Abstract
Observations of bacteria at the single-cell level have revealed many instances of phenotypic heterogeneity within otherwise clonal populations, but the selective causes, molecular bases, and broader ecological relevance remain poorly understood. In an earlier experiment in which the bacterium Pseudomonas fluorescens SBW25 was propagated under a selective regime that mimicked the host immune response, a genotype evolved that stochastically switched between capsulation states. The genetic cause was a mutation in carB that decreased the pyrimidine pool (and growth rate), lowering the activation threshold of a preexisting but hitherto unrecognized phenotypic switch. Genetic components surrounding bifurcation of UTP flux toward DNA/RNA or UDP-glucose (a precursor of colanic acid forming the capsules) were implicated as key components. Extending these molecular analyses—and based on a combination of genetics, transcriptomics, biochemistry, and mathematical modeling—we show that pyrimidine limitation triggers an increase in ribosome biosynthesis and that switching is caused by competition between ribosomes and CsrA/RsmA proteins for the mRNA transcript of a positively autoregulated activator of colanic acid biosynthesis. We additionally show that in the ancestral bacterium the switch is part of a program that determines stochastic entry into a semiquiescent capsulated state, ensures that such cells are provisioned with excess ribosomes, and enables provisioned cells to exit rapidly from stationary phase under permissive conditions.
Keywords: experimental evolution, phenotypic heterogeneity, microbiology, genetics
Introduction
Phenotypic variation between isogenic cells can have adaptive consequences, by allowing populations to survive unpredictable environmental change or promoting interactions between different cell types (Ackermann 2015; van Boxtel et al. 2017). Evidence of adaptive significance derives from various sources (Cohen 1966; Slatkin 1974; Meyers and Bull 2002; Simons 2011; Grimbergen et al. 2015), but particularly from a previous study where stochastic phenotype switching was observed to evolve de novo in experimental bacterial populations propagated under a selective regime that mimicked essential features of the host immune response (Beaumont et al. 2009; Libby and Rainey 2011; Rainey et al. 2011). To escape extinction, populations of Pseudomonas fluorescens SBW25 were required to repeatedly alter colony phenotype in order to passage through a single-cell bottleneck en route to an opposing environment. In two replicate lineages (from of a total of 12), genotypes evolved the capacity to stochastically switch between two distinct (opaque and translucent) colony morphologies. Although stochastic colony switching incurred a cost in Malthusian fitness, switching types were shown to be adaptive in the face of unpredictable environmental change. Long-term success was achieved through production of cells capable of generating variation in colony morphology. Such variation increased the likelihood that a subset of cells expressed a phenotype that was adaptive in a future environment.
While defining ecological conditions that promote the evolution of stochastic switching (Libby and Rainey 2011), the de novo evolution of switching behavior necessarily prompted investigation into cellular and molecular origins. For each of the two focal switching genotypes, 1B4 (from Line 1) and 6B4 (from Line 6), variation in colony morphology is determined by variation in the capacity of individual cells to produce capsules (Beaumont et al. 2009): Cap+ cells produce a colanic acid-like polymer (CAP) and give rise to opaque colonies, whereas Cap− cells produce no capsule and generate translucent colonies.
In both 1B4 and 6B4, the mechanism of switching is epigenetic and involves identical regulatory circuitries (this paper and the accompanying paper by Gallie et al. [2019]); however, the mutations generating the switching state are quite different. The switch-causing mutation in 1B4 is a single nonsynonymous mutation in carB (c2020t), which encodes carbamoyl phosphate synthase, involved in de novo pyrimidine biosynthesis. In 6B4, the causal mutation resides in the housekeeping sigma factor rpoD (t1682c). To date attention has focused on the mechanistic bases of switching in 1B4 (Gallie et al. [2015] and this paper), whereas an accompanying paper (Gallie et al. 2019) shows how the two different causal mutations realize a single phenotypic outcome.
The switch between capsulation states is bistable (Beaumont et al. 2009; Gallie et al. 2015, 2019) with both Cap+ and Cap− cells coexisting in any sufficiently large population (Gallie et al. 2015). As expected of a bistable switch, cells of each capsulation state maintain memory of their past phenotype (there is hysteresis), without which distinct opaque and translucent colonies would not manifest. Nonetheless, close investigation of each colony type shows that they are composed of both Cap+ and Cap− cells: with a certain probability each type switches to the other, and yet maintains memory of the new state for sufficient generations to produce distinct lineages of cell types (Beaumont et al. 2009). This is apparent in, for example, the formation of opaque sectors within otherwise translucent colonies (Beaumont et al. 2009; Gallie et al. 2015), but particularly evident in studies of switching in 1B4 at the single cell level (Gallie et al. 2015).
In the first study of switching, which centered on 1B4 (Gallie et al. 2015), attention focused on genes and regulatory circuits underpinning bifurcation of UTP (the end point of the pyrimidine biosynthetic pathway) into CTP (a precursor for DNA and RNA metabolism) and UDP-glucose (required for synthesis of cellular building blocks including polysaccharides such as CAP). This led to a model in which switching was viewed as integral to the cell cycle and particularly to the decision-making process surrounding partitioning of UTP, with parallels seen in sporulation of Bacillus subtilis and persister formation in Escherichia coli (Balaban et al. 2004; Rocco et al. 2013; Siebring et al. 2014). Implicit was the notion that the switch exists in the ancestral genotype with presumed relevance in nature, with the threshold of activation having been lowered by the carB mutation favored during the course of the selection experiment. Indeed, Gallie et al. (2015) confirmed that switching behavior does exist in the ancestral genotype, which while not evident at the level of colonies, is nonetheless apparent at the level of individual cells (Cap+ cells arise from Cap− cells at a rate of about 1 in every 1,000 cells).
Here, we extend previous work (Gallie et al. 2015) and provide new understanding of the genetic circuitry underpinning switching between capsulation states. The starting point is an experiment that showed the primary signaling molecule linking status of the environment to likelihood of switching is not UDP-glucose as originally hypothesized. This led to reassessment of transcriptome data, recognition that ribosome levels in switching genotypes are elevated, and to experiments that show that switching depends upon competition between ribosomes and GacA-regulated CsrA/RsmA proteins that together determine translation of pflu3655, a positively autoregulated activator of the colanic acid biosynthetic locus. In addition to detailing the mechanistic basis of capsule switching, we show that the switch is central to a program that 1) determines stochastic entry into a semiquiescent capsulated state (as shown by Gallie et al. [2015]), 2) ensures that such cells are provisioned with excess ribosomes, and 3) enables provisioned cells to exit rapidly from stationary phase under permissive conditions.
Results
Capsulation Is Not Induced by Depletion of UDP-Glucose
In previous work that analyzed the switcher genotype 1B4 (Beaumont et al. 2009) and the link between pyrimidine limitation (caused by a defect in CarB), growth and heterogeneous expression of capsules (Gallie et al. 2015), attention was given to bifurcation of UTP flux toward DNA/RNA versus UDP-glucose (a precursor of colanic acid from which capsules are synthesized). Extensive analyses showed that capsule production was tied to entry into a semiquiescent state triggered by reduction of flux through the pyrimidine biosynthetic pathway. The primary signaling molecule was not identified, but it was hypothesized to be a product of the pyrimidine biosynthetic pathway (Gallie et al. 2015), with UDP-glucose being a prime candidate given its role in regulation of cell size and bacterial growth (Vadia and Levin 2015).
To test the hypothesis that UDP-glucose underpins the switch to capsule production, we used a translational reporter for PFLU3655 expression. PFLU3655 is the primary transcriptional activator of the wcaJ–wzb operon and its production was previously shown to be restricted to capsulated cells (see Gallie et al. [2015] and below). The translational reporter (Ppflu3655-GFP) contains ∼500 nucleotides upstream of the PFLU3655 start codon with the first 39 coding nucleotides being fused in frame to GFP. It was introduced into the chromosome of a galU mutant of 1B4. The galU mutant is unable to convert UTP to UDP-glucose and is therefore Cap− (Gallie et al. 2015). The proportion of cells expressing the Ppflu3655-GFP reporter was reduced in the mutant compared with the 1B4 switching genotype (supplementary fig. 1, Supplementary Material online). This finding was inconsistent with the prediction that low UDP-glucose is the signal that increases the chance of switching to the capsulated state. Accordingly, the mechanistic links between pyrimidine limitation, growth, and the heterogeneous production of capsules were reassessed.
Ribosomes Are Overproduced in carB Mutants
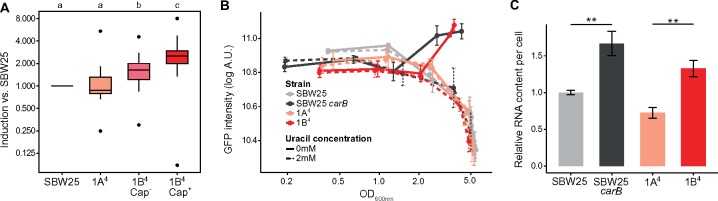
Transcriptomic data published previously (Gallie et al. 2015) were interrogated to identify signaling pathways displaying different levels of activity as a result of the causal carB mutation. These data were derived from experiments performed in strains SBW25, 1A4, and 1B4 (with Cap− and Cap+ cells analyzed separately). SBW25 is the wild-type (ancestral) genotype of P. fluorescens SBW25. Genotype 1A4 is the immediate ancestor of 1B4 from which it differs by a single nonsynonymous mutation in carB. Compared with SBW25, there are eight mutations in 1A4, all targeting components of the c-di-GMP signaling network underpinning repeated phenotypic reversals as required by the original selection experiment (Beaumont et al. 2009). KEGG-enrichment analyses showed overrepresentation of ribosomal components among the genes expressed at least 2-fold more in the capsulated subpopulation of 1B4 (1B4 Cap+) compared with its (noncapsulated) ancestor 1A4 (supplementary table 1, Supplementary Material online). By extracting raw expression values from the available RNA-seq data sets, it was found that the average expression levels of ribosomal protein genes increased in 1B4 (in both Cap− and Cap+) compared with 1A4 or ancestral SBW25 (fig. 1A). Such a finding is surprising because ribosome production is usually proportional to growth rate (Dennis et al. 2004; Paul et al. 2004) and was expected to be reduced in the slower growing strain 1B4 (supplementary fig. 2, Supplementary Material online).
Fig. 1.
Increased ribosome production in carB mutants. (A) Transcriptional induction of ribosomal protein genes in SBW25, 1A4, 1B4 Cap−, and 1B4 Cap+ cells. Absolute expression levels of ribosomal protein genes (KEGG pathway “0310-Ribosomes,” n = 26) were extracted from a previous RNA-seq data set (Gallie et al. 2015) and normalized to SBW25. Boxplots represent the distribution of expression ratios. Bold segments inside rectangles show the median, lower and upper limits of the box represent first and third quartiles, respectively. Whiskers extend up to 1.5 times the interquartile range and dots represent outliers, if present. Letter groups indicate statistical significance, P < 0.05, Kruskall–Wallis test with Dunn’s post hoc correction. (B) Expression kinetics of the PrrnB-GFP transcriptional reporter. Fluorescence in individual cells was measured by flow cytometry. Mean fluorescence of bacterial populations ± SD over biological replicates is shown, n = 4. Data are representative of three independent experiments. (C) Total RNA content in bacterial cells during exponential phase (OD600nm = 0.5–0.6). Values were normalized to SBW25 control within each experiment. Means ± SD are shown, n = 6. Data are pooled from four independent experiments. **P < 0.01, two-tailed t-test.
Bacteria adjust ribosome concentration to match nutrient availability in order to maximize growth rate. They do so by modulating transcriptional activity at ribosomal RNA (rrn) operon promoters (Dennis et al. 2004; Paul et al. 2004) causing the production of ribosomal proteins to match available rRNA (Keener and Nomura 1996). It was plausible that overexpression of ribosomal protein genes in 1B4 may reflect transcriptional upregulation at rrn promoters. Using a chromosomally integrated reporter, PrrnB-GFP transcriptional activity was monitored during bacterial growth (fig. 1B). No effect of the carB mutation was detected during exponential phase, but an increase in PrrnB-GFP transcription was apparent when cultures reached OD600nm ∼2, a density at which 1B4 cultures undergo a noticeable increase in capsulation (supplementary fig. 3, Supplementary Material online). Supplementing growth media with 2 mM uracil—a treatment known to suppress capsulation (Gallie et al. 2015)—restored wild-type PrrnB-GFP expression in strains carrying the mutant carB allele (fig. 1B). Measurement of cellular RNA content also showed higher levels in carB mutants (fig. 1C). These measurements were not affected by cell size (supplementary fig. 4, Supplementary Material online) indicating a bona fide increase in ribosome concentration. Together, these results show that pyrimidine limitation leads to an increase in the pool of ribosomes.
High Ribosome Levels Are Required for Capsulation
The counterintuitive effect of the carB mutation on ribosome levels suggested a causal connection between ribosome concentration and capsulation. In support of this hypothesis, a previous transposon-mutagenesis screen found that insertions in ribosome- or translation-associated genes (prfC, rluB, rluC, glu/gly tRNA) decreased or abolished capsulation in 1B4 (Gallie et al. 2015). We set out to directly manipulate ribosome concentration in 1B4 in order to test whether ribosome abundance affects capsulation.
Pseudomonas fluorescens SBW25 harbors five copies of rrn (rrnA–E). Because deletion of a single rrn operon can often be compensated by overexpression of those remaining (Condon et al. 1993; Bollenbach et al. 2009; Gyorfy et al. 2015), capsulation was quantified in both single and double rrn deletion mutants using the chromosomally inserted translational reporter Ppflu3655-GFP (Gallie et al. 2015). Although single mutants were not significantly affected in capsulation status, double mutants produced fewer capsulated cells (fig. 2A and supplementary fig. 5, Supplementary Material online). Growth rate was only marginally affected in certain mutant combinations (supplementary fig. 6, Supplementary Material online), but a significant reduction in total RNA content was observed in three out of the six rrn double mutants (supplementary fig. 7, Supplementary Material online). Together, these results suggest that capsulation is positively affected by increased ribosome abundance, which is itself a response to pyrimidine starvation.
Fig. 2.
Genetic bases of capsulation. (A) Capsulation in rrn deletion mutants. The Ppflu3655-GFP reporter was introduced in 1B4 bacteria and derived rrn mutants. Capsulation was measured by quantifying the proportion of GFP-positive cells by flow cytometry at the onset of stationary phase (OD600nm = 1–2). Means ± SEM are shown, n = 8 (1B4 ΔrrnB) or n = 11 (all other strains). Data are pooled from four independent experiments. **P < 0.01, ***P < 0.001, Kruskall–Wallis test with Dunn’s post hoc correction, comparison to 1B4. (B) Capsulation in gac/rsm mutants. Means ± SEM are shown, n = 12 (1B4), n = 15 (1B4 ΔgacA), or n = 9 (all other strains). Data are pooled from three independent experiments. *P < 0.05, **P < 0.001, ***P < 0.001, Kruskall–Wallis test with Dunn’s post hoc correction, comparison to 1B4. (C) Model for capsulation in 1B4. See text for details.
A Ribosome–Rsm Competition Model for the Control of Capsulation
Next, we asked how ribosome abundance influences production of capsulated cells. Results from a previous transposon mutagenesis screen showed that the Gac/Rsm two-component signaling pathway is required for expression of CAP capsules (Gallie et al. 2015). The Gac/Rsm signaling pathway controls important ecological traits in many Gram-negative bacteria, including secretion of exoproducts, the transition between biofilm and planktonic cells, and pathogenicity (Lapouge et al. 2008; Vakulskas et al. 2015; Valentini et al. 2018). Its activity is mediated through posttranscriptional regulators of the CsrA/RsmA family that prevent translation of mRNA targets by binding to sites adjacent to or overlapping ribosome-binding sites (RBS; Lapouge et al. 2008; Vakulskas et al. 2015). Upon perception of unknown extracellular signals (Valentini et al. 2018), the sensor kinase GacS activates the cognate response regulator GacA and the transcription of small noncoding RNAs rsmY and rsmZ. Binding of these sRNAs to CsrA/RsmA proteins antagonizes their translation inhibition activity. Three CsrA/RsmA homologs are present in SBW25 and were named rsmA1 (PFLU4746), rsmA2 (PFLU4324) and rsmE (PFLU4165). The phenotypic effect of the Gac/Rsm pathway was investigated by creating deletion mutants for the response regulator gacA and the two CsrA/RsmA homologs rsmA1 and rsmE. Capsulation was completely abolished in a gacA deletion strain, confirming the transposon-mutagenesis results (fig. 2B). Deletion of rsmA1 or rsmE increased the production of capsulated cells, consistent with their typical inhibitory role in Gac/Rsm signaling pathways.
We postulated that variations in the relative concentration of free ribosomes and RsmA/E may determine translational output of a positive regulator of capsule biosynthesis (fig. 2C). While searching for such a regulator, attention turned to pflu3655, the first gene of a putative operon (pflu3655–3657) located just upstream of the colanic acid biosynthetic operon. PFLU3655 is annotated as a hypothetical protein carrying a two-component response regulator C-terminal domain (PFAM PF00486) and is among the most highly upregulated genes in 1B4 Cap+ cells. Moreover, transposon insertions in the promoter region were shown to abolish capsulation in 1B4 (Gallie et al. 2015).
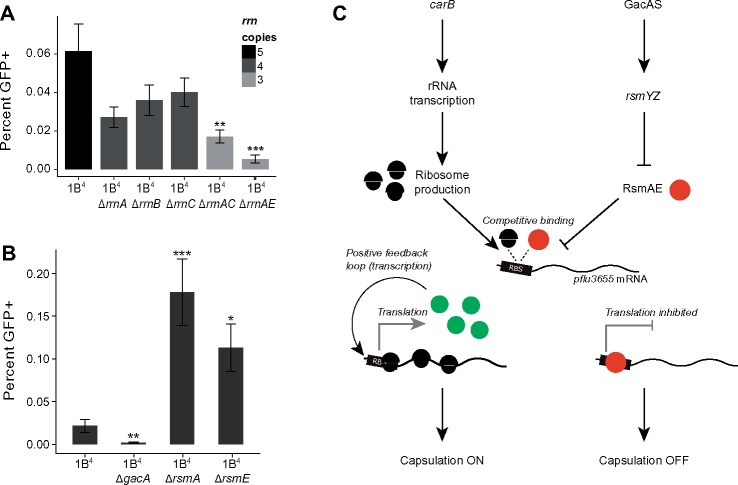
To test the role of pflu3655, a nonpolar deletion of pflu3655 was made in 1B4. The corresponding mutant produced no capsulated cells, whereas complementation of the mutant with an IPTG-inducible copy of pflu3655 on a low copy number plasmid (pME6032) restored capsulation (fig. 3A). Overexpression of pflu3655 in SBW25 and 1B4 led to high capsulation levels, showing that PFLU3655 is a positive regulator of colanic acid biosynthesis, the expression of which is sufficient for capsulation. Additionally, ectopic expression of pflu3655 induces expression from its own promoter, as measured using the chromosomally encoded Ppflu3655-GFP translational fusion (fig. 3B). This shows that pflu3655 expression can generate a positive feedback loop, a motif often underpinning bistable gene expression (Veening et al. 2008; Norman et al. 2015).
Fig. 3.
PFLU3655 is required for capsulation. (A) Capsulation in SBW25, 1B4 Δpflu3655, and 1B4 strains carrying the pME6032-pflu3655 plasmid or the empty vector (EV) after induction with 1 mM IPTG. Phase-contrast microscopy images of bacterial suspensions counterstained with Indian ink. White halos around cells indicate capsulation. Scale bar = 10 µm. (B) PFLU3655 establishes a positive feedback loop. GFP fluorescence from the Ppflu3655-GFP reporter in SBW25 (left), 1B4 Δpflu3655 (middle), or 1B4 (right) cells carrying the pME6032-pflu3655 plasmid. pflu3655 expression was induced with IPTG at indicated concentration and fluorescence was measured by flow cytometry. Data are representative of three independent experiments (A, B).
Existence of a positive feedback loop does not guarantee bistability. Often required is the additional presence of a mechanism to convert small input deviations (typically, molecular noise) into large output differences (Ferrell and Ha 2014). When signaling components are present in large numbers, ultrasensitive responses and threshold effects can arise through molecular titration where a molecule (RNA or protein) is sequestered and inhibited by another protein (Buchler and Louis 2008; Buchler and Cross 2009; Mukherji et al. 2011; Ferrell and Ha 2014). This led to recognition that titration of pflu3655 by RsmA/E may be involved in phenotypic bistability.
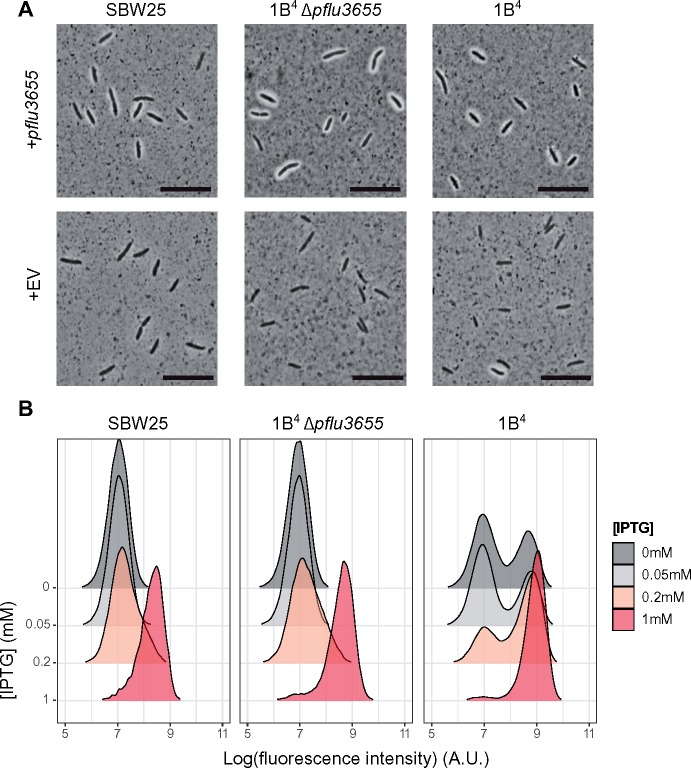
Two putative Rsm binding sites are located in the promoter and 5′ region of the coding sequence of pflu3655 (fig. 4A), indicating that pflu3655 mRNA could be a direct target of RsmA/E. The Ppflu3655-GFP capsulation reporter was used to test this hypothesis (the two putative RsmA/E binding sites are conserved in this synthetic construct). Using site-directed mutagenesis, nucleotides located in the putative RsmA/E binding sites were substituted and effects on GFP expression in the Ppflu3655-GFP reporter strain determined (fig. 4A and B). The G-8A point mutation increased GFP production, consistent with the expected effect arising from reduction in binding of an inhibitor. By contrast, the A33T mutation had a minor effect. This second putative binding site might be of lesser functional importance, in agreement with the fact that most CsrA/RsmA binding sites are located in 5′-UTR regions of target genes (Holmqvist and Vogel 2018). Altering the putative RBS (GG-7AC) completely abolished GFP expression (fig. 4B), which is expected given the need for translation.
Fig. 4.
RsmA/E binding sites in pflu3655 control capsulation. (A) Schematic diagram of pflu3655 5′-region. Two putative RsmA/E binding sites (orange squares) are located in the promoter and 5′-region of the gene. Numbers indicate nucleotide positions relative to start codon (not to scale). Sequences of putative RsmA/E binding sites are shown, the putative RBS is underlined. Gray box: Point mutations introduced in the different sequences by site-directed mutagenesis. (B) Expression of the Ppflu3655-GFP reporter carrying the different point mutations in the 1B4 background. GFP fluorescence was measured by flow cytometry. Data are representative of three independent experiments. (C, D) Mutations in putative RsmA/E binding sites affect capsulation in 1B4 (C) and SBW25 (D). Individual point mutations were reintroduced into 1B4 and SBW25 carrying the wild-type Ppflu3655-GFP reporter and the proportion of GFP positive cells in late exponential phase (OD600nm = 1–2) was measured by flow cytometry. Means ± SEM are shown, n = 9 (1B4) or n = 7 (SBW25). Data are pooled from three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, Kruskall–Wallis test with Dunn’s post hoc correction, comparison to 1B4. (E) Specific interaction between RsmA1 and pflu3655 mRNA. EMSA experiments were performed with 6.25 nM biotin-labelled oligonucleotides, either using a wild-type pflu3655 sequence (WT RNA; left) or a version carrying both G-8A and A33T point mutations (mut RNA; right). An increasing concentration of purified His6-RsmA1 (RsmA1) or a high concentration of proteins purified from SBW25 expressing a non-His6-tagged version of RsmA1 (Control protein) was added to the reactions.
To determine effects on capsulation, the individual point mutations were reintroduced at the native locus in 1B4 and SBW25. The G-8A mutation, but not the A33T mutation, increased the proportion of capsulated cells in both genotypes (fig. 4C and D), mirroring the difference observed on expression of GFP. While consistent with the region defined by G-8A being required for binding of RsmA/E, an alternate possibility is that this mutation creates a stronger RBS on pflu3655 mRNA, thus directly increasing translation. To test whether pflu3655 mRNA is the target of RsmA/E, we performed electrophoretic mobility shift assays using His6-tagged RsmA1 purified from P. fluorescens SBW25 (supplementary fig. 8A, Supplementary Material online) and biotin-labeled RNA oligonucleotides. A shift in RNA migration occurred with the wild-type oligonucleotide (spanning both putative RsmA/E binding sites) and was detected at a lower RsmA1 concentration than with an oligonucleotide carrying both G-8A and A33T mutations (fig. 4E). No shift was observed using proteins extracted from a strain expressing untagged RsmA1, indicating that the shift is specific to this protein. Shifted bands disappeared when an excess of nonlabeled oligonucleotide was added to the reaction mix (supplementary fig. 8B, Supplementary Material online). These results demonstrate that the identified RsmA/E binding sites in pflu3655 mRNA, although not detected by a sequence-based bioinformatics algorithm (Kulkarni et al. 2014; J. Gallie, personal communication), are required for binding of RsmA1.
Together, these data support the model proposed earlier (fig. 2C). Given this model, it is likely that other RsmA/E targets are overexpressed in carB mutants. A list of genes that were differentially expressed in an SBW25 gacS mutant (Cheng et al. 2013) was extracted and their expression levels were compared using the RNA-seq data set. On average, genes that were upregulated in the gacS mutant were expressed at lower levels in 1B4 (both Cap− and Cap+) than in SBW25 or 1A4 (supplementary fig. 9, Supplementary Material online). Genes that were downregulated in gacS showed a slight bias toward higher expression in 1B4 Cap+ but this difference was not statistically significant. These results are consistent with the opposing effects of gacS inactivation (leading to constitutive activation of RsmA/E) and carB-dependent increase in ribosome concentration on RsmA/E targets.
A Qualitative Mathematical Model of Posttranscriptional Control of a Positively Regulated Gene
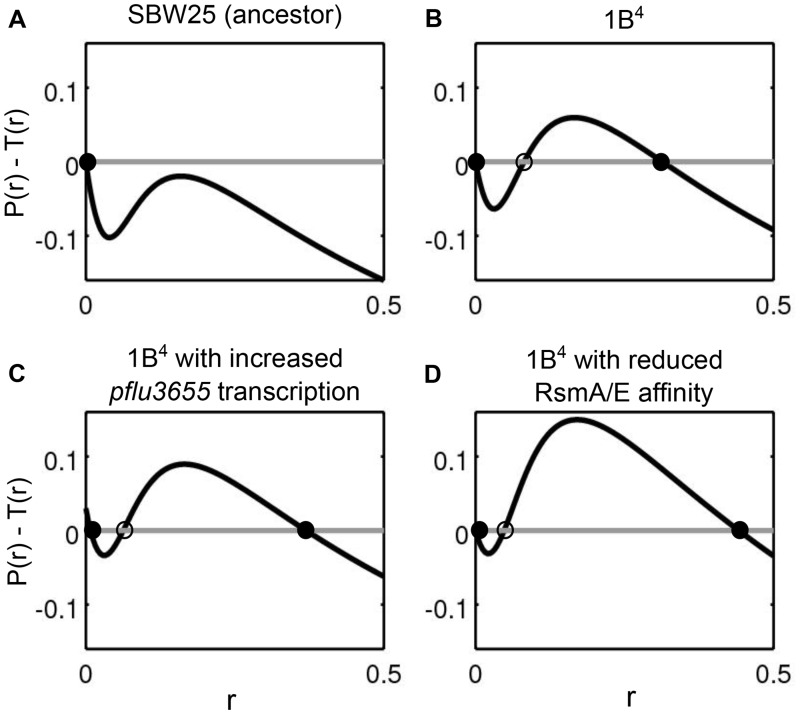
Mathematical models are a useful way of testing whether experimental observations of genes and regulatory circuitry are sufficient to explain experimental observations. Here, we explore whether the nonlinearities induced by positive feedback and molecular titration can give rise to bistable states compatible with experimental observations (Buchler and Louis 2008; Buchler and Cross 2009; Mukherji et al. 2011; Ferrell and Ha 2014). An ordinary differential equation for the concentration of mRNA transcribed from pflu3655 formalizes the observation that transcription is positively regulated by PFLU3655 protein concentration, and that ribosomes and the regulator RsmA/E compete for binding to pflu3655 mRNA (see supplementary note, Supplementary Material online, for detailed information on the model). The model describes the dynamics of PFLU3655 production and predicts the effect of variation in total ribosome concentration, basal rate of production of PFLU3655, and RsmA/E binding efficiency on capsulation (fig. 5). An increase in the concentration of ribosomes shifts the monostable system (fig. 5A) to a bistable regime, with the appearance of a second stable, positive equilibrium point in PFLU3655 concentration corresponding to the capsulated state (fig. 5B). When either a second unregulated source of mRNA production is added (fig. 5C), or the binding affinity of the regulator is reduced (fig. 5D), both the capsulation probability (indicated by the position of the unstable equilibrium) and the protein levels in capsulated cells increase (cf. figs. 3B and 4B). The consistency between the qualitative outputs of the model and experimental results shows that the regulatory circuit as depicted in figure 2C is sufficient to generate bistable capsulation states. Given that the unstable equilibrium represents a threshold at which mRNA translation overcomes sequestration by RsmA/E, a corollary of the model is that capsulated cells are expected to have an increased average ribosome content with respect to noncapsulated cells. Indeed, RNA-seq data indicate that ribosomal protein genes are expressed at higher levels in 1B4 Cap+ compared with 1B4 Cap− (fig. 1A).
Fig. 5.
A mathematical model of the capsulation regulatory network based on the experimental observations (see supplementary note, Supplementary Material online) explains the capsulation phenotype as the result of a bistable switch. Alternative states of transcription of pflu3655 are points (circles) where the net increase in mRNA production (production P(r) minus post-transcriptional binding T(r) of pflu3655; y axis), plotted against pflu3655 mRNA concentration (r; x axis), is null. Empty and filled circles indicate unstable and stable equilibria, respectively. Two qualitatively different scenarios are possible: (A) monostability of the Cap− state (SBW25) and (B–D) bistability between Cap− and Cap+ states (1B4 and its mutants). In the bistable cases, the position of the unstable equilibrium delimits the basins of attraction of the Cap− (left) and Cap+ (right) states. Assuming that the transition between the two alternative states takes place due to stochastic processes at the molecular level, extension of the basin of attraction of either stable equilibrium can be taken as a proxy of the probability of observing cells in the corresponding state. The proportion of capsulated cells is thus expected to increase when either the basal mRNA production is enhanced (C), or RsmA/E binding efficiency is reduced (D). See supplementary note, Supplementary Material online, for details on parameter values used in the different panels.
Consequences of Elevated Ribosome Concentration on Growth Resumption in Capsulated Cells
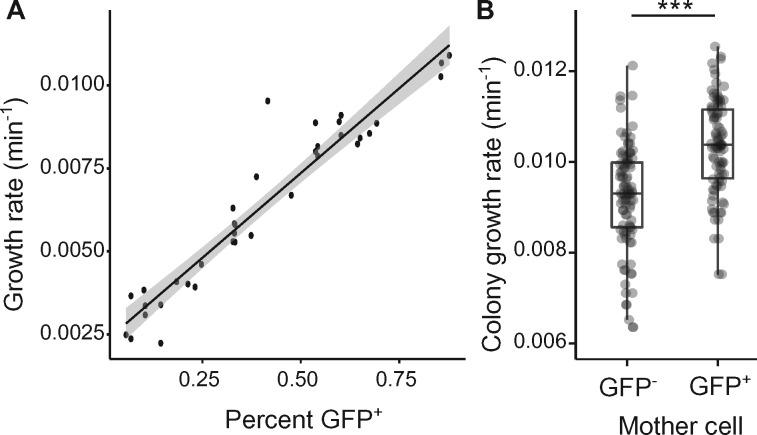
When the quality and/or quantity of nutrients rises abruptly, differences in ribosome abundance can significantly affect fitness (Mori et al. 2017; Kohanim et al. 2018). Reaching higher ribosome concentrations—required for maximal growth rate after nutrient upshift—is time-consuming and introduces a time-delay between environment change and future growth (Ehrenberg et al. 2013). Cells with higher ribosome concentrations before nutrient upshift might be considered as having been provisioned for rapid acclimation to the new conditions. If it is true that ribosomes promote growth resumption after nutrient upshift, then capsulated cells should have an average growth advantage under these conditions. To test this prediction, 1B4 cells were grown to late exponential phase (OD600nm ∼ 1) and cell suspensions enriched in Cap− or Cap+ cells were used to inoculate fresh cultures. The initial growth rate after nutrient upshift in batch cultures was positively correlated with the proportion of capsulated cells (fig. 6A). Time-lapse microscopy on solid agar pads confirmed that colonies founded by GFP+ (capsulated) cells grew ∼10% faster than those originating from GFP− (noncapsulated) cells (fig. 6B).
Fig. 6.
Capsulation and growth in 1B4. (A) Initial growth rate after nutrient upshift is correlated with the proportion of capsulated cells in 1B4 populations. Data points are pooled from two independent experiments. n = 36, r2 = 0.91. Shaded area indicates 95% confidence interval. (B) Growth rate of microcolonies founded by Cap− (GFP−) or Cap+ (GFP+) cells measured by time-lapse microscopy. n = 97 (GFP−) or n = 94 (GFP+). Data are pooled from seven independent experiments. ***P < 0.001, two-tailed t-test.
Capsulation in Ancestral P. fluorescens SBW25
Identification of the mechanism promoting production of CAP capsules in the derived 1B4 switcher genotype raises questions as to the role and conditions for expression of colanic acid in ancestral SBW25. In bacteria, extracellular capsules are important for bacterial pathogenicity (Roberts 1996) and are associated with broader environmental versatility (Rendueles et al. 2017). Recent work has also demonstrated a role for CAP capsules in the positioning of cells at the surface of bacterial colonies, providing access to oxygen and a fitness advantage (Kim et al. 2014, 2016).
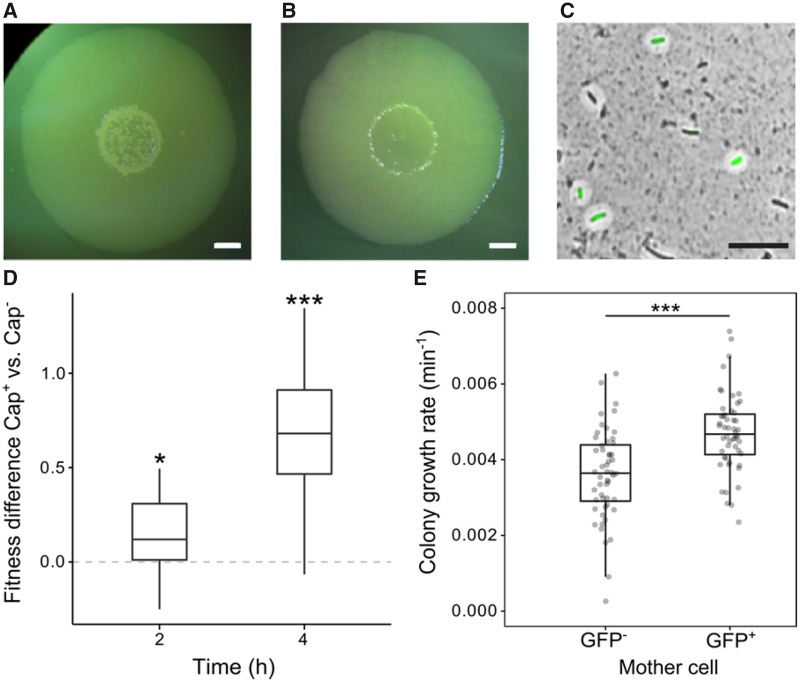
In order to test whether capsulation occurs in ancestral SBW25 colonies, bacterial suspensions were spot-inoculated on agar plates with incubation at 28 °C for several days. Mucoid outgrowths (papillations) were observed in the center of colonies from 5 days post-inoculation and both their size and number increased over time (fig. 7A). Such protrusions were not observed in colonies derived from a colanic acid mutant and their appearance was delayed when 2 mM uracil was added to agar plates (fig. 7B). In ancestral SBW25, cells sampled from papillated regions showed a high proportion of capsulated cells, ∼60% of which expressed the Ppflu3655-GFP reporter (fig. 7C). When streaked on new agar plates, no phenotypic difference was observed in colonies arising from papillated versus nonpapillated regions (data not shown), indicating that papillations comprised of capsulated cells are not caused by de novo mutation in aging colonies. To test whether capsulation status in ancestral SBW25 is also associated with higher average ribosome content, expression of the ribosomal protein gene rpsL was measured in capsulated and noncapsulated cells originating from aged ancestral SBW25 colonies by reverse-transcription quantitative polymerase chain reaction (RT-qPCR). We found an expression ratio of 1.67 (±0.35 SD, n = 5, two-tailed t-test: P = 0.012) in Cap+ versus Cap− cells, suggesting that SBW25-capsulated cells contain on average more ribosomes than their noncapsulated counterparts.
Fig. 7.
Capsulation and growth in SBW25. SBW25 colony grown on KB agar plate (A) or KB agar plate supplemented with 2 mM uracil (B) for 7 days. Scale bar = 2 mm. (C) SBW25 cells carrying the Ppflu3655-GFP reporter and sampled from a 7-day-old colony were counterstained with Indian ink to detect the presence of CAP capsules. A GFP image is overlaid with the phase-contrast image. Scale bar = 10 µm. Images are representative of at least three independent experiments (A–C). (D) Competitive fitness difference between SBW25 Cap− and Cap+ cells. Boxplot of the differences in Malthusian parameters between cultures enriched in Cap+ versus Cap− cells is shown, n = 12. Data are pooled from two independent experiments. *P < 0.05,***P < 0.001, comparison to 0 with two-tailed t-test. (E) Initial growth rate of microcolonies founded by GFP− or GFP+ cells measured by time-lapse microscopy. n = 65. Data are pooled from four independent experiments. ***P < 0.001, Wilcoxon test.
Given previous results showing a growth advantage to capsulated (ribosome-provisioned) 1B4 cells, we reasoned that capsulated ancestral SBW25 cells might similarly show a growth advantage upon transference to growth permissive conditions (fresh medium). Cells were collected from 7-day-old colonies and enriched in capsulated or noncapsulated cells by centrifugation. When transferred to fresh batch cultures, cell suspensions enriched in capsulated cells showed a faster initial growth rate (supplementary fig. 10, Supplementary Material online). To directly measure the fitness effect of capsulation during growth resumption, cellular suspensions from colonies of ancestral SBW25 or its isogenic variant carrying a neutral lacZ marker (Zhang and Rainey 2007) were collected. SBW25 Cap− cells were mixed with SBW25-lacZ Cap+ cells, and vice versa, to initiate competition experiments. A significant fitness advantage of capsulated cells was detected after 2- and 4-h growth in King’s Medium B (KB) (fig. 7D). Finally, time-lapse microscopy experiments revealed that microcolonies founded by GFP+ cells display an initial growth rate significantly higher than colonies arising from GFP− cells (fig. 7E).
Overall, the results from experiments carried out in ancestral (wild-type) P. fluorescens SBW25 are consistent with the capsulation model proposed for 1B4. During “long-term” (i.e., 7 days) starvation, nutrient limitation (possibly triggered by a reduction in flux through the pyrimidine pathway) causes a shift toward increased ribosome content. Cells with higher ribosome levels have an enhanced chance of flipping to the capsulated state where growth remains slow and cells enter a semiquiescent state. Even though giving up competition for nutrients in stationary phase, these cells stand primed for rapid growth upon nutrient upshift.
Discussion
Studies of adaptive phenotypes derived from selection experiments are by nature multifaceted, but two parallel lines of inquiry are of particular importance. The first concerns the nature of the adaptive phenotype, including its selective and molecular causes. The second concerns the ecological significance of the traits affected by adaptive evolution, prior to the occurrence of the adaptive mutation(s). A satisfactory answer to the first requires understanding of the latter.
Early characterization of the 1B4 switching genotype (Beaumont et al. 2009) showed the behavior to be a consequence of a single nonsynonymous nucleotide substitution in carbamoyl phosphate synthase (carB). In and of itself discovery of the causal switch-generating mutation shed no light on the adaptive phenotype. Understanding began to emerge only upon recognition that the carB mutation had altered the activation threshold of a preexisting switch (Gallie et al. 2015). Here, we have substantially extended knowledge of the mechanistic bases of phenotypic switching in the derived 1B4 genotype.
Experiments examining function of the bistable behavior in a galU mutant of the 1B4 switcher showed that previous conclusions concerning the central role of UTP or related molecules required revision. Although pyrimidine biosynthesis and the UTP decision point are important components of the pathways leading to capsulation, the mechanism of switching resides elsewhere. Data presented here have led to formulation of a compelling new model. Central to the model (fig. 2C) is competition between RsmA/E and ribosomes for pflu3655 mRNA, a positive regulator of capsulation. That translation initiation and/or efficiency may affect signaling through the GacAS two-component system has been suggested previously (Kitten and Willis 1996; Blumer and Haas 2000).
Importantly, in our model, titration of pflu3655 mRNA by RsmA/E has the potential to generate bistability. An increase in ribosome production resulting from the carB mutation leads the system to a two-state regime. Small fluctuations in ribosome and/or in RsmA/E activities beyond an activation threshold initiate a positive feedback loop leading to capsulation. The position of the threshold and the consequent probability of switching are affected by even small changes in parameters of the system. A significant change in these parameters occurs at the onset of stationary phase, as evidenced by increased capsulation frequency (supplementary fig. 3, Supplementary Material online). This effect is most likely explained by the increase in rsmY/Z expression (inhibiting RsmA/E activity) during this phase of growth (Valverde et al. 2003) and amplified by the fact that ribosome levels do not decrease in the carB mutants. The prevalence of inhibitory interactions in signaling networks offers a common route for the evolution (or evolutionary tuning) of bistable switches through relatively minor changes to the basal level or the interaction affinity of threshold-defining components (Buchler and Cross 2009; Rotem et al. 2010; Zhang et al. 2013). Bimodal expression of GacAS-regulated genes has been observed in P. aeruginosa (Broder et al. 2017) with the relevance of ribosome-mediated heterogeneity for numerous two-component signaling pathways involving RNA-binding proteins warranting further attention. Such regulatory strategies would enable bacterial cells to couple their internal metabolic and physiological status to multiple signaling outputs. A recent paper reports that heterogeneity in persister resuscitation in E. coli correlates with ribosome content (Kim et al. 2018) and provides a further example of the association between ribosomes and phenotypic heterogeneity.
A remaining open question is the mechanistic link between pyrimidine starvation and ribosome biosynthesis. Although the stringent response is thought to tune ribosome production to cellular needs (Bosdriesz et al. 2015), a phenomenon of ribosome overcapacity was previously reported in slow growing bacteria (Koch 1971; Dai et al. 2017; Li et al. 2018). This phenomenon occurs concomitantly with a reduction in the rate of translation and/or accumulation of inactive ribosomes. Although bearing similarity to what we describe as “ribosome provisioning,” the mechanistic basis appears to differ. RelA-dependent production of ppGpp is necessary for ribosome accumulation under nitrogen starvation (Li et al. 2018), but in unpublished work on 1B4 switching was unaffected in a relA spoT mutant (G.C. Ferguson, unpublished data). It is possible that in 1B4 an imbalance in the nucleotide pool—a factor known to influence ribosome production in E. coli (Gaal et al. 1997)—may directly alter ribosome production.
In vitro selection experiments can shed light on hitherto unrecognized aspects of bacterial physiology (Hall 1976; Clarke 1983; Hindre et al. 2012; Blank et al. 2014; Laan et al. 2015; Rainey et al. 2017). Understanding the evolutionary origin of switching in capsulation states in 1B4 requires knowledge of the function and ecological significance of the switch in the ancestral genotype. Previous work showed that switching to the capsulated type was accompanied by a reduction in growth rate and that the probability of switching to the capsulated state was more likely in starved cells (particularly in cells starved of pyrimidines). This was understood as a mechanism that allowed cells entering starvation conditions to hedge their bets in the face of uncertainty surrounding the future state of the environment (Gallie et al. 2015). Discovery that capsulated cells, despite slow growth, are replete in ribosomes was perplexing, but caused attention to focus on exit from the semiquiescent state. Just as cells entering a slow growing phase stand to be outcompeted by conspecific types that remain in the active growth phase should the environment unexpectedly return to one conducive for growth, cells exiting from a slow growth state stand to be outcompeted by types that are already actively growing unless they can rapidly resume “life in the fast lane.” This phenomenon that we refer to as “ribosome provisioning” has parallels with recent reports of ribosome dynamics in exponentially growing cells (Metzl-Raz et al. 2017; Kohanim et al. 2018). In environments where resource availability fluctuates, it appears that the control of ribosome biosynthesis is subject to a trade-off between maximizing growth rate during nutrient limitation and growth resumption upon nutrient upshift. This trade-off may be solved at the single-cell level, where heterogeneity in ribosome activity may optimize long-term geometric mean fitness.
Although the subject of little attention in the microbiological world (but see Joers and Tenson [2016]; Metzl-Raz et al. [2017]; Mori et al. [2017]; Kohanim et al. [2018]), ideas concerning the provisioning of future generations with resources sufficient to aid their establishment is a component of life-history evolution theory (Stearns 1992). It has been particularly well developed in the context of seed dormancy and the evolution of postgermination traits (Evans and Cabin 1995). It is not difficult to conceive that bacterial cells entering a slow or nongrowing state, such as persisters, or the capsulated cells of SBW25, will, through evolutionary time, experience selection for mechanisms that facilitate rapid reentry to active growth. Our data here are suggestive of such an evolutionary response.
Materials and Methods
Bacterial Strains and Growth Conditions
Bacterial strains used in this study are listed in supplementary table 2, Supplementary Material online. Pseudomonas fluorescens strains were cultivated in King’s Medium B (KB; King et al. 1954) at 28 °C. Escherichia coli DH5α λpir was used for cloning and was grown on Lysogeny Broth at 37 °C. Bacteria were plated on their respective growth medium containing 1.5% agar. Antibiotics were used at the following concentrations: ampicillin (50–100 μg ml−1), gentamicin (10 μg ml−1), tetracycline (10 μg ml−1), kanamycin (25 or 50 μg ml−1 for E. coli or P. fluorescens, respectively), and nitrofuorantoin (100 μg ml−1). Uracil (Sigma-Aldrich) was added to culture medium at 2 mM final concentration when indicated. For competition experiments, 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal) was used at a concentration of 60 mg l−1 in agar plates.
For capsulation assays, precultures were inoculated from precalibrated dilutions of frozen glycerol aliquots in order to reach an OD600nm of 0.3–0.5 after overnight culture.
For colony assays with SBW25, 5 μl of cell suspensions was spot-inoculated on KB agar plates and incubated for 7 days at 28 °C. Cells from the center of these colonies were resuspended in phosphate-buffered saline (PBS) or Ringer’s solution for growth and competition assays and time-lapse microscopy, or in RNAlater solution (Invitrogen) for RT-qPCR.
Molecular Techniques
Oligonucleotides and plasmids used in this study are listed in supplementary tables 3 and 4, Supplementary Material online, respectively. Standard molecular biology techniques were used for DNA manipulations (Sambrook et al. 1989). DNA fragments used to generate promoter fusions and gene deletion constructs were prepared by splicing by overhang extension polymerase chain reaction (SOE-PCR; Ho et al. 1989). All DNA fragments generated by SOE-PCR were first cloned into the pGEM-T easy vector (Promega) and their fidelity was verified by Sanger sequencing (Macrogen, Seoul). Plasmids were introduced into P. fluorescens by triparental conjugations with the helper plasmid pRK2013 (Ditta et al. 1980), carrying the tra and mob genes required for conjugation. Tn7-based plasmids were mobilized into recipient strains with the additional helper plasmid pUX-BF13 (Bao et al. 1991).
To generate deletion mutants (rrn operons, gacA, rsmA1, rsmE, and pflu3655), regions flanking the genes or operons of interest were amplified from SBW25 genomic DNA and assembled by SOE-PCR. Deletion cassettes were inserted into the pUIC3 plasmid (Rainey 1999) as SpeI fragments and mutants were obtained following the two-step allelic exchange protocol described previously (Zhang and Rainey 2007). Deletion mutants were checked by PCR. To check rrn copy number after rrn deletions, quantitative PCR was performed using a protocol described previously (Farr et al. 2017).
For complementation and overexpression studies, pflu3655 was amplified and cloned into pME6032 (Heeb et al. 2002) as an EcoRI/XhoI restriction fragment, downstream of the Ptac promoter.
To generate the Tn7-PrrnB-GFP reporter, a ∼600-bp fragment upstream of the rrnB operon (pflur7–11) was amplified from SBW25 genomic DNA and fused by SOE-PCR to gfpmut3 sequence containing the T0 terminator previously amplified with oPR152/FluomarkerP2 from the miniTn7(Gm)-PrrnB1-gfpmut3 plasmid (Lambertsen et al. 2004). The resulting fragment was cloned into pUC18R6K-mini-Tn7T-Gm (Choi et al. 2005) as a SpeI restriction fragment.
Site-directed mutagenesis of putative RsmA/E binding sites in pGEMT easy-Ppflu3655-GFP plasmid was performed using the Quick Change mutagenesis kit (Stratagene) according to manufacturer’s instructions. Mutagenized fragments were then cloned into pUC18R6K-miniTn7T-Gm (Choi et al. 2005) as a SpeI restriction fragment. In order to reengineer point mutations in RsmA/E into the P. fluorescens genome, a 1.5-kb fragment spanning equal length on each side of the target sites was amplified and cloned into pGEM-T easy vector. Site-directed mutagenesis was performed on this plasmid as described above, and the resulting DNA fragments were cloned in pUIC3 as SpeI restriction fragments and introduced into the P. fluorescens genome via the two-step allelic exchange protocol.
For electrophoretic mobility shift assay (EMSA) experiments, rsmA1 was amplified and cloned into pME6032. A His6-tag was introduced immediately after the start codon of rsmA1 in the expression plasmid pME6032-rsmA1 by site-directed mutagenesis using primers DWR037 and DWR038 following the protocol described by Li et al. (2011).
RNA Extractions and RT-qPCR
For quantification of RNA concentration in bacterial cultures, cells were harvested from 1 ml of cultures at an OD600nm of 0.5–0.6 and resuspended in 200 μl of RNAlater solution (Invitrogen). For total RNA quantification, we followed the method described by You et al. (2013), except that, before processing, cells cultures were resuspended in RNAlater (Invitrogen) instead of being fast frozen on dry ice. To normalize total RNA concentrations, the relationship between cell density and OD600nm was established for each strain by counting cells with a hemocytometer in five independent cultures of similar OD to those used for RNA extractions. For rrn double mutants, no significant difference in the cell/OD600nm ratio was found when compared with 1B4, so RNA quantities were normalized with OD600nm values.
RT-qPCR was performed as described previously (Farr et al. 2017), using gyrA as an internal control. Oligonucleotide primers used for RT-qPCR are listed in supplementary table 3, Supplementary Material online.
Purification of RsmA1
Induction of RsmA1 expression was performed based on the protocol of Romero et al. (2018). Overnight cultures of strains SBW25(pME6032-His6-rsmA1) and SBW25(pME6032-rsmA1) were transferred to 500 ml of prewarmed KB and grown for 2 h to an OD600 of ∼0.35, when Isopropyl β-d-thiogalactoside (IPTG) was added to a final concentration of 1 mM. Cultures were incubated for 6 additional hours at 28 °C. Cells were then harvested by centrifugation at 4,000 × g for 20 min, washed in PBS, and flash frozen in liquid nitrogen for overnight storage at −80 °C. Pulldowns were performed using the QIAexpress Ni-NTA Fast Start kit (QIAGEN) following the manufacturer’s instructions for purification of His6-tagged proteins under native conditions (both lysozyme and Benzonase Nuclease were included in the native lysis buffer). Proteins were eluted in two 1-ml aliquots of native elution buffer, combined, and concentrated to 200 μl using 3 kDa Amicon Ultra-4 centrifugal filters (Millipore) then split into 5× 35 μl aliquots (stored at −80 °C) for use in EMSA and 2× 5 μl aliquots which were mixed with 5 μl 2× Laemlli sample buffer (Bio-Rad) containing 5% β-mercaptoethanol and stored at −20 °C for use in Western blots.
Western Blot
Samples stored in Laemlli sample buffer containing 5% β-mercaptoethanol were thawed on ice and then incubated at 99 °C for 5 min. Samples (9 μl) were loaded onto an 8–16% Mini-PROTEAN TGX gel (Bio-Rad) and run for 80 min at 90 V, followed by transfer to a polyvinylidene fluoride (PVDF) membrane using a Trans-Blot SD semidry transfer cell (Bio-Rad) at 15 V for 15 min. Blots were blocked using 3% (w v-1) bovine serum albumin (BSA) in PBS containing 0.1% (v v-1) Tween 20 (PBST) for 1 h at room temperature, then incubated overnight at 4 °C with 1 μgml-1 His6 Tag Mouse Monoclonal IgG2b antibody (HIS.H8; MA1-21315, ThermoFisher) diluted in 1% BSA in PBST. Membranes were washed 3 × 10 min in PBST, then incubated for 1 h at room temperature with horse radish peroxidase-conjugated goat antimouse IgG cross-adsorbed secondary antibody (A16072, ThermoFisher) diluted 1:2,500 in 1% BSA in PBST, followed by 3 × 10 min washes in PBST. Protein bands were detected using Clarity Western ECL Substrate (Bio-Rad) and imaged using a FluorChem digital imager (Alpha Innotech).
Preparation of RNA Oligonucleotides
3′-Biotin-TEG-labeled HPLC-purified 66-nt-long oligonucleotides corresponding to the wild-type 5′-region of PFLU3655 (from nucleotide position −19 to +47: 5′-AUGCCGGAAAAGGGAGUAGGUGAUGCAUUUUUCCAACGUCCUCGCUAUUGCACGGACUCAAUCGAA-BIOTEG-3′) and the double mutant containing the G-8A and A33T substitutions (5′-AUGCCGGAAAAAGGAGUAGGUGAUGCAUUUUUCCAACGUCCUCGCUAUUGCUCGGACUCAAUCGAA-BIOTEG-3′), were commercially prepared (Eurofins). An unlabeled version of the wild-type oligo was generated by in vitro transcription using the HiScribe T7 In Vitro Transcription Kit (NEB) following the manufacturer’s instructions for reactions with short RNA transcripts. The template was prepared by PCR amplification of the 66-nt wild-type sequence from SBW25 genomic DNA using Phusion High-Fidelity DNA polymerase (Thermo Scientific) and primers DWR100/101, incorporating the T7 promoter sequence to the 5′-end of the PCR product. RNA was transcribed from 10 μl unpurified PCR product during a 16-h incubation at 37 °C and purified using the RNA Clean & Concentrator Kit (Zymo Research) including an on-column DNase treatment using RNase-free DNase Set (Qiagen). RNA was quantified using a NanoDrop 1000 Spectrophotometer (Thermo Scientific) and the correct size was confirmed by polyacrylamide gel electrophoresis compared with the commercially prepared oligos.
Electrophoretic Mobility Shift Assays
Assays were performed using the LightShift Chemiluminescent RNA EMSA Kit (Thermo Scientific). Binding reactions (20 μl) contained 1× REMSA binding buffer, 5% glycerol, 2 μg nonspecific competitor tRNA, and 6.25 nM Biotin-TEG-labelled RNA oligonucleotide. Variable amounts of concentrated protein eluate from the Ni-NTA Fast Start kit were also included in the binding reactions: 1×, 5× and 25× correspond to 0.2, 1 and 5 µl of protein eluate, respectively. We have reported relative amounts of protein because our pulldown contained multiple proteins (supplementary fig. 8A, Supplementary Material online) and it was therefore not possible to accurately quantify the amount of RsmA1 in the eluate. Unlabeled wild-type RNA oligos were included in some control reactions at a concentration 200× or 1,000× that of the Biotin-TEG-labelled oligo. Reactions were incubated for 30 min at room temperature before the addition of loading buffer. Samples were loaded into a prerun 5% Mini-PROTEAN TBE gel (Bio-Rad) and run for 60 min at 100 V under nondenaturing conditions. Bands were transferred to a Biodyne B membrane (Thermo Fisher) using a Trans-Blot SD semidry transfer cell (Bio-Rad) at 20 V for 60 min. RNA was crosslinked to the membrane using a Hoefer UVC 500 Crosslinker (Amersham Biosciences) at 120 mJ cm−2 for 60 s and visualized using streptavidin–horseradish peroxidase conjugate and Luminol/Enhancer solution as described in the instructions of the Chemiluminescent Nucleic Acid Detection Module of the LightShift kit. Images were recorded using a FluorChem digital imager (Alpha Innotech).
Capsulation and Gene Expression Assays
For capsulation test, cells were grown from standardized glycerol aliquots stored at −80 °C. Aliquots were diluted in KB and precultures were grown overnight in order to reach an OD of 0.3–0.5 in the morning. For gene expression studies, overnight precultures were grown to saturation. In both cases, precultures were diluted to OD600nm = 0.05 in KB and incubated at 28 °C. IPTG was added to a final concentration of 0.1–1 mM when indicated. Samples were taken at different time points for flow cytometry and OD measurements. GFP fluorescence in bacterial populations was measured with a BD FACS Canto II flow cytometer. Cell suspensions were diluted to a density of ∼105 cells ml−1 in filter-sterilized PBS and at least 20,000 cells were analyzed by flow cytometry. Cellular debris was filtered using side- and forward-scatter channels. GFP fluorescence was detected with a 488-nm laser with 530/30 bandwidth filter. Laser intensity was set to 600 V, except for PrrnB-GFP analyses where intensity was lowered to 300 V. Flow cytometry data files were analyzed in R (R Core Team 2018) using the “flowCore” package (Ellis et al. 2018). For capsulation experiments, the relative sizes of GFP positive and negative subpopulations were measured after manual thresholding of GFP intensity. The distributions of expression intensities in figure 3B were plotted using the R package “ggridges” (Wilke 2018) and were smoothed using the “KernSmooth” package (Wand 2015) for figure 4B.
Growth Curves in Microplate Reader
Overnight precultures were adjusted to an OD600nm of 0.05 and 200 μl KB cultures were grown in 96-well plates. Cultures were incubated in a Synergy 2 microplate reader (Biotek) for at least 24 h at 28 °C with constant shaking and OD600nm was read every 5 min.
To measure growth rates of cultures enriched in capsulated or noncapsulated cells, cells were harvested from late-exponential phase (OD600nm ∼ 1; 1B4) or 7-day-old colonies (SBW25) and centrifuged (1 min, 3,000 rpm). Supernatants and pellets were collected, representing subpopulations enriched in capsulated and noncapsulated cells, respectively. Cell suspension density was adjusted to an OD600nm of 0.05 in KB to start growth curves. Initial growth rates were calculated by performing a linear regression on the logarithm of the measured OD values during the first 2 h of growth.
Microscopy
Microscopy experiments were performed with an Olympus BX61 upright microscope equipped with an F-View II monochrome camera, a motorized stage, and a temperature-controlled chamber set at 28 °C. Devices were operated by the Cell^P or CellSens software (Olympus). Phase-contrast images were acquired with an oil-immersion 100×/N.A. 1.30 objective. GFP fluorescence images were acquired with the following filter set: excitation (460–480 nm), emission (495–540 nm), and dichroic mirror (DM485).
Indian ink staining of capsulated cells was performed as described previously (Gallie et al. 2015). To determine cell size, exponentially growing bacteria were diluted 1:10 in KB and transferred on 1% agarose-KB gel pads. Cell sizes were determined from phase-contrast images with MicrobeJ (Ducret et al. 2016). For time-lapse microscopy, bacteria were harvested from late exponential phase (1B4, OD600nm ∼ 1–2) or from 7-day-old colonies (SBW25), diluted 1:1,000 in KB and 2 μl of the resulting suspension was immediately transferred on a gel pad (1% agarose KB) located on a glass slide within an adhesive frame (GeneFrame, Thermo-Fisher). When dry, a cross-section of the pad was removed with a razor blade in order to allow gas exchanges to occur; the preparation was sealed with a glass cover-slip and transferred to the microscope incubation chamber preheated to 28 °C. One GFP image was taken before starting the experiment in order to determine the capsulation status of each cell and phase-contrast images were then recorded every 10 min.
Phase-contrast images were segmented with Fiji (Schindelin et al. 2012) and individual colony areas were extracted. For each colony analyzed, the capsulation status of the founding cell was determined manually based on GFP signal. The logarithm of the growth rate of individual 1B4 colonies was then fitted using a linear regression. For SBW25, segmented linear regressions were found to better fit the data and the slope of the first line was reported.
RNA-Seq Analyses
RNA-seq data were published previously (Gallie et al. 2015). KEGG orthology terms for the SBW25 genome were downloaded from the KEGG Orthology database (www.genome.jp; accessed in April 2016). KEGG enrichment statistics were computed with a hypergeometric test and were performed separately for up- and downregulated genes. Data from the transcriptome analysis of SBW25 gacS mutant were retrieved from supplementary table 3 from Cheng et al. (2013).
Competition Experiments
SBW25 or SBW25-lacZ cell suspensions were spot-inoculated on separate KB agar plates and grown for 7 days. Cells from the center of colonies were harvested, resuspended in Ringer’s solution, and gently centrifuged to enrich suspensions in capsulated or noncapsulated cells. Capsulated SBW25 cells were mixed with noncapsulated SBW-lacZ cells, and vice versa. Mixed suspensions were diluted 1:100 in KB medium and grown at 28 °C with orbital shaking for 4 h. Appropriate dilutions of the cultures were plated on KB + X-gal plates at 0, 2, and 4 h postinoculation in order to measure the ratio of white–blue colonies. The difference in Malthusian parameters (Lenski et al. 1991) was used as a measure of relative fitness.
Data Availability
Data and material are available from corresponding authors upon reasonable request.
Supplementary Material
Acknowledgments
The authors thank Jenna Gallie and Camille De Almeida for discussion, Xue-Xian Zhang for the generous gift of pUIC3-ΔgacA deletion plasmid, Heather Hendrickson, Nicolas Desprat and Peter Lind for help with microscopy and flow cytometry, and Elena Denisenko for assistance with KEGG enrichment analyses. This work was supported in part by the Marsden Fund Council and a James Cook Research Fellowship from government funding administered by the Royal Society of New Zealand. S.D.M. has received support under the program “Investissements d’Avenir” launched by the French Government and implemented by ANR with the references ANR-10‐LABX‐54 MEMOLIFE and ANR‐10‐IDEX‐0001‐02 PSL* Research University.
Author Contributions
P.R. and P.B.R. designed the study; P.R., G.C.F., E.M.C., and D.W.R. performed experiments and analyzed the data; S.D.M. designed and analyzed the mathematical model; P.R. and P.B.R. wrote the paper with contributions from all authors.
References
- Ackermann M. 2015. A functional perspective on phenotypic heterogeneity in microorganisms. Nat Rev Microbiol. 13(8): 497–508. [DOI] [PubMed] [Google Scholar]
- Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S.. 2004. Bacterial persistence as a phenotypic switch. Science 305(5690): 1622–1625. [DOI] [PubMed] [Google Scholar]
- Bao Y, Lies DP, Fu H, Roberts GP.. 1991. An improved Tn7-based system for the single-copy insertion of cloned genes into chromosomes of gram-negative bacteria. Gene 109(1): 167–168. [DOI] [PubMed] [Google Scholar]
- Beaumont HJE, Gallie J, Kost C, Ferguson GC, Rainey PB.. 2009. Experimental evolution of bet hedging. Nature 462(7269): 90–93. [DOI] [PubMed] [Google Scholar]
- Blank D, Wolf L, Ackermann M, Silander OK.. 2014. The predictability of molecular evolution during functional innovation. Proc Natl Acad Sci U S A. 111(8): 3044–3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumer C, Haas D.. 2000. Multicopy suppression of a gacA mutation by the infC operon in Pseudomonas fluorescens CHA0: competition with the global translational regulator RsmA. FEMS Microbiol Lett. 187(1): 53–58. [DOI] [PubMed] [Google Scholar]
- Bollenbach T, Quan S, Chait R, Kishony R.. 2009. Nonoptimal microbial response to antibiotics underlies suppressive drug interactions. Cell 139(4): 707–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosdriesz E, Molenaar D, Teusink B, Bruggeman FJ.. 2015. How fast-growing bacteria robustly tune their ribosome concentration to approximate growth-rate maximization. FEBS J. 282(10): 2029–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broder UN, Jaeger T, Jenal U.. 2017. LadS is a calcium-responsive kinase that induces acute-to-chronic virulence switch in Pseudomonas aeruginosa. Nat Microbiol. 2:16184. [DOI] [PubMed] [Google Scholar]
- Buchler NE, Cross FR.. 2009. Protein sequestration generates a flexible ultrasensitive response in a genetic network. Mol Syst Biol. 5:272.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchler NE, Louis M.. 2008. Molecular titration and ultrasensitivity in regulatory networks. J Mol Biol. 384(5): 1106–1119. [DOI] [PubMed] [Google Scholar]
- Cheng X, de Bruijn I, van der Voort M, Loper JE, Raaijmakers JM.. 2013. The Gac regulon of Pseudomonas fluorescens SBW25. Environ Microbiol Rep. 5(4): 608–619. [DOI] [PubMed] [Google Scholar]
- Choi KH, Gaynor JB, White KG, Lopez C, Bosio CM, Karkhoff-Schweizer RR, Schweizer HP.. 2005. A Tn7-based broad-range bacterial cloning and expression system. Nat Methods. 2(6): 443–448. [DOI] [PubMed] [Google Scholar]
- Clarke PH. 1983. Experimental evolution In: Bendall DS, editor. Evolution from molecules to men. Cambridge: Cambridge University Press; p. 235–252. [Google Scholar]
- Cohen D. 1966. Optimizing reproduction in a randomly varying environment. J Theor Biol. 12(1): 119. [DOI] [PubMed] [Google Scholar]
- Condon C, French S, Squires C, Squires CL.. 1993. Depletion of functional ribosomal RNA operons in Escherichia coli causes increased expression of the remaining intact copies. EMBO J. 12(11): 4305–4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai XF, Zhu ML, Warren M, Balakrishnan R, Patsalo V, Okano H, Williamson JR, Fredrick K, Wang YP, Hwa T.. 2017. Reduction of translating ribosomes enables Escherichia coli to maintain elongation rates during slow growth. Nat Microbiol. 2:16231.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis PP, Ehrenberg M, Bremer H.. 2004. Control of rRNA synthesis in Escherichia coli: a systems biology approach. Microbiol Mol Biol Rev. 68(4): 639–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditta G, Stanfield S, Corbin D, Helinski DR.. 1980. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci U S A. 77(12): 7347–7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducret A, Quardokus EM, Brun YV.. 2016. MicrobeJ, a tool for high throughput bacterial cell detection and quantitative analysis. Nat Microbiol. 1(7): 16077.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenberg M, Bremer H, Dennis PP.. 2013. Medium-dependent control of the bacterial growth rate. Biochimie 95(4): 643–658. [DOI] [PubMed] [Google Scholar]
- Ellis B, Haaland P, Hahne F, Le Meur N, Gopalakrishnan N, Spidlen J, Jiang M.. 2018. flowCore: basic structures for flow cytometry data. R package version 1.48.1. https://bioconductor.org/packages/release/bioc/html/flowCore.html
- Evans AS, Cabin RJ.. 1995. Can dormancy affect the evolution of post-germination traits? The case of Lesquerella fendleri. Ecology 76(2): 344–356. [Google Scholar]
- Farr AD, Remigi P, Rainey PB.. 2017. Adaptive evolution by spontaneous domain fusion and protein relocalization. Nat Ecol Evol. 1(10): 1562–1568. [DOI] [PubMed] [Google Scholar]
- Ferrell JE, Ha SH.. 2014. Ultrasensitivity part II: multisite phosphorylation, stoichiometric inhibitors, and positive feedback. Trends Biochem Sci. 39(11): 556–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaal T, Bartlett MS, Ross W, Turnbough CL, Gourse RL.. 1997. Transcription regulation by initiating NTP concentration: rRNA synthesis in bacteria. Science 278(5346): 2092–2097. [DOI] [PubMed] [Google Scholar]
- Gallie J, Bertels F, Remigi P, Ferguson GC, Nestmann S, Rainey PB.. 2019. Repeated phenotypic evolution by different genetic routes: the evolution of colony switching in Pseudomonas fluorescens SBW25. Mol. Biol. Evol.36(5): 1071–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallie J, Libby E, Bertels F, Remigi P, Jendresen CB, Ferguson GC, Desprat N, Buffing MF, Sauer U, Beaumont HJE, et al. 2015. Bistability in a metabolic network underpins the de novo evolution of colony switching in Pseudomonas fluorescens. PLoS Biol. 13(3): e1002109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimbergen AJ, Siebring J, Solopova A, Kuipers OP.. 2015. Microbial bet-hedging: the power of being different. Curr Opin Microbiol. 25:67–72. [DOI] [PubMed] [Google Scholar]
- Gyorfy Z, Draskovits G, Vernyik V, Blattner FF, Gaal T, Posfai G.. 2015. Engineered ribosomal RNA operon copy-number variants of E. coli reveal the evolutionary trade-offs shaping rRNA operon number. Nucleic Acids Res. 43(3): 1783–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall BG. 1976. Experimental evolution of a new enzymatic function. Kinetic analysis of ancestral (ebg0) and evolved (ebg+) enzymes. J Mol Biol. 107(1): 71–84. [DOI] [PubMed] [Google Scholar]
- Heeb S, Blumer C, Haas D.. 2002. Regulatory RNA as mediator in GacA/RsmA-dependent global control of exoproduct formation in Pseudomonas fluorescens CHA0. J Bacteriol. 184(4): 1046–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindre T, Knibbe C, Beslon G, Schneider D.. 2012. New insights into bacterial adaptation through in vivo and in silico experimental evolution. Nat Rev Microbiol. 10(5): 352–365. [DOI] [PubMed] [Google Scholar]
- Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR.. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77(1): 51–59. [DOI] [PubMed] [Google Scholar]
- Holmqvist E, Vogel J.. 2018. RNA-binding proteins in bacteria. Nat Rev Microbiol. 16(10): 601–615. [DOI] [PubMed] [Google Scholar]
- Joers A, Tenson T.. 2016. Growth resumption from stationary phase reveals memory in Escherichia coli cultures. Sci Rep. 6:24055.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keener J, Nomura M.. 1996. Regulation of ribosome synthesis In: Neidhardt FC, Curtiss R III, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger H.E., editors. Escherichia coli and Salmonella: cellular and molecular biology. Washigton (DC: ): American Society for Microbiology; p. 1417–1431. [Google Scholar]
- Kim JS, Yamasaki R, Song S, Zhang WW, Wood TK.. 2018. Single cell observations show persister cells wake based on ribosome content. Environ Microbiol. 20(6): 2085–2098. [DOI] [PubMed] [Google Scholar]
- Kim W, Levy SB, Foster KR.. 2016. Rapid radiation in bacteria leads to a division of labour. Nat Commun. 7:10508.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W, Racimo F, Schluter J, Levy SB, Foster KR.. 2014. Importance of positioning for microbial evolution. Proc Natl Acad Sci U S A. 111(16): E1639–E1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King EO, Ward MK, Raney DE.. 1954. Two simple media for the demonstration of pyocyanin and fluorescin. J Lab Clin Med. 44(2): 301–307. [PubMed] [Google Scholar]
- Kitten T, Willis DK.. 1996. Suppression of a sensor kinase-dependent phenotype in Pseudomonas syringae by ribosomal proteins L35 and L20. J Bacteriol. 178(6): 1548–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch AL. 1971. The adaptive responses of Escherichia coli to a feast and famine existence. Adv Microb Physiol. 6:147–217. [DOI] [PubMed] [Google Scholar]
- Kohanim YK, Levi D, Jona G, Towbin BD, Bren A, Alon U.. 2018. A bacterial growth law out of steady state. Cell Rep. 23(10): 2891–2900. [DOI] [PubMed] [Google Scholar]
- Kulkarni PR, Jia T, Kuehne SA, Kerkering TM, Morris ER, Searle MS, Heeb S, Rao J, Kulkarni RV.. 2014. A sequence-based approach for prediction of CsrA/RsmA targets in bacteria with experimental validation in Pseudomonas aeruginosa. Nucleic Acids Res. 42(11): 6811–6825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laan L, Koschwanez JH, Murray AW.. 2015. Evolutionary adaptation after crippling cell polarization follows reproducible trajectories. Elife 4:e09638.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambertsen L, Sternberg C, Molin S.. 2004. Mini-Tn7 transposons for site-specific tagging of bacteria with fluorescent proteins. Environ Microbiol. 6(7): 726–732. [DOI] [PubMed] [Google Scholar]
- Lapouge K, Schubert M, Allain FHT, Haas D.. 2008. Gac/Rsm signal transduction pathway of gamma-proteobacteria: from RNA recognition to regulation of social behaviour. Mol Microbiol. 67(2): 241–253. [DOI] [PubMed] [Google Scholar]
- Lenski RE, Rose MR, Simpson SC, Tadler SC.. 1991. Long-term experimental evolution in Escherichia coli. 1. Adaptation and divergence during 2,000 generations. Am Nat. 138(6): 1315–1341. [Google Scholar]
- Li CK, Wen AY, Shen BC, Lu J, Huang Y, Chang YC.. 2011. FastCloning: a highly simplified, purification-free, sequence- and ligation-independent PCR cloning method. BMC Biotechnol. 11(1): 92.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SHJ, Li ZY, Park JO, King CG, Rabinowitz JD, Wingreen NS, Gitai Z.. 2018. Escherichia coli translation strategies differ across carbon, nitrogen and phosphorus limitation conditions. Nat Microbiol. 3(8): 939–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libby E, Rainey PB.. 2011. Exclusion rules, bottlenecks and the evolution of stochastic phenotype switching. Proc R Soc Lond B Biol Sci. 278(1724): 3574–3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzl-Raz E, Kafri M, Yaakov G, Soifer I, Gurvich Y, Barkai N.. 2017. Principles of cellular resource allocation revealed by condition-dependent proteome profiling. Elife 6:e28034.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers LA, Bull JJ.. 2002. Fighting change with change: adaptive variation in an uncertain world. Trends Ecol Evol. 17(12): 551–557. [Google Scholar]
- Mori M, Schink S, Erickson DW, Gerland U, Hwa T.. 2017. Quantifying the benefit of a proteome reserve in fluctuating environments. Nat Commun. 8(1): 1225.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherji S, Ebert MS, Zheng GXY, Tsang JS, Sharp PA, van Oudenaarden A.. 2011. MicroRNAs can generate thresholds in target gene expression. Nat Genet. 43(9): 854–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman TM, Lord ND, Paulsson J, Losick R.. 2015. Stochastic switching of cell fate in microbes. Annu Rev Microbiol. 69:381–403. [DOI] [PubMed] [Google Scholar]
- Paul BJ, Ross W, Gaal T, Gourse RL.. 2004. rRNA transcription in Escherichia coli. Annu Rev Genet. 38:749–770. [DOI] [PubMed] [Google Scholar]
- R Core Team. 2018. R: a language and environment for statistical computing. Vienna (Austria: ): R Foundation for Statistical Computing; http://www.R-project.org/. [Google Scholar]
- Rainey PB. 1999. Adaptation of Pseudomonas fluorescens to the plant rhizosphere. Environ Microbiol. 1(3): 243–257. [DOI] [PubMed] [Google Scholar]
- Rainey PB, Beaumont HJE, Ferguson GC, Gallie J, Kost C, Libby E, Zhang XX.. 2011. The evolutionary emergence of stochastic phenotype switching in bacteria. Microb Cell Fact. 10(Suppl 1): S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainey PB, Remigi P, Farr AD, Lind PA.. 2017. Darwin was right: where now for experimental evolution? Curr Opin Genet Dev. 47:102–109. [DOI] [PubMed] [Google Scholar]
- Rendueles O, Garcia-Garcera M, Neron B, Touchon M, Rocha EPC.. 2017. Abundance and co-occurrence of extracellular capsules increases environmental breadth: implications for the emergence of pathogens. PLoS Pathog. 13(7): e1006525.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts IS. 1996. The biochemistry and genetics of capsular polysaccharide production in bacteria. Annu Rev Microbiol. 50:285–315. [DOI] [PubMed] [Google Scholar]
- Rocco A, Kierzek AM, McFadden J.. 2013. Slow protein fluctuations explain the emergence of growth phenotypes and persistence in clonal bacterial populations. PLoS One 8(1): e54272.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero M, Silistre H, Lovelock L, Wright VJ, Chan KG, Hong KW, Williams P, Camara M, Heeb S.. 2018. Genome-wide mapping of the RNA targets of the Pseudomonas aeruginosa riboregulatory protein RsmN. Nucleic Acids Res. 46(13): 6823–6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotem E, Loinger A, Ronin I, Levin-Reisman I, Gabay C, Shoresh N, Biham O, Balaban NQ.. 2010. Regulation of phenotypic variability by a threshold-based mechanism underlies bacterial persistence. Proc Natl Acad Sci U S A. 107(28): 12541–12546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T.. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor (NY: ): Cold Spring Harbor Laboratory. [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods. 9(7): 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebring J, Elema MJH, Vega FD, Kovacs AT, Haccou P, Kuipers OP.. 2014. Repeated triggering of sporulation in Bacillus subtilis selects against a protein that affects the timing of cell division. ISME J 8(1): 77–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons AM. 2011. Modes of response to environmental change and the elusive empirical evidence for bet hedging. Proc R Soc Lond B Biol Sci. 278(1712): 1601–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatkin M. 1974. Hedging one's evolutionary bets. Nature 250(5469): 704–705. [Google Scholar]
- Stearns SC. 1992. The evolution of life histories. Oxford: Oxford University Press. [Google Scholar]
- Vadia S, Levin PA.. 2015. Growth rate and cell size: a re-examination of the growth law. Curr Opin Microbiol. 24:96–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakulskas CA, Potts AH, Babitzke P, Ahmer BMM, Romeo T.. 2015. Regulation of bacterial virulence by Csr (Rsm) systems. Microbiol Mol Biol Rev. 79(2): 193–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentini M, Gonzalez D, Al Mavridou D, Filloux A.. 2018. Lifestyle transitions and adaptive pathogenesis of Pseudomonas aeruginosa. Curr Opin Microbiol. 41:15–20. [DOI] [PubMed] [Google Scholar]
- Valverde C, Heeb S, Keel C, Haas D.. 2003. RsmY, a small regulatory RNA, is required in concert with RsmZ for GacA-dependent expression of biocontrol traits in Pseudomonas fluorescens CHA0. Mol Microbiol. 50(4): 1361–1379. [DOI] [PubMed] [Google Scholar]
- van Boxtel C, van Heerden JH, Nordholt N, Schmidt P, Bruggeman FJ.. 2017. Taking chances and making mistakes: non-genetic phenotypic heterogeneity and its consequences for surviving in dynamic environments. J R Soc Interface. 14(132): 20170141.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veening JW, Smits WK, Kuipers OP.. 2008. Bistability, epigenetics, and bet-hedging in bacteria. Annu Rev Microbiol. 62:193–210. [DOI] [PubMed] [Google Scholar]
- Wand M. 2015. KernSmooth: functions for kernel smoothing supporting Wand & Jones (1995).
- Wilke CO. 2018. ggridges: ridgeline plots in ‘ggpot2’. R package version 0.5.1. https://CRAN.R-project.org/package=ggridges.
- You CH, Okano H, Hui S, Zhang ZG, Kim M, Gunderson CW, Wang YP, Lenz P, Yan DL, Hwa T.. 2013. Coordination of bacterial proteome with metabolism by cyclic AMP signalling. Nature 500(7462): 301–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Bhattacharya S, Andersen ME.. 2013. Ultrasensitive response motifs: basic amplifiers in molecular signalling networks. Open Biol. 3(4): 130031.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XX, Rainey PB.. 2007. Construction and validation of a neutrally-marked strain of Pseudomonas fluorescens SBW25. J Microbiol Methods. 71(1): 78–81. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data and material are available from corresponding authors upon reasonable request.