SUMMARY
O-GalNAc glycosylation is initiated in the Golgi by glycosyltransferases called GALNTs. Proteomic screens identified >600 O-GalNAc-modified proteins, but the biological relevance of these modifications has been difficult to determine. We have discovered a conserved function for GALNT3 in trophoblast stem (TS) cells, blastocyst trophectoderm, and human mammary epithelial cells (HMECs). The loss of GALNT3 expression in these systems reduces O-GalNAc glycosylation and induces epithelial-mesenchymal transition. Furthermore, Galnt3 expression is reduced in aggressive, mesenchymal claudin-low breast cancer cells. We show that GALNT3 expression controls the O-GalNAc glycosylation of multiple proteins, including E-cadherin in both TS cells and HMECs. The loss of GALNT3 results in the intracellular retention of E-cadherin in the Golgi. Significantly, re-expression of GALNT3 in TS cells increases O-GalNAc glycosylation and restores the epithelial state. Together, these data demonstrate the critical biological role of GALNT3 O-GalNAc glycosylation to promote the epithelial phenotype in TS cells, blastocyst trophectoderm, and HMECs.
In Brief
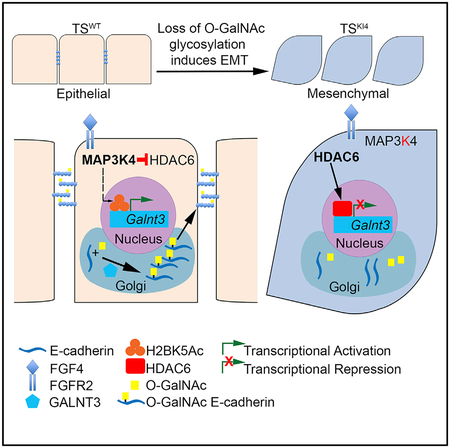
Raghu et al. demonstrate that O-GalNAc glycosylation is critical for epithelial state maintenance in trophoblast stem cells and HMECs. MAP3K4 promotes GALNT3 O-GalNAc modification of E-cadherin. Loss of GALNT3 results in the retention of E-cadherin in the Golgi. GALNT3 re-expression restores cell surface localization of E-cadherin, protecting the epithelial state.
Graphical Abstract
INTRODUCTION
Epithelial cell-cell adhesion is promoted by both proteins located at the cell surface and those secreted into the extracellular matrix. These proteins are heavily modified by the addition of carbohydrates controlling their localization, stability, secretion, and proteolytic processing. Carbohydrate modifications occur in two ways: N-glycosylation, carbohydrates attached to amide groups of asparagine (N), and O-glycosylation, carbohydrates attached to hydroxyl groups of serine and threonine (S/T). Proteomic screens suggest the majority of membrane and secreted proteins are glycosylated, but the biological roles of these modifications remain mostly unknown.
Disruption of epithelial cell-cell adhesion occurs during epithelial-to-mesenchymal transition (EMT) when epithelial cells with tight cell-cell adhesion and apical-basal polarity convert to motile, mesenchymal cells with front-back polarity (Yang and Weinberg, 2008). EMT is critical during mammalian development, including implantation, gastrulation, and neural crest formation (Thiery et al., 2009). Importantly, EMT is reversible through a mesenchymal–to-epithelial transition (MET) in which motile, mesenchymal cells are restored to a non-motile, epithelial state (Yang and Weinberg, 2008). In addition to its role in development, EMT is reactivated during tumor progression and cancer metastasis. Aberrant O-glycosylation has been implicated in cancer EMT (Chia et al., 2016). However, its role in developmental EMT is poorly understood.
In mammals, the first developmental EMT occurs in trophoblast stem (TS) cells during implantation, where epithelial TS cells in the trophectoderm (TE) transition into invasive giant cells (Thiery et al., 2009). Multipotent TS cells isolated from pre-implantation blastocysts can be cultured indefinitely in the presence of fibroblast growth factor 4 (FGF4) (Tanaka et al., 1998). Upon FGF4 withdrawal, TS cells differentiate forming all the mature trophoblast subtypes of the placenta (Tanaka et al., 1998). Wild-type TS (TSWT) cells are epithelial with tight cell-cell adhesion and apical-basal polarity. In contrast, TS cells isolated from mice with a targeted mutation that inactivates the kinase activity of MAP3K4 (TSKI4 cells) exhibit a mesenchymal morphology with front-back polarity (Abell et al., 2011). Embryos with inactivated MAP3K4 display developmental disorders, including neural tube, skeletal, and implantation defects, that are due to perturbations in EMT (Abell et al., 2009; Abell et al., 2005). TSKI4 cells show key characteristics of EMT, including reduced expression of epithelial markers, such as E-cadherin, with increased expression of the mesenchymal markers Vim and Cdh2 and the EMT-inducing transcription factors Twist1 and Snai2 and increased invasiveness (Abell et al., 2011). Importantly, TSKI4 cells and mesenchymal claudin-low (CL) breast cancer cells share gene expression profiles and display properties of stemness and EMT (Abell et al., 2011).
Using DNA microarray data from Abell et al. (2011) and Neve et al. (2006), we have identified a gene, Galnt3, whose expression is reduced in both TSKI4 cells and CL breast cancer cells. Uridine-diphosphate (UDP)-GalNAc transferase 3 (GALNT3) is a member of a large family of homologous genes called GALNTs, each displaying selective tissue and target specificities (Beaman and Brooks, 2014). GALNTs initiate O-GalNAc glycosylation in the Golgi through the transfer of N-acetylgalactosamine (GalNAc) onto serine and threonine residues, forming a GalNAc-S/T (Tn) antigen on specific protein substrates (Bennett et al., 2012). Abnormalities in the activity of GALNTs have been implicated in several pathologies. The role of specific GALNTs in developmental EMT has been difficult to determine using GALNT-specific knockout mice, due in part to the potential functional redundancy of this large family of glycosyltransferases. GALNT3 initiates O-GalNAc glycosylation, but its function in stem cells is unknown. Reduced expression of GALNT3 in mesenchymal TSKI4 cells relative to TSWT cells and in CL breast cancer cells relative to human mammary epithelial cells (HMECs) suggested that GALNT3 may be important in promoting an epithelial phenotype in these cell types.
Herein, we show significant O-GalNAc glycosylation at the cell surface of the TE in embryonic day 3.5 (E3.5) blastocysts and in cultured TS cells. The loss of GALNT3 expression in TS cells, blastocyst TE, and HMECs results in the acquisition of the mesenchymal state and reduced total O-GalNAc glycosylation. Importantly, GALNT3 re-expression in the mesenchymal TSKI4 cells restores both O-GalNAc glycosylation and the epithelial phenotype. Our findings show that GALNT3 promotes initiation of O-GalNAc on multiple proteins, including E-cadherin, resulting in the cell surface localization of E-cadherin, linear adherens junction (AJ) assembly, and epithelial state maintenance. The loss of GALNT3 expression in TSKI4 cells reduces O-GalNAc glycosylation of E-cadherin and increases the intracellular retention of E-cadherin in the Golgi. GALNT3 re-expression restores E-cadherin localization to the plasma membrane and leads to an epithelial state. Together, our findings demonstrate that GALNT3 protects the epithelial phenotype in TS cells and HMECs by promoting O-GalNAc glycosylation of proteins, including E-cadherin.
RESULTS
Loss of GALNT3 Is Associated with Acquisition of the Mesenchymal State
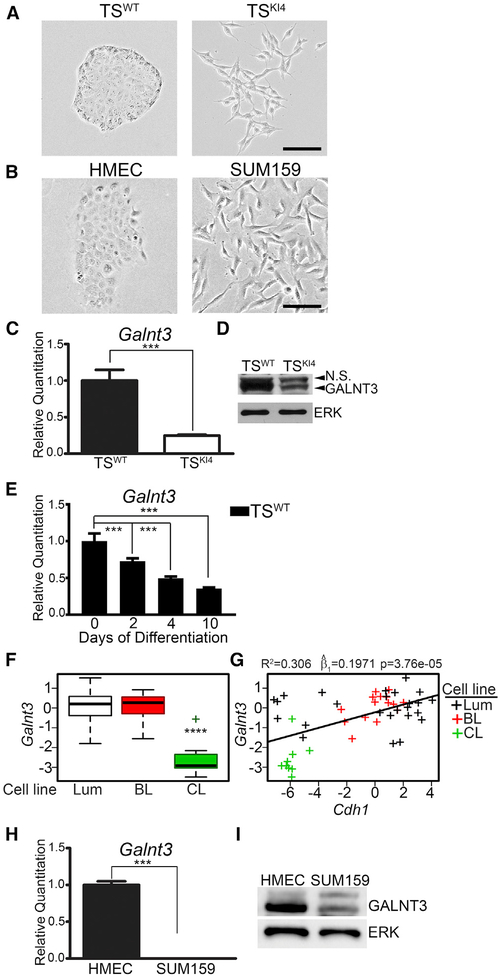
We previously demonstrated that TSKI4 cells lacking MAP3K4 kinase activity exist in a metastable state, having reduced epithelial features and increased mesenchymal characteristics while maintaining stemness (Abell et al., 2011). Unlike TSWT cells that displayed an epithelial morphology with tight cell-cell adhesion and apical-basal polarity, TSKI4 cells displayed a mesenchymal-like morphology with the loss of cell-cell adhesion and gain of front-back polarity (Figure 1A). Similar to TSKI4 cells, CL breast cancer cells display properties of both EMT and stemness (Abell et al., 2011). Unlike HMECs that have an epithelial morphology, the CL breast cancer cell line SUM159 displayed a mesenchymal morphology, consistent with the EMT features of CL breast cancer cells (Figure 1B).
Figure 1. Loss of GALNT3 Correlates with the Gain of a Mesen chymal State.
(A and B) Phase microscopy images of TS cells (A) and HMEC and SUM159 cells (B) are representative of three independent experiments. Black bar represents 100 μm.
(C) Reduced Galnt3 transcripts in mesenchymal TSKI4 cells relative to TSWT cells. qPCR data normalized to Actb are expressed as a fold-change relative to TSWT cells and are the mean ± range of two independent experiments.
(D) Western blots are representative of three independent experiments. N.S., Non-Specific.
(E) Decreased Galnt3 transcripts with differentiation of TSWT cells by FGF4 withdrawal for the indicated number of days. qPCR data normalized to Actb are expressed as a fold-change relative to undifferentiated TSWT cells (0 day) and are the mean ± range of two independent experiments.
(F) Galnt3 expression is significantly reduced in claudin-low (CL) breast cancer cell lines. Luminal (Lum), basal-like (BL), and CL breast cancer subtypes. p value was calculated using Student’s t test and Bonferroni’s correction for multiple comparisons. ****p < 0.0001.
(G) Loss of Galnt3 is significantly correlated with reduced Cdh1 levels. Plot shows the correlation between Galnt3 and Cdh1 transcripts in breast cancer subtypes. Each “+” represents a specific breast cancer line. p value was calculated using linear regression and, represents the estimate of the slope of linear regression line.
(H) Galnt3 transcripts are reduced in mesenchymal SUM159 cells relative to HMECs. qPCR data normalized to Gapdh are expressed as a fold-change relative to HMECs and are the mean ± SEM of three independent experiments.
(I) Western blots are representative of three independent experiments.
***p < 0.001; Student’s t test.
An analysis of DNA microarray data from Abell et al. (2011) suggested that Galnt3 expression is reduced in TSKI4 cells relative to TSWT cells. The Galnt3 transcript and protein were reduced in TSKI4 cells relative to TSWT cells, as measured by real-time qPCR and western blotting, respectively (Figures 1C and 1D). The induction of EMT through differentiation of TS cells also decreased Galnt3 transcript levels (Figure 1E). An examination of Galnt3 expression in the breast cancer cell line microarray dataset from Neve et al. (2006) revealed that the loss of Galnt3 expression was strongly associated with the CL form of breast cancer (p < 0.0001) (Figure 1F). E-cadherin levels are lowest in the CL form of breast cancer as compared to all other breast cancer subtypes (Prat et al., 2010). A comparison of Cdh1 and Galnt3 expression in breast cancer cells from the Neve et al. (2006) dataset showed that the Galnt3 loss was associated with the loss of Cdh1 (p < 0.0001), suggesting a strong correlation between Galnt3 and Cdh1 expression (Figure 1G). The Galnt3 transcript and protein were reduced in mesenchymal SUM159 cells relative to epithelial HMECs (Figures 1H and 1I). These data showed that GALNT3 expression was decreased in both mesenchymal-like TSKI4 cells and CL breast cancer cells, suggesting that the loss of GALNT3 is associated with the loss of the epithelial state in TS cells and CL breast cancer.
Transcriptional Regulation of Galnt3 Expression by MAP3K4 and HDAC6
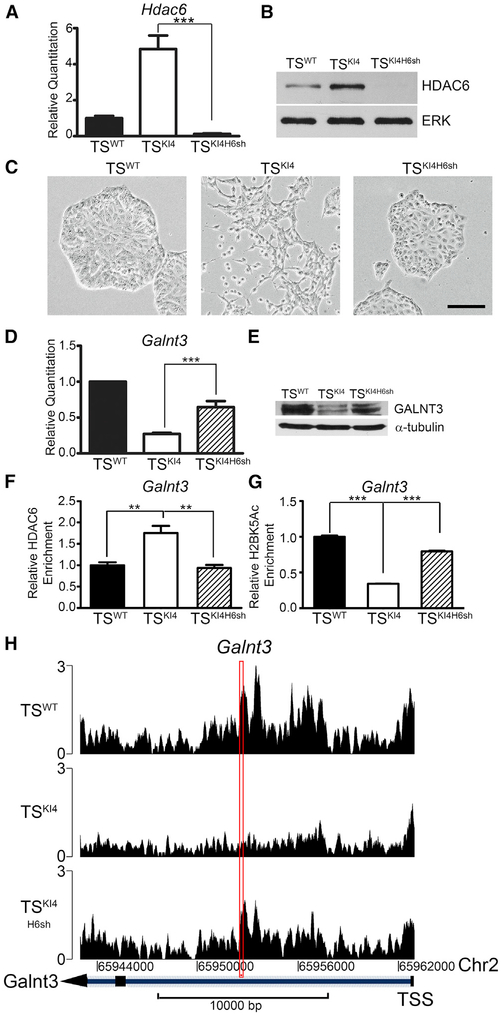
We have previously demonstrated that MAP3K4 inhibits HDAC6 expression and activity (Mobley et al., 2017). HDAC6 induces EMT in TS cells by binding to the promoters of epithelial maintenance genes, deacetylating histone H2BK5, and repressing gene expression. RNA sequencing (RNA-seq) data from Mobley et al. (2017) suggested that Galnt3 expression was HDAC6 dependent. HDAC6 expression at both transcript and protein levels was elevated in TSKI4 cells relative to TSWT cells (Figures 2A and 2B). The HDAC6 protein was also elevated in the nuclei of SUM159 cells relative to HMECs (Figure S1A). Knockdown of HDAC6 in TSKI4 cells with a short hairpin RNA (shRNA) (TSKI4H6sh cells) reduced HDAC6 transcript and protein levels and restored an epithelial morphology (Figures 2A–2C). Galnt3 transcripts were increased in TSKI4H6sh cells relative to TSKI4 cells, resulting in a 50% increase in the GALNT3 protein (Figures 2D and 2E). These data suggested that HDAC6 inhibits the expression of Galnt3.
Figure 2. Galnt3 Expression Is Co-regulated by MAP3K4 and HDAC6.
(A) Hdac6 transcripts analyzed by qPCR in TSWT cells and TSKI4 cells expressing control shRNA or TSKI4 cells expressing Hdac6 shRNA (TSKI4H6sh) are shown. qPCR data normalized to Actb are expressed as a fold-change relative to TSWT cells and are the mean ± SEM of three independent experiments.
(B) Western blots of whole-cell lysates are representative of three independent experiments.
(C) Knockdown of HDAC6 in TSKI4 cells restores an epithelial morphology. Phase microscopy images are representative of three independent experiments. Black bar represents 100 μm.
(D) Hdac6 knockdown increases Galnt3 transcripts in TSKI4H6sh cells relative to TSKI4 cells. qPCR data normalized to Actb are expressed as a fold-change relative to TSWT cells and are the mean ± SEM of five independent experiments.
(E) GALNT3 protein expression increases with knockdown of HDAC6 in TSKI4H6sh cells. Western blots are representative of two independent experiments.
(F) Anti-HDAC6 ChIP-PCR shows increased HDAC6 enrichment on the Galnt3 promoter in TSKI4 cells relative to TSWT cells. Data shown are the mean ± SEM of four independent experiments.
(G) Decreased H2BK5 acetylation on the Galnt3 promoter in TSKI4 cells relative to TSWT cells, as measured by anti-H2BK5Ac ChIP-PCR. Data shown are the mean ± SEM of six independent experiments.
(H) Knockdown of HDAC6 increases H2BK5 acetylation on the Galnt3 promoter in TSKI4H6sh cells relative to TSKI4 cells. H2BK5Ac ChIP-seq read density plots at the Galnt3 transcription start site + 20 kb are shown.
Red rectangle indicates the region amplified in ChIP PCR reactions in (F) and (G). Data are expressed as the normalized read count of the immunoprecipitate (IP) divided by the normalized read count of the TSWT cell input. **p < 0.01; ***p < 0.001; Student’s t test.
See also Figure S1.
We predicted that HDAC6 regulates the expression of Galnt3 by directly binding the Galnt3 promoter and deacetylating H2BK5. As measured by anti-HDAC6 chromatin immunoprecipitation coupled to qPCR (ChIP-PCR), HDAC6 was enriched on the Galnt3 promoter in TSKI4 cells relative to TSWT cells (Figures 2F and S1B). Anti-H2BK5Ac ChIP-PCR and published ChIP sequencing (ChIP-seq) data from Mobley et al. (2017) showed reduced H2BK5Ac on the Galnt3 promoter in TSKI4 cells relative to TSWT cells (Figures 2G, 2H, and S1C). Knockdown of HDAC6 in TSKI4 cells increased H2BK5Ac on the Galnt3 promoter (Figures 2G and 2H). Similar to TS cells, anti-HDAC6 ChIP-PCR showed enrichment of HDAC6 on the Galnt3 promoter in SUM159 cells relative to HMECs (Figures S1D and S1E). Furthermore, anti-H2BK5Ac ChIP-PCR showed a concomitant decrease in H2BK5Ac on the Galnt3 promoter in SUM159 cells (Figure S1F). These data suggested that HDAC6 is bound to the Galnt3 promoter in mesenchymal-like TSKI4 cells and CL SUM159 breast cancer cells, actively deacetylating H2BK5 and repressing Galnt3 expression.
Knockdown of GALNT3 in TSWT Cells Induces EMT
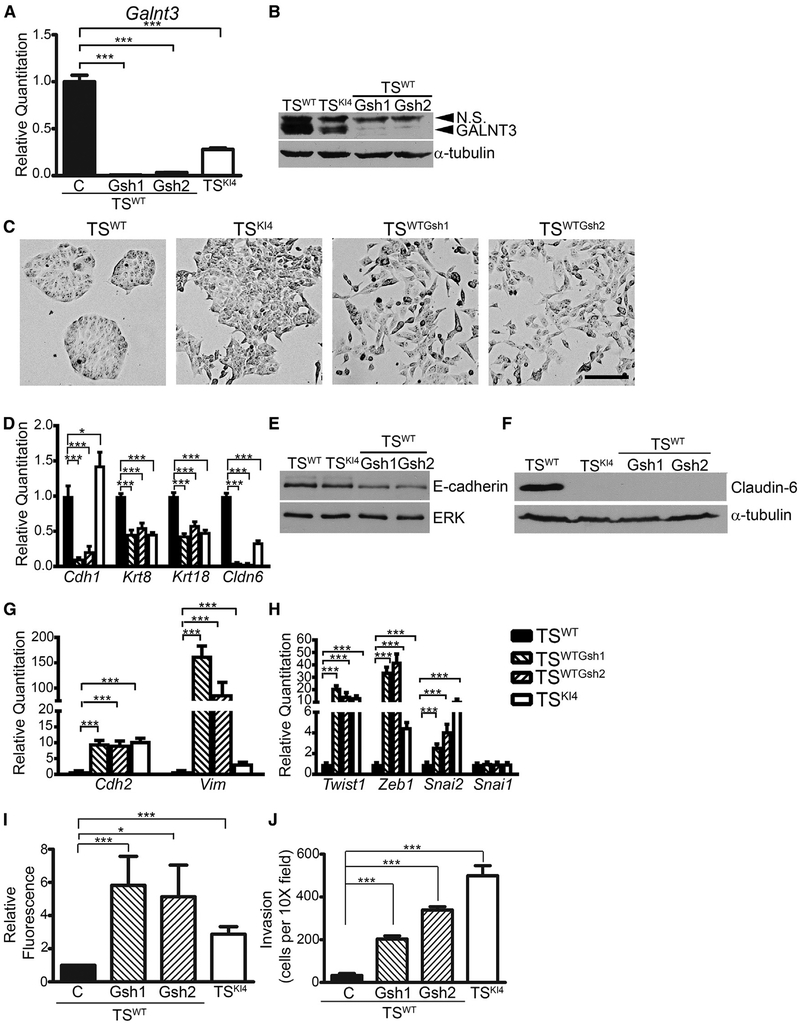
To define the role of GALNT3 in TS cell EMT, we infected TSWT cells with lentiviruses expressing two independent Galnt3 shRNAs (TSWTGsh1 and TSWTGsh2 cells). GALNT3 expression was reduced by >98% at both message and protein levels in TSWTGsh1 and TSWTGsh2 cells relative to TSWT cells (Figures 3A and 3B). The loss of GALNT3 expression in TSWTGsh1 and TSWTGsh2 cells induced a mesenchymal morphology similar to TSKI4 cells (Figure 3C). Transcripts of epithelial genes Cdh1, Krt8, Krt18, and Cldn6 were reduced in TSWTGsh1 and TSWTGsh2 cells relative to TSWT cells (Figure 3D). E-cadherin and Claudin-6 protein levels were also diminished with GALNT3 knockdown (Figures 3E and 3F). In addition to the loss of epithelial markers, TSWTGsh1 and TSWTGsh2 cells gained the expression of mesenchymal markers Cdh2 and Vim and the EMT-inducing transcription factors Twist1, Zeb1, and Snai2 (Figures 3G and 3H). Consistent with changes in Claudin-6 transcript and protein levels, immunofluorescence confocal microscopy showed reduced Claudin-6 expression and a loss of co-localization with ZO1 at cell-cell junctions in TSKI4 cells and TSWTGsh cells (Figure S2A). Furthermore, fluorescent dye exclusion assays showed increased permeability in TSKI4 cells and TSWTGsh cells relative to TSWT cells (Figure 3I). These data suggested that the loss of GALNT3 reduces epithelial features and disrupts barrier formation.
Figure 3. Knockown of GALNT3 in TSWT Cells Induces EMT.
(A) Transcripts were measured using qPCR in TSWT and TSKI4 cells expressing control shRNA (C) or TSWT cells expressing two independent Galnt3 shRNAs (TSWTGsh1 and TSWTGsh2).
(B) Western blots of whole-cell lysates are representative of three independent experiments. N.S., Non-Specific.
(C) GALNT3 knockdown in TSWT cells induces a mesenchymal morphology. Representative phase microscopy images from three independent experiments are shown. Black bar represents 100 μm.
(D) GALNT3 knockdown in TSWT cells reduces epithelial marker expression in TSWTGsh1 and TSWTGsh2 cells.
(E and F) Western blots of E-cadherin (E) and Claudin-6 (F) are representative of three independent experiments.
(G and H) Increased transcripts of mesenchymal markers (G) and EMT-inducing transcription factors (H) with reduced Galnt3 expression are shown.
(I) Impaired barrier formation in TSWTGsh1 and TSWTGsh2 cells with reduced GALNT3 expression. Diffusion of fluorescent dye across a confluent monolayer of cells is expressed as a fold-change in fluorescence relative to TSWT cells. Data shown are the mean ± SEM of three independent experiments.
(J) Loss of GALNT3 increases invasiveness in TSWTGsh1 and TSWTGsh2 cells through growth-factor-reduced Matrigel-coated transwells. Data show cells per 103 field and are the mean ± range of two independent experiments performed in triplicate.
(A, D, G, and H) qPCR data normalized to Actb are expressed as a fold-change relative to TSWT cells and are the mean ± SEM of three independent experiments. *p < 0.05; ***p < 0.001; Student’s t test.
See also Figure S2.
A key feature of EMT is the acquisition of cell motility and/or invasiveness. Cell movement assessed through live cell imaging showed TSWT cells remaining in a static position while rapidly dividing (Video S1). In contrast, TSKI4 cells showed frequent, rapid movements throughout the field (Video S2). Knockdown of GALNT3 in TSWT cells increased motility similar to TSKI4 cells (Video S3). In addition to examining motility, we measured invasiveness using growth-factor-reduced Matrigel-coated transwell assays. TSKI4, TSWTGsh1, and TSWTGsh2 cells displayed increased invasiveness relative to TSWT cells, suggesting that the loss of GALNT3 increases invasiveness (Figure 3J). Although the loss of GALNT3 induced EMT, cells lacking GALNT3 retained the expression of stemness markers and the ability to differentiate into all trophoblast subtypes, suggesting that stemness features were maintained in the absence of GALNT3 (Figures S2B–S2I). These data showed that TS cells lacking GALNT3, either through the loss of MAP3K4 activity in TSKI4 cells or with shRNA knockdown of GALNT3 in TSWT cells, expressed both stemness and EMT features. Together, these data suggested that the loss of GALNT3 in TS cells induces EMT.
We also examined the role of GALNT3 in HMECs by transiently expressing two independent Galnt3 shRNAs. We were unable to achieve a stable reduction of GALNT3 expression in HMECs, suggesting that GALNT3 performs key functions. Transient expression of Galnt3 shRNAs resulted in the partial reduction of Galnt3 transcripts and morphological changes that were similar to TSKI4 cells (Figures S2J and S2K). Control-infected HMECs formed epithelial colonies, whereas GALNT3 knockdown cells were individual with front to back polarity and long cytoplasmic extensions (Figure S2K). Although total E-cadherin protein levels were unchanged with the knockdown of GALNT3 in HMECs, cell movement was altered (Figure S2L; Videos S4 and S5). Control-infected HMECs moved as sheets of cells (Video S4). In contrast, HMECs with GALNT3 knockdown moved more as individual cells with front to back polarity that was similar to SUM159 cells (Video S5; data not shown). Together, these data suggested that GALNT3 is important for maintaining the epithelial state in both TS cells and HMECs, as reduced expression of GALNT3 induced EMT characteristics.
Re-expression of GALNT3 in Mesenchymal-like TSKI4 Cells Restores the Epithelial State
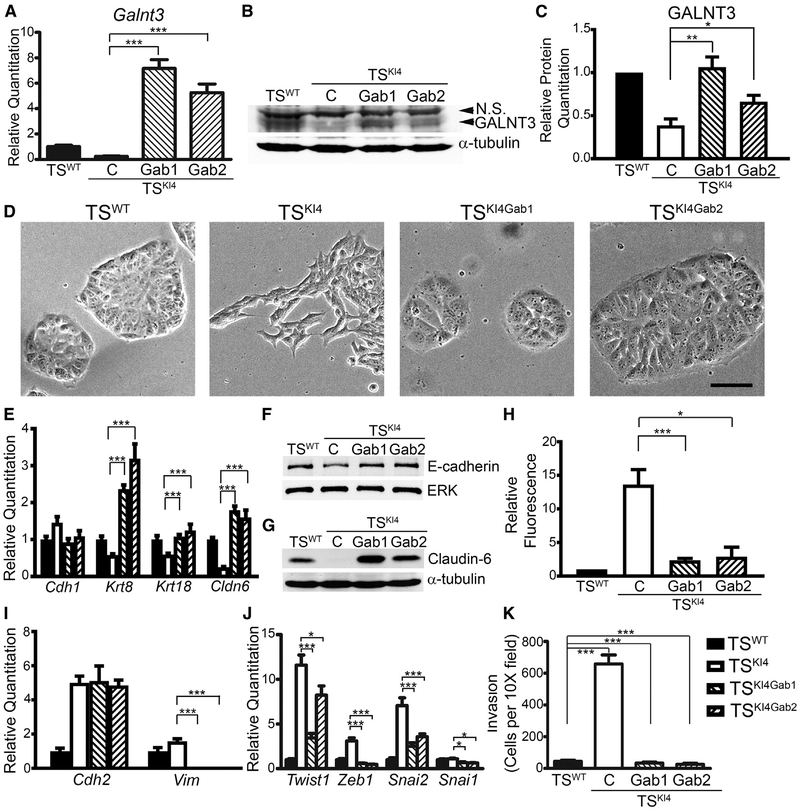
Using a lentiviral construct for human Galnt3, we re-expressed GALNT3 in TSKI4 cells (TSKI4Gab1 and TSKI4Gab2 cells). Galnt3 transcripts were increased in TSKI4Gab1 and TSKI4Gab2 cells relative to TSKI4 cells, as measured with primers detecting both human and mouse Galnt3 (Figure 4A). Similarly, GALNT3 protein levels in TSKI4Gab1 and TSKI4Gab2 cells were also increased rela tive to TSKI4 cells (Figures 4B and 4C). Importantly, re-expression of GALNT3 in TSKI4Gab cells restored an epithelial morphology in TSKI4 cells that was similar to TSWT cells (Figure 4D). In addition, transcripts of epithelial markers Krt8, Krt18, and Cldn6 were also increased in TSKI4Gab1 and TSKI4Gab2 cells relative to TSKI4 cells (Figure 4E). Furthermore, E-cadherin and Claudin-6 proteins were expressed in TSKI4Gab1 and TSKI4Gab2 cells at levels similar to TSWT cells (Figures 4F and 4G). Fluorescent dye flux was reduced in TSKI4Gab cells relative to TSKI4 cells, suggesting partial restoration of tight junctions and barrier function with re-expression of GALNT3 (Figure 4H). Together, these data suggested that GALNT3 re-expression in TSKI4 cells restored epithelial features.
Figure 4. GALNT3 Re-expression in TSKI4 Cells Restores an Epithelial Phenotype.
(A) Increased Galnt3 transcripts in TSKI4 cells expressing human GALNT3. Transcripts were measured using qPCR in TSWT and TSKI4 cells infected with a control lentiviral construct (C) or TSKI4 cells infected with a lentiviral construct expressing human Galnt3 (TSKI4Gab1 and TSKI4Gab2).
(B and C) Re-expression of GALNT3 in TSKI4Gab1 and TSKI4Gab2 cells was measured by western blotting.
(B) Blots are representative of three independent experiments.
(C) Densitometry was used to quantify three independent experiments.
(D) GALNT3 re-expression restores an epithelial morphology. Representative phase microscopy images from three independent experiments are shown. Black bar represents 100 μm.
(E) Increased transcripts of epithelial markers with re-expression of GALNT3.
(F and G) Western blots of E-cadherin (F) and Claudin-6 (G) are representative of three independent experiments.
(H) GALNT3 re-expression promotes barrier formation. Diffusion is expressed as a fold-change in fluorescence relative to TSWT cells. Data shown are the mean ± SEM of three independent experiments.
(I and J) Re-expression of GALNT3 reduces mesenchymal markers (I) and EMT-inducing transcription factors (J).
(K) Re-expression of GALNT3 in TSKI4 cells reduces invasiveness through growth-factor-reduced Matrigel. Data show cells per 10× field and are the mean ± range of two independent experiments performed in triplicate.
(A, E, I, and J) qPCR data normalized to Actb are expressed as a fold-change relative to TSWT cells and are the mean ± SEM of three independent experiments. *p < 0.05; **p < 0.01; ***p < 0.001; Student’s t test.
See also Figure S3.
Re-expression of GALNT3 in TSKI4 cells reduced the expression of mesenchymal markers and EMT-inducing transcription factors. Although Cdh2 expression remained elevated, TSKI4Gab1 and TSKI4Gab2 cells showed reduced expression of the mesenchymal marker Vim and EMT-inducing transcription factors Twist1, Zeb1, and Snai2 relative to TSKI4 cells (Figures 4I and 4J). Importantly, TSKI4Gab cells had reduced cell motility, as measured by live cell imaging and reduced invasiveness to TSWT cell levels (Video S6; Figure 4K). Altogether, these data suggested that re-expression of GALNT3 in TSKI4 cells induces MET, restoring the epithelial phenotype in TS cells.
In contrast to TS cells, re-expression of GALNT3 in SUM159 cells at levels greater than HMECs failed to alter cell morphology (Figures S3A–S3C). Although GALNT3 colocalized with the Golgi marker Giantin, epithelial proteins such as E-cadherin and claudins were not expressed, and mesenchymal markers and EMT-inducing transcription factors remained elevated (Figure S3D; data not shown). These data suggested that unlike TS cells, GALNT3 re-expression was insufficient to restore the epithelial state in SUM159 cells.
GALNT3 Controls the Localization of E-Cadherin
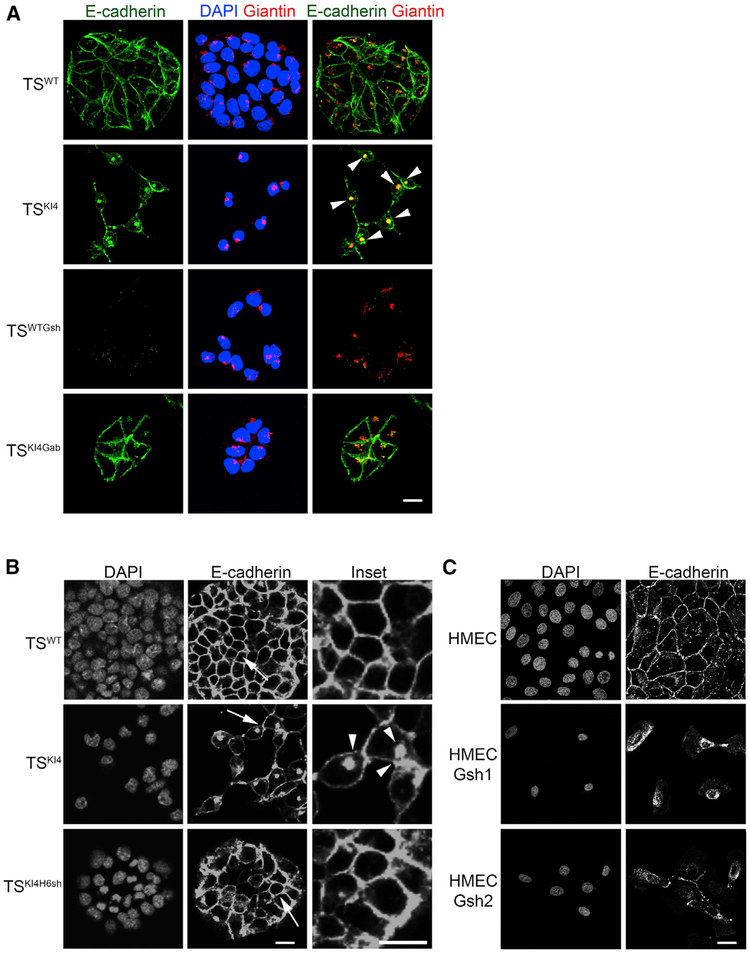
E-cadherin expression at both transcript and protein levels was similar in TSWT and TSKI4 cells (Figures 3D and 3E). However, confocal immunofluorescence microscopy showed a change in E-cadherin localization from the plasma membrane in TSWT cells to the Golgi in TSKI4 cells, as indicated by a 54% increase in co-localization of E-cadherin with a Golgi marker, Giantin (Figure 5A). Importantly, cell surface E-cadherin localization was restored in TSKI4Gab cells, suggesting that GALNT3 controls E-cadherin localization (Figure 5A). Similarly, knockdown of HDAC6, a deacetylase that normally represses Galnt3 expression, restores E-cadherin localization to the plasma membrane (Figure 5B). Knock down of GALNT3 in HMECs resulted in E-cadherin retention in the Golgi (Figure 5C; data not shown). Together, these data suggested that the loss of GALNT3 in TSKI4 cells and HMECs with GALNT3 knockdown results in the intracellular retention of E-cadherin in the Golgi. Re-expression of GALNT3 in TSKI4 cells through either HDAC6 knockdown or a GALNT3 lentiviral construct restored the localization of E-cadherin to the plasma membrane.
Figure 5. Loss of GALNT3 Results in the Intracellular Retention of E-Cadherin in the Golgi.
(A) Reduced GALNT3 expression in TSKI4 cells results in the retention of E-cadherin in the Golgi. Confocal images in TSWT and TSKI4 cells infected with a control virus, TSWT cells infected with Galnt3 shRNA (TSWTGsh) and TSKI4 cells expressing human GALNT3 (TSKI4Gab) are shown. 4′,6-Dia midino-2-phenylindole (DAPI; blue), E-cadherin (green), and Giantin (Golgi) (red). Arrowheads indicate co-localization of E-cadherin and Giantin in the Golgi.
(B) HDAC6 knockdown in TSKI4 cells results in the partial restoration of E-cadherin to the cell surface. Confocal images in TSWT cells and TSKI4 cells expressing control shRNA or TSKI4 cells expressing Hdac6 shRNA (TSKI4H6sh) are shown. Arrows show the area of enlarged insets. Arrowheads indicate punctate intracellular E-cadherin localization.
(C) GALNT3 knockdown in HMECs results in the intracellular trapping of E-cadherin. Confocal images for HMECs infected with a control shRNA or two independent Galnt3 shRNAs (HMECGsh1 and HMECGsh2) are shown.
(A–C) Images are representative of three independent experiments. White bar represents 50 μm.
Loss of GALNT3 Disrupts the Assembly of Adherens Junctions
The loss of E-cadherin often leads to disruption of the AJ assembly (Gumbiner, 1996). TSKI4, TSKI4Gab, and TSKI4H6sh cells expressed α-catenin and δ-catenin protein at levels similar to TSWT cells (Figures S4A and S4B). In addition, transient knockdown of GALNT3 in HMECs resulted in modest changes in α-catenin and δ-catenin protein levels relative to control-infected HMECs (Figure S4C). Although total protein expression was unchanged in TSKI4 cells, localization of AJ proteins was altered (Figure S4D). Unlike TSWT cells with AJ components E-cadherin, δ-catenin, and α-catenin co-localizing at cell-cell junctions, cell surface expression of all AJ components was reduced in TSKI4 cells, resulting in the loss of co-localization (Figure S4D; data not shown). Re-expression of GALNT3 in TSKI4Gab cells restored cell surface co-localization of AJ components (Figure S4D). In TSWTGsh and SUM159 cells having a near complete loss of GALNT3, the expression of δ-catenin and α-catenin were significantly reduced (Figures S4D–S4F). Together, these data suggested that AJ assembly and stability in TS cells and HMECs are GALNT3 dependent.
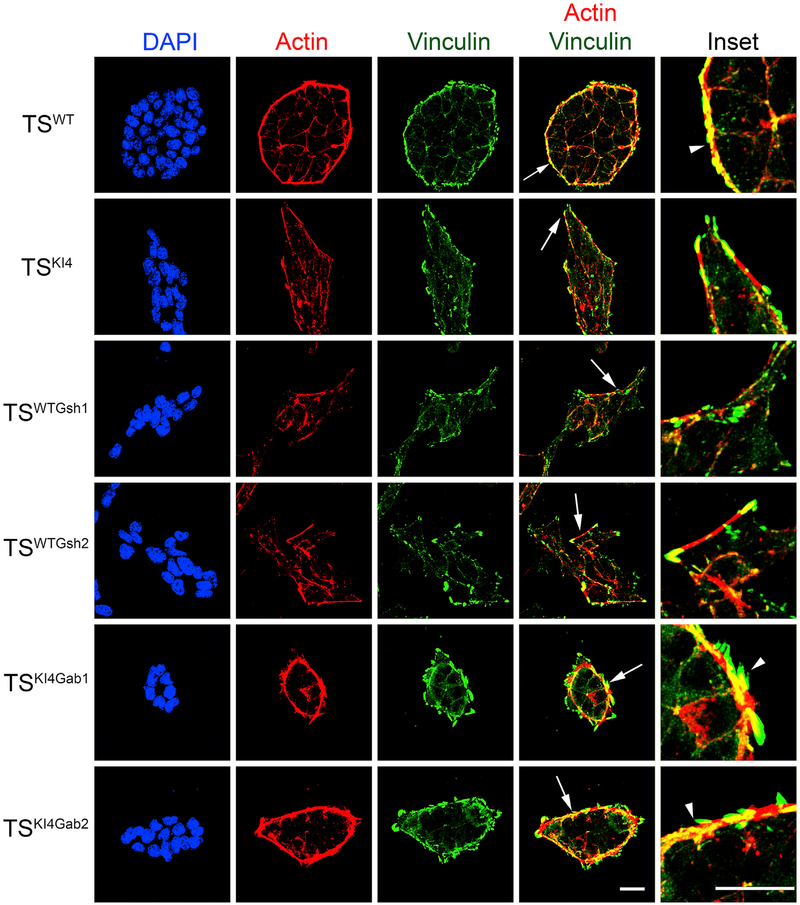
AJs can be classified as two main forms, namely, linear or punctate, based on their association with actin (Takeichi, 2014). TSWT cells displayed linear cortical actin filaments that ran parallel to the cell borders, forming static, linear AJs (Figures 6 and S5A). In TSKI4 cells, actin stress fibers were positioned perpendicular to cell borders, suggesting the presence of dynamic, punctate AJs (Figures 6 and S5A). The re-expression of GALNT3 in TSKI4Gab cells restored cortical actin that ran parallel to the cell borders (Figures 6 and S5A). AJs are often linked to the actin cytoskeleton through the actin-binding protein Vinculin (Takeichi, 2014). Total Vinculin expression did not change significantly under any conditions tested (Figure S4). In TSWT cells, AJs contained small Vinculin-positive puncta with larger Vinculin-positive adhesions positioned continuously around the entire TSWT colony (Figures 6 and S5A). In TSKI4 and TSWTGsh cells, Vinculin was present in discontinuous, finger-like projections (Figures 6 and S5A). The re-expression of GALNT3 in TSKI4Gab cells restored continuous Vinculin-positive adhesions around the colony (Figures 6 and S5A). However, these Vinculin-positive focal adhesions were more finger-like than those found in TSWT colonies. Western blotting showed that shRNA knockdown or re-expression of GALNT3 did not alter HDAC6 levels (Figures S5B–S5D). The retention of elevated HDAC6 expression in TSKI4Gab cells may explain the retention of finger-like Vinculin projections. Altogether, these data suggested that the loss of GALNT3 leads to active remodeling and disassembly of AJs, and GALNT3 re-expression promotes stable AJ assembly.
Figure 6. GALNT3 Promotes Adherens Junction Assembly and Maintenance.
Confocal images in TSWT and TSKI4 cells infected with a control virus, TSWT cells infected with Galnt3 shRNAs (TSWTGsh1 and TSWTGsh2), and TSKI4 cells expressing human GALNT3 (TSKI4Gab1 and TSKI4Gab2) are shown. DAPI (blue), Actin (red), and Vinculin (green). Arrows show the area of enlarged insets. Arrowheads indicate Actin and Vinculin co-localization. Images are representative of three independent experiments. White bar represents 50 μm.
See also Figures S4 and S5.
Decreased O-GalNAc Glycosylation with Loss of GALNT3
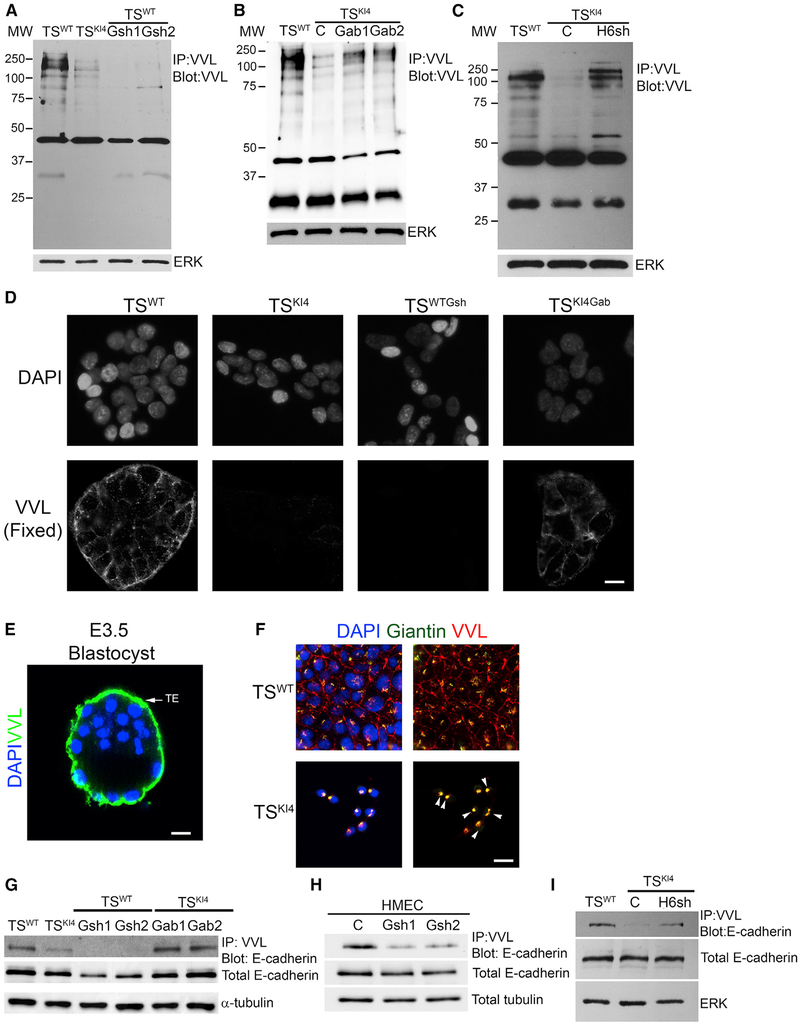
GALNT3 is an O-GalNAc glycosyltransferase localized to the Golgi that modifies specific serine and threonine with the sugar residue GalNAc. In most cell types, O-GalNAc is further modified by additional enzymes that extend the sugar, adding galactose, GlcNAc, and/or sialic acid (Bennett et al., 2012). Vicia villosa lectin (VVL) binds with high affinity to unmodified O-GalNAc residues. Using this lectin, we measured the amount of unextended O-GalNAc-labeled proteins in TSWT cells relative to TSKI4 cells. Whole-cell lysates were immunoprecipitated with agarose-bound VVL, and blots were probed with biotinylated VVL. Unmodified O-GalNAc-labeled proteins were detected in TSWT cells at higher levels relative to TSKI4 cells that have reduced GALNT3 expression (Figure 7A). Furthermore, shRNA knockdown of Galnt3 in TSWT cells resulted in the near complete loss of O-GalNAc-labeled proteins (Figure 7A). Importantly, re-expression of GALNT3 in TSKI4 cells and in SUM159 cells increased the levels of O-GalNAc-labeled proteins (Figures 7B and S6A). Knockdown of HDAC6 in TSKI4 cells also increased O-GalNAc to levels similar to TSWT cells (Figure 7C). VVL binding was also reduced in HMECs expressing Galnt3 shRNAs and in CL SUM159 cells relative to HMECs (Figures S6B and S6C). These data suggested that EMT in TS cells and HMECs was associated with the loss of O-GalNAc-labeled proteins.
Figure 7. Reduced O-GalNAc Glycosylation in Cells Lacking GALNT3 that Is Restored by Re-expression of GALNT3.
(A) Total protein O-GalNAc glycosylation is reduced with the loss of GALNT3 in TSWTGsh1 and TSWTGsh2 cells. IP with VVL agarose and blotting with biotinylated VVL are shown. Western blots of TSWT and TSKI4 cells expressing control shRNA (C) or TSWT cells expressing two independent Galnt3 shRNAs (TSWTGsh1 and TSWTGsh2) are representative of three independent experiments.
(B) Re-expression of GALNT3 in TSKI4 cells (TSKI4Gab1 and TSKI4Gab2) increases protein O-GalNAc glycosylation. Experiments were performed as in (A). Western blots of TSWT and TSKI4 cells expressing a control lentiviral vector (C) or or TSKI4 cells infected with a lentiviral vector expressing human Galnt3 (TSKI4Gab1 and TSKI4Gab2) are representative of three independent experiments.
(C) Knockdown of HDAC6 in TSKI4 cells (TSKI4H6sh) increases total protein O-GalNAc glycosylation similar to TSWT cells. Experiments were performed as in (A). Western blots of TSWT cells and TSKI4 cells expressing control shRNA (C) or TSKI4 cells expressing HDAC6 shRNA (TSKI4H6sh) are representative of two independent experiments.
(D) Absence of cell surface O-GalNAc glycosylation in TSKI4 and TSWTGsh cells with reduced GALNT3. VVL immunofluorescence staining in non-permeabilized TSWT and TSKI4 cells infected with a control virus, TSWT cells infected with Galnt3 shRNA (TSWTGsh), and TSKI4 cells re-expressing GALNT3 (TSKI4Gab) are shown. Images are representative of three independent experiments.
(E) O-GalNAc glycosylation of the trophoectoderm (TE) of intact wild-type E3.5 hatched blastocyst is shown by confocal microscopy using biotinylated VVL. Image is representative of seven blastocysts. DAPI (blue) and VVL (green).
(F) Reduced GALNT3 expression in TSKI4 cells induces retention of O-GalNAc glycosylated proteins in the Golgi. DAPI (blue), VVL (red), and Giantin (Golgi) (green) staining of Triton-permeabilized cells. Arrowheads show co-localization between VVL and Giantin in the Golgi. Images are representative of three independent experiments.
(G and H) O-GalNAc glycosylation of E-cadherin is reduced in cells lacking GALNT3. IP with VVL agarose and blotting with anti-E-cadherin antibody are shown for TS cells (G) and HMECs (H). Western blots are representative of three independent experiments.
(I) Knockdown of HDAC6 increases E-cadherin O-GalNAc glycosylation in TSKI4H6sh cells. Experiments were performed as in (G and H). Western blots are representative of two independent experiments.
IP, immunoprecipitation; MW, molecular weight in kDa; white bar represents 50 μm.
See also Figures S6 and S7.
We also used biotinylated VVL and immunofluorescence to examine the localization of O-GalNAc-labeled proteins in intact or detergent-permeabilized cells. Surprisingly, TSWT cells showed strong cell surface expression of VVL-labeled proteins in non-permeabilized cells that was weaker in TSKI4 cells or undetectable in TSWTGsh cells lacking GALNT3 (Figures 7D and S6D). TSKI4Gab cells showed increased cell surface VVL staining relative to TSKI4 cells (Figure 7D). Quantitation revealed that >90% of TSWT cells were positive for cell surface VVL staining, compared to 45% of TSKI4 cells and 0.5% of TSWTGsh cells (Figure S6E). Similar to cultured TS cells, the TE layer of intact wild-type blastocysts displayed significant VVL staining, suggesting the presence of unextended O-GalNAc-modified proteins (Figure 7E and S6F). Detergent permeabilization revealed intracellular VVL staining that was tightly localized to puncta positioned near the nucleus in all cell conditions (Figure S6G). However, total VVL staining intensity in permeabilized cells was reduced in TSKI4 cells and nearly absent in TSWTGsh cells, consistent with western blotting data (Figures 7A and S6D–S6G). Co-localization of VVL and the Golgi marker Giantin was greater in TSKI4 cells relative to TSWT cells (Figure 7F). Together, these data suggested that the loss of GALNT3 reduced O-GalNAc-labeled proteins at the cell surface.
The presence of low levels of O-GalNAc-labeled proteins in TSKI4 cells and TSWTGsh cells suggested that other GALNTs may be alternatively expressed after the loss of GALNT3. TSKI4 cells showed a modest increase in several of the GALNTs relative to TSWT cells that was reduced by re-expression of GALNT3 in TSKI4Gab cells (Figure S6H). Importantly, knockdown of GALNT3 in TSWTGsh cells did not induce transcripts of other GALNTs except Galnt7 and Galnt12 (Figure S6I). Together, these data suggested that GALNT3 is the dominant O-GalNAc glycosyltransferase in TS cells.
To further define the role of GALNT3 in TS cells, we examined the impact of the loss of GALNT3 in pre-implantation blastocysts. Mid-stage E3.5 blastocysts were hatched and infected with lentiviruses expressing either control shRNA or Galnt3 shRNA. This approach has previously been shown to only infect the TE layer of these intact blastocysts (Georgiades et al., 2007). Using semiquantitative PCR, we validated that infected blastocysts expressed transcripts for the puromycin resistance gene encoded by these viruses (Figure S7A). shRNA knockdown of Galnt3 in the TE of intact blastocysts resulted in premature attachment and outgrowth compared to control shRNA infections (Figures S7B and S7C). The attachment of blastocysts with Galnt3 knockdown occurred eight hours earlier than either control-shRNA-infected or uninfected blastocysts, suggesting that the loss of Galnt3 leads to premature EMT (Figure S7B). Furthermore, cell surface VVL staining of the TE was reduced by 57% in blastocysts infected with Galnt3 shRNAs compared to control shRNA infections (Figures S7D and S7E). Examination of VVL staining of uninfected blastocysts during attachment and outgrowth revealed the reduction of VVL staining with attachment and outgrowth, suggesting that the loss of cell surface O-GalNAc-labeled proteins on the TE may represent a normal process during blastocyst development (Figure S7F). Together, these data demonstrate the role of Galnt3 in EMT of the TE, which is the origin of TS cells.
E-Cadherin O-GalNAc Glycosylation Is Dependent on GALNT3
The loss of GALNT3 reduced the total levels of O-GalNAc glycosylated proteins in TS cells, suggesting that GALNT3 promotes the O-GalNAc glycosylation of multiple proteins in TS cells (Figures 7A–7C). Examination of VVL blots revealed reduced O-GalNAc glycosylation of a protein of ~140 kDa in TSKI4 and TSWTGsh cells that is restored in TSKI4Gab cells and TSKI4H6sh cells (Figures 7A–7C). Although the calculated mass of E-cadherin is 97.5 kDa, E-cadherin migrates around 135 kDa on polyacryl-amide gels. In addition, E-cadherin was recently shown by mass spectrometry to be modified with a single O-GalNAc on the first extracellular cadherin domain (EC1) at threonine position 63, but the function of this modification and the O-GalNAc glycosyltransferases were undefined (Vester-Christensen et al., 2013). Based on changes in O-GalNAc levels at ~140 kDa and the change in E-cadherin localization with the loss of GALNT3 in TS cells, we predicted that GALNT3 may be required for O-GalNAc glycosylation of E-cadherin. Immunoprecipitation of lysates with VVL agarose and probing with an anti-E-cadherin antibody revealed O-GalNAc-labeled E-cadherin in TSWT cells that was lost in TSKI4 cells and in TSWTGsh cells (Figure 7G). Immunoprecipitation with anti-E-cadherin antibody and probing with VVL biotin also showed O-GalNAc glycosylation of E-cadherin in TSWT cells that was diminished in TSKI4 and TSWTGsh cells (Figure S7G). A similar loss of O-GalNAc-labeled E-cadherin was observed with knockdown of GALNT3 in HMECs and in SUM159 cells relative to HMECs (Figures 7H and S7H). Knockdown of HDAC6 in TSKI4 cells to levels that restore GALNT3 expression and GALNT3 re-expression in TSKI4Gab cells increased O-GalNAc modification of E-cadherin (Figures 7G, 7I, and S7G). Furthermore, substitution of threonine 63 to alanine in E-cadherin resulted in a 50% decrease in O-GalNAc glycosylation of E-cadherin relative to wild-type E-cadherin (Figures S7I and S7J). Together, these data suggest that GALNT3 promotes O-GalNAc glycosylation of E-cadherin in TS cells and HMECs and this O-GalNAc glycosylation correlates with the correct localization of E-cadherin.
DISCUSSION
Herein, we define a critical role of O-GalNAc glycosylation in epithelial TS cells derived from the preimplantation blastocyst. We demonstrate that O-GalNAc glycosylation of E-cadherin in TS cells is dependent on GALNT3. The loss of GALNT3 results in the retention of E-cadherin in the Golgi, disruption of AJs, and induction of EMT. Importantly, re-expression of GALNT3 in mesenchymal-like TSKI4 cells restores E-cadherin localization to the cell surface and the epithelial state. Furthermore, we identify a conserved role for GALNT3 in HMECs, where the loss of GALNT3 is associated with the gain of the mesenchymal state. Our work defines a key role for GALNT3 in promoting the epithelial state in TS cells and HMECs. This study identifies the role of a specific O-GalNAc glycosyltransferase in TS cells of the early blastocyst.
E-cadherin is a transmembrane protein localized to AJs that mediates calcium-dependent adhesion. The loss of E-cadherin is a key step in EMT associated with tumor progression and resistance to conventional drug therapies (Pattabiraman et al., 2016). Previous studies have focused on the transcriptional regulation of E-cadherin. However, the mechanisms regulating the folding and localization of E-cadherin are poorly understood. Using mass spectrometry, Vester-Christensen et al. (2013) identified an O-GalNAc site on the EC1 domain of E-cadherin, but the functionality of the site was undefined. Our data show that a MAP3K4-dependent gene, Galnt3, regulates EMT by controlling the O-GalNAc glycosylation of multiple proteins, including E-cadherin, and the localization of E-cadherin. Differences in absolute levels of GALNT3 in TSKI4 cells and TSWTGsh cells may result in a partial versus full EMT and differences in E-cadherin levels. Although O-GlcNAc and N-glycosylation of E-cadherin results in cytoplasmic retention and disruption of AJs, we show that O-GalNAc modification of E-cadherin correlates with cell surface localization and proper AJ assembly, including α and δ-catenin localization (Jamal et al., 2009; Zhu et al., 2001). Together, these data demonstrate that GALNT3 promotes the epithelial TS cell phenotype, in part, by mediating O-GalNAc modification of E-cadherin.
In many cancers, heterogeneity in protein structures due to aberrant glycosylation results in a loss of cell-cell adhesion and gain of invasiveness. Changes in protein glycosylation occur in two ways: incomplete synthesis and neo-synthesis. The loss of expression of initiating glycosyltransferases results in truncated glycans and disrupted protein structures. In contrast, synthesis of glycosyltransferases not normally expressed can lead to new carbohydrate structures on proteins that may trigger a tumorigenic response (Pinho and Reis, 2015). Thus, transcriptional changes in glycosyltransferases are a key factor associated with different types of cancers. We show GALNT3 expression is nearly absent in CL SUM159 cells, and knockdown of GALNT3 in HMECs results in phenotypic changes related to EMT. Changes in GALNT3 expression have been implicated during cancer EMT. In poorly differentiated pancreatic cancers, GALNT3 expression is reduced (Maupin et al., 2010). These same mesenchymal-like pancreatic cancers have a loss of E-cadherin. Furthermore, knockdown of GALNT3 in differentiated pancreatic cancers induces EMT (Chugh et al., 2016). In contrast, GALNT3 expression is increased in ovarian cancers, including A2780s cells (Wang et al., 2014). shRNA knockdown of GALNT3 in A2780s cells reduced proliferation and invasiveness and increased E-cadherin. The authors suggested that these effects were related to GALNT3-dependent MUC1 glycosylation (Wang et al., 2014). Importantly, CL SUM159 cells that lack GALNT3 have a >90% decrease in MUC1 expression relative to HMECs, suggesting MUC1 glycosylation does not promote EMT in CL SUM159 cells (D. Raghu, unpublished data). Differences in the impact of altered GALNT3 expression may be due to the functions of GALNT3 in normal versus cancerous tissues and different stages and grades of cancer. Furthermore, the absence or presence of GALNT3 targets, such as MUC1, or differential activation of pathways triggering GALNT3 mislocalization might lead to different responses in specific cancers (Chia et al., 2016). Compensatory changes in the expression of other GALNTs may also result in different outcomes. Wang et al. (2014) showed that Galnt14 expression was strongly reduced with GALNT3 knockdown in A2780s cells. Re-expression of GALNT3 in TSKI4 cells did not increase the expression of other GALNTs, but their activity may be affected. Our data emphasize a key role for GALNT3 in the maintenance of the epithelial state in TS cells, but its role in cancer cells may be dependent on cell-type-specific expression of GALNT3 and its targets.
GALNTs comprise a large evolutionarily conserved family of O-GalNAc glycosyltransferases that include 20 isoforms in humans, 19 in mice, and 12 in Drosophila. Due to their functional redundancies that result in compensatory mechanisms, it has been difficult to identify the essential functions of GALNTs in developmental EMT. However, specific roles for a few O-GalNAc glycosyltransferases in development have been demonstrated. For example, pgant35A is required for apical-basal polarity, tight junctions, and barrier formation in Drosophila lung epithelium (Tian and Ten Hagen, 2007). In mice, Galnt1 deficiency reduces growth of the submandibular gland during development due to decreased proliferation and basement membrane secretion (Tian et al., 2012). The deletion of Galnt3 in mice with a mixed C57B6/129/SvEv background results in males with growth retardation and infertility, but these mice do not display ectopic calcifications found in humans (Ichikawa et al., 2009). In contrast, mice with a Trp589Arg mutation in Galnt3 in a mixed C57B6/C3H background are infertile and also display ectopic calcifications (Esapa et al., 2012). Dissimilarities in phenotypes of Galnt3-deleted versus mutated mice may reflect differences in genetic background and/or altered compensation mechanisms that produce other outcomes. Both male and female MAP3K4 kinase-inactive mice in a pure 129/SvEv background are infertile, growth retarded, and also have defective decidualization induced by TS cell hyperinvasiveness (Abell et al., 2009; Abell et al., 2005). Knockdown of Galnt3 in the TE of blastocysts resulted in premature attachment and outgrowth and loss of O-GalNAc on the TE, suggesting that the loss of Galnt3 promotes these early events. Interestingly, TSKI4 cells were more invasive than TSWTGsh cells with Galnt3 knockdown, suggesting that the loss of MAP3K4 activity is more detrimental than the Galnt3 loss alone. Our findings highlight the need for additional models and approaches to dissect GALNT functions.
The role of O-GalNAc glycosylation in stem cells is poorly understood. Unextended O-GalNAc-modified proteins have been detected in early preimplantation blastocysts, but the specific GALNT(s) that initiate this glycosylation were unknown (Poirier and Kimber, 1997). Differentiation of mouse embryonic stem cells to embryoid bodies or extraembryonic endoderm (XEN) cells increases the expression of GALNTs, including Galnt3. These findings suggest that O-GalNAc glycosyltransferases may play key roles in early differentiation (Nairn et al., 2012). Here, we demonstrate GALNT3-dependent O-GalNAc glycosylation on the cell surface of the TE of E3.5 blastocysts. Cell surface O-GalNAc-modified proteins are dramatically reduced in TSKI4 cells and TSWTGsh cells lacking GALNT3 relative to TSWT cells. Total O-GalNAc glycosylation levels in TS cells correlate with the absolute expression levels of GALNT3. For example, TSKI4 cells with a 70% decrease in GALNT3 show reduced total O-GalNAc glycosylation relative to TSWT cells. Furthermore, >98% shRNA knockdown of GALNT3 in TSWT cells results in a complete loss of total O-GalNAc glycosylation. In addition, the loss of GALNT3 in blastocysts resulted in decreased total O-GalNAc levels. Importantly, re-expression of GALNT3 in TSKI4 cells increases total O-GalNAc glycosylation. Although GALNT3 strongly controls total O-GalNAc glycosylation in TS cells, alterations in GALNT3 levels in pancreatic and ovarian cancers result in more modest changes in total O-GalNAc glycosylation (Chugh et al., 2016; Maupin et al., 2010; Wang et al., 2014). Together, these data demonstrate that GALNT3-dependent O-GalNAc glycosylation maintains the epithelial TS cells of the TE during early development.
A key feature of EMT is the loss of tight junctions and AJs. shRNA knockdown of GALNT3 completely disrupts AJ and tight junction assembly. Importantly, re-expression of GALNT3 in the TSKI4 cells increases AJ assembly and restores tight junction protein expression and barrier function. We have previously shown that HDAC6 directly deacetylates H2BK5 on the promoter of a key tight junction protein, Cldn6, repressing its expression (Mobley et al., 2017). Similar to Cldn6, we show that HDAC6 represses Galnt3 expression by directly deacetylating H2BK5 on the Galnt3 promoter. These data suggest that HDAC6 controls the expression of several key genes that promote the epithelial state and repression of these genes results in EMT. Interestingly, HDAC6 levels remain elevated in TSKI4 cells re-expressing GALNT3, providing an explanation for the more dynamic AJs in these cells. However, the expression and localization of Claudin-6 is restored in TSKI4Gab cells. This HDAC6-independent restoration of Claudin-6 suggests that GALNT3 may regulate other key targets that control the expression and localization of Claudin-6. Together, these data demonstrate that the MAP3K4, HDAC6, and H2BK5Ac regulated gene Galnt3 plays a key role in regulating tight junction and AJ assembly.
In summary, we define a post-translational mechanism of EMT regulation through GALNT3-dependent O-GalNAc glycosylation of proteins, including E-cadherin. Importantly, GALNT3 regulates the localization of tight junction and AJ proteins, indicating that O-GalNAc glycosylation promotes TS cell-cell adhesion. We predict that GALNT3 O-GalNAc glycosylates additional targets that are important in regulating EMT in TS cells, and future work will focus on identifying these targets. In addition, we detected Tn antigens at the surface of wild-type TS cells and the TE of E3.5 blastocysts, suggesting a possible role for unextended O-GalNAc glycosylation during early development. Further investigation will determine the identity of O-GalNAcmodified proteins and their roles in early development. Together, our work provides insights into O-GalNAc glycosylation events occurring during developmental EMT that may be reactivated during cancer progression.
STAR⋆METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Amy N. Abell (anabell@memphis.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
Kinase-inactive MAP3K4 (KI4) mice having a point mutation of Lysine 1361 to Arginine were created as described in Abell et al. (2005). These mice were maintained on a pure 129/SvEv background as described in Abell et al. (2009). Because this mutation is embryonic lethal, mice heterozygous for the mutation were used to maintain the line. Mice were housed under standard animal house conditions. Wild-type mice used in these experiments were generated from heterozygous crosses of the KI4 mice. Male and female wild-type mice ages 2–6 months were used in timed breedings to generate blastocysts. Female mice were euthanized 3.5 days post mating and blastocysts were isolated as described under the headings Blastocyst analysis and Blastocyst immunostaining. The procedures and the project were approved by and performed in accordance with the Institutional Animal Care and Use Committee (IACUC) following NIH guidelines.
Cell lines and culture conditions
Mouse extraembryonic TSWT cells (male) and TSKI4 cells (female) were isolated as previously described from heterozygote crosses of mice with a targeted mutation in MAP3K4 (K1361R) that inactivates MAP3K4 kinase activity (Abell et al., 2009). TS cells were cultured in the absence of feeders in 30% TS media (RPMI 1640, 20% heat inactivated fetal bovine serum (FBS), 1% penicillin and streptomycin (PS), 1% L-glutamine, 1% sodium pyruvate, and 100 μM β-mercaptoethanol) and 70% TS media conditioned by mitotically inactivated mouse embryonic fibroblasts (MEF-CM). TS cells were supplemented with FGF4 (37.5 ng/ml) and Heparin (1 μg/ml) to maintain their stemness. In differentiation experiments, TS cells were cultured in TS media lacking growth factors and MEF-CM. The human cell lines HMECs (adult female), SUM159 cells (adult female), and HEK293T (fetal female) were a kind gift from Dr. Gary Johnson (UNC, Chapel Hill). HMEC cells were cultured in HuMEC ready medium (Thermo Fisher Scientific) containing 5% FBS, 1% PS with HuMEC supplement and bovine pituitary extract. SUM159 cells were cultured in Ham’s F12 medium (Thermo Fisher Scientific) supplemented with 5% FBS, 1% PS with 5 μg/ml insulin and 1 mg/ml hydrocortisone. Dulbecco’s modified essential medium supplemented with 10% FBS and 1% PS was used to culture HEK293T cells. All cell lines were cultured in a humidified atmosphere at 37°C containing 7% CO2 (mouse) or 5% CO2 (human).
METHOD DETAILS
Plasmids
Lentiviral pLKO, TRCN0000055098 and TRCN0000055099 plasmids (Open Biosystems) were used to create control and murine GALNT3 knockdown cells. TRCN0000035456 and TRCN0000035458 plasmids (Dharmacon) were used to create human GALNT3 knockdown cells. To create a human FLAG-GALNT3 and E-cadherin lentiviral constructs, we used clone # 55179 BCII3565 in pDONR 223 (Vidal human ORFeome (Version 5.1)) and pENTR-Cdh1 plasmid # 49776 Gateway entry vectors respectively. These inserts were cloned into a lentiviral FLAG tagged destination vector (Jordan et al., 2013) using LR clonase II enzyme mix (Invitrogen). pENTR-Cdh1 was a gift from Jamie Davies (Addgene plasmid#49776; http://addgene.org/49776; RRID:Addgene_49776).
Site directed mutagenesis
Q5 Site-Directed Mutagenesis Kit (NEB) was used to create single substitutions in pENTR-Cdh1 at threonine at positions 40, 63 and 68 to alanine. Briefly, PCR amplification was performed with custom designed mutagenesis primers for substitutions created using NEBaseChanger software (NEB). The PCR amplified product was treated with Kinase-Ligase-DpnI (KLD) enzyme mix for 5 min to degrade the parent templates. The KLD product was transformed into high-efficiency NEB 5-alpha competent E.coli. The transformed bacteria were grown for 16 hours and plasmids were purified using GeneJet miniprep kit (Thermo Fisher Scientific). The plasmids were validated for substitution mutations through Sanger sequencing at the Molecular Resource Center at the University of Tennessee Health Science Center. The custom designed primers are listed in the Key Resources Table.
Lentiviral production and infection
HEK293T cells (fetal female) were used to produce replication incompetent lentivirus as previously described (Abell et al., 2011). Briefly, HEK293T cells were cotransfected with either pLKO, shRNAs (Open Biosystems) or FLAG-GALNT3 constructs in combination with pMD2.G and psPAX2 (Addgene) using calcium phosphate. After 48–72 hours, viral supernatants were harvested by ultra-centifugation and viral pellets were resupended in 100 μL of TS media with growth factors or HuMEC media or Ham’s F12 media. Infection of TS cells was performed as previously described (Abell et al., 2011). Transduced cells were selected using puromycin (2 μg/ml).
Real-time quantitative PCR
Isolation of RNA from TS cells was performed using RNeasy Plus minikit (QIAGEN). High-Capacity reverse transcription kit (Thermo Fisher Scientific) was used to prepare cDNA from 3 μg RNA. The Bio-Rad CFX96 Touch qPCR system with iTaq (Bio-Rad) or SsoAdvanced (Bio-Rad) was used to measure gene expression changes. Expression levels were measured using 2 ΔΔCt method and were normalized to mouse Actb or human Gapdh or human Tbp. Primers used are specified in Key Resources Table.
Western blot analysis
Whole-cell, cytoplasmic and nuclear lysates of TS cells were harvested as previously described (Abell et al., 2009; Abell et al., 2005). Briefly, whole-cell lysates were harvested in buffer A (20 mM Tris pH 7.4, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA and 1% Triton X) with protease inhibitors (1 mM PMSF and 17 μg/ml aprotinin) and phosphatase inhibitors (1 mM sodium vanadate and 1 mM sodium fluoride). Cytoplasmic lysates were harvested in 0.5% Triton in 1X phosphate buffered saline with protease and phosphatase inhibitors described above. Nuclear lysates were harvested in RIPA buffer (buffer A, 0.1 mg/ml sodium dodecyl sulfate and 0.1 mg/ml sodium deoxycholate) with protease and phosphatase inhibitors described above. Lysates were probed with the indicated antibodies specified in the Key Resources Table.
Immunofluorescence staining
Immunostaining was performed as previously described (Abell et al., 2009). Briefly, cells were cultured on glass coverslips for two or three days. Cells were fixed with 3% paraformaldehyde in 1X PBS for 10 min and then washed three times with 1X PBS. Cells were permeabilized with 0.1% Triton for 3 min. Cells were blocked with 10% fetal bovine serum for 1 hour at RT. Coverslips were incubated with primary antibody overnight at 4C. Next, the cells were washed for 30 min and then incubated with DAPI (0.1 μg/ml), Dy-Alexa 488 (1:500) (Thermo Fisher Scientific), Alexa 594 (1:250) (Cell Signaling Technology) for one hour at RT. Coverslips were washed and mounted on slides with mounting media (90% glycerol and 10% 1mM Tris pH 7.5). Coverslips were imaged using an EVOS epifluorescence microscope or a Nikon A1 laser scanning confocal microscope.
Immunofluorescence imaging
Immunofluorescence imaging was performed using Nikon A1 laser scanning confocal microscopy and a 60X Plan Apochromat 1.4 numerical-aperture (NA) oil objective with lasers at 408, 488 and 594 nm. Individual Z stacks and the 2D images were obtained by either Maximum Intensity Projection (MIP) or Extended Depth of Field (EDF) for all wavelengths. Confocal images for E-cadherin/Giantin (Figure 5A) were obtained using MIP. Confocal images for Claudin6/ZO1 (Figures S2A), E-cadherin (Figure 5C), E-cadherin/δ-catenin (Figure S4D), and Actin/Vinculin (Figure 6) were obtained using EDF. Confocal 3D images shown in Figure S5A were obtained using NIS elements analysis software. Images displayed are a reconstruction of all the z stacks with a step size of 0.413 μm. The pixel resolution of each section is 512 × 512. Imaging of in vitro biotinylated VVL staining experiments were performed using EVOS epifluorescence microscope at 20X and 40X magnification for 408, 488 and 594 wavelengths. (Figures 7D, 7F, S6F, S6G, S7D, and S7F).
Vicia villosa lectin assays
For VVL pull down assays, cells lysed in buffer A plus protease and phosphatase inhibitors were incubated for 2 hours with VVL agarose beads (Vector Laboratories). Immunoprecipitates were washed four times with buffer A + PMSF and then boiled in 2X Laemmli buffer for 7 min. Immunoprecipitates were separated on SDS-PAGE and proteins were transferred onto nitrocellulose. Samples were then probed with the indicated antibodies detailed in Key Resources Table. For E-cadherin immunoprecipitations, cells lysed in buffer A plus protease and phosphatase inhibitors were incubated for 1 hour with anti-E-cadherin antibody (Abcam). Then, lysates were incubated for 2 hours with Protein A Sepharose beads (Thermo Fisher Scientific). Immunoprecipitates were washed four times with buffer A + PMSF and then boiled in 2X Laemmli buffer for 7 min. Immunoprecipitates were separated by SDS-PAGE and proteins were transferred onto nitrocellulose overnight. Next day, membranes were washed in distilled water for 15 min and blocked with 1X carbo-free block (Vector Laboratories) for 30 min at room temperature. Biotinylated VVL (5 μg/ml) was used to probe membranes for 30 min at room temperature. Phosphate buffered saline (1X) with 0.5% Tween was used to wash membranes for 30 min. Membranes were probed with streptavidin HRP (1:500) (Thermo Fisher Scientific) for 1 hour at room temperature in the dark. Clarity ECL (Bio-Rad) treated membranes were imaged and quantified using a Bio-Rad ChemiDoc and Image lab software respectively.
VVL immunostaining
For VVL immunofluorescence staining, TS cells were cultured on glass coverslips for two days. For both intact and detergent permeabilized VVL staining, cells were fixed with 3% paraformaldehyde in 1X PBS for 10 min and then washed three times with 1X PBS. Coverslips for permeabilized VVL staining were treated with 0.1% Triton for 3 mins and then washed three times with 1X PBS. Intact and permeabilized VVL coverslips were blocked with 1X carbo-free block (Vector laboratories) for 30 min at RT. Then, coverslips were incubated with 5 mg/ml of biotinylated VVL (Vector Laboratories) in 1X PBS for 30 min at RT. Next, cells were washed five times with 1X PBS-Tween (0.1%) and then incubated with DAPI (0.1 μg/ml) and 1:500 of Streptavidin-Alexa 488 (Thermo Fisher Scientific) or Streptavidin-Alexa 594 for one hour at RT. Coverslips were washed and mounted on slides with mounting media and imaged. For co-localization of Golgi and VVL, cells were first incubated with anti-Giantin antibody (Abcam) overnight at 4C and the next day VVL staining was performed.
Blastocyst analysis
Wild-type blastocysts were isolated from timed matings of pure 129/SvEv males with females in the pure 129/SvEv and 129/SvEv/C57B6 mixed backgrounds. Mid-stage blastocysts were hatched by transfer into drops of Acid-tyrodes (Millipore Sigma) for two min. The trophectoderm layer of the hatched, intact blastocysts was infected as previously described (Georgiades et al., 2007). Briefly, 100 μL drops of lentiviruses encoding shControl or shGalnt3 resuspended in KSOM-AA (Millipore Sigma) were used to infect hatched blastocysts for 6 hours. Post infection, blastocysts were transferred into KSOM-AA for 24 hours. Attachment of the blastocysts was assessed after transfer of infected blastocysts into 70% TS medium supplemented with FGF4.
For measurement of puromycin resistance gene expression, RNA was isolated five days post infection from attached and differentiated trophoblast cells in shControl and shGalnt3 using RNeasy micro kit (QIAGEN). High-Capacity reverse transcription kit (Thermo Fisher Scientific) was used to prepare cDNA. PCR reactions for puromycin resistance gene encoded by the lentiviruses were performed. PCR products were run on a 2% agarose gels and imaged using Bio-Rad ChemiDoc.
Blastocyst immunostaining
Wild-type 129/SvEv mice were euthanized 3.5 days post-mating. The uterus was isolated and washed in KSOM-HEPES buffer (Milli-pore Sigma). The utero-tubule junctions were cut and the uterine horns were flushed using a 25 gauge needle filled with 0.4 mL of M2 medium (Millipore Sigma). Using a mouth controlled Pasteur pipette, blastocysts were collected and transferred into a drop of PBS. Blastocysts were hatched by serial passage through drops of Acid Tyrodes (Millipore Sigma). Hatched blastocysts were transferred into drops of PBS, and then fixed in 3% paraformaldehyde and VVL immunofluorescence staining was performed as described under Vicia villosa lectin assays. Blastocyst images were obtained using EVOS epifluorescence and Nikon A1 laser scanning confocal microscope. Immunostaining of the trophectoderm of intact, infected blastocysts was performed 48 hours post-infection. The shControl or shGalnt3 infected blastocysts were fixed with 3% paraformaldehyde and blocked for 30 min using 1X carbofree block. Blastocysts were stained for 1 hour using VVL-FITC (Vector Laboratories) at room temperature. The VVL-FITC stained blastocysts were imaged at 20X using EVOS microscopy.
Chromatin IP coupled to qPCR
ChIP was performed as previously described (Abell et al., 2011). Sonicated samples were immunoprecipitated (IP) with 5 μg of anti-HDAC6 antibody (Cell Signaling Technology) or 5 mg of anti-H2BK5Ac antibody (Abcam). The Bio-Rad CFX96 Touch qPCR system with SsoFast (Bio-Rad) was used to measure promoter enrichment. Primers used for ChIP coupled to qPCR (ChIP-PCR) are specified in Key Resources Table.
H2BK5Ac ChIP-seq read density plot analysis
Anti-H2BK5Ac ChIP-seq data from GEO: GSE92426 aligned to the mm9 reference genome was visualized as in Mobley et al. (2017) using Easeq. Aligned data files were deposited as Datasets and annotated using the mouse mm9 genome Geneset. The FillTrack utility in Easeq software was used to display the read density relative to input at the Galnt3 transcription start site + 20 kb. The units of the y axis are normalized, binned read counts calculated as specified in Lerdrup et al (2016).
Transwell solute flux assay
Barrier assays were performed as previously described (Mobley et al., 2017). Briefly, TS cells were plated in 24 well 0.4 μm pore size
PET membrane Transwells (TSKI4Gab cells) or polycarbonate membrane transwells (TSWTGsh cells) (Corning Life Sciences). Cells were grown to confluence for three days and then dye diffusion was measured using the Synergy H1MD plate reader (BioTek).
Invasion assay
Invasion assays were performed as previously described (Abell et al., 2011). Briefly, TS cells were plated on growth factor reduced Matrigel coated 8 μm pore transwell (TW) chambers. After 48 hours, invasion assays were terminated. Non-invading cells from the top of the TW were removed using swabs and 1X PBS washes. Invasive cells at the bottom of the TW were fixed in 3% paraformaldehyde for 10 min and stained with DAPI (2 μg/ml) for 30 mins. Five 10X fields for each TW were imaged using EVOS epifluorescence microscope. These images were manually counted and graphs were plotted using Prism7 (version 7.0, GraphPad Prism software).
Live cell movies
Cells were cultured in 6-well tissue culture dishes. After 48 hours, cells were fed and live cell imaging was performed using a Lionheart FX automated live cell imager (BioTek). Beacons were set for each cell type using Gen5 software. The LionHeart FX captured 3 × 3 20X montages at 5 min intervals for 21–28 hours for each beacon. Using BioTek Gen5 (version 3.02) software, montages were stitched together and movies were created.
QUANTITATION AND STATISTICAL ANALYSIS
Statistical methods
The statistical details of the experiments including statistical tests used, value of n, and what n represents, can be found in the figure legends. A *p value < 0.05, **p value < 0.01, ***p value < 0.001, and ****p value < 0.0001 were considered statistically significant. Softwares used to calculate statistical analyses are detailed below.
For qPCR data, the Bio-Rad CFX Maestro software (version 3.0) was used to perform a Student’s t test corrected for multiple comparisons. For all other statistics shown, Prism7 (version 7.0, GraphPad software) was used to perform a Student’s t test corrected for multiple comparisons or ANOVA with Bonferroni’s Multiple Comparison test was used to analyze VVL quantitation.
Breast cancer data statistical analysis
For the analysis of breast cancer cell line data from Neve et al. (2006) the molecular subtypes were assigned by Prat et al. (2010) based on gene expression signatures. Of the 52 cell lines analyzed, 49 were classified into molecular subtypes by hierarchical clustering by Prat et al. (2010): 25 as luminal, 15 as basal-like, and 9 as claudin-low. Using the “stats” package in “R,” the following analyses were performed on the 49 classified cell lines (R Core Development Team, 2013). The significance of differences in expression of Galnt3 between luminal, basal-like, and claudin-low subtypes was assessed using the Student’s t test with Bonferroni’s correction for multiple comparisons (pairwise.t.test()). Linear regression analysis (lm()) was used to model the relationship between Galnt3 and Cdh1 gene expression across the 49 classified breast cancer cell lines.
VVL staining quantitation
TS cells were plated for two days and cells were lifted using enzyme free PBS based cell dissociation solution (Millipore Sigma). Cells were centrifuged at 1000 rpm for 3 min and cell pellets were washed in 1X PBS. For both intact and detergent permeabilized cells, VVL staining was performed as described under Immunofluorescence staining. However, all procedures were performed on ice. The stained cells were resuspended in 400 μL 2% FBS in 1X PBS and 100 μL of the resuspended cells were plated in a 96 well plate. Immunofluorescence staining for cell surface and permeabilized VVL (green) was measured using a LionHeart FX automated live cell imager (BioTek). For quantitation, beacons were set for each cell type using BioTek Gen5 (version 3.02) software. The LionHeart FX captured 3 × 3 20X montages for DAPI (Blue) and VVL (Green) for each cell type. Using BioTek Gen5 (version 3.02) software, montages were stitched together and cellular analysis was performed. Parameters used for quantitation included total cell count, integrated green intensity (Total VVL intensity) and percentage of cells positive for green (VVL). Data were plotted using GraphPad Prism7.
NIS-Elements
For E-cadherin and Golgi quantitation, immunofluorescence images were captured using Nikon A1 laser scanning confocal microscopy. NIS-Elements Advanced Research software (version 4.51.00) was used to measure the intensity of E-cadherin (green) in Golgi (red). To measure the intensity of E-cadherin in the Golgi, the Region of Interest Tool was used to select the Golgi area for each cell. Then, the Perform Measurements Tool was used to determine the intensity of E-cadherin (green) in Golgi (red) for each cell. Mean intensity of E-cadherin in Golgi per cell was analyzed and plotted using GraphPad Prism7 software.
For blastocyst VVL-FITC quantitation, NIS analysis elements software was used to measure the intensity of VVL-FITC (green) in the trophectoderm. To measure the intensity of VVL, the Region of Interest Tool was used to select the trophectoderm area and the Perform Measurements Tool was used to determine the intensity. The VVL-FITC intensity for the blastocysts was analyzed and plotted using GraphPad Prism7 (version 7.0).
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Alexa Fluor Phalloidin 594 | Thermo Fisher Scientific | Cat#A12381; RRID: AB_2315633 |
| Alexa Fluor 488-conjugated Streptavidin | Jackson ImmunoResearch Labs | Cat#016-540-084; RRID: AB_2337249 |
| Donkey anti-mouse peroxidase conjugate | Jackson ImmunoResearch Labs | Cat#715-035-151; RRID: AB_2340771 |
| Donkey anti-rabbit peroxidase conjugate | Jackson ImmunoResearch Labs | Cat#711-035-152; RRID: AB_10015282 |
| Goat anti-mouse polyclonal Daylight 488 | Thermo Fisher Scientific | Cat#35503; RRID: AB_1965946 |
| Goat anti-rabbit polyclonal Daylight 488 | Thermo Fisher Scientific | Cat#35553; RRID: AB_1965947 |
| Mouse monoclonal anti-claudin-6 | Santa Cruz Biotechnology | Cat#sc-393671 |
| Mouse monoclonal anti-E-cadherin | BD Biosciences | Cat#610181; RRID: AB_397580 |
| Mouse monoclonal anti-Flag | Thermo Fisher Scientific | Cat#MA1–91878; RRID: AB_1957945 |
| Mouse monoclonal anti-GAPDH | Thermo Fisher Scientific | Cat#AM4300; RRID: AB_2536381 |
| Mouse monoclonal anti-Tubulin | Santa Cruz Biotechnology | Cat#sc-53646; RRID: AB_630403 |
| Mouse monoclonal anti-Tubulin | Sigma-Aldrich | Cat#T6793; RRID: AB_477585 |
| Mouse monoclonal anti-Vinculin | Abcam | Cat#ab18058; RRID: AB_444215 |
| Rabbit polyclonal Alexa 594 | Cell Signaling Technology | Cat#8889; RRID: AB_2716249 |
| Rabbit monoclonal anti-Alpha 1 catenin | Abcam | Cat#ab51032; RRID: AB_868700 |
| Rabbit monoclonal anti-Delta 1 catenin | Abcam | Cat#ab92514; RRID: AB_10565040 |
| Rabbit monoclonal anti-E-cadherin | Abcam | Cat#ab212059 |
| Rabbit monoclonal anti-H2BK5Ac | Active Motif | Cat#39123; RRID: AB_2615079 |
| Rabbit monoclonal anti-HDAC6 | Cell Signaling Technology | Cat#7612; RRID: AB_10889735 |
| Rabbit monoclonal anti-IgG | Abcam | Cat#ab172730; RRID: AB_2687931 |
| Rabbit monoclonal anti-Lamin B1 | Abcam | Cat#ab133741; RRID: AB_2616597 |
| Rabbit polyclonal anti-ERK2 | Santa Cruz Biotechnology | Cat#sc-154; RRID: AB_2141292 |
| Rabbit polyclonal anti-GALNT3 | Abgent | Cat#AP9208C; RRID: AB_10612485 |
| Rabbit polyclonal anti-Giantin | Abcam | Cat#ab24586; RRID: AB_448163 |
| Rabbit polyclonal anti-HDAC6 | Millipore Sigma | Cat#07–732; RRID: AB_441966 |
| Rabbit polyclonal anti-IgG | Abcam | Cat#ab171870; RRID: AB_2687657 |
| Rabbit polyclonal anti-ZO-1 | Thermo Fisher Scientific | Cat#40–2200; RRID: AB_2533456 |
| Biotinylated Vicia Villosa Lectin | Vector Laboratories | Cat#B-1235; RRID: AB_2336855 |
| Peroxidase-streptavidin | Jackson ImmunoResearch Labs | Cat#016-030-084; RRID: AB_2337238 |
| Bacterial and Virus Strains | ||
| Mouse Galnt3 shRNA (Gsh1) | Open Biosystems | TRCN0000055098 |
| Mouse Galnt3 shRNA (Gsh2) | Open Biosystems | TRCN0000055099 |
| Human Galnt3 shRNA (Gsh1) | Dharmacon | TRCN0000035456 |
| Human Galnt3 shRNA (Gsh2) | Dharmacon | TRCN0000035458 |
| Entry Vector Human Galnt3 | Vidal Human ORFeome (Version 5.1) | Clone# 55179 BCII3565 in pDONR223 |
| pENTR-Cdh1 | Cachat et al., 2014 | Addgene Plasmid#49776; RRID: Addgene_49776 |
| Lentiviral FLAG tagged destination vector | Kind Gift from Dr. Gary Johnson (Jordan et al., 2013) | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| 2-mercaptoethanol (BME) | Thermo Fisher Scientific | Cat#31350010 |
| Agarose bound Vicia Villosa Lectin beads | Vector Laboratories | Cat#AL-1233; RRID: AB_2336854 |
| Enzyme Free Cell Dissociation Solution PBS Based (1X) | Millipore Sigma | Cat#S-014-C |
| Fetal Bovine Serum-GIBCO | Thermo Fisher Scientific | Cat#10437–028 |
| Hyclone Glutamine | Thermo Fisher Scientific | Cat#SH3003401 |
| Heparin | Sigma-Aldrich | Cat#H3149–10KU; CAS: 9041-08-1 |
| Hyclone Penicillin-Streptomycin | Thermo Fisher Scientific | Cat#SV30010 |
| Recombinant Human FGF4 | Peprotech | Cat#100–31 |
| Recombinant Protein A Sepharose 4B | Thermo Fisher Scientific | Cat#101141 |
| Sodium pyruvate | Thermo Fisher Scientific | Cat#SH30239.01 |
| Trypsin (0.05%) | Thermo Fisher Scientific | Cat#SH30236.01 |
| Insulin, Human Recombinant Zinc Solution | Thermo Fisher Scientific | Cat#12585014 |
| Critical Commercial Assays | ||
| iTaq Universal SYBR Green Supermix | Bio-Rad | Cat#1725125 |
| Rneasy Plus Mini Kit | QIAGEN | Cat#74134 |
| Rneasy Micro Kit | QIAGEN | Cat#74004 |
| High capacity cDNA Reverse Transcription Kit | Thermo Fisher Scientific | Cat#4368813 |
| Gateway LR Clonase II Enzyme mix | Invitrogen | Cat#11791 −020 |
| Corning BioCoat Matrigel Invasion Chamber: With GFR Matrigel Matrix | Thermo Fisher Scientific | Cat#08-774-193 |
| SsoAdvanced Universal SYBR Green Supermix | Bio-Rad | Cat#1725274 |
| GeneJet Plasmid Miniprep Kit | Thermo Fisher Scientific | Cat#K0502 |
| Transwells 6.4mm with 0.4 mm pore polycarbonate | Corning | Cat#3413 |
| Transwells 6.4mm with 0.4 mm pore polyethylene terephthalate | Corning | Cat#353095 |
| Q5 Site-Directed Mutagenesis Kit | New England BioLabs | Cat#E0554S |
| Experimental Models: Cell Lines | ||
| Human: HEK293T | Kind Gift from Dr. Gary Johnson | N/A |
| Human: Human Mammary Epithelial cells | Kind Gift from Dr. Gary Johnson | N/A |
| Human: SUM159 Claudin-low breast cancer cells | Kind Gift from Dr. Gary Johnson | N/A |
| Mouse: MAP3K4 KI4 Trophoblast stem cells | Abell et al., 2009 | N/A |
| Mouse: Wild-type Trophoblast stem cells | Abell et al., 2009 | N/A |
| Experimental Models: Organisms/Strains | ||
| 129/SvEv strain Wild-type Females | Abell et al., 2005, 2009 | N/A |
| Oligonucleotides | ||
| See Table S1 for primer sequences | This paper | Table S1 |
| Software and Algorithms | ||
| Bio-Rad CFX Maestro | CFX Manager (Version 3) | http://www.bio-rad.com/en-us/product/cfx-maestro-software-for-cfx-real-time-pcr-instruments?ID=OKZP7E15 |
| BioTek Lionheart FX (Live cell and VVL quantitation) | BioTek Gen5 (Version 3.2) | https://www.biotek.com/products/software-robotics/ |
| Easeq | Lerdrup et al., 2016 | https://easeq.net/ |
| GraphPad Prism (Plot Graphs and Statistical analysis) | Graph Pad Prism Software (Version 7) | https://www.graphpad.com/scientific-software/prism/; RRID: SCR_002798 |
| NIS-Elements Advanced Research | NIS-Elements AR software (Version 4.51.00) | https://www.microscope.healthcare.nikon.com/products/software; RRID: SCR_014329 |
| Stats package in R | R Development Core Team (2013) | http://www.R-project.org/; RRID: SCR_001905 |
| Other | ||
| EmbryoMax Acidic Tyrode’s Solution | Millipore Sigma | Cat#MR-004-D |
| Hyclone Dulbecco’s Modified Eagles Medium | Thermo Fisher Scientific | Cat#SH3024301 |
| Ham’s F12 Nutrient Mix | Thermo Fisher Scientific | Cat#11765047 |
| HuMEC Basal Serum-Free Medium (1X) | Thermo Fisher Scientific | Cat#12753018 |
| HuMEC Supplement Kit | Thermo Fisher Scientific | Cat#12755013 |
| KSOM Powdered Embryo Culture Medium | Millipore Sigma | Cat#MR-020P-5D |
| EmbryoMax KSOM+AA with D-Glucose and Phenol Red | Millipore Sigma | Cat#M-121-D |
| EmbryoMax M2 Medium (1X), liquid, with Phenol Red | Millipore Sigma | Cat#MR-015-D |
| Hyclone RPMI-1640 medium (1X) | Thermo Fisher Scientific | Cat#SH3002701 |
Highlights.
TS cells express high levels of cell surface-unextended O-GalNAc-modified proteins
HDAC6 represses the expression of GALNT3
Loss of GALNT3 in TS cells, blastocyst trophectoderm, and HMECs induces EMT
GALNT3 promotes O-GalNAc modification and cell surface localization of E-cadherin
ACKNOWLEDGMENTS
A.N.A. is supported by the Memphis Research Consortium and by an NIH grant (GM116903). D.R. is supported by the Dr. Bill Simco Graduate Student Scholarship. We thank Dr. Omar Skalli and the Integrated Microscopy Center for use of Nikon A1 confocal microscope.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found with this article online at https://doi.org/10.1016/j.celrep.2019.02.093.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Abell AN, Rivera-Perez JA, Cuevas BD, Uhlik MT, Sather S, Johnson NL, Minton SK, Lauder JM, Winter-Vann AM, Nakamura K, et al. (2005). Ablation of MEKK4 kinase activity causes neurulation and skeletal patterning defects in the mouse embryo. Mol. Cell. Biol 25, 8948–8959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abell AN, Granger DA, Johnson NL, Vincent-Jordan N, Dibble CF, and Johnson GL (2009). Trophoblast stem cell maintenance by fibroblast growth factor 4 requires MEKK4 activation of Jun N-terminal kinase. Mol. Cell. Biol 29, 2748–2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abell AN, Jordan NV, Huang W, Prat A, Midland AA, Johnson NL, Granger DA, Mieczkowski PA, Perou CM, Gomez SM, et al. (2011). MAP3K4/CBP-regulated H2B acetylation controls epithelial-mesenchymal transition in trophoblast stem cells. Cell Stem Cell 8, 525–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaman EM, and Brooks SA (2014). The extended ppGalNAc-T family and their functional involvement in the metastatic cascade. Histol. Histopathol 29, 293–304. [DOI] [PubMed] [Google Scholar]
- Bennett EP, Mandel U, Clausen H, Gerken TA, Fritz TA, and Tabak LA (2012). Control of mucin-type O-glycosylation: a classification of the poly-peptide GalNAc-transferase gene family. Glycobiology 22, 736–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cachat E, Liu W, Hohenstein P, and Davies JA (2014). A library of mammalian effector modules for synthetic morphology. J. Biol. Eng 8, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia J, Goh G, and Bard F (2016). Short O-GalNAc glycans: regulation and role in tumor development and clinical perspectives. Biochim. Biophys. Acta 1860, 1623–1639. [DOI] [PubMed] [Google Scholar]
- Chugh S, Meza J, Sheinin YM, Ponnusamy MP, and Batra SK (2016). Loss of N-acetylgalactosaminyltransferase 3 in poorly differentiated pancreatic cancer: augmented aggressiveness and aberrant ErbB family glycosylation. Br. J. Cancer 114, 1376–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esapa CT, Head RA, Jeyabalan J, Evans H, Hough TA, Cheeseman MT, McNally EG, Carr AJ, Thomas GP, Brown MA, et al. (2012). A mouse with an N-Ethyl-N-nitrosourea (ENU) Induced Trp589Arg Galnt3 mutation represents a model for hyperphosphataemic familial tumoural calcinosis. PLoS One 7, e43205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiades P, Cox B, Gertsenstein M, Chawengsaksophak K, and Rossant J (2007). Trophoblast-specific gene manipulation using lentivirus-based vectors. Biotechniques 42, 317–318, 320,, 322–315. [DOI] [PubMed] [Google Scholar]
- Gumbiner BM (1996). Cell adhesion: the molecular basis of tissue architecture and morphogenesis. Cell 84, 345–357. [DOI] [PubMed] [Google Scholar]
- Ichikawa S, Sorenson AH, Austin AM, Mackenzie DS, Fritz TA, Moh A, Hui SL, and Econs MJ (2009). Ablation of the Galnt3 gene leads to low-circulating intact fibroblast growth factor 23 (Fgf23) concentrations and hyper-phosphatemia despite increased Fgf23 expression. Endocrinology 150, 2543–2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamal BT, Nita-Lazar M, Gao Z, Amin B, Walker J, and Kukuruzinska MA (2009). N-glycosylation status of E-cadherin controls cytoskeletal dynamics through the organization of distinct β-catenin- and g-catenin-containing AJs. Cell Health Cytoskelet 2009, 67–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan NV, Prat A, Abell AN, Zawistowski JS, Sciaky N, Karginova OA, Zhou B, Golitz BT, Perou CM, and Johnson GL (2013). SWI/SNF chromatin-remodeling factor Smarcd3/Baf60c controls epithelial-mesenchymal transition by inducing Wnt5a signaling. Mol. Cell. Biol 33, 3011–3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerdrup M, Johansen JV, Agrawal-Singh S, and Hansen K (2016). An interactive environment for agile analysis and visualization of ChIP-sequencing data. Nat. Struct. Mol. Biol 23, 349–357. [DOI] [PubMed] [Google Scholar]
- Maupin KA, Sinha A, Eugster E, Miller J, Ross J, Paulino V, Keshamouni VG, Tran N, Berens M, Webb C, and Haab BB (2010). Glycogene expression alterations associated with pancreatic cancer epithelial-mesenchymal transition in complementary model systems. PLoS One 5, e13002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mobley RJ, Raghu D, Duke LD, Abell-Hart K, Zawistowski JS, Lutz K, Gomez SM, Roy S, Homayouni R, Johnson GL, and Abell AN (2017). MAP3K4 Controls the Chromatin Modifier HDAC6 during Trophoblast Stem Cell Epithelial-to-Mesenchymal Transition. Cell Rep 18, 2387–2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nairn AV, Aoki K, dela Rosa M, Porterfield M, Lim JM, Kulik M, Pierce JM, Wells L, Dalton S, Tiemeyer M, and Moremen KW (2012). Regulation of glycan structures in murine embryonic stem cells: combined transcript profiling of glycan-related genes and glycan structural analysis. J. Biol. Chem 287, 37835–37856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, Clark L, Bayani N, Coppe JP, Tong F, et al. (2006). A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell 10, 515–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattabiraman DR, Bierie B, Kober KI, Thiru P, Krall JA, Zill C, Reinhardt F, Tam WL, and Weinberg RA (2016). Activation of PKA leads to mesenchymal-to-epithelial transition and loss of tumor-initiating ability. Science 351, aad3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinho SS, and Reis CA (2015). Glycosylation in cancer: mechanisms and clinical implications. Nat. Rev. Cancer 15, 540–555. [DOI] [PubMed] [Google Scholar]
- Poirier F, and Kimber S (1997). Cell surface carbohydrates and lectins in early development. Mol. Hum. Reprod 3, 907–918. [DOI] [PubMed] [Google Scholar]
- Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI, He X, and Perou CM (2010). Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res 12, R68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeichi M (2014). Dynamic contacts: rearranging adherens junctions to drive epithelial remodelling. Nat. Rev. Mol. Cell Biol 15, 397–410. [DOI] [PubMed] [Google Scholar]
- Tanaka S, Kunath T, Hadjantonakis AK, Nagy A, and Rossant J (1998). Promotion of trophoblast stem cell proliferation by FGF4. Science 282, 2072–2075. [DOI] [PubMed] [Google Scholar]
- R Development Core Team. (2013). R: A language and environment for statistical computing R. Foundation for Statistical Computing. [Google Scholar]
- Thiery JP, Acloque H, Huang RY, and Nieto MA (2009). Epithelialmesenchymal transitions in development and disease. Cell 139, 871–890. [DOI] [PubMed] [Google Scholar]
- Tian E, and Ten Hagen KG (2007). A UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase is required for epithelial tube formation. J. Biol. Chem 282, 606–614. [DOI] [PubMed] [Google Scholar]
- Tian E, Hoffman MP, and Ten Hagen KG (2012). O-glycosylation modulates integrin and FGF signalling by influencing the secretion of basement membrane components. Nat. Commun 3, 869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vester-Christensen MB, Halim A, Joshi HJ, Steentoft C, Bennett EP, Levery SB, Vakhrushev SY, and Clausen H (2013). Mining the O-mannose glycoproteome reveals cadherins as major O-mannosylated glycoproteins. Proc. Natl. Acad. Sci. USA 110, 21018–21023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZQ, Bachvarova M, Morin C, Plante M, Gregoire J, Renaud MC, Sebastianelli A, and Bachvarov D (2014). Role of the polypeptide N-acetylgalactosaminyltransferase 3 in ovarian cancer progression: possible implications in abnormal mucin O-glycosylation. Oncotarget 5, 544–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, and Weinberg RA (2008). Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev. Cell 14, 818–829. [DOI] [PubMed] [Google Scholar]
- Zhu W, Leber B, and Andrews DW (2001). Cytoplasmic O-glycosylation prevents cell surface transport of E-cadherin during apoptosis. EMBO J 20, 5999–6007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.