Abstract
INF2 is a member of the formin family of actin assembly factors. Dominant mis-sense mutations in INF2 link to two diseases: focal segmental glomerulosclerosis (FSGS), a kidney disease; and Charcot-Marie-Tooth disease (CMTD), a neuropathy. All disease mutations map to the autoinhibitory Diaphanous Inhibitory Domain (DID). Curiously, purified INF2 is not autoinhibited, suggesting the existence of additional cellular inhibitors. We purified an INF2 inhibitor from mouse brain, and identified it as a complex between lysine-acetylated actin (KAc-actin) and cyclase-associated protein (CAP). Inhibition of INF2 by CAP/KAc-actin requires INF2 DID. Treatment of CAP/KAc-actin with histone deacetylase 6 (HDAC6) releases INF2 inhibition, while HDAC6 inhibitors block cellular INF2 activation. INF2 disease mutants are poorly inhibited by CAP/KAc-actin, suggesting that FSGS and CMTD result from reduced CAP/KAc-actin binding. These findings reveal a role for lysine-acetylated actin in the regulation of an actin assembly factor by a mechanism which we call facilitated auto-inhibition.
Keywords: cyclase-associated protein, FSGS, CMTD, HDAC6, facilitated autoinhibition, tubastatin A
INTRODUCTION
Most cellular processes are controlled by regulation of key proteins, with two common regulatory mechanisms being autoinhibition and post-translational modification (PTM). Many well-known regulatory proteins are autoinhibited1, requiring mechanisms to release the autoinhibitory interaction. A variety of PTM can trigger major changes in protein activity or location. Frequently, both autoinhibition and PTM serve to regulate the same protein1, 2.
Members of the formin family of actin assembly factors are key players in diverse cellular functions, including cytokinesis, cell motility, phagocytosis and organelle dynamics. Several of the 15 mammalian formns are known or predicted to be autoinhibited3–5, with 10 mammalian formins6, 7 containing two auto-regulatory sequences, the N-terminal Diaphanous Inhibitory Domain (DID) and the C-terminal Diaphanous Auto-regulatory Domain (DAD). The auto-inhibitory mechanism of one formin, mDia1, has been described in detail8. Binding of GTP-charged RhoA to mDia1’s GTPase Binding Domain (GBD) activates the formin by disrupting the DID/DAD interaction8, 9. Atomic structures have been solved of both DID alone and DID/DAD complex, and a single DID point mutation in the DAD binding site eliminates autoinhibition9, 10.
Inverted Formin 2 (INF2) is a mammalian formin containing DID and DAD sequences. Biochemically, INF2 has multiple effects on actin11, 12, as well as having both direct and indirect effects on microtubules13–16. In cells, INF2-mediated actin polymerization plays roles in mitochondrial fission, ER-to-mitochondrial calcium transfer, vesicle trafficking, and Golgi structure17–21.
Dominant mis-sense mutations in INF2 are linked to two diseases, focal segmental glomerulosclerosis (FSGS), a kidney disease22; and Charcot Marie Tooth disease (CMTD), a peripheral neuropathy23. Interestingly, all of the >35 disease-linked mutations map to the DID, suggesting regulatory effects. However, few disease mutations are in the DAD binding site. In addition, three results suggest that INF2 does not engage in canonical autoinhibition. First, INF2’s DID/DAD interaction is >10-fold weaker than that of mDia111, 24, 25. Second, actin monomers bind to INF2’s DAD, and compete with DID24. Third, purified INF2 is constitutively active in biochemical actin polymerization assays24. While these results suggest that INF2 is not regulated by autoinhibition, cellular studies show that INF2 is tightly regulated in a manner dependent on DID/DAD interaction19, 24.
These results suggest that an additional factor may mediate INF2’s DID/DAD interaction. In this work, we identify this factor as a complex between cyclase-associated protein (CAP) and lysine-acetylated actin. INF2 mutants linked to FSGS and CMTD are not inhibited by CAP/actin, suggesting a mechanism for disease.
RESULTS
INF2 is constitutively active in vitro but inhibited by DID/DAD interaction in cells
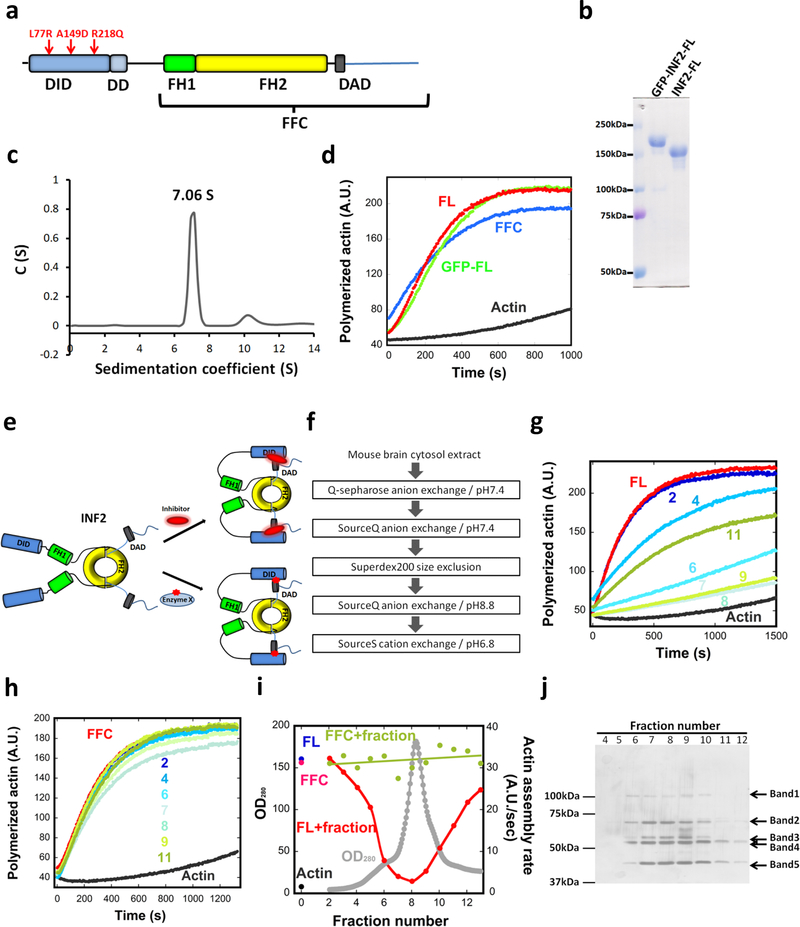
INF2 has two splice variants differing in C-terminal sequence: ER-bound INF2-CAAX18, 19, and cytosolic INF2-nonCAAX17, 20, 21. To test regulatory mechanisms, we used INF2-nonCAAX, expressing the human full-length protein (INF2-FL, Fig. 1a) in mammalian HEK293 cells as an N-terminally tagged fusion with GFP and Strep-tag II (GFP-Strep), and purified the protein to apparent homogeneity either with or without GFP-Strep (Fig. 1b). By sedimentation velocity analytical ultracentrifugation (vAUC), INF2 is a dimer (Fig. 1c). Next, we examined the effect of purified INF2 on actin dynamics, comparing INF2-FL to a truncated INF2 construct lacking the DID (INF2-FFC). Similar to INF2-FL purified from insect cells24, HEK293-purified INF2-FL accelerates actin polymerization from monomers with comparable potency to INF2-FFC (Fig. 1d). These results show that purified INF2 is not autoinhibited.
Figure 1. Constitutive activity of purified INF2, and inhibition by a brain factor.
(a) Schematic diagram of human INF2-nonCAAX (1240 residues). The A149D mutation disrupts DID/DAD interaction. The R218Q and L77R link to FGSG and CMTD, respectively. Domain boundaries: DID, 32–261; DD, 263–342; FH1, 421–520; FH2, 554–940; DAD, 971–1000. FFC construct: 469–1240. (b) Coomassie-stained SDS-PAGE of purified human INF2-nonCAAX, with and without the GFP-Strep tag. Experiment performed five times with similar results. (c) Sedimentation velocity analytical ultracentrifugation of INF2-nonCAAX (3.8 μM). Mass: 270.3 kDa (sedimentation), 134.6 kDa (sequence). Experiment performed once. (d) Pyrene actin polymerization assay (2μM actin) using 20nM INF2 (with or without GFP-Strep) or INF2 FFC. Experiment performed six times with similar results. (e) Schematic of two possible mechanisms for facilitated autoinhibition of INF2. Upper: direct inhibitor binding. Lower: INF2 post-translational modification (red star). (f) Flow chart of chromatography steps for inhibitor purification. (g) and (h) Representative pyrene-actin assays (2μM actin) containing 20nM INF2-FL (g) or 20nM INF2-FFC (h) with or without indicated BIF fractions (fractions 2–11, from Fig. 2i). Experiment performed once. (i) Graph showing protein elution profile from the final SourceS column (OD280 nm, gray) as well as activities of 20 nM INF2-FL (red) or FFC (green) in pyrene-actin assays on addition of the eluted fraction. Activity of INF2-FL alone (blue), INF2 FFC alone (magenta) and actin alone (black) shown on left side. Experiment performed once. (j) Silver stained SDS-PAGE of indicated fractions from figure 2i. Bands identified as follows: Band1, Exportin1/7; Band 2, VATA and HSP7C; Band 3, CAP1, CAP2, VATB2; Band 4, CAP1, CAP2, VATA; and Band 5, actin. Experiment performed once.
We also tested the effect of GFP-INF2-nonCAAX on cytosolic actin polymerization in U2OS cells (Fig. S1a). Wild-type INF2 causes no apparent increase in cytoplasmic actin filaments, while expression of a DID mutant predicted to disrupt DID/DAD interaction (INF2-A149D) increases cytoplasmic actin filaments significantly (Fig. S1b,c). These results agree with previous findings24, and suggest that INF2 is inhibited through DID/DAD interaction in cells, but that this interaction is insufficient to inhibit purified INF2. One explanation could be that a cellular factor facilitates DID/DAD interaction. In this work, we identify such a factor.
Identification of an INF2 inhibitory protein
Two possible mechanisms by which an inhibitor could enhance DID/DAD interaction are: (1) direct binding; or (2) post-translational modification (Fig. 1e). We used a biochemical approach to identify either mechanism, mixing chromatographic fractions from mouse brain with purified INF2-FL, then assaying actin polymerization rate. We made two assumptions: 1) the inhibitor would be cytosolic, because both INF2-CAAX and INF2-nonCAAX are inhibited in cells; and 2) the inhibitor would be abundant, because even cells strongly over-expressing WT-INF2 do not display aberrant actin polymerization (Fig. S1b, c).
We used five chromatographic steps in the purification (Fig. 1f), assessing INF2-FL inhibition by pyrene-actin assay (Fig. 1g,h, Fig. S2a–c). In the last step, inhibitory activity eluted with the major protein peak (Fig. 1i). We refer to this fraction as Brain Inhibitory Fraction (BIF). Importantly, BIF does not inhibit actin polymerization by INF2-FFC (Fig. 1h,i) or mDia1-FFC (Fig. S2d), demonstrating that it is not acting as a general actin polymerization inhibitor. The inability of the BIF to inhibit INF2-FFC also demonstrates that DID is required for inhibition. BIF does not cause significant INF2 proteolysis (Fig. S2e), suggesting that the inhibitor is not a protease. BIF loses all activity upon boiling (Fig. S2f). These experiments reveal a cytosolic protein inhibitor of INF2 that requires INF2’s DID for inhibition.
CAP/actin complex is an INF2 inhibitor
BIF contains five visible bands by SDS-PAGE and silver staining (Fig. 1j), which were identified as: Band 1, Exportin1/7; Band 2, Heat shock cognate 71kDa protein (HSP7C) and Vacuolar ATPase subunit A (VATA); Band 3, cyclase associated proteins 1 and 2 (CAP1, CAP2) and Vacuolar ATPase subunit B2 (VATB2); Band 4, CAP2, CAP1 and VATA; and Band 5, actin (Supplementary Table 1). We immuno-depleted individual proteins from BIF and tested INF2-FL inhibition by these depleted fractions. Depletion of CAP2 eliminates INF2 inhibition (Fig. 2a,b), whereas depletion of VATA, VATB2 or HSP7C has no effect (Fig. S3). Available antibodies against CAP1 and exportin proved unsuitable for depletion.
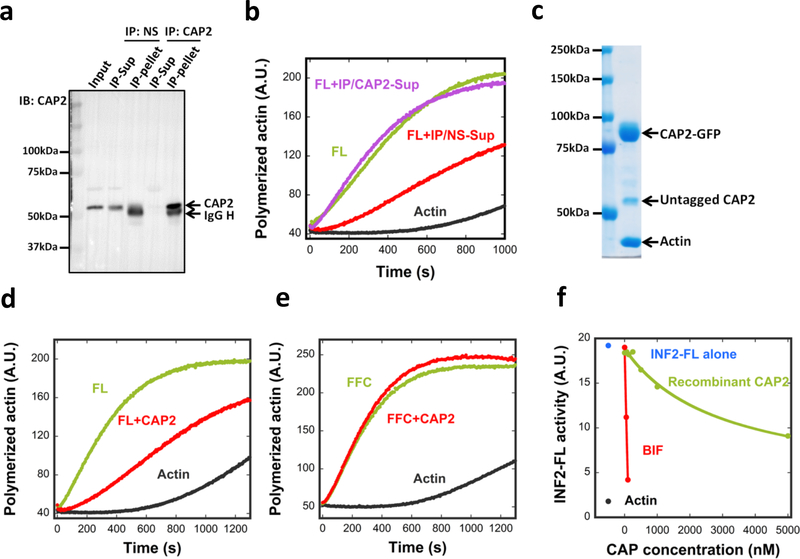
Figure 2. CAP/actin is an INF2 inhibitor.
Immuno-depletion of CAP2. An INF2-inhibiting fraction from mouse brain (Fraction 38–40 pool from SourceQ#1 (Fig. S2a)) was incubated with either a nonspecific AB (NS) or anti-CAP2 AB followed by precipitation using Protein A beads. CAP2 protein in input, supernatant and pellet detected by western blot. AB heavy chain, IgG H. Experiment performed once. (b) Pyrene-actin polymerization assay (2μM actin monomer, 5% pyrene) testing inhibitory activity of immune-depleted supernatants on 20nM INF2-FL. IP/NS-Sup: non-specific antibody supernatant. IP/CAP2-Sup: anti-CAP2 supernatant. Experiment performed once in triplicate. (c) Coomassie stained SDS-PAGE of co-expressed CAP1-GFP/CAP2-GFP purified from HEK293 cells. Experiment performed 24 times with similar results. (d) and (e) Pyrene-actin polymerization assay (2μM actin monomer, 5% pyrene) testing effect of co-expressed CAP1/2 (5 μM, untagged) on 20nM INF2-FL (d) or on 20nM INF2 FFC (e). Experiment performed five times with similar results. (f) Comparing inhibitory activities of recombinant CAP (green) and CAP in BIF (red) on INF2-FL (20nM) in pyrene-actin assays. Assembly rate of actin alone (black) and with INF2-FL (blue) also shown. CAP concentration in BIF determined as described in Methods. Experiment performed five times with similar results for recombinant CAP, and once for BIF.
These results suggest that CAP2 is required for INF2 inhibition by BIF. Since additional inhibitory peaks were identified in the first two purification steps (Fig. S2a,b), we used western blotting to ask whether CAP1 or CAP2 were present in these fractions. Both CAP1 and CAP2 were detected in inhibitory fractions from anion exchange chromatography (Fig. S2a) and size exclusion chromatography (Fig. S2b). These results suggest that endogenous CAP complexes are heterogeneous in size, but that all CAP-containing peaks can inhibit INF2.
To further test CAP as an INF2 inhibitor, we purified recombinant human CAP, containing C-terminal GFP and Strep-tag II (CAP-GFP), in HEK293 cells. We purified three versions of CAP: CAP1, CAP2, and co-expressed CAP1/CAP2. When analyzed by SDS-PAGE, all CAP-GFP fusions contain two additional bands: untagged CAP and actin (Fig. 2c, Fig. S4a). The mean molar ratio of untagged CAP to CAP-GFP is 0.16 in all cases, whereas the mean actin:CAP-GFP ratio is 0.88 (Table 1). Quantification of the protein bands in BIF reveals an actin:CAP ratio of ~1:1 (Table 1). CAP elutes as a high mass particle by gel filtration (Fig. S4b), and CAP-GFP sediments as a 14.5 S particle by vAUC, with a calculated mass of 726 kDa (Fig. S4c). Considering past studies showing CAP hexamerization26, 27, we interpret our purified CAP-GFP to be a hexamer with a 5:1 ratio of CAP-GFP to untagged CAP (likely endogenous HEK293 CAP), and actin monomer bound to a mean of five CAP subunits.
Table 1.
Molar ratios in the CAP/actin complex.
| Mole percentage of indicated protein compared to CAP* | CAP1 | CAP2 | CAP1&2 | BIF | |
|---|---|---|---|---|---|
| Actin | Avg | 82.5% | 88.1% | 90.5% | 106.8% |
| STD | 9.4% | 11.7% | 10.7% | 4.3% | |
| N | 6 | 24 | 2 | 3 | |
| Untagged CAP | Avg | 16.8% | 16.3% | 16.5% | NA |
| STD | 14.3% | 12.4% | 15.0% | NA | |
| N | 2 | 20 | 3 | NA | |
Calculated from densitometry of Coomassie-stained SDS-PAGE. CAP1, CAP2, and CAP1&2 = purified GFP-fusion proteins from HEK293 cells. BIF = purified brain inhibitory fraction.
represents CAP-GFP (for CAP1, CAP2, and CAP1&2) or CAP (BIF) set to 100%.
N = number of independent protein preparations analyzed.
Recombinant CAP/actin inhibits actin polymerization by INF2-FL (Fig. 2d) but not by INF2-FFC (Fig. 2e, Fig. S4f). However, the inhibitory potency of recombinant CAP/actin is significantly lower than that of BIF (Fig. 2f). The IC50 for CAP in BIF is 69 nM, whereas recombinant CAP does not reach its IC50 at the maximum concentration tested. We obtain similar results for recombinant CAP1/actin and CAP2/actin, both with and without GFP (Fig. S4d). In the absence of INF2, CAP/actin has little effect on actin polymerization at the highest concentration tested (Fig. S4e). These results suggest that CAP is a component of the INF2 inhibitory factor, although recombinant CAP does not inhibit maximally.
Actin is an important component of CAP-mediated INF2 inhibition
We reasoned that the difference in inhibition between recombinant CAP and BIF might be due to differences in the bound actin. Therefore, we attempted to exchange actin in our recombinant CAP/actin (heretofore called CAP/293A) with other types of actin. We first tested actin exchange on CAP by mixing immobilized CAP/293A with fluorescently-labeled actin (TMR-actin) and monitoring bead-bound fluorescence (Fig. S5a). Appreciable fluorescent actin associates with the CAP/293A-bound beads but not with control GFP beads (Fig. S5b). The ratio of actin:CAP is not changed significantly compared with mock-exchanged CAP (Fig. S5b). These results show that actin monomers can exchange onto CAP.
We exchanged CAP with total brain actin (BA, Fig. S5c). CAP/BA displays significantly more INF2 inhibition (IC50 54 nM) than does CAP/293A (Fig. 3a,b), but does not inhibit INF2-FFC (Fig. 3c). We next exchanged CAP with two types of skeletal muscle actin, from chicken (CSKA) or rabbit (RSKA). Interestingly, CAP/CSKA inhibits INF2-FL with an IC50 approaching CAP/BA (254 nM Fig. 3d,e), but does not inhibit INF2-FFC (Fig. S4f). In contrast, CAP/RSKA is a poor INF2 inhibitor, similar to CAP/293A (Fig. 3d,e). These results show that the source of the CAP-bound actin significantly affects INF2 inhibition.
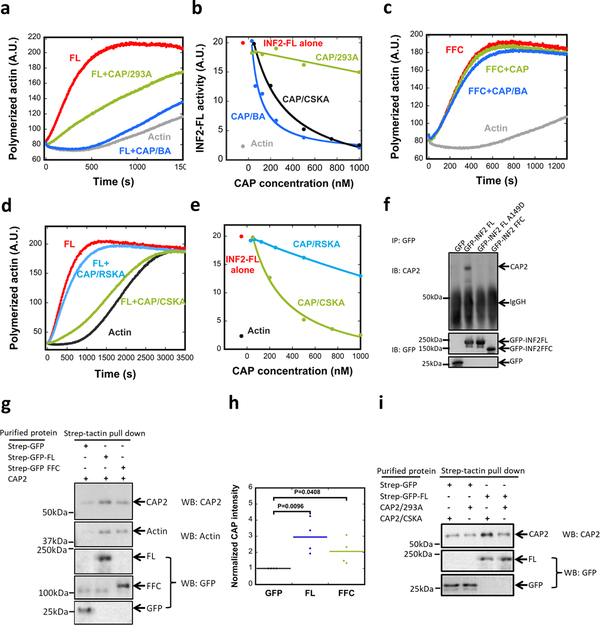
Figure 3. Actin-dependent differences in INF2 inhibition by CAP/actin.
(a) Pyrene-actin polymerization assay (2μM actin monomer, 5% pyrene) containing 20nM GFP-INF2-FL in the presence or absence of 5μM purified CAP2-GFP (CAP/293A) or 250nM CAP2-GFP exchanged with brain actin (CAP/BA). Experiment conducted once in triplicate. (b) Concentration dependence of INF2 activity inhibition by CAP/293A, CAP/BA, or by CAP exchanged with chicken muscle actin (CAP/CSKA). Experiment conducted once in triplicate. (c) Pyrene-actin polymerization assay (2μM actin monomer, 5% pyrene) containing 20nM GFP-INF2 FFC in the presence or absence of 250nM CAP/293A or CAP/BA. Experiment conducted once in triplicate. (d) Pyrene-actin polymerization assay (2μM actin monomer, 5% pyrene) containing 20nM GFP-INF2 full-length in the presence or absence of 1μM CAP/CSKA or CAP/RSKA (rabbit muscle actin). Experiment conducted three (CAP/CSKA) or four times (CAP/RSKA) in triplicate. (e) Concentration dependence of INF2 inhibition by recombinant CAP2 exchanged with CAP/CSKA or CAP/RSKA. Experiment conducted once in triplicate. (f) Co-immunoprecipitation assay of endogenous CAP2 with transfected GFP-fusion proteins: GFP alone, GFP-INF2-FL, GFP-INF2FL A149D mutant, and GFP-INF2-FFC. Top panel: anti-CAP2 western on precipitated samples. Bottom panel: anti-GFP western on same samples. Experiment conducted two times. (g) Strep-tactin pull-downs performed with 1μM bead-bound strep-tagged GFP (control), strep-tagged GFP-INF2 full-length, or strep-tagged GFP-INF2 FFC. These were mixed with 2μM untagged CAP exchanged with chicken muscle actin (CAP/CSKA). Bound samples were analyzed by immunoblotting with anti-CAP2, anti-actin and anti-GFP. Experiment conducted three times. (h) Quantification of bound CAP2 from pull-downs as in Fig. 3g. CAP2 intensity from immunoblots normalized to strep-GFP pulldown. Bars, mean. Error bars, S.E.M. (n = 4 experiments). P values, one-sided student’s t-test. (i) Strep-tactin pull-downs performed with 1μM bead-bound strep-tagged GFP (control), or strep-tagged GFP-INF2 full-length. These were mixed with 2μM untagged CAP exchanged with chicken muscle actin (CAP/CSKA) or w/o exchange (CAP/293A). Bound samples analyzed by immunoblotting with anti-CAP2 and anti-GFP. Experiment conducted two times.
We next asked whether INF2 binds CAP/actin directly. First, we used a cellular co-IP approach, transfecting U2OS cells with GFP-INF2 then conducting anti-GFP IP and probing for CAP. CAP2 co-precipitates with GFP-INF2-FL but not with GFP-INF2-FFC nor with the INF2-FL-A149D (Fig. 3f). We also analyzed INF2/CAP interaction using purified proteins, by incubating immobilized GFP-INF2-FL or GFP-INF2-FFC with CAP/CSKA in solution, then analyzing the bead-associated CAP. CAP associates with INF2-FL to a greater extent than INF2-FFC (Fig. 3g,Fig. 3g,h). These results suggest that CAP/actin inhibits INF2 by direct interaction. In addition, CAP/CSKA binds INF2 better than does CAP/293A (Fig. 3i), consistent with their different inhibitory potencies. These results suggest that DID/DAD interaction enhances INF2 binding to CAP/actin, and that INF2/CAP interaction requires a specific type of actin.
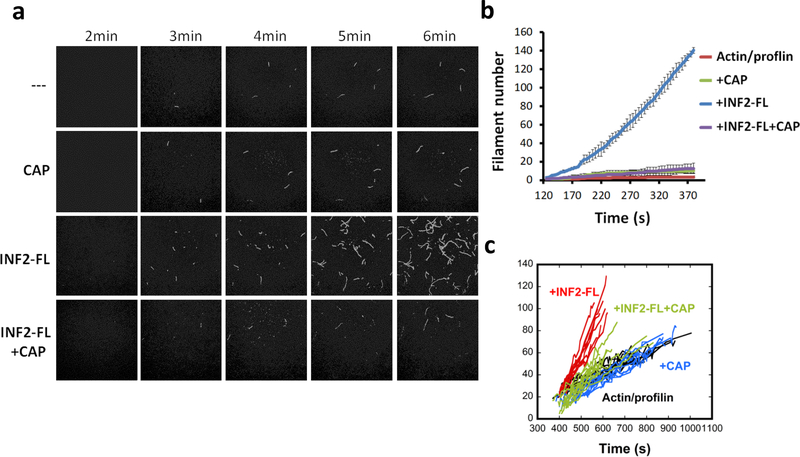
Profilin is an abundant cytosolic actin-binding protein, and a significant proportion of cytosolic actin monomer is profilin-bound28. In addition, profilin binds CAP29. We tested the effect of profilin on INF2 inhibition by CAP/CSKA. By pyrene-actin polymerization, CAP/CSKA still potently inhibits INF2 in the presence of profilin (Fig. S5d), even though profilin slows overall actin polymerization significantly. We also conducted total internal reflection (TIRF) microscopy assays of INF2-mediated actin polymerization in the presence of profilin, distinguishing between effects on actin nucleation and elongation. INF2 significantly increases nucleation rate (Fig. 4a,b), and increases the filament elongation rate approximately 2-fold (Fig. 4c). CAP/CSKA eliminates the INF2 effect on nucleation, while not significantly affecting nucleation of profilin/actin alone (Fig. 4a,b). CAP/CSKA has little effect on profilin/actin elongation rate, but inhibits profilin/actin elongation in the presence of INF2 (Fig. 4c). These results show that INF2 inhibition by CAP/actin can occur in the presence of profilin.
Figure 4. CAP inhibits INF2 mediated nucleation observed by TIRF microscopy.
Time-lapse TIRF microscopy images showing actin polymerization (1μM actin monomer, 10% TAMRA-actin) containing 3μM profilin in the presence or absence of 1nM INF2-FL and 50nM CAP2 exchanged with CSKA. (b) Quantification of filament number over time for TIRF assays. Three microscope fields analyzed for INF2 FL, and five fields for other conditions. Centre value represents mean and error bars are standard error of the mean. (c) Measurement of filament length over time for TIRF assays. 10 filaments for each condition.
Actin acetylation increases INF2 inhibition by CAP/actin
We next probed the mechanism behind the differing inhibitory potencies of CAP/CSKA and CAP/RSKA. Actin is highly conserved, with CSKA and RSKA being 100% identical in amino acid sequence. One possible explanation for inhibitory differences is differential actin post-translational modification. By mass spectroscopy analysis, we identified acetylation on several lysines of actin for CAP/CSKA: including K50, K61, K326 and K328 (Supplementary Table 2). We therefore tested whether lysine acetylation of actin was required for INF2 inhibition.
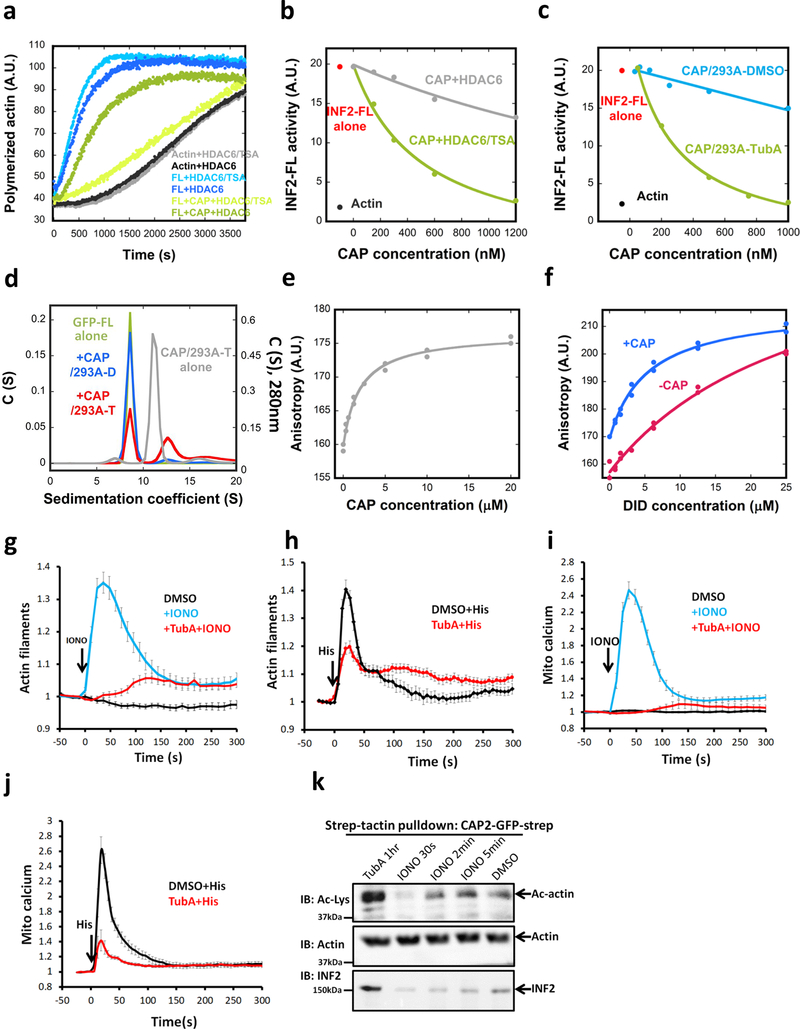
First, we attempted to deacetylate CAP/CSKA. HDAC6 is a major cytosolic lysine deacetylase30. We expressed and purified FLAG-tagged HDAC6 (Fig. S5e), which contains substantial deacetylase activity on brain tubulin (Fig. S5f). In pyrene-actin assays, HDAC6 strongly reduces CAP/CSKA inhibition of INF2, while minimally affecting polymerization of actin alone or in the presence of INF2 (Fig. 5a). Pre-treatment of HDAC6 with trichostatin A (TSA), an HDAC inhibitor, abolishes its effect on CAP/CSKA (Fig. 5a). The concentration curves of CAP/CSKA treated with HDAC6 alone versus HDAC6+TSA show a dramatic difference in inhibitory activities (Fig. 5b), suggesting that lysine acetylation on actin is necessary for CAP/actin inhibition of INF2.
Figure 5. Acetylation-induced regulation of INF2 biochemically and in cells.
Pyrene actin polymerization assay (2μM actin monomer) containing 20nM GFP-INF2-FL in the presence or absence of 1μM CSKA-exchanged CAP2 (CAP2/CSKA) after pre-incubation with either HDAC6/TSA (HDAC6 with its inhibitor, TSA) or HDAC6 alone. Experiment conducted twice in triplicate. (b) Concentration dependence of CAP2/CSKA inhibition of INF2-FL after pre-treatment with HDAC6/TSA or HDAC6 alone. Experiment conducted twice. (c) Concentration dependence of INF2 inhibition by CAP2/293A purified from cells pretreated with DMSO or 50μM Tubastatin A for 1hr prior to lysis and purification. Experiment conducted four times. (d) Sedimentation velocity analytical ultracentrifugation of GFP-INF2-FL (1.8μM), with or w/o 18μM CAP2/293A-D or CAP2/293A-T. Mass of GFP-INF2-FL: 325.2 kDa (from sedimentation), 161.3 kDa (from sequence). Left axis: 490 nm (GFP-INF2), right axis, 280 nm (CAP). Experiment conducted once. (e) Fluorescence anisotropy for CAP2/293A-T binding by INF2-Cterm (100 nM). Experiment conducted once. (f) Fluorescence anisotropy for INF2-Nterm binding to INF2-Cterm (100 nM) in presence or absence of CAP2/293-T (20 μM). Additional values at 48 μM INF2-Nterm, not present on graph but accounted for in the fit curves, are 217 for -CAP and 211 for +CAP. Experiment conducted once. (g) Ionomycin-induced changes in actin filaments in U2OS cells pretreated with either DMSO or 50μM TubA. Mean and S.E.M. shown. N = 40, 38 and 48 independent cells for DMSO, IONO, and IONO+TubA, repectively. (h) Histamine-induced changes on actin filaments in U2OS cells pretreated with either DMSO or 50μM TubA. Mean and S.E.M. shown. N = 15 independent cells for both conditions. (i) Ionomycin-induced changes on mitochondrial calcium levels in U2OS cells pretreated with either DMSO or 50μM TubA. Mean and S.E.M. shown. N = 29, 28, and 22 independent cells for DMSO, IONO, and IONO+TubA, respectively. (j) Histamine-induced changes on mitochondrial calcium in U2OS cells pretreated with either DMSO or 50μM TubA. Mean and S.E.M. shown. N = 15 independent cells for both conditions. (k) Strep-tactin pull-downs performed with cell lysates of U2OS cells overexpressing CAP2-GFP-2xStrep. Cells treated identically prior to lysis. CAP2-GFP was isolated, and CAP2-bound actin and probed by western using anti-actin and anti-Ac-Lys antibodies. Experiment conducted once.
As a second approach to test HDAC6’s effect, we purified CAP/actin from 293 cells in which HDAC6 activity is inhibited. Cells were treated either with DMSO or with the HDAC6-specific inhibitor tubastatin A (TubA), before cell harvesting and purification. Two tests indicate the effectiveness of TubA treatment. First, acetylated tubulin in cell extracts increases dramatically (Fig. S5g). Second, the amount of acetylated lysine detected on actin in purified CAP/293A complex increases (Fig. S5h). TubA-treated CAP/293A (CAP/293A-T) inhibits INF2-mediated actin polymerization with an IC50 of 263nM, which is significantly more potent that the DMSO-treated sample (CAP/293A-D, Fig. 5c). These results suggest that 1) HDAC6 de-acetylates actin in cells, and 2) increased actin acetylation increases CAP/actin inhibitory potency.
We used the higher inhibitory potency of CAP/293A-T versus CAP/293A-D to analyze the direct interaction between INF2 and CAP/actin in more detail. By vAUC, CAP/293A-T causes a portion of GFP-INF2-FL to shift to a 12.6 S species (Fig. 5d, red curve), which is greater than that of the un-tagged CAP/293A-T complex (11.2 S, gray curve), suggesting INF2 binding to the hexameric CAP complex. CAP/293A-D does not produce this magnitude of shift in GFP-INF2 (Fig. 5d, blue curve), suggesting lower affinity. In addition, CAP/293A-T causes no shift GFP-INF2-FFC (Fig. S5i), suggesting that DID is required for high-affinity interaction.
We also used fluorescence polarization of fluorescently-labeled INF2-Cterm (containing DAD) to examine its binding to INF2-Nterm (containing DID) in the presence or absence of CAP/293A-T. INF2-Cterm displays some affinity for CAP/293A-T, with a Kdapp of 1.82 μM (Fig. 5e). Intriguingly, the affinity of INF2-Cterm for INF2-Nterm increases >5-fold in the presence of CAP/293A-T (Fig. 5f), with the Kdapp of DID/DAD estimated at 28 μM and 5.4 μM in the absence and presence of CAP/293A-T, respectively (see Methods for Kdapp determination). These results suggest that CAP/actin complex increases the affinity between DID and DAD.
We used mass spectroscopy to compare actin acetylation patterns for CAP/293A-T and CAP/293A-D samples. Four acetylation sites on actin are appreciably enriched in CAP/293A-T (K50, K68, K215, and K315), while three sites display no clear acetylation change (K191, K326, K328, Supplementary Table 3). These results suggest that the increase in CAP/actin inhibitory potency might be linked to acetylation of K50, K68, K215 and/or K315.
The ability of HDAC6 to abolish CAP/actin inhibition of INF2 suggests that HDAC6 might be required for cellular INF2 activation. Previously, we and others have shown that cell stimulation with ionomycin or histamine causes an INF2-dependent cytosolic actin “burst”18, 31–33. Here, we show that TubA pre-treatment strongly reduces the both ionomycin- and histamine-induced actin burst in U2OS cells (Fig. 5g,h), and the down-stream consequence of increased mitochondrial calcium18 (Fig. 5i,j). These results show that HDAC6 can activate INF2 both biochemically and in cells.
We also tested whether ionomycin treatment alters KAc-actin levels in U2OS cells. Since western blotting of Ac-K from cell lysates resulted in high background, we took an alternate approach in which we expressed 2xStrep-tagged CAP2, then affinity-purified the CAP2 and examined Ac-K levels on the bound actin at various time points after treatment. Mock treatment with DMSO results in measurable KAc-actin, and treatment with TubA strongly increases KAc-actin (Fig. 5k). Ionomycin treatment causes a significant drop in KAc-actin at 30 sec, with a return to control levels at 2 min (Fig. 5k). This time course correlates with that of the ionomycin-induced actin burst (Fig. 5g). In addition, endogenous INF2 co-purifies with CAP2 in DMSO-treated and the TubA-treated samples, but co-purification is greatly reduced at 30 sec post-ionomycin treatment (Fig. 5k). These results suggest that ionomycin causes a transient decrease in lysine acetylation of actin, resulting in decreased INF2 binding.
INF2 disease mutants display reduced inhibition by CAP/actin
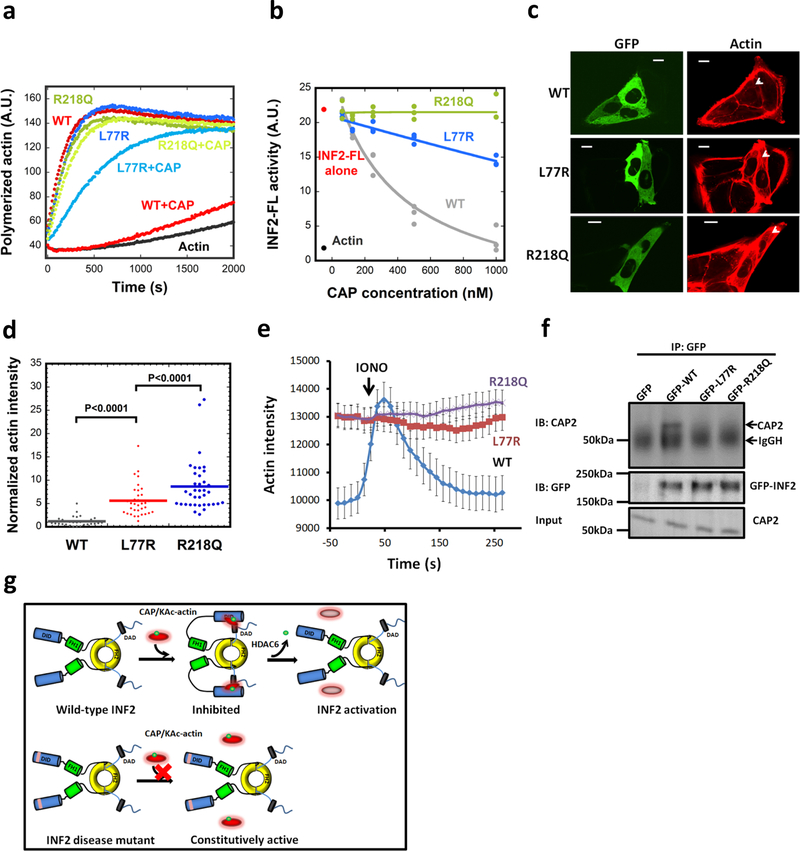
Although multiple mutations in INF2 DID are linked to FSGS and CMTD, the molecular mechanism of disease progression is unknown. We therefore asked whether CAP/actin could inhibit two INF2 mutants: R218Q, a common FSGS mutant22; and L77R, a CMTD mutant23. R218 is located near the DAD-binding site but is not predicted to make direct contact with DAD22, while L77 is localized on the opposite face of the DID from the DAD binding site. CAP/CSKA displays reduced inhibition of both mutants biochemically, with R218Q being particularly resistant (Fig. 6a,b), suggesting that INF2 disease mutants are not efficiently regulated by CAP/KAc-actin.
Figure 6. INF2 disease mutants display decreased regulation by CAP/actin.
(a) Pyrene-actin polymerization assay (2μM actin monomer, 5% pyrene) containing 20nM GFP-INF2 wildtype (WT) or mutants (FSGS mutant R218Q, CMTD mutant L77R) in presence or absence of 1μM CAP2/CSKA. Experiment conducted two times in triplicate. (b) Concentration dependence of CAP2/293A-T inhibition of INF2-FL and disease mutants, in pyrene-actin assays conducted as in panel a. Experiment conducted once (1000 nM CAP points conducted twice) in triplicate. (c) Representative GFP and TRITC-phalloidin confocal microscopy images showing cytosolic actin polymerization by exogenously expressed GFP-INF2 constructs (WT, L77R, or R218Q) in INF2-KO U2OS cells. Cells were fixed and stained with TRITC-phalloidin to label actin filaments. Images are maximum intensity projections of three confocal imaging planes in the middle z region of the cell. Bar, 10 μm. (d) Normalized fluorescence intensity of TRITC-phalloidin was quantified from images similar to (c). Lines represent mean (WT: n=31 cells, mean=1.19, STD=1.02; L77R: n=34 cells, mean=5.60, STD=3.40; R218Q: n=37 cells, mean=8.63, STD=5.47). p values determined by one-sided student’s t-test. (e) Time course of ionomycin-induced changes in actin filaments in INF2-KO U2OS cells expressing GFP-fusions of INF2 WT or the indicated mutants along with mApple-Ftractin. Mean and S.E.M. shown. N = 34, 35, and 33 cells for WT, L77R and R218Q, respectively. (f) Co-immunoprecipitation assay of endogenous CAP2 with transfected GFP-fusion proteins: GFP alone, GFP-INF2-FL, GFP-INF2FL L77R mutant, and GFP-INF2FL R218Q mutant. Top panel: anti-CAP2 western on precipitated samples (IgGH, IgG heavy chain from the IP). Middle panel: anti-GFP western showing GFP-INF2 in precipitated samples. Bottom panel: anti-CAP2 western of input samples. Experiment conducted once. (g) Schematic model of INF2 facilitated autoinhibition by CAP/KAc-actin, and activation by HDAC6-mediated actin de-acetylation. INF2 mutants linked with FSGS and CMTD are defective in CAP/KAc-actin binding, leading to constitutive activity.
We also tested regulation of L77R- and R218Q-INF2 in cells, by transfecting mutant nonCAAX constructs into INF2-KO U2OS cells. Fixed-cell analysis shows that both mutants display significantly higher cytosolic actin filaments than WT-INF2 (Fig. 6c,d). Live-cell analysis shows that cytoplasmic actin filament levels are significantly higher for both mutants than for WT-INF2, and that ionomycin stimulation does not induce increased actin filaments for the mutants (Fig. 6e). We also tested the interaction between INF2 mutants and CAP2 in cells, and found no apparent co-immunoprecipitation, in contrast to INF2-WT (Fig. 6f). Finally, we tested the effect of TubA treatment on cytoplasmic actin filament levels in L77R-INF2 or R218Q-INF2 expressing cells, and found no decrease in these levels, suggesting that increased lysine acetylation of actin does not inhibit these mutants efficiently (Fig. S5j,k). These results show that two INF2 disease mutants are poorly regulated in cells.
DISCUSSION
We reveal a cellular role for lysine-acetylated actin (KAc-actin): inhibition of INF2 while complexed with CAP. We refer to the mechanism as ‘facilitated autoinhibition’ because an additional molecule is required to mediate INF2’s autoinhibitory interaction. Actin de-acetylation through HDAC6 relieves INF2 inhibition. Cellular HDAC6 inhibition blocks stimulus-induced INF2 activation. Conversely, INF2 mutants linked to two human diseases, FSGS and CMTD, are poorly inhibited by CAP/KAc-actin and are constitutively active in cells.
A variety of actin post-translational modifications have previously been identified34. Actin acetylation research has mainly focused on the N-terminus, where all actins are modified35. Lysine acetylation on actin has been identified34 but its significance has been unclear. Our demonstration that KAc-actin inhibits INF2 adds to known acetylation events for other cytoskeletal proteins. Two actin-binding proteins have been shown to be acetylated: cortactin36, and the formin mDia237. In both cases, acetylation inhibits interaction with actin. Interestingly, formin proteins can influence tubulin acetylation through a variety of mechanisms15, 38, 39.
The presence of CAP in the INF2 inhibitory complex is intriguing. CAP was originally identified in budding yeast as an interacting protein for adenylyl cyclase40, 41, but this interaction is not conserved in mammals. CAP binds both actin monomers and filaments42–45. Purified CAP stimulates actin filament disassembly in the presence of cofilin or twinfilin, but CAP alone has little effect on actin disassembly27, 46. The two mammalian CAPs, CAP1 and CAP2, are ~60% identical and are differentially expressed47, 48. We show that both CAP1 and CAP2 inhibit INF2, and that CAP/actin directly interacts with INF2.
CAP/actin is a high molecular weight complex, consistent with previous reports of CAP hexamerization26, 27. Interestingly, additional INF2-inhibitory fractions from mouse brain also contain CAP1 or CAP2. Gel filtration profiles of these fractions suggest that cellular CAP is heterogeneous, and might include some non-hexameric species. Alternately, CAP might associate with additional proteins. CAP can interact with several actin-binding proteins, including cofilin, profilin, twinfilin and ABP127, 46, 49. Our purified BIF fraction contains several co-eluting proteins, including subunits of vacuolar ATPase and exportin. These proteins might provide additional properties.
Auto-inhibition through DID/DAD interaction is a major regulatory mechanism for most metazoan formins3–5. In addition, some formins not containing canonical DID or DAD are still subject to direct auto-inhibition through N-terminal/C-terminal interaction50. Other formins are regulated by protein binding in trans51–57. Our results demonstrate facilitation of formin auto-inhibition by another protein. The fact that a mutation known to disrupt DID/DAD interaction results in constitutive INF2 activity (Fig. S1) suggests that DID/DAD interaction is required. However, we cannot rule out the possibility that CAP/actin serves as a bridge between DID and DAD, with DAD binding KAc-actin directly11, 24. The fact that CAP can associate with INF2 suggests that CAP is part of the inhibitory complex and not merely delivering KAc-actin. Our schematic model of an inhibitory complex between CAP/actin and INF2 (Fig. 6g) does not distinguish between direct DID/DAD interaction or the CAP/actin bridge model, nor does it depict CAP hexamerization, two issues to be addressed in future work.
The acetylation positions identified on actin provide insight into the inhibitory mechanism of the CAP/KAc-actin complex. Four lysines display acetylation increases in the inhibitory CAP/actin complex: K50, K68, K215 and K315. These residues are all on the surface of the actin monomer (Fig. S6a), although K315 likely contributes to interactions between sub-domains. These residues do not make significant inter-subunit contacts in the filament (Fig. S6b), so acetylation may not disrupt polymerization. One acetylation site, K68, might alter CAP binding affinity, since it is at the binding interface in the dimeric structure of CAP’s CARP domain with actin (Fig. S6c)58. Current models of FH2/actin structure suggest little interaction of these residues with the FH2 (Fig. S6d).
Several questions arise regarding cellular INF2 regulation through this mechanism. First, what percentage of actin must be acetylated? We have previously determined total cytosolic actin to be ~200 μM in U2OS cells59, consistent with values in other cell types28, whereas INF2 levels are likely to be < 1 μM. Therefore, acetylation of <1% of cytosolic actin could suffice for INF2 inhibition.
A second question concerns the cellular signal triggering actin deacetylation and INF2 activation. It is clear that increased cytoplasmic calcium activates INF218, 31–33, but the link to decreased actin acetylation is unclear. One possibility is through calcium-mediated metabolic changes60. Lysine acetylation is governed by the balance between lysine acetyltransferase (KAT) and deacetylase activities, and the appreciable cellular acetylation/deacetylation flux30 could be due changes in either process. Several cytosolic members of the ~20 mammalian KAT family could mediate actin acetylation30. An intriguing possibility is that fluctuating levels of the acetyl donor, acetyl-CoA, could regulate KAT activity61, perhaps relating to INF2’s role in mitochondrial division18, 19 and the relationship between mitochondrial division and metabolism62. Alternately, increased calcium might increase deacetylase activity, with HDAC6 being a prime candidate. Interestingly, HDAC6 activity has been shown to cause an increase cellular actin polymerization36, 63. Alternately, calmodulin might participate in calcium-mediated INF2 activation. An interaction between INF2 and calcium-bound calmodulin has been shown in cell lysates33.
Our results have exciting implications for disease mechanisms of both FSGS and CMTD. All disease-linked INF2 mutations map to DID, but few are at the DAD-binding site, suggesting that they do not affect this interaction directly. In addition, all disease mutations are dominant, and expression of select mutants causes aberrant cellular actin polymerization22, suggesting that both diseases are due to increased INF2 activity. We show that two disease mutants are poorly inhibited by CAP/KAc-actin, suggesting that this interaction is affected. Interestingly, recent studies show that aberrant HDAC6 activity can lead to CMTD64–66. Overall, our study provides a framework for understanding both actin regulation and disease progression in a different way.
METHODS:
Plasmids
Human INF2 full-length nonCAAX cDNA was made from human INF2-CAAX clone (Origene SC313010, NM_022489) followed by replacement of the CAAX exon with the nonCAAX exon, correction of a 12-base deletion in the purchased clone, and codon optimization. This construct was subsequently cloned into the EcoR1/Xho1 sites of a modified eGFP-C1 vector (Clonetech Inc) containing Strep-tag II (IBA Life Sciences) and an HRV3C cleavage site N-terminal to the INF2 start codon. INF2 mutations (A149D, L77R, R218Q) were generated in the above-mentioned INF2 full-length nonCAAX construct by site-directed mutagenesis. Human CAP1 and CAP2 cDNAs were purchased from NovoPro (710829–5 (NM-006367) and 710470–11 (NM-006366)), then the entire ORF was PCR-amplified and cloned into a modified eGFP-N1 vector (Clonetech) containing an HRV3C cleavage site followed by the GFP ORF, followed by Strep-tag II, using Xho1 and Kpn1 sites. FLAG tagged human HDAC6 plasmid was purchased from Addgene (30482). Mito-R-GECO1 (Kd= 0.48 μM for calcium) construct, as well as GFP-F-tractin and mApple-F-tractin constructs have been described previously18.
Recombinant protein expression and purification
For protein expression in mamalian cells, 2.24 × 106 FreeStyle 293-F cells (Life Tech R790–07) growing in 1L FreeStyle 293 expression medium (Life Tech 12338–018) were transfected with 0.5mg DNA and 1.5mg sterile 25kDa linear PEI mixed in Opti-MEM reduced-serum medium (Life Tech 31985070). Proteins were expressed at 37 °C, 8% CO2 with shaking at 125 rpm for 2 days (INF2) or 3 days (CAP1, CAP2 and HDAC6). For the Tubstatin A treatment of CAP2-transfected cells, cells were centrifuged at 300xg at 4°C after 3 days of transfection, and resuspended in Dulbecco modified Eagle media (DMEM) without serum. DMSO or 50 μM Tubastatin A (Selleckchem S2627) was added and cells were cultured for an additional 1hr before harvest.
For strep-tagged protein purification from FreeStyle 293 cells, all following steps were performed at 4°C or on ice. Cells were pelleted at 300xg for 15min, and pellet was resuspended in 45mL EB-S (100mM Hepes pH7.4, 500mM NaCl2, 5mM EDTA, 1mM DTT, 1% v/v TritonX-100, 2μg/mL leupeptin, 10μg/mL aprotinin, 2μg/mL pepstatin A, 1μg/mL calpeptin, 1μg/mL calpain inhibitor 1, 1mM bezamidine, 1:1000 dilution of universal nuclease (Thermo Fisher Inc 88702) per 5mL cell pellet followed by 30min end-over-end mixing at 4°C. For tubastatin A-treated cells, EB-S also contained 500 nM tricostatin A (Selleck S1045) and 10 mM sodium butyrate (Sigma-Aldrich 303410). The cell debris was removed by ultracentrifugation at 185,000xg for 1 hr (Ti45 rotor, Beckman), and the supernatant was blocked with avidin (20μg/mL Sigma-Aldrich 189725) then applied to Strep-Tactin Superflow resin (2-1206-025; IBA, Göttingen, Germany) equilibrated in EB-S. After thorough wash with WB (10mM Hepes pH7.4, 150mM NaCl2, 1mM EDTA, 1mM DTT), column was either: 1) treated with HRV3C protease in WB (at 1:50 enzyme to substrate molar ratio) for 16hrs at 4°C followed by 3 column volume WB wash to obtain untagged protein; or 2) eluted with strep elution buffer (10mM Hepes pH7.4, 150mM NaCl, 1mM EDTA, 1mM DTT, 2.5mM desthiobiotin) to obtain strep-GFP fusion protein. Protein was concentrated with 30,000 MWCO Amicon Ultra-15 filter unit (Millipore) before further purification through Superdex 200 16/60 gel-filtration column (GE Bioscience) equilibrated with10mM Hepes pH7.4, 50mM KCl, 1mM MgCl2, 1mM EGTA, 1mM DTT. Protein was further concentrated to >10 μM before being aliquoted, frozen in liquid nitrogen, and stored at −80°C. N.B. – in all cases, Hepes pH given is at 23°C.
For FLAG-tagged protein purification from Freestyle 293 cells, cells were harvested the same way as described above, and lysed in EB-F (50 mM Hepes 7.4, 150 mM NaCl, 1 mM EDTA, 1 mM DTT, 1% v/v TritonX-100, 2μg/mL leupeptin, 10μg/mL aprotinin, 2μg/mL pepstatin A, 1μg/mL calpeptin, 1μg/mL calpain inhibitor 1, 1mM bezamidine, 1:1000 dilution of universal nuclease). Cell lysate was clarified by ultracentrifugation at 185,000xg for 1 hr (Ti45), and supernatant loaded onto Anti-FLAG M2 affinity gel equilibrated with EB-F. Column washed with WB-F (10 mM Hepes 7.4, 50 mM KCl, 1mM MgCl2, 1 mM EGTA, 1 mM DTT) thoroughly, and FLAG-tagged protein was eluted with FLAG elution buffer (10 mM Hepes 7.4, 50 mM KCl, 1mM MgCl2, 1 mM EGTA, 1 mM DTT, 100μg/mL FLAG octapeptide (Sigma F3290)). Protein was gel filtered on Superdex 200, concentrated and stored as described above.
Actin preparation
Details described in16. Rabbit skeletal muscle actin (fresh muscle from Fox and Crow Farm, Barnstead NH) or chicken skeletal muscle actin (frozen chicken breast from Trader Joe’s) was purified from acetone powder. Rabbit muscle actin was labeled with pyrenyliodoacetamide, TAMRA NHS ester (Molecular probes; C1171) or Oregon488 NHS ester (Thermo Fisher; O6185). Both labeled and unlabeled actin were gel-filtered on Superdex 75 (GE Biosciences) and stored in G-buffer (2 mM Tris-HCl pH 8, 0.5 mM DTT, 0.2 mM ATP, 0.1 mM CaCl2, 0.01% w/v sodium azide) at 4°C.
For brain actin purification, all steps were at 4°C. Mouse brains (50g) homogenized using polytron (Kinematica PT3100) in EB (50mM Hepes pH7.4, 1mM MgCl2, 1mM EGTA, 2mM DTT, 2μg/mL leupeptin, 10μg/mL aprotinin, 2μg/mL pepstatin A, 1μg/mL calpeptin, 1μg/mL calpain inhibitor 1, 1mM bezamidine). Cell debris removed by ultracentrifugation at 185,000xg (Ti45), and supernatant filtered through 0.45μm filters. Supernatant was supplemented with ATP (4mM), MgCl2 (4mM), and sodium phosphate (NaPO4, pH 7.0, 20 mM), followed by end-over-end mixing at 4°C for 16hrs. Polymerized brain actin was pelleted by centrifugation for 3hrs at 185,000xg, and the pellet was washed by resuspension-pelleting cycle sequentially with 2xWB1 (500mM KCl, 0.5% v/v thesit, 10mM NaPO4 pH7.0, 2mM MgCl2, 2mM ATP, 1mM DTT, 2μg/mL leupeptin), and 2xWB2 (50mM KCl, 10mM NaPO4 pH7.0, 2mM MgCl2, 2mM ATP, 1mM DTT, 2μg/mL leupeptin). Pellet was then washed with G-buffer with minimal disturbance and then resuspended in G-buffer before shearing through 30G 1/2 needle. Sheared actin was dialyzed in G-buffer for 16hrs before ultracentrifugation 185,000xg (Ti70.1 rotor, Beckman), then concentration to >10μM.
BIF purification
Conducted at 4°C. 100g mouse brains (Pel-Freez Biologicals, Rogers Arkansas). were homogenized using polytron in EB. Cell debris was removed by ultracentrifugation at 185,000xg (Ti45 rotor), and supernatant filtered through 0.45μm filters then supplemented with KCl to 100mM before loading onto 90mL Q-sepharose column (GE Biosciences). BIF containing fraction was eluted with Q350 (350mM KCl, 50mM Hepes pH7.4, 1mM MgCl2, 5mM EGTA, 1mM DTT, 2μg/mL leupeptin, 10μg/mL aprototin, 2μg/mL pepstatin A, 1μg/mL calpeptin, 1μg/mL calpain inhibitor 1). Eluate was diluted 3-fold with EB before loading onto 50mL Source Q column (GE Biosciences). Linear ionic strength gradient elution was performed with Q100 (100mM KCl, 10mM Hepes pH7.4, 1mM MgCl2, 1mM EGTA, 1mM DTT) and Q350 (Q100 + 250 mM KCl). Fractions were dialyzed into 1.5xKMEH+DTT (15mM Hepes pH7.4, 75mM KCl, 1.5mM MgCl2, 1.5mM EGTA, 1mM DTT) and tested for inhibition of INF2 full-length activity by pyrene actin polymerization assay (20 nM INF2, 2 μM actin (5% pyrene)). Inhibitory peak fractions were pooled and passed over Superdex 200 16/60 gel filtration column equilibrated in 1.5xKMEH+DTT. Inhibitory peak fractions were pooled and purified through 8mL Source Q column (GE Biosciences) at pH8.8 with gradient from 100–300 mM KCl (in 10mM Tris pH8.8, 1mM MgCl2, 1mM EGTA, 1mM DTT). Fractions dialyzed into 1.5xKMEH+DTT and assayed. Peak inhibitory fractions were pooled and dialyzed into Q0 (10mM Hepes pH6.8 at 4°C, 1mM MgCl2, 1mM EGTA, 1mM DTT) before applied to 1mL SourceS column (GE Biosciences) with gradient elution from from 0–100 mM KCl (in 10mM Hepes pH6.8 at 4C, 1mM MgCl2, 1mM EGTA, 1mM DTT). Fractions were dialyzed into 1.5xKMEH+DTT, assayed for inhibitory activity, analyzed by silver-stained SDS-PAGE and pooled.
For CAP concentration determination in BIF, samples from Source S column fractions (Fig. 1i) were resolved along with actin standards with known mass by 7.5% SDS-PAGE. Gel was stained with Colloidal Blue stained SDS-PAGE (Thermo Fisher), and CAP concentration was analyzed by densitometry (ImageJ), using actin as a standard. For determination of IC50 for CAP in BIF, linear fit was applied to activity of BIF in Fig. 2F, and activity of actin was used as baseline.
TIRF Microscopy
Detailed methods described elsewhere17. Briefly, glass flow chambers assembled using VWR micro cover glasses (22×22 and 18×18 mm, No 1.5) with double-stick Scotch tape to hold 10μL volume. Before assembly, cover glasses washed in acetone (50 min), ethanol (10 min), and MilliQ water (1 min), then incubated in 1:2 ratio of 30% H2O2:H2SO4 for 1 h. Glasses then rinsed with water, followed by 0.1M KOH, followed by water again, and dried with inert gas before silanization overnight in 0.0025% dichlorodimethyl silane (Sigma-Aldrich 85126) in chloroform, washed with methanol, dried with inert gas, and stored in clean sealed containers. Chambers were incubated with 1% w/v Pluronic F127 (Sigma-Aldrich P2443) in BRB80 (80mM PIPES/KOH, pH 6.9, 1 mM EGTA, 1 mM MgCl2) for 1 min and then equilibrated in TIRF buffer (10mM imidazole pH7, 50mM KCl, 1mM MgCl2, 1mM EGTA, 1mM DTT, 100 mM DTT, 0.2 mM ATP, 15 mM glucose, 0.5% methylcellulose, 0.01mg/ml catalase [Sigma C3515], 0.05mg/ml glucose oxidase [Sigma G6125], and 0.1% BSA). Nucleation-elongation assays were conducted as previously described16. Briefly, unlabeled rabbit skeletal muscle actin monomers were mixed with 20% TAMRA or Oregon-labeled actin monomers (2μM) in G buffer, diluted with profilin (3μM) in 2xTIRF buffer in the presence or absence of 1 nM untagged INF2-FL and/or 100nM CAP/CKSA, and introduced into the flow chamber. The filaments were visualized on a Nikon Eclipse Ti-E inverted microscope with 488- or 561-nm lasers and driven by Nikon Elements software. Single-color images were acquired every 5 s at 100 ms exposure with TIRF objective (60X1.49 N.A.) and Andor electron-multiplying charge-coupled device camera with an Andor TuCam adapter using Perfect Focus. Filament number and length measured manually from frame to frame using Nikon NIS-Elements 4.40.00. For display of TIRF images, original images were adjusted using Photoshop CS5 (Adobe Inc). Curve function was used to lock the intensity of the filaments and then reduce the background haze. All panels were adjusted as a single image.
Pyrene actin polymerization assay
Rabbit skeletal muscle actin in G-buffer (6 μM actin, 5% pyrene) was converted to Mg2+ salt by addition of 1 mM EGTA and 0.1 mM MgCl2 (from 10x stock) for 2 min at 23°C immediately prior to polymerization. Polymerization was induced by addition of 2 volumes of formin with or without other proteins (INF2 and/or inhibitory fraction) in 1.5xpolymerization buffer (75mM KCl, 1.5mM MgCl2, 1.5mM EGTA, 15 mM Hepes pH 7.4, 2mM DTT, 2 mM Tris-HCl, 0.2 mM ATP, 0.1 mMCaCl2, and 0.01% w/v NaN3). Pyrene fluorescence (365/410 nm) was monitored in a 96-well fluorescence plate reader (Infinite M1000; Tecan, Mannedorf, Switzerland) within 1min of mixing actin and formin or other proteins/fraction. Slope of curve at 1/2 max is plotted to represent INF2 activity.
Velocity Analytical Ultracentrifugation
Analytical ultracentrifugation was conducted using a Beckman Proteomelab XL-A and an AN-60 rotor. For sedimentation velocity analytical ultracentrifugation, untagged INF2 full-length nonCAAX (3.8μM), CAP2-GFP (7μM), or GFP-INF2 (1.8μM) in K50MEHD (10mM Hepes pH7.4, 50mM KCl, 1mM MgCl2, 1mM EGTA, 1mM DTT) was centrifuged at 25,000 rpm with monitoring at 490 nm. Data analyzed by Sedfit to determine sedimentation coefficient, frictional ratio, and apparent mass. Sedimentation coefficient reported is that of the major peak (at least 80% of the total analyzed mass) at OD280 for untagged INF2 or CAP2 or OD490 for GFP-INF2 or CAP2-GFP.
Antibodies
Vacuolar ATPase subunit A (rabbit anti-ATP6V1A, Abcam, ab199325), Vacuolar ATPase subunit B2 (rabbit anti-ATP6V1B2, Abcam, ab73404), heat shock cogate 71kDa protein (rat anti-Hsc70, Abcam, ab51052), CAP2 (goat polyclonal, Santa Cruz, sc-167378), CAP1 (rabbit polyclonal, Abcam ab96354), GFP (rabbit polyclonal, gift from William Wickner (Dartmouth)), actin (mouse monoclonal, clone C4, Millipore, MAB1501) alpha-tubulin (mouse monoclonal, clone DM1A, Sigma-Aldrich, T9026), acetyl-lysine (rabbit polyclonal, Cell Signaling Technologies, 9441), anti-acetylated tubulin (mouse monoclonal, clone 6–11B-1, Sigma-Aldrich, T7451). Anti-INF2 was described previously (27; 941–1249 antibody used, rabbit polyclonal). Details on antibodies are provided in Supplementary Table 5.
Immuno-depletion
The initial SourceQ inhibitory fraction pool from mouse brain prep (fractions 38–40, Fig. S2A) was incubated with antibody against Vacuolar ATPase subunit A, Vacuolar ATPase subunit B2, heat shock cogate 71kDa protein, CAP2, or control IgG (rabbit anti-GFP), respectively at 4°C. 10μL of each antibody was added into 500μL fraction followed by 16hrs incubation with end-over-end rmixing at 4°C. The antigen-antibody complex was precipitated by protein A or protein G agarose beads (GE Biosciences). Supernatant recovered and tested for INF2 inhibition. Supernatants and pellets probed for the depleted protein.
CAP bound actin exchange assay
CAP2-GFP-2xstrep protein purified from 293 cells (also containing bound actin) was immobilized on strep-tactin beads, then washed with G-buffer. The indicated species of actin (in G-buffer) was incubated with CAP2-GFP strep-tactin beads at 4°C with end-over-end mixing for 12hrs. Concentrations of added actin and bead-bound CAP2-GFP in exchange reactions were 5μM and 1μM, respectively. Beads were washed with 20 volumes G-buffer. Exchanged CAP2-GFP removed from beads by either: eluting with G-buffer containing 2.5mM dethiobiotin, or 2) cleavage from beads by HRV3C protease (1:50, overnight at 4°C).
Protein binding assays
For strep-tactin pulldown of purified proteins, 1μM strep-GFP tagged protein (GFP, INF2FL or INF2FFC) was incubated with 2μM untagged CAP (with or w/o actin exchange) in IPB (50mM KCl, 1mM MgCl2, 1mM EGTA, 10 mM Hepes pH 7.4, 1mM DTT, 1% v/v thesit) with end-over-end mixing at 4°C for 8hrs. Separately, 0.2 volumes of 50% Strep-Tactin Superflow bead slurry was blocked with 10mg/mL avidin and washed in PD before adding to protein mix. After 8hr incubation at 4°C, beads were washed with IPB (now containing 0.2% v/v thesit). The bead pellet was probed by western blot with anti-GFP, anti-CAP2, or anti-actin antibodies.
For strep-tactin pulldown from cell extracts, Wild-type U2OS cells expressing CAP2-GFP were treated with: 50μM Tubstatin A for 1hr, DMSO for 15min, or 4μM ionomycin (in serum-containing medium) for the indicated length of time prior to lysis in prechilled PDEB (50mM KCl, 1mM MgCl2, 5mM EGTA, 10 mM Hepes pH 7.4, 1mM DTT, 3% v/v thesit, 2μg/mL leupeptin, 10μg/mL aprototin, 2μg/mL pepstatin A, 1μg/mL calpeptin, 1μg/mL calpain inhibitor 1, 1mM bezamidine, 500nM Tricostatin A, 10mM sodium butyrate) in the dish (immediately after removal of medium) and then incubated with shaking at 4°C for 15min. Lysate centrifuged at 100K rpm in TLA120 rotor (Beckman) at 4°C for 20min, and supernatant was blocked with avidin (20μg/mL Sigma-Aldrich 189725) then mixed with Strep-Tactin Superflow resin (prewashed with PDEB) with end-over-end mixing for 3hrs at 4C. Beads were washed with PDEB twice and 1xPBS twice. The pellet was resolved by SDS-PAGE, and western blot probed with anti-actin, anti-acetyl-lysine or anti-INF2 antibodies.
For immunoprecipitation of purified proteins, 0.5μM strep-GFP tagged protein (GFP, INF2-FL or INF2-FFC) was incubated with 2μM untagged CAP (with or w/o exchange) in IPB for 3hrs at 4°C. 50μg anti-GFP antibody was added and incubated 12hrs with end-over-end mixing at 4°C. Protein A beads were blocked with 10mg/mL avidin and washed in IPB before incubating with protein mix for 3hrs. Beads were washed with IPB (now containing 0.2% v/v thesit). The bead pellet was resolved by SDS-PAGE, and the western blot was probed with anti-GFP, anti-CAP2, or anti-actin antibodies. For statistical analysis, results from three independent experiments were quantified. CAP2 band intensity normalized against GFP control using ImageJ.
For immuno-precipitation from cell extracts, INF2KO U2OS cells expressing GFP, GFP-INF2 full-length nonCAAX WT, GFP-INF2FFC or GFP-INF2 full-length nonCAAX A149D, GFP-INF2 full-length nonCAAX L77R, GFP-INF2 full-length nonCAAX R218Q mutants were trypsinized, centrifuged at 300xg for 5 min, and the cell pellet washed with PBS. Cell pellet was lysed with prechilled IPEB (50mM KCl, 1mM MgCl2, 5mM EGTA, 10 mM Hepes pH 7.4, 1mM DTT, 3% v/v thesit, 2μg/mL leupeptin, 10μg/mL aprototin, 2μg/mL pepstatin A, 1μg/mL calpeptin, 1μg/mL calpain inhibitor 1, 1mM bezamidine), then treated with latrunculin A (Sigma-Aldrich, L5163) at 20μM for 15min on ice. Cell debris removed by ultracentrifugation 100,000 rpm TLA-100 rotor). Supernatant incubated with 10μg anti-GFP with end-over-end mixing at 4°C for 12hrs. Protein A beads (20μL) pre-blocked with 10mg/mL avidin and washed in IPEB before incubating with lysate for 3hr. Beads washed with IPW (50mM KCl, 1mM MgCl2, 1mM EGTA, 10 mM Hepes pH 7.4, 1mM DTT, 0.2% v/v thesit), bound proteins resolved by SDS-PAGE western, probed with anti-GFP and anti-CAP2.
Fluorescence polarization anisotropy measurements conducted using INF2-Cterm (amino acids 941–1249 of INF2-CAAX, expressed in bacteria) labeled with tetramethylrhodamine-succinimide (TMR), as previously described24. TMR-INF2-Cterm (100 nM) mixed with CAP/293A-T and/or INF2-Nterm (amino acids 1–420, expressed in bacteria as described24) in 50mM KCl, 1mM MgCl2, 5mM EGTA, 10 mM Hepes pH 7.4, 1mM DTT at 23°C, and fluorescence anisotropy measured in the Tecan M1000 plate reader. Concentration of CAP/293A-T was held constant at 20 μM in some assays. Binding curves fit using standard hyperbolic saturation fitting67. In the case of the INF2-Cterm/INF2-Nterm interaction in the absence of CAP/293A-T, saturation was not obtained and had to be assumed based on extrapolation of the curve, which adds uncertainty to the Kdapp value of 28 μM. For this reason, we state that CAP/293A-T increases the affinity of INF2-Nterm for INF2-Cterm at least 5-fold, which is a conservative estimate of this increase.
Mass spectrometry
Samples were separated by SDS-PAGE, bands excised, destained, and digested with trypsin in 50 mM Ammonium bicarbonate overnight at 37°C. Peptides were extracted using 5% formic acid/50% ACN and dried. Peptides analyzed on a Fusion Orbitrap mass spectrometer (ThermoScientific) equipped with an Easy-nLC 1000 (ThermoScientific). Raw data were searched using COMET in high resolution mode68 trypsin enzyme specificity with up to three missed cleavages, and carbamidomethylcysteine as fixed modification. Oxidized methionine, and acetylated lysine were searched as variable modifications. Quantification of LC-MS/MS spectra was performed using MassChroQ69. Peptides were corrected based on protein amount, ratios CAP TubA / DMSO were calculated on a per charge state and per peptide basis and averaged. Additional analyses were performed at the Taplin Mass Spectrometry Facility at Harvard.
Live cell imaging
Wild-type human osteosarcoma U2OS cells (American Type Culture Collection HTB96) or INF2 KO U2OS cells (described in18) were grown in DMEM (Invitrogen) supplemented with 10% calf serum (Atlanta Biologicals). Cells tested every 6 months for mycoplasma contamination using LookOut PCR detection kit (Sigma-Aldrich). Cells seeded at 4×105 cells/well in 6-well dish ~16 hours prior to transfection. Plasmid transfections performed in OPTI-MEM media (Invitrogen) with 2 μL Lipofectamine 2000 (Invitrogen) and 500 ng GFP-Ftractin and/or mito-R-GECO1, or GFP-INF2 full-length nonCAAX, GFP-INF2 full-length nonCAAX L77R mutant or GFP-INF2 full-length nonCAAX R218Q mutant with mApple-Ftractin per well for 6 hours, followed by trypsinization and re-plating onto glass bottom MatTek dishes (P35G-1.5–14-C) at ~2×105 cells per well. Cells imaged in live in DMEM (GIBCO, #21063–029) supplemented with 10% NCS (HyClone, #SH30118.03) ~16–24 hours after transfection. For histamine or ionomycin treatments, 100 μM histamine (Sigma Aldrich H7125, from 100 mM stock in DMSO), 4 μM ionomycin (Sigma Aldrich I0634, from 2 mM stock in DMSO), or DMSO alone was added at 1 min after commencement of imaging, and imaging continued for 5–10 min. Medium was pre-equilibrated for tempurature and CO2 content before use. For tubastatin A treatments, cells were pretreated with either 50 μM tubastatin A (from 10 mM stock in DMSO) or equal volume DMSO (control) for 60 min prior to addition of histamine or ionomycin. Note: Tubastatin A-containing media was made immediately prior to addition to the cells, individually for each plate. Imaging was conducted on a Dragonfly 302 spinning disk confocal (Andor Technology Inc, Belfast UK) on a Nikon Ti-E base and equipped with an iXon Ultra 888 EMCCD camera, and a Tokai Hit stage-top incubator. Lasers: solid state 405 smart diode 100 mW, solid state 488 OPSL smart laser 50 mW, solid state 560 OPSL smart laser 50 mW, solid state 637 OPSL smart laser 140 mW. Objective: 100× 1.4 NA CFI Plan Apo (Nikon). Images acquired using Fusion software (Andor). Imaging was conducted in a medial region of the cell, approximately 2 μm above the basal surface.
Fix cell imaging
Cells were transfected with GFP, GFP-INF2 full-length nonCAAX, GFP-INF2 full-length nonCAAX A149D mutant, GFP-INF2 full-length nonCAAX L77R mutant or GFP-INF2 full-length nonCAAX R218Q mutant, and split onto MatTek dishes, in the same way as for live cells. Cells were then treated with DMSO or 50 μM Tubastatin A for 1hr prior to fixation with 4% paraformaldehyde (Electron Microscopy Sciences Inc) in PBS for 20min at 23°C. After washing with PBS, the cells were permeabilized on ice with 0.25% Triton X-100 in PBS for 15min. Cells were then washed with PBS prior to blocking with 10% calf serum in PBS for 1hr at 23°C. Actin was stained with 100nM TRITC-phalloidin (Sigma-Aldrich) and DNA was stained with 0.1mg/L w/v DAPI (Calbiochem 268298) for 10min at 23°C in dark followed by PBS wash. Cells were imaged in PBS using the Andor Dragonfly microscope and Zyla 4.2 Mpixel sCMOS camera. Z stacks with 0.2μm step size were acquired. Images from middle three z planes were used for quantification. Cells selected based on GFP expression, then imaged for both GFP and phalloidin staining without first examining the degree of phalloidin staining.
Measurements of actin intensity in fixed cells, and calcium changes and actin burst in live cells
TRITC-phalloidin intensity of two ROIs in the perinuclear region were taken per cell, and was normalized by the intensity of 2 ROIs taken from nuclear region (which lack measurable phalloidin staining).
Measurements of mitochondrial calcium and actin burst in live cells were conducted as described previously18. Mean fluorescence was calculated for each cell using ImageJ (NIH). Fluorescence values for each time point after drug treatment (F) were normalized by the average initial fluorescence (first 5–6 frames prior to stimulation-F0) and plotted against time as F/F0. For calcium measurements, ll cells in the imaging field were analyzed. Where cells were closely juxtaposed, calcium changes were analyzed in the same ROI. For actin “burst” measurements, mean Fluorescence values for each time point were calculated from 4 ROI/cell selected in the peri-nuclear region using ImageJ. Only cells fully in the imaging field were analyzed. Cells were excluded from analysis for reasons: they were rounded up, or a significant concentration of basal stress fiber signal in the field contributed to abnormally high initial signal. Excluded cell percentages as follows. Fig. 5g: 15%, 16% and 19% for DMSO, +Iono and +TubA+Iono, respectively. Fig. 5h: 32% for both DMSO+histamine and TubA+histamine.
Statistical Analysis and Reproducibility
Error (standard error of the mean) was calculated using Excel (Microsoft, version 16.16.1). Comparisons by one-sided T-test (paired) were conducted using GraphPad. Most experiments were reproduced three or more times (given in figure legends) with similar results. One exception was the BIF purification procedure, that was only performed in full one time. All purification steps, however, were optimized in small-scale pilot experiments, typically two or three for each chromatographic column. The purified brain actin was only tested for INF2 inhibition one time.
Data Availability
The mass spectrometry proteomics data (Supplementary Tables 1,2, and 3) have been deposited to the ProteomeXchange Consortium70 through the PRIDE partner repository. PX accession number PXD010484. Raw numerical data for the results in Figure 4b, Figure 5g–j, and Figure 6e are provided in Supplementary Table 4. Unprocessed blots and gels for all relevant figures are in Supplementary Figure 7.
Supplementary Material
ACKNOWLEDGEMENTS
We thank A. Hatch for getting the project started, Z. Svindrich and A. Lavanway for help with imaging and image analysis, T.Y. Chang for a generous supply of neurons, W. Wickner for excellent advice on protein purification, M. Pollak for in-put on FSGS, J. McLellan and M. Ragusa for invaluable comments and support, and C. Detteyala for modifying our behavior. This work was supported by NIH R01 GM069818 and R35 GM122545 to HNH, R01 DK088826 to M. Pollak (HNH sub-contract), R35 GM119455 to AK, and P20 GM113132 to the BioMT COBRE.
Footnotes
COMPETING INTERESTS STATEMENT
The authors declare that they have no financial and non-financial competing interests.
REFERENCES
- 1.Pufall MA & Graves BJ Autoinhibitory domains: modular effectors of cellular regulation. Annu Rev Cell Dev Biol 18, 421–462 (2002). [DOI] [PubMed] [Google Scholar]
- 2.Torres E & Rosen MK Contingent phosphorylation/dephosphorylation provides a mechanism of molecular memory in WASP. Mol Cell 11, 1215–1227 (2003). [DOI] [PubMed] [Google Scholar]
- 3.Campellone KG & Welch MD A nucleator arms race: cellular control of actin assembly. Nature Reviews Molecular Cell Biology 11, 237–251 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goode BL & Eck MJ Mechanism and Function of Formins in Control of Actin Assembly. Annu Rev Biochem (2007). [DOI] [PubMed] [Google Scholar]
- 5.Higgs HN Formin proteins: a domain-based approach. Trends Biochem Sci 30, 342–353 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Higgs HN & Peterson KJ Phylogenetic analysis of the formin homology 2 domain. Molecular biology of the cell 16, 1–13 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pruyne D Revisiting the Phylogeny of the Animal Formins: Two New Subtypes, Relationships with Multiple Wing Hairs Proteins, and a Lost Human Formin. PLoS One 11, e0164067 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li F & Higgs HN The mouse formin mDia1 is a potent actin nucleation factor regulated by autoinhibition. Curr Biol 13, 1335–1340 (2003). [DOI] [PubMed] [Google Scholar]
- 9.Otomo T, Otomo C, Tomchick DR, Machius M & Rosen MK Structural basis of Rho GTPase-mediated activation of the formin mDia1. Mol Cell 18, 273–281 (2005). [DOI] [PubMed] [Google Scholar]
- 10.Lammers M, Rose R, Scrima A & Wittinghofer A The regulation of mDia1 by autoinhibition and its release by Rho*GTP. Embo J 24, 4176–4187 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chhabra ES & Higgs HN INF2 is a WH2 motif-containing formin that severs actin filaments and accelerates both polymerization and depolymerization. J. Biol. Chem 281, 26754–26767 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Gurel PS, Hatch AL & Higgs HN Connecting the cytoskeleton to the endoplasmic reticulum and Golgi. Current biology : CB 24, R660–672 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andres-Delgado L et al. INF2 promotes the formation of detyrosinated microtubules necessary for centrosome reorientation in T cells. The Journal of cell biology 198, 1025–1037 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bartolini F et al. An mDia1-INF2 formin activation cascade facilitated by IQGAP1 regulates stable microtubules in migrating cells. Mol Biol Cell 27, 1797–1808 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fernandez-Barrera J et al. The actin-MRTF-SRF transcriptional circuit controls tubulin acetylation via alpha-TAT1 gene expression. J Cell Biol 217, 929–944 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gurel PS et al. INF2-mediated severing through actin filament encirclement and disruption. Current biology : CB 24, 156–164 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andres-Delgado L, Anton OM, Madrid R, Byrne JA & Alonso MA Formin INF2 regulates MAL-mediated transport of Lck to the plasma membrane of human T lymphocytes. Blood 116, 5919–5929 (2010). [DOI] [PubMed] [Google Scholar]
- 18.Chakrabarti R et al. INF2-mediated actin polymerization at the ER stimulates mitochondrial calcium uptake, inner membrane constriction, and division. J Cell Biol 217, 251–268 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Korobova F, Ramabhadran V & Higgs HN An actin-dependent step in mitochondrial fission mediated by the ER-associated formin INF2. Science 339, 464–467 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Madrid R et al. The formin INF2 regulates basolateral-to-apical transcytosis and lumen formation in association with Cdc42 and MAL2. Developmental Cell 18, 814–827 (2010). [DOI] [PubMed] [Google Scholar]
- 21.Ramabhadran V, Korobova F, Rahme GJ & Higgs HN Splice variant-specific cellular function of the formin INF2 in maintenance of Golgi architecture. Molecular biology of the cell 22, 4822–4833 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brown EJ et al. Mutations in the formin gene INF2 cause focal segmental glomerulosclerosis. Nature genetics 42, 72–76 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boyer O et al. INF2 mutations in Charcot-Marie-Tooth disease with glomerulopathy. The New England journal of medicine 365, 2377–2388 (2011). [DOI] [PubMed] [Google Scholar]
- 24.Ramabhadran V, Hatch AL & Higgs HN Actin monomers activate inverted formin 2 by competing with its autoinhibitory interaction. The Journal of biological chemistry 288, 26847–26855 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun H, Schlondorff JS, Brown EJ, Higgs HN & Pollak MR Rho activation of mDia formins is modulated by an interaction with inverted formin 2 (INF2). Proceedings of the National Academy of Sciences of the United States of America 108, 2933–2938 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chaudhry F et al. Srv2/cyclase-associated protein forms hexameric shurikens that directly catalyze actin filament severing by cofilin. Mol Biol Cell 24, 31–41 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jansen S, Collins A, Golden L, Sokolova O & Goode BL Structure and mechanism of mouse cyclase-associated protein (CAP1) in regulating actin dynamics. J Biol Chem 289, 30732–30742 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pollard TD, Blanchoin L & Mullins RD Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu Rev Biophys Biomol Struct 29, 545–576 (2000). [DOI] [PubMed] [Google Scholar]
- 29.Makkonen M, Bertling E, Chebotareva NA, Baum J & Lappalainen P Mammalian and malaria parasite cyclase-associated proteins catalyze nucleotide exchange on G-actin through a conserved mechanism. J Biol Chem 288, 984–994 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Drazic A, Myklebust LM, Ree R & Arnesen T The world of protein acetylation. Biochim Biophys Acta 1864, 1372–1401 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Ji WK, Hatch AL, Merrill RA, Strack S & Higgs HN Actin filaments target the oligomeric maturation of the dynamin GTPase Drp1 to mitochondrial fission sites. eLife 4, e11553 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shao X, Li Q, Mogilner A, Bershadsky AD & Shivashankar GV Mechanical stimulation induces formin-dependent assembly of a perinuclear actin rim. Proc Natl Acad Sci U S A 112, E2595–2601 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wales P et al. Calcium-mediated actin reset (CaAR) mediates acute cell adaptations. Elife 5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Terman JR & Kashina A Post-translational modification and regulation of actin. Curr Opin Cell Biol 25, 30–38 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arnesen T, Marmorstein R & Dominguez R Actin’s N-terminal acetyltransferase uncovered. Cytoskeleton (Hoboken) (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang X et al. HDAC6 modulates cell motility by altering the acetylation level of cortactin. Mol Cell 27, 197–213 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li X et al. Histone deacetylase 6 regulates cytokinesis and erythrocyte enucleation through deacetylation of formin protein mDia2. Haematologica 102, 984–994 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Destaing O et al. A novel Rho-mDia2-HDAC6 pathway controls podosome patterning through microtubule acetylation in osteoclasts. J Cell Sci 118, 2901–2911 (2005). [DOI] [PubMed] [Google Scholar]
- 39.Thurston SF, Kulacz WA, Shaikh S, Lee JM & Copeland JW The ability to induce microtubule acetylation is a general feature of formin proteins. PLoS One 7, e48041 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Colicelli J et al. Mutational mapping of RAS-responsive domains of the Saccharomyces cerevisiae adenylyl cyclase. Mol Cell Biol 10, 2539–2543 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fedor-Chaiken M, Deschenes RJ & Broach JR SRV2, a gene required for RAS activation of adenylate cyclase in yeast. Cell 61, 329–340 (1990). [DOI] [PubMed] [Google Scholar]
- 42.Balcer HI et al. Coordinated regulation of actin filament turnover by a high-molecular-weight Srv2/CAP complex, cofilin, profilin, and Aip1. Curr Biol 13, 2159–2169 (2003). [DOI] [PubMed] [Google Scholar]
- 43.Gottwald U, Brokamp R, Karakesisoglou I, Schleicher M & Noegel AA Identification of a cyclase-associated protein (CAP) homologue in Dictyostelium discoideum and characterization of its interaction with actin. Mol Biol Cell 7, 261–272 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moriyama K & Yahara I Human CAP1 is a key factor in the recycling of cofilin and actin for rapid actin turnover. J Cell Sci 115, 1591–1601 (2002). [DOI] [PubMed] [Google Scholar]
- 45.Normoyle KP & Brieher WM Cyclase-associated protein (CAP) acts directly on F-actin to accelerate cofilin-mediated actin severing across the range of physiological pH. J Biol Chem 287, 35722–35732 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Johnston AB, Collins A & Goode BL High-speed depolymerization at actin filament ends jointly catalysed by Twinfilin and Srv2/CAP. Nat Cell Biol 17, 1504–1511 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bertling E et al. Cyclase-associated protein 1 (CAP1) promotes cofilin-induced actin dynamics in mammalian nonmuscle cells. Mol Biol Cell 15, 2324–2334 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Swiston J, Hubberstey A, Yu G & Young D Differential expression of CAP and CAP2 in adult rat tissues. Gene 165, 273–277 (1995). [DOI] [PubMed] [Google Scholar]
- 49.Ono S The role of cyclase-associated protein in regulating actin filament dynamics - more than a monomer-sequestration factor. J Cell Sci 126, 3249–3258 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bor B, Vizcarra CL, Phillips ML & Quinlan ME Autoinhibition of the formin Cappuccino in the absence of canonical autoinhibitory domains. Mol Biol Cell 23, 3801–3813 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chesarone M, Gould CJ, Moseley JB & Goode BL Displacement of formins from growing barbed ends by bud14 is critical for actin cable architecture and function. Developmental cell 16, 292–302 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chesarone-Cataldo M et al. The myosin passenger protein Smy1 controls actin cable structure and dynamics by acting as a formin damper. Dev Cell 21, 217–230 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eisenmann KM et al. Dia-interacting protein modulates formin-mediated actin assembly at the cell cortex. Curr Biol 17, 579–591 (2007). [DOI] [PubMed] [Google Scholar]
- 54.Garabedian MV et al. Integrated control of formin-mediated actin assembly by a stationary inhibitor and a mobile activator. J Cell Biol (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Graziano BR, Jonasson EM, Pullen JG, Gould CJ & Goode BL Ligand-induced activation of a formin-NPF pair leads to collaborative actin nucleation. The Journal of cell biology 201, 595–611 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Quinlan ME, Hilgert S, Bedrossian A, Mullins RD & Kerkhoff E Regulatory interactions between two actin nucleators, Spire and Cappuccino. J Cell Biol 179, 117–128 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vizcarra CL et al. Structure and function of the interacting domains of Spire and Fmn-family formins. Proc Natl Acad Sci U S A 108, 11884–11889 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kotila T et al. Structural basis of actin monomer re-charging by cyclase-associated protein. Nat Commun 9, 1892 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hatch AL, Ji WK, Merrill RA, Strack S & Higgs HN Actin filaments as dynamic reservoirs for Drp1 recruitment. Mol Biol Cell 27, 3109–3121 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Denton RM Regulation of mitochondrial dehydrogenases by calcium ions. Biochim Biophys Acta 1787, 1309–1316 (2009). [DOI] [PubMed] [Google Scholar]
- 61.Menzies KJ, Zhang H, Katsyuba E & Auwerx J Protein acetylation in metabolism - metabolites and cofactors. Nat Rev Endocrinol 12, 43–60 (2016). [DOI] [PubMed] [Google Scholar]
- 62.Mishra P & Chan DC Metabolic regulation of mitochondrial dynamics. The Journal of cell biology 212, 379–387 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ran J, Yang Y, Li D, Liu M & Zhou J Deacetylation of alpha-tubulin and cortactin is required for HDAC6 to trigger ciliary disassembly. Sci Rep 5, 12917 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Benoy V et al. HDAC6 is a therapeutic target in mutant GARS-induced Charcot-Marie-Tooth disease. Brain 141, 673–687 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.d’Ydewalle C et al. HDAC6 inhibitors reverse axonal loss in a mouse model of mutant HSPB1-induced Charcot-Marie-Tooth disease. Nat Med 17, 968–974 (2011). [DOI] [PubMed] [Google Scholar]
- 66.Mo Z et al. Aberrant GlyRS-HDAC6 interaction linked to axonal transport deficits in Charcot-Marie-Tooth neuropathy. Nat Commun 9, 1007 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hulme EC & Trevethick MA Ligand binding assays at equilibrium: validation and interpretation. Br J Pharmacol 161, 1219–1237 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Eng JK, Jahan TA & Hoopmann MR Comet: an open-source MS/MS sequence database search tool. Proteomics 13, 22–24 (2013). [DOI] [PubMed] [Google Scholar]
- 69.Valot B, Langella O, Nano E & Zivy M MassChroQ: a versatile tool for mass spectrometry quantification. Proteomics 11, 3572–3577 (2011). [DOI] [PubMed] [Google Scholar]
- 70.Vizcaino JA et al. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat Biotechnol 32, 223–226 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data (Supplementary Tables 1,2, and 3) have been deposited to the ProteomeXchange Consortium70 through the PRIDE partner repository. PX accession number PXD010484. Raw numerical data for the results in Figure 4b, Figure 5g–j, and Figure 6e are provided in Supplementary Table 4. Unprocessed blots and gels for all relevant figures are in Supplementary Figure 7.