Abstract
C2H2 zinc finger protein (ZFP) genes have been extensively studied in many organisms and can function as transcription factors and be involved in many biological processes including plant growth and development and stress responses. In the current study, a comprehensive genomics analysis of the C2H2-ZFP genes in B. rapa was performed. A total of 301 B. rapa putative C2H2-ZFP (BrC2H2-ZFP) genes were identified from the available Brassica genome databases, and further characterized through analysis of conserved amino acid residues in C2H2-ZF domains and their organization, subcellular localization, phylogeny, additional domain, chromosomal location, synteny relationship, Ka/Ks ratio, and expression pattern. We also analyzed the expression patterns of eight B. rapa C2H2-ZFP genes under salt and drought stress conditions by using qRT-PCR technique. Our results showed that about one-third of these B. rapa C2H2-ZFP genes were originated from segmental duplication caused by the WGT around 13 to 17 MYA, one-third of them were highly and consecutively expressed in all tested tissues, and 92% of them were located in nucleus by prediction supporting then their functional roles as transcription factors, of which some may play important roles in plant growth and development. The Ka/Ks ratios of 264 orthologous C2H2-ZFP gene pairs between A. thaliana and B. rapa were all, except two, inferior to 1 (varied from 0.0116 to 1.4919, with an average value of 0.3082), implying that these genes had mainly experienced purifying selection during species evolution. The estimated divergence times of the same set of gene pairs ranged from 6.23 to 38.60 MY, with an average value of 18.29 MY, indicating that these gene members have undergone different selective pressures resulting in different evolutionary rates during species evolution. In addition, a few of these B. rapa C2H2-ZFPs were shown to be involved in stress responses in a similar way as their orthologs in A. thaliana. Comparison between A. thaliana and B. rapa orthologous C2H2-ZFP genes showed that the majority of these C2H2-ZFP gene members encodes proteins with conserved subcellular localization and functional domains between the two species but differed in their expression patterns in five tissues or organs. Thus, our study provides valuable information for further functional determination of each C2H2-ZFP gene across the Brassica species, and may help to select the appropriate gene targets for further in-depth studies, and genetic engineering and improvement of Brassica crops.
Introduction
The zinc finger proteins (ZFPs) are one of the most common groups of proteins widely existing in all eukaryotic organisms. Zinc finger proteins were known to be involved in many cellular processes including DNA-binding, RNA-binding, protein-protein interaction, and protein folding, etc. [1–3], and play important roles in plant growth and development as well as response to various environmental stresses [4–7]. The zinc finger (ZF) structure is maintained by the zinc ion, which coordinates conserved cysteine and/or histidine residues in different combinations. According to the type and order of these zinc coordinated residues, the ZFs can be classified into different types, such as, C2H2, C2C2, and C3H1 domains coordinating one zinc ion, whereas C3HC4 RING finger, PHD and LIM domains coordinating two zinc ions [8–10]. As high as 30 types of ZFs were approved in human genome [9].
The C2H2-ZFs, also called classical ZFs, were the first class to be characterized among all the types of ZFs [9]. The C2H2-ZF domain was defined as about 30 amino acids with two conserved Cys and two His residues which bound to one Zn2+ atom tetrahedrally and form a structure as C-X2-4–C-X3–F-X5-L-X2–H-X3-5–H, where X represents any amino acid [2,11]. Plant C2H2 zinc finger proteins differ from other eukaryotic organisms by a long and diverse space between two zinc fingers, while in animal, the spaces between two tandem repeated zinc fingers are often short (seven amino acid residues) and known as the HC links [8], as it was found in Drosophila and yeast [12]. Additionally, the α-helix in the plant zinc finger domain has a very conserved sequence, QALGGH, which is absent in other organisms, and probably the result of evolution through natural selection for a specific regulatory process unique in plants [7,13]. C2H2-ZFP family has been reported in several plant genomes, including 179 members in Arabidopsis thaliana [14], 189 in rice [15], 321 in soybean [16], 211 in maize [17], 124 in foxtail millet [18], 122 in durum wheat [19], 118 in tobacco [20], and 109 in poplar [21].
Recently, extensive studies showed that most of C2H2-ZFPs functioned as transcription factors and were involved in various biological processes, including plant growth and development, hormone signaling and stress responses in many plants species [4,18,22,23]. For example, in rice ZFP179 is involved in salt tolerance [24], and ZFP245 can increase the tolerance to cold, drought and oxidative stresses [25]. In soybean, SCOF-1 is involved in low-temperature tolerance [26]. In Brassica napus, BnLATE is involved in silique shattering [27]. In A. thaliana, RHL41 is involved in light stress [24]. In Capsicum annuum, CAZFP1 is involved in pathogen defense [28]. ZFP252 can enhance drought and salt tolerance in rice [29]. SIZF2 can influence flower and leaf shape in A. thaliana, and enhance salt tolerance in tomato [30]. BcZAT12 can improve tolerance to drought in tomato [31]. ZFP1 positively regulates tolerance to both cold and drought stress in A. thaliana [32], GsZFP1 is involved in ABA signaling, by reduced ABA sensitivity and decreased stomata size in transgenic A. thaliana plants [33]. GmWRKY49 is a positive regulator of salinity tolerance in soybean [34]. SIZF3 can significantly increase the level of AsA and enhance salt stress tolerance [35], GhSTOP1 can accelerate root growth and is essential for aluminum and proton stress tolerance in A. thaliana [36]. SGR5 is involved in primary events of geotropism in inflorescence stems in A. thaliana [37]. SUPERMAN (SUP), controls the boundary between the stamen and carpel whorls [38–40], prevents class B gene expression and promotes stem cell termination in the fourth whorl of A. thaliana flowers [41]. TRANSPARENT TESTA1 (TT1) controls seed endothelium differentiation in A. thaliana [42].
Brassica genus includes numerous economically important species with notable morphological diversity due to long term evolution [43], providing human nutrition in the form of vegetables, oil, condiments, dietary fiber, and vitamin C [44]. Among the Brassica species, Brassica rapa (n = 10, AA genome, 529 Mb genome size) is one of the most economically important diploid progenitor species which contributes the ‘A’ genome to the allopolyploid oilseed crops, B. napus (n = 19, AACC) and B. juncea (n = 18, AABB) [45]. B. rapa also serves as a model plant for studies related to genomic or evolution due to small genome size (about 529 Mbp) [46] and exhibits very close relation with the model plant A. thaliana. Additionally, it experienced a whole-genome triplication (WGT) event since its divergence from A. thaliana about 13 to 17 million years ago (MYA) [47]. The availability of whole B. rapa genome sequences offers an unprecedented opportunity for genome-wide identification, evolution and functional analysis of various important gene families in this species.
In this study, we performed the genome-wide identification of the C2H2-ZFP gene family in B. rapa. Comprehensive analyses were carried out based on phylogeny, chromosome location, and syntenic relationship between B. rapa and A. thaliana. Furthermore, expression analysis was carried out based on RNA-seq data of this gene family in different tissues, and the expression patterns of a few members under salt and drought stress treatments were also investigated through qRT-PCR. Our results provide a solid foundation for further functional characterization of C2H2-ZFP genes and provide helpful information in the field of genetics and the evolution of this model species.
Materials and methods
Identification and classification of C2H2-ZFPs in B. rapa
We identified the C2H2-ZFPs in B. rapa using two different approaches. First, all 176 known C2H2-ZFPs in A. thaliana [14] were retrieved from the TAIR database (http://www.arabidopsis.org/) and used as queries to BLASTp against the whole B. rapa genome annotation data deposited at the Brassica Database (BRAD, ver. 1.5, http://brassicadb.org/brad/). Second, each type of representative A. thaliana C2H2-ZF domains was used as queries to BLASTp against the same database in order to fully identify the C2H2-ZFPs. In both cases, the retrieved irredundant sequences were submitted to SMART database (http://smart.embl-heidelberg.de/) with the chosen option of Pfam domains to confirm the presence of C2H2 domains, combined by manual inspection of each C2H2 zinc finger domain sequence [48]. We determined the C2H2 type for each identified B. rapa C2H2 domain, according to the plant-specific amino acid residues and distances between two∼nine C2H2-ZF domains, as have been previously adopted in soybean [16]. C2H2-ZFPs containing two∼nine C2H2 domains, and every two nearby domains were linked by less than 12 amino acid residues, were classified as Tandem ZFs with two subsets assigned as Br-t1-SF and Br-t2-SF, respectively; while C2H2-ZFPs containing a single and/or two∼four C2H2 domains and every two nearby domains alienated by more than 11 amino acid residues were classified as dispersed C2H2-ZFPs. Furthermore, different types of C2H2-ZFs domains were classified based on the variation of the plant-specific conserved amino acid sequence “QALGGH” and distances between metal ligands [2,21,49]. For example, C2H2-ZF Q-type domains were defined as X2-C-X2-C-X7-QALGGH-X3-H. M-type (M1∼M5) domains were defined for those with one∼five degraded amino acids in the plant-specific conserved amino acid sequence “QALGGH” and certain modifications in the spacing between the two cysteine’s and two histidines. The Z-type domains were defined as those with more than 12 (Z1) and less than 12 (Z2) in their spacing between the second cysteine and the first histidine. The D-type domains were defined as those missing the second histidine in the C2H2-ZF domain compared with the other three types. According to these defined C2H2-ZF types, C2H2-ZFPs containing a single ZF domain were further classified into four clearly distinguishable subsets (Br-1i-Q-SF, Br-1i-M-SF, Br-1i-Z-SF, and Br-1i-D-SF). C2H2-ZFPs containing two Q-types, two M-types or two Z-types of C2H2-ZF domains were defined as Br-2i-Q-SF, Br-2i-M-SF or Br-2i-Z-SF, respectively, while the C2H2-ZFPs containing two different types of C2H2-ZF domains were classified as Br-2i-Mix-SF. All C2H2-ZFPs containing three or four dispersed ZFs were classified into the subsets of Br-3i-SF or Br-4i-SF, respectively. For each identified B. rapa C2H2-ZFP, their size, amino acid properties, charge, molecular weight (kDa) and isoelectric points (pI) were determined using the online available ProtParam tool (http://web.expasy.org/protparam/) [50], and their subcellular localization was predicted by using the online cello2go software (http://cello.life.nctu.edu.tw/cello2go/) in combination with the WoLF PSORT program (https://www.genscript.com/wolf-psort.html).
Multiple sequence alignments, and phylogenetic analysis
The C2H2-ZF domains retrieved from proteins were aligned by using program Clustal W and manually edited by BioEdit software to specify metal ligand positions. Based on B. rapa C2H2-ZFP sequences, phylogenetic trees were generated by using the MEGA 7.0 software with the Maximum Likelihood (ML) method and a bootstrap analysis of 1000 replicates.
Chromosome location of C2H2-ZFPs protein genes in B. rapa
The chromosome location data of each B. rapa C2H2-ZFP genes were downloaded from the BRAD database. Those genes that were assigned to unassembled genomic scaffolds (absence of chromosomal position) were removed from the dataset while the remaining genes were mapped into the specific chromosomes of B. rapa by using Map chart 2.3v software. Tandem repeated and segmentally duplicated genes were indicated by different color lines.
Syntenic relationships between B. rapa and A. thaliana C2H2 zinc finger protein genes
For each predicted A. thaliana and B. rapa C2H2-ZFP, we used the Search Syntenic Gene function of the BRAD database to find out its A. thaliana ortholog (if existed). On the other hand, for each known A. thaliana C2H2-ZFP gene, we used the same function to find out its B. rapa ortholog(s) (if existed). In each query, the information about the corresponding orthologous gene name(s) in A. thaliana and B. rapa, their localization on tPCK (Translocation Proto-Calepineae Karyotype) chromosomes and ancestral chromosome blocks, LF (least fractioned), MF1 (medium fractionated) and MF2 (most fractionated) subgenomes [51–53], as well as the eventual tandem repeats in the two species were recorded. The C2H2-ZFP full-length amino acid sequences were aligned by using program Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/) and then the synonymous rate (Ks), non-synonymous rate (Ka), and evolutionary constriction (Ka/Ks) were calculated by using the PAL2NAL by using the codeml program in PAML (http://www.bork.embl.de/pal2nal/index.cgi?example=Yes#RunP2N) [54,55]. The divergence time was calculated through formula T = Ks/2R, where T mentions to divergence time, Ks mentions to the synonymous substitutions per site, R is intended for the divergence rate of nuclear genes from plants, R-value is considered as 1.5 × 10−8 synonymous substitutions per site per year in case of dicotyledonous plants [56].
Expression pattern of C2H2-ZFP genes in B. rapa
The RNA-seq data of gene expression of six tissues (callus, root, stem, leaf, flower, and silique) of the B. rapa accession Chiifu-401–42 was retrieved from the Gene Expression Omnibus (GEO) database of NCBI (http://www.ncbi.nlm.nih.gov/geo/) using the accession number GSE43245 [57]. The expression values (Fragments Per Kilobase of exon model per Million mapped, FPKM) of identified B. rapa C2H2-ZFP genes were extracted from the dataset and submitted to clustering analysis using Cluster software v3.0 (http://bonsai.hgc.jp/mdehoon/software/cluster/) with log2-transformed FPKM values, Euclidean distances, and the average linkage clustering method. The clustering tree together with the gene expression heat map was generated by using the Java Tree view software (Version1.1.5r2, http://jtreeview.sourceforge.net/).
Preparation of plant material and qRT-PCR analyses
For plant samples preparation, we cultivated B. rapa accession Chiifu-401–42 seeds were sterilized and sown in a Petri dish with moisture-absorbent filter papers and incubated at 25°C. The germinated seeding was then transferred into plastic pots containing growth medium with vermiculite and peat 3:1 grown in a greenhouse at 22 °C with a photoperiod of 16/8 h for light/dark. 21-days-old seedlings were used for different abiotic treatments. For salinity and PEG treatments, the plants were irrigated with 200 mM NaCl and 20% (w/v) polyethylene glycol (PEG 6000). Then, leaves (third, fourth leaves) from control and stressed plants were harvested after 0, 1, 3, and 24 h of treatments and immediately immersed in liquid nitrogen, and stored at -80 °C until RNA extraction use. For each treatment, three biological replicates were prepared to decrease the error rate.
Total RNA was isolated from the frozen leaves of each sample of B. rapa using a Plant RNA extraction Kit, according to the manufacturer’s instructions (OMEGA, China). The quality of RNA was checked by agarose gel electrophoresis, and also by using the NanoDrop 2000 Spectrophotometer (Thermo Fisher Scientific, Inc., Waltham, MA, USA). First-strand cDNA was synthesized using 1μg of total RNA per sample with the cDNA Synthesis Kit from TaKaRa Bio Inc. (Dalian, China). The reverse transcription products were diluted 20-fold and stored at −20 °C prior to analysis. Gene-specific primers for the selected B. rapa C2H2-ZFP genes were designed using Primer3Plus software (http://www.primer3plus.com/). The B. rapa Actin-2 gene (GenBank accession number XM_018658258) was used as an internal reference gene. The primers used for qRT-PCR and their expected amplification product size are summarized in S6 Table. The qRT-PCR analysis was performed on an ABI 7500 Fast Real-time PCR amplification system (Applied Biosystems, Foster, CA, USA). The analysis was carried out in a total volume of 20 μL containing 2 μL template of cDNA, 0.8 μL of the forward and reverse primers (10 μM), 10μL of SYBR Green PCR Master (ROX) (Roche, Shanghai, China), 6.4 μL of sterile distilled water. The PCR amplification parameters were as following: 95 °C for 1 minute, followed by 40 cycles of 95 °C for 15 s, and 60 °C for 70 s. For each sample, three replicates were run to compute the average Ct values. The data were analyzed using the 2−ΔΔCt methods [58]. Relative gene expression levels were normalized against the expression of the housekeeping gene BrActin-2. The significance of differences with P<0.05 among relative expression levels of the genes were statistically analyzed (Tukey HSD test) by using IBM SPSS version 22.
Results
Genome-wide identification and classification of C2H2-ZFPs in B. rapa
To identify the C2H2-ZFPs, we searched the Brassica genome database and further confirmed through Smart database and manual inspection. A total of 301 putative C2H2-ZFPs containing 534 C2H2 domains were identified in B. rapa genome, and summarized in S1 Table together with their relative information. Furthermore, the 534 C2H2-ZF domains were classified into four categories (named Q, M, Z and D) based on the variation of the plant-specific conserved amino acid sequence “QALGGH” and distances between metal ligands (Table 1, S2 Table). S1 Fig illustrated the multiple sequence alignments of all the identified 534 C2H2-ZFs in B. rapa.
Table 1. The types and sub-types of C2H2-ZF domains and their characteristics in Brassica rapa.
X represents any amino acid and the number represents the consensus spacing between the conserved amino acid residues.
| Type of ZF | Sub-type | Conserved motif sequence description | Conserved spacing |
|---|---|---|---|
| Q | - | QALGGH | X2-C-X2-C-X7-QALGGH-X3-H |
| M | M1 | 1 degraded amino acid in QALGGH | X2-C-X2-C-X12-H-X(3,4)-H |
| M2 | 2 degraded amino acids in QALGGH | X2-C-X2-C-X12-H-X(3,4)-H | |
| M3 | 3 degraded amino acids in QALGGH | X2-C-X(2,4)-C-X12-H-X(3,4,5)-H | |
| M4 | 4 degraded amino acids in QALGGH | X2-C-X(1,2,4)-C-X12-H-X(1,3,4,5,7)-H | |
| M5 | 5 degraded amino acids in QALGGH | X2-C-X(2,4)-C-X12-H-X(3,4,5,8)-H | |
| Z | Z1 | - | X2-C-X(2,3,4)-C-X(>12)-H-X(2,3,4,5)-H |
| Z2 | - | X2-C-X2-C-X(<12)-H-X(3,4)-H | |
| D | - | - | X2-C-X(2,4)-C-X(12,13)-H-X2 |
Among the identified 301 putative C2H2-ZFPs, fifty (16.6%) were classified as tandem C2H2-ZFPs, including two subsets, i.e., Br-t1-SF (48 ZFPs) and Br-t2-SF (two ZFPs). Among the Br-t1-SF subset, 37 C2H2-ZFPs contained three C2H2 domains, 5 had four C2H2 domains, four had two C2H2, and two possessed five C2H2 domains while other subset Br-t2-SF (two ZFPs) possessed nine C2H2 domains (Fig 1). Furthermore, 251 (83.38%) ZFPs were identified as isolated ZFPs, which were further divided into ten subsets: Br-1i-Q-SF (51, 20.31%), Br-1i-M-SF (92, 36.65%), Br-1i-Z-SF (14, 5.57%), Br-1i-D-SF (2, 0.8%), Br-2i-Q-SF (37, 14.74%), Br-2i-M-SF (15, 5.98%), Br-2i-Z-SF (3, 1.19%), Br-2i-Mix-SF (19, 7.57%), Br-3i-SF (12, 4.78%), and Br-4i-SF (6, 2.39%) (Fig 1). Among the 251 isolated ZFPs, 159 contained a single C2H2 domain, 74 contained two, 12 contained three C2H2 domains, and six contained four C2H2 domains (Fig 1, S2 Table). Furthermore, 63 contained a single Q-type C2H2-ZF domain (51 in Br-1i-Q-SF, seven in Br-2i-Mix-SF, and five in Br-3i-SF), followed by 46 containing two Q-type C2H2-ZF domains (37 in Br-2i-Q-SF, six in Br-3i-SF and three in Br-4i-SF, respectively) and one containing four Q-type C2H2-ZF domains (Br-4i-SF) (S2 Table).
Fig 1. Classification of C2H2-ZFP genes in Brassica rapa genome.
Numbers of C2H2-ZFPs in the 12 different subsets and containing 1–5 and 9 C2H2-ZF domains were shown by different colors. For the detailed information, see S2 Table.
Analysis of the 301 B. rapa C2H2-ZFP sequences showed that their length of amino acid ranged from 81 to 1673 aa, with computed molecular weight ranged from 9.34 kDa to 190.32 kDa, and isoelectric points (pIs) ranged from 4.42 to 10.46. The predictions of subcellular localization showed that 277 (92%) members of B. rapa C2H2-ZFPs were localized in nucleus, 11 (3.65%) C2H2-ZFs were localized in cytoplasm, and 13 (4.31%) were located in different organelles (S1 Table).
Phylogenetic analysis of the classified B. rapa C2H2-ZFP genes
In order to investigate the phylogenetic relationships of 301 C2H2-ZFPs in B. rapa, a phylogenetic tree was constructed based on the multiple sequence alignment of all identified C2H2-ZFPs by using the ClustalW program implemented in MEGA 7.0 software (Fig 2). The phylogenetic relationships of 12 main C2H2-ZFP types (Br-t1-SF, Br-t2-SF, Br-1i-Q-SF, Br-1i-M-SF, Br-1i-Z-SF, Br-1i-D-SF, Br-2i-Q-SF, Br-2i-M-SF, Br-2i-Z-SF, Br-2i-Mix-SF, Br-3i-SF and Br-4i-SF) were clearly illustrated by the tree in Fig 2. The C2H2-ZFPs that had been classified into a same subset according to their contained C2H2-ZFs (Fig 1) tended to be also clustered together in the phylogenetic tree, although most of the main branches have often low bootstrap values (<50%) (Fig 2). Interestingly, a few exceptions were also observed as shown in Fig 2 where the differently colored points were occasionally found among a given C2H2-ZFP type.
Fig 2. Phylogenetic tree analysis of the classified Brassica rapa C2H2-ZFPs.
The tree was determined by using MEGA7.0 software with the Neighbor–Joining (NJ) algorithm and a bootstrap analysis of 1,000 replicates. The 12 classified B. rapa C2H2-ZFP subsets were clustered and indicated by different colors, respectively.
Chromosomal distribution and gene duplication
To obtain a global view about the distribution of C2H2-ZFP genes on the B. rapa genome, we retrieved the chromosomal location data of each identified C2H2-ZFP gene from the BRAD database (S1 Table) and constructed a physical map of C2H2-ZFP genes in B. rapa (Fig 3). Our results showed that 295 out of 301 (98.0%) B. rapa C2H2-ZFP genes were mapped into the 10 chromosomes (Fig 3), while the remaining six were located actually on scaffolds, and not mapped to any specific chromosome (S1 Table). As illustrated by Fig 3, these C2H2-ZFP genes were unequally distributed across the 10 chromosomes of B. rapa. The number of C2H2-ZF protein genes is 24, 42, 42, 10, 24, 30, 35, 17, 35 and 26 for A01, A02, A03, A04, A05, A06, A07, A08, A09 and A10, respectively. There existed an obvious tendency of clustering of C2H2-ZFP genes on some regions of B. rapa genome. 183 out of 301 (60.8%) C2H2-ZFP genes were involved in segmental duplication, and 16 pairs of C2H2-ZFP genes were involved in tandem duplication (Fig 3).
Fig 3. Distribution of 295 C2H2-ZFP genes on 10 chromosomes of Brassica rapa.
The 295 C2H2-ZFP genes unevenly located on each conserved collinear blocks of the chromosomes. Chromosome number (A01-A10) is indicated at the top of each chromosome. Gene name is indicated on the right side of each chromosome. The physical position (Mb) of each mapped gene is indicated on the left side of each chromosome. The genes located on duplicated chromosomal segments are framed by same colors and connected by same color lines between the two relevant chromosomes. The tandem repeated genes are framed by red color on the chromosomes.
Syntenic relationships between B. rapa and A. thaliana
Brassica species diverged from a common ancestor with the Arabidopsis lineage ~17 million years ago (MYA), and have all undergone a whole genome triplication (WGT) event ~15.9 MYA after their divergence from Arabidopsis [59–61]. B. rapa is a paleohexaploid and contains three subgenomes designated as least fractionalized (LF), moderately fractionized (MF1) and most fractionized (MF2) according to the extent of gene loss [51,60,61]. For each B. rapa C2H2-ZFP gene, we identified its syntenic paralogs on three subgenomes of B. rapa as well as its orthologs in A. thaliana from BRAD database (http://brassicadb.org/brad/). On the other hand, for each known Arabidopsis C2H2-ZFP gene, we checked if there existed the corresponding orthologous C2H2-ZFP genes on three B. rapa subgenomes. The syntenic relationships between the C2H2-ZFP genes of B. rapa and A. thaliana were summarized in S3 Table. All the 301 B. rapa C2H2-ZFP genes were assigned to one of the three subgenomes: 130 out of 301 (43.19%) on LF, 104 out of 301 (34.55%) on MF1, and 67 out of 301 (22.26%) on MF2. In 20 cases, the three copies were well conserved on the three subgenomes, while in 56 cases, two of the three copies were present, and in 112 cases, only one copy was present. 263 out of 301 (87.37%) B. rapa C2H2-ZFP genes found to their corresponding 164 syntenic orthologs in A. thaliana, which were derived from 23 blocks of 7 ancestral chromosomes of translocation Proto-Calepineae Karyotype (tPCK) [51,52,62,63]. According to the actual version of BRAD database, 38 out of 301 (12.63%) B. rapa C2H2-ZFP genes didn’t find their corresponding syntenic orthologs in A. thaliana (S3 Table), while 32 out of 196 (16.33%) A. thaliana C2H2-ZFP genes didn’t find their corresponding orthologs in B. rapa.
To have an idea about the selective pressure of these B. rapa C2H2-ZFP genes, the Ka, Ks and Ka/Ks values, as well as the divergence times were calculated for the different A. thaliana and B. rapa orthologous C2H2-ZFP gene pairs and summarized in S4 Table. The Ka/Ks ratios varied from 0.0116 to 1.4919, with an average value of 0.3082. Two orthologous gene pairs, AT1G04445-Bra030571 and AT5G61470-Bra029315, have Ka/Ks ratios (1.0174 and 1.4919, respectively) superior to 1, implying that they experienced positive selection, while all other 262 gene pairs have Ka/Ks ratios inferior to 1, implying that they experienced negative selection in the process of species evolution. The estimated divergence times for orthologous gene pairs between A. thaliana and B. rapa C2H2-ZFP genes ranged from 6.23 to 38.60 MY, with an average value of 18.29 MY, signifying that these C2H2-ZFP genes had evolved with different rates during species evolution [64].
Additional domain analysis outside of the C2H2-ZF domain
In order to better classify the identified 301 B. rapa C2H2-ZFPs, we inspected the full-length protein sequence of each C2H2-ZFP by Smart tool analysis for additional known domain besides the C2H2-ZF domains. A total of 57 other known domains outside the C2H2-ZF domains were identified. Based on the presence or not of these additional domains and their organization, the 301 B. rapa C2H2-ZNPs were classified into 34 major groups and subgroups (S5 Table). The first major group includes 205 members (205/301 = 68.1%) containing only C2H2-ZF domains with no any other additional domain detected. The second major group includes 15 members (15/301 = 4.98%) containing each a Coiled-coil domain in addition to the C2H2-ZF domain(s). While the rest of 32 groups includes, 1–10 members with 1–7 additional known domains predicted to be involved in various molecular functions, such as protein-protein interaction (ANK, Ald_Xan_dh_C2, coiled-coil, PWWP, JmjC), protein binding (SET, PUG, CactinC_cactus, UBX), nucleic acid binding (KH, Ald_Xan_Dh_C2, tRNA_anti-codon, G_patch, DEXDc, tRNA-synt_2, HELICc, PPR, HA2, Kin17 curved), Histone binding/phosphorylation/methylation (POST SET, JmjN VEFS-Box, zf-TRM13_CCCH, MTS, Inositol_P domain), and Zinc ion binding (RING, AN1, IBR, Znf UBA, Znf C4HC3), etc. Other known domains such as C1, Transmembrane, NYN, P4HC, UFD1, zf-LYAR domains were also found to be associated with the C2H2-ZFP domain. All these additional known domains were also found in other plant species including A. thaliana, Medicago truncatula, Nicotiana tobacum, Oryza sativa, Vitis vinifera, Glycine max, Glycine Soja, Solanum lycopersicum, Malus Domestica, Zea mays, etc.
Expression analysis of B. rapa C2H2-ZFP genes in different tissues
To gain insights into the expression patterns of individual B. rapa C2H2-ZFP genes in different tissues, we used a publicly available RNA-seq transcriptomic dataset generated by deep sequencing of mRNA from six different tissues (callus, root, stem, leaf, flower and silique) of B. rapa (GSE43245). Except 17 genes, the expression data of 284 B. rapa C2H2-ZFP genes were available from the dataset, of which four genes showed an expression value of zero for all the six tissues and were then excluded from the analysis, while the remaining 280 genes were expressed in at least one of the six tissues. Based on the log2-transformed fragments per kilobase of transcript per million fragments mapped (FPKM) values of the dataset, a clustered heat map (Fig 4) was generated to displaying the expression patterns of B. rapa C2H2-ZF protein genes in callus, root (root_1 and root_2), stem, leaf (leaf_1 and leaf_2), flower and silique. The 280 B. rapa C2H2-ZFP genes were clustered into seven groups, The group I includes 9 genes, of which 88.8% were preferentially expressed both in root and stem. The group II includes 12 genes, which were all preferentially expressed in the stem, 83% in leaf, 66% in silique and 58% in flower. The group III includes 33 C2H2-ZFP genes, of which 72% were preferentially expressed in flower. The group IV includes 65 genes which were almost all highly and consecutively expressed in all the six tissues. The group V includes 11 genes, of which 100% were highly expressed in five tissues except in flower (54%). The group VI includes 128 genes, which were almost all very lowly (or not) expressed in all the tested tissues, except 31(24%) genes which were preferentially expressed in one or more tissues with relatively higher expression levels. The group VII includes 22 C2H2-ZFP genes, of which more than 63% were preferentially expressed in the root.
Fig 4. Expression profiles of 280 Brassica rapa C2H2-ZFP genes in different tissues or organs revealed by clustering analysis of RNA-seq data.

The 280 genes were divided into seven major groups (I-VII) based on the log2-transformed fragments per kilobase of transcript per million fragments mapped (FPKM) values. The scale bar indicates relative expression level as shown on the top left side. The different tissue types are shown on the top side. The individual gene names are indicated on the right side.
Table 2 summarized the distribution of 280 C2H2-ZFP genes in different expression groups of Fig 4 in relation to their classification by C2H2-ZF domain types in Fig 1. We can observe that the Br-t1-SF members were mainly distributed into the expression group VI (33.3%) and IV (31.2%), while the Br-1i-Q-SF into group VI (65.9%) and VII (19.1%), the Br-2i-Q-SF into group VI (47.1%) and V (20.6%), the Br-1i-M-SF into group IV (38.2%) and VI (38.2%), etc.
Table 2. Distribution of 280 Brassica rapa C2H2-ZFP genes in the different expression groups of Fig 4 in relation to the classification of their encoded proteins in Fig 1.
Values in parentheses indicate the percentages of genes per total genes of each C2H2 finger type in each expression group.
| Expression group | No. of genes | Br-t1-SF | Br-t2-SF | Br-1i-Q-SF | Br-2i-Q-SF | Br-1i-M-SF | Br-2i-M-SF | Br-1i-D-SF | Br-1i-Z-SF | Br-2i-Z-SF | Br-2i-Mix-SF | Br-3i-SF | Br-4i-SF |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| I | 9 | 2 (4.1)b | - | 4 (8.5) | 3 (8.8) | - | - | - | - | - | - | - | - |
| II | 12 | 8 (16.7) | - | - | - | 4 (4.5) | - | - | - | - | - | - | - |
| III | 33 | 7 (14.6) | 1 (50.0) | 2 (4.2) | 3 (8.8) | 12 (13.5) | 4 (28.6) | 1 (50.0) | 2 (18.2) | - | - | 1 (10.0) | - |
| IV | 65 | 15 (31.2) | 1 (50.0) | - | 1 (2.9) | 34 (38.2) | 5 (35.7) | 1 (50.0) | 4 (36.4) | 1 (25.0) | 1 (7.1) | 1 (10.0) | 1 (20.0) |
| V | 11 | - | - | 1 (2.1) | 7 (20.6) | 1 (1.1) | - | - | - | - | 1 (7.1) | - | 1 (20.0) |
| VI | 128 | 16 (33.3) | - | 31 (65.9) | 16 (47.1) | 34 (38.2) | 5 (35.7) | - | 4 (36.4) | 3 (75.0) | 8 (57.1) | 8 (80.0) | 3 (60.0) |
| VII | 22 | - | - | 9 (19.1) | 4 (11.8) | 4 (4.5) | - | - | 1 (9.1) | - | 4 (28.6) | - | - |
| Total No. | 280 | 48 | 2 | 47 | 34 | 89 | 14 | 2 | 11 | 4 | 14 | 10 | 5 |
Expression analysis of the B. rapa C2H2-ZFP genes under abiotic stresses
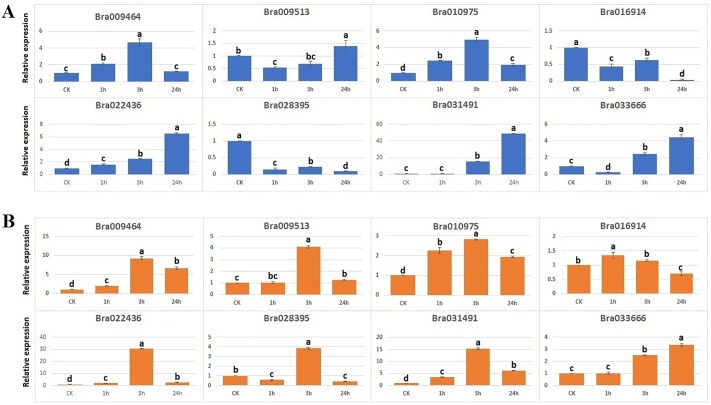
To gain some information about the response of C2H2-ZFP genes to abiotic stresses, we examined the expression response of eight B. rapa C2H2-ZFP genes (S6 Table) to salt (200 mM NaCl) (Fig 5A) and drought (20% (w/v) PEG6000) (Fig 5B) stresses in the leaves of three weeks seedlings by using qRT-PCR technique. The eight genes were selected based on their high expression levels in most of the tested tissues. The results showed that all the eight B. rapa C2H2-ZFP genes were responsive to the two abiotic stress treatments.
Fig 5. qRT-PCR expression patterns of eight Brassica rapa C2H2-ZFP genes under salt (A) and drought (B) treatments.
The time points represent by x-axis and the scale of relative expression shown by y-axis. Tukey tests were used to determine differences among effects on different time courses under salt (A) and drought (B) treatments and different letters indicate the statistical significant deference’s with p ≤ 0.05.
In the case of salt treatment, six B. rapa C2H2-ZF protein genes were up-regulated and two down-regulated compared to control (CK) after 1 h, 3 h or 24 h of salt treatment, respectively (Fig 5A). The most remarkable case is the gene Bra031491 (corresponding to AT1G60640) which was induced by more than 40-fold under salt treatment at 24 h. Furthermore, the gene Bra022436 was progressively inducted along with the time and reached 6-fold at 24 h compared to the control.
In the case of drought treatment, all the six B. rapa C2H2-ZFP genes were responsive to the treatment compared to control (CK) after 1 h, 3 h or 24 h of treatment, respectively (Fig 5B). The highest induction was recorded for Bra022436, (corresponding to AT3G19580) which showed >30-fold up-regulation at 3h and moderately expressed at 24 h compared to the control. Furthermore, the genes Bra009464, Bra009513, Bra010975, and Bra031491 were progressively inducted along with the time and reached 2.5 to 15-fold at 3 h compared to the control and retained the expression more than 1 to 7-fold at 24 hours of treatment compared to the control.
Comparison between A. thaliana and B. rapa orthologous C2H2-ZFP genes
In order to gain more insights into the degree of conservation between B. rapa and A. thaliana C2H2-ZFP genes, we compared the subcellular localization, number of C2H2 domain(s), number and type of additional domain(s), and expression pattern between their corresponding orthologs (S7 and S8 Tables). Our results showed that the subcellular localizations were globally well conserved for all of the 264 A. thaliana—B. rapa orthologous gene pairs, with some differences observed for 40 out of 264 (15.2%) cases where one or two additional localization(s) were detected for one of the two members. In 23 out of 264 (8.7%) cases, the number of C2H2-ZF domains contained by the compared orthologous C2H2-ZFPs was different between B. rapa and A. thaliana, with six cases of increase and 17 cases of decrease in the number of C2H2-ZF domains in B. rapa compared to A. thaliana. In 20 out of 264 (7.6%) cases, the type or/and number of additional known domains differed between B. rapa and A. thaliana (S7 Table).
For comparison of expression pattern, we resembled the expression data of five tissues or organs (root, stem, leaf, flower and silique) for 191 out of 264 A. thaliana–B. rapa orthologous C2H2-ZFP gene pairs, and summarized them in S8 Table. The Pearson’s correlation coefficient (P) of expression levels (sum of values from five tissues or organs) between B. rapa and A. thaliana orthologous C2H2-ZFP genes was calculated and equaled 0.68. In 32 out of 191 (16.8%) cases (gene pairs), the correlation coefficients of expression patterns (different expression levels in five tissues or organs) between B. rapa and A. thaliana orthologous genes were superior to 0.8. Based on the data resembled in S8 Table, an expression heatmap was generated and presented in Fig 6, giving a global view of expression patterns of 191 B. rapa—A. thaliana orthologous C2H2-ZFP gene pairs. We can observe that while some C2H2-ZFP gene members tend to conserve similar expression patterns between B. rapa and A. thalian, many other members showed different expression patterns across the two species.
Fig 6. Heatmap comparison of expression patterns in five tissues or organs (root, stem, leaf, flower and silique) between Brassica rapa and Arabidopsis thaliana orthologous C2H2-ZFP genes (191 gene pairs).
The map was constructed using Cluster software v3.0 (http://bonsai.hgc.jp/mdehoon/software/cluster/) with the option of “center genes”. The expression data of B. rapa C2H2-ZFP genes were extracted from the dataset GSE43245 (accession number) of NCBI Gene Expression Omnibus (GEO) database, while those of A. thaliana C2H2-ZFP genes were extracted from TAIR database (https://www.arabidopsis.org/) using the Arabidopsis eFP Browser. The A. thaliana gene names are indicated on the left side, while the B. rapa gene names (followed by their subgenome localisation) are indicated on the right side of the map. The scale representing the relative signal values is shown on the lower right side. The detail values based on which the map was constructed are summarized in S8 Table.
Discussion
C2H2-ZF transcription factor family are widely existing in all eukaryotic organisms and involved in many biological processes, including plant growth, development, hormone signaling, and stress response [4,18,22,33]. To date, C2H2-ZFP genes have been investigated in multi-plant species, such as A. thaliana, rice, foxtail millet, durum wheat, tobacco, and poplar [14,15,18–21]. In the present study, a total of 301 C2H2-ZFP genes were identified with 533 C2H2-ZF domains in B. rapa genomes. The 301 C2H2-ZFPs were further subdivided into 12 different classes (Fig 1), based on the variation of the plant-specific conserved amino acid sequence “QALGGH” and distances between metal ligands within C2H2-ZF domains (Table 1). This classification was well supported by phylogenetic analysis with only a few exceptions (Fig 2). Majority of B. rapa C2H2-ZFPs contain one or more C2H2 domains with the QALGGH motif, and were identified as Q-type C2H2-ZFPs (Fig 1, S2 Table) [65]. The Q-type C2H2-ZFPs are plant specific, and have not been reported in any other organisms suggesting that these proteins may be involved in plant-specific life processing [8]. The complexity of these C2H2-ZF domains (Fig 1, S2 Table) plus the presence of additional known domain(s) in some C2H2-ZFP members (S5 Table) implies the high functional diversity of these C2H2-ZFPs in plant growth and development. Our classifications of B. rapa C2H2-ZFPs provide then helpful information for further functional characterization of this gene family among Brassica species.
It has been reported, that the model plant A. thaliana, has experienced three WGD events: a γ event shared with most dicots and two following genome duplications (α and β) shared with other associates of Brassicales [66]. B. rapa shares a common ancestor with A. thaliana, and has also experienced the three WGD events. In addition, the ancestor of B. rapa underwent an additional WGT event around 13 to 17 MYA followed by genome diploidization involving substantial genome reshuffling and gene losses in duplicated genomic blocks [47,67]. In the current study, the number of C2H2-ZFPs identified in B. rapa (301) was 1.54 times of that in A. thaliana (196), and 264 syntenic C2H2-ZFP orthologous gene pairs involving 148 A. thaliana C2H2-ZFP genes were identified between the two species (S3 and S4 Tables), showing the important gene losses of C2H2-ZFPs in B. rapa genome following the WGT event. Our syntenic relationship analysis (S3 Table) showed that in 20 cases, the three triplicated C2H2-ZFP gene copies were well conserved on the three subgenomes, while in 56 other cases, two of the three copies were maintained in B. rapa genome. Thus, about one-third of C2H2-ZFP gene members were increased due to the segmental chromosomal duplication caused by the WGT event. The tandem duplication is another central mode of gene expansion. For example, 1135, 1569, 1751 and 2137 tandemly duplicated gene clusters were discovered in Thellungiella parvula, A. thaliana, B. rapa, and Arabidopsis lyrata, respectively [68]. Among the 301 B. rapa C2H2-ZFP genes, 16 tandem arrays involving 32 C2H2-ZFP genes were identified (Fig 3), indicating that the tandem duplication has also contributed to the expansion of C2H2-ZFP gene family in the B. rapa genome. These duplicated C2H2-ZFP gene members are the results of natural and artificial selections during species evolution, and constitute then interesting candidates for studying the functional diversification of duplicated genes in Brassica crops [69, 70]
Our analysis of Ka/Ks values of the 264 orthologous C2H2-ZFP gene pairs between A. thaliana and B. rapa (S4 Table) indicated that almost all tested gene pairs (except AT1G04445-Bra030571 and AT5G61470-Bra029315) have the Ka/Ks ratios inferior to 1, implying that these genes have experienced purifying selection during species evolution [71,72]. AT1G04445 and AT5G61470 each encodes a putative protein of 172 aa and 304 aa in size, respectively, and have not been investigated in the model plant A. thaliana, constituting then the interesting targets together with their B. rapa orthologs for further functional study in both species. The estimated divergence time of each of the 264 orthologous C2H2-ZFP gene pairs between A. thaliana and B. rapa ranged from 6.23 to 38.60 MY, with an average value of 18.29 MY, showing that these gene members have undergone different selective pressures resulting in different evolutionary rates during species evolution [64]. The most slowly evolved gene pair is AT3G23130-Bra023759 with an estimated divergence time of 6.23 MY and a Ka/Ks ratio of 0.5723, while the most rapidly evolved gene pair is AT5G16470-Bra023567 with an estimated divergence time of 38.60 MY and a Ka/Ks ratio of 0.0115 (S4 Table). AT3G23130 (also named as FLO10, FON1 or SUPERMAN) is characterized as a flower-specific gene controlling the boundary of the stamen and carpel whorls (https://www.arabidopsis.org/servlets/TairObject?id=37697&type=locus), while AT5G16470 (also named as MBS2) is characterized as required for induction of singlet oxygen-dependent gene expression and involved in ROS signaling mediated stress responsive pathway in A. thaliana (https://www.arabidopsis.org/servlets/TairObject?id=134268&type=locus). We can expect that those gene pairs with a low estimated divergence time value may maintain a conserved function between the two species, while those with a high divergence time value may generate new functions as have been suggested in other similar studies [73–75].
Many previous studies reported that most of C2H2-ZFPs function as transcription factors and play important roles in plant growth and development [23,36,37,40,76]. Our subcellular localization prediction showed that 92% of B. rapa C2H2-ZFPs were located in nucleus (S1 Table), which supported their functional roles as transcription factors in nucleus. Our expression pattern analysis showed that 280 of 301 B. rapa C2H2-ZFP genes were expressed in at least one of the six tested tissues, of which about one-third was almost always highly and consecutively expressed in all the tested tissues, one-third preferentially expressed in one to three tissues, and one-third very lowly expressed in all the tested tissues, illustrating the functional diversity of this gene family in B. rapa plant growth and development (Fig 4). Several members displaying tissue-specifically expressed patterns may be the good candidates for further in-depth studies for their molecular functions and potential applications on genetic improvement of Brassica species.
Our expression pattern analysis of eight B. rapa C2H2-ZFP genes in response to salt (Fig 5A) and drought (Fig 5B) stresses showed that all of them were responsive to the treatments of two abiotic stresses with different expression levels (Fig 5). One of the eight tested genes, Bra009464, orthologous to A. thaliana gene AT5G04340 or AtZAT6, was significantly up-regulated at 1h (1-fold) and 3h (up to 4-fold) under salt stress (Fig 5A). Interestingly, AtZAT6 was also up-regulated at 1h and 3h after treatment of Col-0 plants by 300 mM NaCl as demonstrated in a previous study [77], suggesting that the functions of two orthologous genes were probably well conserved in both species. Moreover, overexpression of AtZAT6 positively regulated the cadmium tolerance in Arabidopsis through the glutathione-dependent pathway [78], and showed pleiotropic phenotypes with curly leaves, small sized plant at vegetative stage and reduced size of floral organs and siliques at the reproductive stage [77]. Similarly, the orthologous gene of Bra009513 in A. thaliana is AT5G03740 or HD2C, which was shown to be involved in ABA, salt [79], and heat stress responses [80]. The orthologous gene of Bra022436 in A. thaliana is AT3G19580 or AZF2, which was shown to function as a transcriptional repressor involved in the inhibition of plant growth under abiotic stress conditions [24,81]. These results indicated that all the eight tested genes were more or less involved in stress response in B. rapa, in a similar way as their orthologs in A. thaliana. Further stress responsive expression analysis of a larger number of B. rapa C2H2-ZFP genes, including those lowly expressed ones, would allow to identify the most actives members involved in stress tolerance in Brassica crops.
Our comparative analysis between A. thaliana and B. rapa corresponding orthologous C2H2-ZFP genes (264 gene pairs) showed that the majority of them encode proteins with conserved subcellular localization, C2H2-ZF domain(s) and additional known domain(s) between the two species (S7 Table). This means that most of the C2H2-ZFP members may conserved their basic cellular functions across Arabidopsis and Brassica species, while the other minor portion (7~15%) of C2H2-ZFP genes may have deviated from their initial functions or gained new functions during the evolution of plant species. Comparison of expression pattern between A. thaliana and B. rapa corresponding orthologous C2H2-ZFP genes (191 gene pairs) showed that there existed a good positive correlation of expression levels (sum of values from five tissues or organs) between Arabidopsis and B. rapa C2H2-ZFP genes, with a P value equals 0.68 (S8 Table), reflecting somewhat the degree of functional conservation of these C2H2-ZFP genes between the two species. However, only about one-third of A. thaliana—B. rapa orthologous C2H2-ZFP gene pairs displayed P values ranging from 0.5 to 1 for their expression patterns in five tissues or organs, while other two-third genes pairs displayed P values ranging from -1 to 0.5 (S8 Table), suggesting that the functional roles in plant growth and development may be modified between the two species for most of these C2H2-ZFP genes by differed expression patterns which were well illustrated in Fig 6.
Conclusions
In the current study, we analyzed the C2H2-ZFP family in B. rapa using the available Brassica genome databases. A total of 301 C2H2-ZFP genes were identified and further characterized through analysis of conserved amino acid residues in C2H2-ZF domains and their organization, subcellular localization, phylogeny, additional domain, chromosomal location, synteny relationship, and expression pattern, etc. We also analyzed the expression patterns of eight B. rapa C2H2-ZFP genes under salt and drought stress conditions by using qRT-PCR technique. Our results showed that about one-third of these B. rapa C2H2-ZFP genes were originated from segmental duplication caused by the WGT around 13 to 17 MYA, one-third of them were highly and consecutively expressed in all tested tissues, and 92% of them were located in nucleus by prediction supporting then their functional roles as transcription factors, of which some may play important roles in plant growth and development. In addition, a few of these B. rapa C2H2-ZFP genes were shown to be involved in stress responses in a similar way as their orthologs in A. thaliana. Comparison between A. thaliana and B. rapa orthologous C2H2-ZFP genes showed that most of these C2H2-ZFP gene members encodes proteins with conserved subcellular localization and functional domains between the two species but differed in their expression patterns in five tissues or organs. Thus, our study provides valuable information for further functional determination of each C2H2-ZFP gene across the Brassica species, and may help to select the appropriate gene targets for further genetic engineering and genetic improvement of Brassica crops.
Supporting information
(PDF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This work was supported by a start-up fund for distinguished scholars of Fujian Agriculture and Forestry University, No. 114120019 to YHL. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Mackay JP, Crossley M. Zinc fingers are sticking together. Trends Biochem Sci. 1998;23: 1–4. [DOI] [PubMed] [Google Scholar]
- 2.Takatsuji H. Zinc-finger proteins: the classical zinc finger emerges in contemporary plant science. PlantMol Biol. 1999;39: 1073–1078. [DOI] [PubMed] [Google Scholar]
- 3.Von Arnim A, Deng XW. Ring finger motif of Arabidopsis thaliana COP1 defines a new class of zinc-binding domain. J Biol Chem. 1993;268: 19626–31. [PubMed] [Google Scholar]
- 4.Liu D, He S, Song X, Zhai H, Liu N, Zhang D, et al. IbSIMT1, a novel salt-induced methyltransferase gene from Ipomoea batatas, is involved in salt tolerance. Plant Cell Tissue Organ Cult. 2015;120: 701–15. [Google Scholar]
- 5.Liu D, He S, Zhai H, Wang L, Zhao Y, Wang B, et al. Overexpression of IbP5CR enhances salt tolerance in transgenic sweet potato. Plant Cell Tissue Organ Cult. 2014;117: 1–16. [Google Scholar]
- 6.Liu D, Yang L, Luo M, Wu Q, Liu S, Liu Y. Molecular cloning and characterization of PtrZPT2-1, a ZPT2 family gene encoding a Cys2/His2-type zinc finger protein from trifoliate orange (Poncirus trifoliata (L.) Raf.) that enhances plant tolerance to multiple abiotic stresses. Plant Sci. 2017;263: 66–78. 10.1016/j.plantsci.2017.07.012 [DOI] [PubMed] [Google Scholar]
- 7.Wang F, Hong Z, An Y, Si Z, He S, Liu Q. Overexpression of IbMIPS1 gene enhances salt tolerance in transgenic sweet potato. J Integr Agric. 2016;15: 271–81. [Google Scholar]
- 8.Takatsuji H. Zinc-finger transcription factors in plants. Cell. Mol. Life Sci.1998; 54: 582–96. 10.1007/s000180050186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cassandri M, Smirnov A, Novelli F, Pitolli C, Agostini M, Malewicz M, et al. Zinc-finger proteins in health and disease. Cell Death Dis. 2017;3: 17071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kosarev P, Mayer KF, Hardtke CS. Evaluation and classification of RING-finger domains encoded by the Arabidopsis genome. Genome Biol. 2002;3: research0016.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pavletich NP, Pabo CO. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science. 1991;252: 809–17. [DOI] [PubMed] [Google Scholar]
- 12.Schuh R, Aicher W, Gaul U, Côte S, Preiss A, Maier D, et al. A conserved family of nuclear proteins containing structural elements of the finger protein encoded by Krüppel, a Drosophila segmentation gene. Cell. 1986;47: 1025–32. [DOI] [PubMed] [Google Scholar]
- 13.Zhang X, Zhang B, Li MJ, Yin XM, Huang LF, Cui YC, et al. OsMSR15 encoding a rice C2H2-type zinc finger protein confers enhanced drought tolerance in transgenic Arabidopsis. J Plant Biol. 2016;59: 271–81. [Google Scholar]
- 14.Englbrecht CC, Schoof H, Böhm S. Conservation, diversification and expansion of C2H2 zinc finger proteins in the Arabidopsis thaliana genome. BMC Genomics. 2004;5: 39 10.1186/1471-2164-5-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Agarwal P, Arora R, Ray S, Singh AK, Singh VP, Takatsuji H, et al. Genome-wide identification of C2H2 zinc-finger gene family in rice and their phylogeny and expression analysis. Plant Mol. Boil. 2007;65: 467–85. [DOI] [PubMed] [Google Scholar]
- 16.Yuan S, Li X, Li R, Wang L, Zhang C, Chen L, et al. Genome-wide identification and classification of soybean C2H2 zinc finger proteins and their expression analysis in legume-Rhizobium symbiosis. Front Microbiol. 2018;9: 126 10.3389/fmicb.2018.00126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wei K, Pan S, Li Y. Functional characterization of maize C2H2 zinc-finger gene family. Plant Mol Biol Rep. 2016;34: 761–76. [Google Scholar]
- 18.Muthamilarasan M, Bonthala VS, Mishra AK, Khandelwal R, Khan Y, Roy R, et al. C2H2 type of zinc finger transcription factors in foxtail millet define response to abiotic stresses. Funct Integr Genomics. 2014;14: 531–43. 10.1007/s10142-014-0383-2 [DOI] [PubMed] [Google Scholar]
- 19.Faraji S, Rasouli SH, Kazemitabar SK. Genome-wide exploration of C2H2 zinc finger family in durum wheat (Triticum turgidum ssp. Durum): insights into the roles in biological processes especially stress response. Biometals. 2018;10: 1007. [DOI] [PubMed] [Google Scholar]
- 20.Yang M, Chao J, Wang D, Hu J, Wu H, Gong D, et al. Genome-wide identification and expression profiling of the C2H2-type zinc finger protein transcription factor family in tobacco. Yi Chuan. 2016;38: 337–49. 10.16288/j.yczz.15-440 [DOI] [PubMed] [Google Scholar]
- 21.Liu Q, Wang Z, Xu X, Zhang H, Li C. Genome-wide analysis of C2H2 Zinc-finger family transcription factors and their responses to abiotic stresses in poplar (Populus trichocarpa). PLoS One. 2015;10: e0134753 10.1371/journal.pone.0134753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Y, Liu D, Hu R, Hua C, Ali I, Zhang A, et al. AtGIS, a C2H2 zinc-finger transcription factor from Arabidopsis regulates glandular trichome development through GA signaling in tobacco. Biochem Biophys Res Commun. 2017;483: 209–15. 10.1016/j.bbrc.2016.12.164 [DOI] [PubMed] [Google Scholar]
- 23.Takatsuji H, Mori M, Benfey P, Ren L, Chua N. Characterization of a zinc finger DNA‐binding protein expressed specifically in Petunia petals and seedlings. EMBO J. 1992;11: 241–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kodaira KS, Qin F, Tran LSP, Maruyama K, Kidokoro S, Fujita Y, et al. Arabidopsis C2H2 zinc-finger proteins AZF1 and AZF2 negatively regulate ABA-repressive and auxin-inducible genes under abiotic stress conditions. Plant Physiol. 2011;111: 182683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang J, Sun SJ, Xu DQ, Yang X, Bao YM, Wang ZF, et al. Increased tolerance of rice to cold, drought and oxidative stresses mediated by the overexpression of a gene that encodes the zinc finger protein ZFP245. Biochem Biophys Res Commun. 2009;389: 556–61. 10.1016/j.bbrc.2009.09.032 [DOI] [PubMed] [Google Scholar]
- 26.Kim JC, Lee SH, Cheong YH, Yoo CM, Lee SI, Chun HJ, et al. A novel cold-inducible zinc finger protein from soybean, SCOF-1, enhances cold tolerance in transgenic plants. Plant J. 2001;25: 247–59. [DOI] [PubMed] [Google Scholar]
- 27.Tao Z, Huang Y, Zhang L, Wang X, Liu G, Wang H. BnLATE, a Cys2/His2-type zinc-finger protein, enhances silique shattering resistance by negatively regulating lignin accumulation in the silique walls of Brassica napus. PloS One. 2017;12: e0168046 10.1371/journal.pone.0168046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim SH, Hong JK, Lee SC, Sohn KH, Jung HW, Hwang BK. CAZFP1, Cys2/His2-type zinc-finger transcription factor gene functions as a pathogen-induced early-defense gene in Capsicum annuum. Plant Mol Biol. 2004;55: 883–904. 10.1007/s11103-004-2151-5 [DOI] [PubMed] [Google Scholar]
- 29.Xu DQ, Huang J, Guo SQ, Yang X, Bao YM, Tang HJ, et al. Overexpression of a TFIIIA-type zinc finger protein gene ZFP252 enhances drought and salt tolerance in rice (Oryza sativa L.). FEBS Lett. 2008;582: 1037–43. 10.1016/j.febslet.2008.02.052 [DOI] [PubMed] [Google Scholar]
- 30.Hichri I, Muhovski Y, Žižková E, Dobrev PI, Franco-Zorrilla JM, Solano R, et al. The Solanum lycopersicum Zinc Finger2 cysteine-2/histidine-2 repressor-like transcription factor regulates development and tolerance to salinity in tomato and Arabidopsis. Plant Physiol. 2014;164: 1967–90. 10.1104/pp.113.225920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rai AC, Singh M, Shah K. Effect of water withdrawal on formation of free radical, proline accumulation and activities of antioxidant enzymes in ZAT12-transformed transgenic tomato plants. Plant Physiol Biochem. 2012;61: 108–14. 10.1016/j.plaphy.2012.09.010 [DOI] [PubMed] [Google Scholar]
- 32.Luo X, Bai X, Zhu D, Li Y, Ji W, Cai H, et al. GsZFP1, a new Cys2/His2-type zinc-finger protein, is a positive regulator of plant tolerance to cold and drought stress. Planta. 2012;235: 1141–55. 10.1007/s00425-011-1563-0 [DOI] [PubMed] [Google Scholar]
- 33.Luo X, Cui N, Zhu Y, Cao L, Zhai H, Cai H, et al. Over-expression of GsZFP1, an ABA-responsive C2H2-type zinc finger protein lacking a QALGGH motif, reduces ABA sensitivity and decreases stomata size. J Plant Physiol. 2012;169: 1192–202. 10.1016/j.jplph.2012.03.019 [DOI] [PubMed] [Google Scholar]
- 34.Xu Z, Raza Q, Xu L, He X, Huang Y, Yi J, et al. GmWRKY49, a salt-responsive nuclear protein, improved root length and governed better salinity tolerance in transgenic Arabidopsis. Front Plant Sci. 2018;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Y, Chu Z, Luo J, Zhou Y, Cai Y, Lu Y, et al. The C2H2 zinc‐finger protein Sl ZF3 regulates AsA synthesis and salt tolerance by interacting with CSN 5B. Plant Biotechnol J. 2018;16: 1201–13. 10.1111/pbi.12863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kundu A, Das S, Basu S, Kobayashi Y, Kobayashi Y, Koyama H, et al. GhSTOP1, a C2H2 type zinc finger transcription factor is essential for Aluminum and proton stress tolerance and lateral root initiation in cotton. Plant Biol. 2018;10: 1111–12895. [DOI] [PubMed] [Google Scholar]
- 37.Morita MT, Sakaguchi K, Kiyose Si, Taira K, Kato T, Nakamura M, et al. A C2H2‐type zinc finger protein, SGR5, is involved in early events of gravitropism in Arabidopsis inflorescence stems. Plant J. 2006;47: 619–28. 10.1111/j.1365-313X.2006.02807.x [DOI] [PubMed] [Google Scholar]
- 38.Dathan N, Zaccaro L, Esposito S, Isernia C, Omichinski JG, Riccio A, et al. The Arabidopsis SUPERMAN protein is able to specifically bind DNA through its single Cys 2–His 2 zinc finger motif. Nucleic Acids Res. 2002;30: 4945–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sakai H, Krizek BA, Jacobsen SE, Meyerowitz EM. Regulation of SUP expression identifies multiple regulators involved in Arabidopsis floral meristem development. Plant Cell. 2000;12: 1607–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sakai H, Medrano LJ, Meyerowitz EM. Role of SUPERMAN in maintaining Arabidopsis floral whorl boundaries. Nature. 1995;378: 199 10.1038/378199a0 [DOI] [PubMed] [Google Scholar]
- 41.Prunet N, Yang W, Das P, Meyerowitz EM, Jack TP. SUPERMAN prevents class B gene expression and promotes stem cell termination in the fourth whorl of Arabidopsis thaliana flowers. Proc Natl Acad Sci USA 2017;114: 7166–71. 10.1073/pnas.1705977114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sagasser M, Lu GH, Hahlbrock K, Weisshaar B. A. thaliana TRANSPARENT TESTA 1 is involved in seed coat development and defines the WIP subfamily of plant zinc finger proteins. Genes Dev. 2002;16: 138–49. 10.1101/gad.212702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kanwal M, Rabbani AM, Iqbal S, Fayyaz L, Afzal M. Genetic diversity in Brassica species and Eruca sativa for yield associated parameters. Genetika. 2014;46: 537–543. [Google Scholar]
- 44.Talalay P, Fahey JW. Phytochemicals from cruciferous plants protect against cancer by modulating carcinogen metabolism. J Nutr. 2001;131: 3027S–33S. 10.1093/jn/131.11.3027S [DOI] [PubMed] [Google Scholar]
- 45.Nagaharu U. Genome analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Jap J Bot. 1935;7: 389–452. [Google Scholar]
- 46.Johnston JS, Pepper AE, Hall AE, Chen ZJ, Hodnett G, Drabek J, et al. Evolution of genome size in Brassicaceae. Ann Bot 2005;95: 229–35. 10.1093/aob/mci016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Town CD, Cheung F, Maiti R, Crabtree J, Haas BJ, Wortman JR, et al. Comparative genomics of Brassica oleracea and Arabidopsis thaliana reveal gene loss, fragmentation, and dispersal after polyploidy. Plant Cell. 2006;18: 1348–59. 10.1105/tpc.106.041665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Letunic I, Doerks T, Bork P. SMART: recent updates, new developments and status in 2015. Nucleic acids Res. 2014;43: D257–D60. 10.1093/nar/gku949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang D, Tong J, Xu Z, Wei P, Xu L, Wan Q, et al. Soybean C2H2-type zinc finger protein GmZFP3 with conserved QALGGH motif negatively regulates drought responses in transgenic Arabidopsis. Front Plant sci. 2016;7: 325 10.3389/fpls.2016.00325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gasteiger E, Hoogland C, Gattiker A, Wilkins MR, Appel RD, Bairoch A. Protein identification and analysis tools on the ExPASy server In: Walker JM, editor. The proteomics protocols handbook, Humana Press; 2005. pp. 571–607 [Google Scholar]
- 51.Cheng F, Wu J, Fang L, Wang X. Syntenic gene analysis between Brassica rapa and other Brassicaceae species. Front Plant Sci. 2012;3: 198 10.3389/fpls.2012.00198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mandáková T, Lysak MA. Chromosomal phylogeny and karyotype evolution in x = 7 crucifer species (Brassicaceae). Plant Cell. 2008;20: 2559–70. 10.1105/tpc.108.062166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lysak MA, Cheung K, Kitschke M, Bureš P. Ancestral chromosomal blocks are triplicated in Brassiceae species with varying chromosome number and genome size. Plant Physiol. 2007;145: 402–10. 10.1104/pp.107.104380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Goldman N, Yang Z. A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol Biol Evol. 1994;11: 725–36. 10.1093/oxfordjournals.molbev.a040153 [DOI] [PubMed] [Google Scholar]
- 55.Suyama M, Torrents D, Bork P. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006;34: W609–W612. 10.1093/nar/gkl315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Koch MA, Haubold B, Mitchell-Olds T. Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis, and related genera (Brassicaceae). Mol Biol Evol. 2000;17: 1483–98. 10.1093/oxfordjournals.molbev.a026248 [DOI] [PubMed] [Google Scholar]
- 57.Tong C, Wang X, Yu J, Wu J, Li W, Huang J, et al. Comprehensive analysis of RNA-seq data reveals the complexity of the transcriptome in Brassica rapa. BMC Genomics. 2013;14: 689 10.1186/1471-2164-14-689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C T method. Nature protoc. 2008;3: 1101. [DOI] [PubMed] [Google Scholar]
- 59.Lysak MA, Koch MA, Pecinka A, Schubert I. Chromosome triplication found across the tribe Brassiceae. Genome Res. 2005;15: 516–25. 10.1101/gr.3531105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang X, Wang H, Wang J, Sun R, Wu J, Liu S, et al. The genome of the mesopolyploid crop species Brassica rapa. Nature Genet. 2011;43: 1035 10.1038/ng.919 [DOI] [PubMed] [Google Scholar]
- 61.Liu S, Liu Y, Yang X, Tong C, Edwards D, Parkin IA, et al. The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nature Commun. 2014;5: 3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schranz ME, Lysak MA, Mitchell-Olds T. The ABC’s of comparative genomics in the Brassicaceae: building blocks of crucifer genomes. Trends Plant Sci. 2006;11: 535–42. 10.1016/j.tplants.2006.09.002 [DOI] [PubMed] [Google Scholar]
- 63.Cheng F, Mandáková T, Wu J, Xie Q, Lysak MA, Wang X. Deciphering the diploid ancestral genome of the mesohexaploid Brassica rapa. Plant Cell. 2013;113: 110486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Woodhouse MR, Cheng F, Pires JC, Lisch D, Freeling M, Wang X. Origin, inheritance, and gene regulatory consequences of genome dominance in polyploids. Proc Natl Acad Sci. 2014;111: 5283–5288. 10.1073/pnas.1402475111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kubo KI, Sakamoto A, Kobayashi A, Rybka Z, Kanno Y, Nakagawa H, et al. Cys2/His2 zinc-finger protein family of petunia: evolution and general mechanism of target-sequence recognition. Nucleic acids Res. 1998;26: 608–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bowers JE, Chapman BA, Rong J, Paterson AH. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 2003;422:433 10.1038/nature01521 [DOI] [PubMed] [Google Scholar]
- 67.Yang YW, Lai KN, Tai PY, Li WH. Rates of nucleotide substitution in angiosperm mitochondrial DNA sequences and dates of divergence between Brassica and other angiosperm lineages. J Mol Evol. 1999;48: 597–604. [DOI] [PubMed] [Google Scholar]
- 68.Fang L, Cheng F, Wu J, Wang X. The impact of genome triplication on tandem gene evolution in Brassica rapa. Front Plant Sci. 2012;3: 261 10.3389/fpls.2012.00261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Alam I, Yang YQ, Wang Y, Zhu ML, Wang HB, Chalhou B, Lu YH. Genome-wide identification, evolution and expression analysis of RING finger protein genes in Brassica rapa. Sci Rep. 2017;7: 40690 10.1038/srep40690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang YQ, Lu YH. Genome-wide survey, characterization, and expression analysis of RING finger protein genes in Brassica oleracea and their syntenic comparison to Brassica rapa and Arabidopsis thaliana. Genome. 2018;61: 685–697. 10.1139/gen-2018-0046 [DOI] [PubMed] [Google Scholar]
- 71.Stebbins G. Variation and Evolution in Plants. New York: Columbia University press; 1950. [Google Scholar]
- 72.Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290: 1151–5. [DOI] [PubMed] [Google Scholar]
- 73.Zhang H, Cao Y, Shang C, Li J, Wang J, Wu Z, et al. Genome-wide characterization of GRAS family genes in Medicago truncatula reveals their evolutionary dynamics and functional diversification. PloS One. 2017;12: e0185439 10.1371/journal.pone.0185439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen Y, Yang Z, Xiao Y, Wang P, Wang Y, Ge X, et al. Genome-wide analysis of the NF-YB gene family in Gossypium hirsutum L. and characterization of the role of GhDNF-YB22 in embryogenesis. Int J Mol Sci. 2018;19: 483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.He Y, Liu X, Ye L, Pan C, Chen L, Zou T, et al. Genome-wide identification and expression analysis of two-component system genes in tomato. Int J Mol Sci. 2016;17: 1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kim H, Hwang H, Hong J-W, Lee Y-N, Ahn IP, Yoon IS, et al. A rice orthologue of the ABA receptor, OsPYL/RCAR5, is a positive regulator of the ABA signal transduction pathway in seed germination and early seedling growth. J Exp Bot 2011;63: 1013–24. 10.1093/jxb/err338 [DOI] [PubMed] [Google Scholar]
- 77.Shi H, Wang X, Ye T, Chen F, Deng J, Yang P, et al. The Cysteine2/Histidine2-Type transcription factor ZINC FINGER OF ARABIDOPSIS THALIANA 6 modulates biotic and abiotic stress responses by activating salicylic acid-related genes and C-REPEAT-BINDING FACTOR genes in Arabidopsis. Plant Physiol. 2014;165: 1367–79. 10.1104/pp.114.242404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen J, Yang L, Yan X, Liu Y, Wang R, Fan T, et al. Zinc-finger transcription factor ZAT6 positively regulates cadmium tolerance through glutathione-dependent pathway in Arabidopsis. Plant Physiol. 2016;171: 707–719. 10.1104/pp.15.01882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Luo M, Wang Y-Y, Liu X, Yang S, Lu Q, Cui Y, et al. HD2C interacts with HDA6 and is involved in ABA and salt stress response in Arabidopsis. J Exp Bot 2012;63: 3297–306. 10.1093/jxb/ers059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Buszewicz D, Archacki R, Palusiński A, Kotliński M, Fogtman A, Iwanicka-Nowicka R, et al. HD2C histone deacetylase and a SWI/SNF chromatin remodelling complex interact and both are involved in mediating the heat stress response in Arabidopsis. Plant Cell Environ. 2016;39: 2108–22. 10.1111/pce.12756 [DOI] [PubMed] [Google Scholar]
- 81.Drechsel G, Raab S, Hoth S. Arabidopsis zinc-finger protein 2 is a negative regulator of ABA signaling during seed germination. J Plant physiol. 2010;167: 1418–21. 10.1016/j.jplph.2010.05.010 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.