Abstract
Mastitis is a major challenge to bovine health. The detection of sensitive markers for mastitis in dairy herds is of great demand. Suitable biomarkers should be measurable in milk, and should report pathogen specific changes at an early stage to support earlier diagnosis and more efficient treatment. However, the identification of sensitive biomarkers in milk has remained a challenge, in part due to their relatively low concentration in milk. In the present study, we used a selected reaction monitoring (SRM) mass spectrometry approach, which allowed for the first time the absolute quantitation of 13 host response proteins in milk. These proteins were measured during a 54 h time-course upon an in vivo challenge with cell wall components from either gram-negative (lipopolysaccharide from Escherichia coli; LPS) or gram-positive bacteria (peptidoglycan from Staphylococcus aureus; PGN). While our data clearly demonstrate that all challenged animals have consistent upregulation of innate immune response proteins after both LPS and PGN challenge, they also reveal clearly that LPS challenge unleashes faster and shows a more intense host response compared to PGN challenge. Biomarker candidates that may distinguish between gram-negative and gram-positive bacteria include A2M, A1AT, HP, SAA3, CD14, CALGB, CATC, VAN1, LGALS1, LGALS3 and IL-8. Our approach can support further studies of large cohorts of animals with natural occurring mastitis, in order to validate the relevance of these suggested biomarkers in dairy production.
Keywords: mastitis, selected reaction monitoring (SRM), host response proteins, lipopolysaccharide (LPS), peptidoglycan (PGN)
Graphical Abstract
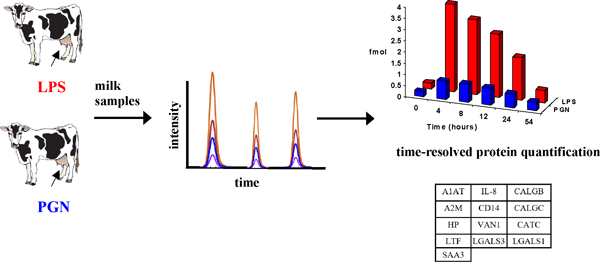
Interpretive summary:
Selected Reaction Monitoring mass spectrometry reveals that regulation of bovine host response proteins depends on the type of invading mastitis pathogen. By Kusebauch et al.
Mastitis is a major challenge to the dairy industry. Markers detectable in milk which support early recognition of causal mastitis pathogens are in great demand. We used a selected reaction monitoring mass spectrometry approach to evaluate changes in 13 candidate markers for mastitis caused by Escherichia coli or Staphylococcus aureus. All proteins in this study increased in concentration after challenge with either LPS (mimicking Escherichia coli mastitis) or PGN (mimicking Staphylococcus aureus mastitis). Eleven of these proteins increased dramatically more after challenge with LPS compared to PGN, and are therefore potential markers for mastitis surveillance in the dairy industry.
INTRODUCTION
Mastitis, an inflammatory reaction of the mammary gland, represents a major challenge to animal health in the dairy farming industry, compromising both animal welfare and economic gain (Hogeveen et al., 2011). Escherichia coli (E. coli) and Staphylococcus aureus (S. aureus) are two of the major causing pathogens in the mammary gland leading different progressions of pathology (Bannerman, 2008, Schukken et al., 2011, Wall et al., 2016a). Mastitis caused by E. coli is usually associated to acute inflammation and severe clinical symptoms whereas S. aureus infections mainly result in chronic and subclinical infections, (Yang et al., 2008, Hernandez-Castellano et al., 2017a). The innate immune system responds to the earliest stages of the infection and relies on cell surface receptors that recognize conserved microbial molecules (Schukken et al., 2011), as well as soluble components such as acute phase proteins and cytokines which are secreted into blood and milk (Aitken et al., 2011).
Regulation of bovine host response proteins is not fully understood, but accumulating evidence suggests that host response is specifically tailored to different types of mastitis causing pathogens (Bannerman, 2008, Griesbeck-Zilch et al., 2008, Yang et al., 2008). Hence, the abundance of specific host response proteins secreted in milk may provide information for early monitoring of mastitis pathogens. Validated markers could be integrated in automated milking systems to provide routine surveillance of animal health, and help to improve detection, diagnosis and treatment of mastitis in dairy herds. Nonetheless, accurate quantification of specific proteins in milk is still technically challenging. Shotgun proteomics has been applied to identify mastitis biomarkers in bovine milk (Danielsen et al., 2010, Boehmer, 2011). However, these studies have failed to report low abundant proteins related to the immune response such as cytokines and their receptors. This is partly due to the stochastic nature of the precursor selection in shotgun mass spectrometry which is biased towards the more abundant proteins in complex biological samples like milk, blood, and tissues (Domon and Aebersold, 2010, Hernández-Castellano et al., 2014; 2015). Selected reaction monitoring (SRM) by MS was developed to provide increased sensitivity, dynamic range and reproducibility of protein analyses (Picotti and Aebersold, 2012). The SRM approach provides absolute quantification of selected protein-specific peptides by paired correlation to stable-isotope labeled standards, and is currently the method of choice for sub-femtomolar quantification of specific protein biomarkers across diverse biological samples including mammalian tissues and body fluids (Ebhardt et al., 2015, Hernández-Castellano et al., 2016, Kusebauch et al., 2016).
Lipopolysaccharides (LPS) have been used in several studies to mimick intra-mammary E. coli mastitis in vivo as well as in cultured mammary epithelial cells (Burvenich et al., 2003, Vels et al., 2009, Wall et al., 2016b). Similarly, peptidoglycans (PGN) have been used to mimick intramammary S. aureus mastitis, but so far only studied in cultured bovine mammary epithelial cells (Mount et al., 2009, Im et al., 2014, Sulabh et al., 2016), and the effect of PGN on expression of host response proteins in live animals remains uncharacterized.
Based on these considerations, the aim of this study was to investigate a time-resolved and parallel monitoring of bovine host response to LPS and PGN, measured by the absolute quantity and concentration dynamics of 13 specific host response proteins in milk.
MATERIAL AND METHODS
Animals and treatments
Six healthy Danish Holstein-Friesian cows in mid-lactation were randomly selected for two parallel intramammary challenge experiments. In this study, three cows were inoculated with LPS (LPS O111:B4, Sigma Aldrich A/S, Brøndby, Denmark) and three cows were similarly inoculated with PGN (Sigma Aldrich A/S). Infusions were performed in one quarter with 10 mL of sterile 0.9 % NaCl containing either 20 µg LPS/mL or PGN/mL (total dose of 200 µg) 3 h after the morning milking following procedures described in Vels et al. (2009). Before the infusions, milk somatic cell counts (SCC) were measured in all quarters, resulting in SCC <100,000 cells/mL. The SCC was determined directly upon sampling according to Fogsgaard et al. (2012) and Torres et al. (2014) using a DeLaval somatic cell counter (DeLaval International AB, Tumba, Sweden).
All procedures and experimental handling of animals were approved by the Danish Animal Experiments Inspectorate (license no 210601–075, J.no. 2001/561–410) and conducted in the dairy barn facilities at the Department of Animal Science, Aarhus University, Denmark.
Milk samples
Milk samples were collected from the infused quarter as well as from an unchallenged, negative control quarter at six different time points from each cow: 0, 4, 8, 12, 24 and 54 h after LPS and PGN inoculations. The SCC was determined directly upon sampling as described above. Milk samples were filtered using a 100 μm nylon mesh (Sefar, AG, Switzerland) cut to fit a 6 cm polystyrene funnel, and stored at −20 °C until further processing. Milk samples were thawed at 37 °C to ensure homogenization of milk fat. The casein proteins were precipitated by adjusting the pH to 4.6 by adding 0.06 % trifluoroacetic acid (TFA) to the milk (9:1), followed by centrifugation at 15,000 x g for 10 min. After that, the milk whey fraction (supernatant) was collected for further analysis. Protein concentration was determined using the Pierce BCA Protein Kit (VWR - Bie & Berntsen A/S, Herlev, Denmark) with bovine serum albumin (BSA) as standard, according to manufacturer’s instructions.
Digestion of milk proteins
Each whey sample (200 µg protein) was precipitated using ice-cold acetone and resuspended in 19 µL digestion buffer (0.5 M Triethylammonium bicarbonate (TEAB), 0.1 % (v/v) SDS). Protein disulfide bonds were reduced with 1 μl of 50 mM Tris (2-carboxyethyl)phosphine hydrochloride (TCEP-HCl) to reach a final concentration of 2.5 mM TCEP-HCl, and incubated at 60 ºC for 1 h. Proteins were then alkylated by adding 1 μl of a 200 mM iodo acetic acid (IAA) and samples incubated at room temperature for 10 min in the dark. Proteins were digested with trypsin (1:10 W/W) (SCIEX, Framingham, MA, USA) at 37 ºC, 16 h. Samples were passed through a centrifugal filter (pore size 0.2 µm) (VWR International, West Chester, PA, USA) for 10 min at 10,000 x g and dried down in a vacuum centrifuge.
Mixed-mode cation exchange (MCX) purification of milk peptides for SRM analysis
Dried peptides (50 µg) were dissolved in 2 % (v/v) acetonitrile (ACN), 0.1 % (v/v) formic acid (pH < 3) and purified with mixed-mode ion-exchange. An Oasis MCX µElution Plate (Waters, Milford, MA, USA) was preconditioned with 800 µL methanol and equilibrated with 3 × 800 µL 0.1 % (v/v) TFA in water before peptide samples were loaded on the plate, followed by a washing step with 2 × 800 µL 0.1 % TFA in water and 2 × 80 % (v/v) ACN/0.1 % (v/v) TFA in water. Peptides were eluted with 2 × 800 µL 10 % (v/v) NH4OH/90 % (v/v) methanol. The eluate was evaporated to dryness in a speed vacuum centrifuge and stored at −80 °C.
SRM assays
The SRM method used in this study was previously developed in our laboratory with the aim to quantify candidate markers for host response directly from crude tissue and body fluid samples as described in detail by Bislev et al. (2012). A panel of 13 host response proteins was selected: five acute phase proteins including lactoferrin (LTF), alpha-2 macroglobulin (A2M), alpha-1 antitrypsin (A1AT), haptoglobin (HP) and serum amyloid A3 (SAA3), and eight proteins previously associated with the onset of bovine host response to mastitis including cluster of differentiation 14 (CD14), calgranulin B (CALGB, S100-A9), calgranulin C (CALGC, S100-A12), cathepsin C (CATC), vanin-1 (VAN1), galectin-1 (LGALS1), galectin-3 (LGALS3) and interleukin 8 (IL-8). In the current study, we measured two proteotypic tryptic peptides from each protein. Detailed criteria for selecting the target peptides are described in Bislev et al. (2012). For the present study we used isotope labelled reference peptides generated by chemical synthesis. These peptides have the same physiochemical properties as the reference peptides generated by the QconCAT approach used in Bislev et al (2012), the origin of reference peptides does not affect the quantification method.
Isotope labeled heavy standard peptides
Isotope heavy labeled standard peptides corresponding to the 26 targeted proteotypic peptides were purchased as Heavy Peptides AQUA QuantPro™ incorporating stable-isotope-labeled amino acids, either [13C6/15N2]Lys or [13C6/15N4]Arg (Thermo Scientific, Waltham, MA, USA) with a concentration of 5 pmol/µL in 5 % (v/v) ACN/water. Peptides were pooled to prepare an internal standard solution of 125 fmol/µL of each peptide.
Liquid Chromatography (LC) and SRM analysis of milk samples
The digested and dried protein extracts were reconstituted in 2 % (v/v) ACN, 0.1 % (v/v) formic acid to 2 µg/µL and mixed with the heavy labeled internal standard solution in a 1:1 ratio. For each LC-SRM analysis 2 µL of sample were injected, corresponding to 125 fmol of each standard peptide and representing the peptide equivalents of 2 µg milk protein. Samples were analyzed with a 5500 QTRAP LC-MS system equipped with a NanoSpray Source III and an Eksigent Tempo™ nano MDLC system (Sciex Framingham, MA, USA). Peptides were loaded on a peptide captrap column (Microm Bioresources Inc., Auburn, CA, USA) in 0.1 % (v/v) formic acid in water for 5 min at a flow rate of 5 µL/min and separated by a 70 min linear gradient from 2–25 % (v/v) acetonitrile, 0.1 % (v/v) formic acid in water at a flow rate of 300 nL/min with an Acclaim PepMap 100 C18 analytical column (0.75 × 150 mm, 3 µm particles, Dionex, Sunnyvale, CA, USA). Peptides were analyzed in scheduled SRM mode with Q1 and Q3 set to unit resolution, a 2 s cycle time, a 360 s retention time window, a declustering potential of 70 and an interface heater temperature of 150 °C. As described in Bislev et al. (2012), each peptide sequence was targeted by ten transitions, five transitions for the endogenous light peptide and five transitions for the heavy labeled standard. All 36 milk samples were measured in triplicate (technical replicates), 108 LC-SRM analyses total.
SRM data analysis
SRM data were processed using Skyline (ver. 2.5) (https://skyline.gs.washington.edu, MacLean et al., 2010). Data were manually inspected to ensure correct peak detection and accurate integration. SRM data and transitions used to measure these proteins were deposited in the PASSEL data repository and are available at http://www.peptideatlas.org/PASS/PASS01014. Individual protein concentrations were calculated as the mean of the three signal intensities from the two peptides from each protein (six intensities per protein).
Statistical analysis
All results are presented as least squares means ± SEM. Statistical analyses were performed using SAS software (version 9.4, SAS Institute Inc., Cary, NC). The MIXED procedure was used to evaluate changes during the experimental period and differences between the experimental groups. The final model included treatment (LPS vs. PGN), experimental period (0, 4, 8, 12, 24 and 54 h) and the interaction between both as dependent variables and cow as repeated subject. The concentration of each of the 13 targeted proteins μg/mL or mg/mL) in the original milk samples was calculated from the relative signal intensities (light/heavy), given by spiking 125 fmol of each AQUA standard peptide into each of the 36 prepared milk samples. Finally, a Tukey-Kramer test was used to evaluate differences between groups. Values were considered significant when P < 0.05.
RESULTS
Somatic cell counts increase in quarters challenged with LPS and PGN
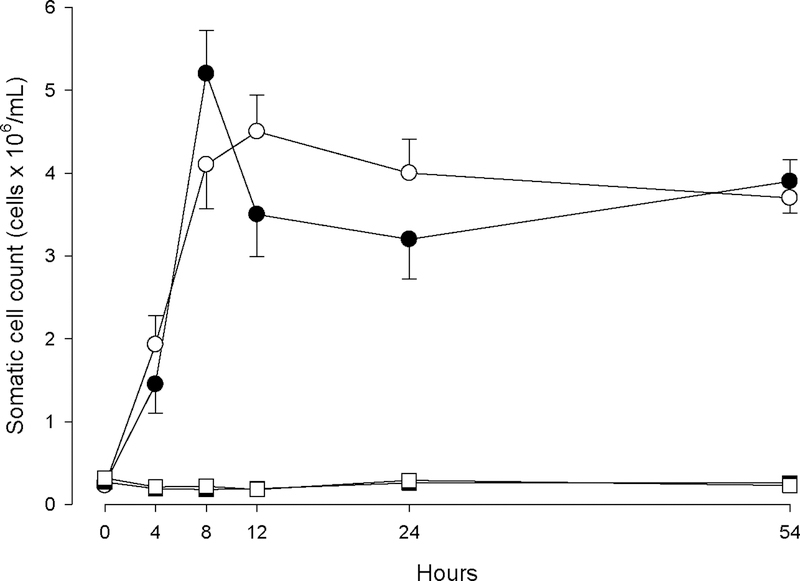
Somatic cell counts were measured in order to monitor extent and timing of host response to LPS and PGN challenge. Figure 1 shows the rise of SCC in challenged quarters with LPS or PGN, and also clearly demonstrates that the respective control quarters remain healthy. Milk SCC was significantly increased at 4 h (P<0.05) in all quarters challenged with either LPS or PGN, and showed the highest SCC at 8 h for LPS (P<0.05) and at 12 h for PGN (P<0.05). During the entire experimental period control quarters remained unchanged (P>0.05).
Figure 1.
Somatic cell count of quarters treated with either LPS (n=3; ●) or PGN (n=3; ○), as well as the control-LPS (n=3;■) and control-PGN (n=3; □) quarters during the experimental period.
SRM analysis of milk samples
All 26 isotope labeled standard peptides (two peptides per protein) were readily detected in milk and therefore support quantification of the 13 targeted host response proteins. Ten of these 13 proteins were robustly quantified in all 36 milk samples by the detection of the two selected proteotypic peptides, while three proteins, namely CALGC, A2M and LGALS1, could only be quantified by one peptide sequence due to the very low endogenous signal intensity of the second targeted peptide from each of these three proteins. The 13 targeted proteins are listed in Table 1 together with their corresponding peptides and measured peptide ions. Supplementary Table 1 provides the concentration of the 13 host response proteins measured in milk that has been collected at 0, 4, 8, 12, 24, and 54 h post challenge.
Table 1.
Target proteins, their corresponding two proteotypic peptides and transitions used in the SRM analysis. Q3 ions that passed the manual verification step and used in this study for quantification are highlighted in bold. Q3 ions are named according to the precursor charge state and fragment ion position in the y ion series; hence 2y6 refers to a y6 fragment ion derived from the doubly charged precursor ion.
| Protein name | Protein abbreviation | Peptide sequence | Transitions |
|---|---|---|---|
| Alpha-1 antitrypsin | A1AT | ADLSGITK | 2y5;2y6;2y7;3y3;3y4 |
| Alpha-1 antitrypsin | A1AT | LSISETYDLK | 2y5;2y6;2y7;2y8;2y9 |
| Alpha 2 macroglobulin | A2M | IQHHTLLASPVR | 2y8;3y5;3y6;3y7;3y8 |
| Alpha 2 macroglobulin | A2M | LPPNVVEESAR | 2y6;2y7;2y8;2y9;3y5 |
| Calgranulin B (S100A9) | CALGB | ELPNFLK | 2y4;2y5;2y6;3y3;3y4 |
| Calgranulin B (S100A9) | CALGB | LGHYDTLIQK | 2y6;2y7;3y4;3y5;3y6 |
| Calgranulin C (S100A12) | CALGC | DQPTIDK | 2y4;2y5;2y6;3y3;3y4 |
| Calgranulin C (S100A12) | CALGC | VGHFDTLNK | 2y5;2y6;3y3;3y4;3y5 |
| Cathepsin C | CATC | NVHGINFVTPVR | 2y7;2y9;3y4;3y5;3y6 |
| Cathepsin C | CATC | TGNTSENVNVNTAR | 2y10;2y11;2y7;2y8;2y9 |
| CD14 | CD14 | LGAAQVPAQLLVAVLR | 2y11;2y8;3y6;3y7;3y8 |
| CD14 | CD14 | VQPQSLDLSHNSLR | 3y5;3y6;3y7;3y8;3y9 |
| Haptoglobin | HP | NQLVEVEK | 2y4;2y5;2y6;2y7;3y3 |
| Haptoglobin | HP | VGYVSGWGR | 2y5;2y6;2y7;2y8;3y3 |
| Interleukin 8 | IL-8 | THSTPFHPK | 2y5;2y7;3y3;3y4;3y5 |
| Interleukin 8 | IL-8 | VVQVFVK | 2y4;2y5;2y6;3y3;3y4 |
| Galectin 1 | LGALS1 | DAGAWGAEQR | 2y5;2y6;2y7;2y8;3y3 |
| Galectin 1 | LGALS1 | LPDGYEFK | 2y4;2y5;2y6;2y7;3y3 |
| Galectin 3 | LGALS3 | GNDVAFHFNPR | 2y5;2y7;3y4;3y5;3y6 |
| Galectin 3 | LGALS3 | IQVLVEPDHFK | 2y7;2y9;3y5;3y6;3y7 |
| Lactotransferrin | LTF | GEADALNLDGGYIYTAGK | 2y10;2y11;3y6;3y7;3y8 |
| Lactotransferrin | LTF | YYGYTGAFR | 2y6;2y7;2y8;3y3;3y4 |
| Serum amyloid A3 | SAA3 | ADQFANEWGR | 2y5;2y6;2y7;2y8;2y9 |
| Serum amyloid A3 | SAA3 | ETIQGITDPLFK | 2y6;2y8;3y4;3y5;3y6 |
| Vanin-1 (Pantetheinase) | VAN1 | DSAPNTLSDLTTQALR | 2y10;2y8;2y9;3y5;3y6 |
| Vanin-1 (Pantetheinase) | VAN1 | NLDLLEGAVTSASK | 2y10;2y11;2y12;2y9;3y6 |
Acute phase proteins measured in milk
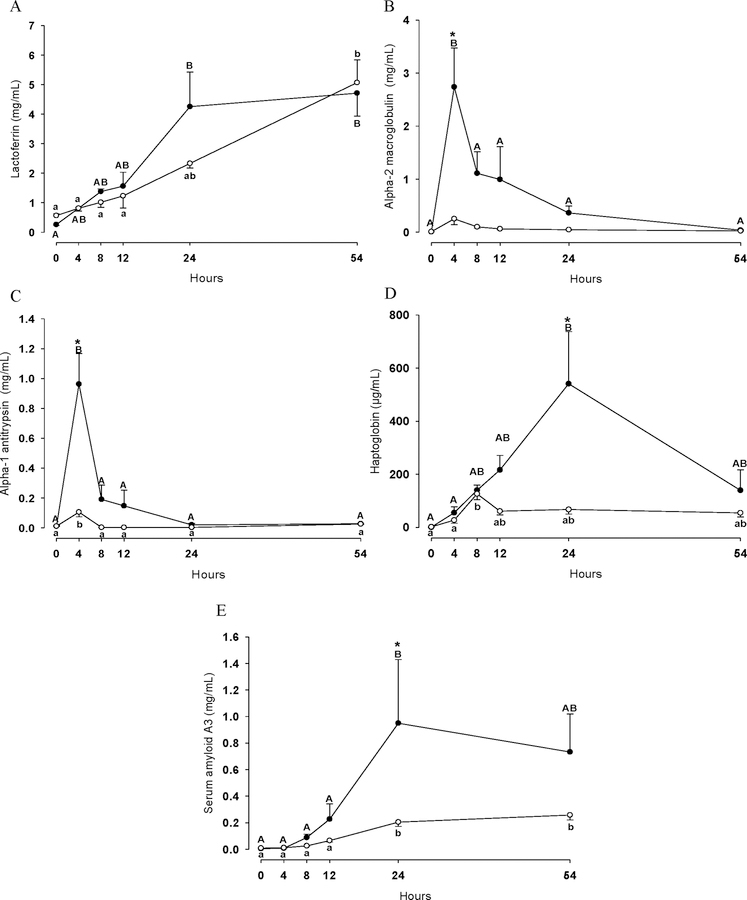
Figure 2 shows concentration changes of the acute phase proteins LTF (Figure 2A), A2M (Figure 2B), A1AT (Figure 2C), HP (Figure 2D) and SAA3 (Figure 2E) after LPS and PGN challenge over the time course of 54 h. The concentration of these five acute phase proteins increased in milk in the LPS and PGN group (P<0.05) with the exception of A2M which remained unchanged over time in the PGN group (P>0.05). In the LPS group, A2M, A1AT, HP and SAA3 increased at both 4 h (A2M and A1AT) and 24 h (HP and SAA3) compared to the PGN group (P<0.05). Lactoferrin shows a remarkable increase over time, but a significant difference between LPS and PGN challenge could not be correlated to the LTF levels in milk throughout the entire experimental period (P>0.05).
Figure 2.
Lactoferrin (A), alpha-2 macroglobulin (B), alpha-1 antitrypsin (C), haptoglobin (D) and serum amyloid A3 (E) concentrations in quarters treated with either LPS (n=3; ●) or PGN (n=3; ○) during the experimental period. Different uppercase letters (A-B) indicate significant differences between time points within LPS group (P < 0.05). Different lowercase letters (a-b) indicate significant differences between time points within PGN group (P < 0.05). Consecutive time points labelled with the same letters (e.g. A-A and a-a) denote that protein changes were not significantly different (P>0.05) between these two time points. * Stands for significant differences between LPS and PGN groups at the marked time point (P < 0.05).
Cellular host response proteins measured in milk
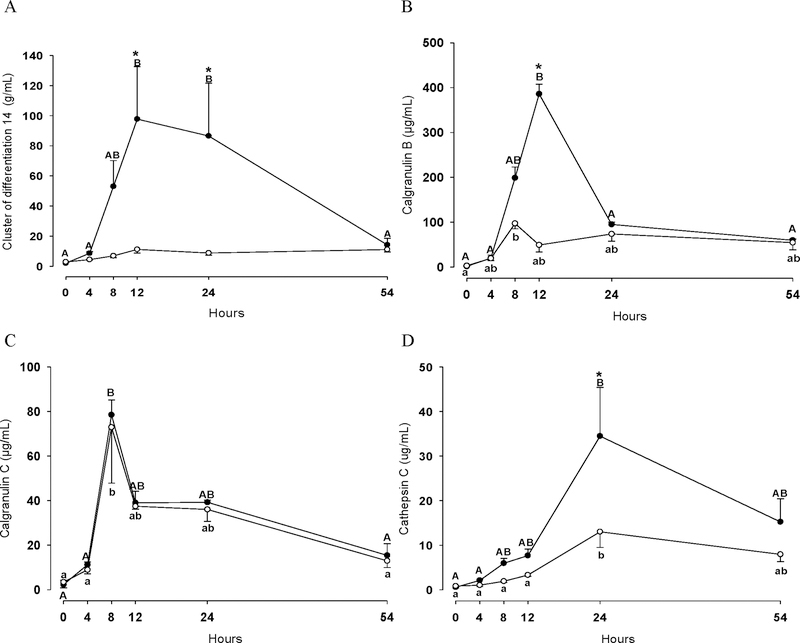
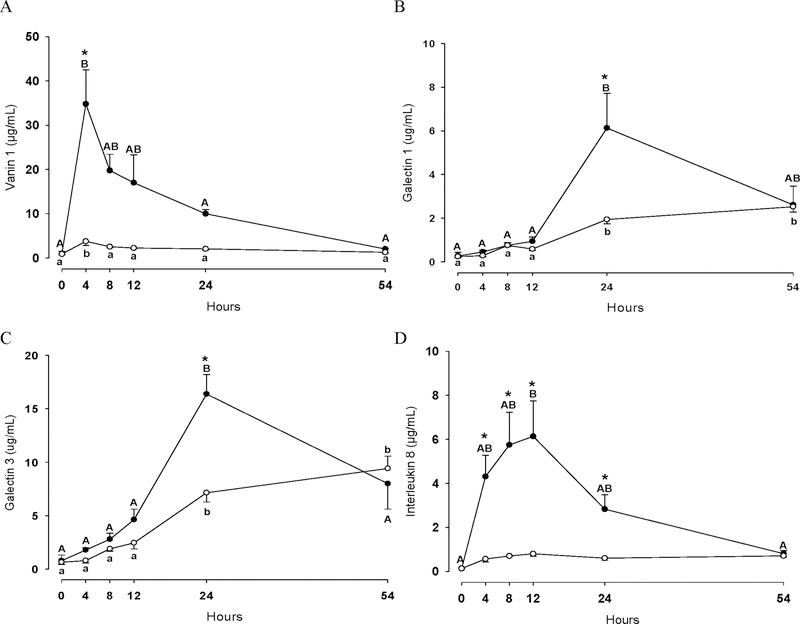
Figure 3 and 4 show concentration changes of CD14 (Figure 3A), CALGB (Figure 3B), CALGC (Figure 3C), CATC (Figure 3D), VAN1 (Figure 4A), LGALS1 (Figure 4B), LGALS3 (Figure 4C) and IL-8 (Figure 4D) in quarters infused either with LPS or PGN. Overall, the concentration of these eight proteins significantly increased in both groups (P<0.05), LPS and PGN, with the exception of CD14 and IL-8 which remained unchanged in the PGN group during the entire experimental period (P>0.05). Notably, the majority of the studied proteins increased significantly in greater extent in the LPS group at 4 h (VAN1 and IL-8), 8 h (IL-8), 12h (CD14, CALGB and IL-8) and 24 h (CD14, CATC, LGALS1, LGALS3 and IL-8) compared to the PGN group (P<0.05). For VAN1 a relatively low concentration change over the time-course in the PGN group was observed, although VAN1 significantly increased at 4 h in this group. For CALGC, no differences in concentration was observed between the two groups over the time course (P>0.05).
Figure 3.
Cluster of differentiation 14 (A), calgranulin B (B), calgranulin C (C) and cathepsin C (D) concentrations in quarters treated with either LPS (n=3; ●) or PGN (n=3; ○) during the experimental period. Different uppercase letters (A-B) indicate significant differences between time points within LPS group (P < 0.05). Different lowercase letters (a-b) indicate significant differences between time points within PGN group (P < 0.05). Consecutive time points labelled with the same letters (e.g. A-A and a-a) denote that protein changes were not significantly different (P>0.05) between these two time points. * Stands for significant differences between LPS and PGN groups at the marked time point (P < 0.05).
Figure 4.
Vanin 1 (A), Galectin 1 (B), galectin 3 (C) and interleukin 8 (D) concentrations in quarters treated with either LPS (n=3; ●) or PGN (n=3; ○) during the experimental period. Different uppercase letters (A-B) indicate significant differences between time points within LPS group (P < 0.05). Different lowercase letters (a-b) indicate significant differences between time points within PGN group (P < 0.05). Consecutive time points labelled with the same letters (e.g. A-A and a-a) denote that protein changes were not significantly different (P>0.05) between these two time points. * Stands for significant differences between LPS and PGN groups at the marked time point (P < 0.05).
DISCUSSION
The activation of bovine host response to mastitis is tailored to fight the different invading mammary pathogenic bacteria (Wellnitz et al., 2013). While LPS has been widely used to simulate E. coli mediated mastitis by both in vivo and in vitro experiments, (Wellnitz et al., 2011, Wall et al., 2016a, Wall et al., 2016b), PGN has so far only been reported for in vitro simulation of mastitis caused by S. aureus (Mount et al., 2009, Im et al., 2014, Sulabh et al., 2016). Based on the increased SCC in both of our experimental groups, we evaluate that PGN is suitable for experimental simulation of mastitis caused by S. aureus. Thus, we present for the first time a parallel study to compare the bovine host response to E. coli and S. aureus mediated challenge, measured by the timed regulation of 13 host response proteins in milk samples collected after intramammary LPS and PGN challenges.
General observations
Generally, all 13 targeted proteins increased their concentration in milk after challenge, and cows challenged with LPS showed a larger increase of these proteins in milk compared to those challenged with PGN. These observations are in agreement with previous transcriptome studies which compared E. coli and S. aureus mastitis in cows (Lee et al., 2006, Mount et al., 2009). Also, ELISA based studies (Riollet et al., 2000, Bannerman et al., 2004, Pyorala et al., 2011) have reported that gram-negative bacteria unleash a more acute and severe host response than gram-positive bacteria. In the present study, six host response proteins, namely CALGC, CATC, VAN1, LGALS1 and LGALS3, were measured in milk after in vivo challenge with either LPS or PGN.
Biomarkers candidates for mastitis detection and diagnosis
Our present study clearly demonstrates that the acute phase proteins A2M and A1AT show very early an increased concentration in cows challenged with LPS compared to those challenged with PGN. Therefore, both proteins are good candidates for early detection of mastitis caused by gram-negative bacteria. Both A2M and A1AT are protease inhibitors synthesized mainly in the liver in response to inflammation, and both are important regulators of the bulk of proteases that are released upon injury (Murata et al., 2004). Both A2M and A1AT are found in milk due to the increased permeability of the blood-milk barrier during inflammation. Our results are in agreement with previous studies, reporting higher A1AT concentrations in bovine milk sampled from coliform mastitis compared to staphylococcal mastitis (Pyörälä and Syväjärvi, 1987). Similar observations about A2M are not available in the literature, thus, our study also present a first time observation on A2M response in challenged cows.
The acute phase proteins SAA3 and HP, which are known to be expressed locally by mammary gland cells (Hiss et al., 2004, Eckersall et al., 2006, Hernandez-Castellano et al., 2017b), also revealed different response patterns after LPS versus PGN challenge. As described above, our data indicate that LPS triggers a more intense reaction of the innate immune system than PGN, thus both SAA3 and HP are interesting candidates for detection and diagnosis of mastitis. In the study from Pyorala et al. (2011), cows infected with E. coli showed increased HP and SAA concentrations (243.5 mg/L and 275.5 mg/L, respectively), which are in agreement to the concentrations showed in the present study (216.5 ± 61.0 µg/mL and 226.6 ± 11.2 µg/mL in cows 12 h post-challenge with LPS, respectively). Additionally and also in agreement with our results, Pyorala et al. (2011) showed low concentrations for HP (33 mg/L) and SAA (16.4 mg/L) in cows infected with S. aureus (our concentrations for HP and SAA in cows 12 h post-challenge with PGN were 54.36 ± 11.04 µg/mL and 65.12 ± 1.54 µg/mL, respectively). Therefore our observations are in agreement with the results from Pyorala et al. (2011), who also suggested that HP and SAA3 can reflect the severity of clinical mastitis.
Despite LTF has been considered as a suitable marker for mastitis, its concentration increased but did not differ between the LPS and the PGN group. Although there is some controversy about the specific LTF concentration during mastitis, several authors have reported similar increased LTF concentrations during mastitis caused either by E. coli or S. aureus (Kawai et al., 1999, Griesbeck-Zilch et al., 2008).
In addition to acute phase proteins, several immune cells (and their secreted substances) are involved in the host response during mastitis. It is well known that bacteria invading the mammary gland can cause pathogen-specific differential permeability of the blood-milk barrier (Wellnitz et al., 2016), which affect the transfer of blood components into the milk (Wellnitz et al., 2011). Pathogens like E. coli have a greater effect on the blood-milk barrier, and consequently a larger transfer of blood proteins to milk occurs in comparison to mastitis caused by S. aureus (Bannerman et al., 2004). Therefore, proteins secreted by immune cells during the host response to mastitis causing pathogens could be considered as useful biomarkers, which additionally can provide information about the severity and stage of mastitis infections (Bannerman et al., 2004). In our study, diverse proteins related to the immune response such as CD14, CALGB, CATC, LGALS1, LGALS3 and IL-8 were increased in cows challenged with LPS compared to those challenged with PGN. These observations support previous ELISA based studies by Persson Waller et al. (2003) and Bannerman et al. (2004), who reported different increase patterns of CD14 and IL-8 in milk from cows with mastitis caused by either E. coli or S. aureus. Additionally, Bislev et al. (2012) showed increased concentrations of CD14, CALGB, CATC, LGALS1, LGALS3 and IL-8 in mammary gland tissue as a consequence of the intramammary infection with Streptococcus uberis, however, the present study shows for the first time that these proteins can be directly measured in milk. Consequently, we suggest that also the seven immune related proteins we report here (CD14, CALGB, CALGC, CATC, LGALS1, LGALS3 and IL-8), are interesting candidates for their use as clinical markers for mastitis detection. Thus, these protein patterns may be used for the identification of the type of causing mastitis bacteria (gram-negative vs. gram-positive). For example, we observed that CALGC is increased in mastitis caused by either gram-negative or gram-positive bacteria, however CD14 is only increased in mastitis caused by gram-negative bacteria. The fact that we can detect and quantify IL-8 and CD14, and other immune related proteins such as CALGB, CALBC, CATC, LGALS1 and LGALS3 by targeted proteomics, reflects the strength and potential of the SRM methods for further validation and research in large scale animal experiments. Additionally, VAN1 showed a pathogen-dependent response during the mastitis challenge in our study. Despite its relatively low concentration in milk, we robustly quantified concentration changes of this protein in all 36 milk samples, having a remarkable response already 4 h after LPS challenge. Vanin-1 is a GPI-anchored cell-specific surface pantetheinase, which, upon pantetheine (coenzyme A precursor) hydrolysis, provides cysteamine to tissues. This protein has been described to have a role in inflammation, oxidative stress and cell migration (Kaskow et al., 2012). In addition, a previous study in human gut demonstrated a pro-inflammatory role of VAN1 in relation to colitis (Berruyer et al., 2006). In a previous publication from our group, we showed increased VAN1 concentrations in bovine mammary gland tissue infected with Streptococcus uberis (Bislev et al., 2012). The present study shows for the first time the quantification of VAN1 in milk.
The relevance of biomarkers in the dairy industry
From the 13 proteins we have presented in this study, eleven proteins showed increased concentrations upon challenge, and seven of them, namely A2M, CALGB, CALBC, CATC, LGALS1, LGALS3, VAN1, are described for the first time in either LPS or PGN challenged cows. We suggest that A2M, A1AT, VAN1 and IL-8 are the most pertinent candidates for future investigations regarding their relevance in mastitis detection and diagnosis. Our considerations are based on their early (A2M, A1AT and VAN1) or long-term (IL-8) increased concentration after LPS challenge compared to PGN challenge. However, monitoring fluctuations of all eleven potential biomarkers may become valuable as early indicators of mastitis caused by gram-negative bacteria, particularly if fast and accurate assays can be implemented in routine on-line detection during milking in automatic milking robots. It should be noted that all biomarker candidates described here must be further validated in a large scale animal experiment in order to fully evaluate their potential as biomarkers for mastitis detection and diagnosis. Based on our results and due to the high throughput and low cost, our SRM approach can provide the means to measure large sets of milk samples including both biological replicates and frequent sampling times in order to further investigate the potentials and general value of these markers.
CONCLUSIONS
We clearly demonstrate a more extent increase in concentration of eleven host response proteins in cows challenged by LPS, reflecting a more acute inflammatory response in mastitis caused by E. coli compared to PGN challenge. Likewise, our observations of an impaired inflammatory response to PGN may contribute to the well-known ability of S. aureus to establish chronic intra-mammary infections. Potential biomarker candidates for distinguishing gram-negative mastitis from gram-positive mastitis include A2M, A1AT, HP, SAA3, CD14, CALGB, CATC, VAN1, LGALS1, LGALS3 and IL-8. The targeted quantitative SRM mass spectrometry approach described here can be used as a validation method of these non-invasive biomarkers in large scale experiments.
Supplementary Material
Supplementary file S1 shows individual concentrations for each of the 13 host response proteins in milk from cows inoculated with either LPS (group 1, mimicking E. coli mastitis) or PGN (group 2, mimicking S. aureus mastitis). Milk was collected at 0, 4, 8, 12, 24, and 54 h post challenge.
ACKNOWLEDGEMENTS
This work was carried out in a BIOSENS Consortium project and was financed by the Danish Ministry of Food Agriculture and Fisheries, Lattec I/S, The Milk Levy Fund, the Faculty of Agriculture Sciences and the Graduate School of Agriculture, Food and Environment at Aarhus University. Ulrike Kusebauch was supported by a fellowship of the German Academic Exchange Service, National Institutes of Health, National Institute for General Medical Sciences (grant R01 GM087221) and Center for Systems Biology (grant 2P50 GM076547), the National Human Genome Research Institute and the American Recovery and Reinvestment Act (ARRA) (grant RC2 HG005805), and the National Science Foundation MRI (grant 0923536).
REFERENCES
- Aitken SL, Corl CM, and Sordillo LM. 2011. Immunopathology of mastitis: insights into disease recognition and resolution. J. Mammary Gland Biol. Neoplasia 16(4):291–304. [DOI] [PubMed] [Google Scholar]
- Bannerman DD 2008. Pathogen-dependent induction of cytokines and other soluble inflammatory mediators during intramammary infection of dairy cows. J. Anim. Sci 87:10–25. [DOI] [PubMed] [Google Scholar]
- Bannerman DD, Paape MJ, Lee JW, Zhao X, Hope JC, and Rainard P. 2004. Escherichia coli and Staphylococcus aureus elicit eifferential innate immune responses following intramammary infection. Clin. Diagn. Lab. Immunol 11(3):463–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berruyer C, Pouyet L, Millet V, Martin FM, LeGoffic A, Canonici A, Garcia S, Bagnis C, Naquet P, and Galland F. 2006. Vanin-1 licenses inflammatory mediator production by gut epithelial cells and controls colitis by antagonizing peroxisome proliferator-activated receptor γ activity. J. Exp. Med 203(13):2817–2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bislev SL, Kusebauch U, Codrea MC, Beynon RJ, Harman VM, Rontved CM, Aebersold R, Moritz RL, and Bendixen E. 2012. Quantotypic properties of QconCAT peptides targeting bovine host response to Streptococcus uberis. J. Proteome Res 11(3):1832–1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehmer JL 2011. Proteomic analyses of host and pathogen responses during bovine mastitis. J. Mammary Gland Biol. Neoplasia 16(4):323–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burvenich C, Van Merris V. r., Mehrzad J, Diez-Fraile A, and Duchateau L. 2003. Severity of E. coli mastitis is mainly determined by cow factors. Vet. Res 34(5):521–564. [DOI] [PubMed] [Google Scholar]
- Danielsen M, Codrea MC, Ingvartsen KL, Friggens NC, Bendixen E, and Rontved CM. 2010. Quantitative milk proteomics - Host responses to lipopolysaccharide-mediated inflammation of bovine mammary gland. Proteomics 10(12):2240–2249. [DOI] [PubMed] [Google Scholar]
- Domon B and Aebersold R. 2010. Options and considerations when selecting a quantitative proteomics strategy. Nat. Biotechnol 28(7):710–721. [DOI] [PubMed] [Google Scholar]
- Ebhardt HA, Root A, Sander C, and Aebersold R. 2015. Applications of targeted proteomics in systems biology and translational medicine. Proteomics 15(18):3193–3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckersall PD, Young FJ, Nolan AM, Knight CH, McComb C, Waterston MM, Hogarth CJ, Scott EM, and Fitzpatrick JL. 2006. Acute phase proteins in bovine milk in an experimental model of Staphylococcus aureus subclinical mastitis. J. Dairy Sci 89(5):1488–1501. [DOI] [PubMed] [Google Scholar]
- Fogsgaard KK, Røntved CM, Sørensen P, and Herskin MS. 2012. Sickness behavior in dairy cows during Escherichia coli mastitis. J. Dairy Sci 95(2):630–638. [DOI] [PubMed] [Google Scholar]
- Griesbeck-Zilch B, Meyer HH, Kuhn CH, Schwerin M, and Wellnitz O. 2008. Staphylococcus aureus and Escherichia coli cause deviating expression profiles of cytokines and lactoferrin messenger ribonucleic acid in mammary epithelial cells. J. Dairy Sci 91(6):2215–2224. [DOI] [PubMed] [Google Scholar]
- Hernandez-Castellano LE, Wall SK, Stephan R, Corti S, and Bruckmaier R. 2017a. Milk somatic cell count, lactate dehydrogenase activity, and immunoglobulin G concentration associated with mastitis caused by different pathogens: A field study. Schweiz Arch. Tierheilkd 159(5):283–290. [DOI] [PubMed] [Google Scholar]
- Hernandez-Castellano LE, Hernandez LL, Sauerwein H, and Bruckmaier RM. 2017b. Endocrine and metabolic changes in transition dairy cows are affected by prepartum infusions of a serotonin precursor. J. Dairy Sci 100(6):5050–5057. [DOI] [PubMed] [Google Scholar]
- Hernández-Castellano LE, Almeida AM, Ventosa M, Coelho AV, Castro N, and Arguello A. 2014. The effect of colostrum intake on blood plasma proteome profile in newborn lambs: low abundance proteins. BMC Vet. Res 10:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández-Castellano LE, Arguello A, Almeida AM, Castro N, and Bendixen E. 2015. Colostrum protein uptake in neonatal lambs examined by descriptive and quantitative liquid chromatography-tandem mass spectrometry. J. Dairy Sci 98(1):135–147. [DOI] [PubMed] [Google Scholar]
- Hernández-Castellano LE, Ferreira AM, Nanni P, Grossmann J, Arguello A, Capote J, Cai G, Lippolis J, Castro N, and Almeida AM. 2016. The goat (Capra hircus) mammary gland secretory tissue proteome as influenced by weight loss: A study using label free proteomics. J. Proteomics 145: 60–69 [DOI] [PubMed] [Google Scholar]
- Hiss S, Mielenz M, Bruckmaier RM, and Sauerwein H. 2004. Haptoglobin concentrations in blood and milk after endotoxin challenge and quantification of mammary Hp mRNA expression. J. Dairy Sci 87(11):3778–3784. [DOI] [PubMed] [Google Scholar]
- Hogeveen H, Huijps K, and Lam T. 2011. Economic aspects of mastitis: New developments. N. Z. Vet. J 59(1):16–23. [DOI] [PubMed] [Google Scholar]
- Im J, Lee T, Jeon JH, Baik JE, Kim KW, Kang S-S, Yun C-H, Kim H, and Han SH. 2014. Gene expression profiling of bovine mammary gland epithelial cells stimulated with lipoteichoic acid plus peptidoglycan from Staphylococcus aureus. Int. Immunopharmacol 21(1):231–240. [DOI] [PubMed] [Google Scholar]
- Kaskow BJ, Michael Proffit J, Blangero J, Moses EK, and Abraham LJ. 2012. Diverse biological activities of the vascular non-inflammatory molecules – The Vanin pantetheinases. Biochem. Biophys. Res. Commun 417(2):653–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai K, Hagiwara S, Anri A, and Nagahata H. 1999. Lactoferrin concentration in milk of bovine clinical mastitis. Vet. Res. Commun 23(7):391–398. [DOI] [PubMed] [Google Scholar]
- Kusebauch U, Campbell DS, Deutsch EW, Chu CS, Spicer DA, Brusniak MY, Slagel J, Sun Z, Stevens J, Grimes B, Shteynberg D, Hoopmann MR, Blattmann P, Ratushny AV, Rinner O, Picotti P, Carapito C, Huang CY, Kapousouz M, Lam H, Tran T, Demir E, Aitchison JD, Sander C, Hood L, Aebersold R, and Moritz RL. 2016. Human SRMAtlas: a resource of targeted assays to quantify the complete human proteome. Cell 166(3):766–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J-W, Bannerman DD, Paape MJ, Huang M-K, and Zhao X. 2006. Characterization of cytokine expression in milk somatic cells during intramammary infections with Escherichia coli or Staphylococcus aureus by real-time PCR. Vet. Res 37(2):219–229. [DOI] [PubMed] [Google Scholar]
- MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, Kern R, Tabb DL, Liebler DC, and MacCoss MJ. 2010. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26(7):966–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mount JA, Karrow NA, Caswell JL, Boermans HJ, and Leslie KE. 2009. Assessment of bovine mammary chemokine gene expression in response to lipopolysaccharide, lipotechoic acid plus peptidoglycan, and CpG oligodeoxynucleotide 2135. Can. J. Vet. Res 73(1):49–57. [PMC free article] [PubMed] [Google Scholar]
- Murata H, Shimada N, and Yoshioka M. 2004. Current research on acute phase proteins in veterinary diagnosis: an overview. Vet. J 168(1):28–40. [DOI] [PubMed] [Google Scholar]
- Persson Waller K, Colditz IG, Lun S, and Östensson K. 2003. Cytokines in mammary lymph and milk during endotoxin-induced bovine mastitis. Research in Veterinary Science 74(1):31–36. [DOI] [PubMed] [Google Scholar]
- Picotti P and Aebersold R. 2012. Selected reaction monitoring-based proteomics: workflows, potential, pitfalls and future directions. Nat. Methods 9(6):555–566. [DOI] [PubMed] [Google Scholar]
- Pyorala S, Hovinen M, Simojoki H, Fitzpatrick J, Eckersall PD, and Orro T. 2011. Acute phase proteins in milk in naturally acquired bovine mastitis caused by different pathogens. Vet. Rec 168(20):535. [DOI] [PubMed] [Google Scholar]
- Pyörälä S and Syväjärvi J. 1987. Bovine Acute Mastitis Part I. Clinical Aspects and Parameters of Inflammation in Mastitis Caused by Different Pathogens. J. Vet. Med., Series B 34(1–10):573–584. [DOI] [PubMed] [Google Scholar]
- Riollet C, Rainard P, and Poutrel B. 2000. Differential induction of complement fragment C5a and inflammatory cytokines during intramammary infections with Escherichia coli and Staphylococcus aureus. Clin. Diagn. Lab. Immunol 7(2):161–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schukken YH, Günther J, Fitzpatrick J, Fontaine MC, Goetze L, Holst O, Leigh J, Petzl W, Schuberth HJ, Sipka A, Smith DGE, Quesnell R, Watts J, Yancey R, Zerbe H, Gurjar A, Zadoks RN, and Seyfert HM. 2011. Host-response patterns of intramammary infections in dairy cows. Vet. Immunol. Immunopathol 144(3–4):270–289. [DOI] [PubMed] [Google Scholar]
- Sulabh S, Bhushan B, Panigrahi M, Verma A, Baba NA, and Kumar P. 2016. Differential response of immune-related genes to peptidoglycan and lipoteichoic acid challenge in vitro. Vet. World 9(9):983–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres A, Hernández-Castellano LE, Morales-delaNuez A, Sánchez-Macías D, Moreno-Indias I, Castro N, Capote J, and Argüello A. 2014. Short-term effects of milking frequency on milk yield, milk composition, somatic cell count and milk protein profile in dairy goats. J. Dairy Res 81(3):275–279. [DOI] [PubMed] [Google Scholar]
- Vels L, Røntved CM, Bjerring M, and Ingvartsen KL. 2009. Cytokine and acute phase protein gene expression in repeated liver biopsies of dairy cows with a lipopolysaccharide-induced mastitis. J. Dairy Sci 92(3):922–934. [DOI] [PubMed] [Google Scholar]
- Wall SK, Hernandez-Castellano LE, Ahmadpour A, Bruckmaier RM, and Wellnitz O. 2016a. Differential glucocorticoid-induced closure of the blood-milk barrier during lipopolysaccharide- and lipoteichoic acid-induced mastitis in dairy cows. J. Dairy Sci 99(9):7544–7553. [DOI] [PubMed] [Google Scholar]
- Wall SK, Wellnitz O, Hernandez-Castellano LE, Ahmadpour A, and Bruckmaier RM. 2016b. Supraphysiological oxytocin increases the transfer of immunoglobulins and other blood components to milk during lipopolysaccharide- and lipoteichoic acid-induced mastitis in dairy cows. J. Dairy Sci 99(11):9165–9173. [DOI] [PubMed] [Google Scholar]
- Wellnitz O, Arnold ET, and Bruckmaier RM. 2011. Lipopolysaccharide and lipoteichoic acid induce different immune responses in the bovine mammary gland. J. Dairy Sci 94(11):5405–5412. [DOI] [PubMed] [Google Scholar]
- Wellnitz O, Arnold ET, Lehmann M, and Bruckmaier RM. 2013. Short communication: differential immunoglobulin transfer during mastitis challenge by pathogen-specific components. J. Dairy Sci 96(3):1681–1684. [DOI] [PubMed] [Google Scholar]
- Wellnitz O, Zbinden C, Huang X, and Bruckmaier RM. 2016. Short communication: Differential loss of bovine mammary epithelial barrier integrity in response to lipopolysaccharide and lipoteichoic acid. J. Dairy Sci 99(6):4851–4856. [DOI] [PubMed] [Google Scholar]
- Yang W, Zerbe H, Petzl W, Brunner RM, Günther J, Draing C, von Aulock S, Schuberth H-J, and Seyfert H-M. 2008. Bovine TLR2 and TLR4 properly transduce signals from Staphylococcus aureus and E. coli, but S. aureus fails to both activate NF-κB in mammary epithelial cells and to quickly induce TNFα and interleukin-8 (CXCL8) expression in the udder. Molecular Immunol 45(5):1385–1397. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary file S1 shows individual concentrations for each of the 13 host response proteins in milk from cows inoculated with either LPS (group 1, mimicking E. coli mastitis) or PGN (group 2, mimicking S. aureus mastitis). Milk was collected at 0, 4, 8, 12, 24, and 54 h post challenge.