Abstract
Current HPV vaccines target a subset of the oncogenic human papillomavirus (HPV) types. If HPV types compete during infection, vaccination may trigger replacement by the non-targeted types. Existing approaches to assess the risk of type replacement have focused on detecting competitive interactions between pairs of vaccine and non-vaccine types. However, methods to translate any inferred pairwise interactions into predictors of replacement have been lacking. In this paper, we develop practical predictors of type replacement in a multi-type setting, readily estimable from pre-vaccination longitudinal or cross-sectional prevalence data. The predictors we propose for replacement by individual non-targeted types take the form of weighted cross-hazard ratios of acquisition versus clearance, or aggregate odds ratios of coinfection with the vaccine types. We elucidate how the hazard-based predictors incorporate potentially heterogeneous direct and indirect type interactions by appropriately weighting type-specific hazards and show when they are equivalent to the odds-based predictors. Additionally, pooling type-specific predictors proves to be useful for predicting increase in the overall non-vaccine-type prevalence. Using simulations, we demonstrate good performance of the predictors under different interaction structures. We discuss potential applications and limitations of the proposed methodology in predicting type replacement, as compared to existing approaches.
This article is part of the theme issue ‘Silent cancer agents: multi-disciplinary modelling of human DNA oncoviruses’.
Keywords: pathogen-type interactions, vaccination, prediction, type replacement, hazard ratio, odds ratio
1. Introduction
Predicting the impact of vaccination against a pathogen can be challenging if the pathogen consists of many, potentially interacting (sub)types. When the vaccine targets only a subset of the pathogenic types, it is particularly relevant to evaluate the risk of replacement by the non-targeted types. In this paper, we expand existing methodology for predicting type replacement for multi-type pathogens with the focus on the human papillomavirus (HPV).
HPV is one of the most common oncogenic DNA viruses in humans. Persistent infection with HPV can cause cancer in various parts of the body [1]. In particular, 12–15 HPV types are classified as high-risk or probable high-risk due to their association with cervical cancer [2]. Currently, three HPV vaccines are available, covering two, four and nine HPV types. All three vaccines protect against HPV 16 and 18, two high-risk types that together account for approximately 70% of all cervical cancers in unvaccinated populations. HPV 31, 33 and 45, accounting for an additional 10–15% of cases [3], are among the cross-protective types of the bivalent and quadrivalent vaccines and are included in the nonavalent vaccine [4].
In countries where HPV vaccination has been implemented with high coverage, circulation of both the vaccine and cross-protective HPV types has decreased considerably [5,6]. However, concerns have been raised that the ecological niche vacated by the targeted types could be taken over by the non-targeted high-risk HPV types [7–9]. Thus far, post-implementation surveillance data have been reassuring, as only sporadic increases in the prevalence of infection with some non-vaccine types have been observed without a clear signal of type replacement [4,10–12]. While waiting for the long-term impact of HPV vaccination to become apparent, evaluation of the potential for type replacement remains crucial.
Whether HPV vaccination will trigger type replacement depends on the existence and strength of competitive interactions between HPV types. HPV types may compete during coinfection, by either diminishing each other’s opportunities to establish a productive infection or by promoting viral clearance (e.g. through activation of antigen-presenting cells) [13–17]. Owing to such competitive mechanisms, the hazard of acquiring (or clearing) a given HPV type may be reduced (or increased, respectively) by infection with other types. In epidemiological terms, different type interactions can be conveniently quantified in terms of appropriately defined hazard ratios [18–20].
We have previously shown, in a simple model of one vaccine and one non-vaccine type, that the cross-hazard ratio of acquisition versus clearance can be used to predict type replacement, provided that the two types interact symmetrically and there is no long-lasting cross-immunity [21]. The latter assumption seems plausible for HPV as even the existence of natural homologous immunity is still debated [22,23]. Given susceptible–infected–susceptible (SIS) transmission dynamics, the appropriate cross-hazard ratio is, moreover, equivalent to an odds ratio of coinfection, and thus estimable from cross-sectional prevalence data [21]. Here, the odds ratio is defined as the odds of infection with one type in the presence versus absence of coinfection with the other type.
With more than two interacting types, not only does prediction of replacement require inference of interactions between multiple types but also an adequate way of combining them. To predict how vaccination will affect the prevalence of a given non-vaccine type, one needs to take account of its direct interactions with the vaccine types as well as any indirect interactions via other non-vaccine types. Meaningful prediction should also incorporate possible heterogeneities in strength or direction (competition versus synergy) of type interactions.
Previous studies evaluating the potential of HPV-type replacement have focused on inferring interactions between pairs of vaccine and non-vaccine types [7,24–27]. In these studies, for each vaccine type, pairwise odds ratios have been compared to pooled odds ratios (pooled across the non-vaccine types) to identify likely candidates for type replacement. With this approach, however, each non-vaccine type is evaluated multiple times according to its interactions with different vaccine types, while the occurrence of type replacement is determined by all these interactions jointly. In addition, the pooled odds ratio, which has been interpreted as the tendency of the vaccine type in question to be involved in coinfection, lacks a clear interpretation for prediction of type replacement.
In this paper, we consider prediction of replacement following vaccination in a setting with an arbitrary number of interacting vaccine and non-vaccine types. We propose predictors of type-specific replacement, i.e. increase in the prevalence of individual non-vaccine types, and pooled predictors of increase in the overall non-vaccine-type prevalence. The predictors are initially defined in terms of steady-state hazards of type-specific acquisition and clearance. Using a mechanistic SIS model, we explain how the predictors relate to the underlying mode of type interactions and show under which interaction structures these hazard-based predictors can be estimated as odds ratios from cross-sectional prevalence data. Finally, we evaluate the performance of the proposed predictors by means of numerical simulations.
2. Material and methods
(a). Prediction framework
We derive predictors of type replacement for a pathogen consisting of many potentially interacting types. The prediction method applies to any pathogen for which naturally acquired immunity is short-lived, so that the transmission dynamics of each type can be approximated by an SIS model. The predictors are constructed in terms of the following data collected at the pre-vaccination steady state:
-
—
prevalence of each (co)infection state, estimable from cross-sectional data;
-
—
type-specific hazards (per capita rates) of acquisition and clearance, estimable from longitudinal data.
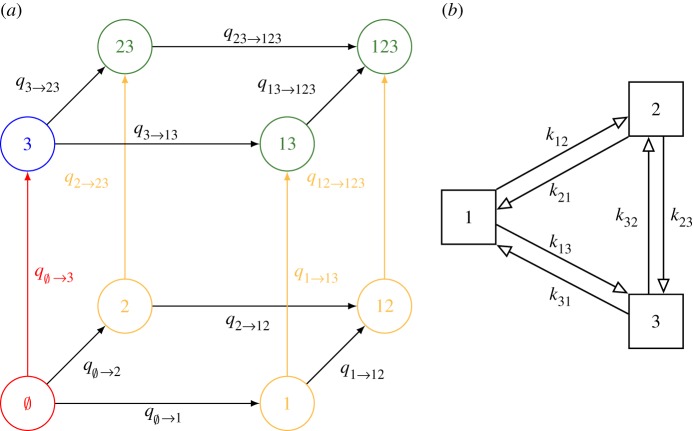
Figure 1a shows the eight (co)infection states and the respective transitions for a pathogen with three types. Type interaction is assumed to operate through current (co)infection, with one or multiple types modifying acquisition hazards of other types, or modifying clearance hazards of coinfecting types. Interactions between different types are allowed to be either competitive or synergistic and to vary in strength. Furthermore, we focus on predicting replacement by non-vaccine types that are not cross-protected by the vaccine, as these types are the most salient for evaluating the potential for replacement.
Figure 1.
(a) The eight infection states and transitions pertaining to acquisitions in a three-type system. For convenience, the reverse transitions (i.e. clearances) are not shown. The colours indicate the aggregate states for vaccine type = {1, 2}, i = 3 and the corresponding type-3 acquisitions; in yellow, in green, in blue, in red. (b) Graphical representation of the interaction parameters under the symmetric multiplicative structure or pairwise-asymmetric multiplicative structure in a three-type system. Under the pairwise-symmetric multiplicative structure, pairs of reverse interaction parameters would moreover be identical (i.e. kij = kji).
(b). Type-specific and overall replacement
We consider replacement, here defined as increase in the prevalence of non-vaccine-type infection once the vaccine types are eliminated in the post-vaccination steady state, at two levels:
-
—
type-specific replacement, defined as increase in the prevalence of a given non-vaccine type i. This occurs when , where I′X and IX denote the post- and pre-vaccination steady-state prevalence of infection state X, respectively. The sums here are taken over all states containing type i, e.g. states {3, 13, 23, 123} in a three-type system when considering i = 3;
-
—
overall replacement, defined as increase in the overall non-vaccine-type prevalence. This occurs when , where NVT is the set of all non-vaccine types, and denotes the number of types in a given set. The sums are taken over all states containing at least one non-vaccine type, e.g. states {2, 3, 23, 12, 13, 123} in a three-type system when considering NVT = {2, 3}.
(c). Predictors in a two-type setting
Previously, we have shown that the following pairwise odds ratio is an exact predictor of type replacement in a simple SIS model of one vaccine type (type 1) and one non-vaccine type (type 2), provided the two types interact symmetrically [21]:
| 2.1 |
This pairwise odds ratio compares the odds of non-vaccine-type infection between those infected and uninfected with the vaccine type.
Whenever the pre-vaccination steady-state value of OR1,2 is less than one, replacement will occur. This correspondence follows from OR1,2 being equivalent to the following pairwise cross-hazard ratio:
| 2.2 |
where qX→Y denotes the transition hazard from state X to Y. The numerator (denominator) of expression (2.2) is the hazard ratio of acquiring (clearing) the non-vaccine type in those infected versus those uninfected with the vaccine type and thus quantifies interaction in acquisition (clearance). In other words, the numerator (denominator) quantifies to what extent vaccine-type infection accelerates or decelerates acquisition (clearance) of non-vaccine-type infection. As a whole, the ratio captures the joint effects of interactions both in acquisition and clearance on the occurrence of non-vaccine-type infection.
Of note, the above pairwise odds ratio (2.1) has been used for inferring interactions between HPV genotypes [7,25–27], whereas the pairwise cross-hazard ratio (2.2) has been used for describing competition between Streptococcus pneumoniae serotypes [28].
(d). Predictors in a multi-type setting
(i). Type-specific cross-hazard ratio as a predictor of type-specific replacement
We now generalize the pairwise cross-hazard ratio (2.2) to a setting with an arbitrary number of interacting vaccine and non-vaccine types. To predict replacement by a given non-vaccine type i, we first aggregate the infection states into the four disjoint collections of states given in table 1 (figure 1a).
Table 1.
Four disjoint collections of infection states constructed for the definition of the type-specific cross-hazard ratio (2.3). Here, VT denotes the set of vaccine types.
 |
Based on the transitions between these four aggregate states, we propose the following generalization of the four hazards in the pairwise cross-hazard ratio (2.2):
| 2.3 |
where denotes the relative prevalence of state X conditioned within aggregate state . Each term in expression (2.3) is a weighted average of hazards as it aggregates hazards of acquiring or clearing type i with weights given by the conditional steady-state prevalence of the states from which the transitions occur. In effect, this weighting adjusts for the time each individual spends being at risk for the respective transitions [29]. Similar to the pairwise cross-hazard ratio (2.2), the numerator and denominator of the type-specific cross-hazard ratio (2.3) are hazard ratios of acquiring and clearing type i in those infected with at least one vaccine type () versus those uninfected with any of the vaccine types (), respectively. In short, the type-specific cross-hazard ratio (2.3) combines interactions by the vaccine types on the non-vaccine type of interest.
Example with multiple vaccine types. Assume that VT = {1, 2}, NVT = {3}, and there is interaction only in acquisition (figure 1a), and consider the type-specific cross-hazard ratio (2.3) as a predictor for replacement by type 3. As the denominator of expression (2.3) now equals one, the predictor reduces to
| 2.4 |
where I1|{1,2,12} = I1/C, I2|{1,2,12} = I2/C, I12|{1,2,12} = I12/C, and C = I1 + I2 + I12.
(ii). Type-specific odds ratio as a predictor of type-specific replacement
The type-specific cross-hazard ratio HRVT,i as given in (2.3) requires estimation of type-specific acquisition and clearance hazards from longitudinal data. However, collecting such data may be cumbersome and expensive due to repeated observations of the infection states in the same study subjects. It would thus be advantageous if the cross-hazard ratio HRVT,i could be approximated using steady-state cross-sectional (i.e. prevalence) data only, in a similar way as in the two-type setting using the pairwise odds ratio (2.1).
In a setting with more than two types, the pairwise odds ratio (2.1) can be generalized to the following type-specific odds ratio:
| 2.5 |
which is the odds of infection with non-vaccine type i in the presence versus absence of coinfection with any of the vaccine types. The approximation of HRVT,i by ORVT,i is exact given detailed balance, i.e. when IXqX→Y = IYqY→X for any pair of states X, Y (see electronic supplementary material, section A for the proof). In reality, whether this property holds depends on the underlying structure of type interactions.
Example continued. In the previous example where VT = {1, 2}, NVT = {3} (figure 1a), the type-specific odds ratio (2.5) becomes
| 2.6 |
(iii). Overall odds and cross-hazard ratios as predictors of overall type replacement
Pooling across pairwise odds ratios has been used to summarize interactions of all non-vaccine types with each vaccine type separately [7,25]. However, this approach lacks a clear interpretation for predicting type replacement. Here, we propose the following overall odds ratio for predicting overall replacement:
| 2.7 |
The use of a pooled odds ratio for prediction can be justified by considering the ratio of the odds of overall non-vaccine-type infection in the pre- versus post-vaccination steady states. This odds ratio is essentially the true impact of vaccination with value less than one characterizing overall replacement. Assuming mutual independence between all non-vaccine types, this true odds ratio of overall infection can be approximated by the product of true odds ratios of type-specific infections (see electronic supplementary material, section B for the derivation). As each true type-specific odds ratio can be predicted by the corresponding ORVT,i, we envision their product (2.7) to be a predictor for overall replacement.
Owing to the relation between HRVT,i and ORVT,i, we also propose the following overall cross-hazard ratio as a predictor for overall replacement:
| 2.8 |
(e). Simulated structures of type interactions
Thus far, we have made no assumptions about the structure of type interactions, i.e. no constraints on kXi and hXi in defining and . To investigate the performance of the proposed predictors and their robustness against different interaction structures, we considered the following three alternative structures in simulations (see electronic supplementary material, section C for their exact descriptions).
The first interaction structure is pairwise-symmetric and multiplicative so that each (co)infecting type contributes multiplicatively to the acquisition hazard of an incoming type, i.e. , or to the clearance hazard of a coinfecting type, i.e. . Here kji and hji describe how (co)infection with type j modifies acquisition and clearance of type i, respectively. Values of kji and 1/hji equal to, less and larger than one correspond to independent, competitive and synergistic interactions, respectively. In other words, competition decreases the acquisition rate, whereas synergy decreases the clearance rate. Furthermore, pairs of interaction parameters are assumed to be identical, i.e. kij = kji and hij = hji, resulting in symmetric interaction matrices. With n types, the numbers of parameters governing the interactions in acquisition and clearance are both reduced from n(2n−1 − 1) to n(n − 1)/2 (figure 1b). This pairwise-symmetric multiplicative structure preserves the detailed balance property (see electronic supplementary material, section D for the proof), so that HRVT,i and ORVT,i are equivalent.
Departure from the above structure may disrupt this equivalence of HRVT,i and ORVT,i. Two alternative structures are the pairwise-asymmetric multiplicative and the groupwise-symmetric multiplicative structure. The pairwise-asymmetric multiplicative structure relaxes the symmetry constraint kji = kij. The groupwise-symmetric multiplicative structure assumes the multiplicativity to act per groups of types instead of individual types. For example, in a four-type system with groups A = {1, 2}, B = {3, 4}, and interaction parameters kA, kB (within groups), kAB = kBA (between groups), the hazards of acquiring type 1 from state 23 and 234 are both .
(f). Performance analysis
To evaluate the performance of the proposed predictors, we simulated the effect of vaccination using a deterministic SIS transmission model (see electronic supplementary material, section C for details of the model). We investigated how the performance of the predictors depends on the numbers of vaccine and non-vaccine types and their interaction structure. For each setting, different sets of model parameters, including interaction parameters and type-specific transmissibility, were randomly generated.
The interaction parameters of the two symmetric structures were uniformly generated on a log scale in the interval (1/3, 3), ranging from competitive to synergistic interactions. To examine the effect of increasing asymmetry under the pairwise-asymmetric structure, asymmetric interaction parameters (k′ij, k′ji) were obtained by either perturbing the generated parameters of the pairwise-symmetric structure (kij), or by generating new k′ij, k′ji independently of each other. Perturbation of the pairwise-symmetric parameters was done by adding deviations on a log scale, i.e. log (k′ij) = log (kij) + ε/2 and log (k′ji) = log (kij) − ε/2. In effect, is the ratio between a pair of reverse interaction parameters, k′ij/k′ji with increasing ε inducing more asymmetry. When generating k′ij, k′ji independently, there was no constraint on their ratio.
For each parameter set, prediction of replacement was made at the simulated pre-vaccination steady state and compared to the ‘true’ outcome (replacement/no replacement) at the post-vaccination steady state. For each setting, the performance of each predictor was defined as the proportion of correct predictions among all generated parameter sets (see electronic supplementary material, section E for the exact simulation procedure).
3. Simulation results
(a). Performance under the pairwise-symmetric multiplicative structure
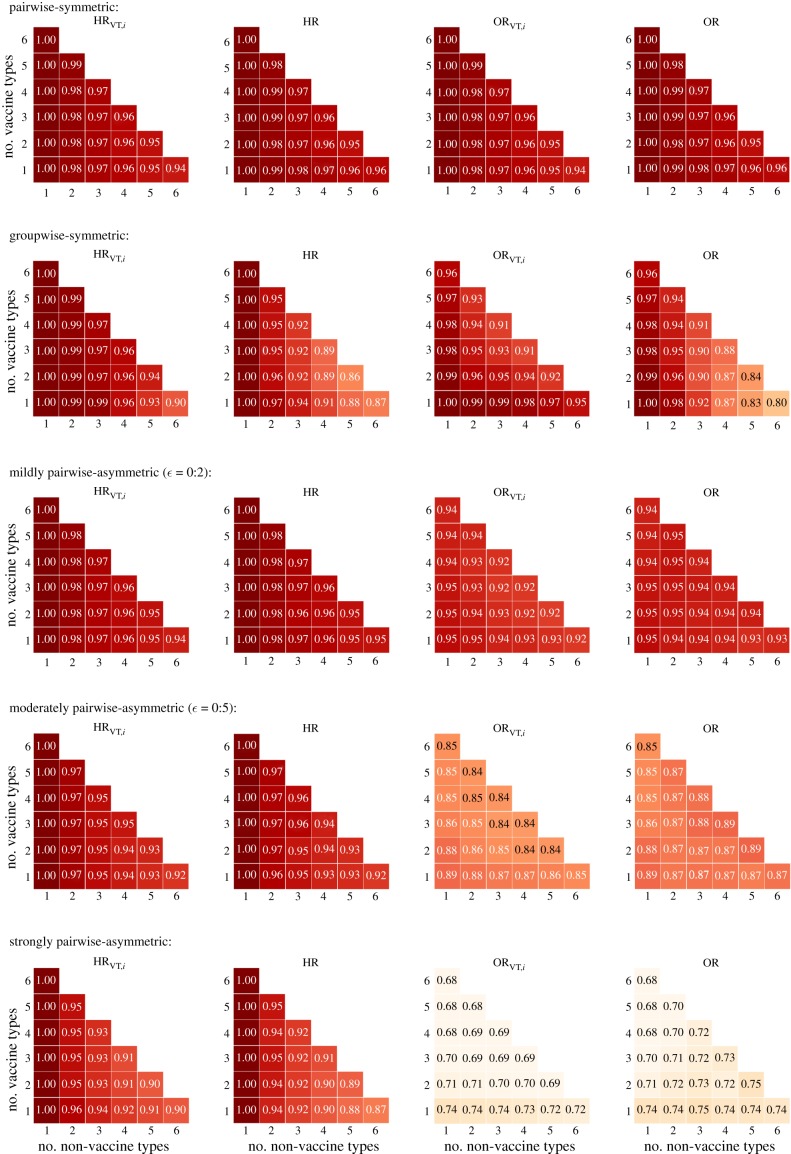
As the pairwise-symmetric multiplicative structure obeys detailed balance, the hazard-based and odds-based predictors are equivalent (HRVT,i = ORVT,i and HR = OR) and indeed performed identically (row 1 of figure 2).
Figure 2.
Performance (proportion of correct predictions among all generated parameter sets) of predictors HRVT,i, HR, ORVT,i, and OR under alternative multiplicative structures for interaction in acquisition (and no interaction in clearance). Row 1: pairwise-symmetric. Row 2: groupwise-symmetric. Rows 3–5: increasing pairwise-asymmetric with increasing values of ε for more asymmetry between pairs of reciprocal interaction parameters. Performance of HRVT,i and ORVT,i were obtained by averaging over the performance of each non-vaccine type i.
With one non-vaccine type, the predictors (HRVT,i = ORVT,i = HR = OR) were always correct, even when some vaccine types would interact synergistically while others would compete with the non-vaccine type. Thus, the predictors correctly captured the balance between the opposing forces of competition and synergy.
With multiple non-vaccine types, prediction of both type-specific and overall replacement became more difficult due to interactions between the non-vaccine types. For example, in a system with VT = {1}, NVT = {2, 3}, if k13 = 1, k12 > 1 and k23 < 1 (figure 1b), vaccination indirectly triggered replacement by type 3 by decreasing type-2 infection and thus increasing type-3 infection. Rewriting expression (2.3) shows that this indirect effect is indicated by the second factor in
| 3.1 |
In particular, the expression in the brackets is less than one when either k12 or k23 (but not both) is less than one (see electronic supplementary material, section F for the derivation of (3.1)). However, if the direct and indirect effects acted in opposite directions, the predictors did not always adequately capture their combined effect (see electronic supplementary material, section G for an example). Indeed, with multiple non-vaccine types, the performance decreased as the number of non-vaccine types increased (follow each subfigure in figure 2 across the diagonals). Nonetheless, the performance of all predictors remained well above 94%. In addition, when the overall predictor failed with a value close to one, the overall prevalence of non-vaccine-type infection usually changed only modestly (see electronic supplementary material, section G for an example).
(b). Performance under alternative interaction structures
Under the alternative interaction structures, the hazard-based and odds-based predictors were no longer equivalent but still performed comparably, except when the asymmetry was strong (figure 2). With one non-vaccine type, the hazard-based predictions remained almost perfect, unlike the odds-based ones. Under the groupwise-symmetric structure, the predictive performance decreased more rapidly as the number of non-vaccine types increased, as compared to the pairwise-symmetric structure. However, it was still above 80% in simulations with up to seven types (row 2 of figure 2).
Under the pairwise-asymmetric structure, the performance of both sets of predictors decreased with increasing asymmetry (row 3–5 of figure 2). Nevertheless, the performance of the hazard-based predictors remained above 85% even when pairs of kij and kji were generated independently, inducing strong asymmetry. The decreasing performance was more vivid for the odds-based predictors, which went down to 70% in the corresponding settings. The superiority of the hazard-based predictors can be illustrated in the following example of two types. Suppose that the vaccine type competes with the non-vaccine type (k12 < 1) while the reverse interaction is synergistic (k21 > 1). Then HR1,2 = k12 < 1 predicts replacement correctly, whereas the symmetrically defined OR1,2 may not because it averages over k12 and k21.
The above results were based on simulations in which interactions occurred only in acquisition. Additional simulations that allowed interactions in both acquisition and clearance showed almost identical performance of both the hazard-based and odds-based predictors (compare figure 2 to electronic supplementary material, section H).
4. Discussion
This paper aims to develop methodology for predicting type replacement for pathogens with many, potentially interacting types using pre-vaccination epidemiological data. We have proposed a predictor of replacement by individual non-vaccine types (i.e. increase in type-specific prevalence) and another predictor of the increase in the overall non-vaccine-type prevalence. Both predictors are initially defined in terms of hazards of acquiring and clearing the non-vaccine types and thus, in principle, require estimation from longitudinal data. In addition, we have derived alternative predictors that can be readily estimated from cross-sectional prevalence data as odds ratios. All proposed predictors demonstrated satisfactory performance (mostly above 85%) across a range of simulated structures of type interactions, except when the interactions were strongly asymmetric.
The predictor of replacement by a given non-vaccine type is constructed as a weighted cross-hazard ratio of acquiring versus clearing infection with the non-vaccine type in question, comparing those infected and uninfected with any of the vaccine types. The odds-based alternative is defined as the odds of infection with that non-vaccine type in presence versus absence of infection with any of the vaccine types.
We note the similarity between type interaction and vaccine efficacy of multi-valent vaccines as both are defined in terms of cross ratios of weighted hazards in a multi-type setting. In both cases, it proves useful to weigh hazards with the steady-state prevalence, conditioned on appropriate risk sets. In defining estimators for vaccine efficacy against one or more vaccine types, the risk sets are those from which acquisitions of the vaccine types may occur [29]. Likewise, in the current paper, the risk sets are those from which acquisitions (or clearances) of the non-vaccine type may occur (see the type-specific cross-hazard ratio (2.3)).
The predictors of overall replacement take form as products of either the hazard-based or the odds-based type-specific predictors. In effect, the overall predictors are obtained by pooling the type-specific predictors on a logarithmic scale. In various studies that have used pairwise odds ratios to evaluate HPV-type interactions, pooling has been performed across the pool of non-vaccine types for each vaccine type separately [7,25,26]. These pooled odds ratios have been interpreted as the affinity of a given vaccine type to be involved in coinfection with any of the non-vaccine types, but their relevance for predicting type replacement has remained elusive. Our results substantiate the predictive value of pooled odds ratios regarding overall replacement, but suggest that pooling is better performed on the entire set of vaccine types, instead of for each vaccine type separately. Furthermore, the pairwise odds ratios have been compared to the pooled ones to identify likely candidates for type replacement [7,25,26]. Accordingly, the potential for replacement by a given non-vaccine type has been assessed separately for each vaccine type, while our type-specific predictors capture interactions with all vaccine types in a comprehensive way.
Our simulation study revealed how the applicability of the new predictors depends on the underlying structure of type interactions. The predictors performed best under the pairwise-symmetric multiplicative structure, in which the hazard-based and odds-based predictors are equivalent. The predictors were mostly able to capture the opposing forces of competition and synergy as well as the interplay between interactions in acquisition and clearance. Under other simulated structures, the hazard-based predictors still performed well, while the odds-based predictors performed fairly up to a reasonable degree of asymmetry. As a rule of thumb, symmetric interactions facilitate prediction of type replacement, while complex and heterogeneous interactions may necessitate more sophisticated predictors that capture details and directions of interactions. For HPV, little is known about the structure of interactions between genotypes as the very existence of interaction is already difficult to determine. For instance, any imbalance between type-specific prevalence may mask possible asymmetric interactions, and although pooling of multiple types may increase the power of detecting interaction, it may also obscure type-specific patterns. Nevertheless, our simulation study demonstrated that the new predictors are robust against various interaction structures.
We cannot ensure good performance of the proposed predictors under mechanisms of interactions other than the ones we considered. For instance, if the pathogen types interact through natural cross-immunity that is long-lasting, infections with different types may be positively associated (i.e. as given by odds ratios greater than one) while there is a risk of type replacement [30,31]. This shortcoming is inherent to the difficulty to distinguish between susceptible individuals and those who have acquired natural immunity, as is the case for HPV infection which induces only a weak antibody response [32]. Nevertheless, if immunity is confined to be type-specific, the predictors work equally well in a two-type model [21], and we envision the proposed predictors to remain applicable also in a multi-type setting. Another mechanism that we did not consider is competition for transmissibility, e.g. through reduction of viral load during coinfection [13–15]. Although competition for transmissibility is not captured by the proposed predictors, it is likely correlated with competition in clearance, as HPV persistence is also determined by viral load [16]. Hence, the predictors may retain good predictive ability even when not all mechanisms of interactions are captured.
Despite the theoretical appeal of the proposed predictors and their promising performance in simulations, some challenges still need to be addressed. First, more extensive simulations are needed to investigate how well the predictors perform with high numbers of pathogen types. Here, simulations were performed with at most seven types, while HPV consists of up to 15 high-risk types. Second, our method assumes elimination of all vaccine-targeted types, although some vaccine types or cross-protective types may persist, especially when vaccination coverage is low [4,5]. If persistence of targeted types would mostly prevent or limit the extent of type replacement, our method may still provide good predictions as a worst-case scenario. However, formal analysis would be required to sharpen this intuition. Third, it remains to be investigated whether the hazard-based and odds-based predictors can be accurately estimated from limited data. In particular, although the hazard-based predictors performed better in simulations, it is not straightforward which ones are more suitable for the empirical setting. In general, hazard-based estimates are more robust against different sources of bias (e.g. due to population entry and unobserved heterogeneity) [31] but require a larger sample size than estimation of type-specific point prevalence. In addition, statistical methods need to be developed to deal with possible confounding due to common risk factors, which were neglected in our analysis. In particular, multivariate statistical methods (e.g. generalized estimating equations and random effect models) may be adapted to adjust for observed heterogeneity or control for unobserved heterogeneity when estimating the new predictors, which are essentially odds and hazard ratios.
For HPV, most epidemiological studies have concluded a low risk of type replacement based on a lack of systematic patterns of negative associations in the co-occurrence of vaccine and non-vaccine types [12,25]. The methodology presented in this paper may help to translate the observed patterns into explicit prediction of the overall risk of replacement. Additionally, application of our type-specific predictors could discover hitherto hidden potential for type replacement, as this potential is not only determined by direct interactions with the vaccine types but also shaped though indirect interactions with other non-vaccine types. As the circulation of vaccine types will be further reduced in the near future, more data will become available to validate any predictions about the long-term impact of HPV vaccination. The proposed methodology could also be used to give better insights into the underpinnings of type competition in Streptococcus pneumoniae, a pathogen for which replacement has been widely observed after the introduction of the pneumococcal conjugate vaccination [33,34].
To conclude, we developed novel methodology for predicting type replacement in a setting of many interacting types. We did so by relating available epidemiological data to the underlying mechanisms of how pathogen types may interact. The proposed predictors may help to better anticipate and understand the impact of vaccination against pathogens with many coexisting (sub)types.
Supplementary Material
Data accessibility
Simulation code: https://github.com/irene-man/Prediction-method-type-replacement.git.
Competing interests
We declare we have no competing interests.
Funding
This study was supported by grant S/113005/01/PT (Prometheus project) through the Strategic Programme from the National Institute for Public Health and the Environment (RIVM) of The Netherlands.
References
- 1.de Martel C, Plummer M, Vignat J, Franceschi S. 2017. Worldwide burden of cancer attributable to HPV by site, country and HPV type. Int. J. Cancer 141, 664–670. ( 10.1002/ijc.v141.4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Muñoz N, Bosch FX, De Sanjosé S, Herrero R, Castellsagué X, Shah KV, Snijders PJ, Meijer CJ. 2003. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N. Engl. J. Med. 348, 518–527. ( 10.1056/NEJMoa021641) [DOI] [PubMed] [Google Scholar]
- 3.de Sanjose S. et al. 2010. Human papillomavirus genotype attribution in invasive cervical cancer: a retrospective cross-sectional worldwide study. Lancet Oncol. 11, 1048–1056. ( 10.1016/S1470-2045(10)70230-8) [DOI] [PubMed] [Google Scholar]
- 4.Mesher D. et al. 2016. Population-level effects of human papillomavirus vaccination programs on infections with nonvaccine genotypes. Emerg. Infect. Dis. 22, 1732–1740. ( 10.3201/eid2210.160675) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kavanagh K, Pollock K, Potts A, Love J, Cuschieri K, Cubie H, Robertson C, Donaghy M. 2014. Introduction and sustained high coverage of the HPV bivalent vaccine leads to a reduction in prevalence of HPV 16/18 and closely related HPV types. Br. J. Cancer 110, 2804–2811. ( 10.1038/bjc.2014.198) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drolet M. et al. 2015. Population-level impact and herd effects following human papillomavirus vaccination programmes: a systematic review and meta-analysis. Lancet Infect. Dis. 15, 565–580. ( 10.1016/S1473-3099(14)71073-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tota JE, Ramanakumar AV, Jiang M, Dillner J, Walter SD, Kaufman JS, Coutlée F, Villa LL, Franco EL. 2013. Epidemiologic approaches to evaluating the potential for human papillomavirus type replacement postvaccination. Am. J. Epidemiol. 178, 625–634. ( 10.1093/aje/kwt018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lloyd-Smith JO. 2013. Vacated niches, competitive release and the community ecology of pathogen eradication. Phil. Trans. R. Soc. B 368, 20120150 ( 10.1098/rstb.2012.0150) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Safaeian M, Rodriguez AC. 2014. Invited commentary: multiple human papillomavirus infections and type replacement—anticipating the future after human papillomavirus vaccination. Am. J. Epidemiol. 180, 1076–1081. ( 10.1093/aje/kwu265) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saccucci M, Franco EL, Ding L, Bernstein DI, Brown D, Kahn JA. 2018. Non-vaccine-type human papillomavirus prevalence after vaccine introduction: no evidence for type replacement but evidence for cross-protection. Sex Transm. Dis. 45, 260–265. ( 10.1097/OLQ.0000000000000731) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kahn JA, Brown DR, Ding L, Widdice LE, Shew ML, Glynn S, Bernstein DI. 2012. Vaccine-type human papillomavirus and evidence of herd protection after vaccine introduction. Pediatrics 130, e249–e256. ( 10.1542/peds.2011-3587) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tota JE, Ramanakumar AV, Villa LL, Richardson H, Burchell AN, Koushik A, Mayrand MH, Coutlée F, Franco EL. 2015. Evaluation of human papillomavirus type replacement postvaccination must account for diagnostic artifacts: masking of HPV52 by HPV16 in anogenital specimens. Cancer Epidemiol. Biomarkers Prev. 24, 286–290. ( 10.1158/1055-9965.EPI-14-0566) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xi LF, Edelstein ZR, Meyers C, Ho J, Cherne SL, Schiffman M. 2009. Human papillomavirus types 16 and 18 DNA load in relation to coexistence of other types, particularly those in the same species. Cancer Epidemiol. Biomarkers Prev. 18, 2507–2512. ( 10.1158/1055-9965.EPI-09-0482) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roden RB, Stern PL. 2018. Opportunities and challenges for human papillomavirus vaccination in cancer. Nat. Rev. Cancer 18, 240–254. ( 10.1038/nrc.2018.13) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Biryukov J, Meyers C. 2018. Superinfection exclusion between two high-risk human papillomavirus (HPV) types during a co-infection. J. Virol. 92, e01993-17 ( 10.1128/JVI.01993-17) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marra E. et al. 2018. Anal HPV 16 and 18 viral load: a comparison between HIV-negative and -positive MSM and association with persistence. J. Med. Virol. 90, 76–83. ( 10.1002/jmv.v90.1) [DOI] [PubMed] [Google Scholar]
- 17.Sofonea MT, Alizon S, Michalakis Y. 2017. Exposing the diversity of multiple infection patterns. J. Theor. Biol. 419, 278–289. ( 10.1016/j.jtbi.2017.02.011) [DOI] [PubMed] [Google Scholar]
- 18.Thomas KK, Hughes JP, Kuypers JM, Kiviat NB, Lee SK, Adam DE, Koutsky LA. 2000. Concurrent and sequential acquisition of different genital human papillomavirus types. J. Infect. Dis. 182, 1097–1102. ( 10.1086/jid.2000.182.issue-4) [DOI] [PubMed] [Google Scholar]
- 19.Rousseau MC, Pereira JS, Prado JC, Villa LL, Rohan TE, Franco EL. 2001. Cervical coinfection with human papillomavirus (HPV) types as a predictor of acquisition and persistence of HPV infection. J. Infect. Dis. 184, 1508–1517. ( 10.1086/jid.2001.184.issue-12) [DOI] [PubMed] [Google Scholar]
- 20.Merikukka M, Kaasila M, Namujju PB, Palmroth J, Kirnbauer R, Paavonen J, Surcel HM, Lehtinen M. 2011. Differences in incidence and co-occurrence of vaccine and nonvaccine human papillomavirus types in Finnish population before human papillomavirus mass vaccination suggest competitive advantage for HPV33. Int. J. Cancer 128, 1114–1119. ( 10.1002/ijc.25675) [DOI] [PubMed] [Google Scholar]
- 21.Man I, Wallinga J, Bogaards JA. 2018. Inferring pathogen type interactions using cross-sectional prevalence data: opportunities and pitfalls for predicting type replacement. Epidemiology 29, 666–674. ( 10.1097/EDE.0000000000000870) [DOI] [PubMed] [Google Scholar]
- 22.Gravitt PE. 2011. The known unknowns of HPV natural history. J. Clin. Invest. 121, 4593–4599. ( 10.1172/JCI57149) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ranjeva SL, Baskerville EB, Dukic V, Villa LL, Lazcano-Ponce E, Giuliano AR, Dwyer G, Cobey S. 2017. Recurring infection with ecologically distinct HPV types can explain high prevalence and diversity. Proc. Natl Acad. Sci. USA 114, 13 573–13 578. ( 10.1073/pnas.1714712114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tota JE. et al. 2016. Epidemiologic evaluation of human papillomavirus type competition and the potential for type replacement post-vaccination. PLoS ONE 11, e0166329 ( 10.1371/journal.pone.0166329) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chaturvedi AK. et al. 2011. Human papillomavirus infection with multiple types: pattern of coinfection and risk of cervical disease. J. Infect. Dis. 203, 910–920. ( 10.1093/infdis/jiq139) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mollers M. et al. 2014. Population- and type-specific clustering of multiple HPV types across diverse risk populations in the Netherlands. Am. J. Epidemiol. 179, 1236–1246. ( 10.1093/aje/kwu038) [DOI] [PubMed] [Google Scholar]
- 27.Beachler DC, Jenkins G, Safaeian M, Kreimer AR, Wentzensen N. 2015. Natural acquired immunity against subsequent genital human papillomavirus infection: a systematic review and meta-analysis. J. Infect. Dis. 213, 1444–1454. ( 10.1093/infdis/jiv753) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mehtälä J, Antonio M, Kaltoft MS, O’Brien KL, Auranen K. 2013. Competition between Streptococcus pneumoniae strains: implications for vaccine-induced replacement in colonization and disease. Epidemiology 24, 522–529. ( 10.1097/EDE.0b013e318294be89) [DOI] [PubMed] [Google Scholar]
- 29.Auranen K, Rinta-Kokko H, Halloran ME. 2013. Estimating strain-specific and overall efficacy of polyvalent vaccines against recurrent pathogens from a cross-sectional study. Biometrics 69, 235–244. ( 10.1111/j.1541-0420.2012.01826.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Durham DP, Poolman EM, Ibuka Y, Townsend JP, Galvani AP. 2012. Reevaluation of epidemiological data demonstrates that it is consistent with cross-immunity among human papillomavirus types. J. Infect. Dis. 206, 1291–1298. ( 10.1093/infdis/jis494) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malagón T, Lemieux-Mellouki P, Laprise JF, Brisson M. 2016. Bias due to correlation between times-at-risk for infection in epidemiologic studies measuring biological interactions between sexually transmitted infections: a case study using human papillomavirus type interactions. Am. J. Epidemiol. 184, 873–883. ( 10.1093/aje/kww152) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vink MA, Berkhof J, van de Kassteele J, van Boven M, Bogaards JA. 2016. A bivariate mixture model for natural antibody levels to human papillomavirus types 16 and 18: baseline estimates for monitoring the herd effects of immunization. PLoS ONE 11, e0161109 ( 10.1371/journal.pone.0161109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feikin DR. et al. 2013. Serotype-specific changes in invasive pneumococcal disease after pneumococcal conjugate vaccine introduction: a pooled analysis of multiple surveillance sites. PLoS Med. 10, e1001517 ( 10.1371/journal.pmed.1001517) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Flasche S, de Waroux OLP, O’Brien KL, Edmunds WJ. 2015. The serotype distribution among healthy carriers before vaccination is essential for predicting the impact of pneumococcal conjugate vaccine on invasive disease. PLoS Comput. Biol. 11, e1004173 ( 10.1371/journal.pcbi.1004173) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Simulation code: https://github.com/irene-man/Prediction-method-type-replacement.git.