Abstract
We developed a computational approach called Domain-architecture Aware Inference of Orthologs (DAIO) for the analysis of protein orthology by combining phylogenetic and protein domain-architecture information. Using DAIO, we performed a systematic study of the proteomes of all human Herpesviridae species to define Strict Ortholog Groups (SOGs). In addition to assessing the taxonomic distribution for each protein based on sequence similarity, we performed a protein domain-architecture analysis for every protein family and computationally inferred gene duplication events. While many herpesvirus proteins have evolved without any detectable gene duplications or domain rearrangements, numerous herpesvirus protein families do exhibit complex evolutionary histories. Some proteins acquired additional domains (e.g., DNA polymerase), whereas others show a combination of domain acquisition and gene duplication (e.g., betaherpesvirus US22 family), with possible functional implications. This novel classification system of SOGs for human Herpesviridae proteins is available through the Virus Pathogen Resource (ViPR, www.viprbrc.org).
Keywords: Herpesviridae, Protein domain, Ortholog, Protein family, Phylogenetics, Evolution, Gene duplication, Domain architecture, Comparative genomics, Nomenclature
1. Introduction
1.1. Human herpesviruses
Herpesviruses comprise a large and diverse order (Herpesvirales) of double stranded DNA viruses that infect humans and a wide range of other hosts (Pellet and Roizman, 2007, Virus Taxonomy: The Classification and Nomenclature of Viruses The Online (10th) Report of the ICTV, 2017). Human diseases caused by herpesviruses range from vesicular rashes to cancer. The order Herpesvirales is subdivided into three families, including the Herpesviridae, which is further subdivided into three subfamilies, the Alpha-, Beta-, and Gammaherpesvirinae. Within subfamilies, groups of related herpesvirus species are classified into genera. The nine species of human herpesviruses are distributed across the three subfamilies and several genera ( Table 1); these viruses are the main focus of this work. Prior studies found that the Beta- and Gammaherpesvirinae are more closely related to each other than to Alphaherpesvirinae (Montague and Hutchison, 2000). In contrast to some other human viruses, the human herpesviruses have a long evolutionary history, with evidence suggesting that the primordial herpesvirus diverged into the Alpha-, Beta, and Gammaherpesvirinae approximately 180 million to 220 million years ago (McGeoch et al., 1995). Coupled with their genome complexity and the availability of numerous complete genome sequences, this deep evolutionary history makes herpesviruses a tractable and informative model to study virus genome evolution at the levels of gene duplication and protein domain rearrangement.
Table 1.
Classification and properties of the human herpesviruses.
| Subfamily | Genus | Species | Common name | Genome length (kb) | RefSeq Accession | Number of annotated proteinsa |
|---|---|---|---|---|---|---|
| Alphaherpesvirinae | ||||||
| Simplexvirus | Human alphaherpesvirus 1 | Herpes simplex 1 (HSV1) | 152 | NC_001806 | 77 | |
| Simplexvirus | Human alphaherpesvirus 2 | Herpes simplex 2 (HSV2) | 155 | NC_001798 | 77 | |
| Varicellovirus | Human alphaherpesvirus 3 | Varicella-zoster virus (VZV) | 125 | NC_001348 | 73 | |
| Betaherpesvirinae | ||||||
| Cytomegalovirus | Human betaherpesvirus 5 | Human cytomegalovirus (HCMV) | 236 | NC_006273 | 169 | |
| Roseolovirus | Human betaherpesvirus 6A | Human herpesvirus 6A (HHV-6A) | 159 | NC_001664 | 88 | |
| Roseolovirus | Human betaherpesvirus 6B | Human herpesvirus 6B (HHV-6B) | 162 | NC_000898 | 104 | |
| Roseolovirus | Human betaherpesvirus 7 | Human herpesvirus 7 (HHV-7) | 153 | NC_001716 | 86 | |
| Gammaherpesvirinae | ||||||
| Lymphocryptovirus | Human gammaherpesvirus 4 | Epstein-Barr Virus (EBV) | 172 | NC_007605 | 94 | |
| Rhadinovirus | Human gammaherpesvirus 8 | Kaposi sarcoma-associated herpesvirus (KSHV); Human herpesvirus 8 (HHV-8) | 138 | NC_009333 | 86 |
Protein numbers are based on CDS entries in the associated RefSeq.
1.2. Phylogenomics
Homologs are genes that are evolutionarily related, regardless of the mechanism. Orthologs were defined by Fitch in 1970 as homologous genes in different species that diverged from a common ancestral gene by speciation. Genes that, either in the same or different species, diverged by a gene duplication have been termed paralogs (Fitch, 2000, Fitch, 1970). While the terms ortholog and paralog have no consistent functional implications (Jensen, 2001), orthologs are oftentimes considered more functionally similar than paralogs at the same level of sequence divergence. This has been termed the “ortholog conjecture”, which remains a topic of active research (Altenhoff et al., 2012, Chen and Zhang, 2012, Nehrt et al., 2011, Rogozin et al., 2014), due to its importance for computational sequence functional analysis (Eisen, 1998, Zmasek and Eddy, 2002) and the significance of gene duplications for biological evolution (Zhang, 2003).
Orthologs (or groups/clusters of orthologs) have often been inferred by indirect methods based on (reciprocal) pairwise highest similarities [e.g. (Remm et al., 2001, Tatusov et al., 1997)]. In this work, we used explicit phylogenetic inference combined with comparison to a trusted species tree for orthology inference, as this approach is likely to yield more accurate results (Zmasek and Eddy, 2002, Zmasek and Eddy, 2001).
1.3. Protein domains and domain architectures
Many eukaryotic proteins, and by extension, proteins of eukaryotic viruses, are composed of multiple domains, components that can each have their own evolutionary history and functional implications. The architecture of a protein is a product of the ordered arrangement of its several domains and their overall tertiary structure. Evolutionarily, individual domains can combine with other partner domains, enabling formation of a vast number of domain combinations, even within the same species (Moore et al., 2008). Assembling multiple domains into a single protein creates a distinct entity that can be more than the sum of its constituent parts. The emergence of proteins with novel combinations of duplicated and then diverged domains is considered to be a major mechanism for rapid evolution of new functionality in eukaryotic genomes (Itoh et al., 2007, Peisajovich et al., 2010). It is especially important in the evolution of pathways, where novel linkages between existing domains may result in the rearrangement of pathways and their behaviors in the cell (Peisajovich et al., 2010). The modular structure of eukaryotic proteins provides a mechanism that enables evolutionarily-rapid differentiation and emergence of a multitude of novel protein functions from an initially limited array of functional domains. Proteins can gain (or lose) new domains via genome rearrangements, creating (or removing) domain combinations, in addition to modification of domains themselves by small-scale mutations (Patthy, 2003, Ye and Godzik, 2004).
Here we present a systematic classification of proteins catalogued in the NCBI RefSeq entries for each of the nine human herpesviruses plus selected comparisons with homologs from non-human herpesviruses based on phylogenetic inferencing and domain architecture analysis using Domain-Architecture Aware Inference of Orthologs (DAIO). This analysis resulted in the classification of proteins into “Strict Ortholog Groups” (SOGs), in which all proteins are orthologous to each other (related by speciation events) and exhibit the same domain architecture. The SOG classification also enabled the development of an informative name convention for each SOG that includes information about the protein's function (if known) and a suffix indicating the taxonomic distribution of the protein. For example, an “aBG” suffix would indicate that proteins of this group are found in some (but not all) human Alphaherpesvirinae species (lowercase “a”), and all human Beta- and Gammaherpesvirinae species (uppercase “B” and “G”). Such suffixes allow for the quick understanding of presumed conserved protein function and minimal common genome across the Herpesviridae family. The SOG classification results have been made publicly available through the Virus Pathogen Resource (ViPR) (Pickett et al., 2012) at https://www.viprbrc.org.
2. Results and discussion
For this analysis, we developed a rational, phylogeny- and domain architecture-aware classification approach for human herpesvirus proteins, the Domain-architecture Aware Inference of Orthologs (DAIO) method, which produces Strict Ortholog Groups (SOGs) of proteins. Before we present genome-wide findings, we show results for a few instructive SOG examples, including protein groups that have evolved in a “simple” manner, recapitulating the Herpesviridae evolutionary tree without gene duplications or domain rearrangements, and protein groups in which domain rearrangements (domain gains) and/or gene duplications have occurred.
Table 2 lists the 23 SOGs common to all nine human herpesviruses. For every SOG, a suggested name is provided, composed of a protein names and a suffix indicating the taxonomic distribution (A, B, G: present in all human members of the Alpha-, Beta-, Gammaherpesvirinae, respectively; a, b, g: present in some but not all human members of the Alpha-, Beta-, Gammaherpesvirinae, respectively). Gene names/symbols (a forward slash is either part of the accepted gene name or is used to separate multiple gene names) and Pfam domain architecture names are also included. The table is organized into three sections. The first section lists protein families that have apparently evolved without gene duplication or domain rearrangements [e.g., uracil DNA glycosylase and the capsid scaffolding protein protease (CSPP)]; the second section lists proteins that have evolved with domain rearrangements and or duplications [e.g., glycoprotein B (gB), DNA polymerase, and multifunctional regulator of expression proteins (mRE)], and the third section lists proteins that share some function (and even genome region) but have been formed from distantly or unrelated domains (e.g., gL, gN, and DNA polymerase processivity factor).
Table 2.
Names of Herpesviridae Proteins Common to All 9 Herpesviruses Based on Strict Ortholog Groups.
| Suggested Name |
Alpha |
Beta |
Gamma |
DA (Pfam domains) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
HSV-1/2 |
VZV | CMV | HHV-6A | HHV-6B | HHV-7 | EBV | KSHV | |||
| UL/US | Other | |||||||||
| Uracil-DNA glycosidase_ABG | UL2 | ORF59 | UL114 | U81 | U81 | U81 | BKRF3 | ORF46 | UDG | |
| Helicase-primase ATPase subunit_ABG | UL5 | ORF55 | UL105 | U77 | U77 | U77 | BBLF4 | ORF44 | Herpes_Helicase | |
| Glycoprotein M_ABG | UL10 | ORF50 | UL100 | U72 | U72 | U72 | BBRF3 | ORF39 | Herpes_glycop | |
| Alkaline deoxyribonuclease_ABG | UL12 | ORF48 | UL98 | U70 | U70 | U70 | BGLF5 | ORF37 | Herpes_alk_exo | |
| Serine threonine protein kinase_ABG | UL13 | ORF47 | UL97 | U69 | U69 | U69 | BGLF4 | ORF36 | UL97 Pfam domain (Beta) | |
| Terminase_ABG | UL15 | ORF42 | UL89 | U66/U60 | U66/U60 | U66/U60 | LMP2 | ORF29 | DNA_pack_N––DNA_pack_C | |
| Tegument protein_ABG | UL16 | ORF44 | UL94 | U65 | U65 | U65 | BGLF2 | ORF33 | Herpes_UL16 | |
| Capsid transport tegument protein_ABG | UL17 | ORF43 | UL93 | U64 | U64 | U64 | BGLF1 | ORF32 | Herpes_UL17 | |
| Triplex dimer protein_ABG | UL18 | VP23 | ORF41 | UL85 | U56 | U56 | U56 | BDLF1 | ORF26 | Herpes_V23 |
| Major capsid protein_ABG | UL19 | VP5/ICP5 | ORF40 | UL86 | U57 | U57 | U57 | BcLF1 | ORF25 | Herpes_MCP |
| Glycoprotein H_ABG | UL22 | ORF37 | UL75 | U48 | U48 | U48 | BXLF2 | ORF22 | Herpes_glycop_H | |
| UL24 Protein_ABG | UL24 | ORF35 | UL76 | U49 | U49 | U49 | BXRF1 | ORF20 | Herpes_UL24 | |
| Portal capping protein_ABG | UL25 | ORF34 | UL77 | U50 | U50 | U50 | BVRF1 | ORF19 | Herpes_UL25 | |
| Protease-scaffolding protein_ABG | UL26 | ORF33 | UL80 | U53 | U53 | U53 | BVRF2 | ORF17 | Peptidase_S21 | |
| Terminase DNA binding subunit_ABG | UL28 | ORF30 | UL56 | U40 | U40 | U40 | BALF3 | ORF7 | PRTP | |
| Major DNA binding protein_ABG | UL29 | ICP8 | ORF29 | UL57 | U41 | U41 | U41 | BALF2 | ORF6 | Viral_DNA_bp |
| Nuclear egress lamina protein_ABG | UL31 | ORF27 | UL53 | U37 | U37 | U37 | BFLF2 | ORF69 | Herpes_UL31 | |
| Capsid transport nuclear protein_ABG | UL32 | ORF26 | UL52 | U36 | U36 | U36 | BFLF1 | ORF68 | Herpes_env | |
| Terminase binding protein_ABG | UL33 | ORF25 | UL51 | U35 | U35 | U35 | BFRF1A | ORF67A | Herpes_UL33 | |
| Nuclear egress membrane protein_ABG | UL34 | ORF24 | UL50 | U34 | U34 | U34 | BFRF1 | ORF67 | Herpes_U34 | |
| Triplex monomer_ABG | UL38 | VP19c | ORF20 | UL46 | U29 | U29 | U29 | BORF1 | ORF62 | Herpes_VP19C |
| Deoxyuridine 5′-triphosphate nucleotidohydrolase_ABG | UL50 | ORF8 | UL72 | U45 | U45 | U45 | BLLF3 | ORF54 | dUTPase | |
| DNA primase_ABG | UL52 | ORF6 | UL70 | U43 | U43 | U43 | BSLF1 | ORF56 | Herpes_UL52 | |
| Portal protein_ABG.ABG | UL6 | ORF54 | UL104 | U76 | U76 | U76 | LMP2 | ORF43 | Herpes_UL6 | |
| Encapsidation and egress protein_ABG.ABG | UL7 | ORF53 | UL103 | U75 | U75 | U75 | BBRF2 | ORF42 | Herpes_UL7 | |
| Encapsidation and egress protein_ABG.g | ORF43 | Herpes_UL6––Herpes_UL7 | ||||||||
| Helicase primase subunit_ABG.ABg | UL8 | ORF52 | UL102 | U74 | U74 | U74 | BBLF2/BBLF3 | ORF41 | Herpes_HEPA | |
| Helicase primase subunit_ABG.g | ORF40 | Herpes_HEPA––Herpes_heli_pri | ||||||||
| Glycoprotein B_ABG.AbG | UL27 | ORF31 | U39 | U39 | U39 | BALF4 | ORF8 | Glycoprotein_B | ||
| Glycoprotein B_ABG.b | UL55 | HCMVantigenic_N––Glycoprotein_B | ||||||||
| DNA polymerase_ABG.a | UL30 | DNA_pol_B_exo1––DNA_pol_B––DNAPolymera_Pol | ||||||||
| DNA polymerase_ABG.aBG | ORF28 | UL54 | U38 | U38 | U38 | BALF5 | ORF9 | DNA_pol_B_exo1––DNA_pol_B | ||
| Large tegument protein_ABG.A | UL36 | VP1–2 | ORF22 | Herpes_teg_N––Herpes_UL36 | ||||||
| Large tegument protein_ABG.BG | UL48 | U31 | U31 | U31 | BPLF1 | ORF64 | Herpes_teg_N | |||
| Ribonucleotide reductase large subunit_ABG.AG | UL39 | ICP6 | ORF19 | BORF2 | ORF61 | Ribonuc_red_lgN––Ribonuc_red_lgC | ||||
| Ribonucleotide reductase large subunit_ABG.B | UL45 | UL28 | UL28 | UL28 | Ribonuc_red_lgC | |||||
| Multifunctional regulator of expression_ABG.a | UL54 | ICP27 | HHV− 1_VABD––Herpes_UL69 | |||||||
| Multifunctional regulator of expression_ABG.aBG | ORF4 | UL69 | U42 | U42 | U42 | BSLF2/BMLF1 | ORF57 | Herpes_UL69 | ||
| Cytoplasmic egress facilitator-1_A | UL51 | ORF7 | Herpes_UL51 | |||||||
| Cytoplasmic egress facilitator-1_BG | U71 | U44 | U44 | U44 | BSRF1 | ORF55 | Herpes_U44 | |||
| Cytoplasmic egress facilitator-2_A | UL21 | ORF38 | Herpes_UL21 | |||||||
| Cytoplasmic egress facilitator-2_B | UL88 | U59 | U59 | U59 | Herpes_U59 | |||||
| Cytoplasmic egress facilitator-2_G | BTRF1 | ORF23 | Herpes_BTRF1 | |||||||
| Cytoplasmic egress tegument protein_A | UL11 | ORF49 | UL11 | |||||||
| Cytoplasmic egress tegument protein_CMV | UL99 | |||||||||
| Cytoplasmic egress tegument protein_G | BBLF1 | ORF38 | DUF2733 | |||||||
| Cytoplasmic egress tegument protein_R | U71 | U71 | U71 | |||||||
| DNA polymerase processivity subunit_A | UL42 | ORF16 | Herpes_UL42––Herpes_UL42 | |||||||
| DNA polymerase processivity subunit_B | UL44 | U27 | U27 | U27 | Herpes_PAP | |||||
| DNA polymerase processivity subunit_G | BMRF1 | ORF59 | Herpes_DNAp_acc | |||||||
| Tegument protein UL14_A | UL14 | ORF46 | Herpes_UL14 | |||||||
| Tegument protein UL14_BG | UL95 | U67 | U67 | U67 | BGLF3 | ORF34 | Herpes_UL95 | |||
| Glycoprotein L_A.a | ORF60 | Herpes_UL1 | ||||||||
| Glycoprotein L_A.S | UL1 | Herpes_UL1––GlyL_C | ||||||||
| Glycoprotein L_B | UL115 | U82 | U82 | U82 | Cytomega_gL | |||||
| Glycoprotein L_G | BKRF2 | ORF47 | Phage_glycop_gL | |||||||
| Glycoprotein N_A | UL49A | |||||||||
| Glycoprotein N_BG.b | UL73 | UL73_N––Herpes_UL73 | ||||||||
| Glycoprotein N_BG.BG | UL73 | U46 | U46 | U46 | BLRF1 | ORF53 | Herpes_UL73 | |||
| Glycoprotein N_a | ORF9A | Herpes_UL49_5 | ||||||||
| Inner tegument protein UL37_A | UL37 | ORF21 | Herpes_UL37_1 | |||||||
| Inner tegument protein UL37_BG | UL47 | U30 | U30 | U30 | BOLF1 | ORF63 | Herpes_U30 | |||
| Small capsid protein_A | UL35 | VP26 | ORF23 | Herpes_UL35 | ||||||
| Small capsid protein_B | UL48A | U32 | U32 | U32 | HV_small_capsid | |||||
| Small capsid protein_G | BFRF3 | ORF65 | Herpes_capsid | |||||||
Abbreviations.
ICP: infected cell protein.
VP: virion protein.
2.1. Uracil DNA glycosylase and capsid scaffolding protein protease: Evolution of a stable domain architecture without gene duplications
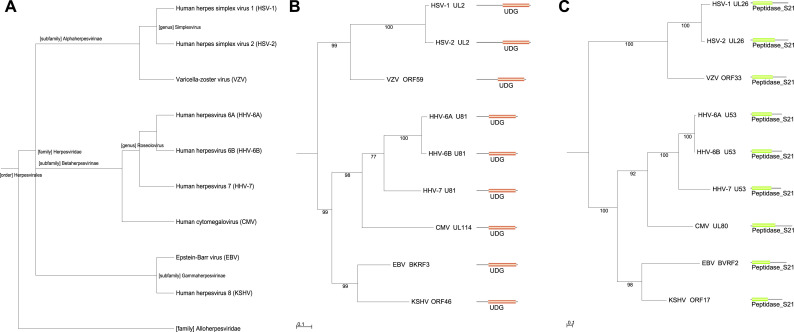
Uracil DNA glycosylases catalyze the first step – removal of the RNA base uracil from DNA – in base excision repair, the mechanism by which damaged bases in DNA are removed and replaced (Krusong et al., 2006). Uracil DNA glycosylases are found in eukaryotes, bacteria, and archaea, as well as in herpesviruses and poxviruses (Chen et al., 2002). Our phylogenomic analysis shows that for all nine human herpesviruses, uracil DNA glycosylase is well conserved and contains one Pfam domain, UDG (uracil DNA glycosylase superfamily). In addition, the gene tree for human herpesvirus uracil DNA glycosylases ( Fig. 1B) precisely recapitulates the herpesvirus species tree (Fig. 1A); therefore, this protein family can be inferred to have evolved from a single common ancestor and without any gene duplications or domain rearrangements (see Table 2 for virus-specific gene names).
Fig. 1.
Proteins with conserved domain architectures that mirror the Herpesvirus species tree. (A) A current view of herpesvirus evolution. The human herpesvirus species tree is based on previous reports (McGeoch et al., 2000, McGeoch et al., 1995, Davison, 2010, Davison, 2002). (B) Maximum likelihood gene tree for uracil DNA glycosylase proteins based on an alignment for UDG Pfam domain amino acid sequences. (C) Maximum likelihood gene tree for capsid scaffolding protein proteases, based on Peptidase_S21 Pfam domain amino acid sequences. For the gene trees, bootstrap values are shown. Branch length distances are proportional to expected changes per site.
Capsid scaffolding protein proteases are essential for herpesvirus capsid assembly and maturation, and have an essential serine protease activity (Liu and Roizman, 1993). These proteins contain one Pfam domain, Peptidase_S21. In contrast to uracil DNA glycosylases, currently available data indicate that protease-scaffolding proteins with a Peptidase_S21 domain are unique to Herpesvirales. Like uracil DNA glycosylases, CSPP evolved without domain architecture rearrangements or gene duplications (Fig. 1C, Table 2).
Other examples of Herpesviridae genes that have evolved without any domain architecture rearrangements or gene duplications are listed in the first section of Table 2.
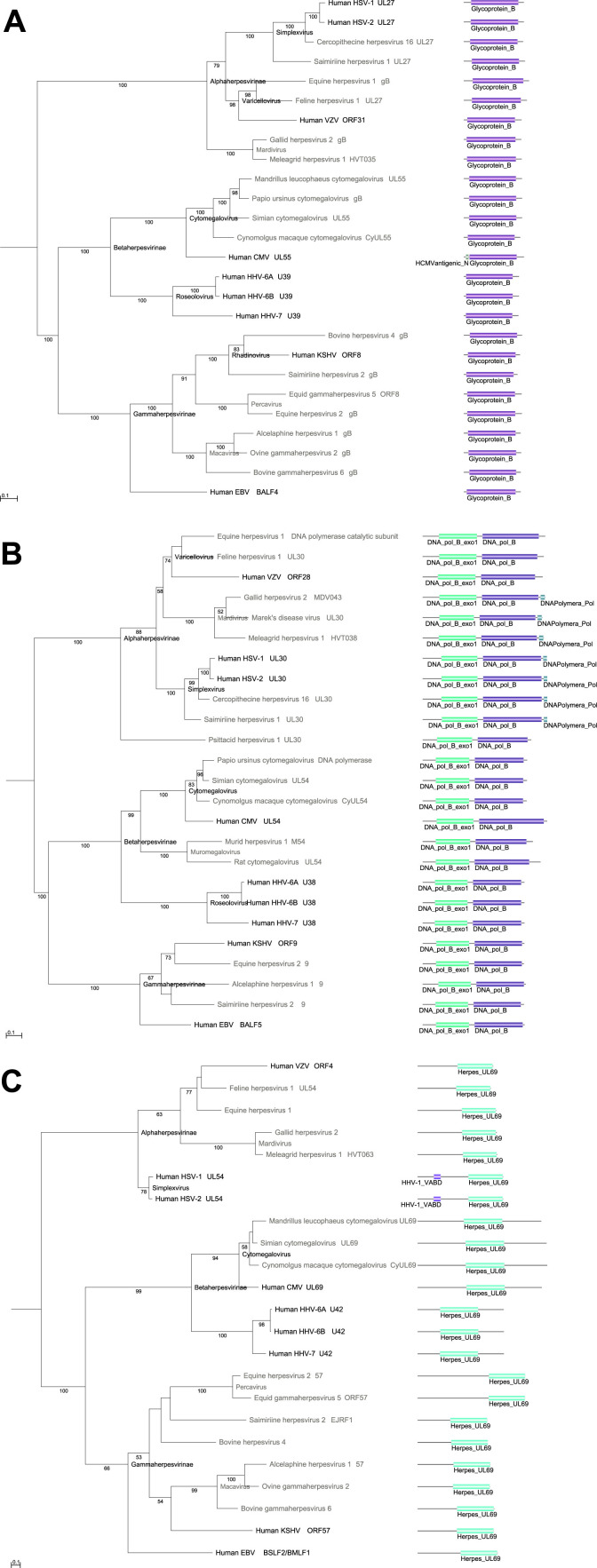
2.2. Molecular evolution of gB: A highly conserved protein required for viral fusion with a recent domain acquisition in one virus lineage
Herpesvirus virions have an envelope that consists of an outer lipid bilayer studded with 12 or more surface glycoproteins (originally defined in HSV). After virion glycoprotein engagement with cell surface receptors, the envelope fuses with the plasma membrane – a process which, for herpes simplex virus 1 (HSV-1), requires four of its 12 envelope glycoproteins, namely glycoproteins gB, gD, gH, and gL (Cai et al., 1988, Forrester et al., 1992, Ligas and Johnson, 1988, Roop et al., 1993, Spear and Longnecker, 2003). In contrast, for other herpesviruses, only glycoproteins gB, gH, and gL have been reported to be required for membrane fusion (AlHajri et al., 2017).
gB and gH are highly conserved across all nine human herpesviruses (Table 2). A protein annotated as gL is also present in all nine human herpesviruses, yet its occurrences in members of the Alpha-, Beta- and Gammaherpesvirinae are homologous within, but not between subfamilies. gLs from different subfamilies contain unrelated protein domains (Pfam: Herpes_UL1, Cytomega_gL, and Phage_glycop_gL). gL is discussed in more detail below.
Detailed phylogenetic analysis of the human herpesvirus gB family ( Fig. 2A), including proteins from selected non-human members of the Herpesviridae, shows a picture of a protein that has evolved without gene duplications (or, at the very least, duplicated genes have not been retained) and with nearly completely conserved domain architectures.
Fig. 2.
Proteins in which an additional domain has been added during the course of evolution. (A) Maximum likelihood gene tree for glycoprotein B proteins based on an alignment for the main glycoprotein_B domain amino acid sequences. (B) Maximum likelihood gene tree for DNA polymerase proteins based on an alignment for DNA_pol_B_exo1––DNA_pol_B domain amino acid sequences. (C) Maximum likelihood gene tree for multifunctional regulator of expression proteins based on an alignment for Herpes_UL69 domain amino acid sequences. Bootstrap values larger than 50 are shown. Branch length distances are proportional to expected changes per site.
The one exception to this is that human cytomegalovirus (HCMV) glycoprotein B (gB) has a short region of about 40 amino acids near its N-terminus that comes in two forms that differ by approximately 50% at the amino acid level. This sequence variant was identified in HCMV strains isolated from Chinese patients (Shiu et al., 1994) and is identified in Pfam as “HCMVantigenic_N domain”. In our global hmmscan analysis (applying the same threshold of E = 10−6 for every Pfam domain) E-value support for presence of this domain in some strains is strong (E < 10–22) and matching over the entire Pfam model while other HCMV strains do not exhibit significant sequence similarity with this domain. It has been suggested that this domain polymorphism may be implicated in HCMV-induced immunopathogenesis, as well as in strain-specific behaviors, such as tissue-tropism and the ability to establish persistent or latent infections (Pignatelli et al., 2004). In our new systemic naming approach (see below) we term the SOG of the protein with HCMVantigenic_N domain “Glycoprotein B_ ABG.b”, whereas all other proteins fall into the “Glycoprotein B_ ABG.AbG” SOG.
2.3. Molecular evolution of DNA polymerase: A highly conserved protein with domain acquisition
All members of the Herpesviridae encode six conserved proteins that play essential roles at the replication fork during viral DNA replication: a single-strand DNA binding protein (major DNA binding protein), a DNA polymerase composed of two independently coded subunits (the catalytic DNA polymerase subunit and a DNA polymerase processivity factor encoded by three distantly related genes in members of the Alpha-, Beta-, and Gammaherpesvirinae, see below), and a three subunit helicase/primase complex (DNA replication helicase, DNA helicase primase complex associated protein, and DNA primase) (Pellet and Roizman, 2007).
Our analysis shows that the catalytic DNA polymerase subunits of all members of the Herpesviridae contain two domains: an N-terminal DNA polymerase family B exonuclease domain, and a C-terminal polymerase domain from DNA polymerase family B (Fig. 2B). Cellular family B DNA polymerases are the main polymerases involved with nuclear DNA replication and repair in eukaryotes and prokaryotes, and include DNA polymerases II and B, and polymerases α, δ, and ε (Garcia-Diaz and Bebenek, 2007). Family B DNA polymerases are also found in other dsDNA viruses, such as the insect Ascoviridae, and members of the Iridoviridae (e.g., fish lymphocystis disease virus) and Phycoviridae (e.g., chlorella virus) (Villarreal and DeFilippis, 2000). In addition to these two large and ubiquitous domains, Simplexvirus (which include human simplex virus 1 and 2) and Mardivirus also possess a small C-terminal domain, called the DNA polymerase catalytic subunit Pol (DNAPolymera_Pol) domain in Pfam (Zuccola et al., 2000), and are longer by about 45 aa on average than DNA polymerase proteins from other Herpesviridae. According to currently available genomic data, DNAPolymera_Pol is found in members of the Simplexvirus genus of the Alphaherpesvirinae. While varicella-zoster virus (Human herpesvirus 3) and other members of the Varicellovirus genus of the Alphaherpesvirinae possesses DNA polymerases that also tend to be longer, similarity of these protein regions to the DNAPolymera_Pol domain is low, using the current Pfam model for DNAPolymera_Pol (Pfam version 31.0). The function of this third domain is to mediate interaction between DNA polymerase and its cognate processivity factor (Bridges et al., 2000, Loregian et al., 2000) based on the observation that a peptide corresponding to the 27 C-terminal amino acids of HSV-1 DNA polymerase has been shown to inhibit viral replication by disrupting the interaction between DNA polymerase and UL42 (Digard et al., 1995, Loregian et al., 1999). In this context, it is interesting to note that the DNA polymerase processivity factors are only distantly-related across the Alpha-, Beta-, and Gammaherpesvirinae (see below). It is therefore conceivable that the interactions of Beta-, and Gammaherpesvirinae DNA polymerase processivity factors with their corresponding DNA polymerases (which lack a DNAPolymera_Pol domain) is different in nature than for Alphaherpesvirinae. As for Varicellovirus it is unclear whether they possess a functional DNAPolymera_Pol domain, and a definitive answer will require similar biochemical assays as have been performed for HSV-1.
Phylogenetic analysis of human herpesvirus DNA polymerase proteins, plus related proteins from selected mammalian herpesviruses, shows that, similar to the glycoprotein B family, DNA polymerases of the Herpesviride evolved without gene duplication. Nonetheless, in contrast to gB, DNA polymerases acquired a new domain early in Alphaherpesvirinae evolution. This domain might have been lost again, or underwent significant mutations, during Varicellovirus evolution. The presence of the longer domain in Varicelloviruses suggests that the longer domain emerged prior to the Varicellovirus/Simplexvirus split.
2.4. Evolution of viral multifunctional regulator of expression (mRE) proteins (homologs of HSV1 ICP27)
Multifunctional regulator of expression (mRE; also known as immediate-early protein IE63, infected cell protein 27, ICP27, and α27) is a protein with homologs in all human herpesviruses (for gene names see Table 2). Multifunctional regulator of expression is a regulatory protein that plays a role in the prevention of apoptosis during HSV1 infection (Aubert and Blaho, 1999). Multifunctional regulator of expression interacts directly with a number of proteins in performing its many roles. In particular, multifunctional regulator of expression protein contributes to host shut‐off by inhibiting pre‐mRNA splicing by interacting with essential splicing factors, termed SR proteins, and affecting their phosphorylation (Sciabica et al., 2003). Furthermore, the mRE protein has been shown to associate with cellular RNA polymerase II holoenzyme in a DNA- and RNA-independent manner and to recruit RNA polymerase II to viral transcription/replication sites (Dai-Ju et al., 2006, Zhou and Knipe, 2002). mRE also competes with some transport receptors, resulting in the inhibition of host pathways while supporting mRNA export factor-mediated transport of HSV-1 mRNAs (Malik et al., 2012).
All of the multifunctional regulator of expression proteins analyzed here have a single copy of a Pfam “Herpesvirus transcriptional regulator family” (Herpes_UL69) domain that is specific to members of the Herpesviridae. In addition to the Herpes_UL69 domain, human Simplexvirus mRE have an additional N-terminal domain, the “Herpes viral adaptor-to-host cellular mRNA binding domain” (HHV-1_VABD) (Tunnicliffe et al., 2011). Besides human Simplexvirus, architectures with C-terminal HHV-1_VABD and N-terminal Herpes_UL69 domains are also found in Chimpanzee herpesviruses (e.g. NCBI Reference Sequence: YP_009011042 (Severini et al., 2013)), while other non-human Simplexviruses lack the HHV-1_VABD domain. Using currently available genomic data, we were unable to detect HHV-1_VABD domains outside of the Simplexvirus genus.
Phylogenetic analysis of human herpesvirus mRE proteins, including proteins from selected herpesviruses of other mammals, shows that multifunctional regulator of expression proteins evolved without observable gene duplications (since this gene tree recapitulates the herpesvirus species tree).
2.5. Different domains performing the same, or similar, functions
Nine groups of human herpesviruses are annotated as performing the same, or very similar function, in the absence of discernable protein sequence similarity (Table 2, Fig. 3).
Fig. 3.
Examples ofHerpesviridaeproteins composed of unrelated or only very distantly related proteins, annotated as performing the same, or very similar function. (A, B, C) Maximum likelihood gene trees for DNA polymerase processivity factor proteins from Alpha-, Beta-, and Gammaherpesvirinae based on alignments for Herpes_UL42 (A), Herpes_PAP (B), and Herpes_DNAp_acc (C) domain amino acid sequences, respectively. The internal domain duplication at the root the Herpes_UL42 tree is shown as a red square. (D, E, F) Maximum likelihood gene trees for gL proteins from human Alpha-, Beta-, and Gammaherpesvirinae based on alignments for Herpes_UL1 (D), Cytomega_gL (E), and Phage_glycop_gL (F) domain amino acid sequences, respectively. Bootstrap support values are shown. Branch length distances are proportional to expected changes per site.
As mentioned above, DNA polymerase processivity factor is one of the six proteins that play essential roles at the replication fork during viral DNA replication. Processivity factors, also called clamp proteins, help to overcome the tendency of DNA polymerase to dissociate from the template DNA, and thus greatly enhance DNA polymerase processivity (Weisshart et al., 1999, Zhuang and Ai, 2010). In contrast to the protein families discussed so far, DNA polymerase processivity factors are only distantly-related across the Alpha-, Beta-, and Gammaherpesvirinae. In the Alphaherpesvirinae, the protein is composed of two tandem Herpes_UL42 domains; Betaherpesvirinae have a single Herpes_PAP domain; Gammaherpesvirinae have a single Herpes_DNAp_acc domain (Fig. 3A, B, C). These three domains are very distant homologs and are members of the DNA clamp superfamily (Pfam clan CL0060).
gL (Fig. 3D, E, F) is another example of a protein function performed by different, probably non-homologous domains present in different Herpesviridae subfamilies (Pfam domains Herpes_UL1, GlyL_C, Cytomega_gL, and Phage_glycop_gL). Interestingly, the open reading frames for these seemingly unrelated proteins are located in analogous conserved genomic contexts, including open reading frame sizes and orientations relative to the surrounding conserved coding regions.
The remaining seven groups with these characteristics are: cytoplasmic egress tegument protein, cytoplasmic egress facilitator-1, cytoplasmic egress facilitator-2, encapsidation chaperone protein, glycoprotein N Pfam clan Herpes_glyco, CL0146), LTP binding protein, and small capsid protein (Table 1 and Supplementary Table 1).
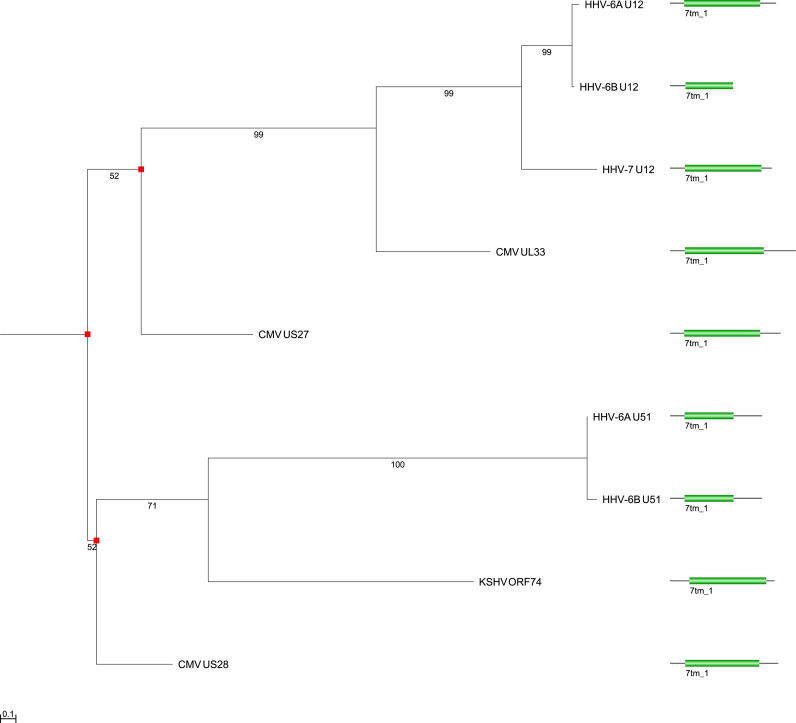
2.6. Gene duplication during viral 7-transmembrane receptor domain protein evolution
In contrast to the protein families discussed so far, the evolutionary history of human Herpesviridae proteins with 7-transmembrane receptor domains is more complex ( Fig. 4) (Spiess et al., 2015). By comparing this gene tree with a species tree for human Herpesviridae (Fig. 1A), we can infer three gene duplication events (marked as red squares in Fig. 4), resulting in four groups of orthologous genes: UL33/U12, US27, U51/ORF74, and US28. In our new nomenclature (see below), we call the first group “G-protein coupled receptor homolog UL33/U12_B” because it is found in all four human Betaherpesvirinae species (uppercase B suffix). The second group is called “G-protein coupled receptor homolog US27_b” as it is found in some human Betaherpesvirinae (lowercase b suffix). The third group is called “G-protein coupled receptor homolog U51/ORF74_bg” because it found in some human Betaherpesvirinae and in some human Gammaherpesvirinae (lowercase “bg” suffix). The fourth group is called “Envelope protein US28_b”. No orthologous genes were found in the human Alphaherpesvirinae. Whenever available, we base our names preferably on (Mocarski, 2007) or the “Recommended name” (under “Protein names”) from the UniProtKB database (Bateman et al., 2017). For reasons of consistency and objectivity, we used an automated approach to root all trees by mid-point rooting. It is possible, that the true root for the 7-transmembrane domain proteins tree is at the base of the U51-ORF74 subtree. In this case there would be only two duplications in the tree, but still the same four ortholog groups: U51/ORF74, US28, US27, UL33/U12. Functionally, all these proteins appear to be hijacked human proteins that are being used by the virus to modulate the host immune system. In particular, many of them appear to act as chemokine (orphan) receptors (Casarosa et al., 2003, Casarosa et al., 2001, Isegawa et al., 1998, Murphy, 2001, Zhen et al., 2005) ( Fig. 5).
Fig. 4.
Gene tree for humanHerpesviridaeproteins with a 7-transmembrane receptor domain. This maximum likelihood tree is based on an alignment of 7tm_1 domain amino acid sequences. Bootstrap values are shown. Branch length distances are proportional to expected changes per site. Red squares indicate gene duplications.
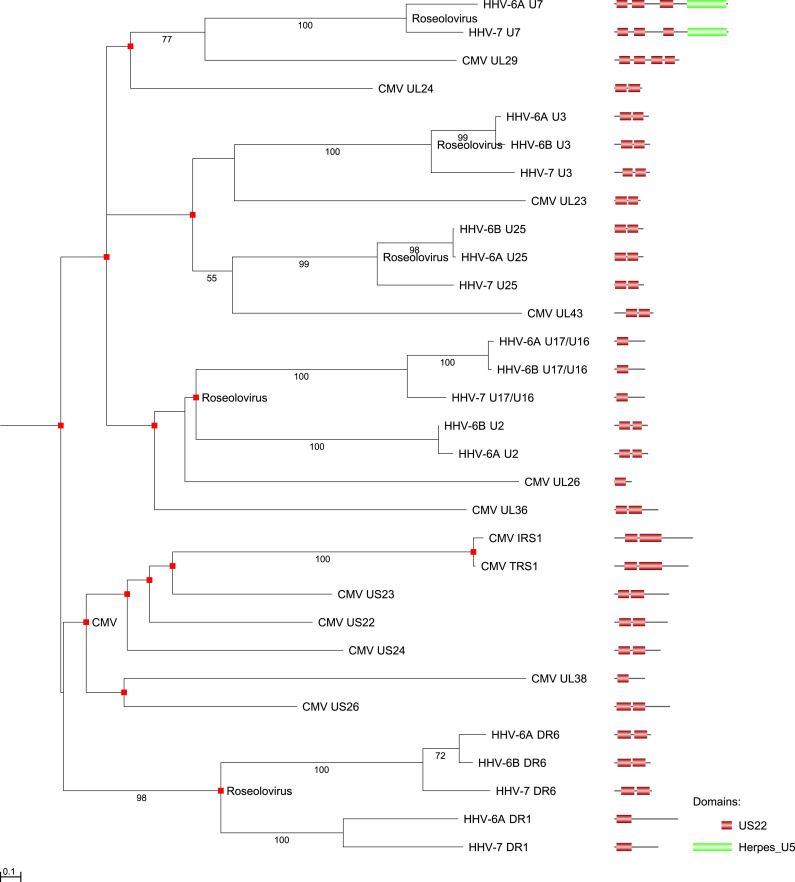
Fig. 5.
Gene tree for humanHerpesviridaeproteins with US22 domain(s). This maximum likelihood tree is based on an alignment of full length protein sequences. Pfam domains are shown with a E = 10−1 cutoff. Bootstrap values larger than 50 are shown. Branch length distances are proportional to expected changes per site. Red rectangles squares indicate the sometimes duplicated US22 domains. Green rectangles indicate the locations of Herpes_U5 domains.
2.7. The complex evolution of US22 domain proteins
Proteins with US22 domains have the most complex evolutionary history of all Herpesviridae proteins, even though among the human herpesviruses, the US22 domain has been found only in betaherpesviruses (Hanson et al., 1999). US22 domain proteins are also present in Gallid herpesvirus 2 (a member of the Alphaherpesvirinae), in members of the Alloherpesviridae family, in other dsDNA viruses (e.g., Poxviridae and Iridoviridae), and in some animal species. Most proteins with US22 domains carry two copies of the domain. US22 is a member of a large group of distantly homologous proteins (the SUKH superfamily, Pfam clan CL0526), which, for example include bacterial Syd proteins. It has been suggested that a function of the US22 family is to act against various anti-viral responses by interacting with specific host proteins (Zhang et al., 2011).
Here we summarize the results of our phylogenetic analysis of US22 domain proteins of the human bataherpesviruses. Unfortunately, the phylogenetic signal across this group of protens is weak, thus some support values are low. Two groups of US22 orthologs span all four human betaherpesviruses: CMV tegument protein UL23 is likely to have orthologs in HHV-6A, HHV-6B, HHV-7 (Roseolovirus) Protein U3 (“Tegument protein UL23/Protein U3_B”). Similarly, CMV Tegument protein UL43 is likely to be orthologous to HHV-6A, HHV-6B, HHV-7 (Roseolovirus) Protein U25 (“Tegument protein UL43/Protein U25_B”). U3 and U25 are paralogous towards each other, as they are connected by a gene duplication, as are HCMV UL23 and 43. Four groups of orthologs specific to Roseolovirus are Tegument protein DR1, Tegument protein DR6, Protein U7, and Protein U17/U16. In U17/U16 proteins, it is unclear whether they possess a second US22 domain, as the similarity to this domain is weak to the point of insignificance. In contrast, U7 proteins possess at least three US22 domains and an additional C-terminal Herpes_U5 domain. Proteins U7 are most closely related to CMV UL29, but differ in their domain architecture (lack of Herpes_U5 domain). Thus CMV UL29 forms its own species-specific group of orthologs. Numerous proteins with US22 domains are specific to CMV (and thus all paralogous to each other) given current data: apoptosis inhibitor UL38, early nuclear protein HWLF1, tegument protein UL26, US24, protein UL24, UL29, UL36, US23, US26, protein IRS1, and protein TRS1.
2.8. The inferred minimal proteomes of the human herpesviruses
As described above, we classified viral proteins into “strict ortholog groups,” requiring that all proteins exhibit the same domain architecture and are orthologous to each other. We attempted to give an informative name for each of these groups including a suffix that indicates the taxonomic distribution of a protein. For example, an “aG” suffix would indicate that proteins of this group are found in some (but not all) members of human alphaherpesvirus species (lowercase “a”), and members of both human gammaherpesvirus species (uppercase “G”).
Families which have a (some) domain(s) in common but differ in their domain architectures, are more difficult to rationally name (we found 17 of these cases). An example of such a family is DNA polymerase. In such cases, the suffix is split by a period into two parts. The first part indicates overall presence of common domain(s) for all members of this SOG, the second part (after the period) relates to specific domain architectures. Thus, “DNA polymerase_ ABG.aBG” refers to the simpler DNA_pol_B_exo1––DNA_pol_B domain architecture present in nearly all Alphaherpesvirinae species. “DNA polymerase_ ABG.a” refers to the DNA_pol_B_exo1––DNA_pol_B—DNAPolymera_Pol DA that is present in a smaller subset of Alphaherpesvirinae species.
The rationale behind this approach for labeling members of protein families that have different domain architectures is that it gives users a choice between “traditional” ortholog groups, which do not consider domain architectures (by ignoring the part after the period), and SOGs (taking the full name into account).
In total, we were able to establish 169 SOGs (Supplementary Table 1). Of these, 40 (23 +8 +9) functionally similar groups (Table 2) are present in all 9 human Herpesviridae species and represent the core proteins of human herpesviruses.
Besides proteins with clearly defined Pfam domains, we found 29 protein families for which Pfam domains have not been defined. Classification of these proteins was based on manual BLAST searches. An example of such a family is the virion host shutoff protein UL41.
Another unusual case is the HSV1 UL13 serine threonine protein kinase. All nine human herpesviruses have homologs of this protein, but its associated Pfam domain UL97 only matches sequences in betaherpesviruses. Extension of the family to alpha- and gammaherpesviruses is thus based on manual BLAST searches.
Finally, two protein families could not be classified due to lack of phylogenetic signal: protein B8 of HHV-6A and HHV-6B (associated gene names U92, U93, HN1, HN92D, B8) and protein UL28/UL29/U8 of HHV-6A, HHV-6B, and HHV-7.
Proteins which are species or strain specific are listed in Supplementary Table 2.
2.9. Dissemination of SOG data through the ViPR database
In order to make the results of DAIO classification available to all Herpesvirus researchers for experimental hypothesis testing, we incorporated SOG data into the Virus Pathogen Resource (ViPR) at https://www.viprbrc.org (Pickett et al., 2012). Through ViPR, scientists can search, sort, and download SOG names (including taxonomic distribution), Pfam domain architecture data, and individual protein sequences belonging to selected SOGs. Fig. 6A shows an example of a search result table, which includes data for some of the protein families discussed above, namely glycoprotein B family members (associated with two distinct SOGs: “Glycoprotein B_ ABG.b” and “Glycoprotein B_ ABG.AbG”), DNA polymerase (“DNA polymerase_ ABG.a” and “DNA polymerase_ ABG.aBG”), and multifunctional regulator of expression (“Multifunctional regulator of expression_ABG.a” and “Multifunctional regulator of expression_ABG.aBG”). By clicking on the “Total # of Proteins” table entries, users can view and download the individual protein sequences belonging to a given SOG. Fig. 6B shows how SOG data, including domain architecture information, is part of protein annotations in ViPR (Simplexvirus “DNA polymerase_ABG.a” example). As new genome sequence data become available, the SOG data in ViPR is continuously updated in order to keep current with the ever expanding universe of Herpesvirus protein sequences. In addition, SOG annotations in ViPR will be expanded to include non-human Herpesviruses in the future. SOG data is also available for Pox- and Coronaviruses in ViPR, and will be applied to other virus families in the future.
Fig. 6.
SOG data in the Virus Pathogen Resource (ViPR,www.viprbrc.org). (A) An example of a protein ortholog group search result is shown. Clicking on the “Total # of Proteins” table entries, allows users to view and download the individual protein sequences belonging to a given SOG. (B) The annotations of an individual protein (Simplexvirus “DNA polymerase_ABG.a” in this example), including SOG name and HMM/Pfam domain architectures, from the Human herpesvirus 1 KOS strain are shown.
3. Conclusions
In this work, we used Domain-architecture Aware Inference of Orthologs (DAIO) to provide a classification for proteins of human herpesviruses, based on domain architecture and phylogenetic history. While the work presented here is limited to human herpesviruses, and thus does not take full advantage of all the sequence data that is currently available, we plan to extend our DAIO approach to all herpesviruses with a known phylogenetic history.
A major contribution of our classification system to herpesvirus biology is that it provides a series of testable hypotheses for further experimental investigations. For example, it informs experimental reconstruction of minimal genome viruses. Such synthesized minimal genomes could prove useful for identification of genes responsible for pathogenic and other biological differences between viruses.
Of particular interest in the field of molecular biology is the relationship between domain architecture and protein function. The detailed analysis of domain architectures presented here suggests studies that investigate the functional effects of removing or swapping domains in viral multidomain protein architectures The fact that Simplexvirus DNA polymerases contain the extra DNAPolymera_Pol domain and that this domain architecture is conserved among Simplexvirus isolates suggests that it may provide some unique function necessary for efficient replication of Simplexviruses. This hypothesis could be explored experimentally. Similarly, what would be the consequence of adding a C-terminal GlyL_C domain to the gL protein of VZV (which contains one Herpes_UL1 domain), and so making it similar to the gL protein found in HSV-1 and HSV-2 (which has a Herpes_UL1––GlyL_C architecture)?
Interestingly, while it has been noted that domain loss is an important mechanism in eukaryote evolution (probably equally—and possibly even more—important than domain gain) (Zmasek and Godzik, 2011); and references therein), in herpesvirus evolution domain loss seems to play a lesser role, as most of the events we were able to detect are domain gains (according to the parsimony principle).
Another implication of this work relates to the observation that in some cases proteins that share the same name are composed of either unrelated (e.g. gL) or very distantly related domains (e.g. DNA polymerase processivity factor) in different herpesvirus species. This raises the question - are such share named truly justified for proteins composed of unrelated domains? And to what extent has their putative shared function been experimentally validated.
Our approach is also expected to facilitate the detection and subsequent experimental study of species- (and strain-) specific proteins (listed in Supplementary Table 2). Whereas HSV1 and HSV2 do not have any species specific proteins given current data, VZV has six, and CMV has by far the most with 130 proteins which are not found in any other species. Interestingly, many of these 130 proteins are specific to one strain (or isolate) of CMV. Unsurprisingly, many of these species- and strain-specific protein do not yet have a Pfam domain (and thus were analyzed by manual BLAST searches in this work). An example of such a protein is the ORF45 protein of KSHV (Zhu and Yuan, 2003). Our automated approach provides a starting point for the systematic computational and experimental study of these species- and strain-specific proteins—studies, which eventually will provide answers to such questions as: Are these species- and strain-specitic proteins essential under certain conditions? Do they result in altered pathology or clinical symptoms? Do they function in host interaction? Do they possess as of yet undiscovered, but shared protein domains?
In summary, we developed a computational approach called Domain-architecture Aware Inference of Orthologs (DAIO) for the classification of viral proteins into groups of orthologous proteins with identical domain architecures (SOGs). In addition, we established a nomenclature for SOGs that provides the user with information about the biological function and taxonomic distribution for the member proteins of a SOG. We applied this classification and nomenclature to the proteomes of all human Herpesviridae species and made the results publicly accessible via the ViPR database. The acquisition and retention of novel domain architectures suggests that some Herpesviridae proteins may have acquired novel functional characteristics, which can now be explored experimentally.
4. Materials and methods
We developed a semi automated software pipeline to analyze amino acid sequences for their protein domain based architectures and to infer multiple sequence alignments and phylogenetic trees for the molecular sequences corresponding to these architectures, followed by gene duplication inference. This pipeline contains the following five major steps: (1) sequence retrival; (2) domain architecture anlysis, including the inference of the taxonomic distributions of domain architectures – each of which corresponding to one preliminary SOG, and manual naming of domain architecures/preliminary SOGs (to be automated in future versions of this pipeline); (3) extraction of molecular sequences corresponding to domain architectures/preliminary SOGs; (4) multiple sequence alignment and phylogenetic inference; (5) gene duplication inference, to determine which preliminary SOGs contain sequences related by gene duplications and thus need to divided in multiple, final SOGs. Links to all custom software programs developed for this work are available here: https://sites.google.com/site/cmzmasek/home/software/forester/daio. In the following the tools and methods used are described in more detail.
4.1. Sequence retrieval
Individual protein sequences were downloaded from the ViPR database (Pickett et al., 2012), while entire proteomes were downloaded from UniProtKB (Bateman et al., 2017).
4.2. Multiple sequence alignments
Multiple sequence alignments were calculated using MAFFT version 7.313 (with “localpair” and “maxiterate 1000” options) (Katoh and Standley, 2013, Kuraku et al., 2013). Prior to phylogenetic inference, multiple sequence alignment columns with more than 50% gaps were deleted. For comparison we also performed the analyses based on alignments for which we only deleted columns with more than 90% gaps.
4.3. Protein domain analysis
Protein domains were analyzed using hmmscan from HMMER v3.1b2 (Eddy, 2011) and the Pfam 31.0 database (Finn et al., 2016).
4.4. Phylogenetic analyses
Phylogenetic trees were calculated for individual domain architectures (not full-length sequences) except for US22 domain proteins, because US22 domain alignments lack phylogeneticly sufficient signal. Distance-based minimal evolution trees were inferred by FastME 2.0 (Desper and Gascuel, 2002) (with balanced tree swapping and “GME” initial tree options) based on pairwise distances calculated by TREE-PUZZLE 5.2 (Schmidt et al., 2002) using the WAG substitution model (Whelan and Goldman, 2001), a uniform model of rate heterogeneity, estimation of amino acid frequencies from the dataset, and approximate parameter estimation using a Neighbor-Joining tree. For maximum likelihood approaches, we employed RAxML version 8.2.9 (Stamatakis et al., 2005) (using 100 bootstrapped data sets and the WAG substitution model). Tree and domain composition diagrams were drawn using Archaeopteryx [https://sites.google.com/site/cmzmasek/home/software/forester]. Rooting was performed by the midpoint rooting method. Unless otherwise noted, Pfam domains are displayed ith a E = 10−6 cutoff. Gene duplication inferences were performed using the SDI and RIO methods (Zmasek and Eddy, 2002, Zmasek and Eddy, 2001). Automated genome wide domain composition analysis was performed using a specialized software tool, Surfacing version 2.002 [Zmasek CM (2012), a tool for the functional analysis of domainome/genome evolution [available at https://sites.google.com/site/cmzmasek/home/software/forester/surfacing]. All conclusions presented in this work are robust relative to the alignment methods, the alignment processing, the phylogeny reconstruction methods, and the parameters used. All sequence, alignment, and phylogeny files are available upon request.
4.5. Phylogenomic analyses and development of novel naming schema using strict ortholog groups
The processes for defining and naming strict ortholog groups were formalized into a set of “rules” and then implemented into a semi-automatic domain-centric phyloinformatics pipeline. Any unique arrangement of single or multiple Pfam domains is considered a domain architecture (DA) (Zmasek and Godzik, 2012, Zmasek and Godzik, 2011). Most proteins of members of the Herpesviridae have DAs consisting of only a single domain. For example, the UDG domain of uracil DNA glycosylase is a single domain DA, whereas the combination of N-terminal DNA_pol_B_exo1 and C-terminal DNA_pol_B (denoted as DNA_pol_B_exo1––DNA_pol_B) of DNA polymerases is a DA with two domains.
In this analysis, we consider a given DA “present” in a given Herpesviridae species S if the DA is present under a set of thresholds in at least one strain of the species S. The rationale for this is that it is possible to miss a DA in a genome, due to incomplete or erroneous sequences, erroneous assembly and gene-predication (false negatives), and even recent, actual gene loss. The opposite (false positive), on the other hand, is far less likely. For this work, we used two thresholds: a minimal domain length of 40% of the length set forth in the Pfam database (domain fragments are unlikely to be functionally equivalent to full length domains) and a hmmscan E-value cutoff of E = 10−6.
For every domain architecture, a set of bootstrap resampled phylogenetic trees (gene trees) was calculated by RAxML (Stamatakis et al., 2005) using protein sequences from one representative for each of the nine human Herpesviridae species. For comparison and validation, we also calculated phylogenetic trees that included non-human hosted Herpesviridae. For illustrations, gene duplications were inferred by comparing the consensus gene trees to the species tree (Fig. 1) for Herpesviridae using the SDI (Speciation Duplication Inference) algorithm (Zmasek and Eddy, 2001). To obtain confidence values on orthology assignments (bootstrap support values), we employed the RIO approach (Resampled Inference of Orthologs) to compare sets of bootstrap resampled phylogenetic trees with the species tree for Herpesviridae (Zmasek and Eddy, 2002).
In this work, we define a strict ortholog group (SOG) as sequences related by speciation events and exhibiting the same domain architecture (based on Pfam domains from Pfam 31.0, a length threshold of 40%, and E-value cutoff of E = 10−6).
Based on this approach for defining SOGs, we developed the following naming syntax.
For protein families such as uracil DNA glycosylase, which exhibit the same DA in all nine human Herpesviridae, and which are related by speciation events only, we base our names on (Mocarski and Edward, 2007) as the base name and add a case-sensitive suffix that indicates the taxonomic distribution - “ABG” in this case, since uracil DNA glycosylase appears in each human Alpha-, Beta-, and Gammaherpesvirinae species. Therefore, the full name is “uracil DNA glycosylase_ABG”. To indicate presence in some, but not all members of a subfamily, we use lower-case suffixes. “Replication origin-binding protein_ Ab” implies that members of this SOG are present in all human Alphaherpesvirinae species (“A”), and in some (but not all) Betaherpesvirinae (“b”).
While most of the human Herpesviridae protein families fall into these basic cases, families which have a (some) domain(s) in common but differ in their DA, are more difficult to rationally name. An example of such a family is glycoprotein B described above. Because members of this family have different DAs, namely “Glycoprotein_B” and “HCMVantigenic_N—Glycoprotein_B”, it is composed of two SOGs (named “Glycoprotein B_ ABG.AbG” and “Glycoprotein B_ ABG.b”). In such cases, we split the suffix into two parts, separated by a period. The first part (“ABG”) indicates overall presence of common domain(s) for all members of this SOG, Glycoprotein_B in this case. The second part (after the period) relates to entire DAs. “. AbG” of “Glycoprotein B_ ABG.AbG” means that the Glycoprotein_B DA is present in all human Alpha- and Gamma-, and some Betaherpesvirinae. “.b” of “Glycoprotein B_ ABG.b” implies that the “HCMVantigenic_N—Glycoprotein_B” DA is present in some Betaherpesvirinae.
Acknowledgements
The authors thank Sanjay Vashee for critical review of the manuscript. We also thank the primary data providers for sharing their data in public archives, including ViPR and UniProtKB. This work was funded by the National Institute of Allergy and Infectious Diseases (NIH/DHHS) under Contract no. HHSN272201400028C to RHS.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.virol.2019.01.005.
Appendix A. Supplementary material
Supplementary materialSupplementary Table 1. Strict Ortholog Groups. This table lists all Herpesviridae proteins that are found in at least two species. For every SOG, a suggested name, including the suffix indicating the taxonomic distribution (A, B, G, S, R, H6: present in all human Alpha-, Beta-, Gammaherpesvirinae, Simplexvirus, Roseolovirus, HHV-6A/HHV-6B respectively; lower-case letter indicate presence in some but not all species of the corresponding group), gene names (a forward slash is either part of the accepted gene name, or is used to separate multiple gene names, a comma indicates a species-specific expansion), the Pfam domain architecture, and the Pfam clan are shown.
Supplementary materialSupplementary Table 2. HumanHerpesviridaeproteins which only appear in one species.
References
- AlHajri S.M., Cunha C.W., Nicola A.V., Aguilar H.C., Li H., Taus N.S. Ovine herpesvirus 2 glycoproteins B, H, and L are sufficient for, and viral glycoprotein Ov8 can enhance, cell-cell membrane fusion. J. Virol. 2017;91 doi: 10.1128/JVI.02454-16. (e02454-16) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altenhoff A.M., Studer R.A., Robinson-Rechavi M., Dessimoz C. Resolving the ortholog conjecture: orthologs tend to be weakly, but significantly, more similar in function than paralogs. PLoS Comput. Biol. 2012;8:e1002514. doi: 10.1371/journal.pcbi.1002514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubert M., Blaho J.A. The herpes simplex virus type 1 regulatory protein ICP27 is required for the prevention of apoptosis in infected human cells. J. Virol. 1999;73:2803–2813. doi: 10.1128/jvi.73.4.2803-2813.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A., Martin M.J., O’Donovan C., Magrane M., Alpi E., Antunes R., Bely B., Bingley M., Bonilla C., Britto R., Bursteinas B., Bye-AJee H., Cowley A., Da Silva A., De Giorgi M., Dogan T., Fazzini F., Castro L.G., Figueira L., Garmiri P., Georghiou G., Gonzalez D., Hatton-Ellis E., Li W., Liu W., Lopez R., Luo J., Lussi Y., MacDougall A., Nightingale A., Palka B., Pichler K., Poggioli D., Pundir S., Pureza L., Qi G., Rosanoff S., Saidi R., Sawford T., Shypitsyna A., Speretta E., Turner E., Tyagi N., Volynkin V., Wardell T., Warner K., Watkins X., Zaru R., Zellner H., Xenarios I., Bougueleret L., Bridge A., Poux S., Redaschi N., Aimo L., ArgoudPuy G., Auchincloss A., Axelsen K., Bansal P., Baratin D., Blatter M.C., Boeckmann B., Bolleman J., Boutet E., Breuza L., Casal-Casas C., De Castro E., Coudert E., Cuche B., Doche M., Dornevil D., Duvaud S., Estreicher A., Famiglietti L., Feuermann M., Gasteiger E., Gehant S., Gerritsen V., Gos A., Gruaz-Gumowski N., Hinz U., Hulo C., Jungo F., Keller G., Lara V., Lemercier P., Lieberherr D., Lombardot T., Martin X., Masson P., Morgat A., Neto T., Nouspikel N., Paesano S., Pedruzzi I., Pilbout S., Pozzato M., Pruess M., Rivoire C., Roechert B., Schneider M., Sigrist C., Sonesson K., Staehli S., Stutz A., Sundaram S., Tognolli M., Verbregue L., Veuthey A.L., Wu C.H., Arighi C.N., Arminski L., Chen C., Chen Y., Garavelli J.S., Huang H., Laiho K., McGarvey P., Natale D.A., Ross K., Vinayaka C.R., Wang Q., Wang Y., Yeh L.S., Zhang J. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017;45:D158–D169. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges K.G., Hua Q., Brigham-Burke M.R., Martin J.D., Hensley P., Dahl C.E., Digard P., Weiss M.A., Coen D.M. Secondary structure and structure-activity relationships of peptides corresponding to the subunit interface of herpes simplex virus DNA polymerase. J. Biol. Chem. 2000;275:472–478. doi: 10.1074/jbc.275.1.472. [DOI] [PubMed] [Google Scholar]
- Cai W.H., Gu B., Person S. Role of glycoprotein B of herpes simplex virus type 1 in viral entry and cell fusion. J. Virol. 1988;62:2596–2604. doi: 10.1128/jvi.62.8.2596-2604.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casarosa P., Bakker R.A., Verzijl D., Navis M., Timmerman H., Leurs R., Smiti M.J. Constitutive signaling of the human cytomegalovirus-encoded chemokine receptor US28. J. Biol. Chem. 2001;276:1133–1137. doi: 10.1074/jbc.M008965200. [DOI] [PubMed] [Google Scholar]
- Casarosa P., Gruijthuijsen Y.K., Michel D., Beisser P.S., Holl J., Fitzsimons C.P., Verzijl D., Bruggeman C.A., Mertens T., Leurs R., Vink C., Smit M.J. Constitutive signaling of the human cytomegalovirus-encoded receptor UL33 differs from that of its rat cytomegalovirus homolog R33 by promiscuous activation of G proteins of the Gq, Gi, and GsClasses. J. Biol. Chem. 2003;278:50010–50023. doi: 10.1074/jbc.M306530200. [DOI] [PubMed] [Google Scholar]
- Chen R., Wang H., Mansky L.M. Roles of uracil-DNA glycosylase and dUTPase in virus replication. J. Gen. Virol. 2002;83:2339–2345. doi: 10.1099/0022-1317-83-10-2339. [DOI] [PubMed] [Google Scholar]
- Chen X., Zhang J. The ortholog conjecture is untestable by the current gene ontology but is supported by RNA sequencing data. PLoS Comput. Biol. 2012;8:e1002784. doi: 10.1371/journal.pcbi.1002784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai-Ju J.Q., Li L., Johnson L.A., Sandri-Goldin R.M. ICP27 interacts with the C-terminal domain of RNA polymerase II and facilitates its recruitment to herpes simplex virus 1 transcription sites, where it undergoes proteasomal degradation during infection. J. Virol. 2006;80:3567–3581. doi: 10.1128/JVI.80.7.3567-3581.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison A.J. Herpesvirus systematics. Vet. Microbiol. 2010;143:52–69. doi: 10.1016/j.vetmic.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison A.J. Evolution of the herpesviruses. Vet. Microbiol. 2002 doi: 10.1016/S0378-1135(01)00492-8. [DOI] [PubMed] [Google Scholar]
- Desper R., Gascuel O. Fast and accurate phylogeny minimum-evolution principle. J. Comput. Biol. 2002;9:687–705. doi: 10.1089/106652702761034136. [DOI] [PubMed] [Google Scholar]
- Digard P., Williams K.P., Hensley P., Brooks I.S., Dahl C.E., Coen D.M. Specific inhibition of herpes simplex virus DNA polymerase by helical peptides corresponding to the subunit interface. Proc. Natl. Acad. Sci. USA. 1995;92:1456–1460. doi: 10.1073/pnas.92.5.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy S.R. Accelerated profile HMM searches. PLoS Comput. Biol. 2011;7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen J.A. Phylogenomics: improving functional predictions for uncharacterized genes by evolutionary analysis. Genome Res. 1998;8:163–167. doi: 10.1101/gr.8.3.163. [DOI] [PubMed] [Google Scholar]
- Finn R.D., Coggill P., Eberhardt R.Y., Eddy S.R., Mistry J., Mitchell A.L., Potter S.C., Punta M., Qureshi M., Sangrador-Vegas A., Salazar G.A., Tate J., Bateman A. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016;44:D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitch W.M. Homology. Trends Genet. 2000;16:227–231. doi: 10.1016/s0168-9525(00)02005-9. [DOI] [PubMed] [Google Scholar]
- Fitch W.M. Distinguishing homologous from analogous proteins. Syst. Zool. 1970;19:99–113. doi: 10.2307/2412448. [DOI] [PubMed] [Google Scholar]
- Forrester A., Farrell H., Wilkinson G., Kaye J., Davis-Poynter N., Minson T. Construction and properties of a mutant of herpes simplex virus type 1 with glycoprotein H coding sequences deleted. J. Virol. 1992;66:341–348. doi: 10.1128/jvi.66.1.341-348.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Diaz M., Bebenek K. Multiple functions of DNA polymerases. CRC Crit. Rev. Plant Sci. 2007;26:105–122. doi: 10.1021/nl061786n.Core-Shell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson L.K., Dalton B.L., Karabekian Z., Farrell H.E., Rawlinson W.D., Stenberg R.M., Campbell A.E. Transcriptional analysis of the murine cytomegalovirus HindIII-I region: identification of a novel immediate-early gene region. Virology. 1999;260:156–164. doi: 10.1006/viro.1999.9796. [DOI] [PubMed] [Google Scholar]
- Isegawa Y., Ping Z., Nakano K., Sugimoto N., Yamanishi K. Human herpesvirus 6 open reading frame U12 encodes a functional beta-chemokine receptor. J. Virol. 1998;72:6104–6112. doi: 10.1128/jvi.77.14.8108-8115.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh M., Nacher J.C., Kuma K., Goto S., Kanehisa M. Evolutionary history and functional implications of protein domains and their combinations in eukaryotes. Genome Biol. 2007;8:R121. doi: 10.1186/gb-2007-8-6-r121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen R.A. Orthologs and paralogs - we need to get it right. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-8-interactions1002. (INTERACTIONS1002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krusong K., Carpenter E.P., Bellamy S.R.W., Savva R., Baldwin G.S. A comparative study of uracil-DNA glycosylases from human and herpes simplex virus type 1. J. Biol. Chem. 2006;281:4983–4992. doi: 10.1074/jbc.M509137200. [DOI] [PubMed] [Google Scholar]
- Kuraku S., Zmasek C.M., Nishimura O., Katoh K. aLeaves facilitates on-demand exploration of metazoan gene family trees on MAFFT sequence alignment server with enhanced interactivity. Nucleic Acids Res. 2013;41:W22–W28. doi: 10.1093/nar/gkt389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligas M.W., Johnson D.C. A herpes simplex virus mutant in which glycoprotein D sequences are replaced by beta-galactosidase sequences binds to but is unable to penetrate into cells. J. Virol. 1988;62:1486–1494. doi: 10.1128/jvi.62.5.1486-1494.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Roizman B. Characterization of the protease and other products of amino-terminus-proximal cleavage of the herpes simplex virus 1 UL26 protein. J. Virol. 1993;67:1300–1309. doi: 10.1128/jvi.67.3.1300-1309.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loregian A., Papini E., Satin B., Marsden H.S., Hirst T.R., Palu G. Intranuclear delivery of an antiviral peptide mediated by the B subunit of Escherichia coli heat-labile enterotoxin. Proc. Natl. Acad. Sci. USA. 1999;96:5221–5226. doi: 10.1073/pnas.96.9.5221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loregian A., Piaia E., Cancellotti E., Papini E., Marsden H.S., Palù G. The catalytic subunit of herpes simplex virus type 1 DNA polymerase contains a nuclear localization signal in the UL42-binding region. Virology. 2000;273:139–148. doi: 10.1006/viro.2000.0390. [DOI] [PubMed] [Google Scholar]
- Malik P., Tabarraei A., Kehlenbach R.H., Korfali N., Iwasawa R., Graham S.V., Schirmer E.C. Herpes simplex virus ICP27 protein directly interacts with the nuclear pore complex through Nup62, inhibiting host nucleocytoplasmic transport pathways. J. Biol. Chem. 2012;287:12277–12292. doi: 10.1074/jbc.M111.331777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGeoch D.J., Cook S., Dolan A., Jamieson F.E., Telford E.A.R. Molecular phylogeny and evolutionary timescale for the family of Mammalian Herpesviruses. J. Mol. Biol. 1995;2:443–458. doi: 10.1006/jmbi.1995.0152. [DOI] [PubMed] [Google Scholar]
- McGeoch D.J., Dolan A., Ralph A.C. Toward a comprehensive phylogeny for Mammalian and Avian Herpesviruses. J. Virol. 2000;74:10401–10406. doi: 10.1128/JVI.74.22.10401-10406.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocarski E.S. Human Herpesviruses: Biology, Therapy and Immunoprophylaxis. Cambridge University Press; 2007. Comparative analysis of herpesvirus-common proteins. [PubMed] [Google Scholar]
- Montague M.G., Hutchison C.A. Gene content phylogeny of herpesviruses. Proc. Natl. Acad. Sci. USA. 2000;97:5334–5339. doi: 10.1073/pnas.97.10.5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore A.D., Björklund Å.K., Ekman D., Bornberg-Bauer E., Elofsson A. Arrangements in the modular evolution of proteins. Trends Biochem. Sci. 2008;33:444–451. doi: 10.1016/j.tibs.2008.05.008. [DOI] [PubMed] [Google Scholar]
- Murphy P.M. Viral exploitation and subversion of the immune system through chemokine mimicry. Nat. Immunol. 2001;2:116–122. doi: 10.1038/84214. [DOI] [PubMed] [Google Scholar]
- Nehrt N.L., Clark W.T., Radivojac P., Hahn M.W. Testing the ortholog conjecture with comparative functional genomic data from mammals. PLoS Comput. Biol. 2011:7. doi: 10.1371/journal.pcbi.1002073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patthy L. Modular assembly of genes and the evolution of new functions. Genetica. 2003;118:217–231. [PubMed] [Google Scholar]
- Peisajovich S.G., Garbarino J.E., Wei P., Lim W.A. Rapid diversification of cell signaling phenotypes by modular domain recombination. Science. (80-.). 2010;328:368–372. doi: 10.1126/science.1182376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellet, P., Roizman, B., 2007. Herpesviridae: a brief introduction. In: Howley, P. (Ed.), Fields Virology. Philadelphia, pp. 2480–2499.
- Pickett B.E., Sadat E.L., Zhang Y., Noronha J.M., Squires R.B., Hunt V., Liu M., Kumar S., Zaremba S., Gu Z., Zhou L., Larson C.N., Dietrich J., Klem E.B., Scheuermann R.H. ViPR: an open bioinformatics database and analysis resource for virology research. Nucleic Acids Res. 2012;40:593–598. doi: 10.1093/nar/gkr859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pignatelli S., Dal Monte P., Rossini G., Landini M.P. Genetic polymorphisms among human cytomegalovirus (HCMV) wild-type strains. Rev. Med. Virol. 2004;14:383–410. doi: 10.1002/rmv.438. [DOI] [PubMed] [Google Scholar]
- Remm M., Storm C.E.V., Sonnhammer E.L.L. Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J. Mol. Biol. 2001;314:1041–1052. doi: 10.1006/jmbi.2000.5197. [DOI] [PubMed] [Google Scholar]
- Rogozin I.B., Managadze D., Shabalina S.A., Koonin E.V. Gene family level comparative analysis of gene expression n mammals validates the ortholog conjecture. Genome Biol. Evol. 2014;6:754–762. doi: 10.1093/gbe/evu051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roop C., Hutchinson L., Johnson D.C. A mutant herpes simplex virus type 1 unable to express glycoprotein L cannot enter cells, and its particles lack glycoprotein H. J. Virol. 1993;67:2285–2297. doi: 10.1128/jvi.67.4.2285-2297.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt H.A., Strimmer K., Vingron M., von Haeseler A. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics. 2002;18:502–504. doi: 10.1093/bioinformatics/18.3.502. [DOI] [PubMed] [Google Scholar]
- Sciabica K.S., Dai Q.J., Sandri-Goldin R.M. ICP27 interacts with SRPK1 to mediate HSV splicing inhibition by altering SR protein phosphorylation. EMBO J. 2003;22:1608–1619. doi: 10.1093/emboj/cdg166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severini A., Tyler S.D., Peters G.A., Black D., Eberle R. Genome sequence of a chimpanzee herpesvirus and its relation to other primate alphaherpesviruses. Arch. Virol. 2013;158:1825–1828. doi: 10.1515/jci-2013-0007.Targeted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu S.Y.W., Chan K.M., Lo S.K.F., Ip K.W.Y., Yuen K.Y., Heath R.B. Sequence variation of the amino-terminal antigenic domains of glycoprotein B of human cytomegalovirus strains isolated from Chinese patients. Arch. Virol. 1994;137:133–138. doi: 10.1007/BF01311179. [DOI] [PubMed] [Google Scholar]
- Spear P.G., Longnecker R. Herpesvirus entry: an update. J. Virol. 2003;77:10179–10185. doi: 10.1128/JVI.77.19.10179-10185.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiess K., Fares S., Sparre-Ulrich A.H., Hilgenberg E., Jarvis M.A., Ehlers B., Rosenkilde M.M. Identification and functional comparison of Seven-transmembrane G-protein-coupled BILF1 receptors in recently discovered Nonhuman primate Lymphocryptoviruses. J. Virol. 2015;89:2253–2267. doi: 10.1128/JVI.02716-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A., Ludwig T., Meier H. RAxML-III: a fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics. 2005;21:456–463. doi: 10.1093/bioinformatics/bti191. [DOI] [PubMed] [Google Scholar]
- Tatusov R.L., Koonin E.V., Lipman D.J. A genomic perspective on protein families. Science (80-.). 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- Tunnicliffe R.B., Hautbergue G.M., Kalra P., Jackson B.R., Whitehouse A., Wilson S.A., Golovanov A.P. Structural basis for the recognition of cellular mRNA export factor REF by herpes viral proteins HSV-1 ICP27 and HVS ORF57. PLoS Pathog. 2011;7:20–22. doi: 10.1371/journal.ppat.1001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villarreal L.P., DeFilippis V.R. A hypothesis for DNA viruses as the origin of eukaryotic replication proteins. J. Virol. 2000;74:7079–7084. doi: 10.1128/jvi.74.15.7079-7084.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virus Taxonomy: The Classification and Nomenclature of Viruses The Online (10th) Report of the ICTV, 2017. [WWW Document]. URL 〈https://talk.ictvonline.org/ictv-reports/ictv_online_report/〉.
- Weisshart K., Chow C.S., Coen D.M. Herpes simplex virus processivity factor UL42 imparts increased DNA-binding specificity to the viral DNA polymerase and decreased dissociation from primer-template without reducing the elongation rate. J. Virol. 1999;73:55–66. doi: 10.1128/JVI.01174-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan S., Goldman N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 2001;18:691–699. doi: 10.1093/oxfordjournals.molbev.a003851. [DOI] [PubMed] [Google Scholar]
- Ye Y., Godzik A. Comparative analysis of protein domain organization. Genome Res. 2004;14:343–353. doi: 10.1101/gr.1610504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Iyer L.M., Aravind L. A novel immunity system for bacterial nucleic acid degrading toxins and its recruitment in various eukaryotic and DNA viral systems. Nucleic Acids Res. 2011;39:4532–4552. doi: 10.1093/nar/gkr036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. Evolution by gene duplication: an update. Trends Ecol. Evol. 2003;18:292–298. doi: 10.1016/S0169-5347(03)00033-8. [DOI] [Google Scholar]
- Zhen Z., Bradel-Tretheway B., Sumagin S., Bidlack J.M., Dewhurst S. The human herpesvirus 6 G protein-coupled receptor homolog U51 positively regulates virus replication and enhances cell-cell fusion in vitro. J. Virol. 2005;79:11914–11924. doi: 10.1128/JVI.79.18.11914-11924.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C., Knipe D.M. Association of herpes simplex virus type 1 ICP8 and ICP27 proteins with cellular RNA polymerase II holoenzyme. J. Virol. 2002;76:5893–5904. doi: 10.1128/JVI.76.12.5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F.X., Yuan Y. The ORF45 protein of Kaposi’s sarcoma-associated herpesvirus is associated with purified virions. J. Virol. 2003;77:4221–4230. doi: 10.1128/JVI.77.7.4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Z., Ai Y. Processivity factor of DNA polymerase and its expanding role in normal and translesion DNA synthesis. Biochim. Biophys. Acta - Proteins Proteom. 2010;1804:1081–1093. doi: 10.1016/j.bbapap.2009.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zmasek C.M., Eddy S.R. RIO: analyzing proteomes by automated phylogenomics using resampled inference of orthologs. BMC Bioinforma. 2002;3:14. doi: 10.1186/1471-2105-3-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zmasek C.M., Eddy S.R. A simple algorithm to infer gene duplication and speciation events on a gene tree. Bioinformatics. 2001;17:821–828. doi: 10.1093/bioinformatics/17.9.821. [DOI] [PubMed] [Google Scholar]
- Zmasek C.M., Godzik A. This Déjà Vu feeling—analysis of multidomain protein evolution in Eukaryotic genomes. PLoS Comput. Biol. 2012;8:e1002701. doi: 10.1371/journal.pcbi.1002701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zmasek C.M., Godzik A. Strong functional patterns in the evolution of eukaryotic genomes revealed by the reconstruction of ancestral protein domain repertoires. Genome Biol. 2011;12:R4. doi: 10.1186/gb-2011-12-1-r4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuccola H.J., Filman D.J., Coen D.M., Hogle J.M. The crystal structure of an unusual processivity factor, herpes simplex virus UL42, bound to the C terminus of its cognate polymerase. Mol. Cell. 2000;5:267–278. doi: 10.1016/S1097-2765(00)80422-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary materialSupplementary Table 1. Strict Ortholog Groups. This table lists all Herpesviridae proteins that are found in at least two species. For every SOG, a suggested name, including the suffix indicating the taxonomic distribution (A, B, G, S, R, H6: present in all human Alpha-, Beta-, Gammaherpesvirinae, Simplexvirus, Roseolovirus, HHV-6A/HHV-6B respectively; lower-case letter indicate presence in some but not all species of the corresponding group), gene names (a forward slash is either part of the accepted gene name, or is used to separate multiple gene names, a comma indicates a species-specific expansion), the Pfam domain architecture, and the Pfam clan are shown.
Supplementary materialSupplementary Table 2. HumanHerpesviridaeproteins which only appear in one species.