (data not shown). In conclusion, this analysis allowed us to assign protein products to four previously hypothetical phage loci (gogB, gogC, gtgE, and sodCIII) and to determine that these loci are normally expressed and regulated in the prophage state.
Figure 2.
Immunodetection of epitope-tagged proteins from Salmonella prophages. Whole-cell bacterial lysates were subjected to electrophoretic separation in a 12% polyacrylamide-SDS gel. Proteins were transferred onto poly(vinylidene difluoride) membrane and probed with anti-FLAG M2 mAb (Sigma). (A) Bacteria grown in LB medium: 1, strain MA7172 (sodCI-30∷3xFLAG KnR); 2, strain MA7180 (gtgE-34∷3xFLAG KnR); 3, strain MA7191 (sodCIII-35∷3xFLAG KnR); and 4, strain MA7192 (gogC-36∷3xFLAG KnR). Expected molecular masses for the 3xFLAG fusion proteins are: SodCI and SodCIII, 19 kDa; GtgE, 29 kDa; and GogC, 22 kDa. (B) Bacteria grown under conditions eliciting SsrB-mediated regulation. Lanes 1–4: strain MA7184 (gogB-33∷3xFLAG KnR ssrB∷ Cm/pBAD ssrB+) grown in LB medium (lane 1), in LB medium supplemented with arabinose (lane 2), in M9-glucose medium (lane 3), and in M9-arabinose medium (lane 4). Lanes 5–8: strain MA7185 (gtgB-37∷3xFLAG KnR ssrB∷Cm/pBAD ssrB+) grown in M9-glucose medium (lane 5), in M9-arabinose medium (lane 6), in LB medium (lane 7), and in LB medium supplemented with arabinose (lane 8). Expected molecular masses for the 3xFLAG fusion proteins are: GtgB, 38 kDa and GogB, 57 kDa. Positions and molecular masses (kDa) of protein standards are indicated on the left.
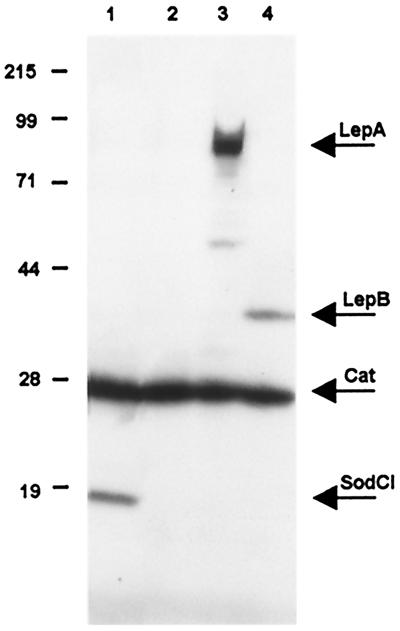
Additional genes that were tagged include the two members of the lep operon, lepA and lepB (37), and the cat gene from a transposon Tn10dTac-cat insertion in the ilvIH operon of Salmonella (38). The lepA gene, which contains the attachment site of the Gifsy-1 prophage at its 5′ end (30), specifies a GTP-binding membrane protein of unknown function (37). The lepB gene specifies signal peptidase 1, a central enzyme in the general secretory pathway of Gram-negative bacteria (39). The normal growth properties of bacteria carrying LepB-tagged construct indicates that the presence of the epitope tail does not adversely affect the catalytic activity of the protein. Likewise, the addition of FLAG or 3xFLAG tags to the Cat protein did not interfere with chloramphenicol resistance. Combining the cat-3xFLAG construct with some of the epitope fusions described above generated doubly tagged strains (Fig. 3).
Figure 3.
Immunodetection of epitope-tagged proteins. Bacterial lysates were subjected to electrophoretic separation in a 10% polyacrylamide-SDS gel. The gel was processed as described in the legend to Fig. 2. 1, strain MA7224 (sodCI-30∷3xFLAG KnR ilvI3305∷Tn10dTac-cat-43∷3xFLAG KnR); 2, strain MA7223 (ilvI3305∷Tn10dTac-cat-43∷3xFLAG KnR); 3, strain MA7227 (lepA-38::3xFLAG KnR ilvI3305∷Tn10dTac-cat-43∷3xFLAG KnR); and 4, strain MA7228 (lepB-39∷3xFLAG KnR ilvI3305∷Tn10dTac-cat-43∷3xFLAG KnR). Expected molecular masses for the 3xFLAG fusion proteins are: SodCI, 19 kDa; Cat, 28 kDa; LepA, 69 kDa; and LepB, 38 kDa; Positions and molecular masses (kDa) of protein standards are indicated on the left. The reasons for the abnormally low mobility of the LepA protein are unknown.
Finally, strains carrying the sodCI gene fused to the HA and 6His epitope were also constructed. The HA-tagged protein was detected with a sensitivity comparable to that of the 3xFLAG version; in contrast, the immunodetection was slightly less efficient and background more pronounced with the 6His-tagged SodCI derivative (data not shown). Still, the latter construct could be useful for the purification of target protein by nickel affinity chromatography.
Detection of Tagged Proteins During Bacterial Infection of Tissue Culture Cells and Animals.
Epithelial cells.
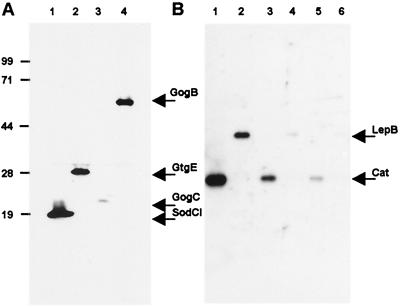
The method described above appeared suitable for the study of gene expression in Salmonella bacteria growing inside eukaryotic cells. Confluent monolayers of epithelial cells (Hep-2) were infected with Salmonella strains carrying FLAG epitope fusions to SodCI, GtgE, GogC, and GogB. Intracellular bacteria were collected 23 h after infection, a period during which their number had increased ≈100-fold. As shown in Fig. 4A, all four epitope-tagged proteins could be detected specifically. The gogB gene appears to be expressed at levels comparable to those of sodCI and gtgE genes. The GogC protein is synthesized in lower amounts. The detection of GogB in intracellular bacteria is consistent with the activation of the gogB gene by the intracellularly induced SsrB regulator.
Figure 4.
Immunodetection of epitope-tagged proteins in intracellular Salmonella bacteria. Hep-2 cells were infected with the indicated strains and bacteria were allowed to multiply intracellularly for 23 h in a gentamicin protection assay (see Materials and Methods). Cells were lysed and extracts were subjected to electrophoretic separation in a 12% SDS-polyacrylamide gel, which was processed as described in the legend to Fig. 2. (A) Prophage proteins: 1, strain MA7172 (sodCI-30∷3xFLAG KnR); 2, strain MA7180 (gtgE-34∷3xFLAG KnR); 3, strain MA7192 (gogC-36∷3xFLAG KnR); and 4, strain MA7084 (gogB-17∷FLAG KnR). (B) Chromosomal proteins. Lanes 1, 3, and 5: strain MA7223 (ilvI3305∷Tn10dTac-cat-43∷3xFLAG KnR) and lanes 2, 4 and 6: strain MA7195 (lepB-39∷3xFLAG KnR). Material loaded corresponds to 106 cfu (lanes 1 and 2), 105 cfu (lanes 3 and 4), and 104 cfu (lane 5 and 6). Positions and molecular masses (kDa) of protein standards are indicated on the left.
To further evaluate the sensitivity and the generality of the method, experiments were performed by using bacteria with tags in genes expected to be expressed constitutively such as the lepB and Tac-cat genes. The results in Fig. 4B set the threshold level of detection at 1 × 105 cfu for the 3xFLAG-tagged derivative of the LepB protein and at as few as 1 × 104 cfu for the same derivative of the Cat enzyme.
Mouse spleen.
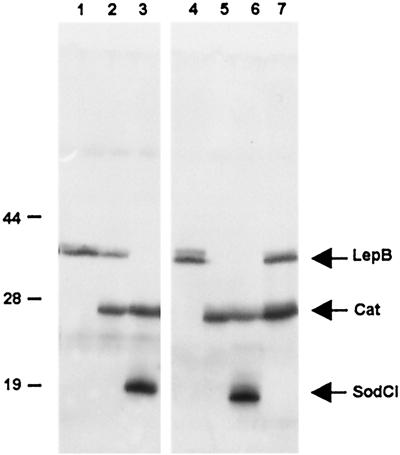
The work above suggested that the tagging procedure could allow the detection of bacterial proteins synthesized during Salmonella infection of an animal host. To test this possibility, two sets of mice were inoculated with strains carrying 3xFLAG tags in sodCI, lepB, and cat genes. Animals were killed 4 days after inoculation, and their spleens were removed and processed for the recovery of bacteria as described (31). Results from representative samples are shown in Fig. 5. Despite the presence of considerable amounts of contaminating material from spleenic tissues, the data in Fig. 5 confirm that the detection of the tagged bacterial products is highly specific and reproducible.
Figure 5.
Immunodetection of tagged bacterial proteins from infected mouse spleens. Bacteria were extracted from spleens and lysed as described in Materials and Methods. Lysates were subjected to electrophoretic separation in a 10% SDS-polyacrylamide gel. Samples are from two groups of mice inoculated i.p. with strain MA7195 (lepB-39∷3xFLAG KnR) (lanes 1 and 4); strain MA7228 (lepB-39∷3xFLAG KnR ilvI3305∷Tn10dTac-cat-43∷3xFLAG KnR) (lanes 2 and 7), and strain MA7224 (sodCI-30∷3xFLAG KnR ilvI3305∷Tn10dTac-cat-43∷3xFLAG KnR) (lanes 3 and 6). One mouse was injected with strain MA7223 (ilvI3305∷Tn10dTac-cat-43∷3xFLAG KnR) (lane 5). Lanes were loaded with material from 5 × 106 cfu.
Discussion
In the present study, we have developed tools enabling the introduction of epitope tails in genes of interest in a bacterium's chromosome. The C-terminal-tagged proteins can be detected immunologically. Although the work was carried out entirely in Salmonella, the technique should be readily extendible to other bacterial species. Here, we used the method to monitor expression and regulation of known or presumptive genes. In this application, the method offers a number of advantages over alternative gene reporter techniques: (i) it does not rely on the enzymatic activity of a reporter gene product; (ii) it is minimally disruptive because of the small size of the epitope tail; (iii) it does not require any cloning steps; and (iv) it is highly sensitive. In addition, the presence of the epitope tag opens the way to a variety of in-depth studies involving immunostaining of cells and tissues, immuno-fluorescence, and protein purification. Included in the latter is the possibility of identifying interacting factors by copurification. For example, immunoaffinity purification of proteins secreted from intracellular bacteria could help identifying proteins targeted by these effectors in the host cytosol. Altogether, the above features make the method particularly suited to the study of genes of intracellular pathogens. Other interesting advantages of the technology proposed here include the possibility of epitope-tagging several proteins within the same bacterial strain. Thus, functional analysis of different genes may be conducted in parallel, a feature of particular interest when studying genes whose activation is expected to be controlled in a coordinated fashion. Epitope-tagged gene(s) expressed constitutively (cat gene, this study) can be used as a positive control and/or as an internal reference. Finally, tagging does not need to be limited to the carboxyl termini of proteins. A set of template plasmids especially designed for the production of internally and N-terminal-tagged proteins was recently constructed in our laboratories and will be available on request.
Some of the results obtained in the present study provide further insight into the involvement of prophage genes in Salmonella pathogenicity. Prophage Gifsy-2 encodes a superoxide dismutase, SodCI, implicated in bacterial protection against macrophage oxidative burst (35, 36) and recently proposed to be involved in the “avirulence” phenotype conferred by another Gifsy-2 gene, grvA (33). Consistent with these findings, we show here that the sodCI gene is highly expressed during active proliferation of Salmonella in epithelial cells (Hep-2) and in murine spleens. Two loci present on the Gifsy-1 prophage, gogB and gogC, are also activated in salmonellae growing inside epithelial cells. The gogB gene encodes a leucine-rich protein of ≈56 kDa with homology to a recently described class of type III secretion substrates (34). We showed that the expression of gogB is under the control of the SsrB transcriptional activator. The latter is part of a two-component regulatory system encoded within Salmonella pathogenicity island 2 (24). We confirmed previous data showing that the gtgB locus of Gifsy-2 [encoding yet another type III secreted protein (34)] is also part of the SsrAB regulatory network (24). Overall, these results indicate that the phage-borne loci evade prophage-specific repression and “plug-in” the regulatory circuitry of the bacterium. Such capacity is likely to be at the basis of the spreading of these sequences within the Salmonella complex.
Acknowledgments
We are indebted to B. Wanner and S. Maloy for the gift of plasmids and strains. We are grateful to M. J. Worley and F. Heffron for the gift of strain MJW141. We thank D. Maloriol for skillful technical assistance. This work was supported by the Centre National de la Recherche Scientifique and by grants from the Ministero dell'Università e della Ricerca Scientifica e Tecnologica, 2001.
Abbreviations
- FRT
FLP recognition target
- HA
hemagglutinin
- cfu
colony-forming unit
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schneider B L, Seufert W, Steiner B, Yang Q H, Futcher A B. Yeast. 1995;11:1265–1274. doi: 10.1002/yea.320111306. [DOI] [PubMed] [Google Scholar]
- 3.Puig O, Rutz B, Luukkonen B G, Kandels-Lewis S, Bragado-Nilsson E, Seraphin B. Yeast. 1998;14:1139–1146. doi: 10.1002/(SICI)1097-0061(19980915)14:12<1139::AID-YEA306>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 4.Knop M, Siegers K, Pereira G, Zachariae W, Winsor B, Nasmyth K, Schiebel E. Yeast. 1999;15:963–972. doi: 10.1002/(SICI)1097-0061(199907)15:10B<963::AID-YEA399>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 5.Russell C B, Thaler D S, Dahlquist F W. J Bacteriol. 1989;171:2609–2613. doi: 10.1128/jb.171.5.2609-2613.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dabert P, Smith G R. Genetics. 1997;145:877–889. doi: 10.1093/genetics/145.4.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Toro C S, Mora G C, Figueroa-Bossi N. J Bacteriol. 1998;180:4750–4752. doi: 10.1128/jb.180.17.4750-4752.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.El Karoui M, Amundsen S K, Dabert P, Gruss A. Nucleic Acids Res. 1999;27:1296–1299. doi: 10.1093/nar/27.5.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Figueroa-Bossi N, Uzzau S, Maloriol D, Bossi L. Mol Microbiol. 2001;39:260–272. doi: 10.1046/j.1365-2958.2001.02234.x. [DOI] [PubMed] [Google Scholar]
- 10.Murphy K C. J Bacteriol. 1998;180:2063–2071. doi: 10.1128/jb.180.8.2063-2071.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poteete A R, Fenton A C. Genetics. 1993;134:1013–1021. doi: 10.1093/genetics/134.4.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Datsenko K A, Wanner B L. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. . (First Published May 30, 2000; 10.1073/pnas.120163297) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murphy K C, Campellone K G, Poteete A R. Gene. 2000;246:321–330. doi: 10.1016/s0378-1119(00)00071-8. [DOI] [PubMed] [Google Scholar]
- 14.Yu D, Ellis H M, Lee E C, Jenkins N A, Copeland N G, Court D L. Proc Natl Acad Sci USA. 2000;97:5978–5983. doi: 10.1073/pnas.100127597. . (First Published May 16, 2000; 10.1073/pnas.100127597) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cherepanov P P, Wackernagel W. Gene. 1995;158:9–14. doi: 10.1016/0378-1119(95)00193-a. [DOI] [PubMed] [Google Scholar]
- 16.Figueroa-Bossi N, Coissac E, Netter P, Bossi L. Mol Microbiol. 1997;25:161–173. doi: 10.1046/j.1365-2958.1997.4451807.x. [DOI] [PubMed] [Google Scholar]
- 17.Prickett K S, Amberg D C, Hopp T P. BioTechniques. 1989;7:580–589. [PubMed] [Google Scholar]
- 18.Xu T, Rubin G M. Development (Cambridge, UK) 1993;117:1223–1237. doi: 10.1242/dev.117.4.1223. [DOI] [PubMed] [Google Scholar]
- 19.Molloy S S, Thomas L, VanSlyke J K, Stenberg P E, Thomas G. EMBO J. 1994;13:18–33. doi: 10.1002/j.1460-2075.1994.tb06231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hernan R, Heuermann K, Brizzard B. BioTechniques. 2000;28:789–793. doi: 10.2144/00284pf01. [DOI] [PubMed] [Google Scholar]
- 21.Miller V L, Mekalanos J J. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herrero M, de Lorenzo V, Timmis K N. J Bacteriol. 1990;172:6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fields P I, Swanson R V, Haidaris C G, Heffron F. Proc Natl Acad Sci USA. 1986;83:5189–5193. doi: 10.1073/pnas.83.14.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Worley M J, Ching K H, Heffron F. Mol Microbiol. 2000;36:749–761. doi: 10.1046/j.1365-2958.2000.01902.x. [DOI] [PubMed] [Google Scholar]
- 25.Maloy S R, Stewart W J, Taylor R K. Genetic Analysis of Pathogenic Bacteria. Plainview, NY: Cold Spring Harbor Lab. Press; 1996. [Google Scholar]
- 26.Miller J H. A Short Course in Bacterial Genetics: A Laboratory Manual and Handbook for Escherichia coli and Related Bacteria. Plainview, NY: Cold Spring Harbor Lab. Press; 1992. [Google Scholar]
- 27.Wilson I A, Niman H L, Houghten R A, Cherenson A R, Connolly M L, Lerner R A. Cell. 1984;37:767–778. doi: 10.1016/0092-8674(84)90412-4. [DOI] [PubMed] [Google Scholar]
- 28.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 29.Lee C A, Jones B D, Falkow S. Proc Natl Acad Sci USA. 1992;89:1847–1851. doi: 10.1073/pnas.89.5.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Figueroa-Bossi N, Bossi L. Mol Microbiol. 1999;33:167–176. doi: 10.1046/j.1365-2958.1999.01461.x. [DOI] [PubMed] [Google Scholar]
- 31.Slauch J M, Mahan M J, Mekalanos J J. Methods Enzymol. 1994;235:481–492. doi: 10.1016/0076-6879(94)35164-3. [DOI] [PubMed] [Google Scholar]
- 32.Stanley T L, Ellermeier C D, Slauch J M. J Bacteriol. 2000;182:4406–4413. doi: 10.1128/jb.182.16.4406-4413.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ho T D, Slauch J M. J Bacteriol. 2001;183:611–620. doi: 10.1128/JB.183.2.611-620.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miao E A, Miller S I. Proc Natl Acad Sci USA. 2000;97:7539–7544. doi: 10.1073/pnas.97.13.7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Farrant J L, Sansone A, Canvin J R, Pallen M J, Langford P R, Wallis T S, Dougan G, Kroll J S. Mol Microbiol. 1997;25:785–796. doi: 10.1046/j.1365-2958.1997.5151877.x. [DOI] [PubMed] [Google Scholar]
- 36.De Groote M A, Ochsner U A, Shiloh M U, Nathan C, McCord J M, Dinauer M C, Libby S J, Vazquez-Torres A, Xu Y, Fang F C. Proc Natl Acad Sci USA. 1997;94:13997–14001. doi: 10.1073/pnas.94.25.13997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.March P E, Inouye M. Proc Natl Acad Sci USA. 1985;82:7500–7504. doi: 10.1073/pnas.82.22.7500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.El Hanafi D, Bossi L. Mol Microbiol. 2000;37:583–594. doi: 10.1046/j.1365-2958.2000.02015.x. [DOI] [PubMed] [Google Scholar]
- 39.March P E, Inouye M. J Biol Chem. 1985;260:7206–7213. [PubMed] [Google Scholar]