Introduction
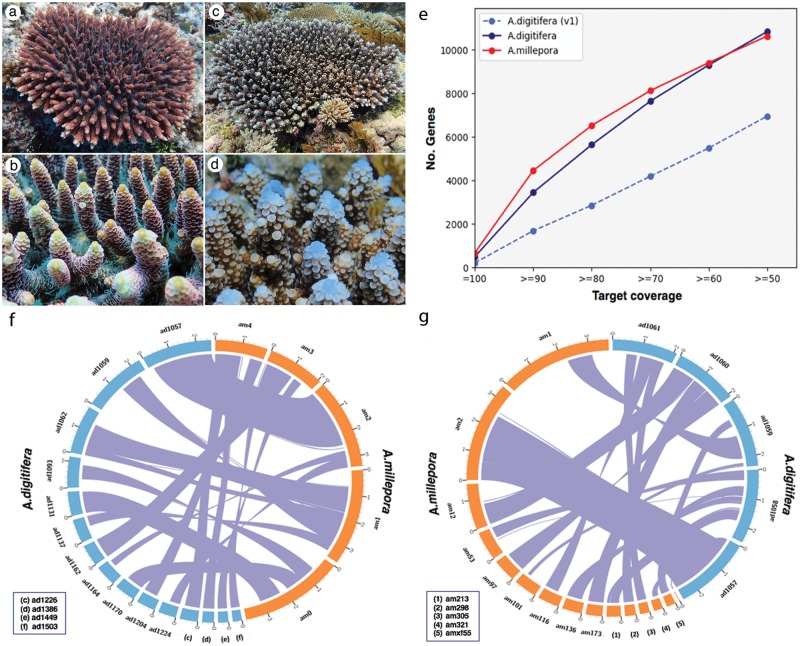
Reef-building corals are iconic animals that are in global decline as a consequence of increasing anthropogenic pressure, but the development of strategies to ensure their conservation is constrained by our limited understanding of the molecular bases of many aspects of coral biology. Some coral genera are particularly sensitive to stress and, among these, Acropora is of particular significance because this is the dominant genus of reef-building corals in the Indo-Pacific. These factors have led to members of this genus often being the subjects of investigation into coral responses to various physical and biological stressors. Fittingly, the first coral genome to be sequenced was Acropora digitifera; the availability of this whole-genome sequence (Shinzato et al. 2011) allowed substantial progress in several areas of coral biology, including the molecular underpinnings of symbiosis and calcification (Hamada et al. 2013; Ramos-Silva et al. 2013). Here we report the whole-genome sequence of a second Acropora species, A. millepora, which has been the most extensively studied Acropora species at the molecular level (reviewed in Miller et al. 2011) by virtue of its wide distribution (Carpenter et al. 2008; Madin et al. 2016) and the ease with which it can be identified in what is a highly speciose genus. Despite being classified on the basis of skeletal characteristics into different species groups sensu Wallace and Wolstenholme (1998), molecular data indicate that A. millepora and A. digitifera are close relatives (e.g., van Oppen et al. 2001) that have diverged since the Oligocene (Santodomingo et al. 2015). The two species are shown in figure 1a–d .
Fig. 1.
—(a–d) Images of the corals whose genomes were compared. (a) An Acropora millepora colony and (b) a close up view of an A. millepora colony. (c) An Acropora digitifera colony and (d) a corresponding close up. Note that the colors are not taxonomically relevant and often vary between colonies. (e) Target coverage of predicted proteins matching to Swiss-Prot database proteins. The A. digitifera (v1) gene model was obtained from Shinzato et al. (2011). (f, g) Circos plots showing relationships between the A. millepora and A. digitifera genomes. The longest five reference scaffolds from (f) A. millepora (orange) or (g) A. digitifera (blue) are arranged around the circumference of the figure. For each reference scaffold, the top three scaffolds containing the most alignments in the other genome are shown. Each purple line crossing the circle represents a unique alignment and the units on the periphery represent 1 Mb. To facilitate display, the scaffold names were shortened as “amil.Sc0000000” to am0, “amil.Sc0000001” to am1, and so on for A. millepora; “NW_015441060.1” to ad1060, “NW_015441061.1” to ad1061, and so on for A. digitifera.
Among corals, early development has been most extensively documented in A. millepora (e.g., Hayward et al. 2002, 2004, 2015) and molecular technologies, including in situ hybridization (e.g., Grasso et al. 2008; Shinzato et al. 2008) and CRISPR/Cas9 (Cleves et al. 2018) are most advanced in their development in this species. Although large RNAseq data sets (Meyer et al. 2011) and a comprehensive transcriptome assembly (Moya et al. 2012) have been available for A. millepora for some time, a genome assembly has not, a situation that is redressed with this publication. In terms of completeness, the genome assembly and associated gene predictions are of similar quality to the recent NCBI-generated version (2.0) of the A. digitifera genome. Despite high heterozygosity (∼2%), the two species show remarkably low divergence at the whole-genome level; average transcript (coding sequence [CDS]) identity was ∼98% and across the whole genome ∼95%. To facilitate access, we have provided a genome browser.
Materials and Methods
DNA Sample Collection, Extraction, and Sequencing
Sperm were collected from a single A. millepora colony at Magnetic Island, Queensland (19°08′S, 146°50′E), snap frozen in liquid nitrogen and stored at −80 °C until needed. High-molecular-weight genomic DNA was prepared using a method based on that described by Blin and Stafford (1976). Paired end (PE) and mate pair (MP) libraries were prepared and sequenced on an Illumina Genome Analyzer IIx at the Australian Genome Research Facility. pFosill vectors were supplied by Andreas Gnirke and MP fosill libraries were constructed as described by Williams et al. (2012). The DNA sequencing libraries with Short Read Archive (SRA) accession numbers are listed in supplementary table S1, Supplementary Material online.
RNA Sample Collection, Extraction, and Sequencing
During the annual spawning event of November 2010, A. millepora embryos were raised at the James Cook University research station on Orpheus Island (18°39′52″S, 146°29′42″E) under GBRMPA permit G09/30327.1. In addition to unfertilized eggs and adult samples, samples from six early life history stages were collected. They were donut (gastrula), sphere (post-gastrula), planula, spindle (late planula), settled, and metamorphosed. Samples were snap frozen in liquid nitrogen and stored at −80 °C until required. RNA was extracted as previously described (Moya et al. 2012). TruSeq stranded mRNA libraries were prepared and sequenced on an Illumina HiSeq2000 by Macrogen Inc., South Korea. The RNA sequencing libraries with SRA accession numbers are listed in supplementary table S2, Supplementary Material online.
Genome Assembly
Prior to genome assembly, quality check and trimming were performed on raw sequencing reads. A preliminary assembly was first conducted using Velvet (Zerbino and Birney 2008). The insert sizes of PE and MP libraries were estimated by read mapping to the selected contigs. This information (supplementary table S3, Supplementary Material online) was used to generate the genome assembly by ALLpath-LG v45633 (Gnerre et al. 2011). We further applied HaploMerger (Huang et al. 2012) and GapCloser v1.12-r6 (Luo et al. 2012) to remove duplicated haplotypes and do scaffolding. The mitochondrial genome sequence was identified from the assembly and compared with known Acropora mitochondrial genomes. Detailed descriptions of assembly methods are provided in Supplementary Materials online.
Genome Annotation
De novo identification of repetitive elements was conducted on the A. millepora genome assembly. To facilitate gene prediction, a high-quality training gene set was produced by transcriptome assembly, Open Reading Frame prediction, filtering, and refinement through a series of criteria. The MAKER2 (Holt and Yandell 2011) annotation pipeline was carried out to generate a protein-coding gene model based on transcript hints, homology, and de novo prediction. Functional annotation was performed by homology searching to match predicted proteins to the PFAM-A protein domain and the Kyoto Encyclopedia of Genes and Genomes (Kanehisa et al. 2017) databases. Detailed description of genome annotation is provided in Supplementary Materials online.
Assessment of the Genome Assembly and Gene Model Data Set
The completeness of the genome assembly and gene model were assessed using the CEGMA (Parra et al. 2009) and BUSCO (Waterhouse et al. 2018) programs. The accuracy of predicted proteins was evaluated by the coverage of matching homologous proteins from the Swiss-Prot database. A detailed description of the assessment process is provided in Supplementary Materials online.
Comparative Genomic Analyses
The updated A.digitifera genome assembly (2.0, NCBI accession GCF_000222465.1) and annotation (NCBI annotation release 100) were downloaded from the NCBI FTP site (ftp://ftp.ncbi.nih.gov/genomes/Acropora_digitifera; last accessed February 28, 2019). Whole-genome alignments between A. millepora and A. digitifera were performed using the NUCmer module of MUMmer v4.0.0beta2 (Marçais et al. 2018) with the default parameters. Alignments with <75% identity were removed. The result was summarized by the dnadiff module of MUMmer and visualized by Circos (Krzywinski et al. 2009).
Results and Discussion
Genome Assembly
The genome of A.millepora (fig. 1 a,b) was sequenced and assembled using a whole-genome shotgun sequencing approach based 140.6 million (PE, MP, and fosmid) paired-end reads (supplementary table S1, Supplementary Material online). The insert sizes ranged between 150 bp and 35 kb. After quality trimming and filtering, a total of 88.5 Gb (∼210× coverage) of sequence data was retained for the final assembly (supplementary table S3, Supplementary Material online). The estimated genome size was between 371 and 454 Mb from k-mer analysis (supplementary table S4, Supplementary Material online), which is in line with the 420-Mb genome size estimate for A. digitifera based on flow cytometry (Shinzato et al. 2011). The genome is highly heterozygous with the estimated SNP rate of ∼2.0% by GenomeScope (supplementary table S4 and fig. S1, Supplementary Material online). The genome assembly statistics (table 1) show better contiguity than the genome assembly by Shinzato et al. (2011) and are close to the NCBI updated version. Unclosed gaps (Ns) comprise 9.72% of the genome sequence and the average GC content is 38.85% (table 1 and supplementary table S5, Supplementary Material online).
Table 1.
Comparison of assembly and annotation statistics for the Acropora millepora and Acropora digitifera genomes
| A. millepora | A. digitifera | ||
|---|---|---|---|
| Assembled genome size (Mb)a | 386.60 | 447.48 | |
| Scaffolds | Number | 3,876 | 2,420 |
| N50 (kb) | 494 | 483 | |
| Largest (kb) | 3,800 | 2,549 | |
| GC% | 38.85 | 39.04 | |
| Number of genesb | 26,615 | 26,060 | |
| Repeatsc | Total repeat (%) | 34.55 | 32.31 |
| Interspersed repeat (%) | 33.46 | 31 | |
| BUSCO: C:P:M (%)d,e | 90:3:7 | 74:11:15 | |
See supplementary table S5, Supplementary Material online, for more detail.
See supplementary table S9, Supplementary Material online, for more detail.
See supplementary table S7, Supplementary Material online, for more detail.
See supplementary table S11, Supplementary Material online, for more detail.
C, P, and M refer to fully represented, partially represented, and missing BUSCO genes.
Anthozoan mitochondrial genomes typically evolve more slowly than the corresponding nuclear genomes (Huang et al. 2008), possibly reflecting DNA repair (Pont-Kingdon et al. 1995). The A. millepora mitochondrial genome was assembled to a single scaffold, whose length is consistent with that from other Acropora spp. (Zhang et al. 2016; supplementary table S6 and fig. S2, Supplementary Material online). Across the genus, levels of nucleotide similarity in the mitochondrial genomes were remarkably high. Of the other Acropora species for which whole mitochondrial sequences are available, A.tenuis is the most divergent (99.32% identity with the A. digitifera reference); this species is always well resolved in molecular phylogenies (see, e.g., van Oppen et al. 2001), and therefore likely to reflect near maximal levels of divergence within the genus (supplementary table S6, Supplementary Material online). For many of the species, apparent differences are below 0.05%. Both the assembled A. digitifera and A. millepora mitochondrial genomes differ from the reference A. digitifera sequence by ∼0.2%, which could reflect assembly artifacts originating from sequencing errors.
Genome Annotation
Prior to gene prediction, de novo repetitive element analyses were carried out on the genome of A. millepora (table 1 and supplementary table S7, Supplementary Material online). A total of 33.44% of the A. millepora genome was made up of interspersed repeats, whereas other repeat classes, including satellite, low complexity and simple repeats account for only 1.11% of the genome sequence. Nearly half of the interspersed repeats (17.85%) could not be explicitly classified by comparison with known repeat databases, and therefore may be cnidarian or coral specific. The classified transposable elements represent over 45 different families and show a slight preference for class I retrotransposons (supplementary table S8, Supplementary Material online).
Based on expressed transcripts, homology and ab initio gene prediction, 26,615 protein-coding genes were annotated in A. millepora (table 1 and supplementary table S9, Supplementary Material online). In total, the annotated genic region comprises 47.59% of the genome, which is close to values typically associated with model organisms (Francis and Wörheide 2017). On average, there are seven exons per gene, and the mean transcript length is 1,818 bp. In total, 16,292 (61.21%) genes have clear homology to proteins in the Swiss-Prot database, with an additional 7,946 (29.86%) genes in the TrEMBL database. Well-defined PFAM-A protein domains were identified in 63.98% of annotated genes, and unambiguous Kyoto Encyclopedia of Genes and Genomes K numbers were assigned to 48.47% of genes. These statistics are in line with the A. digitifera gene models annotated using the sophisticated NCBI Eukaryotic Genome Annotation pipeline (supplementary table S10, Supplementary Material online).
Quality Assessment
To assess the completeness of the conserved core gene set in the genome assembly and gene model data set, the CEGMA and BUSCO pipelines were applied (supplementary table S11, Supplementary Material online). Among the 248 core eukaryotic genes from CEGMA, 65% and 92% of these genes are present in full in the A. millepora genome assembly and predicted transcripts respectively. A larger metazoan gene set containing 978 genes from BUSCO v3 revealed much higher completeness (90.49%) from the assembly perspective, possibly due to the improved searching algorithm. The completeness of putative transcripts is 92.94% which is consistent with the CEGMA outcome. In terms of these statistics, the A. millepora genome data therefore outperform the updated (v2.0) NCBI A. digitifera genome release.
The quality of the gene model data set was further assessed using the manually curated Swiss-Prot protein database, focusing on target coverage only because the query coverage is subject to potential inaccuracies in the lengths of predicted proteins, which are typically unknown. The overall target coverage is similar between putative A. millepora and A. digitifera (2.0) proteins with slightly better performance of A. millepora proteins at the high coverage end (fig. 1e). Meanwhile, the gene model presented here is of considerably higher quality than the v1.0 models provided by Shinzato et al. (2011).
Genome Comparison
Whole-genome alignment of the A. millepora and A. digitifera assemblies confirmed that the two species are closely related. As an initial approach to whole-genome comparison, A. millepora was used as the reference and A. digitifera as the source of query sequences. In total, 98% of A. millepora scaffolds have sequences aligned to all but one of the A. digitifera scaffolds, and 82.35% of A. millepora sequences were aligned to 91.23% of A. digitifera sequences (fig. 1f and g and supplementary table S12, Supplementary Material online). Among these, 82% were aligned uniquely with an average length of 1,550 bp and an average identity of 94.91%. The multiple aligned sequences have a similar average identity of 94.97%. The failure to identify A. digitifera matches for the remaining 17.65% of A. millepora sequences is most likely due to the presence of unclosed gaps (Ns) in both genomes. The reciprocal analyses (i.e., using A. digitifera as the reference and A. millepora as source of query sequences) resulted in very similar outcomes (supplementary table S12, Supplementary Material online).
As another approach to comparing the A. millepora and A. digitifera genome sequences, similarity of protein-coding sequences was evaluated. In total, 16,929 A. millepora CDSs had unambiguous matches to the A. digitifera genome with >100-bp lengths (supplementary table S13, Supplementary Material online), yielding a CDS identity distribution with a mode at 98.38% (supplementary fig. S3, Supplementary Material online). However, the protein alignment on these orthologs based on gene model predictions from each genome suggested much lower levels of similarity that those based on coding sequence and whole-genome alignment. (supplementary fig. S3, Supplementary Material online). This counter intuitive result reflects the generic problems associated with erroneous gene models generated from draft genome assemblies (Zhang et al. 2012; Denton et al. 2014, supplementary fig. S4, Supplementary Material online), resulting in the presence of extensive nonorthologous but aligned regions in the protein alignments. This problem should be thoroughly addressed before assessing gene evolutionary history in future work.
Addendum
During the writing of this article, we became aware of the online availability of an unpublished A millepora draft genome assembly at https://przeworskilab.com/data/, last accessed April 1, 2019.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
This work was supported by the Australian Research Council through the Centre of Excellence for Coral Reef Studies, the Centre for Molecular Genetics of Development and Discovery Grants DP0209460, DP0344483, and DP1095343. The authors also gratefully acknowledge the support of AGRF in providing the sequencing expertise and the resources that enabled the genome assembly. AGRF acknowledges support from Bioplatforms Australia through funding from the National Collaborative Research Infrastructure Strategy. We thank Zoe Richards for the use of the coral photos (fig. 1a–d), and Zoe, Andrew Baird, and Peter Cowman for providing advice on many aspects of coral biology.
Data deposition: This Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession QTZP00000000. The version described in this article is version QTZP01000000. A genome browser is available via http://coralreefgenomes.jcu.edu.au/.
Literature Cited
- Blin N, Stafford DW.. 1976. A general method for isolation of high molecular weight DNA from eukaryotes. Nucleic Acids Res. 3(9):2303–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter KE, et al. 2008. One-third of reef-building corals face elevated extinction risk from climate change and local impacts. Science 321(5888):560–563. [DOI] [PubMed] [Google Scholar]
- Cleves PA, Strader ME, Bay LK, Pringle JR, Matz MV.. 2018. CRISPR/Cas9-mediated genome editing in a reef-building coral. Proc Natl Acad Sci U S A. 115(20):5235–5240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denton JF, et al. 2014. Extensive error in the number of genes inferred from draft genome assemblies. PLoS Comput Biol. 10(12):e1003998.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis WR, Wörheide G.. 2017. Similar ratios of introns to intergenic sequence across animal genomes. Genome Biol Evol. 9(6):1582–1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnerre S, et al. 2011. High-quality draft assemblies of mammalian genomes from massively parallel sequence data. Proc Natl Acad Sci U S A. 108(4):1513–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasso LC, et al. 2008. Microarray analysis identifies candidate genes for key roles in coral development. BMC Genomics. 9:540.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada M, et al. 2013. The complex NOD-like receptor repertoire of the coral Acropora digitifera includes novel domain combinations. Mol Biol Evol. 30(1):167–176. [DOI] [PubMed] [Google Scholar]
- Hayward DC, Grasso LC, Saint RB, Miller DJ, Ball EE.. 2015. The organizer in evolution: gastrulation and organizer gene expression highlight the importance of Brachyury during development of the coral Acropora millepora. Dev Biol. 399:227–247. [DOI] [PubMed] [Google Scholar]
- Hayward DC, Miller DJ, Ball EE.. 2004. Snail expression during embryonic development of the coral Acropora: blurring the diploblast/triploblast divide? Dev Genes Evol. 214(5):257–260. [DOI] [PubMed] [Google Scholar]
- Hayward DC, et al. 2002. Localized expression of a dpp/BMP2/4 ortholog in a coral embryo. Proc Natl Acad Sci U S A. 99(12):8106–8111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt C, Yandell M.. 2011. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics 12:491.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D, Meier R, Todd PA, Chou LM.. 2008. Slow mitochondrial COI sequence evolution at the base of the metazoan tree and its implications for DNA barcoding. J Mol Evol. 66(2):167–174. [DOI] [PubMed] [Google Scholar]
- Huang S, et al. 2012. HaploMerger: reconstructing allelic relationships for polymorphic diploid genome assemblies. Genome Res. 22(8):1581–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K.. 2017. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 45(D1):D353–D361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski MI, et al. 2009. Circos: an information aesthetic for comparative genomics. Genome Res. 19(9):1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo R, et al. 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 1(1):18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madin JS, et al. 2016. The Coral Trait Database, a curated database of trait information for coral species from the global oceans. Sci Data 3:160017.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marçais G, et al. 2018. MUMmer4: a fast and versatile genome alignment system. PLoS Comput Biol. 14(1):e1005944.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer E, Aglyamova GV, Matz MV.. 2011. Profiling gene expression responses of coral larvae (Acropora millepora) to elevated temperature and settlement inducers using a novel RNA‐Seq procedure. Mol Ecol. 20(17):3599–3616. [DOI] [PubMed] [Google Scholar]
- Miller DJ, Ball EE, Forêt S, Satoh N.. 2011. Coral genomics and transcriptomics—ushering in a new era in coral biology. J Exp Mar Biol Ecol. 408(1-2):114–119. [Google Scholar]
- Moya A, et al. 2012. Whole transcriptome analysis of the coral Acropora millepora reveals complex responses to CO2‐driven acidification during the initiation of calcification. Mol Ecol. 21(10):2440–2454. [DOI] [PubMed] [Google Scholar]
- Parra G, Bradnam K, Ning Z, Keane T, Korf I.. 2009. Assessing the gene space in draft genomes. Nucleic Acids Res. 37(1):289–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pont-Kingdon GA, et al. 1995. A coral mitochondrial mutS gene. Nature 375(6527):109–111. [DOI] [PubMed] [Google Scholar]
- Ramos-Silva P, et al. 2013. The skeletal proteome of the coral Acropora millepora: the evolution of calcification by co-option and domain shuffling. Mol Biol Evol. 30(9):2099–2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santodomingo N, Wallace CC, Johnson KG.. 2015. Fossils reveal a high diversity of the staghorn coral genera Acropora and Isopora (Scleractinia: Acroporidae) in the Neogene of Indonesia. Zool J Linn Soc. 175(4):677–763. [Google Scholar]
- Shinzato C, et al. 2008. Sox genes in the coral Acropora millepora: divergent expression patterns reflect differences in developmental mechanisms within the Anthozoa. BMC Evol Biol. 8:311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinzato C, et al. 2011. Using the Acropora digitifera genome to understand coral responses to environmental change. Nature 476(7360):320. [DOI] [PubMed] [Google Scholar]
- van Oppen MJ, McDonald BJ, Willis B, Miller DJ.. 2001. The evolutionary history of the coral genus Acropora (Scleractinia, Cnidaria) based on a mitochondrial and a nuclear marker: reticulation, incomplete lineage sorting, or morphological convergence? Mol Biol Evol. 18(7):1315–1329. [DOI] [PubMed] [Google Scholar]
- Wallace CC, Wolstenholme J.. 1998. Revision of the coral genus Acropora (Scleractinia; Astrocoeniina; Acroporidae) in Indonesia. J Linn Soc. 123(3):199–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse RM, et al. 2018. BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol Biol Evol. 35(3):543–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams LJ, et al. 2012. Paired-end sequencing of Fosmid libraries by Illumina. Genome Res. 22(11):2241–2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino D, Birney E.. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18(5):821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Goodsell J, Norgren RB.. 2012. Limitations of the rhesus macaque draft genome assembly and annotation. BMC Genomics. 13:206.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Yu X, Zhou Z, Huang B.. 2016. The complete mitochondrial genome of Acropora aculeus (Cnidaria, Scleractinia, Acroporidae). Mitochondrial DNA DNA Mapp Seq Anal. 27(6):4276–4277. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.