Summary
Background
The lack of evidence-based outcomes data leads to uncertainty in developing treatment regimens in children newly diagnosed with Ulcerative Colitis (UC). We hypothesized that pre-treatment clinical, transcriptomic, and microbial factors predict disease course.
Methods
We performed an inception cohort study of 428 paediatric UC patients receiving standardised mesalazine or corticosteroids (CS), with pre-established criteria for escalation to thiopurines or anti-TNFα. RNA sequencing (n=206) defined pre-treatment rectal gene expression. 16S sequencing (n=343) characterized rectal/fecal microbiota. The primary outcome was Week 52 CS-free remission (SFR) with no therapy beyond mesalazine.
Findings
Week 52 SFR was achieved in 150/400 (38%) participants; 74/400 (19%) received thiopurines alone, 12¾00 (31%) received anti-TNFα, and 25/400 (6%) colectomy. Lower baseline clinical severity, higher baseline hemoglobin, and Week 4 clinical remission were associated with achieving Week 52 SFR (logistic model AUC:0.70 (95% CI 0.65–0.75), specificity 77% (CI 71–82), n=386). Baseline severity and week 4 remission were validated inan independent cohort of 274 participants. An antimicrobial peptide gene signature (OR:0.6, p=0.002) and Ruminococcaceae (OR:1.4, p=0.04) and Sutterella (OR: 0.8, p=0.05) abundance were independently associated with SFR after adjusting for the clinical predictors. Amongst moderate-to-severe patients, escalation to anti-TNFα was associated with increased baseline clinical severity and decreased hemoglobin, serum 25 (OH) D, and rectal eosinophils (logistic model AUC:0.78 (95% CI 0.72–0.84), specificity 85% (CI 78–93), n=232). A rectal transportgene signature (OR:0.3, p=0.0006) and Oscillospira abundance (OR:0.6, p=0.02) were independently associated with escalation to anti-TNFα after adjusting for the clinical predictors.
Interpretation
Our findings support the utility of using initial clinical activity and treatment response by 4 weeks to predict Week 52 CS-free remission with mesalazine alone in children newly diagnosed with UC. The development of personalized clinical and biological signatures holds the promise of informing UC therapeutic decisions.
Funding
National Institutes of Health
Keywords: ulcerative colitis, children, mesalazine, corticosteroids, eosinophils, microbiome, gene expression
Introduction
Children diagnosed with ulcerative colitis (UC) typically experience a moderate to severe disease course with high rates of corticosteroid (CS) dependency and CS refractory disease driving frequent escalation to immunomodulators and anti-TNFα therapy1,2. Colectomy rates range up to 26% within 5 years.1,3 We hypothesized that in adults and children UC course depends upon initial disease phenotype, therapy offered, and intrinsic patient biology. Initial disease severity, lack of response to early therapy, hypoalbuminemia, anemia, low serum 25 (OH) D, colonic histology, and specific genetic polymorphisms have been associated with worse clinical outcomes2,4–6, though data largely come from studies lacking standardised treatment and many have focused on acutely ill hospitalized patients. In a paediatric observational registry with no standardisation of initial therapy, less than 40% of children newly diagnosed with UC were in CS-free remission without the need for immunomodulators or anti-TNFα therapy one year after diagnosis7.
The PROTECT Study: Predicting Response to Standardised Paediatric Colitis Therapy, was conducted to provide prospective evidence-based data on the course of children newly diagnosed with UC treated with standardised protocols, and identify clinical features and biologic pathways that would improve our understanding of the marked variability in treatment response without the confounding of uncontrolled treatment and retrospective analyses. In PROTECT we characterized rectal gene expression and the intestinal microbiome prior to therapy and in relation to clinical outcomes to develop predictive models of disease course beyond clinical characteristics.
Methods
Study Design and Oversight
We performed a prospective, 29 center North American study enrolling children ages 4–17 years between July 2012 and April 2015 undergoing evaluation for UC. Ileocolonoscopy and esophagogastroduodenoscopy with biopsies were performed with a diagnosis of UC made using established criteria8. Initial therapy was based upon disease severity and included mesalazine or oral/intravenous CS. Standardised treatment guidelines with indications for anti-TNFα therapy or immunomodulators are provided in the Supplementary Appendix and Figure S1.
Clinical and routine laboratory data were collected at diagnosis and weeks 4, 12, and 52 as previously reported9. Disease activity was determined by the Pediatric Ulcerative Colitis Activity Index (PUCAI)10 and Mayo score11. Rectal biopsies were obtained from the most inflamed part of the rectosigmoid with histology evaluated centrally (MHC), and a Histologic Severity Score was defined (Supplementary Appendix)12. Fecal calprotectin was determined centrally as previously described.13 High density DNA genotyping was done using the Affymetrix UK Biobank Axiom Array14. Pretreatment rectal biopsy global pattern of gene expression was determined using RNAseq on the Illumina platform15 and validated on the Lexogen platform16. Rectal and fecal microbiome was profiled using 16S rRNA amplicon sequencing of the V4 region17. Details are noted in the Supplementary Appendix.
Each site’s Institutional Review Board approved the protocol and safety monitoring plan. Informed consent/assent was obtained for each participant. Pentasa® (mesalazine, Shire Pharmaceuticals) was used under IND 111863. This study is registered with ClinicalTrials.gov, NCT 01536535.
Outcomes
The primary outcome was Week 52 steroid free remission (SFR): clinical remission (PUCAI<10) with no CS for ≥4 weeks immediately prior to Week 52, no medical therapy beyond mesalazine, or colectomy. The secondary outcome was escalation to anti-TNFα at any time in the 52 weeks. Additional outcomes included escalation to immunomodulators only, and colectomy. Week 4 remission was defined as PUCAI<10 irrespective of CS status but without immunomodulator, anti-TNFα, or colectomy. We further defined baseline disease severity using a combination of initial PUCAI score and initial therapy as mild (mesalazine, or PUCAI<45 with oral CS) or moderate/severe (PUCAI≥45 with oral CS, or IV CS) based upon previous observations of this cohort9, in which PUCAI < 45 was identified as the only predictor (OR=4.4, p=0.001), other than week 4 remission, of week 12 CS-free remission in the subgroup starting on oral steroids.
Statistical Analysis
Statistical methods were pre-specified in an analysis plan and are fully described in the Supplementary Appendix. This study is reported as per the STROBE statement for observational cohort studies. The planned sample size of 430 patients ensured 90% power to identify an odds ratio (OR) of 2.5 in predictive modeling, a value we expected to be clinically meaningful. Clinical predictors were modeled using the per-protocol population (n=386), excluding protocol violations and early study discontinuations with unknown outcomes (see Consort Diagram, Figure S2). Analysis of biological predictors was limited to a subset (n=177) selected for RNA-sequencing (n=206) and with microbiome (n=343) data. Statistical tests were conducted at a two-sided alpha level of 0.05 with no adjustment for multiple comparisons, unless otherwise specified; results should therefore be interpreted with caution.
An independent prospective inception cohort of children newly diagnosed with UC7 was used to test the clinical model for week 52 SFR. These patients, diagnosed 2002–2010 at 31 centers in North America, prospectively had clinical data obtained at diagnosis, one month post diagnosis, and quarterly. Treatment was by provider dictate, not protocol.
Prediction models for PROTECT clinical data were developed using multiple imputation (MI) multivariable logistic regression with least absolute shrinkage and selection operator (LASSO)18 variable selection to account for missing data and obtain a parsimonious model. Rectal biopsy genes differentially expressed at baseline between moderate/severe patients who achieved or did not achieve Week 52 SFR with fold change (FC) ≥1.5 and false discovery rate (FDR) < 0.05 were defined. Gene set enrichment analyses (GSEA) identified enriched biologic pathways within these genes, and principal components (PC) were built from the gene signatures. Differentially abundant microbial organisms associated with Week 52 SFR and escalation to anti-TNFα therapy at the FDR 0.2 and 0.05 level were identified by multivariable analyses accounting for key clinical and demographic covariates. We tested improvement of clinical prediction models upon the addition of single gene expression PCs and microbial taxa with variables retained at p < 0.05. Identified PCs and microbial taxa were then tested for simultaneous inclusion in the clinical models, where improvement over the clinical model was assessed via comparison of the area under the receiver operator characteristic (ROC) curve (AUC).
Results
Participants
We enrolled 467 participants; 36 were disqualified because of change of diagnosis to Crohn’s disease or other protocol violation; 3 patients/families declined medical therapy, leaving 428 who started medical therapy, 286 (67%) strictly per protocol. (Consort Diagram, Figure S2). Baseline characteristics for the whole cohort and by disease severity are in Table S1. For this group mean age was 12.7±3.3 years, 50% female, 16% non-white. Mild disease presentation (mean PUCAI 31.9±12.1, mean total Mayo score 5.7±1.8) was noted in 178/428 (42%); 136 (76%) initially received mesalazine and 42 (24%) oral CS. Moderate/severe disease presentation (mean PUCAI 62.9±13.2, mean total Mayo score 9.3±1.7) was noted in 250/428 (58%); 102 (41%) initially received oral CS and 148 (59%) received IV CS. For patients taking mesalazine, adherence data were available for 289/383 (75%). Overall 84% of prescribed mesalazine was taken (81% mild vs. 86% moderate/severe, p=0.07). Twenty-eight subjects discontinued before Week 52 without prior colectomy or escalation to IM or anti TNFα leaving 400 (93%) participants evaluable at Week 52. An additional 14 patients were excluded from predictive modeling because of protocol violations.
Outcomes
By Week 52, 150/400 (38%) patients achieved SFR [80/163 (49%) mild disease presentation and 70/237 (30%) moderate/severe disease presentation (Table S2). One hundred forty-five of the 150 were taking mesalazine (97%) and 5 were taking no medication. Amongst the 150 patients achieving Week 52 remission, 98 (65%) supplied stool specimens at that time and fecal calprotectin was <250 mcg/g in 56 (57%). SFR with fecal calprotectin <250 mcg/g was achieved by 56/231 (24%) of those patients who supplied stool for analysis at week 52 (Table S2).
Seventy-four of 400 (19%) used immunomodulators as the only additional medical therapy [18/163 (11%) mild and 56/237 (24%) moderate/severe] and 12¾00 (31%) escalated to anti-TNFα (n=121), CI (n=1) or colectomy (n=1, emergent colectomy prior to anti-TNFα) [28/163 (17%) mild and 95/237 (40%) moderate/severe]. A total of 25/400 (6%) had colectomy [2/163 (1%) mild and 23/237 (10%) moderate/severe]. Figure S3 displays the time-course of treatments.
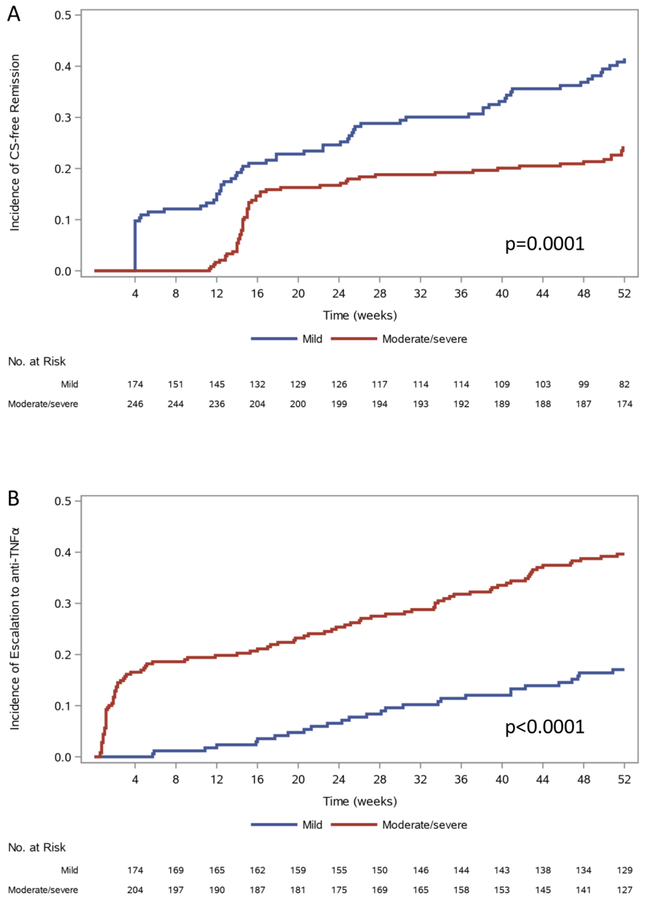
Figure 1 depicts time until SFR and escalation to anti-TNFα by initial disease presentation. Patients with mild disease presentation continued to achieve SFR from week 4 until week 52, while those with moderate/severe disease presentation who achieved SFR had largely done so by week 16. Approximately 50% of patients with moderate/severe disease presentation who escalated to anti-TNFα had done so by week 4. After week 4, mild and moderate/severe patients escalated to anti-TNFα at similar rates. At week 52, 50/105(48%) who escalated to anti-TNFα and had 1-year follow-up were in clinical remission (Table 1). Time to CS-free remission, escalation to immunomodulators, anti-TNFα, or colectomy as a function of Week 4 remission status are shown in Figures S4A–D. By week 16, 21/211(10%) without week 4 remission had achieved SFR; few ultimately reached SFR by week 52 [50/211 (24%). Conversely, 38/211(18%) who achieved week 4 remission escalated to anti-TNFα primarily between weeks 16 and 52.
Figure 1. Time until CS-free Remission (SFR) or Escalation to anti-TNFα by Initial Disease Presentation.
Time until A) CS-free remission (SFR) or B) escalation to anti-TNFa is shown by initial disease presentation of mild (n=178) or moderate-to-severe (n=250). Time until event was censored at last contact. Mild: initiated on mesalazine, or on oral CS with PUCAI < 45. Moderate/Severe: initiated on IV CS, or oral CS with PUCAI ≥ 45. Differences between groups were tested by log-rank test. Data presented represent Kaplan-Meier analysis considering patients lost to follow-up during the first year.
Table 1.
Baseline Clinical and Demographic Characteristics, Week 4 Remission, Week 12 CS-free Remission and Week 52 PUCAI Score Grouped by Week 52 Outcome
| Week 52 F.valliable population (n=400a) |
Week 52 CS-Free Remission (n=150) |
Neither CS-Free Remission nor Additional Therapy (n=53) |
Escalation to Immuno-modulator onlyb (n=74) |
Escalation to Anti-TNFαc (n=123) | p-value |
|---|---|---|---|---|---|
| Age (years) (mean±SD) | 12.3 ± 3.4 | 12.4 ± 3.8 | 12.9 ± 2.9 | 12.9 ± 3.0 | 0.30 |
| Female (%) | 69/150 (46%) | 30/53 (57%) | 36/74 (49%) | 62/123 (50%) | 0.60 |
| Nonwhite race (%) | 22/148 (15%) | 9/53 (17%) | 11/70 (16%) | 20/121 (17%) | 0.98 |
| Clinical Characteristics: | |||||
| Extensive/Pancolitisd | 117 (78%) | 40 (75%) | 65 (88%) | 113 (92%) | |
| % < 45 | 70/150 (47%) | 34/53 (64%) | 16/74 (22%) | 23/123 (19%) | <0.0001 ** |
| ≥65 (Severe) n=133 | 41/133 (31%) | 5/133 (4%) | 18/133 (14%) | 69/133 (52%) | |
| % ≥ 11 | 13 (9%) | 1 (2%) | 8 (11%) | 47 (38%) | <0.0001 ** |
| Mayo endoscopy score (range 0–3) (mean±SD) | N=150 2.1 ± 0.7 |
N=53 1.9 ± 0.6 |
N=74 2.2 ± 0.6 |
N=123 2.5 ± 0.6 |
<0.0001 ** |
| %<10g/dL | 22 (16%) | 7 (14%) | 22 (32%) | 45 (38%) | 0.0001 ** |
| Median (P25, P75) | 19 (11, 34) | 17 (9, 35) | 28 (12, 47) | 30 (17, 46) | 0.0005 ** |
| >ULN | 42 (37%) | 12 (32%) | 24 (47%) | 59 (59%) | 0.0044 * |
| % <3.5 g/dL | 40 (27%) | 15 (28%) | 22 (31%) | 55 (45%) | 0.0155 * |
| ≥ 30 ng/mL | 63 (46%) | 22 (44%) | 31 (46%) | 40 (35%) | |
| % ≥ 1.6 ng/mL | 65 (48%) | 20 (39%) | 21 (32%) | 36 (32%) | 0.045 * |
| Rectal biopsy eosinophilic inflammation (count > 32/hpf) | 75/126 (60%) | 31/46 (67%) | 43/63 (68%) | 45/107 (42%) | 0.0016 * |
| Week 4 Remissione | 100/150 (67%) | 27/53 (51%) | 33/74 (45%) | 38/123 (31%) | <0.0001 ** |
| Week 4 fecal calprotectin (mcg/g) Median (P25, P75) |
N=108 604.5 (144.8, 1519.3) |
N=39 677.6 (233.2, 1597.7) |
N=48 1014.3 (372.0, 2314.2) |
N=78 1576.8 (564.6, 2975.5) |
0.0008 ** |
| Week 12 CS-free remissionf | 79/150 (53%) | 23/53 (43%) | 15/74 (20%) | 19/123 (15%) | <0.0001 ** |
| ≥65 (Severe) | 0 (0%) | 1 (2%) | 2 (3%) | 6 (6%) |
p<0.05,
p < 0.001. P-values comparing groups are from a chi-squared or Fisher’s exact test (noted by #) for categorical variables, a Mantel-Haenszel chi-squared test for ordinal variables, and ANOVA or Kruskal-Wallis test for continuous variables. P25, P75: 25th and 75% percentile Additional characteristics are located in Table S3A
Evaluable population excludes participants who discontinued the study without additional therapy or colectomy
106 patients started on immunomodulators, 32 later escalated to anti-TNFα
Includes one patient starting a calcineurin inhibitor and one patient who received colectomy without escalation to anti-TNFα
Includes patients with fulminant disease with limited colonoscopy
week 4 remission: PUCAI < 10 at week 4 with no prior treatment escalation or colectomy
week 12 CS-free remission: PUCAI < 10 at week 12 and steroid-free for the prior 14 days with no prior treatment escalation or colectomy.
Clinical Factors Associated with Disease Course
Baseline characteristics, Week 4 remission, Week 12 CS-free remission, and Week 52 PUCAI score grouped by Week 52 outcome are shown in Table 1; additional baseline characteristics are in Table S3A. Table S3B shows these same data for the moderate/severe subgroup. Outcome rates did not vary with age, sex, or race. As expected, patients who achieved Week 52 SFR were more likely to have mild clinical and endoscopic severity, to exhibit higher hemoglobin and albumin, lower acute phase reactants, and higher rates of week 4 and week 12 remission. There was substantial variability, however, as 41/133 (31%) of patients with baseline PUCAI≥65 met week 52 CS-free remission, and 13/90 (14%) of subjects with PUCAI <35 were escalated to anti-TNFα by week 52. We observed that the bioavailable fraction of 25 (OH) D was higher in those who achieved Week 52 SFR (median 1.5 ng/mL vs 1.2–1.3 ng/mL, p=0.009). In those with moderate/severe presentation, serum 25 (OH) D and bioavailable fraction were lower in patients escalated to anti-TNFα (median 26.2 and 1.2 ng/mL respectively than other patient groups (median 28.7 – 30.6 and 1.1 – 1.5 ng/mL respectively, both p=0.03). Patients who escalated to anti-TNFα were less likely to exhibit elevated levels of eosinophils on rectal histopathology [45/107(42%) vs 60–68%, p=0.0016]; this was more apparent in those with moderate/severe presentation [31/83 (37%) vs 54–74%,p=0.0004]. Mesalazine adherence was not associated with achieving Week 52 SFR, or escalation to anti-TNFα (Table S7). Week 4 remission and week 12 CS-free remission were both strongly associated with Week 52 outcomes, and exhibited a modest correlation (Spearman correlation = 0.39, n=416, p<0.0001)
Genes Associated with Treatment Responses
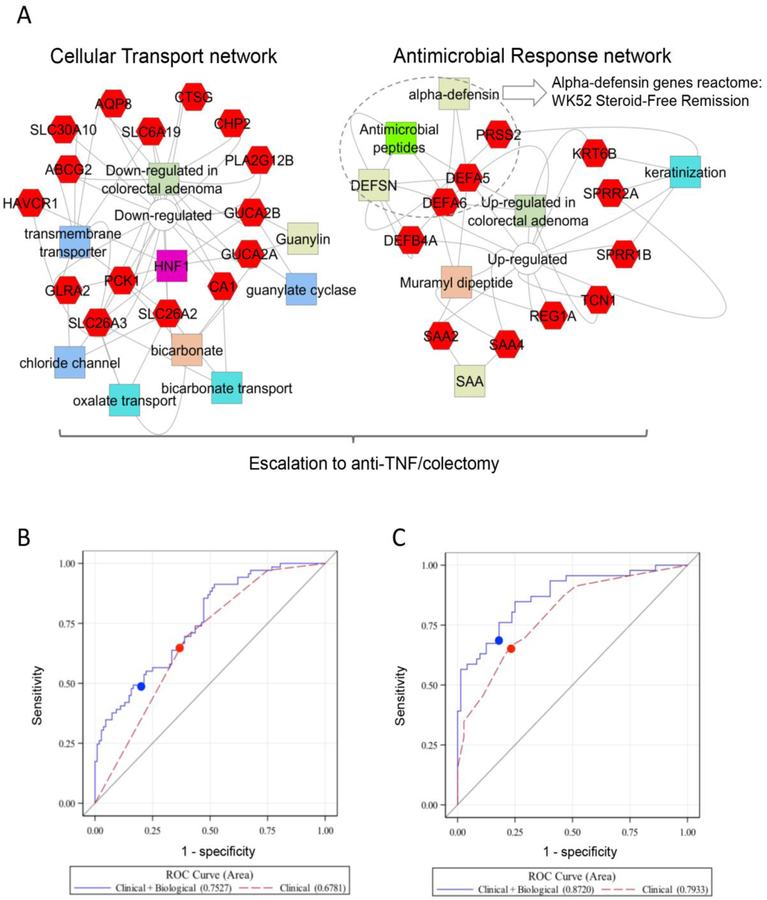
We identified a panel of 33 genes (Figure S5A and Tables S4A–C) which were differentially expressed in the rectum between moderate/severe patients who did or did not achieve Week 52 SFR. Unsupervised hierarchal clustering of the gene panel defined two groups of UC patients (Figure S5A, dendogram branch ii-a and ii-b). Patients who achieved week 52 SFR were more likely to be in block ii-b (52/75, p=0.03) while those that escalated to anti-TNFα were more likely to be in block ii-a (42/64, p=1E−6). Gene set enrichment analysis (GSEA) of the gene panel identified epithelial transporters and channels, and anti-microbial peptides including alpha-defensins (the transport and antimicrobial gene signature, Figure 2A). A specific antimicrobial peptide pathway was induced in UC compared to controls (Figure S6A), but relatively lower in many of the patients who achieved Week 52 SFR (Figure S6B). Consistent with this, a greater frequency of alpha defensin 5 (DEFA5) positive cells was detected in rectal biopsies from UC patients who did not achieve week 52 SFR, compared to UC patients who did achieve week 52 SFR, and controls (Figure S7). By comparison, the epithelial transport gene signature was suppressed in UC compared to controls (Figure S6C), to the greatest degree in patients who escalated to anti-TNFα (Figure S6D). A specific induction of the antimicrobial peptide gene signature (Figure S6E) and suppression of the epithelial transport gene signature (Figure S6F) was confirmed in UC compared to non-IBD controls, and disease controls with Crohn’s disease involving the rectum, in an independent cohort.
Figure 2. Biological Predictors of Disease Outcome.
A. Gene set enrichment analyses (GSEA) of the 33 genes differentially expressed in the rectum [18 increased (left), and 15 decreased (right)] in those that did vs. did not achieve WK52 CS-free remission using ToppGene, ToppCluster, and Cytoscape are shown (See Supplemental Appendix). The increased gene sets were associated with cellular transport and channel functions, while the decreased gene sets were associated with antimicrobial responses. The overall 33 gene set signature was associated with need for escalation to anti-TNFα, while the alpha-defensins reactome (biosystem ID: 1269266) antimicrobial peptides pathway showed a stronger association with the W52 SFR outcome. Genes are indicated in red hexagons, GO annotations in blue colors, drug-associations in orange, and pathways, gene family, interactions, and co-expression in green colors. The full list of GSEA results and p-values are in Table S4. B and C. ROC curves are presented for the logistic regression models in patients with clinical data (red) and clinical+biological data (blue) from the imputed dataset with the median CV AUC for the models for B) CS-free Remission and C) Escalation to Anti-TNFα. The cut-point used to define sensitivity and specificity for each model is indicated by the filled circle. See Table 2 and Table 3 for the AUCs and the p-value comparing the AUC of the two ROC curves averaged across the multiple imputations.
Microbes Associated with Treatment Responses
Microbes from the Clostridiales order have been implicated in maintenance of gut barrier function and immune tolerance19. Multivariable analyses identified taxa associated with week 52 SFR (six at FDR <0.2, with two at FDR<0.05), and escalation to anti-TNFα (56 at FDR <0.2, with 26 at FDR<0.05, Table S5A,B). Almost all of these were increased in patients who achieved SFR, and decreased in those who escalated to anti-TNFα. These included two operational taxonomic units (OTUs) from the Clostridiales order which were increased in patients who achieved week 52 SFR, and 20 Clostridiales OTUs which were decreased in patients who escalated to anti-TNFα therapy (FDR<0.05, Tables S5A,B).
Factors Associated with Week 52 CS-free Remission in Multivariable Regression Models
Mild clinical severity (PUCAI<45, regardless of initial treatment), week 4 remission, and baseline hemoglobin≥10 g/dL in those who did not achieve week 4 remission were associated with achieving Week 52 SFR (Table 2). The multivariable model demonstrated good discriminant power and specificity (Sp), with AUC (95% CI) of 0.70 (0.65, 0.75), cross-validated (CV) AUC 0.63 (95% CI 0.57, 0.69), Sp 0.77 (95% CI 0.71, 0.82), n=386 (Tables S6A, B and Figure S9A). A post-hoc model with Week 12 CS-free remission instead of Week 4 remission had substantially similar discriminant power with AUC of 0.69 (0.64, 0.75), specificity 0.77, (0.71, 0.82), n= 386.
Table 2.
Multivariable Logistic Regression Models of Week 52 CS-free Remission in All Patients
| All patients (N=386a) |
Patients with biological data (N=177) |
||
|---|---|---|---|
| Clinical Model | Clinical Modelb | Clinical + Biological Model | |
| Number of events (%) | 147 (38%) | 69 (39%) | 69 (39%) |
| Baseline predictors: |
Odds Ratio (95% CI) p-value |
Odds Ratio (95% CI) p-value |
Odds Ratio (95% CI) p-value |
| PUCAI <45c | 1.84 (1.18, 2.89) 0.008 |
||
| Hemoglobin ≥10 g/dL (w/o Wk 4 remissiond) | 4.24 (1.46, 12.31) 0.008 |
6.79 (1.45, 31.76) 0.015 |
5.72 (1.16, 28.23) 0.032 |
| Week 4 Remissione | 9.50 (3.39, 26.63) <0.0001 | 15.11 (3.39, 67.44) 0.00037 |
14.92 (3.18, 69.98) 0.00061 |
| Antimicrobial Peptide Gene Signature | 0.57 (0.39, 0.81) 0.0022 |
||
| Ruminococcaceae (560535) OTU log relative abundancef | 1.43 (1.02, 2.00) 0.036 |
||
| Sutterella (589923) OTU log relative abundancef | 0.81 (0.65, 1.00) 0.049 |
||
| Model Characteristics | |||
| R2 | 0.16 | 0.17 | 0.29 |
| Sensitivity (95% CI) | 47% (39%, 55%) | 70% (57%, 80%) | 49% (38%, 61%) |
| Specificity (95% CI) | 77% (71%, 82%) | 60% (50%, 69%) | 78% (70%, 85%) |
| PPV (95% CI) | 56% (47%, 64%) | 53% (42%, 63%) | 58% (46%, 71%) |
| NPV (95% CI) | 70% (65%, 76%) | 76% (65%, 84%) | 71% (62%, 79%) |
| Comparison of AUC | 0.014 | ||
| Hosmer-Lemeshow Goodness of Fit test median MI p-value (% of imputations with p > 0.05) | P = 0.42 (100%) | P = 0.99 (100%) | P=0.25 (100%) |
Week 52 CS-free remission was defined as PUCAI < 10 and no corticosteroids (CS) for 28 days without additional therapy or colectomy.
CV AUC = AUC from leave-one-out cross-validation, PPV=positive predictive value, NPV = negative predictive value
Note: Missing covariate data imputed via multiple imputation. Clinical parameters selected by LASSO. Sensitivity, specificity, PPV, and NPV were based on a predicted probability of 0.50.
The evaluable population excludes participants who discontinued the study without additional therapy or colectomy and without protocol violations.
The clinical model was run on the subset of participants with biological data. Clinical model factors with p > 0.05 in this smaller cohort were removed from the model.
PUCAI < 45, irrespective of initial treatment.
The interaction of hemoglobin ≥10 g/dL and Week 4 Remission was statistically significant (p=0.042) and the combinations of hemoglobin ≥ 10 and week 4 remission were combined into a 3-level variable: 1) neither hemoglobin≥10 and no week 4 remission, 2) hemoglobin≥10 and no week 4 remission, and 3) week 4 remission regardless of hemoglobin. Patients with low hemoglobin not achieving week 4 remission had a very low rate of week 52 cs-free remission, while those reaching week 4 remission had the same chance of week 52 cs-free remission as those who started with higher hemoglobin.
Week 4 remission: PUCAI < 10 at week 4 with no prior treatment escalation or colectomy
Log relative abundance was calculated by taking the log base 10 of the OTU plus half of the minimum value
Compares the clinical model in the subset of patients with biological data against the clinical + biological model
The predicted probability of CS-free remission for someone with PUCAI<45, hemoglobin ≥10 and week 4 remission = exp(−2.449 + 0.612*1 + 1.443*0 + 2.252*1)/(1+ exp(−2.449 + 0.612*1 + 1.443*0 + 2.252*1)) = 0.60; see supplemental Table S6A.
Components of the clinical model were validated in an independent inception cohort of 307 paediatric UC patients enrolled in the prospective Pediatric Inflammatory Bowel Disease Collaborative Research Group Registry7 (Table S10). Baseline clinical and demographic characteristics were quite similar to the PROTECT cohort. Disease activity was classified using Physician Global Assessment (PGA) as enrollment occurred prior to the publication of the PUCAI10. Medical therapy was not standardized, and 28% achieved week 52 SFR. Multivariable analysis in the 274/307 (89%) participants with complete data confirmed that mild baseline clinical disease activity (OR (95% CI): 1.7 (1.0 – 3.0), p=0.056) and week 4 inactive disease (OR (95% CI): 2.8 (1.7–4.8), p=0.0001), were associated with week 52 SFR with similar discriminant power (AUC (95% CI): 0.65 (0.59–0.71). Baseline hemoglobin did not achieve significance, although the direction of effect was consistent with the PROTECT clinical model (continuous hemoglobin OR = 1.08/1-unit increase, p=0.18; hemoglobin≥10 OR=1.55, p=0.19).
The relationship between the probability of achieving week 52 SFR based upon the PROTECT clinical model and the antimicrobial peptide gene signature is shown in Figure S5B. For each level of clinical probability, patients with a lower antimicrobial peptide gene signature PC1 were more likely to achieve SFR. The addition of the antimicrobial peptide gene signature and the relative abundance of Ruminococcaceae and Sutterella organisms to the clinical model improved the model discriminant power with an AUC of 0.75 (95% CI 0.68–0.83; p=0.014, CVAUC 0.72, 95% CI 0.64–0.80, Sp 0.78, 95% CI 0.70–0.85) in the subset with transcriptomic and microbial data (n=177, Figure 2B). Similar results were observed when treating the microbes as present or absent. Patients with lower levels of antimicrobial peptide gene expression and Sutterella organisms and higher relative abundance of Ruminococcaceae were more likely to achieve week 52 SFR.
Factors Associated with Escalation to anti-TNFα in Multivariable Regression Models
As 40% of moderate/severe patients escalated to anti-TNFα (Table S2), we next sought to define factors associated with this outcome. In patients with moderate/severe presentation, a total Mayo score ≥11 and lack of week 4 remission were associated with escalation to anti-TNFα. Notably, baseline hemoglobin < 10 g/dL, lower serum 25 (OH) D, and lower rectal biopsy eosinophil count were also predictive of escalation to anti-TNFα in these very ill patients (Table 3). A model including these factors demonstrated good discriminant power, with AUC (95% CI) of 0.78 (0.72, 0.84), CVAUC= 0.75 (95% CI 0.68–0.82), Sn 0.58 (95% CI 0.47–0.68), Sp 0.85 (95% CI 0.78–0.92), n=232 (Table S8A and Figure S7B). A post-hoc model with Week 12 CS-free remission instead of Week 4 remission had a similar AUC of 0.78 (0.72–0.85), Sn 0.59 (0.48–0.69), Sp 0.85 (0.78–0.92), n= 232. As total Mayo score, serum 25 (OH) D, and rectal biopsy eosinophil count were not collected in the IBD Registry study, we were not able to use these data to validate this secondary model. The relationship between the probability of escalation to anti-TNFα based upon the clinical model and the transport and antimicrobial gene signature is shown in Figure S5C. For each level of clinical probability, patients with a lower transport and antimicrobial gene signature PC1 were more likely to escalate to anti-TNFα Figures S5C and S6). The addition of the transport and antimicrobial gene signature and the abundance of an Oscillospira species to the model improved the model fit with an AUC of 0.88 (0.81, 0.94, p=0.016) CVAUC=0.84 (95% CI 0.76–0.91), Sn 0.66 (95% CI 0.52–0.80), Sp 0.83 (95% CI 0.74–0.91) in the subset with transcriptomic and microbial data (n=118, Figure 2C). Similar results were obtained when treating the microbes as present or absent. Patients with lower levels of transport gene expression and Oscillospira, and higher levels of antimicrobial gene expression, were more likely to escalate to anti-TNFα.
Table 3.
Multivariable Logistic Regression Models of Escalation to Anti-TNFα by Week 52 for Moderate/Severe Patients
| Moderate/severe patients (N=232a) |
Moderate/severe Patients with biological data (N=118b) |
||
|---|---|---|---|
| Clinical Model | Clinical Model | Clinical + Biological Model | |
| Number of events (%) | 94 (41%) | 46 (39%) | 46 (39%) |
| Baseline predictors: |
Odds Ratio (95% CI) p-value |
Odds Ratio (95% CI) p-value |
Odds Ratio (95% CI) p-value |
| Total Mayo score ≥11 | 4.38 (2.25, 8.54) 0.0002 |
||
| Rectal biopsy eosinophil peak count > 32/hpf | 0.48 (0.25, 0.92) 0.027 |
||
| Higher 25 (OH) D per increase in category: 1 = < 20 ng/mL 2 = 20– < 30 ng/mL 3 = ≥ 30 ng/mL |
0.60 (0.37, 0.96) 0.034 |
0.29 (0.11, 0.74) 0.010 |
0.31 (0.14, 0.71) 0.0052 |
| Hemoglobin ≥10 g/dL | 0.50 (0.26, 0.95) 0.033 |
0.39 (0.20, 0.76) 0.0063 |
0.33 (0.11, 0.94) 0.038 |
| Week 4 Remissionc | 0.36 (0.19, 0.68) 0.0015 |
0.16 (0.06, 0.41) 0.00012 |
0.21 (0.07, 0.58) 0.0029 |
| Transport and Antimicrobial Gene Signature | 0.31 (0.16, 0.61) 0.00063 |
||
| Oscillospira (581079) OTU log relative abundanced | 0.64 (0.44, 0.92) 0.018 |
||
| Model Characteristics | |||
| R2 | 0.32 | 0.37 | 0.52 |
| Sensitivity (95% CI) | 58% (47%, 68%) | 63% (49%, 77%) | 66% (52%, 80%) |
| Specificity (95% CI) | 85% (78%, 92%) | 82% (73%, 91%) | 83% (74%, 91%) |
| PPV (95% CI) | 73% (61%, 84%) | 69% (55%, 83%) | 71% (57%, 85%) |
| NPV (95% CI) | 75% (68%, 82%) | 78% (68%, 87%) | 79% (70%, 89%) |
| Comparison of AUC | 0.016 | ||
| Hosmer-Lemeshow Goodness of Fit test median MI p-value (% of imputations with p > 0.05) | P = 0.43 (99%) | P=0.96 (100%) | P=0.53 (100%) |
Moderate/Severe = initiated on IV CS or oral CS with PUCAI ≥ 45. CV AUC = AUC from leave-one-out cross-validation, PPV = positive predictive value, NPV = negative predictive value.
Note: Missing clinical covariate data imputed via multiple imputation. Clinical parameters selected by LASSO. Sensitivity, specificity, PPV, and NPV are based on a predicted probability of 0.5.
Note: Escalation to anti-TNFα includes one patient starting a calcineurin inhibitor and one patient who received colectomy without escalation to anti-TNFα.
The evaluable population excludes participants who discontinued the study without additional therapy or colectomy and with no protocol violations.
The clinical model was run on the subset of evaluable participants with biological data. Clinical model factors with p > 0.05 in this smaller cohort were removed from the model.
Week 4 remission: PUCAI < 10 at week 4 with no prior treatment escalation or colectomy
Log relative abundance was calculated by taking the log base 10 of the OTU plus half of the minimum value
Compares the clinical model in the subset of patients with biological data against the clinical + biological model
The predicted probability of Escalation to Anti-TNFα for someone with mayo ≥11, eos < 32, vitamin D < 20, hemoglobin <10, and no week 4 remission = exp(1.587 + 1.477*1 – 0.738*0 – 0.514*0 – 0.694*0 – 1.014*0) / (1+ exp(1.587 + 1.477*1 – 0.738*0 – 0.514*0 – 0.694*0 – 1.014*0)) = 0.96; see supplemental Table S8A.
Discussion
Despite utilization of standardised initial therapy with mesalazine±CS a minority of children with UC achieved the ideal clinical outcome of CS-free remission with mesalazine alone at one year following diagnosis. Additional medical therapy with thiopurines was common, almost one third received anti-TNFα therapy, and 6% required colectomy within one year of diagnosis. This is consistent with an 8% colectomy rate previously reported1. While more severe clinical presentations were associated with worse outcomes, some severely ill patients went on to do well and some patients with initially mild disease required therapy escalation.
A striking finding of our study was the importance of early response to mesalazine±CS and achieving clinical remission by Week 4 with these therapies as a predictor of Week 52 outcome. Importantly, we replicated this observation in a large independent cohort. A previous retrospective report of children treated in a similar fashion suggested that PUCAI at 3 months was an important predictor of CS-free remission at one year20. Earlier prediction of disease course would be preferable and could lead to more timely introduction of additional medical therapy beyond mesalazine±CS, especially in those moderately to severely ill at presentation. Recent ECCO/ESPGHAN guidelines recommend that children who present with acute severe colitis who respond to IV CS generally be transitioned to thiopurines21. Our data suggest that may not be necessary in all cases if clinical remission is achieved by 4 weeks, and that these children should be given an opportunity to demonstrate their ability to achieve early clinical remission and transition to mesalazine before being routinely placed on thiopurines or anti-TNFα agents.
We also sought to identify biologic predictors of disease course. Patients with pre-treatment levels of 25(OH)D < 20 ng/mL were more likely to escalate to anti-TNFα consistent with a murine study showing beneficial effects of vitamin D upon gut epithelial barrier function and inflammatory immune responses22, and prior data in adults with UC showing worse clinical outcomes associated with low 25 (OH) D levels6. Patients with pre-treatment rectal eosinophils <32/hpf were more likely to escalate to anti-TNFα consistent with adult UC data demonstrating worse outcomes associated with decreased rectal eosinophils23. These data suggest that incorporation of baseline 25(OH)D and rectal eosinophil levels into assessment of the likelihood of escalation to anti-TNFα in UC may be helpful, and may prioritize clinical trials targeting these pathways.
Shifts in the gut microbial community and host intestinal gene expression have been considered in predicting the likelihood of response to the biologic therapies24–27. Data regarding response to initial therapy of mesalazine and CS are lacking. We found that lower expression of genes encoding ion channels and transporters, and higher expression of anti-microbial peptides, was associated with a greater likelihood of escalation to anti-TNFα. The epithelial transport gene signature was enriched for guanylin activity, which ameliorates murine colitis28. While reduced Paneth cell alpha defensin expression has been implicated in the pathogenesis of ileal Crohn’s Disease29, we linked for the first time up-regulated rectal alpha defensin expression to a more refractory course in treatment naïve UC. In this context, alpha defensins may contribute to colonic injury via direct cytotoxic activity, and recruitment of inflammatory leukocytes26. Over two dozen microbial OTUs were linked to the clinical outcomes, with the majority from the Clostridiales order. Lower levels of Clostridiales at diagnosis were associated with escalation to anti-TNFα. This is consistent with recent studies linking these taxa to improved colonic barrier function, and anti-inflammatory regulatory T cell function19. A probiotic for UC therapy based upon Clostridiales species is now in development; our data support its potential clinical utility.
Our study has several limitations and notable strengths. Patient enrollment was not population based and we cannot exclude some degree of selection bias. However, the 29 centers were representative of paediatric UC care in North America. Only 69 (16%) participants were Asian or African-American, and so our results may not be generalizable to a non-white patient population. The complexity and expense of completing this study did not allow us to develop a similar validation cohort with standardised therapy and biological measures. Gene expression and microbiome data were available for only a subset of participants who strictly followed the treatment paradigm; a validation study will be required. A marked strength supporting generalizability of our findings is that our initial standardized treatment paradigm is in harmony with recent ECCO/NASPGHAN guidelines for the treatment of paediatric UC21,30 and we prospectively enrolled a large number of children with varying disease severity. Our primary outcome rate of CS-free remission on mesalazine only is similar to that reported previously from an uncontrolled observational registry of children newly diagnosed with UC7, and we replicated our main clinical model within this group.
Physicians, patients, and families seek early information that facilitates more informed choices when balancing the risks and benefits of additional medical therapy. Our data will have an immediate clinical impact on the treatment of children with UC. By week 4 following initial therapy with mesalazine±CS many children have declared their treatment response. Our validated clinical model including baseline severity coupled with week 4 remission has good specificity; this will minimize the number of false positive results in providing more confidence to guide treatment decisions. Readily available clinical indices of severity, rectal eosinophils and 25(OH)D levels can be used when considering the use of anti-TNFα therapy. Additional studies on rectal gene expression and gut microbial factors may improve prediction of response to initial therapy, identify novel therapeutic targets, and help develop personalized approaches improving outcomes in children with UC.
Supplementary Material
Research in context.
Evidence before the study
The lack of strong evidence-based outcomes data leads to uncertainty in developing treatment regimens in children newly diagnosed with UC, whose course may be unpredictable based upon initial clinical features alone. Robust controlled clinical trial data are not available in children, and current practice is largely based on extrapolation from adult data. We searched PubMed with the MeSH terms “ulcerative colitis”, “corticosteroids”, “5-aminosalicylates”, “treatment”, and “outcomes” for studies published in English before Dec 31, 2017. We identified no systematic prospective studies of initial standardised therapy in treatment-naive children, nor large scale microbiome and transcriptomic studies of stool and rectal tissue obtained at diagnosis to aid in predicting disease outcome at one year. While studies did identify specific clinical factors associated with treatment outcomes, these were limited by small sample sizes, uncontrolled therapies, and lack of replication.
Added value of this study
Our study of 428 treatment naïve children with UC conducted at 29 centers in North America from 2012 to 2016 establishes for the first time prospective controlled one year outcome data following standardised initial therapy with mesalazine±CS. We developed a clinical model for week 52 steroid-free remission with mesalazine alone which includes baseline clinical and laboratory factors and the week 4 response to standardised therapy, and then validated the model in an independent cohort. A similar model with high specificity was developed to predict escalation to anti-TNFα therapy in our primary cohort. We found that rectal gene expression and gut microbial factors not only improved our ability to predict clinical outcomes beyond initial disease severity and laboratory characteristics, but also provided crucial insight into the biological reasons for widely disparate patient courses. These data highlighted more robust host innate immune responses, and loss of microbial Clostridiales taxa, in worse outcomes after controlling for clinical severity.
Implications of all the available evidence
Our study has replicated several prior reports of clinical factors linked to outcomes in UC, and for the first time has developed and validated a clinical model for week 52 steroid-free remission with mesalazine alone in two large paediatric inception cohorts. These results will therefore have an immediate impact on clinical practice by providing specific parameters for initial disease activity, laboratory test results, and histologic features, as well as early response to standardised therapy, to guide therapeutic decisions. The transcriptomic and microbiome data reveal a possible mechanistic basis for success or failure of response to standardised therapy, and the need for treatment escalation. Our data suggest that novel personalized interventions that account for these host:microbe interactions might be beneficial in both guiding more rationale use of current therapies, and developing novel microbial interventions.
Acknowledgements:
The authors thank Frank Hamilton MD and Stephen James, MD from NIDDK for their guidance, Curtis Huttenhower, PhD, Tiffany Poon, and Hera Vlamakis, PhD from The Broad Institute for their assistance, and William Faubion, MD for his role as safety monitor. The study investigators are deeply indebted to Shire Pharmaceuticals for providing Pentasa® for this study, to the research coordinators at the investigative sites for their tireless attention, and to the patients and families who agreed to participate in this important study.
Support for this study was provided by NIDDK 5U01DK095745 and P30 DK078392, Integrative Morphology and Gene Expression Cores, NIDDK DK043351 and AT009708, CSIBD DK043351, Center for Microbiome Informatics and Therapeutics at Massachusetts Institute of Technology; www.protectstudy.com, Clinical trials: NCT 01536535
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
These authors have nothing to disclose:
Alison Marquis, Nathan Gotman, Bradley Saul, Jessie Wang, David Mack, Francisco Sylvester, Jonathan Evans, Keith Benkov, Marian Pfefferkorn, Robert Baldassano, Susan Baker, Brendan Boyle, Stephen Guthery, Boris Sudel, Joshua Noe, Prateek Wali, Suresh Venkateswaran, Vin Tangpricha, Dedrick Moulton, Kevin Hommel, Jose Serrano, Krista Spada, Yael Haberman, Rebekah Karns, Melanie Schirmer, Ramnik Xavier, Angela Mo, Keith Benkov, Urko Marigorta, Greg Gibson
These authors report the following disclosures:
Jeffrey S. Hyams: Advisory Board, Janssen, Consultant, Abbvie, Takeda, Lilly, Boerhinger-Ingelheim, Allergan, Pfizer, Receptos, Astra Zeneca; Sonia Davis Thomas, independent data monitoring committee, Lycera Corporation; Lee A. Denson: Grant Support, Abbvie and Janssen,; Neal LeLeiko: Consultant, Abbvie; Ashish Patel, Speakers Bureau Abbvie, Janssen; James Markowitz,Consultant for Janssen, Celgene, Lilly; Maria Oliva-Hemker: Abbott Immunology—research grant, Janssen –research grant, Hoffman LaRoche—consultant; Anne Griffiths:: Research support Abbvie, Consultant Abbvie, Celgene, Janssen, Lilly, Pfizer, Takeda, Speaker Abbvie, Janssen, Shire; Joel Rosh:Consultant, Abbvie, Celgene, Janssen, Luitpold, Pfizer. Grant Funding Janssen, Abbvie; David Keljo: research support from Genentech and Takeda; Anthony Otley: Advisory Board, Janssen, Abbvie, Research support Lilly, Abbvie, Janssen, Takeda, Celgene; Michael Kappelman:Consultant, Abbvie, Janssen, GlaxoSmithKline, Pfizer; Marla Dubinsky: Consultant, Prometheus Laboratories, Abbvie, Janssen, Takeda, Pfizer, Celgene, UCB, Boehringer Ingelheim, Lilly, Gilead, Allergan; Paul Rufo: Consultant, Shire, Leutpold, Speaker, Abbvie, Research support, TechLab; Cary Sauer, Consultant, Abbvie; Margaret H. Collins, Consultant: Shire, Regeneron, Receptos, Allakos; Contracts with Shire, Regeneron; Subra Kugathasan, Consultant, Janssen, UCB; Jennifer Strople, Consultant and speaker, Abbvie; Melvin Heyman, Research grants Genentech, Abbvie, Shire, Takeda, Mallinkrodt, Janssen, Gilead, David Ziring, Speaker’s bureau: Abbvie, consultant 11th Health and Vitality Biopharma.
References
- 1.Gower-Rousseau C, Dauchet L, Vernier-Massouille G, et al. The natural history of pediatric ulcerative colitis: a population-based cohort study. Am J Gastroenterol 2009;104:2080–8. [DOI] [PubMed] [Google Scholar]
- 2.Turner D, Mack D, Leleiko N, et al. Severe pediatric ulcerative colitis: a prospective multicenter study of outcomes and predictors of response. Gastroenterology 2010;138:2282–91. [DOI] [PubMed] [Google Scholar]
- 3.Hyams JS, Davis P, Grancher K, Lerer T, Justinich CJ, Markowitz J. Clinical outcome of ulcerative colitis in children. J Pediatr 1996;129:81–8. [DOI] [PubMed] [Google Scholar]
- 4.Melson JE, Giusto D, Kwasny M, Eichenseer P, Jakate S, Keshavarzian A. Histopathology predictors of medically refractory ulcerative colitis. Dis Colon Rectum 2010;53:1280–6. [DOI] [PubMed] [Google Scholar]
- 5.Haritunians T, Taylor KD, Targan SR, et al. Genetic predictors of medically refractory ulcerative colitis. Inflamm Bowel Dis 2010;16:1830–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gubatan J, Mitsuhashi S, Zenlea T, Rosenberg L, Robson S, Moss AC. Low Serum Vitamin D During Remission Increases Risk of Clinical Relapse in Patients With Ulcerative Colitis. Clin Gastroenterol Hepatol 2017;15:240–6 e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zeisler B, Lerer T, Markowitz J, et al. Outcome following aminosalicylate therapy in children newly diagnosed as having ulcerative colitis. J Pediatr Gastroenterol Nutr 2013;56:12–8. [DOI] [PubMed] [Google Scholar]
- 8.Bousvaros A, Antonioli DA, Colletti RB, et al. Differentiating ulcerative colitis from Crohn disease in children and young adults: report of a working group of the North American Society for Pediatric Gastroenterology, Hepatology, and Nutrition and the Crohn’s and Colitis Foundation of America. J Pediatr Gastroenterol Nutr 2007;44:653–74. [DOI] [PubMed] [Google Scholar]
- 9.Hyams JS, Davis S, Mack DR, et al. Factors associated with early outcomes following standardised therapy in children with ulcerative colitis (PROTECT): a multicentre inception cohort study. Lancet Gastroenterol Hepatol 2017;2:855–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turner D, Otley AR, Mack D, et al. Development, validation, and evaluation of a pediatric ulcerative colitis activity index: a prospective multicenter study. Gastroenterology 2007;133:423–32. [DOI] [PubMed] [Google Scholar]
- 11.Schroeder KW, Tremaine WJ, Ilstrup DM. Coated oral 5-aminosalicylic acid therapy for mildly to moderately active ulcerative colitis. A randomized study. N Engl J Med 1987;317:1625–9. [DOI] [PubMed] [Google Scholar]
- 12.Boyle B, Collins MH, Wang Z, et al. Histologic Correlates of Clinical and Endoscopic Severity in Children Newly Diagnosed With Ulcerative Colitis. Am J Surg Pathol 2017;41:1491–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burri E, Manz M, Rothen C, Rossi L, Beglinger C, Lehmann FS. Monoclonal antibody testing for fecal calprotectin is superior to polyclonal testing of fecal calprotectin and lactoferrin to identify organic intestinal disease in patients with abdominal discomfort. Clinica chimica acta; international journal of clinical chemistry 2013;416:41–7. [DOI] [PubMed] [Google Scholar]
- 14.Venkateswaran S, Prince J, Cutler DJ, et al. Enhanced Contribution of HLA in Pediatric Onset Ulcerative Colitis. Inflamm Bowel Dis 2018;24:829–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haberman Y, Tickle TL, Dexheimer PJ, et al. Pediatric Crohn disease patients exhibit specific ileal transcriptome and microbiome signature. J Clin Invest 2014;124:3617–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tuerk A, Wiktorin G, Guler S. Mixture models reveal multiple positional bias types in RNA-Seq data and lead to accurate transcript concentration estimates. PLoS Comput Biol 2017;13:e1005515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gevers D, Kugathasan S, Denson LA, et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell host & microbe 2014;15:382–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Friedman J, Hastie T, Tibshirani R. Regularization Paths for Generalized Linear Models via Coordinate Descent. J Stat Softw 2010;33:1–22. [PMC free article] [PubMed] [Google Scholar]
- 19.Atarashi K, Tanoue T, Oshima K, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 2013;500:232–6. [DOI] [PubMed] [Google Scholar]
- 20.Schechter A, Griffiths C, Gana JC, et al. Early endoscopic, laboratory and clinical predictors of poor disease course in paediatric ulcerative colitis. Gut 2015;64:580–8. [DOI] [PubMed] [Google Scholar]
- 21.Turner D, Ruemmele FM, Orlanski-Meyer E, et al. Management of Paediatric Ulcerative Colitis, Part 2: Acute Severe Colitis-An Evidence-based Consensus Guideline From the European Crohn’s and Colitis Organization and the European Society of Paediatric Gastroenterology, Hepatology and Nutrition. J Pediatr Gastroenterol Nutr 2018;67:292–310. [DOI] [PubMed] [Google Scholar]
- 22.Liu W, Chen Y, Golan MA, et al. Intestinal epithelial vitamin D receptor signaling inhibits experimental colitis. J Clin Invest 2013;123:3983–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heatley RV, James PD. Eosinophils in the rectal mucosa. A simple method of predicting the outcome of ulcerative proctocolitis? Gut 1979;20:787–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ananthakrishnan AN, Luo C, Yajnik V, et al. Gut Microbiome Function Predicts Response to Anti-integrin Biologic Therapy in Inflammatory Bowel Diseases. Cell host & microbe 2017;21:603–10 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arijs I, Li K, Toedter G, et al. Mucosal gene signatures to predict response to infliximab in patients with ulcerative colitis. Gut 2009;58:1612–9. [DOI] [PubMed] [Google Scholar]
- 26.Arijs I, De Hertogh G, Lemaire K, et al. Mucosal gene expression of antimicrobial peptides in inflammatory bowel disease before and after first infliximab treatment. PLoS One 2009;4:e7984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.West NR, Hegazy AN, Owens BMJ, et al. Oncostatin M drives intestinal inflammation and predicts response to tumor necrosis factor-neutralizing therapy in patients with inflammatory bowel disease. Nature medicine 2017;23:579–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harmel-Laws E, Mann EA, Cohen MB, Steinbrecher KA. Guanylate cyclase C deficiency causes severe inflammation in a murine model of spontaneous colitis. PLoS One 2013;8:e79180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.VanDussen KL, Liu TC, Li D, et al. Genetic variants synthesize to produce paneth cell phenotypes that define subtypes of Crohn’s disease. Gastroenterology 2014;146:200–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Turner D, Ruemmele FM, Orlanski-Meyer E, et al. Management of Paediatric Ulcerative Colitis, Part 1: Ambulatory Care-An Evidence-based Guideline From European Crohn’s and Colitis Organization and European Society of Paediatric Gastroenterology, Hepatology and Nutrition. J Pediatr Gastroenterol Nutr 2018;67:257–91. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.