Abstract
Type IV pili (Tfp) of Neisseria gonorrhoeae, the Gram-negative etiologic agent of gonorrhea, facilitate colonization of the human host. Tfp are assumed to play a key role in the initial adherence to human epithelial cells by virtue of the associated adhesin protein PilC. To examine the structural and functional basis for adherence in more detail, we identified potential genes encoding polypeptides sharing structural similarities to PilE (the Tfp subunit) within the N. gonorrhoeae genome sequence database. We show here that a fiber subunit-like protein, termed PilV, is essential to organelle-associated adherence but dispensable for Tfp biogenesis and other pilus-related phenotypes, including autoagglutination, competence for natural transformation, and twitching motility. The adherence defect in pilV mutants cannot be attributed to reduced levels of piliation, defects in fiber anchoring to the bacterial cell surface, or to unstable pilus expression related to organelle retraction. PilV is expressed at low levels relative to PilE and copurifies with Tfp fibers in a PilC-dependent fashion. Purified Tfp from pilV mutants contain PilC adhesin at reduced levels. Taken together, these data support a model in which PilV functions in adherence by promoting the functional display of PilC in the context of the pilus fiber.
The adherence of microbes to host tissue represents the initial step in the pathogenesis of infections, with specificity and tissue tropism being defined by precise adhesin-receptor interactions. In most Gram-negative pathogens that colonize mucosal surfaces, attachment is mediated by proteinaceous filaments termed pili or fimbriae (1). The use of these organelles to display adhesive molecules promotes ligand recognition at a distance from the bacterial cell surface. This increase in the effective physical range of interaction allows the pathogen to maintain dense surface structures such as polysaccharide capsules and LPS, which act in immune avoidance.
The structure–function relationships of adhesive pili are best understood for those pili assembled by the chaperone-assisted or chaperone/usher pathway, which accounts for over 30 distinct forms of adhesive organelles in various Gram-negative species (2). These pili are composite structures with a long pilus shaft comprised of the major pilus subunit and a short tip fibrillum made up of pilin-subunit-like molecules, including the actual adhesin and adaptors that link it to the shaft. Type IV pili (Tfp) are a separate unique class of pili expressed by many Gram-negative bacteria of medical, environmental, and industrial importance. Tfp are defined by their shared structural, biochemical, antigenic, and morphological features (3) and a highly conserved biogenesis pathway that resembles that used for the Type II protein secretion pathway (4).
Tfp promote adherence of Neisseria gonorrhoeae, the Gram-negative etiologic agent of gonorrhea, to epithelial cells and therefore play a crucial step in colonization of the human host (5). They also contribute to other phenotypes, including autoagglutination, competence for natural transformation, and twitching motility (6). Tfp-mediated gonococcal adherence appears not to be directly associated with the pilin subunit protein PilE but rather is most strongly correlated with the expression of the less abundant PilC protein, which copurifies with Tfp (7, 8). A similar system appears to operate for the highly related species Neisseria meningitidis. In contrast, Tfp-mediated adherence exhibited by Pseudomonas aeruginosa has been proposed to involve a receptor-binding site within the C terminus of the pilin subunit (9). Tfp also enhance colonization of human epithelium by enteropathogenic strains of Escherichia coli (10) and Vibrio cholerae (11), but is unclear whether specific adhesin–receptor interactions are involved in these species.
The conclusion that PilC is a neisserial Tfp-associated adhesin is based primarily on the findings that (i) purified recombinant PilC imparts human epithelial cell binding activity on latex beads to which it has been coupled and blocks binding of piliated organisms to human epithelial cells (7) and (ii) piliated mutants lacking PilC fail to adhere (12, 13). However, several studies have suggested a role for the pilin subunit PilE in receptor recognition. These include numerous findings showing that different antigenic variant forms of PilE influence epithelial cell adherence and tissue tropism (12, 14–16). However, the structural and functional attributes of PilE accounting for these observations remain unknown. Additionally, PilE itself has been reported to possess human epithelial cell and erythrocyte receptor binding properties (8, 17). The biological significances of the latter in vitro activities to pilus and gonococcal cell adherence remain unclear.
In an effort to understand the molecular biology of neisserial Tfp in more detail, we undertook a project to identify potential genes encoding polypeptides sharing structural similarities to PilE (the Tfp subunit) within the neisserial genome sequence databases. We previously reported on the characterization of one such protein, ComP, which is dedicated to the DNA uptake process associated with Tfp expression (18). Here, we identify and characterize a second protein, termed PilV, which is essential to Tfp-mediated adherence to human epithelial cells but dispensable for organelle biogenesis and other pilus-related phenotypes. The data are most consistent with PilV being a minor constituent of the Tfp fiber, and we propose that it acts indirectly in adherence by promoting the effective display of the associated PilC adhesin.
Materials and Methods
Bacterial Strains, Plasmids, and DNA Manipulations.
Bacterial strains used in this study are described in Table 1. E. coli HB101 was used for plasmid propagation and cloning experiments. Antibiotics were used for selection of gonococcal transformants at the following concentrations: chloramphenicol, 10 μg/ml; erythromycin, 8 μg/ml; kanamycin, 50 μg/ml; nalidixic acid, 1 μg/ml; and tetracycline, 15 μg/ml. Isolation and purification of plasmid DNA were performed by using Qiagen columns according to the manufacturer's specifications (Qiagen, Chatsworth, CA).
Table 1.
Genotype and phenotype of N. gonorrhoeae strains used in this study
| Strain | Parental strain | Relevant genotype | Natural transformation | Twitching motility | Adherence to HCEC* | Reference |
|---|---|---|---|---|---|---|
| N400 | VD300 | recA6(tetM) | + | + | + | (13) |
| N401 | N400 | recA6(kan) | + | + | + | (13) |
| MW4 | N401 | pilTind(tetM) | − | − | + | (13) |
| MW7 | N401 | pilTind, pilC1∷erm, pilC2∷cat | − | − | − | (13) |
| GV1 | N400 | pilVG-1 fs | + | + | − | This work |
| GV2 | N400 | pilV∷kan | + | + | − | This work |
| GV3 | GV2 | pilV∷kan, iga∷pilV | + | + | + | This work |
| GV4 | MW4 | pilV∷kan, pilTind | − | − | − | This work |
| GV5 | GV1 | pilVG-1 fs, iga∷pilV | + | + | + | This work |
| GV6 | MW4 | pilVG-1 fs, pilTind | − | − | − | This work |
| GC1† | N401 | pilC1∷erm | + | + | ND | This work |
| C2† | GC1 | pilC1∷erm, pilVG-1 fs | + | + | ND | This work |
| C3† | C2 | pilC1∷erm, pilVG-1 fs, pilTind | − | − | ND | This work |
| GV7 | C3 | pilC1∷erm, pilVG-1 fs, pilTind, pilC2∷cat | − | − | − | This work |
HCEC, human corneal epithelial cells; +, >100 cfu/cell; −, 0–5 cfu/cell. Results are expressed as the mean values of at least four independent experiments.
These strains were generated in the stepwise creation of GV7.
Based on DNA sequence derived from FA1090 genome project, the pilV locus was amplified by using primers PilV5′ (5′-GTCGAGCTCGAATTCTTTAACCAAACCGATGAAA-3′) and PilV3′ (5′-GTCGAGCTCGGATCCGGGCTTCAGATAAGACCATCC-3′). The resulting 1.2-kb PCR fragment was cut by using the unique flanking BamHI and EcoRI sites (underlined) and forced cloned into pUP6 (19) to yield pPILV1. For the complementation studies, cloning of pilV into p2/16/1 was done by using the same oligonucleotides, but here the resulting product was cut with the flanking SacI sites (shown in italics) and cloned into a unique SacI site in the vector. p2/16/1 is a derivative of pUP6 that allows insertion of DNA fragments into the iga locus (4). The nucleotide sequence of pilV and pilE alleles was determined directly from PCR products at GATC Biotech AG (Konstanz, Germany). A pilV insertion mutant was constructed by cloning a kanamycin resistance gene cassette into the unique XhoI site at codons L +4/E +5 of PilV in pPilV1 to yield pPILV2. A frame-shift mutation was made by PCR by using the oligonucleotide PilV3′ together with PilV-FS (5′-AGCTCGAGCAGCGTAAAGCTTTTGAACGTTTTTCATAGC-3′). This primer overlaps the XhoI site (bold) in PilV, deleted a single base at the G −1 codon and introduced a new restriction enzyme site (HindIII, underlined) that was used for verification of the frame-shift mutation when transformed into gonococcal strains. The PCR product was cut with XhoI and BamHI and ligated into pPilV1 digested with these same enzymes creating the plasmid pPILV3.
Characterization of Twitching Motility and Competence for Natural Transformation.
Twitching motility was assessed by direct visualization of bacteria at the periphery of colonies with a stereomicroscope, as well as by the slide culture method in which bacteria grown on Gc agar slices on microscope slides are covered with a coverslip and viewed in a Zeiss phase microscope by using the ×40 objective (19). Transformation assays were carried out as previously described by using 1 μg/ml plasmid pSY6 DNA (19).
Epithelial Cell Bacterial Adherence and Tfp Binding Assays.
Primary cultures of human corneal epithelial cells were established as described (20). For use in adherence assays, epithelial cells were grown on 12-mm circular glass or thermanox coverslips in 1 ml of medium. Before the start of the infection, the medium was replaced with 1 ml of DMEM supplemented with 5% FCS. Gonococci were grown on GC agar plates (14 h, 5% CO2), suspended in tissue culture medium, and added to the cells (2 × 107 per well). After 1 h incubation (37°C, 10% CO2), the infection was stopped by rinsing the cells three times with 1 ml Dulbecco's PBS (DPBS) to remove unbound microorganisms, followed by fixation (at least 30 min, room temperature) in 0.1% glutaraldehyde/1% paraformaldehyde in DPBS. Specimens were stained with crystal violet (0.007% in distilled water). Infected cells were photographed by using an Olympus (New Hyde Park, NY) BH-2 microscope, and photoprints were directly counted to determine the number of adherent bacteria per cell. In cases where large numbers of adherent bacteria and microcolony formation precluded accurate counting, quantitation of adherence was confirmed as follows: after 1 h, nonadherent bacteria were removed by washing five times with assay media. The monolayers and cell-associated bacteria were then recovered by treatment with 0.25% trypsin for 5 min at 37°C. The recovered bacteria were plated on agar after dilution, and relative adherence was quantified by determining the ratio of cell-associated colony-forming units (cfu) to total cfu of the inoculum. Tfp binding assays were performed similarly, with the exception that the resuspended gonococci (one plate per milliliter of tissue culture medium) were vigorously vortexed (1 min) to shear off Tfp, which were separated from the bacteria by differential centrifugation (13,000 × g, 3 min) and added to the human epithelial cells. After 2 h of incubation at 37°C, the cells were washed and fixed in 2% paraformaldehyde in DPBS (30 min). Bound Tfp were detected by indirect immunofluorescence by using first rabbit serum 2-66 [1/500 dilution in Tris-buffered saline containing 1% BSA (pH 7.2)] raised against purified Tfp from strain N400, followed by staining with Alexa Red (594 nm) conjugated goat anti-rabbit IgG (Molecular Probes; 1/400 dilution in the same buffer). Specimens were mounted in 50% glycerol in DPBS and viewed in a conventional Zeiss fluorescence microscope and by confocal laser microscopy. Images shown correspond to fields that are representative of the overall observations.
Protein Quantitation.
Total cellular protein was extracted by incubating resuspended gonococcal cell pellets in lysis buffer (0.2 M potassium phosphate buffer (pH 7.5), 20% acetone, 40 mM EDTA, and 0.1% Triton X-100) for 15 min on ice. Protein concentrations were determined by using the Bio-Rad Protein Assay and BSA standards according to the manufacturer's instructions.
SDS/PAGE, Immunoblotting, and Staining.
Procedures for SDS/PAGE and immunoblotting have been described previously (13). Quantitative pilus purification was carried out as previously described (13). PilV and PilC were detected by immunoblotting of pilus preps and whole cell lysates by using PilV- and PilC-specific rabbit polyclonal antibodies and alkaline phosphatase-coupled goat anti-rabbit antibodies (Tago). PilC-specific sera were gifts from A.-B. Jonsson (Karolinska Institutet, Stockholm) and T. Rudel (Max Planck Institute for Infection Biology, Berlin). PilV-specific antisera were generated against a synthetic peptide (QNLERYYRQKGTFEKYDSTKL) corresponding to amino acid residues 45–65 of the mature protein (Research Genetics, Huntsville, AL). The pilin subunit from purified pili was detected by Coomassie staining of SDS/PAGE gels. A colloidal silver staining technique was used to detect proteins on poly(vinylidene difluoride) membranes (21).
Electron Microscopy.
Colonies of bacteria grown on GC agar plates (12 h, 37°C, 5% CO2) were gently touched with pioloform-coated grids and air dried. Grids were subsequently stained with 1% ammonium molybdate in water for 2 min, rinsed once with water, air-dried, and viewed in a Hitachi (Tokyo) HU-11E-1 electron microscope. Images shown correspond to fields that are representative of the overall observation.
Results
Identification of a Gonococcal Prepilin-Like ORF.
By using a database search strategy previously successful in the identification of the comP locus (18), we identified a second ORF sharing identity to the N terminus of PilE in the FA1090 gonococcal genome sequence assembled at the University of Oklahoma. To characterize this ORF in more detail, the locus was PCR amplified from strain N400 and sequenced. The derived ORF differed only slightly from the FA1090 ORF and was predicted to encode a polypeptide of 129 aa with a pI of 9.3 and molecular mass of 14.35 kDa. Comparative sequence analysis found homologues in the meningococcal group A (22) and B (23) genome sequences (corresponding to NMA0726 and NMB0547, respectively), with 82% identical sequence over their complete reading frames. The sequence downstream of the predicted translation termination site demonstrated a strong potential to form a stable stem loop RNA structure, and further examination of the sequences upstream and downstream showed sequence similarity to genes encoding an AcrE-like lipoprotein and alcohol dehydrogenase, respectively (data not shown). The organization of this gene cluster is conserved in N. meningitidis strains (22, 23).
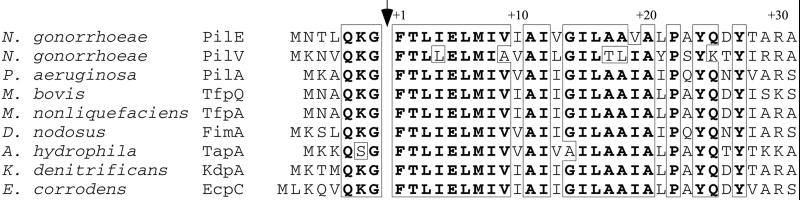
In a blastp search of the GenBank database, the ORF showed the highest identity to several Tfp subunits from various bacterial species (Fig. 1), with the regions of relatedness limited to the first 35 residues. Although the degree of identity within residues −1 through +27 was very high, the ORF did differ from a consensus sequence at residues +4, +9, +17, +18, and +25 (Fig. 1).
Figure 1.
N-terminal aligment of PilV with Tfp prepilins. The arrow indicates the prepilin peptidase processing site of the prepilin proteins and predicted processing site of PilV. Identical residues are boxed and in bold.
PilV Mutants Are Defective in Adherence to Human Epithelial Cells.
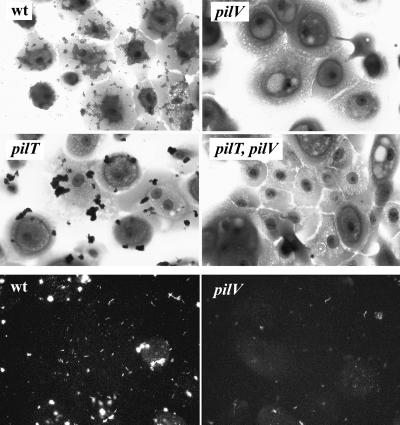
Both a gene disruption and a frameshift mutation (which altered residues carboxyl terminal to E +5 and G −1, respectively; see Materials and Methods) were constructed and introduced into the gonococcal chromosome via transformation. The mutants were indistinguishable from the wild-type parent in Tfp-associated colony morphology, retained the pilus-associated phenotype of twitching motility, and were not quantitatively reduced in competence for transformation relative to the isogenic wild-type parent (Table 1). In contrast, adherence assays showed that the mutants were severely impaired, with the levels of adherence being reduced ≈100-fold relative to the wild-type strains (Fig. 2, Top; Table 1). The defects in binding exhibited by the mutants were successfully complemented by introduction of an intact copy of the ORF region and adjacent 5′ sequences into the iga locus encoding IgA1 protease (Table 1). These results excluded the possibility that the binding deficiency could be due to polarity effects on distal gene expression. To exclude the possibility that the impaired adherence was due to PilE antigenic variation, the pilE genes in the mutants were sequenced and found to be unaltered. Based on these results and findings detailed below, the corresponding gene was named pilV.
Figure 2.
Adherence of gonococcal strains and Tfp to human corneal epithelial cells. (Top and Middle) Adherent cells are stained with crystal violet. wt (N400; recA6(tetM)); pilV (GV2; pilV∷kan); pilT (MW4; pilTind(tetM)); and PilT, pilV (GV4; pilV∷kan, pilTind). (Bottom) Binding of Tfp to human corneal epithelial cells assesed by indirect immunofluorescence. wt (N400; recA6(tetM)); and pilV (GV2; pilV∷kan). All panels are shown at the same level of magnification.
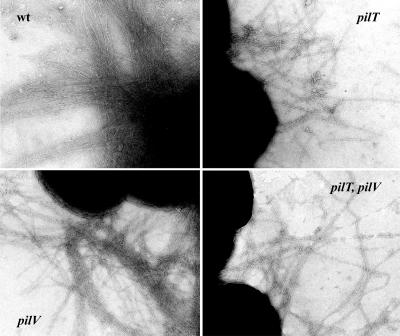
The simplest explanation for the nonadherent mutant phenotype would be a defect in Tfp fiber formation, although the autoagglutinating colony morphology of the pilV mutants suggested that they expressed Tfp at undiminished levels. Examination by transmission electron microscopy confirmed that the pilV mutants retained piliation levels indistinguishable from wild type (Fig. 3 Left). At this level of resolution, no altered pilus fiber morphology or lateral aggregation was discernible. To assess directly relative levels of piliation, a quantitative purification method previously developed was used (13). As shown in Fig. 4B, the yields of pili from the pilV mutants were indistinguishable from those from the wild-type parent and the complemented strains when standardized relative to the weights of whole cells used in purification.
Figure 3.
Piliation of gonococcal strains analyzed by transmission electron microscopy. wt (N400, recA6(tetM)); pilV (GV2, pilV∷kan); pilT (MW4, pilTind (tetM)); and pilT, pilV (GV4, pilV∷kan, pilTind). Micrographs are taken at a magnification of ×90,000.
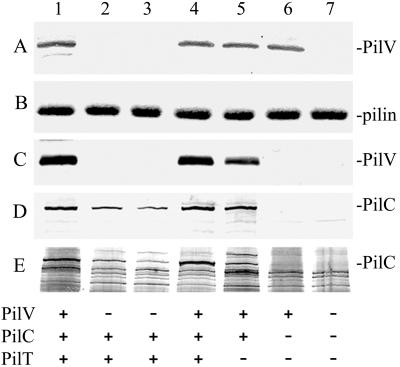
Figure 4.
Quantitative analysis of PilV and PilC in whole cells and purified pili from gonococcal strains. Lanes 1, N401 (wt); 2, GV1 (pilVG-1 fs); 3, GV2 (pilV∷kan); 4, GV3 (pilV∷kan, iga∷pilV); 5, MW4 (pilTind); 6, MW7 (pilTind, pilC1∷erm, pilC2∷cat); and 7, GV7 (pilVG-1 fs,, pilTind, pilC1∷erm, pilC2∷cat). (A) Immunoblotting of whole cell lysates by using rabbit antibodies specific for PilV. (B) Coomassie-stained SDS/PAGE gel showing the relative amounts of PilE in purified pili. (C) Immunoblotting of purified pili by using rabbit antibodies specific for PilV. (D) Immunoblotting of purified pili by using rabbit antibodies specific for PilC. (E) Detection of PilC by colloidal silver staining of purified pilus proteins transferred to a poly(vinylidene difluoride) membrane. The amounts of samples loaded in A were equalized based on the total protein concentration of whole cells. The amounts of purified pili loaded in B–E, lanes 1 through 4, were standardized based on the weight of whole cells from which the preparations were made. The comparable intensities of the Coomassie-stained PilE band in B (reflecting its relative yield) provide a quantitative readout of piliation levels (19). The amounts of pili loaded in lanes 5–7 were adjusted to be equivalent to those used in lanes 1–4.
Gonococcal pili are dynamic structures that undergo both extrusion from the cell and retraction (4, 24). Pilus retraction accounts for the absence of pilus fibers in some classes of biogenesis mutants (4, 13) and has been proposed to be responsible for the dramatic reduction in piliation seen after prolonged interaction of N. meningitidis with a human epithelial cell line (25). It appears then that, under certain circumstances, the equilibrium between pilus growth and retraction can be shifted from one in which extrusion is favored to one in which retraction predominates. It was formally possible then that the adherence defects in pilV mutants might relate to altered kinetics of retraction during interactions with human epithelial cells. We therefore asked whether blocking pilus retraction would rescue the adherence defect in pilV mutants by introducing a nonfunctional allele of pilT, whose product is required for retraction (4) into those backgrounds. These pilT, pilV strains expressed Tfp at the same level as the isogenic pilT strains (data not shown), and no alterations in pilus fiber morphology were seen between the isogenic pairs (Fig. 3 Right). Like the isogenic pilT mutant, the double mutants hyperautoagglutinated (data not shown), did not express twitching motility, and failed to take up DNA during transformation (Table 1). When tested for adherence, however, the pilT, pilV mutants failed to adhere to the human cells whereas the pilT mutant bound efficiently (Fig. 2 Middle; Table 1). Therefore, altered dynamics of pilus retraction do not appear to be responsible for aberrant adherence found for the pilV mutants.
Virji and colleagues (26) reported reduced epithelial cell binding capabilities in N. meningitidis because of a pilus anchorage defect associated with low level expression of PilC. To address whether a related phenomenon might be operating in pilV mutants, crude Tfp preparations (obtained from bacterial suspensions that were subjected to shearing by vortexing and cleared by low speed centrifugation) were incubated with the human epithelial cells. As detected by indirect immunofluorescence, wild-type Tfp bound efficiently to the cell monolayers whereas pilV∷kan Tfp were present in greatly reduced amounts (Fig. 2 Bottom). As such, PilV appears to contribute to the intrinsic epithelial cell adhesiveness of gonococcal Tfp.
PilV Copurifies with Pilus Fibers.
To identify PilV and assess its pattern of expression and localization, polyclonal rabbit serum raised against a synthetic peptide representing residues 45–65 of the mature protein was used in immunoblotting. This serum detects PilV as a Mr 18 kDa polypeptide when expressed in E. coli (data not shown). By using whole cell gonococcal lysates, a similar reactive species migrating with molecular mass of 18 kDa was present in the wild-type background and absent in the pilV mutant strains GV1 (pilVG-1 fs) and GV2 (pilV∷kan; Fig. 4A). Antigen detection was restored in the mutants complemented with pilV expressed from the iga locus. Knowing where PilV migrated on SDS/PAGE, we attempted to localize it in by using Coomassie-stained gels of whole cell lysates, but no specific band was detected (data not shown). Thus, PilV appears to be expressed at relatively low levels.
Yoshida and colleagues (27) previously demonstrated the copurification of the pilin-like PilV protein with thin Tfp encoded by IncI1 plasmids. We used immunoblotting to see whether PilV might also copurify with Tfp fibers and found it present in equivalent levels in pilus preparations from the wild-type and complemented pilV mutant but at approximately 2-fold reduced levels in the pilT mutant preparations (Fig. 4C). We sought to detect PilV in Coomassie-stained gels loaded with the purified pilus preparations but could not detect a corresponding band (data not shown), demonstrating that PilV is present at low levels in purified Tfp preparations.
PilC and PilV Content in Pilus Preparations from Adherence-Defective Mutants.
One potential explanation for the lack of adherence of pilV mutants to human epithelial cells would be that PilV is required for PilC stability, activity, or localization. We first examined the status of PilC antigen in whole cell lysates by immunoblotting but could detect no differences within any of the strains used here save for its absence in the pilC mutants (data not shown). However, when purified Tfp preparations were used, it became clear that the pilV mutants had reduced levels of associated PilC, which was restored to normal levels in the complemented mutant (Fig. 4D). As a negative control, pili from a pilC1, pilC2, pilT triple mutant were included. The inclusion of the pilT mutation was necessary because the pilC1, pilC2 background alone fails to express Tfp (13). Remarkably, this preparation lacked PilV although the protein was present in normal levels in the whole cell lysate sample (Fig. 4 A and C, lane 6). This finding showed that PilC is required for the capacity of PilV to copurify with Tfp and implies that PilV antigen copurification in the other backgrounds is likely to be due to specific interactions and not contamination from other cellular fractions.
Attempts at quantitating the relative levels of PilC in pilus preparations by immunoblotting were complicated by the nonlinear responsiveness associated with the enzyme-amplified immunodetection system used and potential differences in PilC immunogenicity in the pilV mutants. To circumvent these problems, PilC was directly detected by using colloidal silver staining of protein onto blotted poly(vinylidene difluoride) filter membranes (Fig. 4E). Relative quantitation was achieved by comparison of the band intensity of the mutants to that seen for serial dilutions of Tfp preparations from the wild-type strain mixed with preparations from the pilC, pilT mutant (data not shown). This method demonstrated that, relative to wild-type Tfp, PilC levels were reduced between 4- to 6-fold in the pilVG-1 fs mutant, 8- to 10-fold in the pilV∷kan mutant, and 2- to 4-fold in the pilT mutant. There was therefore a clear discrepancy between the marginal differences in copurifying PilC levels in the pilT and pilVG-1 fs mutants and the dramatic differences in their adherence proficiencies (Table 1).
Discussion
N. gonorrhoeae Tfp play a critical role in colonizing the human host by promoting adherence to epithelial tissues. For most pilus colonization factors, the filamentous appendages appear to function primarily in the display of adhesin molecules in forms capable of binding specific host cell receptors. Accordingly, gonococcal mutants failing to express Tfp fibers, as well as piliated mutants lacking the Tfp-associated adhesin PilC, both have severe defects in binding to cells of human origin (5, 12, 13). Here, we discovered that the pilin subunit-like protein PilV is essential for Tfp-mediated epithelial cell adherence but dispensable for pilus expression. In addition, pilV mutants do not express reduced levels of piliation as assessed by quantitative techniques. Moreover, because free Tfp fibers from mutants failed to bind to human epithelial cells and because mutants carrying a concurrent mutation in pilT failed to adhere, the defect associated with the loss of PilV cannot be attributed directly to defects in pilus anchoring to the bacterial cell, fiber fragility, or unstable expression because of organelle retraction. These observations demonstrate that PilV promotes the intrinsic adhesive properties of Tfp fibers for human cell receptors.
The mechanism by which PilV contributes to adherence remains obscure, particularly because both the pilin subunit PilE and PilC proteins have been shown to possess human epithelial cell-binding activities. In contrast to the situation with PilC, however, there is currently no direct evidence that the PilE activity has relevance to pilus-mediated bacterial adherence. We chose then to focus this discussion with the assumption that PilC is a pilus-associated adhesin, a position we feel is warranted by current literature and the findings made here. It is formally possible that PilV acts as an adhesin and that both it and PilC are required for efficient adherence. This possibility would be compatible with present data in the literature on PilC adhesin activity. Given that purified mutant pili retain associated PilC but at diminished levels, two other mutually nonexclusive explanations can be envisioned. In one case, PilV might function in efficient PilC incorporation or association with Tfp fibers, in which case reduced adherence relates to reduced amounts of associated PilC. Secondly, PilV may be required for proper modification or exposure of PilC within Tfp fibers. This potential explanation is supported by the dramatic differences in adherence found for the pilT and pilVG-1 fs mutants although their isolated pili have only minor differences in levels of associated PilC (Table 1 and Fig. 4 D and E). At present, however, it is impossible to differentiate between these possibilities or to ascribe the adhesive defect to either quantitative or qualitative alterations in PilC. Interestingly, Scheuerpflug et al. (8) reported that addition of purified PilC was able to impart human epithelial cell adherence to inert particles coated with PilC-deficient pili produced in N. gonorrhoeae or P. aeruginosa but noted that binding of complemented gonococcal pili produced in P. aeruginosa was detectably weaker. This finding led them to state that they could not “exclude the possibility that additional factors in gonococcal pili facilitated the proper presentation of adhesive PilC.”
The concept that pilus adhesin activity might be influenced by ancillary factors physically associated with the organelle has clear precedents. In the E. coli Pap fimbrial system, PapE and PapF are pilin-like molecules, associated with the pilus as components of the tip fibrillum, that are required for proper localization and, thus, function of the PapG adhesin. Mutant cells lacking PapE retain binding ability whereas pili prepared from this background lack it and PapF mutants are devoid of both bacterial cell and purified pilus receptor binding activities (28). Although PilV copurifies with Tfp during repeated rounds of solubilization and precipitation, we have as yet been unable to demonstrate that it is an integral part of the fiber or constitutes a structural subcomponent such as the tip fibrillum. However the fact that copurification of PilV is reliant on PilC, which is a known fiber-associated protein, strongly suggests that PilC, PilV, and Tfp interact with one another and that PilV pilus association is not merely due to leakage from the cells and nonspecific association. Additionally, it has been found in N. meningitidis that one isoform of PilC (PilC2) is associated with a piliated, nonadherent phenotype (29). Because we have shown that PilC is necessary for the association of PilV with Tfp, we speculate that the lack of adherence imparted by that pilC allele may result from reduced levels of pilus-associated PilV.
Adhesin activity might also be influenced by the major pilus subunit as has been seen in some instances. In the Salmonella type 1 fimbrial system, it was reported that receptor specificity of the associated FimH adhesin is influenced by the fimbrial shaft protein subunit (30). Similarly, receptor specificity of the sialic acid E. coli adhesin SfaS appears to be variable depending on the fimbrial subunit with which it is associated (31). Studies in pathogenic neisseria species have suggested a role for the pilin subunit PilE in epithelial cell interaction because changes in its primary structure arising via antigenic variation have been reported to influence adherence proficiencies and tissue tropism (12, 14–16). Given their structural identities in the amino terminal domains thought to be crucial to subunit–subunit contact, one explanation might relate to the differing capacities of altered PilE protein and PilV to interact productively with one another, leading to reduced or defective PilV activity. It is important to note here that, with the exception of studies of Virji and colleagues (26), none of the former studies have examined the levels of Tfp-associated PilC or the intrinsic adherence properties of Tfp alone. Thus, reduced PilC levels in pili or defects in pilus anchoring might account for the findings for the different pilE and pilC alleles. Whatever the case, it would seem important to ascertain whether levels of fiber-associated PilV vary in these backgrounds.
In summary, the PilV protein was identified as a pilin-like molecule involved in gonococcal Tfp-mediated adherence to human epithelial cells. In addition, this molecule copurifies with Tfp and this association requires the PilC adhesin. The paradigm established in enterobacterial chaperone-usher fimbrial systems in which pilin-like molecules contribute to pilus adherence function can therefore be extended to encompass the gonococcal Type IV pilus systems. This situation is particularly remarkable because these two pilus families use highly unrelated pathways and components for biogenesis. Knowing that gonococcal Tfp-mediated adherence to human epithelial cells requires minimal expression of PilE, PilC, and PilV, it will be important to establish how these three components act in concert to create the adhesive phenotype. PilE is highly antigenically variable and encoded by multiple genes, PilC is highly polymorphic and encoded by a two-gene family, whereas pathogenic Neisseria genomes contain only one pilV allele. Given the potentially limited genetic diversity of PilV, elucidation of its structure and function and the nature of its association with both PilC and Tfp may lead to novel strategies for preventing neisserial disease.
Acknowledgments
The gonococcal strain FA1090 genome sequence database was assembled and provided by the Gonococcal Genome Sequencing Project, and B.A. Roe, S. P. Lin, L. Song, X. Yuan, S. Clifton, and D. W. Dyer at the University of Oklahoma. This work was supported by funds from the Norwegian Research Council (to H.W.-L.), the Biotechnology Centre of Oslo (to F.T.H.), and in part by a subcontract of Public Health Service Grant AI 27837, National Institutes of Health (to M.K.).
Abbreviation
- Tfp
Type IV pili
Footnotes
Data deposition: The pilV nucleotide and amino acid sequences derived from this work have been deposited in the GenBank database (accession no. AY063774).
References
- 1.Soto G E, Hultgren S J. J Bacteriol. 1999;181:1059–1071. doi: 10.1128/jb.181.4.1059-1071.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sauer F G, Barnhart M, Choudhury D, Knight S D, Waksman G, Hultgren S J. Curr Opin Struct Biol. 2000;10:548–556. doi: 10.1016/s0959-440x(00)00129-9. [DOI] [PubMed] [Google Scholar]
- 3.Strom M S, Lory S. Annu Rev Microbiol. 1993;47:565–596. doi: 10.1146/annurev.mi.47.100193.003025. [DOI] [PubMed] [Google Scholar]
- 4.Wolfgang M, van Putten J P, Hayes S F, Dorward D, Koomey M. EMBO J. 2000;19:6408–6418. doi: 10.1093/emboj/19.23.6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heckels J E. Clin Microbiol Rev. 1989;2:S66–S73. doi: 10.1128/cmr.2.suppl.s66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tønjum T, Koomey M. Gene. 1997;192:155–163. doi: 10.1016/s0378-1119(97)00018-8. [DOI] [PubMed] [Google Scholar]
- 7.Rudel T, Scheurerpflug I, Meyer T F. Nature (London) 1995;373:357–359. doi: 10.1038/373357a0. [DOI] [PubMed] [Google Scholar]
- 8.Scheuerpflug I, Rudel T, Ryll R, Pandit J, Meyer T F. Infect Immun. 1999;67:834–843. doi: 10.1128/iai.67.2.834-843.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee K K, Sheth H B, Wong W Y, Sherburne R, Paranchych W, Hodges R S, Lingwood C A, Krivan H, Irvin R T. Mol Microbiol. 1994;11:705–713. doi: 10.1111/j.1365-2958.1994.tb00348.x. [DOI] [PubMed] [Google Scholar]
- 10.Bieber D, Ramer S W, Wu C Y, Murray W J, Tobe T, Fernandez R, Schoolnik G K. Science. 1998;280:2114–2118. doi: 10.1126/science.280.5372.2114. [DOI] [PubMed] [Google Scholar]
- 11.Taylor R K, Miller V L, Furlong D B, Mekalanos J J. Proc Natl Acad Sci USA. 1987;84:2833–2837. doi: 10.1073/pnas.84.9.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rudel T, van Putten J P, Gibbs C P, Haas R, Meyer T F. Mol Microbiol. 1992;6:3439–3450. doi: 10.1111/j.1365-2958.1992.tb02211.x. [DOI] [PubMed] [Google Scholar]
- 13.Wolfgang M, Park H S, Hayes S F, van Putten J P, Koomey M. Proc Natl Acad Sci USA. 1998;95:14973–14978. doi: 10.1073/pnas.95.25.14973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nassif X, Lowy J, Stenberg P, O'Gaora P, Ganji A, So M. Mol Microbiol. 1993;8:719–725. doi: 10.1111/j.1365-2958.1993.tb01615.x. [DOI] [PubMed] [Google Scholar]
- 15.Virji M, Heckels J E. J Gen Microbiol. 1984;130:1089–1095. doi: 10.1099/00221287-130-5-1089. [DOI] [PubMed] [Google Scholar]
- 16.Jonsson A B, Ilver D, Falk P, Pepose J, Normark S. Mol Microbiol. 1994;13:403–416. doi: 10.1111/j.1365-2958.1994.tb00435.x. [DOI] [PubMed] [Google Scholar]
- 17.Schoolnik G, Fernandez R, Tai J Y, Rothbard J, Gotschlich E C. J Exp Med. 1984;159:1351–1357. doi: 10.1084/jem.159.5.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wolfgang M, van Putten J P, Hayes S F, Koomey M. Mol Microbiol. 1999;31:1345–1357. doi: 10.1046/j.1365-2958.1999.01269.x. [DOI] [PubMed] [Google Scholar]
- 19.Wolfgang M, Lauer P, Park H S, Brossay L, Hebert J, Koomey M. Mol Microbiol. 1998;29:321–330. doi: 10.1046/j.1365-2958.1998.00935.x. [DOI] [PubMed] [Google Scholar]
- 20.van Putten J P M, Paul S M. EMBO J. 1995;14:2144–2154. doi: 10.1002/j.1460-2075.1995.tb07208.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Oostveen I, Ducret A, Aebersold R. Anal Biochem. 1997;247:310–318. doi: 10.1006/abio.1997.2052. [DOI] [PubMed] [Google Scholar]
- 22.Parkhill J, Achtman M, James K D, Bentley S D, Churcher C, Klee S R, Morelli G, Basham D, Brown D, Chillingworth T, et al. Nature (London) 2000;404:502–506. doi: 10.1038/35006655. [DOI] [PubMed] [Google Scholar]
- 23.Tettelin H, Saunders N J, Heidelberg J, Jeffries A C, Nelson K E, Eisen J A, Ketchum K A, Hood D W, Peden J F, Dodson R J, et al. Science. 2000;287:1809–1815. doi: 10.1126/science.287.5459.1809. [DOI] [PubMed] [Google Scholar]
- 24.Merz A, So M, Sheetz M P. Nature (London) 2000;407:98–102. doi: 10.1038/35024105. [DOI] [PubMed] [Google Scholar]
- 25.Pujol C, Eugene E, Marceau M, Nassif X. Proc Natl Acad Sci USA. 1999;96:4017–4022. doi: 10.1073/pnas.96.7.4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Virji M, Makepeace K, Peak I, Payne G, Saunders J R, Ferguson D J, Moxon E R. Mol Microbiol. 1995;16:1087–1097. doi: 10.1111/j.1365-2958.1995.tb02334.x. [DOI] [PubMed] [Google Scholar]
- 27.Yoshida T, Furuya N, Ishikura M, Isobe T, Haino-Fukushima K, Ogawa T, Komano T. J Bacteriol. 1998;180:2842–2848. doi: 10.1128/jb.180.11.2842-2848.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jacob-Dubuisson F, Heuser J, Dodson K, Normark S, Hultgren S. EMBO J. 1993;12:837–847. doi: 10.1002/j.1460-2075.1993.tb05724.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nassif X, Beretti J L, Lowy J, Stenberg P, O'Gaora P, Pfeifer J, Normark S, So M. Proc Natl Acad Sci USA. 1994;91:3769–3773. doi: 10.1073/pnas.91.9.3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thankavel K, Shah A H, Cohen M S, Ikeda T, Lorenz R G, Curtiss R, 3rd, Abraham S N. J Biol Chem. 1999;274:5797–5809. doi: 10.1074/jbc.274.9.5797. [DOI] [PubMed] [Google Scholar]
- 31.Hacker J, Kestler H, Hoschutzky H, Jann K, Lottspeich F, Korhonen T K. Infect Immun. 1993;61:544–550. doi: 10.1128/iai.61.2.544-550.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]