Abstract
The food we consume feeds not only us, but also a vast and diverse community of microbiota within our gastrointestinal tract. In a process of symbiotic co-evolution, the gut microbiota became essential for the maintenance of the health and integrity of our colon. The advent of next-generation DNA sequencing technology and metabolic profiling have, in the recent years, revealed the remarkable complexity of microbial diversity and function, and that the microbiota produce a wide variety of bioactive products that are not only active at the mucosal surface, but also absorbed and circulated throughout the body, influencing distant organ health and function. As a result, several microbiota compositional patterns and their associations with both health and disease states have been identified. Importantly, a disturbed micro-biota–host relationship, termed dysbiosis, is now recognized to be the root cause for a growing list of diseases, including colorectal cancer (CRC). There is mounting in vitro and in vivo evidence to suggest that diet selects for the microbiota composition and several health promoting and deleterious effects of diet are, in fact, mediated by the microbiota. Recent findings of the feasibility of dietary fiber to boost the colonic microbial synthesis of anti-proliferative and counter carcinogenic metabolites, particularly butyrate, underscores the prerequisite of dietary modification as a key measure to curb the pandemic of CRC in westernized countries. Better understanding of the diet–microbiota interplay and large-scale studies to evaluate the efficacy of dietary modification and gut microbiota modulation in reversing dysbiosis and restoring health could offer novel preventative and/or therapeutic strategies against westernized diseases, which are now considered the chief threat to public health.
Introduction
A large and diverse community of microbiota comprising about a 100 trillion microbes, termed the microbiome, inhabits the human gastrointestinal tract.1 The microbiome thrives on the undigested dietary residues in the intestinal lumen and in return yields a wide array of metabolites, termed the meta-bolome, by conducting a robust network of intricate metabolic functions. The intestinal commensal bacteria resist proliferation of pathogenic organisms by competing for nutrition and producing bactericidal factors, play a crucial role in the immunomodulation of the gut associated lymphoid tissue, reinforce gut mucosal defense barrier, and synthesize vitamins such as biotin, folic acid and vitamin-K serving an excellent paradigm of symbiosis.2–5 Colon, rich in microbiota present at a concentration of 1012 cfu mL−1, can now be perceived as a ‘new’ metabolic organ, with its functional potential matching that of the liver.6
In the present ‘-omic’ era, state-of-the-art techniques such as high throughput gene sequencing and polymerized chain reaction (PCR) of conserved regions of microbial 16S rRNA have enabled identification of the predominantly ‘unculturable’ anerobic gut microbiota.7 Advances in the fields analyz ing the genomic content of the gut microbiota (meta-genomics); elucidating their metabolic functional potential through metabolite profiling (metabolomics); and studying their interactions and influence on our health and disease (metabonomics) have given us a new perspective of the role of microbiota in our health and disease.7,8
After being passed on maternally and shaped by factors such as breast-feeding, maternal skin and environmental contact, the gross microbiota composition and distribution of an individual remains quite resilient.9,10 Interestingly, fundamentally distinct microbiomes have been identified among people of different origin that could be categorized into three human fecal ‘enterotypes’ based on the abundance of one of three genera: Bacteroides (enterotype 1), Prevotella (enterotype 2) and Ruminococcus (enterotype 3).11 However, microbiome is a dynamic entity that is affected by several factors besides diet such as exposure to antibiotics, and gastrointestinal diseases and surgery that can disturb the composition and metabolic activity of gut microbiota resulting in a state of disturbed host–microbiota homeostasis, termed dysbiosis.12,13 In addition, several studies have shown that microbiome changes in composition and diversity as we age, but specific or consistent patterns have not been recognized to date. However, the existing evidence supports a heightened inflammatory state in the elderly hypothesized to be secondary to a decline in gut immune system (‘immunosenescence’) and perpetuation of a chronic low grade inflammation (‘inflamm’aging’) in association with enrichment of facultative anerobes or pathobionts and decrease in symbiotic species such as Faecalibacterium prauznitzii that have anti-inflammatory effect.14,15 Dysbiosis has been identified as the root cause for a growing list of medical problems including allergy and autoimmune disorders, irritable bowel syndrome, inflammatory bowel disease, diabetes, obesity, clostridium difficile diarrhea, and colon cancer.16–21
Research in this field had produced a wealth of information in the recent years and several reviews have summarized a plethora of observational, in vitro, and in vivo studies describing the role of the microbiota in health and disease.22 In this review, we will focus on the interplay between diet and micro-biota with regards to its influence on risk of CRC.
Colorectal cancer – association with diet and microbiota
Colorectal cancer (CRC) is the most common gastrointestinal cancer, and ranks globally as the third most common cancer by incidence, and fourth most common in mortality, with over a million people affected annually, nearly half of who die based on GLOBOCAN 2012 estimates.23 Over 90% of CRC cases are sporadic, where a complex interplay between genetic and environmental factors determines neoplastic transformation to colorectal carcinogenesis. Seminal analyses by Doll and Peto of the geographic variation in CRC and its association with diet composition in the 80s had shown that over 90% of the gastrointestinal cancers could be attributed to dietary habits.24 Alcohol, smoking, obesity, and inflammatory bowel disease are also recognized risk factors of CRC. Importantly, dietary factors like higher red and processed meat consumption and deficiency of fiber, calcium, vitamin D and folate are well-recognized factors associated with a higher CRC risk.25 Western nations bear up to 60% of the global disease burden, which has been attributed to the high red meat, high fat, and low fiber content of their diet.26,27
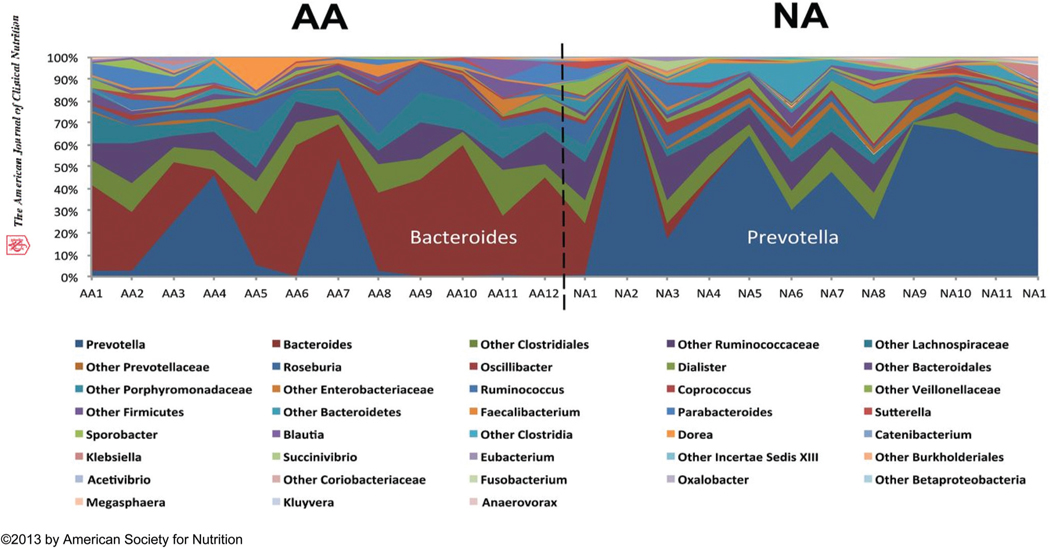
The link between diet and colon cancer is perhaps best exemplified by migration studies. Japanese residents traditionally had a low incidence of CRC, but within one generation of change to a western diet after migration to Hawaii, their incidence increased to levels similar to local Hawaiians.28 In keeping with a microbiota-mediated explanation for the geographic variation of CRC, fundamentally distinct microbiota composition has been identified in different populations. De Filippo et al. noted that the gut microbial composition of European children had higher Enterobacteriaceae representation, whereas the rural African children who consumed higher fiber diet had higher Bacteroidetes and lower Firmicutes counts.29 We confirmed that these differences persisted into adulthood. In our studies into reasons why colon cancer is most common in African Americans (65 : 100 000) in the USA, and rare in rural Africans (<5 : 100 000), we showed that the genotype Prevotella was predominant in native Africans while Bacteroides was in African Americans (Fig. 1).30
Fig. 1.
Distinct colonic microbial composition of African Americans (high CRC risk group) and native rural South Africans (low CRC risk group). Microbial composition was dominated by Bacteroides in the African Americans, which indicated that they belonged to enterotype 1, and was dominated by Prevotella in the native rural South Africans, which categorized them as enterotype 2 (10). AA, African American; CRC, colorectalcancer; NA, native African. Adapted with permission from “Junhai Ou, et al. Am. J. Clin. Nutr., 2013, 98, 111–120, American Society for Nutrition.”
Recent research has produced a wealth of information highlighting the key role of microbiota in mediating the dietary risk of CRC. The fact that colon has the highest microbial concentration and cancer rates along the gastrointestinal tract combined with the observation from gnotobiological studies such as detection of colonic adenocarcinoma in 70% of conventionalized, but not the germ-free TCRbeta and p53 double-knockout (TCRbeta−/− p53−/−) mice, accentuates the crucial role of microbiota in causing CRC.31 Metagenomic studies have now identified specific bacterial species that colonize the tumor as well as non-tumor colonic sites and characterized individualized oncogenic microbiome.21 Association of Streptococcus gallolyticus (previously S. bovis) with CRC was one of the earliest, although the causative link has not been established.32 In one study, fecal samples of CRC patients had reduced butyrate-producing Eubacterium rectale and Faecali-bacterium prausnitzii and increased populations of Enterococcus faecalis, when compared with healthy volunteers.33 High abundance of Fusobacterium necroforum, an oral pathogen causing periodontitis, was recently identified in the CRC tumor tissue compared to normal mucosa.34 Whether this represents cause or effect remains unknown, but as one potential causative mechanism, its adherence to colonic epithelium through FadA adhesion has been shown to stimulate of E-cadherin/β-catenin signaling, a mechanism known to promote carcinogenesis.35 Sobhani et al. compared the fecal microbiota composition in endoscopic mucosal biopsies between healthy subjects and CRC patients.36 They noted significant higher fecal populations of the Bacteroides–Prevotella group in CRC patients with identification of increased representation of IL-17 immunoreactive cells in the healthy subjects, suggesting immunological functional consequences from the dysbiosis.
Overall, review of multiple studies reporting associations between specific microbiota and healthy, adenoma, and CRC populations suggests that there is substantial evidence to date suggestive of microbiotal role in CRC although further research is warranted to establish firm causative links.37 Moreover, it seems clear that it’s not one specific microorganism that is responsible for CRC, but an abundance of a group of bacteria whose detrimental actions surpass those of the beneficial commensals. A recent systematic review of 31 original human and animal studies on association between microbiota and CRC found conclusive evidence of role of microbiota and dysbiosis in CRC.38 It was noted that certain bacteria (such as Fusobacteria, Alistipes, Porphyromonadaceae, Coriobacteridae, Staphylococcaceae, Akkermansia spp. and Methanobacteriales) were consistently augmented, while some other (such as Bifido-bacterium, Lactobacillus, Ruminococcus, Faecalibacterium spp., Roseburia, and Treponema) were underrepresented in CRC. Of note, reduced butyrate and elevated amino acid metabolites were identified throughout colonic carcinogenesis.
Substantial in vitro and in vivo evidence supports the role of microbiota in causing DNA damage and chromosomal instability that lead to mutations responsible for carcino-genesis. Microbiota contribute to neoplastic transformation of the colonic epithelium by instigating a state of chronic inflam mation mediated by signaling pathways such as induction of Toll-like receptors, up-regulation of cyclooxygenase-2 (COX-2), and activation of mitogen-activated protein kinases (MAPK) that promote epithelial proliferation and genetic mutations.39,40 Also, microbiota implement multiple other mechanisms including biotransformation of dietary procarcinogens, production of reactive oxygen and nitrogen species, and genotoxins.41 Contrarily, cancer-protective factors such as folate and biotin that are essential for DNA synthesis and repair are also synthesized by microbiota, such as Bifidobacterium.42
Mechanisms of diet–microbiota mediated CRC risk
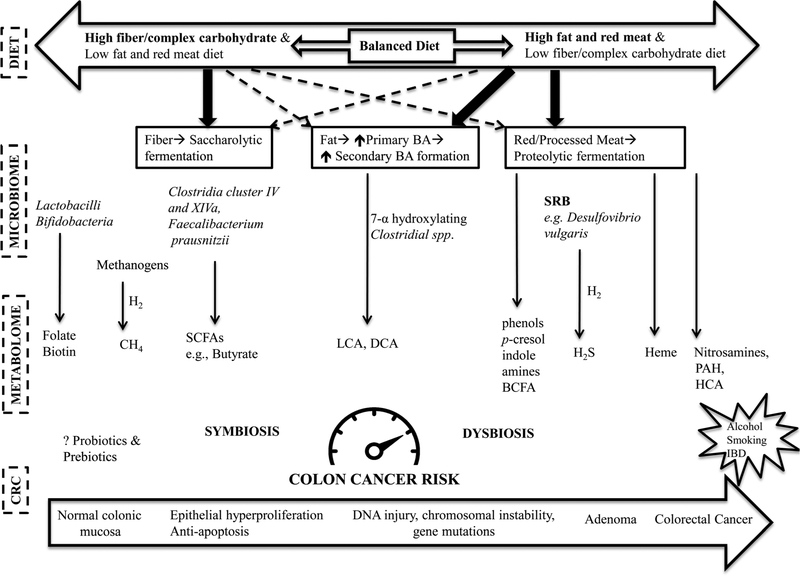
The principal dietary components, i.e., carbohydrates, protein, and fat are essential for the energy and metabolism as well as structural maintenance and repair of all aspects of human functioning. However, their disproportionate consumption or aberrant metabolism predisposes to dysbiosis. Generation of pro-inflammatory and toxic metabolites detrimental to colonic mucosa surmounting the anti-inflammatory defensive meta-bolites seems to be an essential precursor for carcinogenesis. Here, we will discuss the role of the well-recognized diets and microbiota implicated in either aggravating (red meat and fat) or mitigating (fiber) CRC risk (Fig. 2).
Fig. 2.
Dietary risk of CRC is mediated by dysbiosis of gut microbiota and their metabolites. Dietary fiber/complex carbohydrates promote saccharolytic fermentation yielding anti-inflammatory and antiproliferative SCFAs, such as butyrate, whereas, red meat generates inflammatory and genotoxic metabolites by promoting proteolytic fermentation, H2S production from its sulfur-rich amino acid content, and exposing colonic mucosa to other carcinogenic constituents such as heme, nitrosamines, HCA, and PAH. High dietary fat promotes excess primary BA secretion and their conversion to pro-carcinogenic secondary bile acids (LCA, DCA). Dysbiosis, an imbalance between the ‘protective’ and ‘detrimental’ microbiota composition and their metabolic end-products results determines the risk of CRC. BA (bile acids), BCFA (branched-chain fatty acids), CH4 (methane), DCA (deoxycholic acid), HCA (heterocyclic amines), H2 (hydrogen), H2S (hydrogen sulfide), LCA (lithocholic acid), PAH (polyaromatic hydrocarbons), short-chain fatty acids (SCFAs). (Original work).
Fiber
Working in East Africa in the 50s, Burkitt noted the association between the traditional African diet, which contained 50–100 g fiber per day, and absence of non-inflammatory bowel diseases, particularly colon cancer, being one of the first to suggest the protective role of fiber.43 Up to 20% of potentially digestible starch plus resistant starches enter the colon and undergo saccharolytic fermentation by most members of the microbiota, but especially Bacteroides spp., Lactobacillus spp., and Bifidobacterium spp.44 This process yields end-products such as short-chain fatty acids (SCFAs); gases such as carbon dioxide (CO2), hydrogen (H2), methane (CH4); and ethanol. Acetate, propionate, and butyrate constitute the three major SCFAs.
Whilst all three SCFAs have health-promoting effects on the colonic mucosa, butyrate is the most potent with respect to cancer protection. It is produced predominantly by Clostridia clusters XIVa and IV of the genus Firmicutes.45 Butyrate is the chief energy source for colonocytes and regulator of epithelial proliferation.46 Multiple potential mechanisms have been identified suggesting the protective action of butyrate against colorectal carcinogenesis.47 Butyrate exhibits anti-proliferative activity by activation of the apoptosis cascade and arresting the growth of tumors by histone hyperacetylation.48 It suppresses tumors by potentiating p53 gene expression and transforming growth factor-β (TGF-β) signaling, and increasing the immunogenicity of cancer cells.49,50 Its anti-inflammatory properties are mediated by suppressing nuclear factor-kB activation, a transcription factor controlling the expression of genes encoding proinflammatory responses and inflammatory mediators like tumor necrosis factor-α (TNF-α) and nitric oxide.51,52 On the other hand, acetate and propionate are largely absorbed systemically and play key roles in glucose and lipid metabolism.53,54
Red and processed meat
Red meat and processed meat contain an array of procarcinogenic constituents that act either directly or via their colonic end-metabolites.55 High protein and heme content of the red meat have been shown to be deleterious to the colonic mucosal health. Proteolytic fermentation of the meat residues by the colonic microbiota such as Bacteroides spp., and Clostridium spp. generates inflammatory and procarcinogenic end products such as nitrosamines, branched-chain fatty acids, phenolic (phenols and p-cresol) and indolic compounds from aromatic amino acids.56 Heme has been shown to cause colonocyte injury, inhibit apoptosis, and enhance crypt hyperplasia and thereby promoting colonic epithelial proliferation.57
In addition, several carcinogenic chemicals such as poly-cyclic-aromatic hydrocarbons (PAH) and heterocyclic amines (HCA) are formed when cooking meat at very high temperatures and importantly through bacterial enzymatic transformation via decarboxylation of aromatic amino acids and N-nitrosation.58,59 Their metabolic derivatives can cause DNA base alkylation and formation of DNA adducts which pre-dispose to mutations and carcinogenesis.60
Red meat is also rich in sulfur containing amino acids, which promote the growth of sulfur-reducing bacteria (SRBs, e.g., Desulfovibrio vulgaris) that convert the H2 gas produced during saccharolytic fermentation into an inflammatory and genotoxic end-product, hydrogen sulfide (H2S).61 H2S impairs cytochrome oxidase, inhibits mucin synthesis, suppresses butyrate utilization, and promotes methylation of DNA by generating free radicals.62 Also, SRBs suppress the growth of methanogens such as Methanogenic archaea (e.g., Methanobrevibacter smithii) that detoxify the H2 into non-toxic methane gas that may be exhaled in the breath.63
Fat
In a murine model, Devkota et al. have noted that a high fat diet (37% of total calories) increased hepatic synthesis of sulfur-rich taurine conjugates of bile acids that promoted the growth of Bilophila wadsworthia in the colon, which generated H2S causing acute colitis in Il10−/− mice.64 High dietary fat stimulates secretion, enterohepatic circulation, and colonic escape of primary bile acids, cholic acid (CA) and chenodeoxycholic acid (CDA).65 Subsequently, high colonic concentrations of CA and CDA promote 7-α hydroxylating Clostridial spp. that carry out dehydrogenation, sulfation, and 7α-dehydroxylation yielding the secondary bile acids, deoxycholic acid (DCA) and lithocholic acid (LCA).66 Secondary bile acids have been shown to be anti-apoptotic and genotoxic via oxidative stress through generation of reactive oxygen species and suppression of p53 response to DNA damage causing cellular proliferation.67 Fur thermore, Bernstein et al. demonstrated that the addition of secondary bile acids to the drinking water of rats induced spontaneous carcinogenesis that could be blocked by simultaneous administration of antioxidants.68
The role of diet and microbiota in the mediation of CRC risk
There is substantial in vitro and in vivo evidence to suggest that dietary patterns shape the dynamic composition and diversity of colonic microbiota. In a series of studies, we tested our hypothesis that microbiota mediate the dietary risk of CRC by analyzing the dietary patterns, colonic mucosal biopsies, fecal and colonic microbiota (measured by colonic evacuation), breath contents of hydrogen and methane, colonic secondary bile acids and SCFAs concentrations among subjects belonging to different CRC risk populations, African Americans (high risk, incidence of ~65 : 100 000), Caucasian Americans (moderate risk, incidence ~40 : 100 000), and native Africans in rural South Africa (low CRC incidence of ~5 : 100 000).26,30,42,69,70
African Americans were noted to consume significantly (p < 0.01) higher protein (94 ± 9.3 g d−1 vs. 58 ± 4.1 g d−1) and fat (114 ± 11.2 vs. 38 ± 3.0 g d−1), meat, saturated fat, cholesterol, as well as higher (p < 0.05) calcium, vitamin A, and vitamin C, and similar amount of fiber intake when compared to native Africans.42 And they had significantly higher breath H2, fecal colony counts of 7-alpha dehydroxylating bacteria, and colonic crypt cell proliferation rates – a biomarker of cancer risk. On the other hand, native Africans who consumed a diet high in complex carbohydrates and resistant starch had higher breath methane and fecal colony counts of Lactobacilli, colonic SCFAs and butyrate concentrations when compared with Caucasian Americans and African Americans. And, the American groups had a significantly higher colonic content of 2-methylvalerate, a branched-chain fatty acid derived from protein fermentation.70 Interestingly, native Africans, despite their lower dietary consumption of folate, vitamin B12, and biotin, had high colonic contents, indicating de novo synthesis by the microbiota. In terms of dietary fat intake and fecal secondary bile acids, African Americans had 3–4 times higher (p < 0.05) colonic LCA, DCA, and CA content than the native Africans, but similar when compared to Caucasian Americans.70 We have already discussed the major phylogenetic differences in fecal and colonic microbiota above, with African Americans conforming to ‘enterotype 1’ and Africans to ‘enter-otye 2’, with greater abundances of starch degraders, such as Dialister, Oscilispera, Succinovibrio, Xylanibactcter, and butyrate producers.30 Dietary factors seem to play a key role in shaping the gut microbiota as evidenced by strong association of long-term consumption of protein and animal fat rich diet with Bacteroides predominance (enterotype 1) and that of carbohydrates with Prevotella predominance (enterotype 2).71 In a study comparing the fecal microbiota, the rural African children were noted to have a unique abundance of bacteria belonging to the genii Prevotella and Xylanibacter (absent in their European counterparts) that had genes for fermentation of cellulose and xylan hypothesized to be a consequence of high fiber diet in order to allow maximal metabolic energy extraction from ingested plant fiber.29 In keeping with the earlier findings, most of our adult rural South African subjects rural were also found to have predominance of Prevotella and starch degraders (Succinivibrio and Oscillospira) reflecting their high consumption of plant based polysaccharide diet. Equally impressive was the differences in diversity of fecal methanogenic archaea (MA) and SRB. Native Africans were found to have a higher proportion and diversity of MA and distinct SRB populations when compared their American and European counterparts.72 We concluded that it was the colonic milieu rather than an individual microbe, metabolite, nutrient, or component, that determines the overall colon cancer risk.73
Dietary intervention to modulate colonic microbiota and influence CRC risk
Dietary changes have been shown to result in swift shifts in microbial composition. In a germ-free mouse model colonized with human microbiota, switching the diet from a low fat to high fat altered the microbial diversity within a single day.74 In healthy volunteers, David et al. examined how dietary change altered human gut communities within 5 days when the diet was switched from a ‘plant-based’ (rich in grains, legumes, fruits and vegetables) to an ‘animal-based’ (rich in meats, eggs, and cheese) regimen.75 The animal-based diets increased the abundance of microorganisms such as Bilophila wadworthia, which is linked to dietary fat, bile acids, and mucosal inflammation and decreased the levels of saccharolytic Firmicutes. In their 10-day controlled-feeding study on effect of diet on human fecal microbiota, Wu et al. used diet inventories and 16S rDNA sequencing of fecal microbiota and noted the enterotype clustering of Bacteroides predominantly in subjects on protein and animal fat rich diets and Prevotella in those on carbohydrate and simple sugar rich diets.71 Interestingly, changes in microbiome composition were identified when switched between high-fat/low-fiber or low-fat/high-fiber diets, but the enterotype clustering remained stable suggesting it takes long-term dietary changes to allow microbial adaptation and significant shift in their compositions.
In summary, we have substantial human and experimental evidence that change to a western diet explains the increase in colon cancer. However, westernization results in changes in many other aspects of the environment that might also increase exposure to luminal carcinogens. In an attempt to investigate the role of diet alone, we switched the diets of Americans and rural Africans for a 2 week period and measured the changes in the fecal and colonic microbiota, their metabolites, and inflammatory and cancer biomarkers in colonic biopsies. Global analysis of the microbiota was done using 16S rRNA gene phylogenetic microarray (Human Intestinal Tract Chip (HITChip)), which covers over 1000 of the cur rently known bacterial species from the human intestine and has been demonstrated to provide highly concordant results concerning the microbiota composition when compared to 16S or metagenome sequencing.76 HITChip allows deep profiling of phylotypes at high resolution, down to <0.1% relative abundance; corresponding to a duplicated set of 100 000 pyro-sequencing reads per sample with very high reproducibility (>98%) and at considerably lower cost.77–79 In general, targeted analysis of microbes known to be related to the area of interest is best pursued first, followed by complete microbial genome sequencing (e.g. ‘shotgun’ sequencing) which can identify, or predict the presence of unknown microbes and their metabolic potentials.80 The choice of sequencing method, which is evolving daily, very much depends upon budget, as the newer techniques can cost as much as $1000 per sample, making them inappropriate for epidemiological studies. Much of the development is now dependent on advanced computerized integration of the massive amounts of data generated from these techniques, and so the cost of interpretation will exceed the analytical costs. For our study functional gene analysis was performed using real-time quantitative PCR. Fecal short-chain fatty acids were measured by gas chromatography and bile acids by liquid chromatography-mass spectrometry.
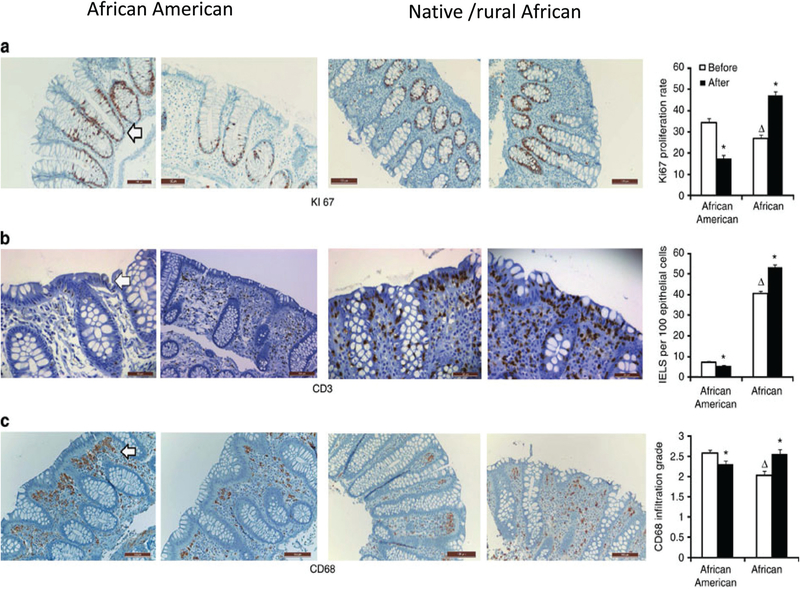
All volunteers were housed for the 2 weeks, African Americans in our Clinical Translational Research Center, Africans in a rural lodge, and their meals were cooked on site and delivered under strict supervision.27 The rural South Africans were fed a western-style diet (51% fat, 27% protein, 20% carbohydrate, 3 g fiber per 1000 kcal), and African Americans were fed a traditional African style diet (17% fat, 15% protein, 68% carbohydrate, 21 g fiber per 1000 kcal). On switching to a high fiber/low fat diet, the butyrate increased by 2.5-fold and secondary bile acids decreased by 70% in African Americans. On the other hand, switching to low fiber/high fat diet in native Africans resulted in reduction of colonic butyrate content by 50% and increased in secondary bile acids by 400%. Similarly, in our functional gene analysis to capture the functional ability of the microbiome (as a whole as several microbes can share the same functional ability), native Americans who initially had greater expression of the bcoA gene that codes for the enzyme butyryl-CoA:acetate CoA-transferase, which is responsible for the last step in butyrate synthesis and mcrA gene for the enzyme responsible for methanogenesis at baseline, had a reduction in these gene expressions after the diet switch. And, baiCD gene reflecting the 7-α-dehydroxylating enzyme responsible for secondary bile acid production was low initially and increased on higher fat diet after switch. Reciprocal changes were noted among the African Americans. Most importantly, these reciprocal changes in aspects of the microbiome and metabolome were associated with reciprocal changes in mucosal biomarkers of cancer risk: namely mucosal epithelial proliferation rate (Ki67 staining) and immunohistochemical markers of inflammation (CD3+ intraepithelial lymphocytes and CD68+ lamina propria macrophages), which were suppressed in Americans by the ‘African’ diet, and increased in Africans by the ‘western’diet (Fig. 3). These findings are excit ing as they show not only that colonic microbial metabolism responds rapidly to dietary modification, but also that these microbial-metabolic changes are accompanied by changes in mucosa known to increase or decrease the susceptibility to neo-plastic change and cancer risk within 2 weeks.
Fig. 3.
Colonic mucosal immunohistochemistry of proliferative and inflammatory biomarkers. Immunohistochemical analysis of colonic mucosal biopsies taken at colonoscopy with their associated quantitative analysis on the right panel. Decreased expression of the cancer biomarker – Ki67 staining of epithelial crypt cells (a) and inflammatory biomarkers – CD3+ staining (b) and CD68+ macrophages in the lamina propria (c) in an African American (the first and second panels) and reciprocal changes in a native/rural African (the third and 4th panels left to right) before and after dietary switch. The bar graphs on the far right summarize the group mean ± S.E. results in 20 African Americans and 12 rural Africans. The two-tailed Mann–Whitney U-test was used for comparisons for non-paired samples and the Wilcoxon rank sum test for paired samples, with Bonferroni correction for multivariate comparisons. Triangles indicate significant (P < 0.05) baseline differences and stars indicate significant changes induced by diet switch. Adapted with permission from Nat. Commun., 20156(Apr 28), 6342, DOI: 10.1038/ncomms7342”.
Conclusion and future direction
Our knowledge of the microbiota–host interactions and their impact on health outcomes is still in infancy as we are transitioning our research efforts from ‘observing the association(s)’ to ‘understanding the causation’. The new ‘organ status’ of the microbiota has attracted great attention as a target for prophylactic or therapeutic intervention. Interestingly, a recent study has identified 20 microbial gene markers that differentiated patients with adenomatous polyps and cancer from healthy control subjects.81 The study also validated 4 of those microbial genes that were enriched in the microbiome of patients, in ethically different (Danish, French, and Austrian) cohorts, with early stage (stage I–II) CRC highlighting the potential role of fecal biomarkers for earlier diagnosis of CRC. There has been tremendous enthusiasm in the field of ‘functional foods’ with promotion of ‘prebiotics’ (indigestible carbohydrates that enhance the population or activity of ‘beneficial’ probiotic intestinal microorganisms), ‘probiotics’ (microorganisms ingested to populate the intestine for health benefits), and ‘synbiotics’ (combination of pre- and probiotics). While their health benefits seem intuitive and the supportive in vitro evidence is promising, it is doubtful that the benefits of prebiotics and prebiotics can outweigh those of a normal ‘balanced’ diet that our genome evolved together with.
Biography
Dr Stephen J. O’Keefe explores the field of nutritional gastroenterology by conducting translational research into the physiological and pathophysiological responses to dietary intake and interventional feeding. He is currently examining microbiota-metabolomic mechanisms, with the support of National Institute of Health, that explain why the chief determinant of colon cancer is diet, and why African Americans have the highest risk of colon cancer in the USA, whilst rural South Africans from KwaZulu-Natal rarely get the disease. Dr Kishore Vipperla is his research associate with interest in the fields of short gut syndrome and modulation of colon cancer risk by dietary modification.
References
- 1.Huttenhower C, et al. , Human Microbiome Project Consortium, Structure, function and diversity of the healthy human microbiome, Nature, 2012, 486(7402), 207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wardwell LH, Huttenhower C and Garrett WS, Current concepts of the intestinal microbiota and the pathogenesis of infection, Curr. Infect. Dis. Rep, 2011, 13(1), 28–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cebra JJ, et al. , Development and maintenance of the gut-associated lymphoid tissue (GALT): the roles of enteric bacteria and viruses, Dev. Immunol, 1998, 6(1–2), 13–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tremaroli V and Backhed F, Functional interactions between the gut microbiota and host metabolism, Nature, 2012, 489(7415), 242–249. [DOI] [PubMed] [Google Scholar]
- 5.O’Keefe SJD, et al. , Products of the Colonic Microbiota Mediate the Effects of Diet on Colon Cancer Risk, J. Nutr, 2009, 139(11), 2044–2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O’Keefe SJ, The colon as a metabolic organ, S. Afr. Med. J, 1994, 84(7), 376–377. [PubMed] [Google Scholar]
- 7.Schloss PD and Handelsman J, Metagenomics for studying unculturable microorganisms: cutting the Gordian knot, Genome Biol., 2005, 6(8), 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nicholson JK and Lindon JC, Systems biology: Metabonomics, Nature, 2008, 455(7216), 1054–1056. [DOI] [PubMed] [Google Scholar]
- 9.Mackie RI, Sghir A and Gaskins HR, Developmental microbial ecology of the neonatal gastrointestinal tract, Am. J. Clin. Nutr, 1999, 69(5), 1035S–1045S. [DOI] [PubMed] [Google Scholar]
- 10.Mueller NT, et al. , The infant microbiome development: mom matters, Trends Mol. Med, 2015, 21(2), 109–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arumugam M, et al. , Enterotypes of the human gut micro-biome, Nature, 2011, 473(7346), 174–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Modi SR, Collins JJ and Relman DA, Antibiotics and the gut microbiota, J. Clin. Invest, 2014, 124(10), 4212–4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li JV, et al. , Metabolic surgery profoundly influences gut microbial-host metabolic cross-talk, Gut, 2011, 60(9), 1214–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Biagi E, et al. , Aging of the human metaorganism: the microbial counterpart, Age, 2012, 34(1), 247–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rajilic-Stojanovic M, et al. , Development and application of the human intestinal tract chip, a phylogenetic micro-array: analysis of universally conserved phylotypes in the abundant microbiota of young and elderly adults, Environ. Microbiol, 2009, 11(7), 1736–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marsland BJ and Salami O, Microbiome influences on allergy in mice and humans, Curr. Opin. Immunol, 2015, 36, 94–100. [DOI] [PubMed] [Google Scholar]
- 17.Collins SM, A role for the gut microbiota in IBS, Nat. Rev. Gastroenterol. Hepatol, 2014, 11(8), 497–505. [DOI] [PubMed] [Google Scholar]
- 18.Hold GL, et al. , Role of the gut microbiota in inflammatory bowel disease pathogenesis: what have we learnt in the past 10 years?, World J. Gastroenterol, 2014, 20(5), 1192–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen J, Obin MS and Zhao L, The gut microbiota, obesity and insulin resistance, Mol. Aspects Med, 2013, 34(1), 39–58. [DOI] [PubMed] [Google Scholar]
- 20.Willing BP, Russell SL and Finlay BB, Shifting the balance: antibiotic effects on host-microbiota mutualism, Nat. Rev. Microbiol, 2011, 9(4), 233–243. [DOI] [PubMed] [Google Scholar]
- 21.Marchesi JR, et al. , Towards the human colorectal cancer microbiome, PLoS One, 2011, 6(5), 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vipperla K and O’Keefe SJ, The microbiota and its metabolites in colonic mucosal health and cancer risk, Nutr. Clin. Pract, 2012, 27(5), 624–635. [DOI] [PubMed] [Google Scholar]
- 23.Ferlay J, Soerjomataram I, Ervik M, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F, GLOBOCAN 2012 v1.0, Cancer Incidence and Mortality Worldwide, IARC CancerBase No. 11 [Online], 2013, http://globocan.iarc.fr, accessed on 30/January/2016.
- 24.Doll R and Peto R, The causes of cancer: quantitative estimates of avoidable risks of cancer in the United States today, J. Natl. Cancer Inst, 1981, 66(6), 1191–1308. [PubMed] [Google Scholar]
- 25.Norat T, et al. , Fruits and vegetables: updating the epidemiologic evidence for the WCRF/AICR lifestyle recommendations for cancer prevention, Cancer Treat. Res, 2014, 159, 35–50. [DOI] [PubMed] [Google Scholar]
- 26.O’Keefe SJ, et al. , Why do African Americans get more colon cancer than Native Africans?, J. Nutr, 2007, 137(1 Suppl), 175S–182S. [DOI] [PubMed] [Google Scholar]
- 27.O’Keefe SJ, et al. , Fat, fibre and cancer risk in African Americans and rural Africans, Nat. Commun, 2015, 6, 6342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Le Marchand L and Kolonel LN, Cancer in Japanese migrants to Hawaii: interaction between genes and environment, Rev. Epidemiol Sante Publique, 1992, 40(6), 425–430. [PubMed] [Google Scholar]
- 29.De Filippo C, et al. , Impact of diet in shaping gut micro-biota revealed by a comparative study in children from Europe and rural Africa, Proc. Natl. Acad. Sci. U. S. A, 2010, 107(33), 14691–14696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ou J, et al. , Diet, microbiota, and microbial metabolites in colon cancer risk in rural Africans and African Americans, Am. J. Clin. Nutr, 2013, 98(1), 111–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kado S, et al. , Intestinal microflora are necessary for development of spontaneous adenocarcinoma of the large intestine in T-cell receptor beta chain and p53 double-knockout mice, Cancer Res, 2001, 61(6), 2395–2398. [PubMed] [Google Scholar]
- 32.Abdulamir AS, Hafidh RR and Abu Bakar F, The association of Streptococcus bovis/gallolyticus with colorectal tumors: the nature and the underlying mechanisms of its ee, J. Exp. Clin. Cancer Res, 2011, 30(11), 1756–9966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Balamurugan R, et al. , Real-time polymerase chain reaction quantification of specific butyrate-producing bacteria, Desulfovibrio and Enterococcus faecalis in the feces of patients with colorectal cancer, J. Gastroenterol. Hepatol, 2008, 23(8 Pt 1), 1298–1303. [DOI] [PubMed] [Google Scholar]
- 34.Castellarin M, et al. , Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma, Genome Res, 2012, 22(2), 299–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rubinstein MR, et al. , Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/beta-catenin signaling via its FadA adhesin, Cell Host Microbe, 2013, 14(2), 195–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sobhani I, et al. , Microbial dysbiosis in colorectal cancer (CRC) patients, PLoS One, 2011, 6(1), 0016393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Keku TO, et al. , The gastrointestinal microbiota and colorectal cancer, Am. J. Physiol. Gastrointest. Liver Physiol, 2015, 308(5), G351–G363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Borges Canha M, Role of colonic microbiota in colorectal carcinogenesis: A systematic review, Rev. Esp. Enferm. Dig, 2015, 107. [DOI] [PubMed] [Google Scholar]
- 39.Terzic J, et al. , Inflammation and colon cancer, Gastroenterology, 2010, 138(6), 2101–2114. [DOI] [PubMed] [Google Scholar]
- 40.Frosali S, et al. , How the Intricate Interaction among Toll-Like Receptors, Microbiota, and Intestinal Immunity Can Influence Gastrointestinal Pathology, J. Immunol. Res, 2015, 2015, 489821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huycke MM, Abrams V and Moore DR, Enterococcus faecalis produces extracellular superoxide and hydrogen peroxide that damages colonic epithelial cell DNA, Carcino-genesis, 2002, 23(3), 529–536. [DOI] [PubMed] [Google Scholar]
- 42.O’Keefe SJ, et al. , Products of the colonic microbiota mediate the effects of diet on colon cancer risk, J. Nutr, 2009, 139(11), 2044–2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Burkitt DP, Diseases of the alimentary tract and western diets, Pathol. Microbiol, 1973, 39(3), 177–186. [DOI] [PubMed] [Google Scholar]
- 44.Cummings JH and Englyst HN, Measurement of starch fermentation in the human large intestine, Can. J. Physiol. Pharmacol, 1991, 69(1), 121–129. [DOI] [PubMed] [Google Scholar]
- 45.Louis P, et al. , Understanding the effects of diet on bacterial metabolism in the large intestine, J. Appl. Microbiol, 2007, 102(5), 1197–1208. [DOI] [PubMed] [Google Scholar]
- 46.Roediger WE, Utilization of nutrients by isolated epithelial cells of the rat colon, Gastroenterology, 1982, 83(2), 424–429. [PubMed] [Google Scholar]
- 47.Fung KY, et al. , A review of the potential mechanisms for the lowering of colorectal oncogenesis by butyrate, Br. J. Nutr, 2012, 108(5), 820–831. [DOI] [PubMed] [Google Scholar]
- 48.Davie JR, Inhibition of histone deacetylase activity by butyrate, J. Nutr, 2003, 133(7 Suppl), 2485S–2493S. [DOI] [PubMed] [Google Scholar]
- 49.Nguyen KA, et al. , Dietary fiber enhances a tumor suppressor signaling pathway in the gut, Ann. Surg, 2006, 243(5), 619–625; discussion 625–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Watson AJ, An overview of apoptosis and the prevention of colorectal cancer, Crit. Rev. Oncol. Hematol, 2006, 57(2), 107–121. [DOI] [PubMed] [Google Scholar]
- 51.Rodriguez-Cabezas ME, et al. , Dietary fiber down-regulates colonic tumor necrosis factor alpha and nitric oxide production in trinitrobenzenesulfonic acid-induced colitic rats, J. Nutr, 2002, 132(11), 3263–3271. [DOI] [PubMed] [Google Scholar]
- 52.Inan MS, et al. , The luminal short-chain fatty acid butyrate modulates NF-kappaB activity in a human colonic epithelial cell line, Gastroenterology, 2000, 118(4), 724–734. [DOI] [PubMed] [Google Scholar]
- 53.Al-Lahham SH, et al. , Biological effects of propionic acid in humans; metabolism, potential applications and underlying mechanisms, Biochim. Biophys. Acta, 2010, 11(83), 4. [DOI] [PubMed] [Google Scholar]
- 54.Puchowicz MA, et al. , Zonation of acetate labeling across the liver: implications for studies of lipogenesis by MIDA, Am. J. Physiol, 1999, 277(6 Pt 1), E1022–E1027. [DOI] [PubMed] [Google Scholar]
- 55.Cross AJ, et al. , A large prospective study of meat consumption and colorectal cancer risk: an investigation of potential mechanisms underlying this association, Cancer Res, 2010, 70(6), 2406–2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hughes R, Magee EA and Bingham S, Protein degradation in the large intestine: relevance to colorectal cancer, Curr. Issues Intest. Microbiol, 2000, 1(2), 51–58. [PubMed] [Google Scholar]
- 57.de Vogel J, et al. , Dietary heme injures surface epithelium resulting in hyperproliferation, inhibition of apoptosis and crypt hyperplasia in rat colon, Carcinogenesis, 2008, 29(2), 398–403. [DOI] [PubMed] [Google Scholar]
- 58.Mirvish SS, et al. , Total N-nitroso compounds and their precursors in hot dogs and in the gastrointestinal tract and feces of rats and mice: possible etiologic agents for colon cancer, J. Nutr, 2002, 132(11 Suppl), 3526S–3529S. [DOI] [PubMed] [Google Scholar]
- 59.Cross AJ and Sinha R, Meat-related mutagens/carcinogens in the etiology of colorectal cancer, Environ. Mol. Mutagen, 2004, 44(1), 44–55. [DOI] [PubMed] [Google Scholar]
- 60.Schut HA and Snyderwine EG, DNA adducts of hetero-cyclic amine food mutagens: implications for mutagenesis and carcinogenesis, Carcinogenesis, 1999, 20(3), 353–368. [DOI] [PubMed] [Google Scholar]
- 61.Gibson GR, Macfarlane GT and Cummings JH, Sulphate reducing bacteria and hydrogen metabolism in the human large intestine, Gut, 1993, 34(4), 437–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Attene-Ramos MS, et al. , Hydrogen sulfide induces direct radical-associated DNA damage, Mol. Cancer Res, 2007, 5 (5), 455–459. [DOI] [PubMed] [Google Scholar]
- 63.Gibson GR, Cummings JH and Macfarlane GT, Competition for hydrogen between sulphate-reducing bacteria and methanogenic bacteria from the human large intestine, J. Appl. Bacteriol, 1988, 65(3), 241–247. [DOI] [PubMed] [Google Scholar]
- 64.Devkota S, et al. , Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10−/− mice, Nature, 2012, 487(7405), 104–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Reddy BS, Diet and excretion of bile acids, Cancer Res, 1981, 41(9 Pt 2), 3766–3768. [PubMed] [Google Scholar]
- 66.Ridlon JM, Kang DJ and Hylemon PB, Bile salt bio-transformations by human intestinal bacteria, J. Lipid Res, 2006, 47(2), 241–259. [DOI] [PubMed] [Google Scholar]
- 67.Bernstein H, et al. , Bile acids as endogenous etiologic agents in gastrointestinal cancer, World J. Gastroenterol, 2009, 15(27), 3329–3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bernstein C, et al. , Carcinogenicity of deoxycholate, a secondary bile acid, Arch. Toxicol, 2011, 85(8), 863–871. Epub 2011 Jan 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.O’Keefe SJ, et al. , Rarity of colon cancer in Africans is associated with low animal product consumption, not fiber, Am. J. Gastroenterol, 1999, 94(5), 1373–1380. [DOI] [PubMed] [Google Scholar]
- 70.Ou J, et al. , Association between low colonic short-chain fatty acids and high bile acids in high colon cancer risk populations, Nutr. Cancer, 2012, 64(1), 34–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wu GD, et al. , Linking long-term dietary patterns with gut microbial enterotypes, Science, 2011, 334(6052), 105–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nava GM, et al. , Hydrogenotrophic microbiota distinguish native Africans from African and European Americans, Environ. Microbiol. Rep, 2012, 4(3), 307–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.O’Keefe SJ, et al. , Products of the Colonic Microbiota Mediate the Effects of Diet on Colon Cancer Risk, J. Nutr, 2009, 139(11), 2044–2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Turnbaugh PJ, et al. , The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnoto-biotic mice, Sci. Transl. Med, 2009, 1(6), 3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.David LA, et al. , Diet rapidly and reproducibly alters the human gut microbiome, Nature, 2014, 505(7484), 559–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.van den Bogert B, et al. , Microarray analysis and barcoded pyrosequencing provide consistent microbial profiles depending on the source of human intestinal samples, Appl. Environ. Microbiol, 2011, 77(6), 2071–2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Claesson MJ, et al. , Comparative analysis of pyrosequencing and a phylogenetic microarray for exploring microbial community structures in the human distal intestine, PLoS One, 2009, 4(8), e6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Le Chatelier E, et al. , Richness of human gut microbiome correlates with metabolic markers, Nature, 2013, 500(7464), 541–546. [DOI] [PubMed] [Google Scholar]
- 79.Lahti L, et al. , Probabilistic analysis of probe reliability in differential gene expression studies with short oligo-nucleotide arrays, IEEE/ACM Trans. Comput. Biol. Bio-inform, 2011, 8(1), 217–225. [DOI] [PubMed] [Google Scholar]
- 80.Fraher MH, O’Toole PW and Quigley EM, Techniques used to characterize the gut microbiota: a guide for the clinician, Nat. Rev. Gastroenterol. Hepatol, 2012, 9(6), 312–322. [DOI] [PubMed] [Google Scholar]
- 81.Yu J, et al. , Metagenomic analysis of faecal microbiome as a tool towards targeted non-invasive biomarkers for colorectal cancer, Gut, 2015, DOI: 10.1136/gutjnl-2015-309800. [DOI] [PubMed] [Google Scholar]