Abstract
Pythium guiyangense, an oomycete from a genus of mostly plant pathogens, is an effective biological control agent that has wide potential to manage diverse mosquitoes. However, its mosquito-killing mechanisms are almost unknown. In this study, we observed that P. guiyangense could utilize cuticle penetration and ingestion of mycelia into the digestive system to infect mosquito larvae. To explore pathogenic mechanisms, a high-quality genome sequence with 239 contigs and an N50 contig length of 1,009 kb was generated. The genome assembly is approximately 110 Mb, which is almost twice the size of other sequenced Pythium genomes. Further genome analysis suggests that P. guiyangense may arise from a hybridization of two related but distinct parental species. Phylogenetic analysis demonstrated that P. guiyangense likely evolved from common ancestors shared with plant pathogens. Comparative genome analysis coupled with transcriptome sequencing data suggested that P. guiyangense may employ multiple virulence mechanisms to infect mosquitoes, including secreted proteases and kazal-type protease inhibitors. It also shares intracellular Crinkler (CRN) effectors used by plant pathogenic oomycetes to facilitate the colonization of plant hosts. Our experimental evidence demonstrates that CRN effectors of P. guiyangense can be toxic to insect cells. The infection mechanisms and putative virulence effectors of P. guiyangense uncovered by this study provide the basis to develop improved mosquito control strategies. These data also provide useful knowledge on host adaptation and evolution of the entomopathogenic lifestyle within the oomycete lineage. A deeper understanding of the biology of P. guiyangense effectors might also be useful for management of other important agricultural pests.
Author summary
Utilization of biocontrol agents has emerged as a promising mosquito control strategy, and Pythium guiyangense has wide potential to manage diverse mosquitoes with high efficiency. However, the molecular mechanisms underlying pathological processes remain almost unknown. We observed that P. guiyangense invades mosquito larvae through cuticle penetration and through ingestion of mycelia via the digestive system, jointly accelerating mosquito larvae mortality. We also present a high-quality genome assembly of P. guiyangense that contains two distinct genome complements, which likely resulted from a hybridization of two parental species. Our analyses revealed expansions of kinases, proteases, kazal-type protease inhibitors, and elicitins that may be important for adaptation of P. guiyangense to a mosquito-pathogenic lifestyle. Moreover, our experimental evidence demonstrated that some Crinkler effectors of P. guiyangense can be toxic to insect cells. Our findings suggest new insights into oomycete evolution and host adaptation by animal pathogenic oomycetes. Our new genome resource will enable better understanding of infection mechanisms, with the potential to improve the biological control of mosquitoes and other agriculturally important pests.
Introduction
Mosquitoes are a major threat to global health since they are vectors of numerous devastating diseases, including malaria, dengue fever, Zika virus and other arboviruses, which together result in hundreds of millions of cases and several million deaths annually [1]. Existing commonly used control methods for reducing disease rely on the application of residual synthetic pesticides. However, intensive and repeated use of pesticides leads to ongoing development of resistance, environmental pollution and toxicity to human and non-target organisms [2]. Control strategies utilizing naturally occurring microbial pathogens have therefore emerged as a promising alternative. A particular focus has been on biological control agents [2]. Among them, the oomycete Lagenidium giganteum and the fungal pathogens, Beauveria bassiana and Metarhizum anisopliae, are well characterized, promising agents for mosquito larvae control, and have been produced commercially for field tests [2–4]. However, so far, available agents for mosquito control are rather limited.
Recently, a new mosquito-pathogenic oomycete, Pythium guiyangense X.Q. Su was isolated from infected larvae of Aedes albopictus from Guizhou, China [5]. It is a virulent pathogen of a wide range of mosquito larvae and is safe to non-target organisms [6, 7]. The pathogen also shows robust adaptability to a variety of natural environments and can be easily mass-produced [7]. All these properties of P. guiyangense make it of interest for practical applications as a potential mosquito control agent. However, little is known about the molecular mechanisms underlying the pathological processes on its mosquito hosts. It belongs to the genus Pythium (kingdom Stramenopila; phylum Oomycota, class Peronosporomycetes) [8]. Within the oomycetes, the genus Pythium is a genetically diverse group with a broad host range. For example, many Pythium species are important plant pathogens causing a variety of diseases [9, 10]. On the other hand, P. undulatum can infect fish and P. insidiosum is a well-known pathogen that is capable of infecting human and other mammals [11]. To date, only P. guiyangense has been proposed for mosquito control [5].
The availability of genome sequences of a variety of pathogenic oomycetes, including Pythium species, provides a unique opportunity for comparative analysis between P. guiyangense and other oomycetes with respect to the evolution of pathogenicity. A number of genome sequences of plant pathogenic oomycetes are now available [12–14]. In addition, the genomes of mycoparasitic Pythium species that infect fungi, the human pathogen P. insidiosum, and the fish pathogens S. parasitica and S. diclina have also been sequenced [15, 16]. Oomycetes secrete an arsenal of effectors into the host to manipulate the host immune system and enable parasitic infection [17]. These effectors have been a central question in the study of plant-oomycete interactions, including extracellular proteins such as toxins and hydrolases, and cell-entering proteins such as the RxLR (Arg-X-Leu-Arg) and Crinkler (CRN) effectors [18]. P. guiyangense may share conserved virulence effectors with other pathogenic oomycetes because of their close evolutionary relationships.
Here, we determined the infection cycle of P. guiyangense and demonstrated an unusual process, in which mycelia were devoured by the mosquito larvae and the mycelia inside digestive system could effectively initialize infection. Then we produced a high-quality genome sequence assembly, which represents the first draft genome from an insect pathogenic oomycete. Based on the transcriptome, effector prediction, and comparative genome analyses with other Pythium species, we investigated its insect-killing mechanisms and finally identified several cytoplasmic effectors having virulence functions in insect cells, thus expanding the roles of oomycete effectors.
Results
Two invasion pathways accelerate mosquito larvae mortality
The P. guiyangense isolate was reported to be highly virulent on mosquito larvae, but infection was not quantitated [5, 19]. We created a quantitative virulence assay by inoculating early second-instar larvae of Aedes albopictus or Culex pipiens pallens with P. guiyangense mycelia. The cumulative survival curves revealed that daily survival of A. albopictus and Cx. pipiens pallens larvae quickly declined from 3~4 days post-inoculation (dpi) onwards, reaching 76% mortality for A. albopictus and 69% for Cx. pipiens pallens by 10 dpi (S1A Fig). A. albopictus larvae died faster than Cx. pipiens pallens larvae, reaching 50% survival by day 6 compared to day 8 (S1A Fig). Furthermore, P. guiyangense could infect all the tested stages of Cx. pipiens pallens, including eggs, larvae, pupae and adults, resulting a visible accumulation of mycelia by 3–4 dpi (S1 Video) and all tissues were fully covered by mycelia (S1B Fig). Together, the results confirmed that P. guiyangense is highly efficacious in killing all life stages of Aedes albopictus and Cx. pipiens pallens.
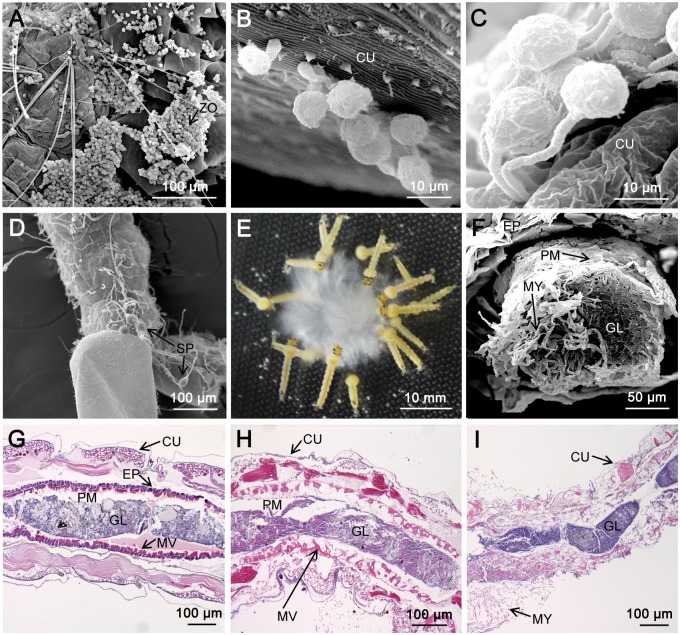
To investigate the infection process of P. guiyangense, early second-instar larvae of Cx. pipiens pallens were incubated with mycelia or swimming zoospores. Our results showed that zoospores attached to almost any part of the mosquito larval cuticle (Fig 1A showing zoospore attachments on the thorax and abdomen). Then, germination of the cysts occurred and appressorium-like swellings appeared at the tip of the germ tubes as visualized using scanning electron microscopy (SEM) (Fig 1B). Penetration hyphae emerging from the appressorium traversed the insect integument (Fig 1C) and invaded the hemocoel of larvae. Eventually the mycelia filled the whole body, then emerged through the inner cuticle and formed sporangia (Fig 1D). We also observed that Cx. pipiens pallens larvae readily ingested P. guiyangense mycelia even in an adequate food environment (Fig 1E, S1C Fig, S2 Video). A thick section of a moribund larva visualized with SEM showed that, following feeding, the midgut was completely packed with mycelia that could initialize infection (Fig 1F). After fixation, embedding in paraffin, and sectioning, microscopic observations showed that the midgut epithelium, muscles, and connective tissues appeared disrupted in the P. guiyangense-infected larvae (Fig 1G and 1H). After 48 hpi, infected larvae appeared almost devoid of internal organs or tissues and the whole body was permeated with mycelia (Fig 1I). Thus invasion through the digestive tract is an effective route of infection by P. guiyangense. Taken together, these observations define two routes of invasion, namely infection through the exterior cuticle and through the digestive tract.
Fig 1. Two infection pathways by P. guiyangense into mosquito larvae.
(A-D) Infection by direct penetration of the mosquito cuticle. (A) Zoospores gathered and adhered to the cuticle of a larva at 4 hpi. (B) Germinating cysts with germ tubes and appressoria at 8 hpi. (C) Germ tubes penetrating larvae at 12 hpi. (D) Sporangia produced at 24 hpi. (E-I) Infection by disruption of the mosquito digestive tract. (E) Mosquito larvae devouring P. guiyangense mycelia. (F) The mosquito larvae midgut packed with mycelia at 2 hpi. (G-H) The mosquito midgut epithelium, muscles, and connective tissues appeared disrupted at 12 hpi and 24 hpi. (I) Complete disruption of the mosquito epithelium and peritrophic membrane after 48 h. ZO: zoospore, MY: mycelia, SP: sporangia, CU: cuticle, EP: epithelium, PM: peritrophic membrane, GL: gut lumen, MV: microvilli.
General features of the genome and transcriptome sequences
A high quality genome sequence of P. guiyangense was generated using a hybrid strategy that combined sequences from Pacific Biosciences long reads and Illumina short reads. The genome assembly indicated in an estimated P. guiyangense genome size of 110 Mb and annotation predicted 30,943 protein-coding genes (Table 1). The assembled genome consisted of 239 contigs with an N50 contig length of 1,009 kb. To assess the completeness of the genome assembly, CEGMA analysis, which identifies orthologs of 248 ultra-conserved core eukaryotic genes (CEGs), was used to identify core genes in the P. guiyangense genome. The results revealed complete matches to 97.6% of CEGs and at least partial matches to 98.4% of CEGs within the P. guiyangense assembly; these results compared to only 78.2–94.4% of complete CEGs and 91.5–95.6% of partial CEGs in other Pythium genomes (S1 Table). Taken together, the comparison with other assembled Pythium genomes including P. insidiosum, P. ultimum, P. aphanidermatum, P. arrhenomanes, P. irregulare and P. iwayamai, revealed that the P. guiyangense assembly represented the best quality among the sequenced Pythium genomes so far.
Table 1. Genome statistics of Pythium genomes.
| P. guiyangense | P. insidiosum | P. ultimum | P. aphanidermatum | P. arrhenomanes | P. irregulare | P. iwayamai | |
|---|---|---|---|---|---|---|---|
| Assembly size (Mb) | 110.1 | 53.2 | 42.8 | 35.9 | 44.7 | 42.9 | 43.3 |
| Contig/Scaffold Number | 239 | 1192 | 975 | 5,667 | 10,978 | 5,887 | 11,542 |
| N50 scaffold Length (Kb) | 1009.1 | 146.3 | 773.5 | 37.4 | 9.8 | 23.2 | 11 |
| G+C content (%) | 58.3 | 57.9 | 49.8 | 51.4 | 54.6 | 53.4 | 53.1 |
| Number of protein-coding genes | 30,943 | 14,962 | 15,297 | 12,305 | 13,805 | 13,804 | 14,875 |
| Average gene length (bp) | 1658 | 2144 | 1299 | 1470 | 1339 | 1495 | 1325 |
| Gene density (genes per Mb) | 281 | 281 | 357 | 343 | 309 | 322 | 344 |
| Introns per gene | 2.0 | 2.4 | 1.6 | 1.3 | 1.6 | 1.7 | 1.8 |
Whole transcriptome sequencing (RNA-seq) was performed using Illumina sequencing of RNAs from P. guiyangense mycelia and from early second-instar larvae 24 hr after inoculation with P. guiyangense mycelia. Transcripts from each individual library matched approximately 69% of the genes, and together matched approximately 74% of the genes (S2 Table). A total of 3,354 genes (10.8%) were differentially expressed (>4-fold expression difference and statistical GFOLD value > 1 or < -1) between the two samples; 1,654 genes were up-regulated in the infection stage while 1,700 genes were down-regulated. Functional enrichment analysis revealed that genes encoding tyrosine kinase-like (TKL) kinases, subtilisin proteases, kazal-type protease inhibitors, and elicitin proteins, were over-represented among the differentially expressed genes (S3 Table). To validate the differentially expressed genes, we selected 18 genes that belonged to the above mentioned over-represented gene families and that were up- or down-regulated based on RNA-Seq data, and then measured their transcript levels by the qRT-PCR assay. The qRT-PCR results showed that the transcriptional patterns of 17 among the 18 genes were consistent with RNA-Seq results (S2 Fig), which further supported the general reliability of the RNA-Seq data.
Interspecies hybridization may contribute to the large size of the P. guiyangense genome
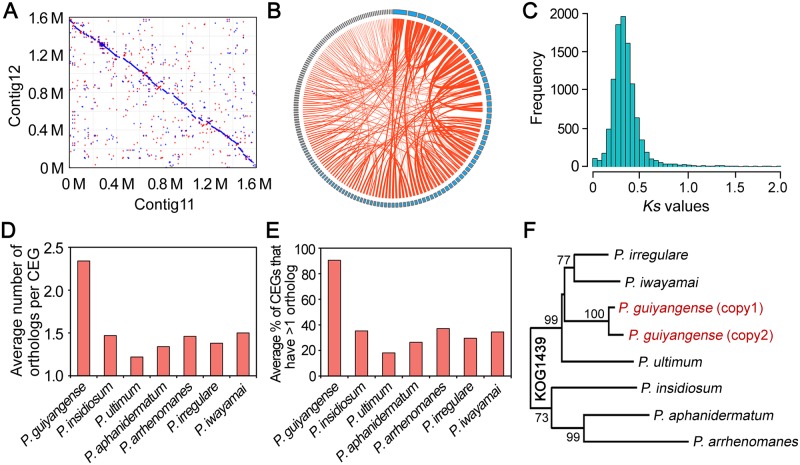
Our comparative genome analysis revealed that the genome size (110 Mb) and predicted gene number of P. guiyangense (30,943 genes) were approximately twice those of the other sequenced Pythium species (Table 1). To explore the potential mechanisms underlying such a large genome size, we initially analyzed the repetitive DNA content within the P. guiyangense genome and found that the repeat sequence content was 6%, similar to that of P. ultimum (7%) which has a genome size of only 43 Mb, thus excluding the possibility that high repeat content was responsible for the large genome size, which was observed in Ph. infestans [14]. Previously, hybridization between two parental species has been reported in yeast and Phytophthora evolution [20, 21]. Most (84%) of the Pythium core genes (present in all the 7 Pythium genomes) were present in two copies in the P. guiyangense genome, consistent with a hybrid origin, which is similar to the yeast [20]. Typically, the copied core genes shared the highest sequence similarity with genes derived from the P. irregulare genome, however, the two copied genes were about 8% different in nucleotide sequence. To further confirm the hypothesis of hybridization, internal synteny is selected as a useful indicator, which could be used to evaluate hybridization between two parental genomes [22, 23]. Therefore, we characterized internal synteny across the P. guiyangense genome using the MCScanX program [24]. In the whole genome, 468 conserved synteny blocks with an average size of 192 kb were identified (Fig 2A, S4 Table). These synteny blocks covered 74% of all the contigs, and together spanned 84% of the genome, suggesting that the P. guiyangense genome could be classified into two subgenomes.
Fig 2. Interspecies hybridization may contribute to the large genome size.
(A) Frequent synteny blocks identified between pairs of contigs in the P. guiyangense genome assembly. Shown is an example of nucleotide alignments of contig 11 (x-axis) versus contig 12 (y-axis). Each point in the plot corresponds to an aligned sequence of >500 bp. The red dots represent the same orientation and the blue dots represent reversed orientations. (B) Representation of homologous genes with conserved synteny in the genome. Each blue rectangle represents one contig. The circle represents all the contigs with synteny, and each line connects two homologous genes in the synteny block. Homologous genes which are found in the same contig were not drawn. The detailed information was listed in S4 Table. (C) The Ks distribution of paralogous gene pairs. (D) Comparison of the average number of orthologs for each CEG among Pythium species. (E) Comparison of the average percentage of detected CEGs that have more than one ortholog among Pythium species. (F) Phylogenetic analysis of one example of CEG named KOG1439 among Pythium species. This CEG had two copies in P. guiyangense while only one copy in each of other Pythium species.
To further compare the genetic relatedness of the two parental subgenomes, the average nucleotide identity (ANI) was calculated, and the ANI between the two subgenomes revealed approximately 91% identity. Notably, a total of 11,068 pairs of homologous genes were identified in these synteny blocks (Fig 2B, S4 Table), consistent with a hybrid origin. We estimated the rates of synonymous substitutions per synonymous site (Ks) of 11,068 pairs of homologous genes. This analysis showed a synonymous site divergence peak of Ks = 0.35 (Fig 2C), indicating that the two subgenomes were relatively diverse. Taken together, we inferred that P. guiyangense was a hybrid genome derived from two distinct parental species.
To further investigate the parental species of P. guiyangense, we systematically analyzed CEGs in the 7 sequenced Pythium genomes. The majority of CEGs were present as two copies in the P. guiyangense genome but only one copy in each of the other Pythium genomes (Fig 2D and 2E). A total of 167 CEGs that contained 2 copies in P. guiyangense and also had orthologs in other Pythium species were utilized for phylogenetic analysis. For each phylogenetic tree, the two copies of the P. guiyangense CEG always clustered together most closely, and then clustered with the orthologs from the other Pythium species (one tree based on the KOG1439 protein is shown in Fig 2F as an example), indicating that the parental species of P. guiyangense were not represented in the data set. In addition, cytochrome oxidase II (cox II) and β-tubulin genes, which have been widely used as phylogenetic maker genes, were available in 35 Pythium species and contained 2 copies in P. guiyangense. Phylogenetic analyses of the two genes showed that the two P. guiyangense orthologs were more similar to one another than the nearest known species (P. orthopogon) (S3A and S3B Fig), indicating that the parental species of P. guiyangense were not represented based on current information. We speculate that the parental species of P. guiyangense are more closely related to each other than to the known Pythium species.
In parallel with the genome analysis, we noticed that an unusual high percentage of P. guiyangense zoospores contained two nuclei rather than one (S3C Fig). Among 500 observed zoospores, nearly 22% of them contained two nuclei in P. guiyangense while in P. aphanidermatum and Ph. capsici, all the spores had only one nucleus (S3C Fig). We then found that the P. guiyangense zoospores containing only one nucleus could also breed similar percent of zoospores containing two nuclei, and PCR amplifications resulted in presence of both of the two copied genes. This observation suggested that P. guiyangense might be a dikaryon, and its relationship with the complex genome is still under investigation.
Evolutionary relationships and species-specific gene families
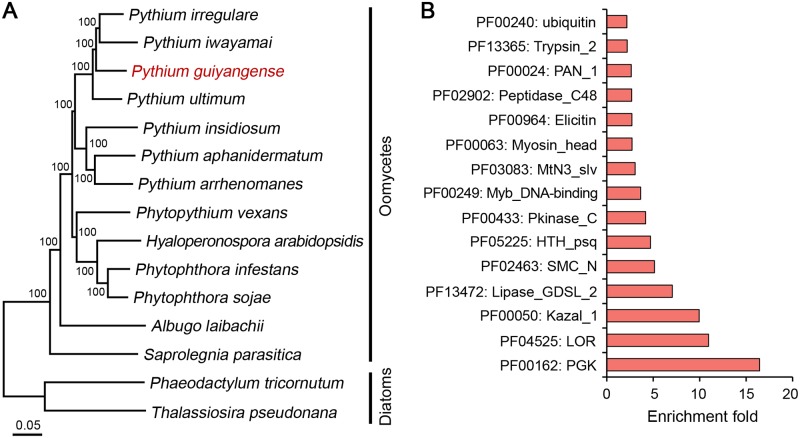
To establish the phylogenetic relationship of P. guiyangense among oomycetes, a phylogenetic tree was constructed based on 248 CEGs from P. guiyangense and other 12 oomycetes, with the diatoms as outgroups (Fig 3A). The tree clearly showed that P. guiyangense was clustered within the clade formed by the plant pathogenic Pythium species, and was distantly related to other genera, including Hyaloperonospora and Phytophthora. This phylogeny was consistent with that in previous publications [15, 25]. These results imply that P. guiyangense, along with the mammalian pathogen P. insidiosum share common ancestors with the plant pathogenic Pythium species (Fig 3A).
Fig 3. Phylogenetic relationships and species-specific genes.
(A) Genome-based phylogenetic relationships of P. guiyangense and other oomycetes. The neighbor-joining tree was constructed from concatenated alignment of 248 core eukaryotic genes (CEGs) identified by CEGMA analysis. (B) Enrichment of PFAM domains in P. guiyangense-specific genes. The fold-enrichment (y-axis) corresponds to the frequency of a given PFAM domain in the species-specific genes divided by the frequency in the rest of the proteome.
To identify the genes responsible for host adaptation in P. guiyangense, the OrthoMCL tool was used to cluster the seven Pythium proteomes on the basis of protein sequence similarity. A total of 25,602 (83%) P. guiyangense genes had orthologs in other Pythium species. Among these, P. guiyangense shared 13,000 core genes with the other Pythium species. In addition, 5,341 genes were identified to be specific to P. guiyangense (S4 Fig). To gain insights into the features of species-specific genes in P. guiyangense, we compared the frequency of occurrence of protein family domains and identified highly over-represented domains included kinase (PF00433), kazal inhibitor (PF00050), elicitin (PF00964), and protease (PF02902) (Fig 3B, S5 Table). These gene families were also enriched among genes differentially expressed during infection.
Expanded protein kinases involved in the infection process
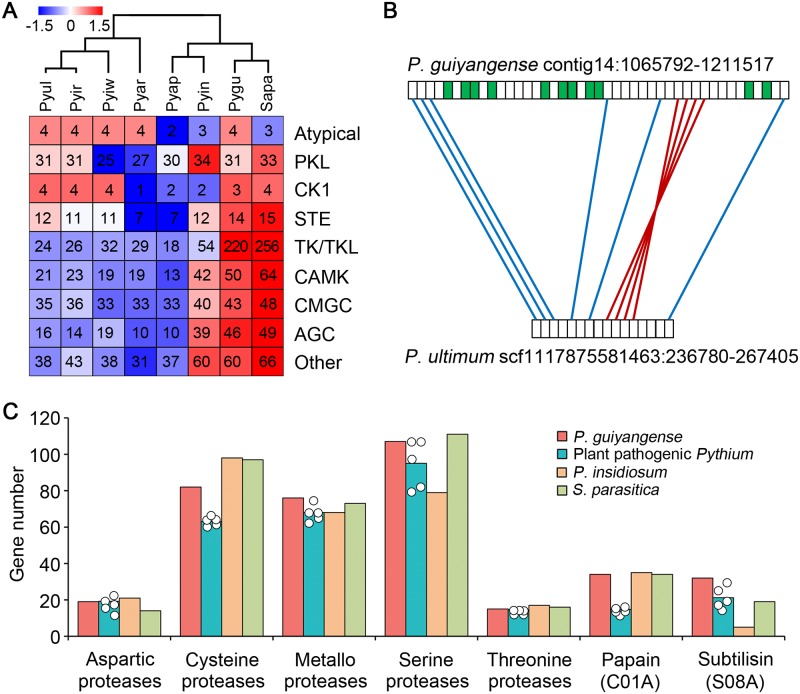
By searching with the HMM profiles of kinase domains derived from KinBase, 471 unique protein kinases (943 kinases in total) were identified in the P. guiyangense genome, greatly surpassing the numbers in plant pathogenic Pythium genomes, which range from 152 to 192 (Table 2). Intriguingly, other two animal pathogenic oomycetes, P. insidiosum and S. parasitica, also have expanded kinomes, coding for 286 and 538 kinases, respectively [15, 16]. We further classified the kinases into 9 families defined by Hanks and Hunter [26]. Five families, including TKL (tyrosine kinase-like), CAMK (calcium/calmodulin-dependent kinase), CMGC [including cyclin-dependent kinases (CDKs), mitogen-activated protein kinases (MAP kinases), glycogen synthase kinases (GSK) and CDK-like kinases], AGC (cAMP-dependent, cGMP-dependent and protein kinase C) and "other" were noticeably expanded in P. guiyangense (Fig 4A). Among the 5 expanded families, a total of 220 unique TKL genes were identified in P. guiyangense kinome. A comparison of the locations of TKL genes in the P. guiyangense and P. ultimum genomes revealed extensive rearrangements, which resulted from species-specific expansions at the locations of these genes (Fig 4B). Forty-six unique kinases belonging to AGC family and 50 unique members of the CAMK family were identified from P. guiyangense. Based on the RNA-Seq data, a total of 92 kinase genes were differentially expressed at the infection stage, including 52 TKL kinase genes (S6 Table). These results suggest that many of the protein kinases may be involved in regulation of infection processes and adaptation to the mosquito hosts.
Table 2. Protein families implicated in pathogen-host interactions.
| P. guiyangensea | P. ultimum | P. irregulare | P. iwayamai | P. aphanidermatum | P. arrhenomanes | P. insidiosum | S. parasitica | |
|---|---|---|---|---|---|---|---|---|
| Kinases | 471 (943) | 185 | 192 | 185 | 152 | 161 | 286 | 538 |
| Proteases | 307 (615) | 284 | 264 | 233 | 272 | 246 | 283 | 311 |
| Serine proteases | 109 (218) | 108 | 97 | 79 | 108 | 83 | 79 | 111 |
| Subtilisin proteases | 32 (64) | 30 | 19 | 18 | 25 | 14 | 5 | 19 |
| Cysteine proteases | 83 (167) | 63 | 66 | 61 | 63 | 62 | 98 | 97 |
| Papain proteases | 34 (68) | 17 | 16 | 14 | 14 | 13 | 35 | 34 |
| Metallo proteases | 79 (159) | 75 | 66 | 63 | 68 | 68 | 68 | 73 |
| Kazal protease inhibitors | 19 (39) | 11 | 10 | 8 | 12 | 9 | 27 | 5 |
| Elicitins | 10 (20) | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| Elicitin-like | 45 (91) | 40 | 25 | 23 | 23 | 29 | 50 | 29 |
| NLP | 1 (2) | 7 | 4 | 4 | 4 | 5 | 0 | 0 |
| CRN | 19 (38) | 46 | 10 | 33 | 45 | 46 | 44 | 0 |
aThe numbers without brackets are numbers of unique genes per haploid genome, while numbers in the brackets are total gene numbers.
Fig 4. Expansion of kinases and proteases in P. guiyangense.
(A) Comparison of kinase gene numbers between P. guiyangense and other oomycetes. The numbers of family members per haploid genome are shown in each case, including P. guiyangense. Overrepresented (white to red) and underrepresented (white to blue) numbers are depicted as Z-scores for each family. Pygu, P. guiyangense; Pyin, P. insidiosum; Pyap, P. aphanidermatum; Pyar, P. arrhenomanes; Pyir, P. irregulare; Pyiw, P. iwayamai; Pyul, P. ultimum; Sapa, S. parasitica. (B) Synteny of TKL kinase genes between P. guiyangense and P. ultimum. The region shown is an example of dense clustering of TKL kinase genes in the P. guiyangense genome assembly. The green boxes represent TKL kinase genes. Red lines join syntenic genes with the same orientation and the blue lines join genes with reversed orientations. (C) Comparison of protease gene numbers between P. guiyangense and other oomycetes. The teal columns represent the average number of genes per haploid genome of the five plant pathogenic Pythium species, while each dot represents the number in an individual plant pathogenic Pythium species. For P. guiyangense, the unique gene number in each haploid genome was used.
A large number of proteases may facilitate cuticle penetration
Since major structural and physiological differences were observed between plant cell walls and insect cuticles, we compared the repertoire of plant cell wall and cuticle degrading enzymes encoded in the P. guiyangense genome to other oomycete genomes. Several groups of plant cell wall degrading enzymes, such as GH53, GH78, CE5, GH10 and GH11, and GH12 were completely absent in P. guiyangense (S7 Table). Genes encoding 12 unique pectin/pectate lyases (PL1, PL3 and PL4), two unique GH28 and 1 unique GH43 involved in pectin backbone degradation were identified in the P. guiyangense genome; however, RNA-Seq data showed that none of these genes exhibited up-regulation during mosquito infection processes.
P. guiyangense had more genes encoding proteases potentially involved in insect cuticle degradation than plant pathogenic Pythium species (Fig 4C). A total of 307 unique proteases (615 genes in total) were encoded in the P. guiyangense genome, compared to an average of 260 proteases in the plant pathogenic Pythium species (Table 2). The two animal pathogen genomes also had large numbers of proteases (Table 2, Fig 4C). Among them, genes encoding cysteine-, metallo- and serine-proteases were particularly highly expanded in P. guiyangense (Fig 4C). The subtilisin serine-protease family had the highest relative expansion with 32 unique genes in P. guiyangense (Table 2, Fig 4C). Phylogenetic analysis revealed that over half of the subtilisin proteases were recently expanded in P. guiyangense due to lineage-specific gene duplications (S5 Fig). Based on the RNA-Seq data, 31% of the total subtilisins were significantly up-regulated during mosquito infection. The peptidase_C1 and carboxypeptidases also exhibited significant expansion in P. guiyangense (Table 2, Fig 4C).
Large family of kazal-type serine protease inhibitors (KPIs) potentially associated with infection
Protease inhibitors regulate various biological and physiological processes in all living systems as modulators of protease activity [27]. Among them, the kazal-type protease inhibitor (KPI) family is one of the best characterized [27]. A total of 19 unique kazal inhibitors were identified in the P. guiyangense genome, which exceeded those in plant pathogenic Pythium species (Table 2). The animal pathogen, P. insidiosum also encoded larger numbers of kazal inhibitors. A phylogenetic tree was constructed using the Pythium kazal inhibitors, and the majority of genes derived from P. guiyangense and P. insidiosum formed clusters that were species-specific (S6A Fig), implying that these genes were retained and diversified independently in these two animal pathogens. During mosquito infection, 25% of the P. guiyangense kazal inhibitors were up-regulated, and four of these exhibited transcript levels over 40 times those in the mycelia sample (S6B Fig). The transcriptional patterns of the 4 kazal inhibitors at three infection time points were analyzed by qRT-PCR, and results revealed that all the 4 genes were up-regulated during the infection process (S6C Fig). Further analysis demonstrated that all of the up-regulated kazal inhibitors contained signal peptides, indicating that they could play important roles in the pathogenesis.
Elicitor genes and host detection
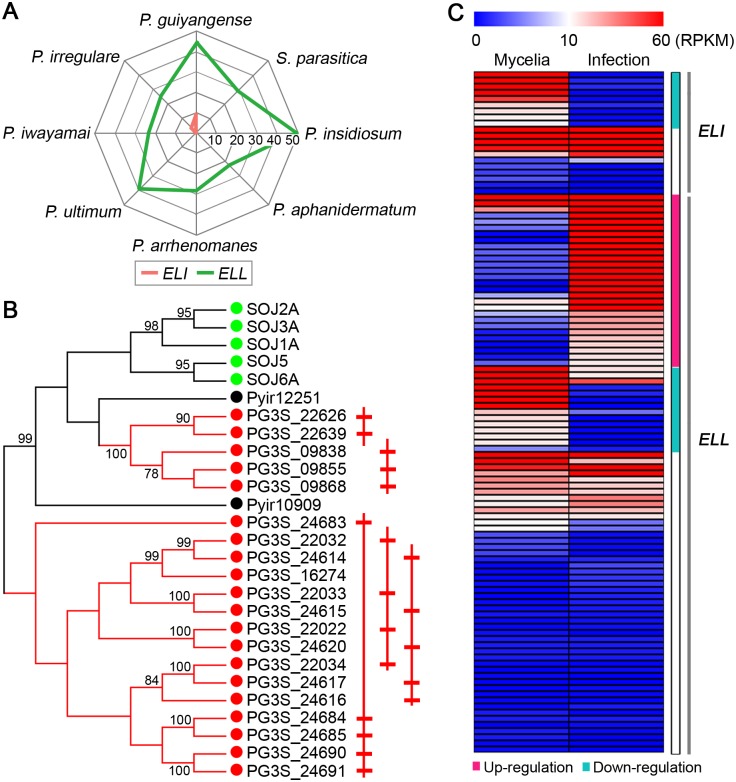
A common feature of many plant pathogenic oomycetes is the secretion of a variety of apoplastic (extracellular) proteins to promote infection, some of which can be detected by the host immune system. These include elicitins (ELIs; lipid-binding proteins), elicitin-like (ELL) proteins and Nep1-like proteins (NLP) [28]. Ten unique ELI genes were identified in the P. guiyangense genome. In contrast, only 2 ELI genes were found in the P. irregulare genome, and none were identified in the other Pythium and S. parasitica genomes (Fig 5A, Table 2). Based on phylogenetic analysis of the elicitin domains, P. guiyangense ELIs were distributed into two clades, and one clade included genes from diverse species while the second clade only contained P. guiyangense ELIs (Fig 5B). Moreover, 19 of the 20 ELI genes were physically clustered in the P. guiyangense genome, suggesting that ELIs were expanded in a species-specific manner. In contrast to the ELI genes, ELL genes were widely distributed in all the detected Pythium genomes. Both P. guiyangense and P. insidiosum had more ELL genes (45 and 50 unique genes) than the plant pathogenic Pythium species (23–40 genes) (Fig 5A, Table 2). Further phylogenetic analysis showed that over half of the ELL genes were distributed in nine clades which were specific to P. guiyangense and contained at least four members; thus many ELL genes were specifically expanded in P. guiyangense. Based on the P. guiyangense transcriptome analysis, 45% of the ELI genes were differentially expressed, and all were down-regulated during infection (Fig 5C). In contrast, 31% of the ELL genes were up-regulated while 15% were down-regulated during infection (Fig 5C). This observation suggested that the diverse ELIs and ELLs had a variety of different functions relative to growth and infection.
Fig 5. Expansion of ELI and ELL genes.
(A) Radar charts showing the numbers of elicitin (ELI) and elicitin-like (ELL) genes found in each Pythium or S. parasitica genome. For P. guiyangense, the unique gene number in each haploid genome was used. Each spoke in the chart represents one species. The concentric circles form a ruler with a primary unit of 10 genes. (B) Phylogenetic tree of the ELI genes, showing species-specific expansion of P. guiyangense ELI genes. Members from P. guiyangense (PG3S, red), P. irregulare (Pyir, black), and Ph. sojae (SOJ, green) are included. On the right the physically linked ELI genes are schematically drawn. (C) Relative transcript abundance of ELI and ELL genes at 24 hpi infection versus mycelia tissue. The teal bar highlights the significantly down-regulated genes, and the pink bar highlights the significantly up-regulated genes.
Another common apoplastic effector family is the necrosis and ethylene-inducing-like proteins (NLP) genes. Many NLPs, but not all, can trigger cell death and defense responses in plants [29]. Only 1 unique NLP gene was found in P. guiyangense and none were found in P. insidiosum (Table 2). This NLP protein belonged to type 1 NLP subfamily with two conserved cysteine residues. Transcriptional analysis revealed no significant change during infection. These observations suggest that NLP proteins may not participate in oomycete-animal interactions.
Some CRN effectors can induce insect cell death
Crinkler (CRN), a large class of cytoplasmic effectors, was first identified in Ph. infestans as a family of proteins that could cause plant cell death and defense responses [30]. A total of 38 CRN candidates were predicted in P. guiyangense (S8 Table), compared to 10–46 predicted CRN proteins in the other Pythium species using the same method (Table 2). Examination of protein alignments of P. guiyangense CRN effectors revealed considerable conservation of the characteristic LxLFLAR/K and HVLVxxP motifs, which were similar to those observed in plant pathogenic Pythium species [13, 25]. Based on five secretion signal predictors, 74% of CRN candidates in P. guiyangense contained a potential signal peptide or non-classical secretion signal (S8 Table), suggesting that the majority of P. guiyangense CRN proteins might be secreted into mosquito hosts.
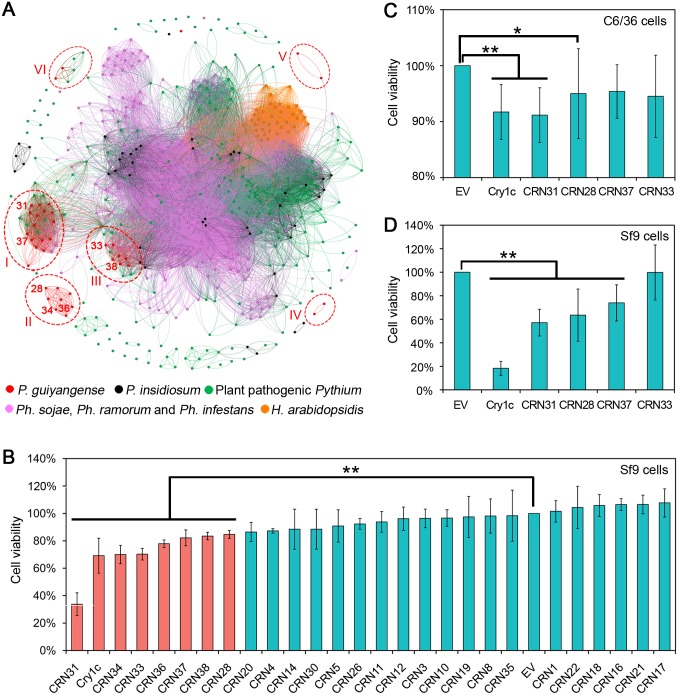
A homology network of the oomycete CRN proteins was generated to investigate the evolutionary relationships between P. guiyangense and other oomycetes. The network is composed of 633 nodes in which each node represents an individual CRN protein. The network contains 34,149 edges that link nodes if the node proteins are homologous based on an all-versus-all BlastP search with an E-value cutoff of 10−10. As shown in Fig 6A, the network was comprised of a crowd of disconnected clusters and a small number of singletons. P. guiyangense CRN proteins were mainly distributed in 3 large and 3 small clusters (cluster I-VI represented by red dotted circles). Cluster I and III were composed primarily of P. guiyangense CRN proteins, with some of these proteins having homology to Pythium proteins. Notably, cluster II, IV and V only contained CRN proteins derived from P. guiyangense, revealing that these CRN proteins did not share significant sequence similarity with other oomycete CRN proteins. Moreover, all of the P. guiyangense CRN proteins showed sequence divergence of at least 50% with the most similar CRN protein in any plant pathogenic Pythium species, indicating that the CRN proteins are highly divergent between insect pathogenic Pythium species and plant pathogenic Pythium species.
Fig 6. Diversification of P. guiyangense CRN proteins and their potential contributions to virulence.
(A) Homology network of oomycete CRN proteins. Each protein is represented by a node. An edge joining two nodes represents sequence similarity shared by the two proteins. The red circled clusters (cluster I-VI) represent P. guiyangense specifically expanded CRN proteins. The number on the left of the red node represents P. guiyangense CRN protein ID. For example, 31 represents CRN31. (B) Evaluation of CRN protein toxicity to Spodoptera frugiperda cells (Sf9). The Sf9 cells were transfected with DNA encoding the CRN proteins. The protein toxicity was measured as the remaining number of viable cells compared to the empty vector (EV) negative control. Protein expression was assessed by western blots (S7A Fig). (C-D) Assay of CRN protein toxicity to insect cells (C, mosquito C6/36 cells; D, Sf9 cells) using CRN proteins produced in E. coli cell extracts. CRN protein production was assessed by western blots (S7C Fig). Differences were assessed for significance using Student’s t-test: *, P<0.05 and **, P<0.01.).
To explore the possible functions of P. guiyangense CRN proteins in insect cells, twenty-six CRN genes were expressed in Spodoptera frugiperda cell (Sf9) lines; successful expression of the proteins was confirmed with western blots or by detecting fluorescence signals under the fluorescence microscope (S7A and S7B Fig). The cell counting Kit-8 assay was used to determine protein toxicity to cells, with the Bacillus thuringiensis Delta-Endotoxin Cry1C as a positive control [31]. The results showed that 7 CRN proteins (CRN31, 33, 34, 36, 37, 38, and 28) significantly decreased the viability of Sf9 cells while the remaining CRN proteins produced responses similar to the negative control (Fig 6B). Notably, CRN31 appeared to be the most toxic to Sf9 cells. To further validate the toxicity of these 7 CRN proteins, a prokaryotic expression system was used to obtain recombinant CRN proteins (S7C Fig). E. coli crude extracts containing the expressed proteins were then incubated with Sf9 cells and with mosquito Aedes albopictus C6/36 cells, respectively, to determine the toxicity using the cell counting Kit-8 assay. The results showed that CRN31 and CRN28 significantly reduced the viability of Sf9 cells and C6/36 cells (Fig 6C and 6D). Since the transcript levels of the CRN31 and CRN28 genes were not elevated during infection at 24 hpi as measured by RNA-seq (S8 Table), we used qRT-PCR to test whether the two CRN genes were significantly up-regulated during earlier infection stages (1–4 hpi) (S7D and S7E Fig). CRN31 exhibited the highest transcript level change with a 65 fold change at 2 hpi while CRN28 showed 4–5 fold changes at 1–3 hpi (S7D and S7E Fig). Together these results suggested that CRN31 and possibly CRN28 might act as cell-killing effectors during insect infection.
Discussion
In this study, we have determined the mode of infection of P. guiyangense on mosquito larvae. In our experiments, P. guiyangense caused up to 76% mortality for A. albopictus and 69% for Cx. pipiens pallens larvae. Analogous to most of the entomopathogenic fungi, P. guiyangense hyphae emerging from germinating zoospore cysts entered their host directly through the exterior cuticle, propagated inside hosts, and produced sporangia to start a new cycle of infection. Another infection route of P. guiyangense was through the ingestion of mycelia by larvae. Mycelia in the digestive tract progressively destroyed internal tissues of the larval midgut, leading to host death. The most common invasion route for aquatic insect pathogens, including Metarhizium anisopliae, Aspergillus clavatus and Beauveria Bassiana, was through ingestion of spores to infect their host [4, 32, 33]. P. guiyangense has evolved a similar strategy to initiate infection in the digestive system. Overall, P. guiyangense utilizes cuticle penetration and ingestion of mycelia into the digestive system to infect mosquito larvae. We speculate that firm adhesion of zoospores to the mosquito larvae epicuticle is critical for the success of the P. guiyangense pathogen which involves a combination of passive hydrophobic and electrostatic forces as well as protein interaction. Hydrophobins found in the outer layer of the spore cell wall of Beauveria Bassiana, mediate adhesion to the arthropod cuticle [34, 35]. Hydrolytic enzymes, Mad1 and Mad2, also assisted in attachment of the fungi to insects [36]. To identify the factors that promote attachment and ingestion of P. guiyangense by the mosquitoes would be interesting to further explore in the future.
To probe the molecular basis underlying the interactions of P. guiyangense with insects, a high-quality genome assembly and transcriptome sequences were generated for P. guiyangense. Our results reveal that P. guiyangense is probably a hybrid genome derived from two parental species. Natural interspecies hybridization events have been described in the genus Phytophthora such as Ph. andina, Ph. nicotianae and Ph. cactorum [37, 38]. It is believed that interspecies hybridization has the potential to create new strains that have a new or expanded host range [21]. Considering the distinct hosts, we speculate that the hybrid feature of P. guiyangense contributes to its adaptation of the mosquito host. The two parental subgenomes of P. guiyangense are approximately 9% different in nucleotide sequence, suggesting that the two parents are relatively diverse, however, the potential parents are still mysterious based on limited Pythium data. We will pay close attention to the new information of Pythium and update the concerns in future study.
The phylogenetic analysis of the currently sequenced oomycete pathogens together with two diatoms demonstrated P. guiyangense is closely related to three plant pathogenic Pythium species (P. irregulare, P. iwayamai and P. ultimum) but has a slightly more distant relationship with the mammalian pathogen, P. insidiosum. This finding suggested that as a facultative mosquito pathogen, P. guiyangense, may have evolved from a common ancestor with the plant pathogens. This result is highly concordant with recent analysis indicating the mosquito oomycete pathogen, L. giganteum has also evolved from a plant pathogen [3].
In conjunction with the transcriptome analysis, oomycete genome comparisons identified several gene families that might contribute to P. guiyangense virulence. In this study, 471 putative unique kinases (943 kinases in total) were identified in the P. guiyangense genome. Comparison with other sequenced oomycete genomes revealed that the genomes of the animal pathogens, P. guiyangense, P. insidiosum and S. parasitica, also encoded significantly more kinases than the plant pathogenic Pythium genomes [15, 16]. Transcriptome analysis revealed that a total of 92 kinases were differentially expressed during infection of P. guiyangense against mosquito larvae, implying that protein kinases may be involved in regulating virulence. We also found that genes involved in insect cuticle degradation were expanded in P. guiyangense while proteins for plant cell wall penetration were absent or lost functions. The P. guiyangense genome encoded a significantly larger number of proteases than plant pathogenic Pythium species, including cysteine-, metallo-, and serine-proteases. Transcript levels of 31% of the total subtilisin-like serine proteases were significantly elevated when P. guiyangense invaded mosquito larvae. Some of these proteases were reported as key virulence determinants in entomopathogenic fungi [39], supporting a potential role of these proteases in P. guiyangense infection.
A large number of Kazal proteinase inhibitors (KPIs) were characterized from P. guiyangense and 25% of these genes were up-regulated during infection of mosquito larvae, suggesting KPIs may be involved in pathogenicity. Our study demonstrated that one invasion route of P. guiyangense was through ingestion of mycelia in the digestive system. The mosquito midgut contains an abundant array of secreted serine proteases for digestion, providing nutrition for development [40, 41]. To aid in colonization in its hosts, P. guiyangense may secrete protease inhibitors, such as KPIs for protection from these proteolytic enzymes. This is consistent with previous studies which show that the animal parasite, Toxoplasma gondii secretes TgPI-1 and TgPI-2, and Hookworm, Ancylostoma ceylanicum, secretes a 8-kDa broad spectrum serine protease inhibitor of the Kunitz family into the host digestive tract to aid in infection [42–44]. Serine proteases are also key components of immune responses and KPIs may manipulate host immune defenses for pathogenicity [45]. A kazal-like serine protease inhibitor was characterized from the plant pathogenic oomycete, Ph. infestans, and it targeted protease P69B to counteract tomato defense responses [46]. Another oomycete pathogen, Ph. palmivora also produced a KPI, PpEP to suppress plant defense [47]. Therefore, we speculate that the large number of KPIs secreted by P. guiyangense may suppress mosquito immune defenses by targeting serine proteases.
In addition to hydrolytic enzymes, plant pathogenic oomycetes deliver a diverse battery of other secreted proteins into host tissue to support infection and interfere with host immune responses, including lipid-binding proteins (elicitins), toxins (e.g. NLPs), and host-cell-entering RxLR and CRN effectors [48, 49]. We found distinct sequence and evolutionary features of these proteins in P. guiyangense. Firstly, no statistically significant evidence for RxLR effectors encoded in the P. guiyangense genome was found, in agreement with previous reports [13, 25]. Secondly, 10 unique ELI genes were present in the P. guiyangense genome whereas these genes were largely absent from the other Pythium species, including P. insidiosum. The P. guiyangense ELI genes appear to have expanded relatively recently to form two species-specific clades, and one clade appears to share a common origin with the Ph. sojae genes. In contrast to ELIs, ELLs have been widely found in all sequenced Pythium species. RNA-Seq data showed that ELI and ELL genes show differential expression patterns in P. guiyangense. ELI genes were typically highly expressed in the mycelia stage while a large number of ELL genes were up-regulated in the infection stage. These results suggested that the two subclasses of elicitins may be involved in different functions. Elicitins have also been reported in another mosquito pathogenic oomycete, L. giganteum [3], suggesting that elicitins in the two aquatic insect oomycetes may be linked to pathogenicity towards the insect hosts.
CRN effectors are considered more ancient cytoplasmic effectors than RxLRs, as they are distributed across a wide range of oomycetes [14, 25] and have also been reported in the fungal animal pathogen, Batrachochytrium dendrobatidis [50] and in arbuscular mycorrhizal fungi [51]. Interestingly, CRN proteins were also detected in the mosquito pathogenic oomycete L. giganteum [3, 52]. They are presumed to enter the host cytoplasm and manipulate cell death and defense responses [30, 53]. It has been widely reported that only a handful of oomycete CRN proteins were predicted to contain canonical signal peptides [13, 14]. In this study, four different signal peptide predictors and one non-classical secretion signal predictor were used, and the majority (74%) of P. guiyangense CRN proteins were predicted to contain potential secretion signals, suggesting that these CRN proteins very likely were secreted into mosquito hosts. Once inside host tissue, the roles of these putative effectors in animal pathogenic oomycetes remains unclear. One investigation detected CRN effectors in an entomopathogenic oomycete, Lagenidium giganteum, but their roles in the mosquito pathogenicity process remained unclear [52]. In this study, twenty-six CRN candidates were characterized in P. guiyangense and insect cell line transformation experiments revealed that CRN31 and CRN28 were toxic to Spodoptera frugiperda (Sf9) cells and to a lesser extent to Aedes albopictus (C6/36) cells. Therefore, we speculate that P. guiyangense has evolved distinct lineages of CRN effectors that are secreted into mosquito cells as virulence factors to induce host cell death.
Overall, we have demonstrated that two infection routes are available for infection of mosquitoes by P. guiyangense. The high-quality genome sequence of P. guiyangense provides new insights into study oomycete evolution and host adaptation because it is an oomycete pathogen that has adapted to mosquitoes. Genome comparisons suggest adaptations to a mosquito-pathogenic lifestyle include loss of plant cell wall degrading enzymes and NLP proteins, and expansions of kinases, proteases, and kazal-type protease inhibitors. Oomycete intracellular CRN effectors were identified and insect cell toxicity was identified in at least one of them, which could serve as a new resource to control agricultural important pests.
Materials and methods
Strain and mosquito source
The P. guiyangense strain Su was kindly provided by Dr. Xiaoqing Su from Guiyang Medical University, Guiyang, China and was maintained on 10% vegetable juice (V8) medium in the dark at 25 ± 1°C. The Nanjing laboratory strains of Aedes albopictus and Cx. pipiens pallens were obtained from Jiangsu Provincial Center for Disease Control and Prevention, Nanjing, China, and were kept at a temperature of 25 ± 1°C in a 16L: 8D photoperiod.
Pathogenicity assays and infection processes of P. guiyangense
For mycelia infection assays, tests were carried out in plastic cups (capacity of 200 mL), each containing 25 early second-instar larvae and 4 agar plugs (10 mm × 10 mm in size) of P. guiyangense mycelia in 100 mL of deionized distilled water. The numbers of dead larvae were recorded every 24 hours for 10 days and each treatment was replicated at least three times. For zoospore infection assays, zoospores were prepared according to the method previously described [54], and then batches of 25 early second-instar larvae were exposed to a concentration of 107 zoospores ml-1 in individual cups to examine the cuticle penetration process. The progress of infection in the larvae was initially documented using light microscope every 2 h for 48 h. For scanning electron microscopy (SEM), representative larvae were collected at 0.5, 2, 4, and 48 hpi and fixed in 2.5% glutaraldehyde solution. The fixed larvae were then rinsed three times in 0.1 M PBS, dehydrated sequentially in 30%, 50%, 70%, 80%, 95% and 100% ethanol, subjected to critical point drying, mounted, and finally gold coated for viewing. To investigate the digestive system infection, larvae were inoculated with 4 agar plugs (10 mm × 10 mm in size) of P. guiyangense mycelia. After different times post-inoculation, larvae were examined with SEM as described above or else embedded in paraffin, sectioned, and stained with haematoxylin and eosin for light microscope observation.
Genome assembly
High-quality genomic DNA of P. guiyangense was prepared and submitted for genome sequencing using the PacBio and Illumina NGS platforms by Berry Genomics Corporation. The 450-bp paired-end libraries were constructed and sequenced on the Illumina HiSeq 2500 platform. The resultant short reads were processed to remove adapter sequences and low-quality sequences, resulting in 10.32 Gb of clean data (approximately 100-fold coverage). Two PacBio 20-kb SMRTcell libraries were constructed and sequenced on the Sequel platform. The generated raw reads were filtered by trimming the adapter sequences and low-quality sequences. This produced 7.55 Gb of cleaned sequences, with an average cleaned read length of 7.12 kb (approximately 70-fold coverage). Both the Illumina and PacBio SMRT sequencing data were used for the genome assembly. The de novo assembly was produced using the PacBio Hierarchical Genome Assembly Process HGAP 3.0 program [55]. First, the PacBio SMRT sequence data were error-corrected using the long filtered read and sub-read cyclic consensus sequences using HGAP error correction. The error-corrected long reads were then assembled using HGAP with default parameters. The Illumina paired-end reads were aligned to the PacBio assembly with BWA [56], and paired-end reads with concordant alignments were selected with SAMtools view for error correction. A final genome assembly error correction was conducted using the Pilon tool [57]. This Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession QXDM00000000.1. The CEMGA pipeline was used to evaluate the completeness and continuity of the genome on the basis of 248 core eukaryotic genes [58].
Gene prediction and functional annotation
The P. guiyangense assembly was masked for low complexity, as well as known transposable elements using RepeatMasker (www.repeatmasker.org). Genes in the repeat-masked genome were predicted using two predictors, AUGUSTUS [59] and SNAP [60]. The P. guiyangense core eukaryotic genes identified by CEGMA analysis were used to train the gene predictor SNAP. The AUGUSTUS predictor was trained using P. ultimum proteins. SNAP and AUGUSTUS were then used as a part of the MAKER software to conduct the gene prediction. Protein sequences from six sequenced Pythium species, plus Ph. sojae, Ph. infestans, H. arabidopsidis and S. parasitica were submitted to MAKER as extrinsic sources of gene structure evidence to improve sensitivity of gene predictions. The transcripts discovered based on the RNA-Seq data were also submitted to MAKER as EST evidence. MAKER was set to filter out short gene models that produce proteins with fewer than 30 amino acids.
All the protein sequences from P. guiyangense were searched against themselves using the BlastP program with the E-value setting to 10−10. Then, the BlastP result file and the GFF file of the P. guiyangense genome were inputted into the MCScanX program to analyze the synteny blocks and homologous genes located in synteny blocks [24]. Circular representations of these homologous genes were produced using Circos program [61]. The Ks values of each pair of homologous genes were calculated using KaKs_Calculator 2.0 [62].
Whole genome protein families were classified by Pfam analysis [63]. The proteomes were screened for CAZymes (carbohydrate active enzymes) using Hmmscan from the HMMER package and the dbCAN HMM profile database [64]. Putative proteases and protease inhibitors were identified by using batch BLAST at the MEROPS server [65]. Protein kinases were classified by Hmmsearch against KinBase (www.kinase.com).
Transcriptome analysis
A sample of 30 early second-instar larvae inoculated with P. guiyangense mycelia was collected at 24 hpi as the infection stage, and another sample of P. guiyangense mycelia was harvested as the control. There were no biological replicates. The total RNAs of the two samples were extracted according to the method previously described [54], and then sequenced by Berry Genomics Corporation using the Illumina 2500 platform. The 150 bp paired-end reads were filtered for quality as described above and aligned to the P. guiyangense genome assembly using Tophat with a maximum of two mismatches [66]. The mapped reads were quantified using the Cufflinks program [67], and the transcript level of each gene was quantified as RPKM (reads per kilobase transcript length per million reads mapped). Differentially expressed genes were identified using the GFOLD algorithm [68]. GFOLD was developed for unreplicated RNA-Seq data and assigns statistics for expression changes based on the posterior distribution of log fold change. Genes with four-fold change and GFOLD > 1 or < -1 were considered differentially expressed between two samples. The reads were also assembled de novo using the Trinity package [69] with default settings to serve as additional evidence for gene prediction.
To detect the transcript levels of particular P. guiyangense genes during infection stages, qRT-PCR assays were performed. The samples of early second-instar larvae inoculated with P. guiyangense mycelia at different infection time points were collected, and another sample of P. guiyangense mycelia was harvested as the control. Then the total RNAs of the above samples were extracted for qRT-PCR assay. qRT-PCR was performed using an ABI Prism 7500 Real-Time PCR system (Applied Biosystems) with SYBR Premix Ex Tag according to the manufacturer’s instructions. The comparative threshold cycle (Ct) method was used to determine relative transcript levels through ABI 7500 System Sequence Detection Software. The relative transcript levels of particular P. guiyangense genes were normalized to the mycelia data using the actin gene as internal standard. At least three biologically independent replicates of the qRT-PCR experiments were carried out.
Orthology and phylogenetic analysis
A phylogenetic analysis was conducted on the core eukaryotic genes identified using the CEGMA pipeline. Multiplex sequence alignments of proteins were made with ClustalW [70] and subsequently concatenated. A neighbor-joining tree was built using MEGA5 with 1000 fold bootstrapping for distance estimation [71]. Orthologous and paralogous groups among the seven Pythium genomes were determined using OrthoMCL with default parameters: BLASTp E-value cutoff of 10−5 and inflation index of 1.5 [72]. The output of OrthoMCL was parsed to separate core, conserved and specific clusters. Pfam domain enrichment analysis was undertaken on genes that were specific to P. guiyangense. The fold-enrichment corresponds to the frequency of a given PFAM domain within a specific gene set divided by the frequency in the rest of the P. guiyangense proteome; a chi-square test with p-value <0.05 was used for significance tests.
Identification of putative elicitins and effectors
The elicitin domain (PF00964) was retrieved from the PFAM database [63], and then used to search against P. guiyangense proteome. Hits with E-value less than 10−5 were considered to be elicitin candidates. To distinguish elicitin (ELI) and elicitin-like (ELL) proteins, the previously known sequence features of the elicitin domain were used [28]. ELIs contain a highly conserved 98-amino acid domain with six cysteine residues and a typical cysteine spacing pattern. ELLs possess shorter or longer elicitin domains and sequences are more diverse.
For CRN effector prediction, well-characterized CRN proteins from Ph. sojae and Ph. infestans were obtained from a previous publication [14], and then were used to construct HMM profiles based on the LFLAK and HVLVVVP motifs. The HMM profiles were used to search against all possible proteins derived from six open reading frames of the genome. The resulting CRN candidates were manually examined for the presence of LFLAK and HVLVVVP motifs. After that, the validated CRN proteins were used to update the HMM profile, which was then used to search the protein database again for new candidates. To determine whether CRN candidates were full length or pseudogenes, we aligned the CRN candidates with previously characterized CRN proteins. If the CRN candidates shared similar sequence with known CRNs at the DNA level, but had an obvious frameshift mutation or earlier stop codon, they were considered to be pseudogenes. To predict secretion signals for CRN proteins, four signal peptide predictors including SignalP 3.0 (http://www.cbs.dtu.dk/services/SignalP-3.0/), SignalP 4.1 (http://www.cbs.dtu.dk/services/SignalP/), iPSORT (http://ipsort.hgc.jp/), PrediSi (http://www.predisi.de/), and one non-classical secretion signal predictor named SecretomeP 2.0 (http://www.cbs.dtu.dk/services/SecretomeP/), were utilized.
Insect cell bioassays
Spodoptera frugiperda sf9 cell lines were cultured with sf-900™ III SFM medium (Gibco) at 27°C and 140 rpm in suspension flasks until they reached 2×106 cells/mL. Sf9 cells were subcultured using fresh medium every 3 days. The mosquito C6/36 cell lines were cultured in Schneider’s Drosophila Medium (Gibco) with 10% Fetal Bovine Serum (Gibco) at 27°C and the culture medium was renewed every 3 days. Cell density was determined using a Countess Automated Cell Counter (Invitrogen) and cell viability was evaluated by staining with trypan blue exclusion dye.
To evaluate CRN protein toxicity, Sf9 cells were transfected with DNAs encoding CRN proteins. The ORFs of CRNs excluding the signal peptide were amplified, and CRNs without the signal peptide predicted by SignalP were amplified by excluding the N-terminal twenty-five amino acids. The PCR products were cloned into pIB/V5-His vector (Invitrogen) using ClonExpress II One Step Cloning Kit (Vazyme). Sf9 cell suspensions were seeded into 96-well plates (100 μL/well). After 24 h of incubation, cells were transfected with 0.2 ug plasmid DNA using the FuGENE HD Transfection Reagent (Promega) as described by the manufacturer, and six parallel wells were used in each group. Cell toxicity was detected by cell counting with the Cell Counting Kit-8 (CCK-8, Dojindo Laboratories Kumamoto, Japan) according to the manufacturer’s instructions. Briefly, after transfection of the Sf9 cells for 60 hours, 10 μL CCK8 solution was added to the cells. After the cells were incubated for 24 hours at 27°C, the absorbance was analyzed at 450 nm with a reference wavelength of 600 nm using SpectraMax M5 microplate reader. The cells receiving empty vector DNA were considered as 100% viable. Then, the cell viability rate was calculated as follows: Cell viability (%) = [(As-Ab)/(Ac-Ab)]×100%, where As represents the test well reading, Ac represents the empty vector well reading, and Ab represents a blank well reading. The data are expressed as the means ± SE based on at least three independent experiments. CRN constructs were compared to empty vector DNA using Student’s t-test. A difference with P < 0.05 was considered to be statistically significant.
To validate the toxicity of specific CRN proteins, the relevant CRN genes excluding the signal peptide were inserted in frame into the pET32a (+) vector (Novagen) by directional cloning between the BamHI and HindIII sites. The pET32a empty vector was used as a negative control. Cell growth and induction of expression were carried out as described in the pET system manual. Briefly, Escherichia coli BL21(DE3) strains were grown at 37°C to an OD600 of 0.6. At that time, 1 mM IPTG was added in order to induce protein expression at 18°C for 18 h. Induced cells were harvested by centrifugation at 4°C and washed three times with PBS buffer. The cells were resuspended in PBS buffer and lysed using short (3 s) ultrasonic bursts separated by 6 s intervals for 6 min. Crude protein extracts were centrifuged for 10 min at 12,000 rpm. Then the extracts were filtered with a 0.22 um filter. Adherent sf9 or C6/36 cell monolayers in 96-well plates were incubated with the protein extracts for 4 hours. Cell viability was assayed using the methods in the above paragraph, and at least three independent repeats were performed.
Western blot analysis
Cells were lysed in ice-cold lysis buffer (Solarbio) for 10 min. Following this, samples were centrifuged at 12,000 g at 4°C for 5 min. The supernatants were collected and boiled with loading buffer at 100°C for 10 min. The samples were separated by 10% SDS-PAGE and transferred onto a polyvinylidene difluoride membrane (Millipore, Billerica, MA, USA). Membranes were blocked with 5% non-fat milk then incubated with anti-His primary antibody for 2 h. The membranes were washed with 0.1% Tween 20 in PBS and probed with IRDye 800CW-conjugated goat (polyclonal) anti-mouse IgG secondary antibodies for 1 h at room temperature. PVDF membranes were visualized using a scanner (LI-COR Odyssey) with excitation at 700 and 800 nm.
Supporting information
(A) The cumulative survival curves of Aedes albopictus (Left) and Culex pipiens pallens (Right) larvae after inoculation with 4 agar plugs (10 mm × 10 mm in size) of P. guiyangense mycelia. (B) P. guiyangense infected egg, larva, pupa and adult stages of mosquitoes. (C) Cx. pipiens pallens larvae prefer to ingest P. guiyangense mycelia even in the adequate food environment. Cx. pipiens pallens larvae were put in the culture dish containing mycelia of P. guiyangense, and P. aphanidermatum (control), V8 medium and mosquito food.
(TIF)
The relative expression levels of the selected 18 genes were verified by qRT-PCR. Error bars represented the SD for three independent experiments. Gene name was shown on the top of each pair of images.
(TIF)
(A) Phylogenetic tree of Cox II genes among 35 Pythium species. (B) Phylogenetic tree of β-tubulin genes among 35 Pythium species. (C) High percentage of zoospores contained two nuclei in P. guiyangense. Observation of nucleus numbers in zoospores by staining with 4',6-diamidino-2-phenylindole (DAPI). The numbers in the brackets represent the percentage of two nuclei observed in one zoospore (a total of 500 zoospores) for each species.
(TIF)
The cladogram was constructed based on the tree shown in Fig 3A. The letter "A" with blue background represents animal host, and the letter "P" with green background represents plant host. The colored bars represent the numbers of core (present in all the seven genomes), conserved (present in two to six genomes) or species-specific (present only in own genome) genes for each species, which were determined using OrthoMCL.
(TIF)
The red branches represent the subtilisin-like proteases from P. guiyangense.
(TIF)
(A) Neighbor-joining tree of kazal protease inhibitors encoded by the seven available Pythium genomes. (B) Relative abundance of P. guiyangense transcripts encoding kazal protease inhibitors at 24 hpi infection versus mycelia. (C) Validation of transcriptional levels of 4 kazal protease inhibitor genes at different infection time points by qRT-PCR. Error bars represented the SD for three independent experiments.
(TIF)
(A) Expression of CRN proteins in Sf9 cells was confirmed with western blot. (B) Expression of CRN proteins in Sf9 cells was confirmed by detecting the fluorescence signals. (C) Prokaryotic expression of selected CRN proteins confirmed by western blot analysis. (D) qRT-PCR analysis of CRN31 transcript levels at early infection time points. (E) qRT-PCR analysis of CRN28 transcript levels at early infection time points. Transcript levels are given relative to the internal standard actin gene. MY, mycelia.
(TIF)
(MP4)
pipiens pallens larvae readily ingested P. guiyangense mycelia.
(MP4)
(DOC)
(DOC)
(DOC)
(XLSX)
(DOC)
(DOC)
(DOC)
(XLSX)
Acknowledgments
We sincerely thank Dr. Xiaoqing Su (Department of Microbiology, Guizhou Medical University, Guiyang, China) for providing the Pythium guiyangense strain and Dr. Hongliang Chu (Jiangsu Provincial Center for Disease Control and Prevention, Nanjing, China) for providing the Culex Pipiens pallens and Aedes albopictus mosquitoes. The authors are also thankful to Dr. Yidong Wu and Shuwen Wu (Entomology Department, Nanjing Agricultural University, and Nanjing, China) for kind collaboration.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This research was supported by grants from the National Natural Science Foundation of China (Grant No. 31770157 to AX, Grant No. 31625023 to DD). URL of the National Natural Science Foundation of China: http://www.nsfc.gov.cn The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Diniz DFA, de Albuquerque CMR, Oliva LO, de Melo-Santos MAV, Ayres CFJ. Diapause and quiescence: dormancy mechanisms that contribute to the geographical expansion of mosquitoes and their evolutionary success. Parasite Vector. 2017;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scholte EJ, Knols BG, Samson RA, Takken W. Entomopathogenic fungi for mosquito control: a review. Journal of insect science. 2004;4:19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Quiroz Velasquez PF, Abiff SK, Fins KC, Conway QB, Salazar NC, Delgado AP, et al. Transcriptome analysis of the entomopathogenic oomycete Lagenidium giganteum reveals putative virulence factors. Applied and environmental microbiology. 2014;80(20):6427–36. 10.1128/AEM.02060-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Butt TM, Greenfield BP, Greig C, Maffeis TG, Taylor JW, Piasecka J, et al. Metarhizium anisopliae pathogenesis of mosquito larvae: a verdict of accidental death. PloS one. 2013;8(12):e81686 10.1371/journal.pone.0081686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Su X. A new species of Pythium isolated from mosquito larvae and its ITS region of rDNA. Mycosystema. 2006;25(4):523–8. [Google Scholar]
- 6.Yu S, Duan S, Su X. A study on induction conditions for a trypsin-like protease (Pr2) of Pythium guiyangense Su. Journal of guiyang medical college. 2008;33(3):221–4. [Google Scholar]
- 7.Zhao J, Su X. The genetic transformation of Pythium guiyangense mediated by Agrobacterium tumefaciens. Mycosystema. 2008;27(4):594–600. [Google Scholar]
- 8.Kamoun S. Molecular genetics of pathogenic Oomycetes. Eukaryot Cell. 2003;2(2):191–9. 10.1128/EC.2.2.191-199.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saunders GA, Washburn JO, Egerter DE, Anderson JR. Pathogenicity of fungi isolated from field-collected larvae of the Western treehole mosquito, Aedes sierrensis (Diptera: Culicidae). Journal of invertebrate pathology. 1988;52(2):360–3. [DOI] [PubMed] [Google Scholar]
- 10.Larkin RP, English JT, Mihail JD. Effects of infection by Pythium spp on root-system morphology of alfalfa seedlings. Phytopathology. 1995;85(4):430–5. [Google Scholar]
- 11.Krajaejun T, Khositnithikul R, Lerksuthirat T, Lowhnoo T, Rujirawat T, Petchthong T, et al. Expressed sequence tags reveal genetic diversity and putative virulence factors of the pathogenic oomycete Pythium insidiosum. Fungal biology. 2011;115(7):683–96. 10.1016/j.funbio.2011.05.001 [DOI] [PubMed] [Google Scholar]
- 12.Lamour KH, Mudge J, Gobena D, Hurtado-Gonzales OP, Schmutz J, Kuo A, et al. Genome sequencing and mapping reveal loss of heterozygosity as a mechanism for rapid adaptation in the vegetable pathogen Phytophthora capsici. Molecular plant-microbe interactions: MPMI. 2012;25(10):1350–60. 10.1094/MPMI-02-12-0028-R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levesque CA, Brouwer H, Cano L, Hamilton JP, Holt C, Huitema E, et al. Genome sequence of the necrotrophic plant pathogen Pythium ultimum reveals original pathogenicity mechanisms and effector repertoire. Genome Biol. 2010;11(7):R73 10.1186/gb-2010-11-7-r73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haas BJ, Kamoun S, Zody MC, Jiang RH, Handsaker RE, Cano LM, et al. Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature. 2009;461(7262):393–8. 10.1038/nature08358 [DOI] [PubMed] [Google Scholar]
- 15.Rujirawat T, Patumcharoenpol P, Lohnoo T, Yingyong W, Kumsang Y, Payattikul P, et al. Probing the phylogenomics and putative pathogenicity genes of Pythium insidiosum by oomycete genome analyses. Sci Rep. 2018;8(1):4135 10.1038/s41598-018-22540-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang RHY, de Bruijn I, Haas BJ, Belmonte R, Lobach L, Christie J, et al. Distinctive expansion of potential virulence genes in the genome of the oomycete fish pathogen Saprolegnia parasitica. Plos Genet. 2013;9(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bozkurt TO, Schornack S, Banfield MJ, Kamoun S. Oomycetes, effectors, and all that jazz. Curr Opin Plant Biol. 2012;15(4):483–92. 10.1016/j.pbi.2012.03.008 [DOI] [PubMed] [Google Scholar]
- 18.Jiang RH, Tyler BM. Mechanisms and evolution of virulence in oomycetes. Annu Rev Phytopathol. 2012;50:295–318. 10.1146/annurev-phyto-081211-172912 [DOI] [PubMed] [Google Scholar]
- 19.Huang S, Su x. Biological studies on Pythium guiyangense, a fungal pathogen of mosquito larvae. Mycosystema. 2007;26(3):380–8. [Google Scholar]
- 20.Braun-Galleani S, Ortiz-Merino RA, Wu Q, Xu Y, Wolfe KH. Zygosaccharomyces pseudobailii, another yeast interspecies hybrid that regained fertility by damaging one of its MAT loci. Fems Yeast Res. 2018;18(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bertier L, Leus L, D’hondt L, de Cock AWAM, Hofte M. Host adaptation and speciation through hybridization and polyploidy in Phytophthora. PloS one. 2013;8(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Louis VL, Despons L, Friedrich A, Martin T, Durrens P, Casaregola S, et al. Pichia sorbitophila, an interspecies yeast hybrid, reveals early steps of genome resolution after polyploidization. G3 (Bethesda). 2012;2(2):299–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choo JH, Hong CP, Lim JY, Seo JA, Kim YS, Lee DW, et al. Whole-genome de novo sequencing, combined with RNA-Seq analysis, reveals unique genome and physiological features of the amylolytic yeast Saccharomycopsis fibuligera and its interspecies hybrid. Biotechnol Biofuels. 2016;9:246 10.1186/s13068-016-0653-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y, Tang H, Debarry JD, Tan X, Li J, Wang X, et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40(7):e49 10.1093/nar/gkr1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adhikari BN, Hamilton JP, Zerillo MM, Tisserat N, Levesque CA, Buell CR. Comparative genomics reveals insight into virulence strategies of plant pathogenic oomycetes. PloS one. 2013;8(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hanks SK, Hunter T. Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J. 1995;9(8):576–96. [PubMed] [Google Scholar]
- 27.Rimphanitchayakit V, Tassanakajon A. Structure and function of invertebrate Kazal-type serine proteinase inhibitors. Dev Comp Immunol. 2010;34(4):377–86. 10.1016/j.dci.2009.12.004 [DOI] [PubMed] [Google Scholar]
- 28.Jiang RHY, Tyler BM, Whisson SC, Hardham AR, Govers F. Ancient origin of elicitin gene clusters in Phytophthora genomes. Molecular biology and evolution. 2006;23(2):338–51. 10.1093/molbev/msj039 [DOI] [PubMed] [Google Scholar]
- 29.Pemberton CL, Salmond GPC. The Nep1-like proteins—a growing family of microbial elicitors of plant necrosis. Mol Plant Pathol. 2004;5(4):353–9. 10.1111/j.1364-3703.2004.00235.x [DOI] [PubMed] [Google Scholar]
- 30.Torto TA, Li S, Styer A, Huitema E, Testa A, Gow NA, et al. EST mining and functional expression assays identify extracellular effector proteins from the plant pathogen Phytophthora. Genome Res. 2003;13(7):1675–85. 10.1101/gr.910003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kwa MSG, de Maagd RA, Stiekema WJ, Vlak JM, Bosch D. Toxicity and binding properties of the Bacillus thuringiensis delta-endotoxin Cry1C to cultured insect cells. Journal of invertebrate pathology. 1998;71(2):121–7. 10.1006/jipa.1997.4723 [DOI] [PubMed] [Google Scholar]
- 32.Bawin T, Seye F, Boukraa S, Zimmer JY, Raharimalala FN, Ndiaye M, et al. Histopathological effects of Aspergillus clavatus (Ascomycota: Trichocomaceae) on larvae of the southern house mosquito, Culex quinquefasciatus (Diptera: Culicidae). Fungal biology. 2016;120(4):489–99. 10.1016/j.funbio.2016.01.002 [DOI] [PubMed] [Google Scholar]
- 33.Lacey CM, Lacey LA, Roberts DR. Route of invasion and histopathology of Metarhizium anisopliae in Culex quinquefasciatus. Journal of invertebrate pathology. 1988;52(1):108–18. [DOI] [PubMed] [Google Scholar]
- 34.Zhang S, Xia YX, Kim B, Keyhani NO. Two hydrophobins are involved in fungal spore coat rodlet layer assembly and each play distinct roles in surface interactions, development and pathogenesis in the entomopathogenic fungus, Beauveria bassiana. Molecular microbiology. 2011;80(3):811–26. 10.1111/j.1365-2958.2011.07613.x [DOI] [PubMed] [Google Scholar]
- 35.Aw KMS, Hue SM. Mode of infection of Metarhizium spp. fungus and their potential as biological control agents. J Fungi (Basel). 2017;3(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Greenfield BP, Lord AM, Dudley E, Butt TM. Conidia of the insect pathogenic fungus, Metarhizium anisopliae, fail to adhere to mosquito larval cuticle. R Soc Open Sci. 2014;1(2):140193 10.1098/rsos.140193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bonants PJ, Hagenaar-de Weerdt M, Man In ’t Veld WA, Baayen RP. Molecular characterization of natural hybrids of Phytophthora nicotianae and P. cactorum. Phytopathology. 2000;90(8):867–74. 10.1094/PHYTO.2000.90.8.867 [DOI] [PubMed] [Google Scholar]
- 38.Goss EM, Cardenas ME, Myers K, Forbes GA, Fry WE, Restrepo S, et al. The plant pathogen Phytophthora andina emerged via hybridization of an unknown Phytophthora species and the Irish potato famine pathogen, P. infestans. PloS one. 2011;6(9):e24543 10.1371/journal.pone.0024543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bidochka MJ, Melzer MJ. Genetic polymorphisms in three subtilisin-like protease isoforms (Pr1A, Pr1B, and Pr1C) from Metarhizium strains. Can J Microbiol. 2000;46(12):1138–44. [DOI] [PubMed] [Google Scholar]
- 40.Terra WR, Ferreira C. Insect digestive enzymes—properties, compartmentalization and function. Comp Biochem Phys B. 1994;109(1):1–62. [Google Scholar]
- 41.Wu DD, Wang GD, Irwin DM, Zhang YP. A profound role for the expansion of trypsin-like serine protease family in the evolution of hematophagy in mosquito. Molecular biology and evolution. 2009;26(10):2333–41. 10.1093/molbev/msp139 [DOI] [PubMed] [Google Scholar]
- 42.Morris MT, Coppin A, Tomavo S, Carruthers VB. Functional analysis of Toxoplasma gondii protease inhibitor 1. J Biol Chem. 2002;277(47):45259–66. 10.1074/jbc.M205517200 [DOI] [PubMed] [Google Scholar]
- 43.Morris MT, Carruthers VB. Identification and partial characterization of a second Kazal inhibitor in Toxoplasma gondii. Mol Biochem Parasitol. 2003;128(1):119–22. [DOI] [PubMed] [Google Scholar]
- 44.Milstone AM, Harrison LM, Bungiro RD, Kuzmic P, Cappello M. A broad spectrum Kunitz type serine protease inhibitor secreted by the hookworm Ancylostoma ceylanicum. J Biol Chem. 2000;275(38):29391–9. 10.1074/jbc.M002715200 [DOI] [PubMed] [Google Scholar]
- 45.Telleria EL, de Araujo APO, Secundino NF, d’Avila-Levy CM, Traub-Cseko YM. Trypsin-like serine proteases in Lutzomyia longipalpis—expression, activity and possible modulation by Leishmania infantum chagasi. PloS one. 2010;5(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tian MY, Huitema E, da Cunha L, Torto-Alalibo T, Kamoun S. A Kazal-like extracellular serine protease inhibitor from Phytophthora infestans targets the tomato pathogenesis-related protease P69B. Journal of Biological Chemistry. 2004;279(25):26370–7. 10.1074/jbc.M400941200 [DOI] [PubMed] [Google Scholar]
- 47.Ekchaweng K, Evangelisti E, Schornack S, Tian M, Churngchow N. The plant defense and pathogen counterdefense mediated by Hevea brasiliensis serine protease HbSPA and Phytophthora palmivora extracellular protease inhibitor PpEPI10. PloS one. 2017;12(5):e0175795 10.1371/journal.pone.0175795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wawra S, Belmonte R, Lobach L, Saraiva M, Willems A, van West P. Secretion, delivery and function of oomycete effector proteins. Curr Opin Microbiol. 2012;15(6):685–91. 10.1016/j.mib.2012.10.008 [DOI] [PubMed] [Google Scholar]
- 49.Kamoun S. A catalogue of the effector secretome of plant pathogenic oomycetes. Annu Rev Phytopathol. 2006;44:41–60. 10.1146/annurev.phyto.44.070505.143436 [DOI] [PubMed] [Google Scholar]
- 50.Rosenblum EB, Poorten TJ, Joneson S, Settles M. Substrate-specific gene expression in Batrachochytrium dendrobatidis, the chytrid pathogen of amphibians. PloS one. 2012;7(11):e49924 10.1371/journal.pone.0049924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kuo A, Kohler A, Martin FM, Grigoriev IV. Expanding genomics of mycorrhizal symbiosis. Front Microbiol. 2014;5:582 10.3389/fmicb.2014.00582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Olivera IE, Fins KC, Rodriguez SA, Abiff SK, Tartar JL, Tartar A. Glycoside hydrolases family 20 (GH20) represent putative virulence factors that are shared by animal pathogenic oomycetes, but are absent in phytopathogens. BMC Microbiol. 2016;16(1):232 10.1186/s12866-016-0856-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu T, Ye W, Ru Y, Yang X, Gu B, Tao K, et al. Two host cytoplasmic effectors are required for pathogenesis of Phytophthora sojae by suppression of host defenses. Plant physiology. 2011;155(1):490–501. 10.1104/pp.110.166470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang C, Shen D, Wang J, Chen Y, Dong Y, Tang Z, et al. An AGC kinase, PgAGC1 regulates virulence in the entomopathogenic oomycete Pythium guiyangense. Fungal biology. 2019;123(1):87–93. 10.1016/j.funbio.2018.11.006 [DOI] [PubMed] [Google Scholar]
- 55.Chin CS, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods. 2013;10(6):563–+. 10.1038/nmeth.2474 [DOI] [PubMed] [Google Scholar]
- 56.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–60. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PloS one. 2014;9(11):e112963 10.1371/journal.pone.0112963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Parra G, Bradnam K, Korf I. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics. 2007;23(9):1061–7. 10.1093/bioinformatics/btm071 [DOI] [PubMed] [Google Scholar]
- 59.Stanke M, Steinkamp R, Waack S, Morgenstern B. AUGUSTUS: a web server for gene finding in eukaryotes. Nucleic Acids Res. 2004;32(Web Server issue):W309–12. 10.1093/nar/gkh379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Korf I. Gene finding in novel genomes. BMC Bioinformatics. 2004;5:59 10.1186/1471-2105-5-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19(9):1639–45. 10.1101/gr.092759.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang D, Zhang Y, Zhang Z, Zhu J, Yu J. KaKs_Calculator 2.0: a toolkit incorporating gamma-series methods and sliding window strategies. Genomics Proteomics Bioinformatics. 2010;8(1):77–80. 10.1016/S1672-0229(10)60008-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016;44(D1):D279–85. 10.1093/nar/gkv1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yin Y, Mao X, Yang J, Chen X, Mao F, Xu Y. dbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2012;40(Web Server issue):W445–51. 10.1093/nar/gks479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rawlings ND, Barrett AJ, Bateman A. MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2012;40(Database issue):D343–50. 10.1093/nar/gkr987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–11. 10.1093/bioinformatics/btp120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):511–5. 10.1038/nbt.1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Feng J, Meyer CA, Wang Q, Liu JS, Shirley Liu X, Zhang Y. GFOLD: a generalized fold change for ranking differentially expressed genes from RNA-seq data. Bioinformatics. 2012;28(21):2782–8. 10.1093/bioinformatics/bts515 [DOI] [PubMed] [Google Scholar]
- 69.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29(7):644–52. 10.1038/nbt.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular biology and evolution. 2011;28(10):2731–9. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li L, Stoeckert CJ Jr., Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13(9):2178–89. 10.1101/gr.1224503 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) The cumulative survival curves of Aedes albopictus (Left) and Culex pipiens pallens (Right) larvae after inoculation with 4 agar plugs (10 mm × 10 mm in size) of P. guiyangense mycelia. (B) P. guiyangense infected egg, larva, pupa and adult stages of mosquitoes. (C) Cx. pipiens pallens larvae prefer to ingest P. guiyangense mycelia even in the adequate food environment. Cx. pipiens pallens larvae were put in the culture dish containing mycelia of P. guiyangense, and P. aphanidermatum (control), V8 medium and mosquito food.
(TIF)
The relative expression levels of the selected 18 genes were verified by qRT-PCR. Error bars represented the SD for three independent experiments. Gene name was shown on the top of each pair of images.
(TIF)
(A) Phylogenetic tree of Cox II genes among 35 Pythium species. (B) Phylogenetic tree of β-tubulin genes among 35 Pythium species. (C) High percentage of zoospores contained two nuclei in P. guiyangense. Observation of nucleus numbers in zoospores by staining with 4',6-diamidino-2-phenylindole (DAPI). The numbers in the brackets represent the percentage of two nuclei observed in one zoospore (a total of 500 zoospores) for each species.
(TIF)
The cladogram was constructed based on the tree shown in Fig 3A. The letter "A" with blue background represents animal host, and the letter "P" with green background represents plant host. The colored bars represent the numbers of core (present in all the seven genomes), conserved (present in two to six genomes) or species-specific (present only in own genome) genes for each species, which were determined using OrthoMCL.
(TIF)
The red branches represent the subtilisin-like proteases from P. guiyangense.
(TIF)
(A) Neighbor-joining tree of kazal protease inhibitors encoded by the seven available Pythium genomes. (B) Relative abundance of P. guiyangense transcripts encoding kazal protease inhibitors at 24 hpi infection versus mycelia. (C) Validation of transcriptional levels of 4 kazal protease inhibitor genes at different infection time points by qRT-PCR. Error bars represented the SD for three independent experiments.
(TIF)
(A) Expression of CRN proteins in Sf9 cells was confirmed with western blot. (B) Expression of CRN proteins in Sf9 cells was confirmed by detecting the fluorescence signals. (C) Prokaryotic expression of selected CRN proteins confirmed by western blot analysis. (D) qRT-PCR analysis of CRN31 transcript levels at early infection time points. (E) qRT-PCR analysis of CRN28 transcript levels at early infection time points. Transcript levels are given relative to the internal standard actin gene. MY, mycelia.
(TIF)
(MP4)
pipiens pallens larvae readily ingested P. guiyangense mycelia.
(MP4)
(DOC)
(DOC)
(DOC)
(XLSX)
(DOC)
(DOC)
(DOC)
(XLSX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.