Abstract
Background:
The purpose of this study was to identify specific pharmacokinetic (PK) and pharmacodynamics (PD) factors that affect the likelihood of treatment remission with a serotonin norepinephrine reuptake inhibitor (SNRI) in depressed patients whose initial selective serotonin reuptake inhibitor (SSRI) failed.
Methods:
Multiple logistic regression modeling of PK and PD variation hypothesized to contribute to SNRI (i.e. duloxetine or venlafaxine) treatment remission in prior SSRI (i.e. citalopram or escitalopram) failure was conducted on 139 subjects from the Pharmacogenomics Research Network (PGRN) and Sequenced Treatment Alternatives to Relieve Depression (STAR*D) studies. Depressive symptoms were assessed with the Quick Inventory of Depressive Symptomatology Clinician-rated (QIDS-C16).
Results:
Venlafaxine-XR remission was associated with a significant interaction between CYP2D6 ultra-rapid metabolizer (URM) phenotype and SLC6A4 5-HTTLPR L/L genotype. A similar significant interaction effect was observed between CYP2D6 URM and SLC6A2 G1287A GA genotype. Stratifying by transporter genotypes, venlafaxine-XR remission was associated with CYP2D6 URM in patients with SLC6A4 L/L (p = 0.001) and SLC6A2 G1287A GA genotypes.
Limitations:
The primary limitation of this post hoc study was small sample size.
Conclusion:
Our results suggest that CYP2D6 ultra-rapid metabolizer status contributes to venlafaxine-XR treatment remission in MDD patients; in particular, there is a PK-PD interaction with treatment remission associated with CYP2D6 URM phenotype and SLC6A4 5-HTTLPR L/L or SLC6A2 G1287A G/A genotype, respectively. These preliminary data are encouraging and support larger pharmacogenomics studies differentiating treatment response to mechanistically different antidepressants in addition to further PK-PD interactive analyses.
Keywords: Venlafaxine-XR, Remission, CYP2D6, SLC6A4, SLC6A2, Pharmacodynamic-pharmacokinetic, interaction
1. Introduction
The World Health Organization (WHO) has ranked depression as the leading cause of medical disability worldwide with an estimated 300 million people living with depression (World Health Organization). In the US, major depressive disorder (MDD) is the leading cause of disability in young people age 15–44 (National Institute Mental Health). While selective serotonin reuptake inhibitors (SSRIs) are considered first line treatment for MDD (Nassan et al., 2016). Only 50% of these patients will respond to initial treatment with SSRIs, and even fewer (~ 30%) patients achieve remission (Sinyor et al., 2010; Connolly and Thase, 2011). Step-wise treatment trials after SSRI non-remission have shown variable response rates when switching to a second SSRI (Joffe et al., 1996; Thase et al., 1997; Thase et al., 2001), to any non-SSRI antidepressant (Fava et al., 2001; Fava et al., 2003a; 2003b), or to a serotonin noradrenergic reuptake inhibitor (SNRI) (de Montigny et al., 1999; Nierenberg et al., 1994; Saiz-Ruiz et al., 2002). A clinical strategy often employed in the setting of SSRI nonresponse is to shift to a second antidepressant with a different mechanism of action. For example, in a study of depressed patients, for whom the majority (65%) failed initial treatment with an SSRI, response and remission rates were significantly higher when patients were subsequently randomized to the SNRI venlafaxine vs a 2nd SSRI paroxetine (Poirier and Boyer, 1999), though other studies do not agree (Rush et al., 2006).
It is increasingly recognized that genetic factors may contribute to inter-individual differences in the overall risk (i.e. side effects) / benefit (i.e. response / remission rate) ratio of antidepressant treatment (Ahmed et al., 2018). Relevant genetic factors include both pharmacokinetic (PK) variation that impacts drug metabolism (i.e. active metabolite) and pharmacodynamic (PD) variation that impacts drug action at the cellular level (Nassan et al., 2016). Several previously published reports have shown that PK (i.e. cytochrome (CYP) 2D6 and 2C19) genetic variation is associated with variation in clinical response to SSRI's (Mrazek et al., 2011; Tsai et al., 2010; Gressier et al., 2015). While there is a suggestion that PD genetic variation in the gene encoding the serotonin transporter (SLC6A4) 5-HTTLPR is associated with MDD (Caspi et al., 2003), there is a larger evidence base that the S/S genotype genetic variation is associated with greater antidepressant side effect burden, including antidepressant-induced mania (Frye et al., 2015) and lower treatment response rate than S/L and L/L genotype (Caspi et al., 2003; Murphy et al., 2004; Yu et al., 2002; Mrazek et al., 2009). Similarly, polymorphisms in the gene encoding for the norepinephrine transporter (NET) (SLC6A2) T-182C and G1287A have been associated with major depression (Inoue et al., 2004; Hahn et al., 2008), but also with decreased SNRI treatment response (T-182C allele) and slower onset of treatment response (G1287A A/A genotype) (Yoshida et al., 2004).
Interaction between pharmacokinetic (PK) and pharmacodynamics (PD) genetic factors (i.e. CYP2D6 and serotonin transporter 5-HTTLPR) and their relationship to treatment response have not been well studied, but they may be of greater value in predicting symptom burden and/or treatment outcome than either genetic factor alone (Suzuki et al., 2006). PK genetic variation contributes to a variable concentration of drug at the site of action, whereas PD genetic variation contributes to a variable quantity or ability of transporter protein to interact with these drugs; therefore, a PK-PD genetic variation interaction may be a meaningful analysis to apply when investigating clinical outcomes.
This study investigated the association between genetic polymorphisms in pharmacokinetic (CYP2D6, CYP2C19), and pharmacodynamic [SLC6A4, SLC6A2 (NET)] genes and treatment remission with SNRIs (duloxetine and venlafaxine-XR) in patients with MDD. This study utilized data from two clinical trials (Mrazek et al., 2014; Rush et al., 2006) of SNRIs in patients with MDD who had previously failed prospective treatment with an SSRI. The questions addressed were: (1) does PK genetic variation in CYP2D6 and CYP2C19 predict SNRI remission, (2) does PD genetic variation in serotonin and norepinephrine transporters (i.e. SLC6A4 and SLC6A2) predict SNRI remission, (3) and does a PK (i.e. CYP2D6 and CYP2C19 metabolizer status) PD (i.e.SLC6A4 and SLC6A2 genetic variation) interaction predict SNRI remission?
2. Methods
2.1. Patient identification and recruitment
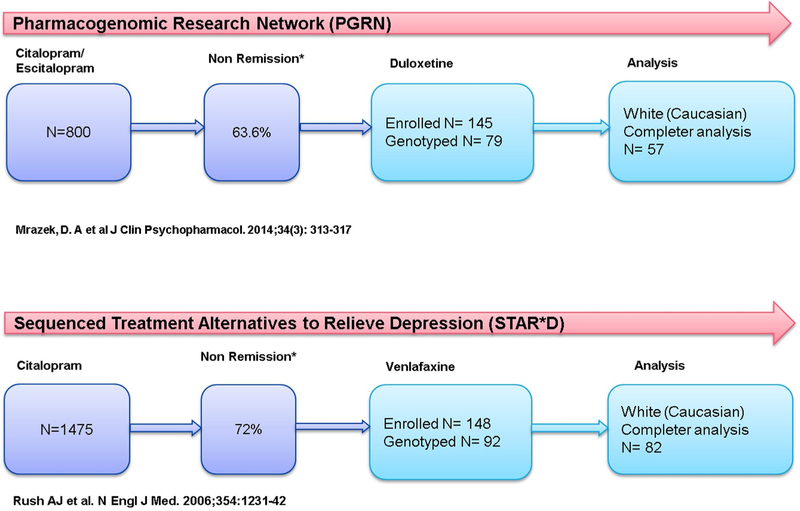
Data sources for these analyses include two clinical trials: the Pharmacogenomic Research Network (PGRN) Antidepressant Medication Pharmacogenomics Study (AMPS) and the Sequenced Treatment Alternatives to Relieve Depression (STAR*D) study (Rush et al., 2006). The PGRN study was an 8-week open label clinical trial which enrolled 800 patients over 4 years who were treated with either citalopram or escitalopram. Patients who did not achieve remission over the course of 8 weeks of treatment were offered subsequent treatment with the SNRI duloxetine (n = 145, mean dose=53.3 mg/day) with enrollment over 2 years. The Sequenced Treatment Alternatives to Relieve Depression (STAR*D) Level 1 was a 12-week open label randomized clinical trial which enrolled 1475 patients to receive citalopram over 3 years. Patients who did not achieve remission at 12 weeks entered Level 2 and were subsequently offered a number of augmentation and switch strategies including the SNRI venlafaxine-XR. The enrollment (N=250) for venlafaxine-XR (mean dose=193.6 mg/day) treatment occurred over 3 years. Both study designs and timelines are depicted in Fig. 1. Given known genetic variation by race and ethnicity, this analysis was conducted on Caucasian patients only [PGRN (N=79) and STAR*D (N=92)].
Fig. 1.
Study and patient enrollment timeline.
2.2. Gene selection
We first focused on metabolizer phenotypes for CYP2D6 and CYP2C19 which included: poor metabolizer (PM), intermediate metabolizer (IM), extensive metabolizer (EM) and ultra-rapid metabolizer (URM). Duloxetine is mainly metabolized to active metabolites (i.e. 4-OH, 5-OH and 6-OH duloxetine) by CYP2D6 and CYP1A2, while venlafaxine-XR is predominately metabolized by CYP2D6 and partially by CYP2C19 to an active metabolite O-desmethylvenlafaxine (ODV) (Gaedigk, 2013). Pharmacodynamically, the initial mechanism of action (MOA) of an SSRI or an SNRI is to block serotonin and norepinephrine reuptake transporters. Therefore, we also investigated allelic variation for SLC6A4, 5-HTTLPR (S/S, L/S and L/L genotypes), SLC6A4 5-HTVNTR (12/12, [9/12, 9/10, 10/12] and 10/10), SIC6A2 G1287A (A/A, G/A and G/G genotypes), and SIC6A2 T182C (C/C, T/C and T/T genotypes) (Caspi et al., 2003; Inoue et al., 2004; Hahn et al., 2008; Lesch, 2001). For the PK investigation, 47/79 (PGRN) and 90/92 (STAR*D) samples were genotyped. For the PD investigation, 77/79 (PGRN) and 90/92 (STAR*D) samples were available for genotyped.
2.3. Statistical analysis
Fig. 1 shows that 57 Caucasian patients completed the entire study duration of 8 weeks with duloxetine (PGRN) and 82 Caucasian patients completed 8 weeks of the 12 week study duration with venlafaxine-XR (STAR*D). There were participants who were genotyped but never started medication (PGRN n = 1, STAR*D n = 6) and participants who were genotyped, but dropped out after starting the medication for either inefficacy or side effects (PGRN n = 21, STAR D n = 4);a secondary intent to treat analysis was attempted by adding the 4 subjects from STAR*D trial to completers analysis.
Remission, defined as QIDS-C16 ≤ 5 at week 8, was the primary outcome measure for both the STAR*D and PGRN clinical trials (Rush et al., 2006; Mrazek et al., 2014) and was used as the primary outcome measure for this pharmacogenomic analysis. Using Chi-square test or Fisher's exact test (if expected frequencies were small), the metabolizer status of CYP2D6 and CYP2C19, as well as the genetic variants SLC6A4 5-HTTLPR, SLC6A4 5-HTVNTR, SLC6A2 G1287A, and SLC6A2 T182C were compared between patients who achieved remission vs patients who did not. Multiple genetic variants and metabolizer status factors were tested concurrently for their association with remission in multiple logistic regression models. To evaluate the effects of metabolizer status of CYP2D6 or CYP2C19, separate models were constructed treating metabolizer status as ordinal (tested via a 1df test) and not ordinal (tested via a 2df test). Separate logistic regression models were used to test pairwise interactions between the effects of metabolizer status and SNPs in PD genes with respect to remission.
A fixed effects model of meta-analysis was performed using the estimates/SE of estimates from both studies for SNPs/metabolizer status modeled univariately versus remission. Pooled odds ratios and 95% confidence intervals are presented for each predictor. To assess heterogeneity across the two studies, we used the I2 statistic (Higgins et al., 2003). Statistical analysis was performed using IBM SPSS Statistics version 22.0 (Armonk, NY: IBM Corp.) or STATA version 14 (StataCorp LP, College Station, Texas).
3. Results
As presented in Table 1, the only significant demographic difference between the two study groups was rate of employment (PGRN=81% vs. STAR*D=60%, p-value=0.009).
Table 1.
Basic demographics, depression ratings, and genotyping outcomes.
| PGRNa Duloxetine N=(57) |
STAR*Da Venlafaxine N=(82) |
P-value | ||
|---|---|---|---|---|
| Demographics | ||||
| Age, yrs (Mean, SD) | 43.8(11.4) | 43.7(11.9) | 0.960 | |
| Gender (N, %) | Female | 31(54.4%) | 48(58.5%) | 0.755 |
| Male | 26(45.6%) | 34(41.5%) | ||
| Marital status (N, %) | Married | 23(40.4%) | 33(40.3%) | 0.989 |
| Not Married | 34(59.6%) | 49(59.7%) | ||
| Employment status (N, %) | Employed | 46(80.7%) | 49(59.8%) | 0.009 |
| Unemployed | 11(19.3%) | 33(40.2%) | ||
| Education, yrs (N, %) | < 12 | 5(8.8%) | 10(12.2%) | 0.379 |
| 12-15 | 26(45.6%) | 44(53.7%) | ||
| ≥16 | 26(45.6%) | 28(34.1%) | ||
| Depression ratings &outcomes | ||||
| QIDS-C16b score(Mean, SD) | Baseline | 12.23(3.86) | 13.27(4.56) | 0.120 |
| Week 8 | 8.91(4.50) | 8.95(5.81) | 0.961 | |
| Remitters (QIDS-C16*score ≥ 5) (N, %) | Yes | 13(22.8%) | 28(34.1%) | 0.210 |
| No | 44(77.2%) | 54(65.9%) | ||
| Genotyping | N (%) | N (%) | ||
| CYP2D6 | PMC | 1(3.6%) | 10(8.4%) | 0.373 |
| IM/EMC | 25(89.3%) | 63(75.9%) | ||
| URMC | 2(7.1%) | 7(12%) | ||
| CYP2C19 | PMC | 2(3.1%) | 1(1.2%) | 0.240 |
| IM/EMC | 29(90.6%) | 74(91.4%) | ||
| URMC | 1(6.3%) | 6(7.4%) | ||
| SLC6A4 5-HTTLPR | L/L | 6(21.4%) | 27(32.9%) | 0.499 |
| L/S | 17(60.7%) | 44(53.7%) | ||
| S/S | 5(17.9%) | 11(13.4%) | ||
| SLC6A4 5-HTVNTR | 12/12 | N/A | 33(41.3%) | N/A |
| 9/12–9/ | N/A | 30(37.5%) | ||
| 10–10/12 | ||||
| 10/10 | N/A | 17(21.3%) | ||
| SLC6A2 G1287A | A/A | 4(7.1%) | 10(12.3%) | 0.613 |
| G/A | 28(50%) | 38(46.9%) | ||
| G/G | 24(42.9%) | 33(40.7%) | ||
| SLC6A2 T182C | C/C | 4(7%) | 5(6.2%) | 0.781 |
| T/C | 24(42.1%) | 39(48.1%) | ||
| T/T | 29(50.9%) | 37(45.7%) |
PGRN, Pharmacogenomic Research Network, START*D, Sequenced Treatment Alternatives to Relieve Depression,
QIDS-C16, Quick Inventory of Depressive Symptomatology Clinician-rated,
PM, poor metabolizer; IM, intermediate metabolizer; EM, extensive metabolizer; URM, ultra-rapid metabolizer;
L, long; S, short allele variants
3.1. CYP2D6 / CYP2C19 metabolizers and SLC6A4 / SLC6A2 genotypes
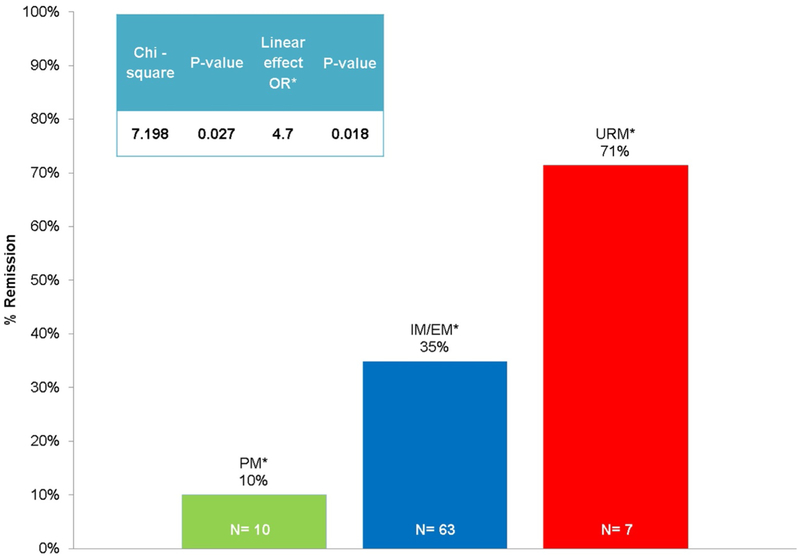
Venlafaxine-XR remission was associated with CYP2D6 metabolism phenotype (p = 0.027). Specifically, remission rates were higher among URM (n = 5, 71.4%) in comparison to CYP2D6 PM (n = 1, 10%). Assuming a linear effect of CYP2D6 metabolizer status on venlafaxine-XR remission, (i.e., increasing level of metabolism: PM, IM/EM and URM), higher metabolism was associated with greater odds of remission (OR = 4.72, p = 0.018) (Table 2 and Fig. 2). An intent to treat analysis did not change the significance of phenotype and remission. We found no significant difference in duloxetine remission rates by CYP2D6 metabolism phenotype. Moreover, we found no significant differences in venlafaxine-XR or duloxetine remission rates by CYP2C19, SLC6A4 5-HTTLPR, SLC6A4 5-HTTVNTR, and by SLC6A2 G1287A genotypes (Table 2)
Table 2.
The association between metabolizer status, SLC6A4 and SLC6A2 allele variants, and SNRIs remission.
| Gene | Drug | Remission rate by genotype | P-valuea | Assuming linear effeetb ORc | P-valued | ||
|---|---|---|---|---|---|---|---|
| CYP2D6 | PMd | IM/EMd | URMd | ||||
| Duloxetine | 0(0%) | 6(24.0%) | 0(0%) | 0.462 | 1.559 | 0.762 | |
| Venlafaxine | 1(10.0%) | 22(34.9%) | 5(71.4%) | 0.027 | 4.727 | 0.018 | |
| CYP2C19 | PMd | IM/EMd | URMd | ||||
| Duloxetine | 1(50.0%) | 7(24.1%) | 0(0%) | 0.559 | 0.266 | 0.332 | |
| Venlafaxine | 0(0%) | 26(35.1%) | 2(33.3%) | 0.650 | 1.194 | 0.825 | |
| SLC6A4e 5-HTTLPR | L/Ld | S/Ld | s/sd | ||||
| Duloxetine | 1(16.7%) | 6(35.3%) | 0(0%) | 0.135 | 0.692 | 0.603 | |
| Venlafaxine | 8(29.6%) | 17(38.6%) | 3(27.3%) | 0.645 | 1.061 | 0.869 | |
| SLC6A4e 5-HTVNTRf | 12/12 | 9/12, 10/12, 9/10 | 10/10 | ||||
| Venlafaxine | 13(39.4%) | 9(30.0%) | 5(29.4%) | 0.671 | 0.777 | 0.423 | |
| SLC6A2e G1287A | AA | GA | GG | ||||
| Duloxetine | 0(0%) | 8(28.6%) | 5(20.8%) | 0.270 | 0.908 | 0.853 | |
| Venlafaxine | 4(40.0%) | 11(28.9%) | 13(39.4%) | 0.604 | 0.880 | 0.715 | |
| SLC6A2e T182C | C/C | T/C | T/T | ||||
| Duloxetine | 1(25.0%) | 9(37.5%) | 3(10.3%) | 0.059 | 2.525 | 0.07 | |
| Venlafaxine | 0(0%) | 17(43.6%) | 11(29.7%) | 0.05 | 1.009 | 0.981 | |
Chi-square test.
By coding different allele variants as continuous variable.
OR, Odds Ratio.
Linear regression model.
PM, poor metabolizer; IM/EM, intermediate metabolizer/extensive metabolizer; URM, ultra-rapid metabolizer;
L, long; S, short allele variants;
SLC6A4, serotonin transporter; SLC6A2, norepinephrine transporter.
VNTR, variable number of tandem repeats.
Fig. 2.
CYP2D6 ultra-rapid metabolizer (URM) phenotype associated with highest rate of remission with venlafaxine. PM=poor metabolizer; IM/EM=intermediate metabolizer/extensive metabolizer; URM=ultra-rapid metabolizer; OR=odds ratio.
3.2. Pharmacokinetic-pharmacodynamic interaction and SNRI remission
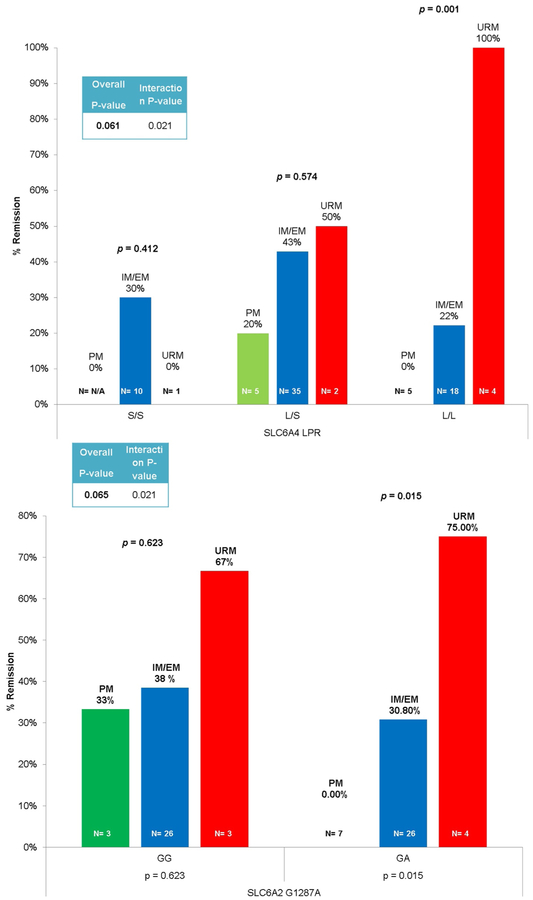
Venlafaxine-XR remission was associated with an interaction between SLC6A4 5-HTTLPR and CYP2D6, when URM were compared with IM/EMs (p = 0.021). A similar interaction effect on remission was observed between CYP2D6 URM and SIC6A2 G1287A (p = 0.021). Post-hoc analysis of venlafaxine-XR remission stratified by SLC6A4 5-HTTLPR genotypes showed that remission was associated with CYP2D6 metabolizer status in L/L genotype carriers (p = 0.001), while, analysis stratified by SLC6A2 G1287A also revealed an association between CYP2D6 and remission in patients with the GA genotype at SLC6A2 G1287A (p = 0.015) (Fig. 3a, b). These stratified analyses suggested that CYP2D6 URM (i.e. those metabolizing more venlafaxine-XR to the active metabolite O-desmethylvenlafaxine) had a higher rate of remission, especially in patients with SLC6A4 5-HTTLPR L/L or SLC6A2 G1287A G/A genotype.
Fig. 3.
(a) CYP2D6 URM phenotype & SLC6A4 L/L genotype associated with highest rate of remission with venlafaxine. SLC6A4=serotonin transporter PM=poor metabolizer; IM/EM=intermediate/extensive metabolizer; URM=ultra-rapid metabolizer. (b) CYP2D6 ultra-rapid metabolism phenotype & SLC6A2 G/A genotype associated with highest rate of remission with venlafaxine. SLC6A2=norepinephrine transporter; PM=poor metabolizer; IM/EM=intermediate/extensive metabolizer; URM=ultra-rapid metabolizer.
3.3. Meta-analysis
In the meta-analysis across the two studies, CYP2D6 URM was significantly associated (p = 0.023) with greater odds of SNRIs remis-sion (OR = 3.9; I2 = 0.0). None of the other genetic variations were associated with SNRI remission in meta-analyses combining the two studies.
4. Discussion
This is one of the first studies to investigate the association between prospectively confirmed SNRI treatment remission and genetic polymorphisms in CYP2D6, CYP2C19, SLC6A4, and SLC6A2 in MDD patients with prospectively confirmed prior SSRI treatment failure. In this study, venlafaxine-XR remission rate was significantly higher in patients with the CYP2D6 URM phenotype. Among patients with the SLC6A4 5-HTTLPR L/L or SLC6A2 G1287A G/A genotypes, those with the CYP2D6 URM phenotype were more likely to achieve remission with venlafaxine-XR in comparison with those with IM/EM phenotypes or PM phenotypes.
We found that the CYP2D6 URM phenotype was associated with greater odds of venlafaxine-XR remission (i.e. increasing level of metabolism from PM to IM/EM to URM). Patients with the URM phenotype were more likely to achieve remission with venlafaxine-XR than those with the IM/EM or PM phenotype. Similarly, Lobello and colleagues showed that patients with the EM phenotype (i.e. “normal” metabolism), in comparison to the PM phenotype, had significantly higher response and remission rates when treated with venlafaxine-XR (Lobello et al., 2010). Unlike our study, Lobello and colleagues did not test URM phenotype in comparison to EM phenotype.
To our knowledge, this is the first PK-PD interaction associated with venlafaxine-XR treatment remission. Among patients with the SLC6A4 5-HTTLPR L/L genotype, those with the CYP2D6 URM phenotype were more likely to achieve remission with venlafaxine-XR in comparison to those with IM/EM or PM phenotypes. Secondly, patients with the combination of the CYP2D6 URM phenotype and SLC6A2 G1287A G/A genotype were more likely to achieve remission with venlafaxine-XR in comparison to the IM/EM or PM phenotype. There is less systematic research on the SLC6A2 G1287A and T182C polymorphisms and pharmacogenomic treatment response in comparison to the well-studied serotonin transporter. Ueda and colleagues, however, found a relationship between the volume of the dorsolateral prefrontal cortex and the SLC6A2 G1287A polymorphism in MDD patients, suggesting an area for future research on disease risk (Ueda et al., 2016). Minn and colleagues (2009) investigated the PD-PD interaction between SLC6A2-T182C and the serotonin transporter in patients with major depression (n = 579) and reported that patients with the combination of SLC6A2-T182C C/C and 5-HTTLPR S/S genotypes (n = 16) had a significantly lower symptom burden of depression in comparison with patients who were T-allele carriers and l-allele carriers respectively (n = 247). In contrast, patients with the 5-HTTLPR L/L and VNTR 12/12 genotypes (n = 21) had a better clinical response to SSRIs treatment in comparison to S-carriers and 10-repeat allele carriers respectively (n = 31) (Min et al., 2009).
Our findings if replicated will have important practical implications. Reliable and valid biological markers that could inform clinical practice with regard to when one should bypass first line treatment with SSRIs for SNRIs as a treatment intervention would have clinical value. This potential biomarker would be an additional factor for inclusion in clinical decision support guidelines, in addition to subtypes of depression (i.e. pain conditions such as fibromyalgia, neuropathy, and musculoskeletal) known to respond to SNRIs.
The primary limitation of this post hoc study was small sample size. That is the main reason that we did not pursue a genome wide association study. Moreover, our multiple stratified analyses were not corrected for multiple testing. While the intent to treat analyses did not differ from the completer analysis, the small sample size, again, may limit the conclusions from these statistical analyses. Additional limitations focus on trial design. While the rating scales and scheduled research visits were the same, the longer 12-week STAR*D trial allowed for a slower titration of study drug in comparison to the shorter 8-week PGRN trial; the maximum dose of venlafaxine-XR was reached at week 8 while the maximum dose for duloxetine was reached by week 4. Moreover, the STAR*D study subjects immediately received SNRI treatment after SSRI treatment failure, whereas the time of receiving SNRIs treatment in the PGRN study subjects varied.
Providing greater precision for antidepressant recommendations for individual patients beyond the large-scale, clinical trials evidence base can potentially reduce side effect toxicity profiles and increase response rates and overall effectiveness. These results underscore the multidimensional aspects of precision medicine as an approach to optimize drug treatment of MDD.
Acknowledgments
The authors of this paper would like to recognize the vision and original contributions to this research of the late Dr. David Mrazek. This publication was made possible by CTSA Grant Number UL1 TR000135 from the National Center for Advancing Translational Sciences (NCAT) as part of Ahmed T Ahmed master's degree thesis. Dr. Ahmed Research reported in this publication was supported by National Institute of General Medical Sciences of the National Institutes of Health under award number T32 GM008685. Dr. Liewei Wang was supported by NIH grants U19 GM61388. Dr. Weinshilboum was supported by NIH grants U19 GM61388 and NIH grants RO1 GM28157.
Disclaimer and conflicts of interest
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Funding: Dr Ahmed Research reported in this publication was supported by National Institute of General Medical Sciences of the National Institutes of Health under award number T32 GM008685.
Dr. Frye has received grant support from AssureRx Health Inc, Myriad, Pfizer Inc, NIMH (R01 MH079261), the National Institute on Alcohol Abuse and Alcoholism (P20AA017830) in the National Institutes of Health at the US Department of Health and Human Services, and the Mayo Foundation. He has been a consultant (for Mayo) to Janssen Global Services, LLC; Mitsubishi Tanabe Pharma Corp; Myriad Genetics, Inc; Sunovion Pharmaceuticals, Inc; and Teva Pharmaceutical Industries Ltd. He has received continuing medical education, travel, and presentation support from American Physician Institute and CME Outfitters.
Dr. Liewei Wang was supported by NIH grants U19 GM61388, U54 GM114838 and NSF164615. Dr. Wang is a cofounder and stockholder in OneOme.
Dr. Weinshilboum was supported by NIH grants RO1 GM28157, U19 GM61388, U54 GM114838 and NSF1624615. Dr. Weinshilboum is a cofounder and stockholder in OneOme.
Mayo Clinic has a financial interest in OneOme.
Dr. Rush has received consulting fees: Akili, Inc, Brain Resource Inc., Compass, Inc., Curbstone Consultant LLC, Emmes Corp, Liva-Nova, MindLinc, Sunovion, Taj Medical, Takeda USA; royalties: Guilford Publications, University of Texas Southwestern Medical Center at Dallas (for the Inventory of Depressive Symptoms and its derivatives); and speaking fees: Liva-Nova.
Dr. Rush is named as co-inventor on two patents: (1) U.S. Patent No. 7,795,033: Methods to Predict the Outcome of Treatment with Antidepressant Medication, Inventors: McMahon FJ, Laje G, Manji H, Rush AJ, Paddock S, Wilson AS. (2) U.S. Patent No. 7,906,283: Methods to Identify Patients at Risk of Developing Adverse Events During Treatment with Antidepressant Medication, Inventors: McMahon FJ, Laje G, Manji H, Rush AJ, Paddock S.
Dr. Gen Shinozaki is supported by NIH grant K23 MH107654, and NSF 1,664,364. Dr. Shinozaki is a cofounder and president of Predelix Medical LLC.
Footnotes
Previous presentation
Presented in part at the Annual Meeting of the National Network of Depression Centers (NNDC) 2017 Ann Arbor, MI and the Annual Meeting of the Society of Biological Psychiatry (SOBP) 2018 NY.
Supplementary materials
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.jad.2018.12.021.
References
- Ahmed A, Weinshilboum R, Frye M, 2018. Benefits of and barriers to pharmacogenomics-guided treatment for major depressive disorder. Clin. Pharmacol. Ther [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi A, Sugden K, Moffitt TE, et al. , 2003. Influence of life stress on depression: moderation by a polymorphism in the 5-HTT gene. Science. 301 (5631), 386–389 July 18. [DOI] [PubMed] [Google Scholar]
- Connolly KR, Thase ME, 2011. If at first you don't succeed: a review of the evidence for antidepressant augmentation, combination and switching strategies. Drugs 71 (1), 43–64 January 01. [DOI] [PubMed] [Google Scholar]
- de Montigny C, Silverstone PH, Debonnel G, et al. , 1999. Venlafaxine in treatment-resistant major depression: a Canadian multicenter, open-label trial. J. Clinic. Psychopharmacol 19 (5), 401–406 October. [DOI] [PubMed] [Google Scholar]
- Fava M, Dunner DL, Greist JH, et al. , 2001. Efficacy and safety of mirtazapine in major depressive disorder patients after SSRI treatment failure: an open-label trial. J. Clinic. Psychiat 62 (6), 413–420 June. [DOI] [PubMed] [Google Scholar]
- Fava M, McGrath PJ, Sheu WP, 2003a. Switching to reboxetine: an efficacy and safety study in patients with major depressive disorder unresponsive to fluoxetine. J. Clinic. Psychopharmacol 23 (4), 365–369 August. [DOI] [PubMed] [Google Scholar]
- Fava M, Papakostas GI, Petersen T, et al. , 2003b. Switching to bupropion in fluoxetine-resistant major depressive disorder. Annal. Clinic. Psychiat. Official J. Am. Acad. Clinic. Psychiat 15 (1), 17–22 March. [DOI] [PubMed] [Google Scholar]
- Frye MA, McElroy SL, Prieto ML, et al. , 2015. Clinical risk factors and serotonin transporter gene variants associated with antidepressant-induced mania. J. Clinic. Psychiat 76 (2), 174–180 February. [DOI] [PubMed] [Google Scholar]
- Gaedigk A, 2013. Complexities of CYP2D6 gene analysis and interpretation. Intern. Rev. Psychiat. (Abingdon, England) 25 (5), 534–553 October. [DOI] [PubMed] [Google Scholar]
- Gressier F, Verstuyft C, Hardy P, et al. , 2015. Response to CYP2D6 substrate antidepressants is predicted by a CYP2D6 composite phenotype based on genotype and comedications with CYP2D6 inhibitors. J. Neur. Trans. (Vienna, Austria: 1996) 122 (1), 35–42 January. [DOI] [PubMed] [Google Scholar]
- Hahn MK, Blackford JU, Haman K, et al. , 2008. Multivariate permutation analysis associates multiple polymorphisms with subphenotypes of major depression. Gene. Brain Behav 7 (4), 487–495 June. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins JP, Thompson SG, Deeks JJ, et al. , 2003. Measuring inconsistency in meta-analyses. BMJ (Clinic. Res. Ed) 327 (7414), 557–560 September 06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K, Itoh K, Yoshida K, et al. , 2004. Positive association between T-182C polymorphism in the norepinephrine transporter gene and susceptibility to major depressive disorder in a Japanese population. Neuropsychobiology 50 (4), 301–304. [DOI] [PubMed] [Google Scholar]
- Joffe RT, Levitt AJ, Sokolov ST, et al. , 1996. Response to an open trial of a second SSRI in major depression. J. Clinic. Psychiat 57 (3), 114–115 March. [PubMed] [Google Scholar]
- Lesch KP, 2001. Serotonergic gene expression and depression: implications for developing novel antidepressants. J. Affect. Disord 62 (1-2), 57–76 January. [DOI] [PubMed] [Google Scholar]
- Lobello KW, Preskorn SH, Guico-Pabia CJ, et al. , 2010. Cytochrome P450 2D6 phenotype predicts antidepressant efficacy of venlafaxine: a secondary analysis of 4 studies in major depressive disorder. J. Clinic. Psychiat 71 (11), 1482–1487 November. [DOI] [PubMed] [Google Scholar]
- Min W, Li T, Ma X, et al. , 2009. Monoamine transporter gene polymorphisms affect susceptibility to depression and predict antidepressant response. Psychopharmacology 205 (3), 409–417 August. [DOI] [PubMed] [Google Scholar]
- Mrazek DA, Rush AJ, Biernacka JM, et al. , 2009. SLC6A4 variation and citalopram response. American journal of medical genetics Part B. Neuropsychiat. Genet. Official Pub. Intern. Soc. Psychiat. Genet. 150b (3), 341–351 April 05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mrazek DA, Biernacka JM, O'Kane DJ, et al. , 2011. CYP2C19 variation and citalopram response. Pharmacogenet. Genom 21 (1), 1–9 January. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mrazek DA, Biernacka JM, McAlpine DE, et al. , 2014. Treatment outcomes of depression: the pharmacogenomic research network antidepressant medication pharmacogenomic study. J. Clinic. Psychopharmacol 34 (3), 313–317 June. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy GM Jr., Hollander SB, Rodrigues HE, et al. , 2004. Effects of the serotonin transporter gene promoter polymorphism on mirtazapine and paroxetine efficacy and adverse events in geriatric major depression. Archive. Gen. Psychiat 61 (11), 1163–1169 November. [DOI] [PubMed] [Google Scholar]
- Nassan M, Nicholson WT, Elliott MA, et al. , 2016. Pharmacokinetic pharmacogenetic prescribing guidelines for antidepressants: a template for psychiatric precision medicine. Mayo Clinic Proceed. 91 (7), 897–907 July. [DOI] [PubMed] [Google Scholar]
- National Institute Mental Health. Major depression statistics. Available at: https://www.nimh.nih.gov/health/statistics/major-depression.shtml.
- Nierenberg AA, Feighner JP, Rudolph R, et al. , 1994. Venlafaxine for treatment-resistant unipolar depression. J. Clinic. Psychopharmacol 14 (6), 419–423 December. [PubMed] [Google Scholar]
- Poirier MF, Boyer P, 1999. Venlafaxine and paroxetine in treatment-resistant depression. Double-blind, randomised comparison. Br. J. Psychiat. J. Ment. Sci 175, 12–16 July. [DOI] [PubMed] [Google Scholar]
- Rush AJ, Trivedi MH, Wisniewski SR, et al. , 2006. Bupropion-SR, sertraline, or venlafaxine-XR after failure of SSRIs for depression. New Eng. J. Med 354 (12), 1231–1242 March 23. [DOI] [PubMed] [Google Scholar]
- Saiz-Ruiz J, Ibanez A, Diaz-Marsa M, et al. , 2002. Efficacy of venlafaxine in major depression resistant to selective serotonin reuptake inhibitors. Prog. Neuro-psycho-pharmacol. Biolog. Psychiat 26 (6), 1129–1134 October. [DOI] [PubMed] [Google Scholar]
- Sinyor M, Schaffer A, Levitt A, 2010. The sequenced treatment alternatives to relieve depression (STAR*D) trial: a review. Can. J. Psychiat. Revue canadienne de psychiatrie 55 (3), 126–135 March. [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Sawamura K, Someya T, 2006. Polymorphisms in the 5-hydroxytryptamine 2A receptor and CytochromeP4502D6 genes synergistically predict fluvoxamine-induced side effects in japanese depressed patients. Neuropsychopharmacol. Official Pub. Am. College Neuropsychopharmacol 31 (4), 825–831 April. [DOI] [PubMed] [Google Scholar]
- Thase ME, Blomgren SL, Birkett MA, et al. , 1997. Fluoxetine treatment of patients with major depressive disorder who failed initial treatment with sertraline. J. Clinic. Psychiat 58 (1), 16–21 January. [DOI] [PubMed] [Google Scholar]
- Thase ME, Feighner JP, Lydiard RB, 2001. Citalopram treatment of fluoxetine non-responders. J. Clinic. Psychiat 62 (9), 683–687 September. [DOI] [PubMed] [Google Scholar]
- Tsai MH, Lin KM, Hsiao MC, et al. , 2010. Genetic polymorphisms of cytochrome P450 enzymes influence metabolism of the antidepressant escitalopram and treatment response. Pharmacogenomics 11 (4), 537–546 April. [DOI] [PubMed] [Google Scholar]
- Ueda I, Kakeda S, Watanabe K, et al. , 2016. Relationship between G1287A of the NET gene polymorphisms and brain volume in major depressive disorder: a voxel-based MRI study. PloS One 11 (3), e0150712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization. Depression and other common mental disorders 3/March/2017. Available at: http://www.who.int/mediacentre/news/releases/2017/world-health-day/en/.
- Yoshida K, Takahashi H, Higuchi H, et al. , 2004. Prediction of antidepressant response to milnacipran by norepinephrine transporter gene polymorphisms. Am. J. Psychiat 161 (9), 1575–1580 September. [DOI] [PubMed] [Google Scholar]
- Yu YW, Tsai SJ, Chen TJ, et al. , 2002. Association study of the serotonin transporter promoter polymorphism and symptomatology and antidepressant response in major depressive disorders. Molecul. Psychiat 7 (10), 1115–1119. [DOI] [PubMed] [Google Scholar]