Abstract
During cell-to-cell communications, the interplay between physical and biochemical cues is essential for informational exchange and functional coordination, especially in multicellular organisms. However, it remains a challenge to visualize intercellular signaling dynamics in single live cells. Here, we report a photonic approach, based on laser microscissors and Förster resonance energy transfer (FRET) microscopy, to study intercellular signaling transmission. First, using our high-throughput screening platform, we developed a highly sensitive FRET-based biosensor (SCAGE) for Src kinase, a key regulator of intercellular interactions and signaling cascades. Notably, SCAGE showed a more than 40-fold sensitivity enhancement than the original biosensor in live mammalian cells. Next, upon local severance of physical intercellular connections by femtosecond laser pulses, SCAGE enabled the visualization of a transient Src activation across neighboring cells. Lastly, we found that this observed transient Src activation following the loss of cell-cell contacts depends on the passive structural support of cytoskeleton but not on the active actomyosin contractility. Hence, by precisely introducing local physical perturbations and directly visualizing spatiotemporal transmission of ensuing signaling events, our integrated approach could be broadly applied to mimic and investigate the wounding process at single-cell resolutions. This integrated approach with highly sensitive FRET-based biosensors provides a unique system to advance our in-depth understanding of molecular mechanisms underlying the physical-biochemical basis of intercellular coupling and wounding processes.
Keywords: high-throughput screening, directed evolution, FRET imaging, laser-induced wounding, highly sensitive FRET-based biosensor SCAGE, transient Src activation, passive structural support of cytoskeleton, active actomyosin contractility
GraphicalAbstract
Cell-to-cell communications are fundamentally pivotal to the differentiation, development, and physiology of metazoans.1–3 Abnormal alterations of ubiquitous and diverse intercellular communications contribute to a wide variety of diseases.4–6 For instance, by disrupting intercellular junctions, wounding causes the loss of cell-cell communications and induces tissue injuries. As a result, wound damages can lead to a high prevalence of chronic disease progression if without appropriate care and healing of wounds.7,8 In order to accomplish effective exchange of information and proper coordination of functions and behaviors between cells, sophisticated repertoires of molecular signals need to be precisely controlled at the intercellular regions.1 Cells exchange various information in different ways, including, but not limited to, biochemical and physical information. For example, direct transfer of molecular information, such as genetic materials and proteins, occurs between cells via the formation of gap junctions or membrane bridges over short distances, or the secretion of extracellular vesicles over long distances.9,10 Tunneling nanotubes provide another long-distance route for intercellular exchange of cellular components and signaling molecules.11 Gap junctions also mediate intercellular exchange of metabolites and ions.9 Besides conveying biochemical information, cells have evolved a variety of communication mechanisms to transmit physical information, especially mechanical signals from extracellular environment and their neighbors.12 For instance, generation and propagation of action potentials in neuronal cells involve mechanosensitive gating of ion channels and plasma membrane deformations, and both synaptic plasticity and dendritic spine plasticity are modulated by mechanical forces.13 Extremely dynamic interfaces of immunological synapses formed between immune cells and antigen-presenting cells provide architectures for cell-cell interactions, and generate mechanical forces to ensure normal physiological functions and effective intercellular communications.14
A group of transmembrane proteins, called cell adhesion proteins, largely participates in different physical-biochemical coupling scenarios, such as force generation, mechanical signaling transduction, and cellular/multicellular response.13–15 Integrins and cadherins, among the most characterized mechanosensitive membrane receptors of three-dimensional adhesive network, regulate cell-cell and cell-extracellular-matrix adhesions as well as transduction of mechanical signals from extracellular matrices and from neighboring cells into downstream intracellular signaling cascades.15,16 Studies suggest that the cell-cell adhesion network endows groups of cells with a number of collective functions and behaviors, such as tissue morphogenesis and regeneration, maintenance of tissue homeostasis and epithelial tissue cohesion.13,15–17 Cadherin-mediated cell-cell adhesions are commonly believed to be involved in these well-orchestrated cellular-level and tissue-level physiological processes. For example, the expression of cadherins is tightly regulated during embryonic development and adult tissue homeostasis and remodeling.16–18 It also correlates with a variety of morphogenetic processes, such as cell sorting, cell rearrangements, and collective cell movements.19–22 Cadherins form physical and functional linkages with different cytoplasmic filaments and serve as a scaffold to mediate mechanical and biochemical signaling transductions.23–27 In fact, the downregulation or loss of cadherin function is often found to be associated with severe pathological situations, such as tumorigenesis, tumor progression, tumor invasion, and metastasis.26,28 It is thus of vital importance to further elucidate the underlying mechanisms responsible for these physiological and pathological scenarios.
Decades of research have revealed multiple mechanisms that underlie the regulation of cadherin-mediated cell-cell contacts.20,29,30 In particular, a wide variety of tyrosine kinases are involved in mediating cadherin-based adhesions, including both receptor and nonreceptor protein kinases.29–31 Among those, Src family kinases (SFKs) were identified as critical regulators.17,32 In fact, SFKs have been shown to concentrate at adherens junctions and correlate with elevated phosphorylation levels of adhesion complex proteins.33 However, there are discrepancies on how SFKs affect cadherin adhesions. For instance, during keratinocyte differentiation, inhibition of SFKs suppresses adhesion formation and strength.34 On the contrary, activation of SFKs by the expression of Src oncogene suppresses cadherin adhesions through direct tyrosine phosphorylation on cadherin/catenin complexes and contributes to invasive and metastatic potential of cells.26,28,35,36 Recently, multiple studies suggested that Src kinase, as the principal member of SFKs, modulates the interaction of epithelial cadherin (E-cadherin) with the cytoskeleton, maintains the integrity of E-cadherin adhesion and supports apical junctional tension.37–39 Meanwhile, with the power of tension sensor module derived from spider flagelliform silk, recent quantitative evidence indicates that E-cadherin not only serves as a mechanosensitive signaling hub through transmitting external mechanical forces, but also senses internal mechanics via enduring constitutive cytoskeleton-dependent tension.40,41 Moreover, E-cadherin tension relaxation and β-catenin nuclear translocation through actomyosin remodeling requires Src-dependent focal adhesion kinase activation.42
Despite these studies, what the dynamics of Src signaling are and how Src signaling is regulated across neighboring cells remain poorly understood, partly because of the technical limitations by conventional biochemical and imaging methods. It is hence of great need to develop novel approaches that can directly enable local physical perturbations and allow the visualization of the dynamics and transmission of subsequent signaling events across adjacent cells, so as to better understand crosstalk and integration of different signaling cascades at cell-cell contacts. Here, we addressed this need by integrating photonic tools with molecular engineering and imaging to study the intercellular signaling dynamics at single-cell resolutions during the wounding process among multicell connections.
A photonic technique, laser microscissors, enables us to ablate subcellular targets of single live cells with high spatiotemporal precision,43‒45 with surrounding regions of the laser focal spot remaining intact and cell viability unaffected. This approach has been applied in a variety of studies to introduce local physical perturbations on different cellular structures with high precision, such as mitochondria and cytoskeletons, and to investigate how cell structures and functions are coupled.46–49
Molecular imaging approaches using Förster resonance energy transfer (FRET) microscopy have been broadly applied in cell biology.50 Numerous genetically encoded biosensors employing fluorescent proteins (FPs) and FRET have been developed for the visualization and quantification of intracellular signaling events with high spatiotemporal resolutions in single live cells.51 However, the empirical optimization processes have hampered the systematic development of highly sensitive and specific FRET-based biosensors. Directed evolution52 employing iterative cycles of mutagenesis to generate libraries coupled with selection to identify target mutants has become a powerful tool for protein engineering in cases where rational approaches are ineffective. Large libraries can be expressed in yeast or bacterial display systems,53,54 which can be screened by the fluorescence-activated cell sorting (FACS) technology to identify desired proteins.
In this study, based on our high-throughput screening platform, we developed a highly sensitive FRET-based biosensor (named SCAGE) for monitoring the activity of Src kinase, which gives a more than 40-fold sensitivity enhancement than the original biosensor. We next sought to combine FRET microscopy and laser scissors technology to investigate how the dynamic activity of Src kinase is regulated between adjacent epithelial cells upon the laser-induced wounding. With SCAGE, we successfully observed a transient Src activation across neighboring cells upon the femtosecond laser-induced disruption of cell-cell contacts, which was not detectable by the original parental Src biosensor.55 Our results provide direct evidence that the transient Src activation across adjoining epithelial cells upon precise physical perturbations depends on the passive structural support of cytoskeleton but not on the active actomyosin contractility.
RESULTS
High-Throughput Screening of Substrate-Binding Affinity toward a Library of c-Src SH2 Domains.
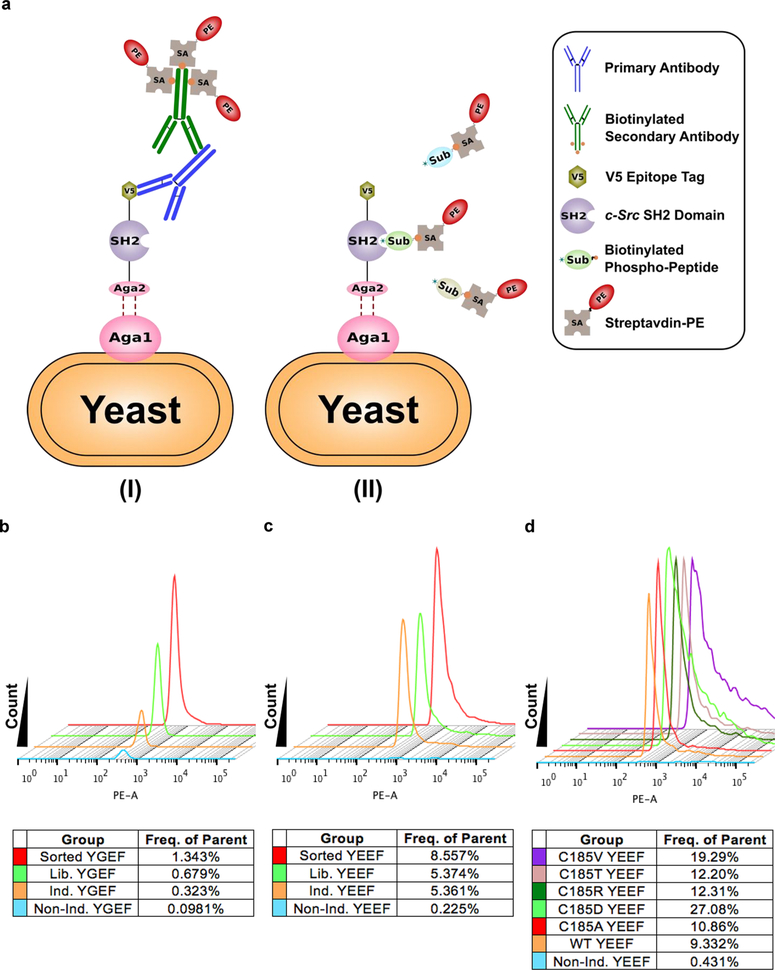
We developed a yeast display system (Figure 1a) to improve the binding between the SH2 domain and the substrate peptide within the Src FRET-based biosensor. Briefly, a library of SH2 mutants was fused with a-agglutinin, an abundant yeast cell wall protein, and displayed outside of the yeast cell, thus allowing the displayed mutant library to be screened for binding activity.54 This system allows a high-throughput screening and identification of optimal SH2 variants and corresponding peptide sequences (Figures 1a and S1). Successful yeast surface display of the recombinant cargo protein was confirmed by the staining of the V5 epitope tag at the C-terminus of the SH2 domain (Figures 1a and S2a). We then screened the buffer conditions and the phosphopeptide concentrations for the binding assay between the expressed SH2 domain and the phosphorylated substrate peptides. The results revealed that the binding buffer containing 0.5% BSA led to consistent staining signals (Figure S2b,c), which was applied for the binding buffers used in the rest of manuscript. We next proceeded to optimize the substrate peptide conditions for yeast binding assays. An ideal substrate sequence in a FRET-based biosensor should have two features: (1) the substrate sequence is favored by the target kinase for phosphorylation; (2) the substrate peptide upon phosphorylation has an optimal binding affinity toward the intramolecular SH2 domain (or its mutant) in the biosensor for FRET changes. It has been shown that EIYGEF and EIYEEF can serve as optimal substrate sequences for c-Src kinase in vitro,56 and a different sequence after phosphorylation pYEEI is preferred for binding by wild-type Src SH2 domain (WT SH2).57 We hence compared these different phosphopeptides (pYGEF, pYEEF, and pYEEI) as well as the unphosphorylatable negative control (FEEI, with phosphorylated tyrosine residue replaced by phenylalanine residue), with respect to their binding toward WT SH2. We stained the yeast cells displaying WT SH2 using these peptides. The results indicate that both pYGEF and pYEEF can bind to WT SH2 proportional to the peptide concentration, with pYEEF clearly demonstrating a stronger binding than the previously identified pYEEI57 (Figure S3a–c). We subsequently utilized pYEEF and pYGEF peptides as the Src favorable substrate sequences (0.2 μg/μL, pYGEF < pYEEI < pYEEF in binding toward WT SH2 as shown in Figure S3a–c) to conduct the binding assay against the yeast cells displaying either WT SH2 or its mutant library. The mutant library was generated by site-saturation mutagenesis58 on the cysteine site (C185) in WT SH2 which is critical for the binding toward phosphotyrosine peptides.59 Compared to yeast cells uniformly expressing WT SH2, the yeast library displaying diversified mutants showed overall stronger binding toward both pYGEF and pYEEF peptides (Figure 1b,c). We further observed stronger binding of both WT SH2 and the mutant library toward pYEEF over pYGEF (Figure 1b,c). We thereafter enriched the yeast cells displaying SH2 mutants with strong binding toward phosphotyrosine peptides (pYEEF and pYGEF) by sorting and repopulating (Figure 1b,c). Among the sorted SH2 variants, C185D, C185R, C185T, C185 V, and C185A mutations were identified by sequencing and verified by yeast staining to show high binding toward the phosphotyrosine peptides, with pYEEF displaying a significantly higher binding capacity than pYGEF toward each mutant (Figures 1d and S4). Our results suggest that yeast display and random mutagenesis can allow the high-throughput screening and identification of efficient interaction pairs consisting of SH2 mutants and phosphotyrosine peptides. This approach is potentially applicable to the engineering and optimization of, in principle, any FRET-based biosensor capable of monitoring tyrosine kinase activities.
Figure 1.
Optimal c-Src SH2 domain variants and tyrosine kinase substrates identified by high-throughput screening. (a) SH2 domains from c-Src kinase were displayed on the yeast cell surface as a fusion protein carrying the V5 epitope tag at the C-terminus. (I) The V5 epitope tag allows the staining of expressed protein cargoes by the primary antibody and the biotinylated secondary antibody, which can then be labeled by streptavidin-R-phycoerythrin (SA-PE) conjugate. (II) Wild-type (WT) and variant SH2 domain mutants bind to the biotinylated phosphotyrosine-containing substrate peptides, which can then be labeled by SA-PE conjugate. (b–d) Identifying the optimal SH2 domain mutants and the corresponding substrate peptides, with peptides EIpYGEF in (b), EIpYEEF in (c), and EIpYEEF together with SH2 domain mutants, as indicated in (d). “Nonind.”, “Ind.”, and “Lib.” represent noninduced yeast group, induced yeast group and yeast library group, respectively, stained with phosphorylated peptide EIpYGEF or EIpYEEF. C185A, C185D, C185R, C185T, and C185 V are different mutations in residue 185 of wild-type c-Src SH2 domain
Characterization of Src Biosensors with Identified Mutations in Live Mammalian Cells.
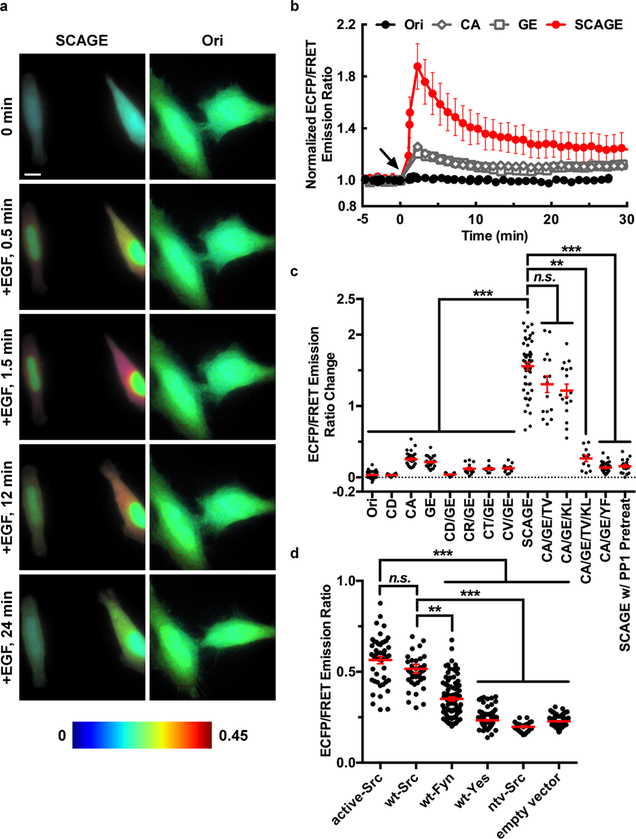
We then constructed FRET-based Src biosensors comprising identified binding pairs of substrates and SH2 mutants, with the expectation that the substrate phosphorylation by Src kinase can cause a conformational change and a reduction of FRET efficiency due to the consequent separation of YPet from ECFP (Figure S5). We replaced EYFP in the original Src biosensor55 with YPet because YPet has been verified to form a better FRET pair with ECFP in enhancing the sensitivities of biosensors.60 We examined the sensitivities and specificities of these Src biosensors in mammalian cells and compared them with the parental biosensor55 used as the template for mutagenesis and directed evolution. In HeLa cells expressing different Src biosensors, epidermal growth factor (EGF) induced a large FRET response (~156%) of SCAGE (SH2 C185A and substrate EIYEEF, with C → A mutation at residue 185 of WT SH2 and G → E mutation in the substrate sequence, Figure 2a–c and Movie S1). Smaller FRET responses were observed in other groups, either the Ori group (original parental biosensor with WT SH2 and substrate sequence EIYGEF, Figure 2a–c and Movie S2) or groups with a single mutation in the SH2 domain or substrate sequence (Figure 2b,c). We further incorporated additional mutations, identified by our earlier publication61 to improve the binding affinity of SH2 domain toward phosphotyrosine peptides, into the SH2 domain of SCAGE and examined their effects. The results indicate that these two additional mutations (SH2 T180 V and K203L) failed to further improve the biosensor sensitivities (Figure 2c). In fact, the CA/GE/TV/KL biosensor group (incorporating triple point mutations C185A, T180 V, K203L in the SH2 domain with the substrate sequence EIYEEF) had a much smaller FRET response (~26.5%, Figure 2c), suggesting that overly high affinity between binding pairs within the biosensor may not enhance the biosensor sensitivity. We further examined the platelet-derived growth factor (PDGF)-induced FRET responses of the Ori, SCAGE, CA (SH2 C185A and substrate EIYGEF, with C → A mutation only at residue 185 of WT SH2) and GE (WT SH2 and substrate EIYEEF, with G → E mutation only in the substrate sequence) biosensors in mouse embryonic fibroblasts (MEFs; Figure S6). Again, the largest FRET response (~108%, Figure S6c) was observed in the SCAGE group, with 15–20% ratio changes for the CA or GE only group and ~5.4% ratio change for the Ori group (Figure S6c). The biosensors in HeLa cells and MEFs had very distinct dynamics (Figures 2b and S6b), with a clear transient time course in HeLa cells but a sustained activity in MEFs, as highlighted by SCAGE. In HeLa cells expressing SCAGE, the EGF-induced FRET response was substantially reduced after pretreatment with PP1, a selective Src family inhibitor (Figure 2c). Ratio change was also reduced markedly when SCAGE was introduced together with a kinase-dead c-Src (ntv-Src) into HeLa cells. To further determine the specificity of SCAGE, we reconstituted Src/Yes/Fyn triple-knockout (SYF −/−) MEFs with active-Src, wt-Src (wild-type c-Src), ntv-Src, wt-Fyn, wt-Yes or an empty vector. Only active-Src and wt-Src restored the ECFP/FRET ratios (Figure 2d). These results suggest that our new SCAGE biosensor can specifically detect Src kinase activity with a high sensitivity. FRET response was absent in HeLa cells expressing the inactive CA/GE/YF (SH2 C185A and EIFEEF, Figure 2c) biosensor in which the substrate tyrosine is mutated to phenylalanine. Therefore, the biosensor response is dependent on the tyrosine phosphorylation of the substrate as we designed (Figure S5b). We further compared SCAGE with our previously developed Src biosensor (WME biosensor),60 which has a relatively high sensitivity but contains two tyrosine residues in the substrate. In HeLa cells, SCAGE showed a significantly higher sensitivity than WME biosensor (~156% vs ~75%, respectively, Figure S7). These results indicate that the interaction pairs identified from our high-throughput screening of libraries successfully enhanced the sensitivities of FRET-based biosensors. In vitro assays of purified biosensor proteins further supported this notion (Figures S8 and S9).
Figure 2.
Characterization of FRET-based Src biosensors in living mammalian cells. (a) Time-lapse FRET imaging in HeLa cells expressing different Src FRET-based biosensors (SCAGE or Ori) stimulated with 50 ng/mL EGF (arrow at 0 min). The color scale at the bottom shows the range of ECFP/FRET emission ratio, with cool and warm colors representing low and high ratios, respectively. Scale bars, 5 μm. (b) Time courses of the normalized ECFP/FRET emission ratio of the Ori (black solid circles, n = 6), CA (open diamonds, n = 7), GE (open squares, n = 8), and SCAGE biosensors (red solid circles, n = 5) in HeLa cells before and after 50 ng/mL EGF stimulation. The ECFP/FRET emission ratios were normalized against the averaged values before EGF stimulation. The error bars represent SEM (the standard error of the mean) and average data are presented as mean ± SEM. (c) Responses of different Src FRET-based biosensors in HeLa cells stimulated with 50 ng/mL EGF. The CA/GE/YF group is the inactive control, the SCAGE and ntv-Src group is the SCAGE biosensor group together with the kinase-dead c-Src (ntv-Src), and the SCAGE w/PP1 Pretreat group is the SCAGE biosensor group pretreated with 10 μM PP1 for 1 h (n = 53, 18, 28, 30, 17, 13, 10, 12, 52, 15, 17, 12, 36, 19, and 113 cells for Ori, CD, CA, GE, CD/GE, CR/GE, CT/GE, CV/GE, SCAGE, CA/GE/TV, CA/GE/KL, CA/GE/TV/KL, CA/GE/YF, SCAGE and ntv-Src and SCAGE w/PP1 Pretreat groups, respectively). (d) ECFP/FRET emission ratios of the SCAGE biosensor in Src/Yes/Fyn triple-knockout (SYF −/−) mouse embryonic fibroblasts (MEFs) cotransfected with various Src family kinases and mutants. SYF −/− MEFs were reconstituted with active-Src (n = 46), wt-Src (n = 44), ntv-Src (n = 24), wt-Fyn (n = 90), wt-Yes (n = 107), and an empty vector (n = 81). (c, d) In scatter plots, the red lines indicate the mean values, the error bars represent SEM and average data are presented as mean ± SEM; ***P < 0.001, **P < 0.01, and “n.s.”P > 0.05 are from Kruskal-Wallis test followed by Dunn’s multiple comparison test.
Observation of Src Signaling Transmission Following the Femtosecond Laser-Induced Wounding.
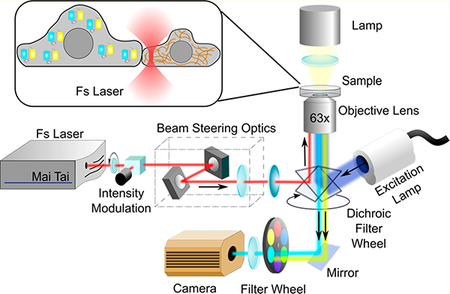
After thoroughly characterizing the specificity and sensitivity of our newly developed Src biosensor, we further decided to use SCAGE to study the dynamics of Src kinase activity during the intercellular signal transmission between neighboring epithelial cells with well-developed cell-cell adhesions. In order to do so, we applied SCAGE for FRET imaging in adult male Potorous tridactylus kidney (PtK2) epithelial cells, which have prominent cell-cell junctions and their associated cytoskeletal structures. As the blow-up schematic drawing shown in Figure 3a, we employed femtosecond near-infrared laser scissors microscopy and selectively ablated cytoskeletal stress fibers along cell-cell junctions to physically induce the loss of cell-cell contacts and mimic the wounding process between adjacent cells with highly controllable precision. Meanwhile, together with SCAGE, we integrated FRET imaging with our laser scissors to visualize the signal transmission and propagation across neighboring cells upon the laser-induced wounding (Figure 3a). Indeed, as shown in Figure 3b (red lines), we can precisely irradiate single actin filaments near cell-cell contact regions within the neighboring cells along the cell of interest, highlighted by an in vivo filamentous actin (F-actin) marker LifeAct-RFP.62 The complete severing operation by the 740 nm scanning laser in Figure 3b took 10.7 s. Consequently, the irradiated neighboring cells quickly retracted and disassociated from the cell of interest in the center (Figure 3b), due to the loss of cell-cell junctions upon the laser ablation of multiple peripheral stress fibers. In response to the loss of cell-cell contacts, the cell of interest also underwent large-scale retraction and transient Src activation (Figure 3c–e and Movie S3). The observed Src activation reached the peak rapidly within 5 min followed by a gradual decrease (Figure 3d). The calculated FRET response of the SCAGE group upon the laser-induced wounding is ~13.7% (Figure 3e,f). Such FRET responses were absent with the disabled CA/GE/RV/YF biosensor (Figure 3f), suggesting that the intramolecular interaction between the binding partners within the biosensor is necessary for the detection of FRET signals. The absence of Src activation in cells residing next to others but not forming stable and physical cell-cell contacts (Figure 3f, Cutting Control) suggests that the observed Src activation requires physical cell-cell contacts, but not the release and diffusion of chemical constituents between neighboring cells. The response from the Ori biosensor was very minor and almost undetectable, clearly highlighting the power and necessity of the new SCAGE Src biosensor in revealing this signal transmission between neighboring cells (Figure S10).
Figure 3.
Regulation of the transient Src activation following the laser-induced wounding. (a) System setup diagram of the laser scissors integrated with an inverted fluorescence microscope for FRET imaging. (b) Time-lapse images of PtK2 cells expressing a filamentous actin (F-actin) marker (LifeAct-TagRFP) upon the laser-induced disruption of cell-cell contacts. RFP-channel F-actin images (upper panel) and phase-contrast images (lower panel) of PtK2 cells at the indicated time points before and after laser ablation are shown. The red lines indicate the position of laser ablation. Scale bars, 5 μm. (c) Time-lapse FRET images of the SCAGE Src biosensor in the representative PtK2 cell before and after the laserinduced disruption of cell-cell contacts. The color scale at the left shows the range of ECFP/FRET emission ratio, with cool and warm colors representing low and high ratios, respectively. Scale bars, 5 μm. (d) Time course of the normalized ECFP/FRET emission ratio of the SCAGE biosensor in the representative PtK2 cell before and after laser ablation. The ECFP/FRET emission ratios were normalized against the averaged values before laser ablation. (e) ECFP/FRET emission ratios of the SCAGE biosensor in PtK2 cells (n = 28) before and after laser ablation. ***P < 0.001 is from the unpaired two-tailed Mann-Whitney test. (f) ECFP/FRET emission ratio changes of Src biosensors as indicated in PtK2 cells before and after laser ablation (n = 28, 6, 11, 14, and 14 cells for SCAGE, CA/GE/RV/YF, cutting control, SCAGE with CytoD Pretreat, and SCAGE with ML-7 Pretreat groups, respectively). A 1 μM cytochalasin D (CytoD) and 10 μM ML-7 were applied to inhibit actin polymerization and actomyosin contractility, respectively. PtK2 cells were pretreated with CytoD for 30 min or with ML-7 for 60 min. **P < 0.01, ***P < 0.001, and “n.s.” P > 0.05 are from Kruskal-Wallis test followed by Dunn’s multiple comparison test. (e, f) In scatter plots, the red lines indicate the mean values, the error bars represent SEM (the standard error of the mean) and average data are presented as mean ± SEM.
Regulation of Src Signaling Activation upon the Laser-Induced Wounding.
Cytoskeletal support and contractile forces are of vital importance in regulating cell-cell adhesions junctions. Physical and functional links between contractile actomyosin bundles and adhesion complexes are widely believed to be the main molecular basis for junctional integrity and functionality.15,17,37,63 In order to better understand the mechanistic regulation of the observed Src transmission upon the laser-induced physical disruption of cell-cell contacts, we decided to investigate the roles of actin cytoskeleton and its related actomyosin-based contractility during the wounding process. We pretreated PtK2 cells with cytochalasin D (CytoD, an inhibitor of actin polymerization) for 30 min to disrupt the actin cytoskeletal structures, or ML-7 (a myosin light chain kinase inhibitor) for 60 min to suppress the active actomyosin contractility. Results of inhibition assays indicate that the transient Src activation upon the laserinduced wounding was completely eliminated with pretreatment by CytoD but not by ML-7 (Figure 3f), although ML-7 clearly suppressed the activity of focal adhesion kinase in PtK2 cells (Figure S11 and Movie S4). We also noticed that ML-7 pretreatment did not affect the Src signaling dynamics (data not shown). Taken together, these results demonstrate that the observed transient Src signaling transmission across neighboring cells following the loss of cell-cell contacts depends on the passive structural support of actin cytoskeleton but not on the active actomyosin contractility.
DISCUSSION
We developed a high-throughput screening method (Figure S1) that allows the systematic development of FRET-based biosensors (Figure S5a). The result is a highly sensitive Src biosensor (SCAGE), which provides a more than 40-fold sensitivity enhancement in live mammalian cells comparing to the parental biosensor template before the directed-evolution-based improvement55 (Figure 2c). This new Src biosensor is also more sensitive than our previously developed Src biosensor based on a rational design with two tyrosine residues in the substrate motif60 (Figure S7). Employing this evolution-optimized biosensor and laser scissors, we further demonstrated that the activation of Src kinase rapidly occurs as a signal transmission between neighboring cells upon the femtosecond laser-induced physical disruption of cell-cell contacts (Figure 3c,d). This transient activation is dependent on the passive structural support of actin cytoskeleton, but not on the active actomyosin contractility (Figure 3f). As such, our high-throughput screening method can be applied to systematically develop highly sensitive FRET-based biosensors, which together with our integrated photonic approach can lead to the revelation of how cells perceive physical perturbations from neighboring cells and trigger biochemical responses, especially from a single-cell perspective.
The development of intramolecular FRET-based biosensors generally suffers from low-throughput optimizations in trial-and-error fashions. Our high-throughput screening method provides a new way for developing sensitive FRET-based biosensors in a systematic fashion. This high-throughput screening platform based on yeast display and directed evolution has a number of following advantages. First, upon induction, the yeast display system can correctly express the protein cargo motifs on the yeast surface, readily accessible by specific binding partners to be screened (Figure S2a). In fact, negative control peptide FEEI showed no significant binding to WT SH2 displayed on the yeast surface even at a high concentration (Figure S3d). Second, this method allows for effective screening and identification of various interaction pairs with better binding affinities, including but not limited to substrate peptides and SH2 domain variants. This method can hence be readily extended to develop, in principle, any FRET-based biosensor for real-time monitoring post-translational modification dynamics (mostly for an enzyme). A potential limitation of our approach is that the yeast, a lower eukaryotic organism, may exhibit different codon usage and give different post-translational modification functions as comparing to mammalian cells. Thus, the binding pairs identified may not directly translate into optimal responses of biosensors in mammalian cells. This may explain why our CD/GE biosensor containing the strongest phospho-substrate binding domain, SH2 C185D, showed significantly lower sensitivity than SCAGE in mammalian cells (Figure 2c). In the future, high-throughput screening approaches based on the full-length biosensor sequences and higher eukaryotic organisms may be needed to further improve the efficiency of biosensor development.
On the other hand, during the biosensor characterization, both our CD/GE biosensor and CA/GE/TV/KL biosensor had remarkably smaller FRET responses compared with the CA/GE biosensor. Since both C185D and the triple (CA/TV/KL) mutant of SH2 domain have higher affinities than the C185A SH2 mutant, merely pursuing stronger binding affinities between binding pairs within the Src biosensors may not necessarily lead to higher sensitivities of biosensors. This is possibly because the CD or triple mutant SH2 domain has a high affinity in binding to weakly phosphorylated substrates under even low Src kinase activities, causing a high basal level of biosensor signals and hence reducing the dynamic range or sensitivity of the biosensors. Indeed, the biosensor containing the triple mutant SH2 domain showed significantly higher basal FRET ratio level before growth factor stimulation and had smaller FRET responses upon the stimulation. Furthermore, biosensors with superbinder SH2 mutants may also bind to nonspecific phosphotyrosine-containing peptides, which may lead to unexpected response and effect of the biosensors. Of note, the triple mutant SH2 domain (T180 V, C185A, K203L) has been reported before to be detrimental to cells due to its unphysiologically strong binding affinity against phosphorylated tyrosine peptides,61 which is consistent with our observation that HeLa cells expressing the CA/GE/TV/KL biosensor were poorly viable in culture.
The biosensor characterization in mammalian cells revealed very distinct dynamics and responses of SCAGE. More specifically, the characterization in mammalian cells revealed that SCAGE showed a transient FRET response upon EGF stimulation in HeLa cells (Figure 2b), but a markedly sustained FRET response upon PDGF stimulation in MEFs (Figure S6b). It is possible that EGF and PDGF receptors involve distinct regulations, such as downregulation of kinase activation and endocytic trafficking of receptor tyrosine kinases.64–67 For instance, EGFR in HeLa cells, but not PDGFR in MEFs, may undergo fast ubiquitination, degradation, or internalization, which could contribute to the transient nature of EGFR signaling. Hence, the high-sensitivity Src biosensor may be applied to reveal the dynamic differences between EGF and PDGF-mediated signaling transductions and shed new lights on the molecular mechanisms underlying their distinct regulations.
Our study on cell-cell contacts using laser scissors, FRET microscopy, and the high-sensitivity Src biosensor (Figure 3a) showed a rapid Src signaling transmission between neighboring cells upon the laser-induced physical disruption of cell-cell contacts mimicking the wounding process. One major advantage of our integrated photonic system over conventional wounding methods is that we employ the laser scissors to precisely introduce physical perturbations on subcellular structures of actin fibers between neighboring cells so as to mimic wounding processes in a precisely controlled manner and simultaneously monitor molecular signaling dynamics at the single-cell level. In addition to visualizing the transient Src activation upon the separation of neighboring cells, we noticed an accompanying retraction occurring on the cell of interest (Movie S3). In fact, the initial Src activation upon laser ablation was accompanied by a cell retraction before a graduate decrease of Src activities for the cell of interest to readapt to its surroundings and re-establish its cellular mechanical stability, while distinct F-actin accumulation happened especially near the original cell-cell junction sites (Figure 3b and Movie S3). This dynamic activation of Src kinase is associated with actin reorganization and mechanical readaption during the period when the cell loses its adhesive contacts with its neighbors. Interestingly, the inhibition assays revealed that this fast and transient Src signaling event depends on the passive cytoskeletal structure but not on the active actomyosin contractility system. These results further highlight that subtle signaling dynamics can become observable with our novel biophotonic approaches and also suggest the physical perturbations on cells introduced by laser pulses may deliver a profound physical impact which can be transmitted afar and across to neighboring cells via the passive structural support without the aid of actomyosin contractility.
In summary, integrated with the micromanipulation photonic system with high precision, our newly developed Src biosensor, SCAGE, reveals how cells react biochemically to photonic-driven physical perturbations from neighboring cells. The combination of the photonic and imaging approaches, laser microscissors and FRET microscopy, establishes a generally applicable and unique platform to allow local physical perturbations with high precision in mimicking the wounding process, and to simultaneously monitor the spatiotemporal transmission of consequent signaling events across neighboring cells. This novel approach will largely complement the conventional biochemical and imaging methods and shed new light on cellular crosstalk and integration of physical and biochemical signals in single live cells, such as molecular mechanisms underlying wounding processes.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Drs. Alice Y. Ting and Michiyuki Matsuda for valuable constructs. The authors are grateful to the members of Dr. Michael W. Berns, Dr. Huimin Zhao, and Dr. Yingxiao Wang’s laboratories for their helpful assistance and input. This work is supported, in part, by Grants from NIH HL121365, GM125379, CA204704, and CA209629 (Y.W.), NSF CBET1360341 (to Y.W.), and by Grants from AFOSR FA9550–08-1–0284, AFOSR FA9550–17-1–0193, and gifts from the Hoag Family Foundation, Huntington Beach CA, the David and Lucille Packard Foundation, Los Altos, CA, and the Beckman Laser Institute Foundation (to M.W.B.). The funding agencies had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACSPublicationswebsite at DOI: 10.1021/acsphotonics.8b00383.
Characterization of the identified Src biosensors in vitro; Supplementary materials and methods; Supplementary figures (PDF).
Movie S1: EGF-induced FRET response in HeLa cells expressing SCAGE Src biosensor (AVI).
Movie S2: EGF-induced FRET response in HeLa cells expressing Ori Src biosensor (AVI).
Movie S3: FRET response in the representative PtK2 cell expressing SCAGE Src biosensor and TagRFP-labeled F-actin subjected to the laserinduced wounding (AVI).
Movie S4: Myosin-inhibitor-induced FRET response in a PtK2 cell expressing Lyn-FAK biosensor (AVI).
The authors declare the following competing financial interest(s): Yingxiao Wang is a scientific co-founder of Cell E&G Inc., and the authors declare competing financial interests in the form of provisional patent application. However, these financial interests do not affect the design, conduct or reporting of this research.
∎ REFERENCES
- (1).Gerhart J 1998 Warkany lecture: signaling pathways in development. Teratology 1999, 60, 226–39. [DOI] [PubMed] [Google Scholar]
- (2).Dale B; Gualtieri R; Talevi R; Tosti E; Santella L; Elder K Intercellular communication in the early human embryo. Mol. Reprod. Dev 1991, 29, 22–8. [DOI] [PubMed] [Google Scholar]
- (3).Van Norman JM; Breakfield NW; Benfey PN Intercellular communication during plant development. Plant Cell 2011, 23, 855–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Niessen CM Tight junctions/adherens junctions: basic structure and function. J. Invest. Dermatol 2007, 127, 2525–32. [DOI] [PubMed] [Google Scholar]
- (5).Lai-Cheong JE; Arita K; McGrath JA Genetic diseases of junctions. J. Invest. Dermatol 2007, 127, 2713–25. [DOI] [PubMed] [Google Scholar]
- (6).Dejana E; Tournier-Lasserve E; Weinstein BM The control of vascular integrity by endothelial cell junctions: molecular basis and pathological implications. Dev. Cell 2009, 16, 209–21. [DOI] [PubMed] [Google Scholar]
- (7).Iizuka M; Konno S Wound healing of intestinal epithelial cells. World J. Gastroenterol 2011, 17, 2161–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Leoni G; Neumann PA; Sumagin R; Denning TL; Nusrat A Wound repair: role of immune-epithelial interactions. Mucosal Immunol. 2015, 8, 959–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Herve JC; Derangeon M Gap-junction-mediated cell-to-cell communication. Cell Tissue Res. 2013, 352, 21–31. [DOI] [PubMed] [Google Scholar]
- (10).Mittelbrunn M; Sanchez-Madrid F Intercellular communication: diverse structures for exchange of genetic information. Nat. Rev. Mol. Cell Biol 2012, 13, 328–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Marzo L; Gousset K; Zurzolo C Multifaceted roles of tunneling nanotubes in intercellular communication. Front. Physiol 2012, 3, 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Ko KS; McCulloch CAG Intercellular mechanotransduction: Cellular circuits that coordinate tissue responses to mechanical loading. Biochem. Biophys. Res. Commun 2001, 285, 1077–1083. [DOI] [PubMed] [Google Scholar]
- (13).Tyler WJ The mechanobiology of brain function. Nat. Rev. Neurosci 2012, 13, 867–78. [DOI] [PubMed] [Google Scholar]
- (14).Basu R; Huse M Mechanical Communication at the Immunological Synapse. Trends Cell Biol. 2017, 27, 241–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).DuFort CC; Paszek MJ; Weaver VM Balancing forces: architectural control of mechanotransduction. Nat. Rev. Mol. Cell Biol 2011, 12, 308–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Weber GF; Bjerke MA; DeSimone DW Integrins and cadherins join forces to form adhesive networks. J. Cell Sci 2011, 124, 1183–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).van Roy F; Berx G The cell-cell adhesion molecule E-cadherin. Cell. Mol. Life Sci 2008, 65, 3756–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Huveneers S; Danen EHJ Adhesion signaling - crosstalk between integrins, Src and Rho. J. Cell Sci 2009, 122, 1059–1069. [DOI] [PubMed] [Google Scholar]
- (19).Takeichi M Cadherin Cell-Adhesion Receptors as a Morphogenetic Regulator. Science 1991, 251, 1451–1455. [DOI] [PubMed] [Google Scholar]
- (20).Gumbiner BM Regulation of cadherin-mediated adhesion in morphogenesis. Nat. Rev. Mol. Cell Biol. 2005, 6, 622–34. [DOI] [PubMed] [Google Scholar]
- (21).Maitre JL; Berthoumieux H; Krens SF; Salbreux G; Julicher F; Paluch E; Heisenberg CP Adhesion functions in cell sorting by mechanically coupling the cortices of adhering cells. Science 2012, 338, 253–6. [DOI] [PubMed] [Google Scholar]
- (22).Heisenberg CP; Bellaiche Y Forces in tissue morphogenesis and patterning. Cell 2013, 153, 948–62. [DOI] [PubMed] [Google Scholar]
- (23).Leckband DE; le Duc Q; Wang N; de Rooij J Mechanotransduction at cadherin-mediated adhesions. Curr. Opin. Cell Biol 2011, 23, 523–530. [DOI] [PubMed] [Google Scholar]
- (24).Schwartz MA; DeSimone DW Cell adhesion receptors in mechanotransduction. Curr. Opin. Cell Biol 2008, 20, 551–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Juliano RL Signal transduction by cell adhesion receptors and the cytoskeleton: functions of integrins, cadherins, selectins, and immunoglobulin-superfamily members. Annu. Rev. Pharmacol. Toxicol 2002, 42, 283–323. [DOI] [PubMed] [Google Scholar]
- (26).Behrens J Cadherins and catenins: role in signal transduction and tumor progression. Cancer Metastasis Rev. 1999, 18, 15–30. [DOI] [PubMed] [Google Scholar]
- (27).Hartsock A; Nelson WJ Adherens and tight junctions: structure, function and connections to the actin cytoskeleton. Biochim. Biophys. Acta, Biomembr 2008, 1778, 660–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Jeanes A; Gottardi CJ; Yap AS Cadherins and cancer: how does cadherin dysfunction promote tumor progression? Oncogene 2008, 27, 6920–6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Daniel JM; Reynolds AB Tyrosine phosphorylation and cadherin/catenin function. BioEssays 1997, 19, 883–91. [DOI] [PubMed] [Google Scholar]
- (30).Gumbiner BM Regulation of cadherin adhesive activity. J. Cell Biol 2000, 148, 399–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Lilien J; Balsamo J The regulation of cadherin-mediated adhesion by tyrosine phosphorylation/dephosphorylation of betacatenin. Curr. Opin. Cell Biol 2005, 17, 459–65. [DOI] [PubMed] [Google Scholar]
- (32).Yeatman TJ A renaissance for SRC. Nat. Rev. Cancer 2004, 4, 470–80. [DOI] [PubMed] [Google Scholar]
- (33).Tsukita S; Oishi K; Akiyama T; Yamanashi Y; Yamamoto T; Tsukita S Specific Protooncogenic Tyrosine Kinases of Src Family Are Enriched in Cell-to-Cell Adherens Junctions Where the Level of Tyrosine Phosphorylation Is Elevated. J. Cell Biol 1991, 113, 867–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Calautti E; Cabodi S; Stein PL; Hatzfeld M; Kedersha N; Dotto GP Tyrosine phosphorylation and src family kinases control keratinocyte cell-cell adhesion. J. Cell Biol 1998, 141, 1449–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Matsuyoshi N; Hamaguchi M; Taniguchi S; Nagafuchi A; Tsukita S; Takeichi M Cadherin-mediated cell-cell adhesion is perturbed by v-src tyrosine phosphorylation in metastatic fibroblasts. J. Cell Biol 1992, 118, 703–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Behrens J; Vakaet L; Friis R; Winterhager E; Van Roy F; Mareel MM; Birchmeier W Loss of epithelial differentiation and gain of invasiveness correlates with tyrosine phosphorylation of the E-cadherin/beta-catenin complex in cells transformed with a temperature-sensitive v-SRC gene. J. Cell Biol 1993, 120, 757–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).McLachlan RW; Kraemer A; Helwani FM; Kovacs EM E-cadherin adhesion activates c-Src signaling at cell–cell contacts. Mol. Biol. Cell 2007, 18, 3214–3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Ren G; Helwani FM; Verma S; McLachlan RW; Weed SA; Yap AS Cortactin is a functional target of E-cadherin-activated Src family kinases in MCF7 epithelial monolayers. J. Biol. Chem 2009, 284, 18913–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Gomez GA; McLachlan RW; Wu SK; Caldwell BJ; Moussa E; Verma S; Bastiani M; Priya R; Parton RG; Gaus K; Sap J; Yap AS An RPTPalpha/Src family kinase/Rap1 signaling module recruits myosin IIB to support contractile tension at apical E-cadherin junctions. Mol. Biol. Cell 2015, 26, 1249–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Grashoff C; Hoffman BD; Brenner MD; Zhou R; Parsons M; Yang MT; McLean MA; Sligar SG; Chen CS; Ha T; Schwartz MA Measuring mechanical tension across vinculin reveals regulation of focal adhesion dynamics. Nature 2010, 466, 263–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Borghi N; Sorokina M; Shcherbakova OG; Weis WI; Pruitt BL; Nelson WJ; Dunn AR E-cadherin is under constitutive actomyosin-generated tension that is increased at cell–cell contacts upon externally applied stretch. Proc. Natl. Acad. Sci. U. S. A 2012, 109, 12568–12573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Gayrard C; Bernaudin C; Dejardin T; Seiler C; Borghi N Src- and confinement-dependent FAK activation causes E-cadherin relaxation and beta-catenin activity. J. Cell Biol 2018, 217, 1063–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Berns MW; Edwards J; Strahs K; Girton J; McNeill P; Rattner J; Kitzes M; Hammer-Wilson M; Liaw L; Siemens A Laser microsurgery in cell and developmental biology. Science 1981, 213, 505–513. [DOI] [PubMed] [Google Scholar]
- (44).Botvinick EL; Berns MW Internet-based robotic laser scissors and tweezers microscopy. Microsc. Res. Tech 2005, 68, 65–74. [DOI] [PubMed] [Google Scholar]
- (45).Berns MW A history of laser scissors (microbeams). Methods Cell Biol. 2007, 82, 1–58. [DOI] [PubMed] [Google Scholar]
- (46).Shen N; Datta D; Schaffer CB; LeDuc P; Ingber DE; Mazur E Ablation of cytoskeletal filaments and mitochondria in live cells using a femtosecond laser nanoscissor. Mech Chem. Biosyst 2005, 2, 17–25. [PubMed] [Google Scholar]
- (47).Kumar S; Maxwell IZ; Heisterkamp A; Polte TR; Lele TP; Salanga M; Mazur E; Ingber DE Viscoelastic retraction of single living stress fibers and its impact on cell shape, cytoskeletal organization, and extracellular matrix mechanics. Biophys. J 2006, 90, 3762–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Wakida NM; Lee CS; Botvinick ET; Shi LZ; Dvornikov A; Berns MW Laser nanosurgery of single microtubules reveals location-dependent depolymerization rates. J. Biomed. Opt 2007, 12, 024022. [DOI] [PubMed] [Google Scholar]
- (49).Tanner K; Boudreau A; Bissell MJ; Kumar S Dissecting regional variations in stress fiber mechanics in living cells with laser nanosurgery. Biophys. J 2010, 99, 2775–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Periasamy A; Day R Molecular imaging: FRET microscopy and spectroscopy; Elsevier, 2011. [Google Scholar]
- (51).Frommer WB; Davidson MW; Campbell RE Genetically encoded biosensors based on engineered fluorescent proteins. Chem. Soc. Rev 2009, 38, 2833–2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Schmidt-Dannert C Directed evolution of single proteins, metabolic pathways, and viruses. Biochemistry 2001, 40, 13125–36. [DOI] [PubMed] [Google Scholar]
- (53).Georgiou G; Stathopoulos C; Daugherty PS; Nayak AR; Iverson BL; Curtiss R 3rd Display of heterologous proteins on the surface of microorganisms: from the screening of combinatorial libraries to live recombinant vaccines. Nat. Biotechnol 1997, 15, 29–34. [DOI] [PubMed] [Google Scholar]
- (54).Cherf GM; Cochran JR Applications of Yeast Surface Display for Protein Engineering. Methods Mol. Biol 2015, 1319, 155–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Ting AY; Kain KH; Klemke RL; Tsien RY Genetically encoded fluorescent reporters of protein tyrosine kinase activities in living cells. Proc. Natl. Acad. Sci. U. S. A 2001, 98, 15003–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Songyang Z; Carraway KL; Eck MJ; Harrison SC; Feldman RA; Mohammadi M; Schlessinger J; Hubbard SR; Smith DP; Eng C; Lorenzo MJ; Ponder BAJ; Mayer BJ; Cantley LC Catalytic Specificity of Protein-Tyrosine Kinases Is Critical for Selective Signaling. Nature 1995, 373, 536–539. [DOI] [PubMed] [Google Scholar]
- (57).Songyang Z; Shoelson SE; Chaudhuri M; Gish G; Pawson T; Haser WG; King F; Roberts T; Ratnofsky S; Lechleider RJ; et al. SH2 domains recognize specific phosphopeptide sequences. Cell 1993, 72, 767–78. [DOI] [PubMed] [Google Scholar]
- (58).Arnold FH; Georgiou G Directed evolution library creation. Methods in Molecular Biology 2003, 231, 231. [Google Scholar]
- (59).Bradshaw JM; Mitaxov V; Waksman G Investigation of phosphotyrosine recognition by the SH2 domain of the Src kinase. J. Mol. Biol 1999, 293, 971–985. [DOI] [PubMed] [Google Scholar]
- (60).Ouyang M; Sun J; Chien S; Wang Y Determination of hierarchical relationship of Src and Rac at subcellular locations with FRET biosensors. Proc. Natl. Acad. Sci. U. S. A 2008, 105, 14353–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Kaneko T; Huang H; Cao X; Li X; Li C; Voss C; Sidhu SS; Li SS Superbinder SH2 domains act as antagonists of cell signaling. Sci. Signaling 2012, 5, ra68. [DOI] [PubMed] [Google Scholar]
- (62).Riedl J; Crevenna AH; Kessenbrock K; Yu JH; Neukirchen D; Bista M; Bradke F; Jenne D; Holak TA; Werb Z; Sixt M; Wedlich-Soldner R Lifeact: a versatile marker to visualize F-actin. Nat. Methods 2008, 5, 605–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Ratheesh A; Yap AS A bigger picture: classical cadherins and the dynamic actin cytoskeleton. Nat. Rev. Mol. Cell Biol 2012, 13, 673–679. [DOI] [PubMed] [Google Scholar]
- (64).Ostman A; Bohmer FD Regulation of receptor tyrosine kinase signaling by protein tyrosine phosphatases. Trends Cell Biol. 2001, 11, 258–66. [DOI] [PubMed] [Google Scholar]
- (65).Chiarugi P; Cirri P; Taddei ML; Talini D; Doria L; Fiaschi T; Buricchi F; Giannoni E; Camici G; Raugei G; Ramponi G New perspectives in PDGF receptor downregulation: the main role of phosphotyrosine phosphatases. Journal of Cell Science 2002, 115, 2219–32. [DOI] [PubMed] [Google Scholar]
- (66).Thien CB; Langdon WY Cbl: many adaptations to regulate protein tyrosine kinases. Nat. Rev. Mol. Cell Biol 2001, 2, 294–307. [DOI] [PubMed] [Google Scholar]
- (67).Goh LK; Sorkin A Endocytosis of receptor tyrosine kinases. Cold Spring Harbor Perspect. Biol. 2013, 5, a017459. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.