Abstract
Incidence of human papillomavirus (HPV) related cancers is increasing, generating substantial interest in understanding how trends in population prevalence of HPV infection are changing. However, there are no direct, population-scale measurements of HPV prevalence prior to 2003. Previous work using models to reconstruct historical trends have focused only on genital infection or seroprevalence (prevalence of antibodies) separately, and the results of these single-measure studies have been hard to reconcile. Here, we develop a mechanistic disease model fit jointly to cervicogential prevalence and seroprevalence in unvaccinated women in the USA using National Health and Nutrition Examination Survey data (2003–2010) and compare it to fits of statistical age–cohort models. We find that including a latent HPV state in our model significantly improves model fit and that antibody waning may be an important contributor to observed patterns of seroprevalence. Moreover, we find that the mechanistic model outperforms the statistical model and that the joint analysis prevents the inconsistencies that arise when estimating historical cohort trends in infection from genital prevalence and seroprevalence separately. Our analysis suggests that while there is substantial uncertainty associated with the estimation of historic HPV trends, there has likely been an increase in the force of infection for more recent birth cohorts.
This article is part of the theme issue ‘Silent cancer agents: multi-disciplinary modelling of human DNA oncoviruses’.
Keywords: human papillomavirus, age–period–cohort model, mechanistic model, seroprevalence, latent infection, waning antibodies
1. Introduction
The human papillomavirus (HPV) is the most common sexually transmitted infection in the USA [1] and is the aetiological agent of many anogenital and oral cancers, including cervical, anal and oropharyngeal. Incidence of HPV-related cancers is increasing [2], generating substantial interest in understanding how trends and patterns of HPV infection in the population are changing. Models connecting HPV prevalence to cancer incidence may allow for the prediction of future trends in HPV-related cancers [3]. Because testing for genital HPV at the population level in the USA did not begin until 2003 for women [4] (and 2013 for men [5]), there are no direct measurements of how HPV incidence and prevalence have changed over the past decades.
Instead, researchers have used mathematical models (e.g. catalytic models connecting seroprevalence (prevalence of presence of antibodies) to the force of infection) or statistical models (e.g. age–period–cohort models) to estimate and reconstruct HPV genital prevalence or seroprevalence over time [6–8]. However, these models paint a complicated picture. Models of seroprevalence in the USA have suggested that antibody prevalence has been increasing since the Sexual Revolution cohort (around 1950) [6,8], consistent with the rising trend in the number of lifetime sexual partners [8]. By contrast, cervicogenital HPV prevalence in women may have decreased over that time period [6]. These results are not inherently contradictory because prevalence and seroprevalence are very different markers. The presence of HPV DNA is a measure of current infection, while seroposivity indicates that a person was infected (or vaccinated) at some time in the past. Seronegativity, moreover, does not indicate that a person has never been infected, as not every infection causes seroconversion and as the body may cease the production of antibodies. Still, it is hard to fully reconcile past modelling results, and the overall picture remains unclear.
To unify these results and develop a more complete picture of HPV infection trends, we jointly and mechanistically model HPV genital infection and seropositivity, comparing and contrasting the results to statistical age–cohort trend analyses of cervicogenital HPV prevalence and seroprevalence data separately.
2. Material and methods
(a). Data
We use data from the US National Health and Nutrition Examination Survey (NHANES), a series of cross-sectional studies administered by the National Center for Health Statistics combining physical examinations, laboratory tests and interviews. Through complex sampling design and participant weighting, NHANES is designed to be a representative sample of the non-institutionalized, civilian US population [9]. Participant sample weights are incorporated in all data analysis. In our analysis, we include data from 6442 female participants between the ages of 18 and 59, who were sampled in 2003–2004, 2005–2006, 2007–2008 or 2009–2010, were tested conclusively and typed for both cervicogenital HPV infection and HPV L1 serum antibodies and had not received any doses of an HPV vaccine. This restricted population did not differ substantially in terms of demographic and sexual and reproductive health behaviour characteristics from the larger population of all 8148 women between ages 18 and 59 sampled in these cycles (see the electronic supplementary material).
The presence of HPV was assessed by a linear array genotyping assay based on L1 consensus polymerase chain reaction amplification of target DNA with biotinylated PGMY primers; genotype-specific prevalence was determined by nucleic acid hybridization of amplicons appropriate to each genotype. Seropositivity was assessed with a multiplexed Luminex assay, where type-specific neutralizing antibodies compete with serum antibodies to bind to conformationally sensitive neutralizing epitopes on the HPV virus-like particles. Detailed laboratory methods are publicly available [9]. Although 37 genotypes were analysed in the full cervicogenital prevalence data, we only consider the four genotypes for which there was serotyping data, namely, 6, 11, 16 and 18. These four genotypes had a population cervicogenital prevalence of 8.5% among unvaccinated women 18–59 in 2003–2010 (compared with 42.3% for any of the 37 genotypes).
For each age 18–59 in each NHANES cycle, we extract the number of people tested for cervicogential HPV or HPV serum antibodies and the weighted number of people who (i) were positive for cervicogenital HPV but had no antibodies, (ii) had antibodies but no current cervicogenital infection, (iii) were positive for cervicogenital HPV and had antibodies and (iv) were negative for cervicogenital HPV and had no antibodies. These four categories are a partition of the population.
(b). Models
(i). Age–cohort model
Age–period–cohort (APC) models are statistical models used to understand how different facets of time—age, period (calendar year) and cohort (birth year)—affect patterns of disease incidence or prevalence in the population [10–13]. Because full APC models suffer from a non-identifiability problem [11], where all three effects cannot be estimated independently from age-specific incidence or prevalence temporal data alone, we use an age–cohort model for the cervicogenital prevalence and seroprevalence of HPV 6, 11, 16 or 18. We choose age–cohort instead of age–period because HPV transmission is driven primarily by sexual contact, and sexual behaviour varies by age and by birth cohort [8]. The model is
| 2.1 |
where P is prevalence, a is age, c is birth cohort, and βA and βC are continuous functions (here, natural splines). Given the data P, one estimates the parameters of βA and βC in a generalized linear regression framework [14]. This model is the same used by Brouwer et al. [6] (though that analysis used the full cervicogential genotype data) and serves as our baseline analysis. Here, we model ages 18–59 over the 2003–2010 period, corresponding to birth cohorts 1944–1991. We allow 4 d.f. to the age splines and five to the cohort splines, corresponding to approximately 1 d.f. per 10-year span, a widely used rule of thumb for APC models.
(ii). Disease model
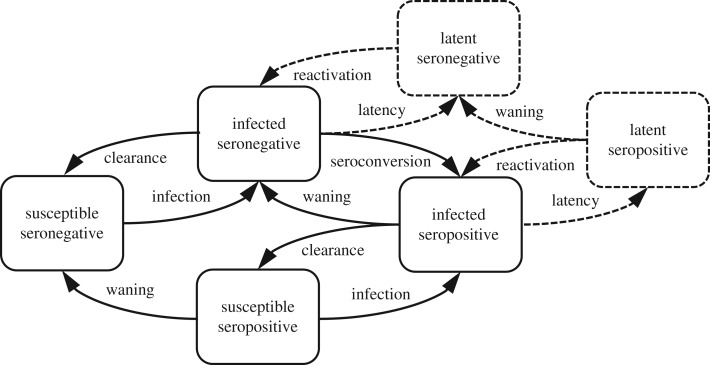
In order to jointly model HPV genital infection and seropositivity, we mechanistically model the fraction of each birth cohort 1944–1991 in each of four disease–sero states (susceptible and seronegative, infected and seronegative, infected and seropositive, and susceptible and seropositive) over time (figure 1). We model the dynamics of each birth cohort over time (i.e. as the cohort ages) separately, assuming no demographic changes. Each birth cohort is simulated starting at age 0 fully susceptible. Because reactivation of latent infections may be a relevant contributor to patterns of prevalence [15], particularly in explaining the higher prevalence in older women, we also include seronegative and seropositive latent states. In this population and with this limited set of genotypes, multiple infections are rare. Here, 0.7% (0.5–1.0%) of women had a genital infection with more than one of the four genotypes (i.e. only 8.4% (5.7–11.1%) of women with a genital infection were positive for more than one of the four genotypes). For simplicity, we do not explicitly model infections with multiple genotypes. We thus modelled six state transitions: infection (see below), clearance (with rate γ), seroconversion (with rate σ), waning antibodies (with rate ω), entering latency (with rate ν) and reactivation of latent infection (with rate μ).
Figure 1.
Schematic of a mechanistic model with cervicogenital HPV infection, HPV sero status (the presence of antibodies to HPV) and potential HPV latency.
Unlike vaccination, which has a substantial protective effect, naturally acquired antibodies are thought to confer at best modest protection for women [16] (while evidence from the HPV in men (HIM) trial suggests that they are not protective in men [17–19]). To model this possible protection in our female population, we include a parameter ρ that represents the attenuation of transmission to seropositive women. Because we model all four genotypes together, this parameter represents a weighted average of same genotype and cross-genotype protection across the four genotypes.
In the usual transmission model framework, the force of infection λ, i.e. the rate at which susceptible individuals become infected, depends on the current prevalence. Because we do not have corresponding age-specific genital HPV prevalence information for men for this time period and as cohort-specific sexual mixing patterns are not well understood, we instead use the simplifying assumption that the force of infection λ is proportional to the rate of partner acquisition. Hence, this model is not, strictly speaking, a transmission model. Analogous to the age–cohort model above, here we model λ(c, a) as the product of a function of the age of the cohort φ(a) and a cohort-dependent coefficient λ0(c),
| 2.2 |
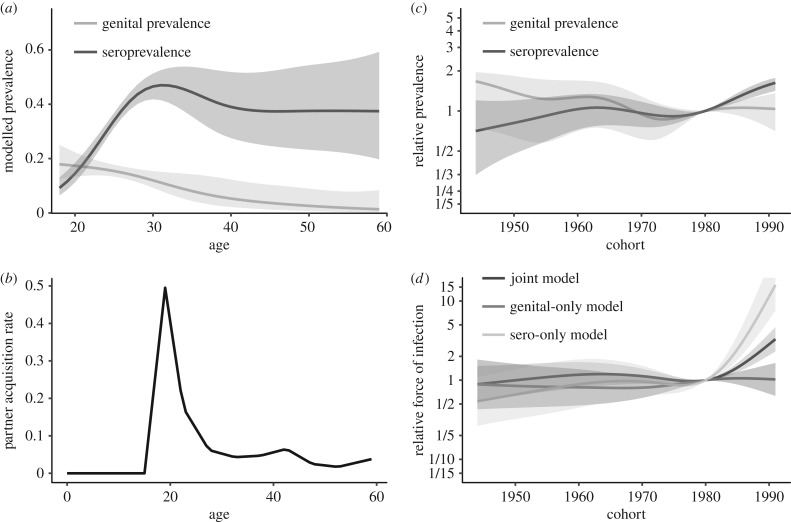
We denote the parameters of the natural spline modelling λ0(c) as ; as above, we use 5 d.f. for the cohorts 1944–1991. We use previous estimates of the age-specific partner acquisition rate as our φ(a) [8] (presented later in figure 3c).
Figure 3.
(a) Age-specific cervicogenital prevalence and seroprevalence of HPV 6, 11, 16 or 18 predicted by an age–cohort model. (b) Age-specific partner acquisition rate, adapted from Ryser et al. [8], used in the mechanistic disease model force of infection. (c) Relative prevalence and seroprevalence by birth cohort (relative to 50% prevalence), as predicted by an age–cohort model. (d) Cohort-specific relative force of infection λ(c) (1980 reference cohort) for the three mechanistic disease models.
The unconstrained model parameters are given in table 1, and the model equations are
| 2.3 |
Here, the dot indicates the derivative with respect to age a. We assume that we observe samples of Ipos and Ineg in the data, as well as samples of the sums Sneg + Lneg and Spos + Lpos since we cannot distinguish between those who test negative because they are not infected and those who test negative because their infections are latent.
Table 1.
Parameters of the joint HPV prevalence and seroprevalence model.
| parameter | definitions |
|---|---|
| λ | force of infection (yr−1) |
| γ | HPV clearance rate (yr−1) |
| σ | seroconversion rate (yr−1) |
| ω | antibody decay rate (yr−1) |
| ρ | attenuation of force of infection when seropositive |
| ν | rate of entering latency (yr−1) |
| μ | rate of reactivation from latency (yr−1) |
(iii). Parameter estimation
We used maximum-likelihood estimation to fit the model to the NHANES data, assuming a multinomial distribution of the data, corresponding to a likelihood
| 2.4 |
where θ is the vector of parameters. Here, n is the sample size at a given age and birth cohort. Because we cannot distinguish between people who are susceptible and those who are latent from the DNA test, Sneg + Lneg represents the modelled proportion of the population who do not have cervicogenital HPV types 6, 11, 16 or 18 and are seronegative (with Ineg, Spos + Lpos, Ipos analogously defined). Then, is the weighted number of individuals who do not have cervicogenital HPV types 6, 11, 16 or 18 and are seronegative in the NHANES data (with , , analogously defined).
The corresponding likelihoods when considering genital HPV infection and seropositivity on their own are, respectively,
| 2.5 |
where I = Ineg + Ipos (and similarly for S and L) for the disease model or the prevalence of those with genital infections predicted by the APC model, and
| 2.6 |
where Apos = Spos + Ipos + Lpos, and similarly for Aneg, for the disease model or the prevalence of those who are seropositive as predicted by the APC model.
We minimize the relative negative log-likelihood (NLL) in R v. 3.4.1 using a Nelder–Mead algorithm in the base package (function optim). Simulation of the differential equation model used the deSolve package. All model parameters are structurally identifiable from the data (see the electronic supplementary material). Questions of practical identifiability are outside of the scope of this analysis but could be addressed in the future. Here, uncertainty quantification for individual parameters and the natural splines was done using the inversion of the Hessian matrix returned by the optimization algorithm.
We compare four hypotheses corresponding to the following model constraints
-
—
Model 0. There are no latent states, and serostatus does not affect infection rates (μ = ν = 0, ρ = 1).
-
—
Model 1. There are no latent states, and serostatus affects infection (μ = ν = 0).
-
—
Model 2. There are latent states, and serostatus does not affect infection rates (ρ = 1).
-
—
Model 3. There are latent states, and serostatus affects infection (no constraints).
Nested models are compared by likelihood-ratio test and by the Aikake information criterion (AIC, +2k, where k is the number of parameters).
3. Results
(a). Model selection
We compare the fits of the joint genital–sero disease model with the four different constraints (table 2). We find that the models that allow latency (non-zero μ and ν), i.e. Models 2 and 3, are able to fit the data much more closely than the models that do not allow latency, i.e. Models 0 and 1. Between the models with latency, the difference in model fit when serostatus was allowed to affect the infection was not sufficient to justify the additional parameter. Hence, we use Model 2, the model with latency but with no effect of serostatus on infection, as our final joint genital–sero model.
Table 2.
Comparison of mechanistic model fits and model selection. We select the model with latency but no effect of serostatus on infection using two model selection criteria. Smaller relative Akaike information criterion (AIC) values indicate better fits after adjusting for the number of parameters. We give the AIC of each model relative to Model 2, the model with the smallest AIC. We also compare models pairwise with the likelihood-ratio test. p-Values are based on the likelihood-ratio test with the noted model and degrees of freedom equal to the difference in number of parameters. A small p-value means that we reject the comparison model.
| model | description | number of parameters | AIC relative to Model 2 | comparison model for likelihood-ratio | p-value |
|---|---|---|---|---|---|
| Model 0 | no latency, no effect of serostatus | 9 | 57.04 | — | — |
| Model 1 | no latency, serostatus affects infection | 10 | 47.97 | Model 0 | 9.7 × 10−4 |
| Model 2 | with latency, no effect of serostatus | 11 | 0 | Model 0 | 4.4 × 10−14 |
| Model 3 | with latency, serostatus affects infection | 12 | 1.93 | Model 2 | 0.77 |
In addition to jointly modelling cervicogenital HPV infection and seropositivity measures, we fit Model 2 to each measure separately, which we call the genital-only and sero-only models. Because ρ = 1 under this model, the genital-only model collapses to a three-compartment model with S = Sneg + Spos, etc., and does not depend on σ and ω (see the electronic supplementary material). The sero-only model uses the full model. We then compare, for each measure, how well each of three models fit the data for that measure: the age–cohort model, the joint genital–sero model (Model 2 with parameters estimated using both measures), the single-measure model (Model 2 with parameters estimated using only the single measure) and the other single-measure model (Model 2 with parameters estimated using only the alternate single measure). The statistical fits of these models to each type of data are compared in table 3. The mechanistic disease models fit substantially better than the statistical age–cohort models, perhaps because the age splines do not have the implicit information about under-18-year-olds that the partner acquisition rate gives the mechanistic model. In each case, the joint genital–sero model fits worse than the single-measure model informed by that measure but not as poorly, at least for the genital data, as the single-measure model informed by the other measure. That is, for example, the sero-only model fits the seroprevalence data better than the joint genital–sero model, but the joint sero-genital model simultaneously fits both the genital prevalence and seroprevalence data better than the sero-only model does. These results suggest that the variability in each dataset allows a single-measure model to fit to each measure in a way that is inconsistent with the other measure. This result underscores the importance of the joint, mechanistic approach to the problem.
Table 3.
Comparison of model fits to the genital HPV prevalence and antibody seroprevalence data separately, comparing the single-measure disease models (models with parameters informed only by the fit to one measure), the joint genital–sero disease model (model with parameters informed by fit to both measures) and the APC model. Smaller relative Akaike information criterion (AIC) values indicate better fits after adjusting for the number of parameters. The ‘dagger’ symbol denotes that there is no natural fit of the genital-only model to the seropositivity data without assuming values for σ and ω.
| measure | model | number of parameters | relative AIC |
|---|---|---|---|
| genital infection | genital-only disease model | 9 | 0 |
| sero-only disease model | 9 | 75.51 | |
| joint genital–sero disease model | 11 | 16.44 | |
| genital APC model | 9 | 941.48 | |
| seropositivity | genital-only disease model | 9 | † |
| sero-only disease model | 11 | 0 | |
| joint genital–sero disease model | 11 | 10.16 | |
| sero APC model | 9 | 482.34 |
(b). Model results
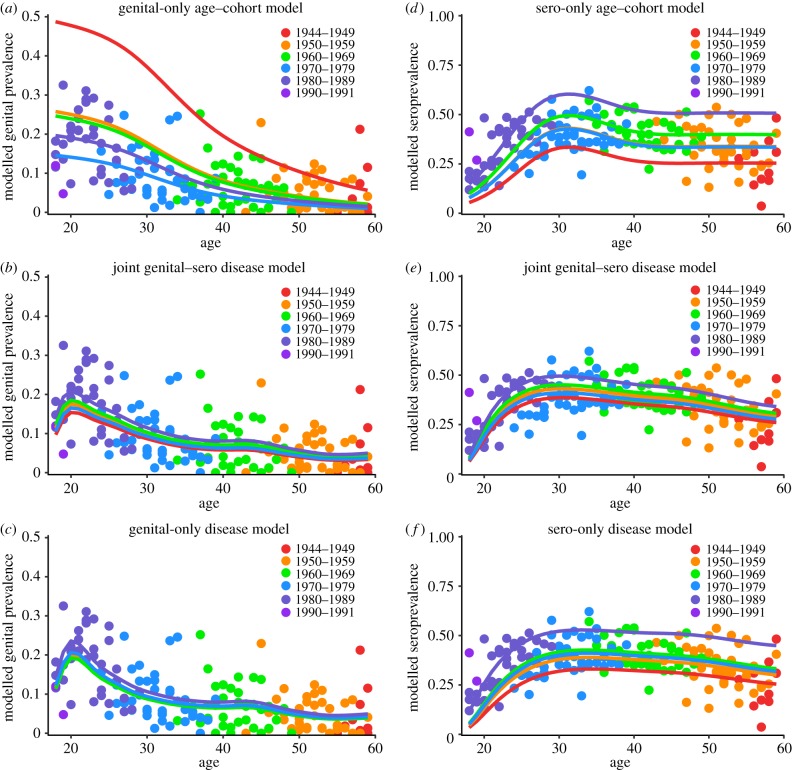
While the age–cohort and disease models predict similar general age-specific cervicogenital prevalence shapes, both achieving their highest values among young people and slowly dropping off with age, the joint genital–sero disease model suggests a different interpretation of the data from the age–cohort model (figure 2a,b). Here, cervicogenital prevalence shows a defined peak in the early 1920s before decreasing for older ages, except for the very slight second bump in the mid-1940s. Without the information about HPV prevalence for women under 18, the APC model does not model the peak that the disease model does (figure 3a); the partner acquisition rate giving the shape of the force of infection is given in figure 3b. Moreover, the age–cohort model predicts substantial spread over the cohorts. By contrast, the disease model predicts a relatively small range of prevalences by cohort. The cohort effects predicted by the two models also differ, with the age–cohort model predicting a decrease between the 1940s and 1970s, followed by a plateau, and the joint genital–sero model predicting a slight peak in the 1960s and a substantial increase starting in the early 1980s (figure 3c,d). The model fitted only to the genital data (figure 2c) predicts a similar shape but less variation by cohort than the joint model. Interestingly, the cohort effects of the genital-only model are almost opposite those estimated by the joint model, which underscores the difficulty in interpreting trends in these data.
Figure 2.
(a) The age–cohort model fit to cervicogenital prevalence of HPV 6, 11, 16 or 18 by birth cohort. (b) The joint genital–sero mechanistic disease model (fit jointly to both measures) plotted on to cervicogenital prevalence. (c) The genital-only mechanistic disease model fit to and plotted on cervicogenital prevalence. (d) The age–cohort model fit to seroprevalence of antibodies to HPV 6, 11, 16 or 18 by cohort. (e) The joint genital–sero mechanistic disease model plotted on seroprevalence. (f) The sero-only mechanistic disease model fit to and plotted on seroprevalence. All mechanistic models are based on Model 2.
The age–cohort and disease models predict similar age-specific seroprevalence shapes for early ages, but they diverge for the older ages. The age–cohort model increases to a peak in the early 1930s but decreases again to a plateau around 1940. The joint genital–sero model similarly predicts a peak in the early 1930s but estimates a gradual decrease in seroprevalence over the rest of the lifetime (figure 2d,e). The two models predict qualitatively similar cohort effects: increasing to a peak in the 1960s, followed by a slight decline in the 1970s, but a larger increase beginning in the 1980s (figure 3c,d). The model fitted only to the seroprevalence data predicts exaggerated cohort variability and an overall flatter profile than the joint model (figure 2f).
Parameter estimates for the joint genital–sero disease model are given in table 4 (parameters for the single-measure models are provided in the electronic supplementary material). The median time between infection and either clearance or latency is 5.7 months. The mean time before antibody waning is 20.8 years. The mean time for reactivation of a latent infection is 1.9 years.
Table 4.
Maximum-likelihood parameter estimates and 95% confidence intervals for the joint HPV prevalence and seroprevalence model.
| parameter | value | 95% CI | definitions |
|---|---|---|---|
| λ0 (1980) | 0.51 | (0.43, 0.59) | force of infection coefficient for 1980 birth cohort |
| γ | 0.41 | (0.29, 0.52) | HPV cervicogenital clearance rate (yr−1) |
| σ | 0.74 | (0.62, 0.86) | seroconversion rate (yr−1) |
| ω | 0.048 | (0.035, 0.061) | rate of waning immunity (yr−1) |
| ν | 1.06 | (0.75, 1.36) | rate of entering latency (yr−1) |
| μ | 0.53 | (0.28, 0.77) | rate of reactivation from latency (yr−1) |
4. Discussion
In this analysis, we developed a mathematical model to jointly describe HPV cervicogenital prevalence and seroprevalence data from women in the USA in NHANES. We compared this analysis to an age–cohort statistical description of the data. Our analysis highlights the difficulty of using available HPV prevalence and seroprevalence to estimate historical trends: the data admit different, sometimes strikingly different, interpretations depending on the model assumptions. This limitation (of not only our study but all similar studies) is a result of the wide intra-cohort variation and relatively small age overlap in the data available for different cohorts; as more population-level data are collected in future NHANES cycles, we will gain confidence in the analyses. Another, more structural limitation is that estimating serostatus based on a cut-off may be prone to misclassification [20]; in general, a mixture model approach could address this problem when viral titres are available, but NHANES currently reports only genotype-specific seropositivity. Finally, we do not consider co-infections, which are relatively rare when considering only the four genotypes, but they may alter the dynamics slightly and will need to be accounted for if larger panels of HPV seroprevalence become available in the future. Despite these limitations, the strength of this analysis lies in the mechanistic construction of the model, which constrains the interpretation to be biologically reasonable, and in its simultaneous treatment of both genital prevalence and seroprevalence, preventing interpretations of trends in one that are inconsistent with trends in the other, as the model fitted to each measure separately demonstrates (figure 3d). All previous analyses of HPV trends, including our own, used statistical models or used only genital or seroprevalence but not both [6–8].
The shape of the force of infection in our analysis was based on the age-specific number of sexual partners modelled by Ryser et al. [8]. However, our estimated trends in the force of infection by birth cohort (figure 3d), estimated by our model, differ somewhat from the cohort trends in the number of sexual partners. In the Ryser et al. analysis, the cohort factor increased steadily from the 1950s birth cohort before peaking in the 1980s cohort and coming down. While that result may seem to conflict with our interpretation of birth cohort trends—a small peak in the mid-1960s dipping to 1980 and increasing substantially in the mid-1980s—the number of sexual partners is only one aspect of the force of infection. The force of infection also depends on the prevalence in the cohorts of the sexual partners (which we do not model) and on the probability of transmission, which is influenced by condom use, for instance. The cohort trends predicted by this model are consistent with increased condom use in response to the HIV/AIDS epidemic and other factors in this time frame [21–23]. However, condom use seems to have largely plateaued since the early 2000s [24,25]. The model suggests that the force of infection for birth cohorts in the late 1980s and early 1990s may be increasing beyond levels seen in previous decades. However, we have comparatively little data on these cohorts, so this prediction should be revised as additional data becomes available. It is unclear what might be driving such an increase: there is no indication of substantial declines in condom use in the USA, and Ryser et al. reported that later cohorts may in fact have lower partner acquisition rates than the preceding cohorts [8]. It may also be the case that HPV genotypes have waxed and waned as fractions of the overall HPV prevalence and that, by considering only the 6, 11, 16 and 18 subset, the trends we estimate here may differ from the trends for infection of any HPV type.
HPV vaccines, which are targeted at pre-adolescents, have been approved for use in certain populations in the USA since 2006 (for only women initially and men later) [26]. Vaccination results in higher antibody titres than produced by infection and appears to offer protection against the vaccine genotypes for a decade or more [27]. Because there were relatively few vaccinated women in the age range for these NHANES cycles and because the effect of seropositivity on infection and clearance is likely different depending on whether the antibodies were produced from infection or vaccination, we decided to exclude vaccinated women from this analysis. Analysis of future NHANES cycles will need to account for vaccination, however, not only because of its effect on disease initiation and progression but because the shape of the force of infection may begin to change as a greater percentage of young people are vaccinated.
Previous analyses of seroprevalence trends in the USA and the UK concluded that antibody waning is minimal and does not substantially contribute to observed patterns of seroprevalence. This interpretation is consistent with our finding that the sero-only analyses predict flatter seroprevalence trajectories (figure 2e,f). Our joint genital–sero analysis, by contrast, demonstrates that slow but meaningful waning (in the order of 20 years) is consistent with observed patterns of both seroprevalence and cervicogenital prevalence. Studies have shown that antibodies to HPV16 tend to be durable [28–30], particularly when individuals have persistent HPV 16 infections. However, other studies have found HPV antibodies to not persist longer than a few years, particularly if the genital infection did not persist or if the infection was a type other than HPV 16 [30,31]. Our results, together with the literature, suggest that waning of natural antibodies could indeed be an important contributor to patterns of seroprevalence.
The estimates of the cervicogenital HPV clearance rate and the rate of infections entering latency in our model correspond to a median time to loss of detectable HPV DNA of approximately six months (and a mean time of approx. eight months). Estimates of the time to clear an HPV infection has varied between studies but appears to have a median of approximately eight to nine months [32–36], suggesting that the model may be overestimating one or both of the relevant parameters (γ or ν).
Many viruses, including cytomegalovirus, Epstein–Barr virus and herpesviruses, are considered ‘silent agents’, whether of carcinogenesis or other diseases, because of their ability to lie latent in the human body for spans of time, only to reactivate later. Although it has been challenging to interpret seroprevalence data in the context of viral latency and reactivation, population-level modelling approaches are offering new insights into how these underlying processes could be driving observed seroprevalence patterns [37–39]. In the context of HPV, it has so far been unclear how important a possible latent infection state is to patterns of HPV prevalence. Although the existence of an immunologically controlled latent state is likely [40–42], the duration of such latency is unknown and likely depends on local irritation or systemic immunological changes. In men, latency, along with autoinnoculation, has been suggested as the mechanism behind successive infections with the same genotype in men [19]. Reactivation after latency in response to menopause-associated changes could also play a role in HPV prevalence peak in older women [43,44], although this peak is also consistent with a so-called second sexual debut around this age. We did not model age-dependent latency here, but we did find that a relatively short latency period (in the order of 2 years) significantly improved the model fit to the data.
It has also so far been unclear how much of an effect cross-reactivity has on the acquisition of new HPV types related to types that one already has antibodies to. Natural immunity has been shown to be largely genotype specific, with limited cross-reactivity in certain species [45]. In this analysis, we found that modelling a reduction in acquisition of strains when one was seropositive did not improve model fit to the data. Given the cross-sectional rather than longitudinal nature of the dataset and the limited number of genotypes we were able to consider, this finding should be considered weak evidence that there is not, at least, a substantial protective effect. This is consistent with previous studies showing that natural infection antibodies are at most modestly protective [16].
Although this model is a mechanistic model of infection in the style of the classic SIR (susceptible–infectious–recovered) model, it is not, strictly speaking, a transmission model as the force of infection does not depend on the model states. Future work may incorporate transmission into this style of model to assess trends, although there are several barriers, not least of which is the need to handle age- and cohort-dependent sexual contact patterns. Nevertheless, this analysis demonstrates that it is possible to include APC frameworks within SIR-style infectious disease models. Furthermore, although this model is structurally identifiable, more future work will need to address the practical identifiability and uncertainty quantification of model parameters from real data, particularly as it relates to possible latent classes. Finally, extending the model to consider the dynamics of distinct types and multiple infections would be a useful direction for future work.
The framework we used here could be modified for other non-persisting viruses. We are not the first to integrate measures of past and current infection for a disease, but this approach is arguably underutilized, perhaps because it is relatively rare to have simultaneous measures of current and past infection in the same population. Nevertheless, increased awareness of methods capable of integrating such data, as well as the increased power we gain for making inferences, may make simultaneous data collection more common in the future. Simultaneous collection will be incredibly useful, for example, when symptoms of infection are rare and asymptomatic shedding is contributing to sustained transmission (as is likely the case for many enteric viruses [46]). Of course, the modelling framework will need to be modified depending on whether the infection is immunizing and antigenically stable (e.g. measles), immunizing but antigenically variable (e.g. influenza, dengue) or non-immunizing (e.g. malaria, HIV) [47].
5. Conclusion
Trend analysis of HPV cervicogenital prevalence and seroprevalence in the USA has been difficult and lacked robustness because of the large intra-cohort variation in prevalence and the relatively small span of the data. Our mechanistic model jointly describing cervicogenital and seroprevalence demonstrated that a latent infection state and waning natural antibodies may both play a role in populational-level prevalence patterns. Moreover, while there are indications that there may be a substantial increase in HPV prevalence in more recent birth cohorts, increasing rates of HPV vaccination may ultimately control adverse HPV-related outcomes, including genital warts and cancer.
Supplementary Material
Supplementary Material
Data accessibility
The datasets supporting this article have been uploaded as part of the electronic supplementary material. Select code is available through a GitHub repository: https://github.com/epimath/joint-genital-sero-HPV-model.
Authors' contributions
Conceived of the work: A.F.B., R.M. and M.C.E.; performed analysis: A.F.B.; drafted manuscript: A.F.B.; and revised and approved manuscript: A.F.B., R.M. and M.C.E.
Competing interests
We have no competing interests.
Funding
All authors were supported by NIH grant no. U01CA182915. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
References
- 1.Centers for Disease Control and Prevention. 2017. Genital HPV infection - fact sheet. See https://www.cdc.gov/std/hpv/stdfact-hpv.htm.
- 2.Jemal A. et al. 2013. Annual report to the nation on the status of cancer, 1975–2009, featuring the burden and trends in human papillomavirus (HPV)-associated cancers and HPV vaccination coverage levels. J. Natl Cancer Inst. 105, 175–201. ( 10.1093/jnci/djs491) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brouwer AF, Eisenberg MC, Meza R. 2018. Case studies of gastric, lung, and oral cancer connect etiologic agent prevalence to cancer incidence. Cancer Res. 78, 3386–3397. ( 10.1158/1538-7445.AM2018-1027) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dunne EF, Unger ER, Sternberg M, McQuillan G, Swan DC, Patel SS, Markowitz LE. 2007. Prevalence of HPV infection among females in the United States. J. Am. Med. Assoc. 297, 813–819. ( 10.1001/jama.297.8.813) [DOI] [PubMed] [Google Scholar]
- 5.Gargano JW, Unger ER, Liu G, Steinau M, Meites E, Dunne E, Markowitz LE. 2017. Prevalence of genital human papillomavirus in males, United States, 2013–2014. J. Infect. Dis. 215, 1070–1079. ( 10.1093/infdis/jix057) [DOI] [PubMed] [Google Scholar]
- 6.Brouwer AF, Eisenberg MC, Carey TE, Meza R, Name MM, Gene L, Pairs B, Temp A. 2015. Trends in HPV cervical and seroprevalence and associations between oral and genital infection and serum antibodies in NHANES 2003–2012. BMC Infect. Dis. 15, 575 ( 10.1186/s12879-015-1314-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Desai S. et al. 2011. Prevalence of human papillomavirus antibodies in males and females in England. Sex. Transm. Dis. 38, 622–629. ( 10.1097/OLQ.0b013e31820bc880) [DOI] [PubMed] [Google Scholar]
- 8.Ryser MD, Rositch A, Gravitt PE. 2017. Modeling of US human papillomavirus (HPV) seroprevalence by age and sexual behavior indicates an increasing trend of HPV infection following the sexual revolution. J. Infect. Dis. 216, 604–611. ( 10.1093/infdis/jix333) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Centers for Disease Control and Prevention. 2018. National health and nutrition examination survey. See http://www.cdc.gov/nchs/nhanes.htm.
- 10.Holford TR. 1983. The estimation of age, period and cohort effects for vital rates. Biometrics 39, 311–324. ( 10.2307/2531004) [DOI] [PubMed] [Google Scholar]
- 11.Holford TR. 1991. Understanding the effects of age, period, and cohort on incidence and mortality rates. Annu. Rev. Public Health 12, 425–457. ( 10.1146/annurev.pu.12.050191.002233) [DOI] [PubMed] [Google Scholar]
- 12.Clayton D, Schifflers E. 1987. Models for temporal variation in cancer rates. II: Age-period-cohort models. Stat. Med. 6, 469–481. ( 10.1002/sim.4780060406) [DOI] [PubMed] [Google Scholar]
- 13.Clayton D, Schifflers E. 1987. Models for temporal variation in cancer rates. I: Age-period and age-cohort models. Stat. Med. 6, 449–467. ( 10.1002/sim.4780060405) [DOI] [PubMed] [Google Scholar]
- 14.Durrleman S, Simon R. 1989. Flexible regression models with cubic splines. Stat. Med. 8, 551–561. ( 10.1002/sim.4780080504) [DOI] [PubMed] [Google Scholar]
- 15.Gravitt PE. 2011. The known unknowns of HPV natural history. J. Clin. Invest. 121, 4593–4599. ( 10.1172/JCI57149) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beachler DC, Jenkins G, Safaeian M, Kreimer AR, Wentzensen N. 2016. Natural acquired immunity against subsequent genital human papillomavirus infection: a systematic review and meta-analysis. J. Infect. Dis. 213, 1444–1454. ( 10.1093/infdis/jiv753) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu B. et al. 2012. Prevalent serum antibody is not a marker of immune protection against acquisition of oncogenic HPV16 in men. Cancer Res. 72, 676–685. ( 10.1158/0008-5472.CAN-11-0751) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pamnani SJ, Nyitray AG, Abrahamsen M, Rollison DE, Villa LL, Lazcano-Ponce E, Huang Y, Borenstein A, Giuliano AR. 2016. Sequential acquisition of anal human papillomavirus (HPV) infection following genital infection among men who have sex with women: the HPV infection in men (HIM) study. J. Infect. Dis. 214, 1180–1187. ( 10.1093/infdis/jiw334) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ranjeva SL, Baskerville EB, Dukic V, Villa LL, Lazcano-Ponce E, Giuliano AR, Dwyer G, Cobey S. 2017. Recurring infection with ecologically distinct HPV types can explain high prevalence and diversity. Proc. Natl Acad. Sci. USA 114, 13 573–13 578. ( 10.1073/pnas.1714712114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vink MA. 2015. Estimating seroprevalence of human papillomavirus type 16 using a mixture model with smoothed age-dependent mixing proportions. Epidemiology 26, 8–16. ( 10.1097/EDE.0000000000000196) [DOI] [PubMed] [Google Scholar]
- 21.Sonenstein FL, Pleck JH, Ku LC. 1989. Sexual activity, condom use and AIDS awareness among adolescent males. Fam. Plan. Perspect. 21, 152–158. ( 10.2307/2135805) [DOI] [PubMed] [Google Scholar]
- 22.Bankole A, Darroch JE, Singh S. 1999. Determinants of trends in condom use in the United States, 1988–1995. Fam. Plan. Perspect. 31, 264–271. ( 10.2307/2991536) [DOI] [PubMed] [Google Scholar]
- 23.Catania JA. et al. 2001. National trends in condom use among at-risk heterosexuals in the United States. J. Acquir. Immune Defic. Syndr. 27, 176–182. ( 10.1097/00126334-200106010-00013) [DOI] [PubMed] [Google Scholar]
- 24.Anderson JE, Warner L, MacAluso M. 2011. Condom use among US adults at last sexual intercourse, 1996–2008: an update from national survey data. Sex. Transm. Dis. 38, 919–921. ( 10.1097/OLQ.0b013e31822545d9) [DOI] [PubMed] [Google Scholar]
- 25.Kahn L, Lowry R, Eaton D, Wechsler H. 2012. Trends in HIV-related risk behaviors among high school students–United States, 1991–2011. Morb. Mortal. Wkly. Rep. 61, 556–560. [PubMed] [Google Scholar]
- 26.U.S. Food and Drug Administration. 2018. Human papillomavirus vaccine. See https://www.fda.gov/biologicsbloodvaccines/vaccines/approvedproducts/ucm172678.htm.
- 27.Pinto LA, Dillner J, Beddows S, Unger ER. 2018. Immunogenicity of HPV prophylactic vaccines: serology assays and their use in HPV vaccine evaluation and development. Vaccine 36, 4792–4799. ( 10.1016/j.vaccine.2017.11.089) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.af Geijersstam V, Wang Z, Koskela P, Pukkala E, Schiller J, Lehtinen M, Dillner J. 1998. Stability over time of serum antibody levels to human papillomavirus type 16. J. Infect. Dis. 177, 1710–1714. ( 10.1086/jid.1998.177.issue-6) [DOI] [PubMed] [Google Scholar]
- 29.Shah KV, Viscidi RP, Alberg AJ, Helzlsouer J, Comstock W, Alberg J, Helzlsouer KJ, Comstock GW. 1997. Antibodies to human papillomavirus 16 and subsequent in situ or invasive cancer of the cervix. Cancer Epidemiol. Biomarkers Prev. 6, 233–237. [PubMed] [Google Scholar]
- 30.Carter JJ, Koutsky LA, Hughes JP, Lee SK, Kuypers J, Kiviat N, Galloway DA. 2000. Comparison of human papillomavirus types 16, 18, and 6 capsid antibody responses following incident infection. J. Infect. Dis. 181, 1911–1919. ( 10.1086/jid.2000.181.issue-6) [DOI] [PubMed] [Google Scholar]
- 31.Ho GY, Studentsov YY, Bierman R, Burk RD. 2004. Natural history of human papillomavirus type 16 virus-like particle antibodies in young women. Cancer Epidemiol. Biomarkers Prev. 13, 110–116. ( 10.1158/1055-9965.EPI-03-0191) [DOI] [PubMed] [Google Scholar]
- 32.Ho GY, Bierman R, Beardsley L, Chang CJ, Burk RD. 1998. Natural history of cervicovaginal papillomavirus infection in young women. N. Engl. J. Med. 338, 423–428. ( 10.1056/NEJM199802123380703) [DOI] [PubMed] [Google Scholar]
- 33.Franco ELL, Villa LLL, Sobrinho JPP, Prado JMM, Rousseau MC, Désy M, Rohan TEE. 1999. Epidemiology of acquisition and clearance of cervical human papillomavirus infection in women from a high-risk area for cervical cancer. J. Infect. Dis. 180, 1415–1423. ( 10.1086/jid.1999.180.issue-5) [DOI] [PubMed] [Google Scholar]
- 34.Giuliano AR, Sedjo RL, Roe DJ, Harris R, Baldwin S, Papenfuss MR, Abrahamsen M, Inserra P. 2002. Clearance of oncogenic human papillomavirus (HPV) infection: effect of smoking (United States). Cancer Causes Control 13, 839–846. ( 10.1023/A:1020668232219) [DOI] [PubMed] [Google Scholar]
- 35.Muñoz N, Méndez F, Posso H, Molano M, van den Brule, Meijer C, Muñoz A. 2004. Incidence, duration, and determinants of cervical human papillomavirus infection in a cohort of Colombian women with normal cytological results. J. Infect. Dis. 190, 2077–2087. ( 10.1086/jid.2004.190.issue-12) [DOI] [PubMed] [Google Scholar]
- 36.Goodman MT. et al. 2008. Prevalence, acquisition, and clearance of cervical human papillomavirus infection among women with normal cytology: Hawaii human papillomavirus cohort study. Cancer Res. 68, 8813–8824. ( 10.1158/0008-5472.CAN-08-1380) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hogea C, Dieussaert I, Effelterre TV, Guignard A, Mols J. 2015. A dynamic transmission model with age-dependent infectiousness and reactivation for cytomegalovirus in the United States: potential impact of vaccination strategies on congenital infection. Hum. Vaccin. Immunother. 11, 1788–1802. ( 10.1080/21645515.2015.1016665) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Boven M, van de Kassteele J, Korndewal MJ, van Dorp CH, Kretzschmar M, van der Klis F, de Melker HE, Vossen AC, van Baarle D. 2017. Infectious reactivation of cytomegalovirus explaining age- and sex-specific patterns of seroprevalence. PLoS Comput. Biol. 13, 1–18. ( 10.1371/journal.pcbi.1005719) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marinelli I, van Lier A, de Melker H, Pugliese A, van Boven M. 2017. Estimation of age-specific rates of reactivation and immune boosting of the varicella zoster virus. Epidemics 19, 1–12. ( 10.1016/j.epidem.2016.11.001) [DOI] [PubMed] [Google Scholar]
- 40.Maglennon GA. 2012. The biology of papillomavirus latency. Open Virol. J. 6(Suppl. 2), 190–197. ( 10.2174/1874357901206010190) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gravitt PE. 2012. Evidence and impact of human papillomavirus latency. Open Virol. J. 6(Suppl 2), 198–203. ( 10.2174/1874357901206010198) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schiffman M, Kjaer SK. 2003. Chapter 2: Natural history of anogenital human papillomavirus infection and neoplasia. J. Natl Cancer Inst. Monogr. 31, 14–19. ( 10.1093/oxfordjournals.jncimonographs.a003476) [DOI] [PubMed] [Google Scholar]
- 43.Rositch AF, Burke AE, Viscidi RP, Silver MI, Chang K, Gravitt PE. 2012. Contributions of recent and past sexual partnerships on incident human papillomavirus detection: acquisition and reactivation in older women. Cancer Res. 72, 6183–6190. ( 10.1158/0008-5472.CAN-12-2635) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gravitt PE, Rositch AF, Silver MI, Marks MA, Chang K, Burke AE, Viscidi RP. 2013. A cohort effect of the sexual revolution may be masking an increase in human papillomavirus detection at menopause in the United States. J. Infect. Dis. 207, 272–280. ( 10.1093/infdis/jis660) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scherpenisse M, Schepp RM, Mollers M, Meijer CJ, Berbers GA, van der Klis FR. 2013. Characteristics of HPV-specific antibody responses induced by infection and vaccination: cross-reactivity, neutralizing activity, avidity and IgG subclasses. PLoS ONE 8, 1–9. ( 10.1371/journal.pone.0074797) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dennehy PH. 2000. Transmission of rotavirus and other enteric pathogens in the home. Pediatr. Infect. Dis. J. 19, S103–S105. ( 10.1097/00006454-200010001-00003) [DOI] [PubMed] [Google Scholar]
- 47.Metcalf CJE, Farrar J, Cutts FT, Basta NE, Graham AL, Lessler J, Ferguson NM, Burke DS, Grenfell BT. 2016. Use of serological surveys to generate key insights into the changing global landscape of infectious disease. Lancet 388, 728–730. ( 10.1016/S0140-6736(16)30164-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets supporting this article have been uploaded as part of the electronic supplementary material. Select code is available through a GitHub repository: https://github.com/epimath/joint-genital-sero-HPV-model.