Abstract
Insect odorant receptor (Or) genes determine the responses of sensory neurons that mediate critical behaviors. The Drosophila melanogaster Or22 locus represents an interesting example of molecular evolution, with high levels of sequence divergence and copy number variation between D. melanogaster and other Drosophila species, and a corresponding high level of variability in the responses of the neuron it controls, ab3A. However, the link between Or22 molecular and functional diversity has not been established. Here, we show that several naturally occurring Or22 variants generate major shifts in neuronal response properties. We determine the molecular changes that underpin these response shifts, one of which represents a chimeric gene variant previously suggested to be under natural selection. In addition, we show that several alternative molecular genetic mechanisms have evolved for ensuring that where there is more than one gene copy at this locus, only one functional receptor is generated. Our data thus provide a causal link between the striking levels of phenotypic neuronal response variation found in natural populations of D. melanogaster and genetic variation at the Or22 locus. Since neuronal responses govern animal behavior, we predict that Or22 may be a key player in underlying one or more olfactory-driven behaviors of significant adaptive importance.
Keywords: odorant receptor, Drosophila melanogaster, olfaction, chimeric gene, natural variants
Introduction
In insects, the responses of many olfactory receptor neurons (ORNs) to ecologically relevant chemicals are determined by the rapidly evolving odorant receptor (Or) gene family (Clyne et al. 1999; Dobritsa et al. 2003; Robertson et al. 2003; Hallem et al. 2004; Hallem and Carlson 2006; McBride et al. 2007; Tunstall and Warr 2012). In Drosophila melanogaster, where these genes were first discovered, there are 61 ligand-binding Ors (Robertson et al. 2003) as well as a single coreceptor, Orco (Larsson et al. 2004). All Or-expressing ORNs express Orco, and the majority of these ORNs coexpress a single ligand-binding Or, with occasionally two (Dobritsa et al. 2003; Hallem et al. 2004; Couto et al. 2005). Within D. melanogaster, the ligand-binding Ors share only 20% identity on average, however, some paralogs show much higher identity levels, up to 78% (Clyne et al. 1999). Similarly, when Or genes are compared across Drosophila species the identity between orthologs is quite variable, ranging from 46% to 91% (Guo and Kim 2007).
Interestingly, even though Or identity between orthologs can be quite low, when ORN response profiles have been compared across Drosophila species they have been found to be in most part quite conserved (Stensmyr, Dekker, et al. 2003; de Bruyne et al. 2010). An exception is the ab3A neuron, which exhibits a high level of response variation across different species (Stensmyr, Dekker, et al. 2003; de Bruyne et al. 2010). The ab3A response is controlled by genes expressed from the Or22 locus, which interestingly shows both high levels of genetic variation and also copy number variation between species (Dobritsa et al. 2003; Guo and Kim 2007; de Bruyne et al. 2010). In the Canton-S laboratory strain of D. melanogaster, and in the genome reference strain, there are two gene copies at this locus, Or22a and Or22b, which share 78% sequence identity (Dobritsa et al. 2003). This level of identity is much higher than for the Or family as a whole and indicates that Or22a and Or22b are likely the result of a relatively recent gene duplication (Aguadé 2009). In the Canton-S strain, both Or22a and Or22b are expressed in the ab3A neurons (Dobritsa et al. 2003). However, when each gene was expressed in ab3A neurons of a strain in which the Or22 locus is deleted (the empty neuron system; Hallem et al. 2004), only Or22a was found to be functional (Dobritsa et al. 2003). Thus, in this strain, Or22a alone determines the response of the ab3A neurons and Or22b is nonfunctional.
Previous studies have identified a naturally occurring variant at the Or22 locus, where a single chimeric gene (called Or22ab) has formed due to a deletion that leaves mostly Or22b sequence fused in-frame with the predicted N terminus from Or22a (Turner et al. 2008; Aguadé 2009). These studies also reported the presence of several nonsynonymous single nucleotide polymorphisms (SNPs) in Or22ab compared with Or22a and Or22b (Turner et al. 2008). The Or22ab allele occurs at different frequencies in natural populations, and at latitudinally-varying frequency in Australia, strongly suggesting the locus-specific action of selection (Turner et al. 2008; Aguadé 2009). However, its effect on olfactory system function is unknown.
Here, we determine the functional consequences of the Or22ab variant on ab3A neuron function. We further identify and characterize a third naturally-occurring variant at this locus, and find that the three variants generate dramatic response differences from the ab3A neurons, including changes in the major ligand. By dissecting the molecular bases of these phenotypes, we determine that three alternative molecular genetic mechanisms have evolved to generate ecologically-relevant functional Or diversity, while ensuring that only one functional Or protein is produced from the Or22 locus. Our findings, together with the known variability of this locus and neuron across Drosophila species, suggest that Or22 may underlie important olfactory-driven behaviors such as oviposition or food seeking.
Results and Discussion
Three ab3A Phenotypic Variants Occur in Natural Populations
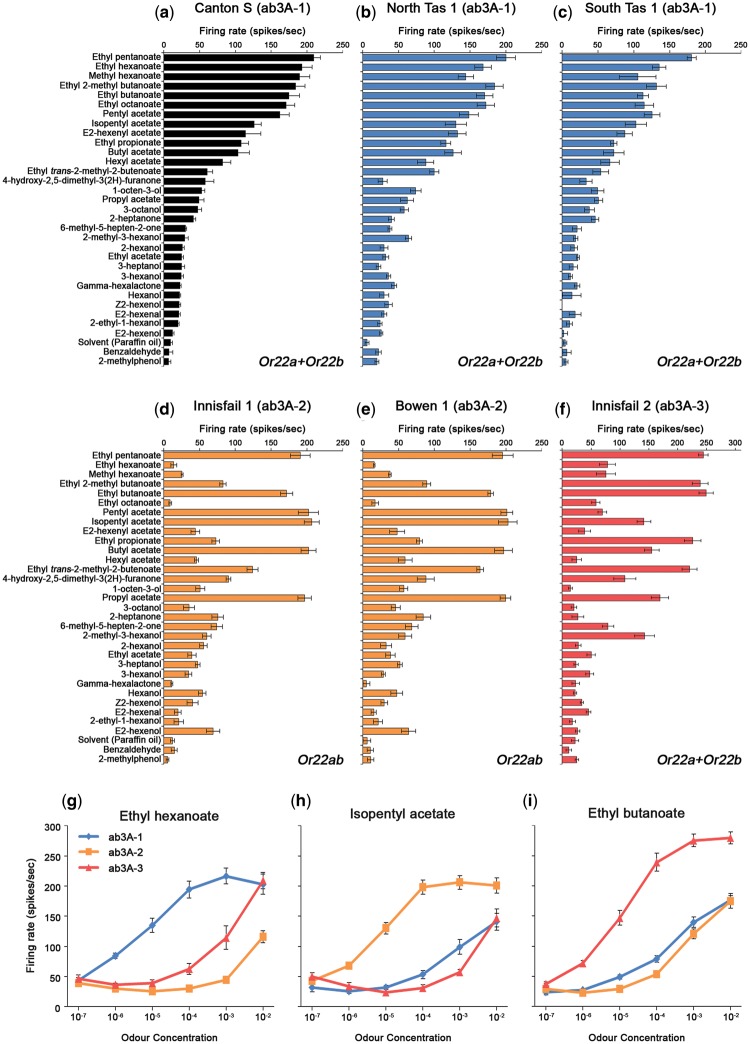
To determine if there are any changes in neuronal responses in flies homozygous for Or22ab, we generated lines that were isogenic for the second chromosome from populations of Drosophila collected from northern regions of eastern Australia. These populations were expected to have relatively high frequencies of Or22ab, with higher frequencies in the north and lower in the south previously observed (Turner et al. 2008). The response profiles of ab3A neurons were measured from individual isogenic lines and the lines were also genotyped for the presence of either Or22ab or both of Or22a and Or22b. In the laboratory strain Canton-S, which has both Or22a and Or22b, this neuron has been shown to respond to a range of esters and alcohols but it is most sensitive to ethyl hexanoate (Stensmyr, Giordano, et al. 2003; Pelz et al. 2006). Several of the isogenic lines showed the same ab3A response profile as Canton-S (fig. 1a–c;supplementary fig. 1, Supplementary Material online), and lines with this phenotype all had both Or22a and Or22b (supplementary fig. 2, Supplementary Material online). Since in the Canton-S strain only Or22a is required for ab3A responses, and Or22b is presumed nonfunctional (Dobritsa et al. 2003), this is likely also to be the case for these isogenic lines.
Fig. 1.
Identification of three ab3A neuronal response phenotypes in natural populations of Drosophila melanogaster. (a) The ab3A response profile of Canton-S; named the ab3A-1 phenotype. (b, c) Two isogenic lines derived from populations from southern Australia (from North and South Tasmania, respectively) show the ab3A-1 phenotype (comparing a and b, Spearman rank correlation, P < 0.001; comparing a and c, P < 0.001). (d, e) Two isogenic lines from northern populations (from Innisfail and Bowen, respectively) exhibit a different ab3A phenotype (comparing a and d, P = 0.380; comparing d and e, P < 0.001); we call this the ab3A-2 phenotype. (f) A second isogenic line derived from the Innisfail population shows an ab3A phenotype distinct from ab3A-1 and -2 (compare a and f, P = 0.096; compare d and f, P = 0.055); named the ab3A-3 phenotype. (g–i) Neuronal dose–response curves for the three ab3A phenotypes to ethyl hexanoate (g), isopentyl acetate (h), and ethyl butanoate (i) showing shifts in major ligand sensitivity. For ease of identification, blue represents the ab3A-1 phenotype, orange the ab3A-2 phenotype, and red the ab3A-3 phenotype. Paraffin oil is the solvent-only control. Data are represented as the mean ± SEM, n = 6 for all except c and e (n =3). For a–f, all odorants were tested at 10−2 except for ethyl and methyl hexanoate (tested at 10−4) due to the known high sensitivity of the ab3A neuron to these odorants (Hallem and Carlson 2006).
Two of the isogenic lines (derived from populations collected from Innisfail and Bowen in northern Australia), however, exhibited a substantially different ab3A response profile (fig. 1d and e;supplementary fig. 2, Supplementary Material online). These lines were found to be homozygous for the Or22ab variant (supplementary fig. 2, Supplementary Material online). In these flies, the responses to ethyl hexanoate and some other esters, such as methyl hexanoate and ethyl 2-methyl butanoate, are strongly reduced. By contrast, the responses to isopentyl-, butyl-, and propyl acetate are strongly increased.
We also identified one isogenic line derived from the Innisfail population which had a different ab3A response profile again (fig. 1f;supplementary fig. 1, Supplementary Material online). Due to multiple ab3A phenotypes being identified, we therefore named the previously published ab3A phenotype from Canton-S the ab3A-1 phenotype, the phenotype associated with Or22ab the ab3A-2 phenotype, and this third phenotype the ab3A-3 phenotype. In ab3A-3 flies, the ab3A neurons display some of the same changes seen in ab3A-2 neurons, such as decreased responses to ethyl- and methyl hexanoate, and an increased response to propyl acetate. However, there are also some major differences such as a decreased response to pentyl acetate and an increased response to ethyl propionate. Amplifying and sequencing the Or22 locus from ab3A-3 flies showed that, as in ab3A-1 flies, both Or22a and Or22b are present in ab3A-3 flies (supplementary fig. 2, Supplementary Material online).
To quantify the shifts in ligand-binding properties in the three types of ab3A neurons, we tested ab3A responses to a range of doses of three odorants. We chose ethyl hexanoate because it is the best known ligand for ab3A-1 neurons (Stensmyr, Giordano, et al. 2003; Hallem and Carlson 2006), and isopentyl acetate and ethyl butanoate because they are the odorants that generated the largest responses in ab3A-2 and ab3A-3 neurons, respectively (fig. 1; supplementary fig. 1, Supplementary Material online). To quantify the differential sensitivities, the expected concentration (EC) resulting in 100 spikes/s was determined for each of the neurons for each of the odorants. These values were compared to determine the magnitude of the differences in sensitivity between the different neurons. Electrophysiological recordings revealed that ab3A-1 neurons are ∼3 log orders (1,000 times) more sensitive to ethyl hexanoate than ab3A-2 neurons, and ∼2 log orders (100 times) more sensitive to ethyl hexanoate than ab3A-3 neurons (ab3A-1 EC = 1.6 × 10−6, ab3A-2 = 7.8 × 10−3, and ab3A-3 = 9.6 × 10−4; fig. 1g). By contrast, ab3A-2 neurons are ∼3 log orders (1,000 times) more sensitive to isopentyl acetate than both ab3A-1 and ab3A-3 neurons (ab3A-1 EC = 4.4 × 10−3, ab3A-2 = 4.1 × 10−6, and ab3A-3 = 4.1 × 10−3; fig. 1h). Finally, ab3A-3 neurons are ∼2 log orders (100 times) more sensitive to ethyl butanoate than both ab3A-1 and ab3A-2 neurons (ab3A-1 EC = 2.6 × 10−4, ab3A-2 = 5.7 × 10−4, and ab3A-3 = 3.2 × 10−6; fig. 1i). The dramatic differences in ab3A neuron sensitivity to these odorants suggest that a major change in ligand specificity has occurred, with large methyl and ethyl esters being the best ligands in ab3A-1, acetate esters in ab3A-2, and smaller ethyl esters in ab3A-3.
Genetic Variation at the Or22 Locus Underlies the Different ab3A Phenotypes
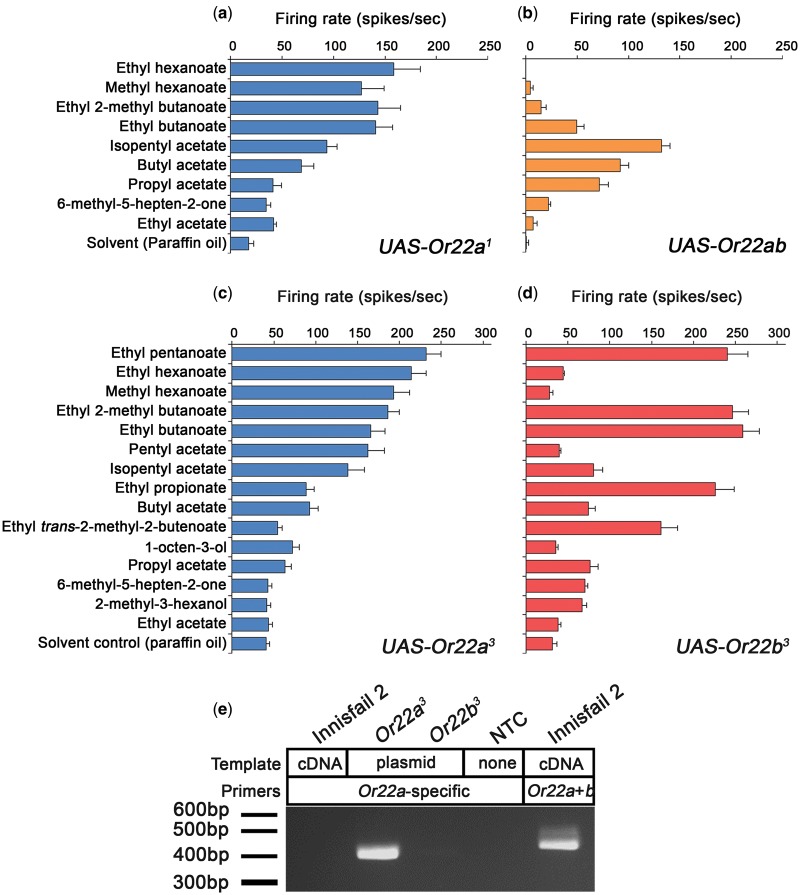
The data from the isogenic lines showed that the presence of the Or22ab variant correlates with an altered ab3A phenotype. However, as there are other genetic background differences between these lines it remained formally possible that the observed effects on the ab3A response were not due to Or22 variation. In this regard, we note that the majority of the sequence of Or22ab is identical to that of Or22b from the Canton-S strain (Aguadé 2009), which, as previously mentioned, does not encode a functional receptor (Dobritsa et al. 2003). Thus, in order to provide evidence of causality, we cloned the Or22ab variant and expressed it in the empty neuron system (Hallem et al. 2004). Expression of the Or22ab transgene in empty ab3A neurons conferred a response profile that was markedly different to that of flies in which Or22a from Canton-S (hereafter designated Or22a1) was expressed (Spearman rank correlation, P = 0.707; fig. 2a). By comparison to flies expressing Or22a1, flies expressing Or22ab have decreased responses to ethyl hexanoate, methyl hexanoate, and ethyl 2-methyl butanoate, as well as increased responses to isopentyl acetate and propyl acetate. Comparing this response profile to that of an isogenic line with the Or22ab genotype revealed that these two profiles are highly similar (P < 0.001; figs. 1d and 2b). Taken together, these data show that Or22ab encodes a functional olfactory receptor that has dramatically different ligand-binding properties to Or22a, and that it is responsible for the ab3A-2 phenotype.
Fig. 2.
Natural variation at the Or22 locus determines the ab3A neuronal response phenotype. (a–d) Response profiles for flies expressing UAS-Or22 transgenes in ab3A neurons lacking their endogenous receptor (Dobritsa et al. 2003). (a) Expression of Or22a1 results in an ab3A-1 phenotype (comparing to fig. 1a, Spearman rank correlation, P < 0.001). (b) Or22ab results in an ab3A-2 phenotype (comparing to fig. 1d, P < 0.001). (c) Or22a3 results in an ab3A-1 phenotype (comparing to fig. 1a, P < 0.001). (d) Or22b3 results in an ab3A-3 phenotype (comparing to fig. 1f, P < 0.001). Data are represented as the mean ± SEM, n = 6 for all recordings. Paraffin oil is the solvent control. (e) Or22a mRNA transcripts are not detected in Innisfail 2 antennal cDNA preparations. Plasmid DNA template control showing that Or22a primers do not amplify Or22b. NTC, no template control for Or22a primers. Or22 primers (not specific to Or22a or Or22b) detect Or22 expression in the same cDNA prep as lane 1.
We next asked how the ab3A-3 response profile is generated, given that there are two gene copies at the Or22 locus, as is also the case in ab3A-1 flies. We first sequenced the two gene copies from the ab3A-3 line. By comparison to their homologs in ab3A-1 flies, Or22a in the ab3A-3 line has two amino acid substitutions, H8Y and I67M, and Or22b also has two amino acid substitutions, V25I and R194M. For clarity, the Or22 alleles associated with the ab3A-3 phenotype will hereafter be called Or22a3 and Or22b3, and the alleles associated with the ab3A-1 phenotype will be called Or22a1 and Or22b1. We then generated transgenic constructs encoding each gene (Or22a3 and Or22b3) and expressed them in empty ab3A neurons. Expressing Or22a3 conferred ab3A function (fig. 2c), however resulted in ab3A neurons with an ab3A-1 phenotype, such as that seen in Canton-S, and not the ab3A-3 phenotype (P < 0.001; figs. 1a and 2c). By contrast, expressing Or22b3 conferred the ab3A-3 phenotype (P < 0.001; fig. 2d).
Given these data show that Or22a3 and Or22b3 both encode functional receptors, yet that Or22b3 alone can confer the ab3A-3 phenotype, we hypothesized that the Or22a3 gene may not be expressed in ab3A-3 flies. To test this idea, we designed primers to regions in the Or22a coding sequence that are variable to the Or22b coding sequence, and as such should specifically amplify Or22a. Specificity was demonstrated by showing the primers amplify a genomic copy of Or22a that had been cloned into the plasmid pGEM but do not amplify Or22b (fig. 2e). We then used these primers to test for Or22a expression in antennal cDNA from the ab3A-3 isogenic line. Consistent with our prediction, no expression of Or22a was detected (fig. 2e). We note that due to the high level of similarity between Or22a and Or22b, we were unable to design primers that amplify Or22b specifically. We therefore used an alternative strategy to confirm that only Or22b is expressed in ab3A-3 flies. Primers that recognize both genes were used to amplify a product from head cDNA, and the resulting polymerase chain reaction (PCR) product was cloned into the plasmid pGEM. We then sequenced 27 subclones to determine which gene(s) were present. All 27 subclones contained Or22b3. Taken together, these data strongly suggest that Or22b3 is the sole contributor to ab3A-3 function, and that Or22a3 is not expressed in ab3A-3 flies.
Specific SNPs Underpin Changes in Or22 Receptor Function
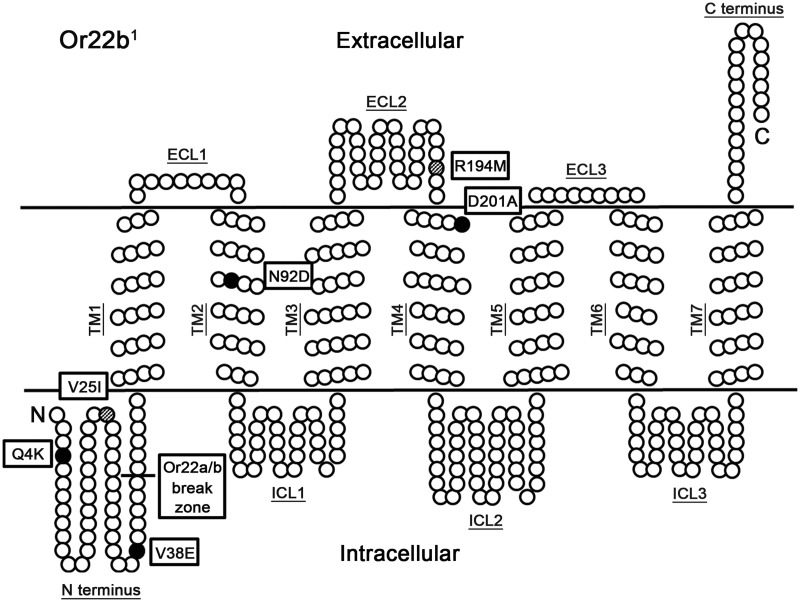
We next aimed to identify which specific amino acid changes between the Or22 copies underlie the functional differences that we had identified. We focused on the Or22b variants as there are too many differences between copies of Or22a and Or22b to enable this approach. Given that Or22b1 is nonfunctional, whereas both Or22ab (composed mostly of the Or22b1 sequence) and Or22b3 are functional receptors, we first asked what amino acid changes between these control this overall change in functionality. Consistent with a previous study, when we sequenced Or22ab from the Innisfail 1 isogenic line we found that the deletion leads to the first 43 amino acids of Or22a1 becoming the first 43 residues of Or22ab, with the remaining 354 amino acids derived from Or22b1 (fig. 3, supplementary fig. 3, Supplementary Material online). Due to the high level of sequence similarity between Or22a1 and Or22b1, this results in only three amino acid substitutions in this region (Q4K, V25I, V38E). We also found three amino acid substitutions present in the remainder of Or22ab when compared with Or22b1. These are: N92D, in the middle of transmembrane (TM) domain 2; R194M in the second extracellular loop (ECL2) and; D201A in TM4. Both R194M and D201A are close to the ECL2-TM4 boundary (fig. 3). As mentioned earlier, there are two amino acid substitutions between Or22b1 and Or22b3 (V25I and R194M) both of which are found in Or22ab. Thus, overall we identified six amino acid changes between Or22b1 and the two functional receptors Or22ab and Or22b3.
Fig. 3.
Amino acid differences between functional Or22b receptors and Or22b1. Schematic representation of the Or22b1 protein including membrane topology and positions of the seven TM regions. The sites of amino acid substitutions found in the Innisfail 1 Or22ab (black and hatched circles) and Innisfail 2 Or22b3 (hatched circles) sequences compared with the Or22b1 sequence are indicated.
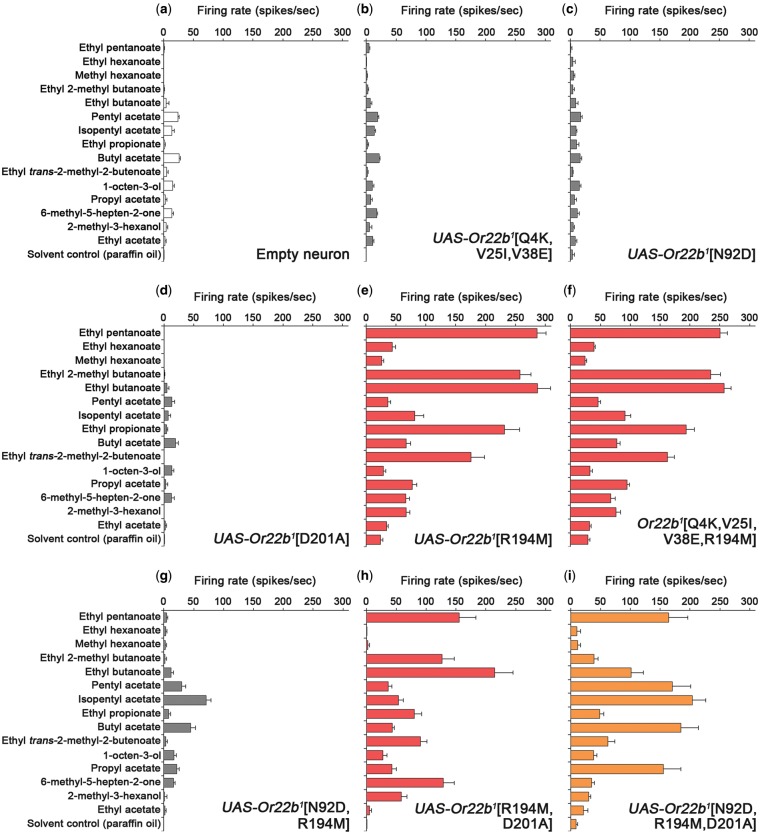
To determine if any of these changes could impart function on Or22b1, we generated four different transgenic constructs that generate changes on an Or22b1 backbone: 1) substitution of the first 43 amino acids of the Or22a1 N terminus in place of the first 43 amino acids of the Or22b1 N terminus, thus introducing three amino acid substitutions, Q4K, V25I, and V38E into Or22b1 (Or22b1[Q4K, V25I, V38E]); 2) the N92D substitution alone (Or22b1[N92D]); 3) the D201A substitution alone (Or22b1[D201A]), and 4) the R194M substitution alone (Or22b1[R194M]). Of the four constructs, one restored ab3A responses when expressed in empty ab3A neurons: Or22b1[R194M] (fig. 4a–e). This demonstrates that Or22b1 lacks function because of the presence of an arginine residue at position 194. In further support of this, we note that of the ∼46 Drosophila Genetic Reference Panel (DGRP; Mackay et al. 2012) lines with the Or22ab variant (thus encoding only one functional receptor), all have a methionine residue at position 194.
Fig. 4.
Molecular basis of altered Or22b receptor function and ligand specificity. (a) The empty neuron control ab3A response profile to a panel of odorants. (b–d) Expression of Or22b1[Q4K, V25I, V38E], Or22b1[N92D], and Or22b1[D201A], respectively, in empty ab3A neurons result in no functional responses. (e) Expression of Or22b1[R194M] results in a functional response profile that closely resembles the ab3A-3 phenotype (comparing to fig. 1f, Spearman rank correlation, P < 0.001). (f) Recordings from ab3A neurons of DGRP line 730, which has Or22b1[Q4K, V25I, V38E, R194M], results in an ab3A-3 phenotype (comparing to fig. 1f, P < 0.001). (g) Expression of Or22b1[N92D, R194M] results in low level ab3A responses. (h) Expression of Or22b1[R194M, D201A] in the empty neuron results in an ab3A-3 phenotype (comparing to fig. 1f, P < 0.001). (i) Expression of Or22b1[N92D, R194M, D201A] results in an ab3A-2 phenotype (comparing to fig. 1d, P < 0.001). Data are represented as the mean ± SEM, n = 6 for all recordings. Paraffin oil is the solvent control.
Interestingly, the response profile of Or22b1[R194M] strongly correlated with the ab3A-3 phenotype (P < 0.001), and not with the ab3A-2 phenotype controlled by Or22ab. We therefore next asked which of the remaining differences between Or22b1 and Or22ab are required in combination with the R194M substitution to generate the ab3A-2 phenotype. We first combined the R194M substitution with the changes that arise from the altered N terminal region (Q4K, V25I, V38E). To do this we took advantage of a DGRP line that had these four changes from Or22b1 but lacked the N92D and D201A substitutions (line 730). We found that this line also showed an ab3A-3 phenotype (P < 0.001; fig. 4f), suggesting that the N terminal exchange is not the cause of the altered ligand-binding properties of Or22ab in ab3A-2 flies. We next combined the two other amino acid substitutions (N92D and D201A) with R194M by generating transgenic constructs encoding these forms of Or22b1. Adding N92D alone into the Or22b1-R194M background generated ab3A neurons which showed very low responses to the tested odorants (fig. 4g). Adding D201A alone into the Or22b1-R194M background did not alter the phenotype from that of ab3A-3 (P < 0.001; fig. 4h). However, when both N92D and D201A were combined with Or22b1-R194M, a response profile remarkably similar to the ab3A-2 phenotype was observed (P < 0.001; fig. 4i). Thus, all three of the N92D, R194M, and D201A substitutions that have occurred between Or22b1 and Or22ab are required to generate the ab3A-2 phenotype. These data further show that amino acid variation at residues 92 and 201 alters the ligand-binding properties of Or22b.
Although the structure of a ligand-binding Or has proven elusive, the structure of an insect Orco homotetramer was recently solved (Butterwick et al. 2018). These authors proposed that the Or-Orco heterotetramer should form an analogous structure, and that the Or ligand-binding pocket is formed within the extracellular leaflet of the plasma membrane between helices TM1–6. In support of this model, mutations in Ors that map to this region have been previously shown to alter ligand specificity (Nichols and Luetje 2010; Leary et al. 2012; Hughes et al. 2014; Yang et al. 2017). Similarly, the residues we found to alter ligand specificity of Or22b, residues 92 and 201, are predicted to lie within this proposed binding pocket. Notably, residue 194, which we found to be essential for Or22b function, lies in ECL2, which in an Orco homoetramer (which does not bind ligands) is proposed to restrict access of odorants to the binding pocket (Xu and Leal 2013; Butterwick et al. 2018). This region is therefore potentially critical for odorant access to the binding pocket in Or22b.
The ab3A-3 Phenotype Is Likely to Be Ancestral to the Simulans Clade
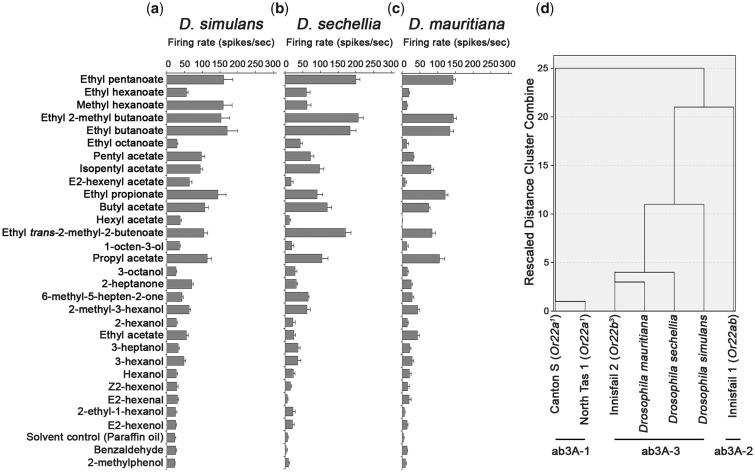
Given that we had identified three different ab3A phenotypes, we were in a position to test which was likely to be the ancestral form of the ab3A response, and how the Or22 variants might have evolved within the D. melanogaster lineage. To investigate this, the ab3A response profiles of the three most closely related species to D. melanogaster (Drosophila simulans, Drosophila sechellia, and Drosophila mauritiana; fig. 5a–c) were determined. Although ab3A responses from these species have been reported previously (Stensmyr, Dekker, et al. 2003; de Bruyne et al. 2010), we needed to test a wider range of odorants to allow us to better compare the responses to the D. melanogaster phenotypes we had found. As previously shown, we found that the responses of these species differed greatly to the previously reported ab3A phenotype of D. melanogaster (ab3A-1). Notably, all three species had reduced responses to ethyl hexanoate, and all but D. simulans had reduced responses to methyl hexanoate. We also observed increased responses to propyl acetate, ethyl propionate, and ethyl trans-2-methyl-2-butenoate, as well as decreased responses to ethyl octanoate, in all three species. Given the observed increased responses to ethyl propionate and ethyl trans-2-methyl-2-butenoate, and decreased response to ethyl hexanoate, as well as no increase in response to isopentyl acetate, these ab3A responses appeared to be most similar to the D. melanogaster ab3A-3 phenotype.
Fig. 5.
The ab3A-3 phenotype is likely to be ancestral to the Drosophila simulans clade. (a–c) Neuronal ab3A responses from D. simulans, Drosophila sechellia, and Drosophila mauritiana, respectively. Data are represented as the mean ± SEM, n = 6 for all recordings. Paraffin oil is the solvent control. (d) Similarity tree shows clustering of the ab3A responses from the tested Drosophila species with the three Drosophila melanogaster ab3A phenotypes. Unlike the ab3A-1 and ab3A-2 phenotypes, the ab3A-3 phenotype of D. melanogaster clusters most closely with the three other Drosophila species.
We tested this idea formally by generating a similarity tree to group the different ab3A responses together (fig. 5d). This showed that the D. sechellia, D. simulans, and D. mauritiana phenotypes cluster with, and therefore most closely resemble, the ab3A-3 phenotype in D. melanogaster. We therefore propose that the ab3A-3 phenotype is likely to be the ancestral ab3A phenotype for the simulans clade.
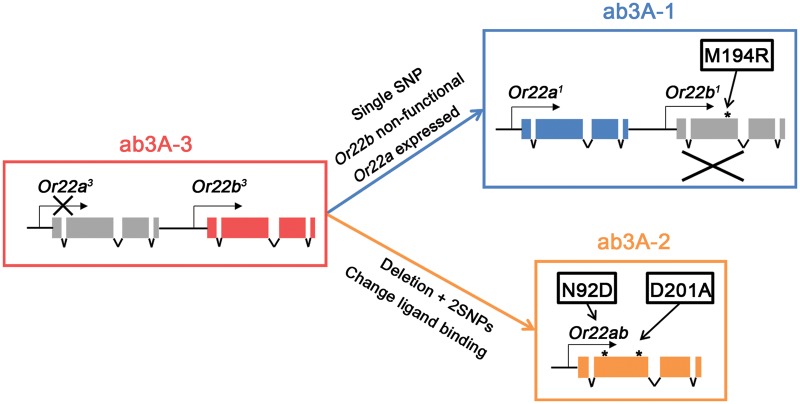
From our findings we can then predict how the D. melanogaster ab3A-1 and ab3A-2 phenotypes may have evolved (fig. 6). From the ab3A-3 situation, in which Or22a is not expressed and Or22b controls neuron function, we suggest that the ab3A-1 phenotype may have arisen via the activation of expression of Or22a together with the M194R substitution in Or22b to render it nonfunctional. To form the chimeric Or22ab receptor and the ab3A-2 phenotype, a deletion between the first intron of Or22a3 and the first intron of Or22b3 took place, together with two amino acid substitutions (N92D and D201A) occurring in the original Or22b portion of the receptor. The two combinations of the changes seem more likely to have arisen independently rather than sequentially because a methionine was found to always occupy position 194 in Or22ab. This suggests that this chimeric gene has evolved from the ancestral Or22a3 and Or22b3, where there is a methionine at residue 194 of Or22b, rather than from Or22a1 and Or22b1, where there is not.
Fig. 6.
A model for the evolution of the three Drosophila melanogaster ab3A phenotypes. The ab3A-3 phenotype is likely the ancestral response profile and is conferred by the Or22a3 and Or22b3 genotype. We therefore propose a model whereby the ab3A-1 and ab3A-2 phenotypes are the result of two distinct molecular genetic mechanisms beginning with the ab3A-3 ancestral state. From here, the ab3A-1 phenotype has arisen via mutations that activated expression of Or22a1 and a SNP that caused the M194R amino acid substitution in Or22b1 to render it nonfunctional. The ab3A-2 phenotype has arisen via the formation of the chimeric gene Or22ab, resulting from an in-frame deletion between the first introns of Or22a3 and Or22b3. Two additional SNPs in Or22ab were further required for the two amino acid substitutions, N92D and D201A, to produce the ab3A-2 phenotype.
In conclusion, we have demonstrated that naturally occurring variation at the Or22 locus causes dramatic changes in the ligand specificity of the ab3A class of olfactory neuron. Our data show that three different molecular mechanisms have evolved to ensure that, despite there sometimes being more than one gene present at the Or22 locus, only one functional receptor is produced. These data further suggest that having only one functional receptor produced from this locus may be of intrinsic importance to the function of the ab3A neuron. Perhaps the ab3A output generated if there is more than one functional receptor might interfere with odor discrimination such that the animal cannot effectively process the chemical stimuli and generate appropriate behaviors. Given that in many other species of Drosophila gene copy number variation at this locus exists (de Bruyne et al. 2010), further support for this idea could be obtained by determining if only one functional receptor type is ever produced in these species.
Are these differences in neuron response profiles likely to cause behavioral differences? The relationship between olfactory neuron physiology and behavior is not straightforward (Stensmyr, Giordano, et al. 2003; Ruebenbauer et al. 2008; Knaden et al. 2012; Richgels and Rollmann 2012). However, naturally-occurring changes in the sequence of some other Or genes have been shown to associate with changes in olfactory behavior (Rollmann et al. 2010; Richgels and Rollmann 2012). In addition, some studies have shown associations between changes in olfactory neuron number and changes in olfactory behavior (Ibba et al. 2010; Dekker et al. 2015).
The rapid molecular evolution at the Or22 locus and accompanying functional diversity in responses of the ab3A neuron, both within D. melanogaster and across Drosophila species, suggest it may be of particular ecological importance compared with other neuron types. This is further supported by the latitudinally-varying frequency of the Or22ab variant in Australia (Turner et al. 2008). Moreover, a recent study reported that wild African D. melanogaster are seasonal specialists on marula fruit, and that flies with the Or22ab allele have increased sensitivity to the odorant ethyl isovalerate (an ester found in high amounts in marula fruit and not other fruits) compared with Canton-S (ab3A-1) flies (Mansourian et al. 2018). Mansourian et al. (2018) also showed that, although the ab3A neuron is not the sole contributor, it plays a role in the ability of flies to locate marula odorants over long distances. It is therefore possible that the Or22 variants investigated here cause changes in behaviors that are critical for individual fitness, and that selective pressure on such behaviors may explain the high level of intra- and inter-specific variation observed at the Or22 locus and in the ab3A neuron response profile.
Materials and Methods
Drosophila Stocks and Maintenance
Four mass-bred populations of D. melanogaster, originally collected from Bowen, Queensland (19°58′S), Innisfail, Queensland (17°31′S), Northern Tasmania (41°S), and Southern Tasmania (43°S), were a kind gift from Carla Sgró, Monash University (Sgrò et al. 2010). Multiple isogenic lines were derived from each mass-bred population by crossing single males to w; If/CyO; MKRS/TM6B virgins, and kept as stocks with balanced second and third chromosomes (w;+/CyO;+/TM6B) from which w;+;+ homozygous flies could be obtained. Species stocks were obtained from the Tucson Drosophila Stock Centre (D. simulans, 14021-0251.169; D. sechellia, 14021-0248.0; D. mauritiana, i.e. 14021-0241.01). All isogenic lines, mass-bred populations and other Drosophila species lines were maintained on standard wheat-based media at 22 °C. To drive expression of olfactory receptor transgenes in the ab3A neuron, we used flies carrying ΔHalo, a small deletion on the second chromosome that removes the Or22 locus as well as 11 other genes (Gross et al. 2003), combined with an Or22a-Gal4 transgene (Dobritsa et al. 2003). The UAS-Or22a transgenic line was a kind gift from John Carlson, Yale University. All crosses using transgenic lines were performed at 25 °C.
Genotyping of Flies
DNA was extracted from isogenic lines using a chloroform–phenol extraction method and genotyped for the presence of either Or22a and Or22b (long variant, 4,542-bp fragment) or Or22ab (short variant, 2,468-bp fragment) by amplification of the region between primers Or22a-F1 (5′-CAA TCA TTT TTC GGT TGC AT-3′) and Or22b-R (5′-CTG TCC CTC TTT TGC ACC AT-3′) using Expand high-fidelity Taq polymerase (Roche).
Or22 Constructs and Generation of Transgenic Lines
The Or22ab gene was amplified and sequenced (UASOr22ab-F 5′-GAA TTC ATG TTA AGC AAG TTT TTT CCC CAC A-3′; UASOr22ab-R 5′-GCG GCC GCT CGT CGA AAG AGA CAA CTG-3′) from genomic DNA extracted from the Innisfail 1 isogenic line using KOD polymerase (Novagen). The amplicon was cloned into pUAST-attB using EcoRI and NotI restriction sites. The Or22a3 and Or22b3 genes were initially amplified and sequenced from genomic DNA extracted from the Innisfail 2 isogenic line using Expand Taq polymerase (Roche) and cloned into pGEM-T (Promega) using primers in the 5′ and 3′ regions of each gene (Or22a3-F1 5′-ATG GTC AAC GCA ATG TCA GA-3′, Or22a3-R1 5′-GGT TCC ATT GAC CAC AAT C-3′, Or22b3-F1 5′-GCA GTT TTT CGC AAA GGA AG-3′, Or22b3-R1 5′-TGT CTC CTA CCC CAG ACC AC-3′). The coding sequences of the Or22a3 and Or22b3 clones were then isolated using PCR (Or22a3-F2 5′-GAA TTC ATG TTA AGC AAG TTT TTT CCC TAC A-3′, Or22a3-R2 5′-GCG GCC GCT AGC AGA GCT CGT CCC TCT C-3′, Or22b3-F2 5′-GAA TTG ATG TTA AGC CAG TTC TTT CCC CAC A-3′, Or22b3-R2 5′-GCG GCC GCA AGC AGA GCT TGC ATA TCT T-3′), and inserted into pUAST-attB using EcoRI and NotI restriction sites. All transgenic constructs with amino acid substitutions (Or22b1[Q4K, V25I, V38E], Or22b1[N92D], Or22b1[R194M], Or22b1[D201A], Or22b1[N92D, R194M], Or22b1[R194M, D201A], and Or22b1[N92D, R194M, D201A]) were synthesized (Genscript) and subcloned into pUAST-attB. All transgenic lines were generated via phi-C31-mediated integration into the ZH-86Fb site (BestGene Inc) with the exception of the Or22b1[N92D, R194M], Or22b1[R194M, D201A], and Or22b1[N92D, R194M, D201A] constructs, which were injected in-house.
cDNA Synthesis and Reverse Transcription-PCR
One hundred antennae were dissected from snap-frozen adults from the Innisfail 2 isogenic line before RNA was isolated using β-mercaptoethanol, chloroform, and phenol extraction. cDNA was synthesized using a Tetro cDNA kit (Bioline) and amplified using GoTaq (Promega) to test for Or22a3 expression using Or22a-specific primers (Or22a-F2 5′-CTC CCA CCT TCG TGG TAA TGA A-3′; Or22a-R 5′-CAA AAA TGG TTC CCG AAA AG-3′). Diluted plasmid DNA was used as template for primer specificity tests. Or22a+b primers (Or22a+b-F 5′-GAG AGA TGC CTT CAT TTA CTT GG-3′; Or22a+b-R 5′-ACC CCA TGA GAA TGA CTT CG-3′) were used to confirm the quality of the Innisfail 2 antennal cDNA preparation. To sequence Or22 transcripts expressed in the Innisfail 2 isogenic line, ten heads were dissected from snap-frozen adults followed by RNA isolation and reverse transcription-PCR using the Or22a+b primers. The amplicon was ligated with pGEM-T (Promega) and 27 clones containing Or22 inserts were sequenced.
Electrophysiological Recordings from ab3A Neurons
Recordings of neuronal responses to odorants were performed as previously described (de Bruyne et al. 2010). Briefly, 4–10-day-old flies were immobilized and glass recording and reference electrodes were inserted into the cuticle at the base of a single sensillum and into the fly eye, respectively. Signals were amplified using an active probe fed into an analog-to-digital (AD) converter with digital amplification (Syntech IDAC 4). All odors were diluted 10−2 v/v in paraffin oil for determining response spectra, except ethyl- and methyl hexanoate (10−4). For the dose–response curves, odorants were diluted further in decadic steps. Volatiles were collected in 5 ml disposable syringes holding 10 µl of the diluted odorant and injected into a clean air stream (Alphagaz Air 1, Air Liquide) delivered to the preparation. Neuronal responses (spikes/s) were calculated from action potential counts during the 500 ms of stimulation after subtracting activity during the 500 ms prior to stimulation. Odor responses were measured from at least six neurons from at least four different flies. Response profiles were compared for similarity using Spearman’s rank correlation in GraphPad Prism (GraphPad Software, Inc.). For analysis of sensitivity in the dilution series, the EC value was calculated by finding the concentration at which a firing rate of 100 spikes/s would be seen. From this, the average EC value could be calculated.
Statistical Analyses
Response profiles were tested for similarities with the ab3A-1, ab3A-2, or ab3A-3 response profiles using a Spearman’s rank correlation in GraphPad Prism (GraphPad Software, Inc.). The similarity tree comparing and grouping response profiles was generated by performing a hierarchical cluster analysis evaluating the squared Euclidian distance to find the average linkage between response profiles using SPSS (IBM SPSS Statistics 23). Sequence alignments and comparisons were performed using MEGA software (supplementary fig. 3, Supplementary Material online).
Supplementary Material
Acknowledgments
We thank the Australian Drosophila Biomedical Research Facility (OzDros) and Jyotika Taneja de Bruyne for technical support, the Bloomington Drosophila Stock Centre, the Tucson Drosophila Stock Centre, John Carlson and Carla Sgró for providing fly stocks, and Ary Hoffman, Stephen McKechnie, Damian Dowling, Charles Robin, and Tim Connallon for valuable discussions and comments on the manuscript. This project was supported by a CSIRO Flagship Collaboration Fund postgraduate award to K.H.S.
Author Contributions
C.G.W. and M.d.B. conceived the experiments, interpreted the data, and co-led the work. K.H.S. conceived the experiments, interpreted the data, and performed the experiments. A.A. and T.K.J. interpreted the data. K.H.S., T.K.J., and C.G.W. wrote the paper.
References
- Aguadé M. 2009. Nucleotide and copy-number polymorphism at the odorant receptor genes Or22a and Or22b in Drosophila melanogaster. Mol Biol Evol. 26(1): 61–70. [DOI] [PubMed] [Google Scholar]
- Butterwick JA, del Mármol J, Kim KH, Kahlson MA, Rogow JA, Walz T, Ruta V.. 2018. Cryo-EM structure of the insect olfactory receptor Orco. Nature 560(7719): 447–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clyne PJ, Warr CG, Freeman MR, Lessing D, Kim J, Carlson JR.. 1999. A novel family of divergent seven-transmembrane proteins: candidate odorant receptors in Drosophila. Neuron 22(2): 327–338. [DOI] [PubMed] [Google Scholar]
- Couto A, Alenius M, Dickson BJ.. 2005. Molecular, anatomical, and functional organization of the Drosophila olfactory system. Curr Biol. 15(17): 1535–1547. [DOI] [PubMed] [Google Scholar]
- de Bruyne M, Smart R, Zammit E, Warr CG.. 2010. Functional and molecular evolution of olfactory neurons and receptors for aliphatic esters across the Drosophila genus. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 196(2): 97–109. [DOI] [PubMed] [Google Scholar]
- Dekker T, Revadi S, Mansourian S, Ramasamy S, Lebreton S, Becher PG, Angeli S, Rota-Stabelli O, Anfora G.. 2015. Loss of Drosophila pheromone reverses its role in sexual communication in Drosophila suzukii. Proc Biol Sci. 282(1804): 20143018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobritsa AA, Van Der Goes Van Naters W, Warr CG, Steinbrecht RA, Carlson JR.. 2003. Integrating the molecular and cellular basis of odor coding in the Drosophila antenna. Neuron 37(5): 827–841. [DOI] [PubMed] [Google Scholar]
- Gross SP, Guo Y, Martinez JE, Welte MA.. 2003. A determinant for directionality of organelle transport in Drosophila embryos. Curr Biol. 13(19): 1660–1668. [DOI] [PubMed] [Google Scholar]
- Guo S, Kim J.. 2007. Molecular evolution of Drosophila odorant receptor genes. Mol Biol Evol. 24(5): 1198–1207. [DOI] [PubMed] [Google Scholar]
- Hallem EA, Carlson JR.. 2006. Coding of odors by a receptor repertoire. Cell 125(1): 143–160. [DOI] [PubMed] [Google Scholar]
- Hallem EA, Ho MG, Carlson JR.. 2004. The molecular basis of odor coding in the Drosophila antenna. Cell 117(7): 965–979. [DOI] [PubMed] [Google Scholar]
- Hughes DT, Wang G, Zwiebel LJ, Luetje CW.. 2014. A determinant of odorant specificity is located at the extracellular loop 2-transmembrane domain 4 interface of an Anopheles gambiae odorant receptor subunit. Chem Senses 39(9): 761–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibba I, Angioy AM, Hansson BS, Dekker T.. 2010. Macroglomeruli for fruit odors change blend preference in Drosophila. Naturwissenschaften 97(12): 1059–1066. [DOI] [PubMed] [Google Scholar]
- Knaden M, Strutz A, Ahsan J, Sachse S, Hansson BS.. 2012. Spatial representation of odorant valence in an insect brain. Cell Rep. 1(4): 392–399. [DOI] [PubMed] [Google Scholar]
- Larsson MC, Domingos AI, Jones WD, Chiappe ME, Amrein H, Vosshall LB.. 2004. Or83b encodes a broadly expressed odorant receptor essential for Drosophila olfaction. Neuron 43(5): 703–714. [DOI] [PubMed] [Google Scholar]
- Leary GP, Allen JE, Bunger PL, Luginbill JB, Linn CE Jr, Macallister IE, Kavanaugh MP, Wanner KW.. 2012. Single mutation to a sex pheromone receptor provides adaptive specificity between closely related moth species. Proc Natl Acad Sci U S A. 109(35): 14081–14086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay TFC, Richards S, Stone EA, Barbadilla A, Ayroles JF, Zhu D, Casillas S, Han Y, Magwire MM, Cridland JM, et al. 2012. The Drosophila melanogaster genetic reference panel. Nature 482(7384): 173–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansourian S, Enjin A, Jirle EV, Ramesh V, Rehermann G, Becher PG, Pool JE, Stensmyr MC.. 2018. Wild African Drosophila melanogaster are seasonal specialists on marula fruit. Curr Biol. 28(24): 3960–3968.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride CS, Arguello JR, O’Meara BC.. 2007. Five Drosophila genomes reveal nonneutral evolution and the signature of host specialization in the chemoreceptor superfamily. Genetics 177(3): 1395–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols AS, Luetje CW.. 2010. Transmembrane segment 3 of Drosophila melanogaster odorant receptor subunit 85b contributes to ligand-receptor interactions. J Biol Chem. 285(16): 11854–11862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelz D, Roeske T, Syed Z, De Bruyne M, Galizia CG.. 2006. The molecular receptive range of an olfactory receptor in vivo (Drosophila melanogaster Or22a). J Neurobiol. 66(14): 1544–1563. [DOI] [PubMed] [Google Scholar]
- Richgels PK, Rollmann SM.. 2012. Genetic variation in odorant receptors contributes to variation in olfactory behavior in a natural population of Drosophila melanogaster. Chem Senses 37(3): 229–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson HM, Warr CG, Carlson JR.. 2003. Molecular evolution of the insect chemoreceptor gene superfamily in Drosophila melanogaster. Proc Natl Acad Sci U S A. 100(2 Suppl): 14537–14542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollmann SM, Wang P, Date P, West SA, Mackay TFC, Anholt RRH.. 2010. Odorant receptor polymorphisms and natural variation in olfactory behavior in Drosophila melanogaster. Genetics 186(2): 687–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruebenbauer A, Schlyter F, Hansson B, Löfstedt C, Larsson M.. 2008. Genetic variability and robustness of host odor preference in Drosophila melanogaster. Curr Biol. 18(18): 1438–1443. [DOI] [PubMed] [Google Scholar]
- Sgrò CM, Overgaard J, Kristensen TN, Mitchell KA, Cockerell FE, Hoffmann AA.. 2010. A comprehensive assessment of geographic variation in heat tolerance and hardening capacity in populations of Drosophila melanogaster from Eastern Australia. J Evol Biol. 23(11): 2484–2493. [DOI] [PubMed] [Google Scholar]
- Stensmyr MC, Dekker T, Hansson BS.. 2003. Evolution of the olfactory code in the Drosophila melanogaster subgroup. Proc R Soc B Biol Sci. 270(1531): 2333–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stensmyr MC, Giordano E, Balloi A, Angioy AM, Hansson BS.. 2003. Novel natural ligands for Drosophila olfactory receptor neurones. J Exp Biol. 206(4): 715–724. [DOI] [PubMed] [Google Scholar]
- Tunstall NE, Warr CG.. 2012. Chemical communication in insects: the peripheral odour coding system of Drosophila melanogaster In: López-Larrea C, editor. Sensing in nature. New York, NY: Springer US; p. 59–77. [DOI] [PubMed] [Google Scholar]
- Turner TL, Levine MT, Eckert ML, Begun DJ.. 2008. Genomic analysis of adaptive differentiation in Drosophila melanogaster. Genetics 179(1): 455–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Leal WS.. 2013. Probing insect odorant receptors with their cognate ligands: insights into structural features. Biochem Biophys Res Commun. 435(3): 477–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K, Huang LQ, Ning C, Wang CZ.. 2017. Two single-point mutations shift the ligand selectivity of a pheromone receptor between two closely related moth species. eLife 6: e29100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.