Abstract
Background
Extracellular vesicles (EV) are shed by tumor cells but little is known about their individual molecular phenotypes and heterogeneity. While exosomes have received considerable attention, much less is known about larger microvesicles. Here we profile single microvesicles (MV) and exosomes from glioblastoma (GB) cells and MV from the plasma of patients.
Methods
EV secreted from mouse glioma GL261 and human primary GBM8 cell lines as well as from the plasma of 8 patients with diagnoses of GB and 2 healthy controls were isolated and processed for single vesicle analysis. EV were immobilized on glass slides and the heterogeneity of vesicle and tumor markers were analyzed at the single vesicle level.
Results
We show that (i) MV are abundant, (ii) only a minority of MV expresses putative MV markers, and (iii) MV share tetraspanin biomarkers previously thought to be diagnostic of exosomes. Using MV capture and staining techniques that allow differentiation of host cell and GB-derived MV we further demonstrate that (i) tumoral MV often present as <10% of all MV in GB patient plasma, and (ii) there is extensive heterogeneity in tumor marker expression in these tumor-derived MV.
Conclusion
These results indicate that single MV analysis is likely necessary to identify rare tumoral MV populations and the single vesicle analytical technique used here can be applied to both MV and exosome fractions without the need for their separation from each other. These studies form the basis for using single EV analyses for cancer diagnostics.
Keywords: cancer, diagnostics, extracellular vesicles, microvesicles
1. Large EV (microvesicles) are abundant in circulation in glioblastoma patients. 2. This study defines the molecular landscape of single microvesicles in circulation.
Key Points.
Large EV (microvesicles) are abundant in circulation in glioblastoma patients.
This study defines the molecular landscape of single microvesicles in circulation.
Importance of the study.
Extracellular vesicles have emerged as important circulating, nano-sized structures with diagnostic and therapeutic potential. These vesicles are shed by a number of cell types in the human body, and analyzing their makeup is essential prior to clinical applications. Despite intense research interests, little is known about the heterogeneity of these vesicles. Here we used a single vesicle analytical technique to profile microvesicles from glioblastoma cells and patients. Our studies detail the landscape of microvesicles in human circulation and provide the basis for future cancer diagnostics.
Many mammalian cells release extracellular vesicles (EV) into circulation. These vesicles differ in ontogeny, size, and molecular composition.1–5 The two most abundant fractions of EV are termed microvesicles (MV; 200–1000 nm), formed by budding of the cell membrane, and exosomes (EX; 50–200 nm), derived from multivesicular bodies. The last few years have seen intense interest in these vesicles given their biological role in cancer6–9 and potential for diagnostic and therapeutic purposes.10–13 To date, vesicles are often differentiated by size and density criteria14–16 and the presence or absence of markers considered “specific” for certain EV subtypes. For example, it is generally accepted that EX fractions are positive for cluster of differentiation (CD)63, CD9, CD81, Alix, and tumor susceptibility gene 101 (TSG101).5 Similarly, it has been reported that MV are positive for integrin beta 1, CD40, vesicle-associated membrane protein 3 (VAMP-3), and ADP-ribosylation factor 6 (Arf6).17–19
Much of the work to date has relied on bulk analysis of vesicle fractions using western blotting20 and microchip analysis of RNA21,22 or protein cargo.4,23,24 Invariably, these data fail to provide granularity on the composition of specific vesicles and thus support coexpression of different markers of EV populations. Yet, it is the protein present on a given vesicle that could ultimately determine its pharmacokinetics and together with its mRNA and miRNA also biological function. For this reason, we have been interested in developing single EV analytical techniques to shed light on the composition of vesicles obtained from patients.
Single EV analytical technologies are slowly emerging for use in cancer22,25–29 and are believed to ultimately provide much needed insight into the molecular makeup and heterogeneity of tumor-derived EV. The majority of emerging technologies have been applied to EV from cell culture lines rather than primary human samples. The goal of the current study was thus to focus on the makeup of MV obtained from peripheral blood of glioblastoma (GB) patients. We adapted a capture/image technology in which EV are immobilized on glass slides and then developed an imaging strategy to separately analyze EV from host and tumor cells based on surface markers. Here we provide a comprehensive single vesicle analysis of large (MV-like) and small (EX-like) fractions of thousands of vesicles. We show unexpected findings of marker expression on MV that have previously been excluded in the diagnostic analysis of plasma EV.
Materials and Methods
Cell Culture
Cell lines in this study were acquired from the Breakefield lab from recent American Type Culture Collection stocks and verified to be mycoplasma free. Mouse glioma GL261 cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal bovine serum (FBS; Sigma) at 37°C in a humidified atmosphere with 5% CO2. Before EV collection, cells were grown in DMEM with 10% exosome-depleted FBS (Thermo Fisher Scientific) for 72 hours. Human primary GBM8 cells were grown as spheroids in Neurobasal media (Thermo Fisher Scientific) with 1.5 mL/100 mL 100x Glutamax (Thermo Fisher Scientific), 0.5 mL/100 mL N-2 supplement (Thermo Fisher Scientific), 2 mL/100 mL B-27 supplement (Thermo Fisher Scientific), 1% penicillin/streptomycin, 20 ng/mL epidermal growth factor (R&D Systems), and 20 ng/mL fibroblast growth factor (Peprotech). Media with supplements were filtered through a 0.22-μm pore size membrane to sterilize. Cells were subcultured by seeding at a 10000–20000 cells/cm2 density and supplied with fresh media every 3 days.
Human Samples
Blood samples were obtained from patients (8 with diagnoses of GB at the Brigham and Women’s Hospital and 2 neurologically healthy controls) per institutional review board–approved protocol with informed consent acquired from each patient. Histological diagnosis of GB was made by a board certified neuropathologist at Brigham and Women’s Hospital. For every patient, the tumor cytogenetic was determined by a comparative genomic hybridization assay. Blood samples were collected in 10 mL EDTA tubes and inverted several times before storing at 4°C. Within 1 hour of blood collection, samples were spun at 400 × g for 10 min at 4°C, and plasma layer was pulled from the top, aliquoted, and stored at −80°C until ready for EV isolation and preparation.30
Preparation of EV
In preparation of EV fractions we adopted a clinically viable method of size separation for analytic purposes. Supernatants from cell culture media and plasma were centrifuged at 300 × g for 5 min followed by centrifugation of the resultant supernatant at 2000 × g for 10 min to remove cell debris. The supernatant from this step was then centrifuged at 10000 × g for 30 min to isolate a fraction of larger EV which we termed a “large (MV-like)” subfraction. The pellet was resuspended in phosphate buffered saline (PBS) and re-spun at 10000 × g for 30 min. Supernatant from the initial isolation spin was centrifuged at 100000 × g for 70 min to isolate a smaller EV population which we termed a “small (EX-like)” fraction. The pellet was washed in PBS and then centrifuged at 100000 × g for 70 min to re-pellet. Size separated EX, and MV fractions were resuspended in 300 μL of PBS and incubated with 333 μM EZ-Link Sulfo-NHS-LC-Biotin (Thermo Fisher Scientific) for 30 min at room temperature. We used a 20-fold molar excess of sulfo-NHS-biotin to EV protein in approximately 0.5 mL volume. Thus about 4–6 biotins were expected to be incorporated per vesicle. Excess biotin was then removed utilizing the Zebra Spin Desalting Column, 7K MWCO (Thermo Fisher Scientific) per the kit instructions. EV were then incubated with 5 µg/mL Cell Tracker CM-DiI membrane dye (Thermo Fisher Scientific) for 30 min at room temperature, and excess dye was removed utilizing the Zebra Spin Desalting Column, 7K MWCO.
Antibody Preparation
Vendor and clone information of all antibodies used are summarized in Supplementary Table 1. All antibodies were validated against positive and negative controls and other published means.31 CD9(VJ1)-CD405M, TSG101, integrin beta 1 (12G10), epidermal growth factor receptor (EGFR)-AF594, CD31-AF647 CD45, epithelial cell adhesion molecule (EpCAM) (AUA1), and Arf6 (poly) antibodies were purchased from Abcam; CD41, CD42b-AF647, CD40 (5C3), CD40 (3/23), CD9 (MZ3), and isocitrate dehydrogenase 1 (IDH1) (O92H9) antibodies were purchased from Biolegend; CD63 (AHN16.1/16-4-5) antibody was purchased from Ancell; Alix, CD81, and VAMP-3 antibodies were purchased from Santa Cruz; IDH1-R132H, Arf6 (EPR8357), and integrin beta 1 (poly) antibodies were purchased from EMD Millipore; CD235a-AF647 antibody was acquired from BioRad; EGFR variant III (EGFRvIII) and IDH1 (D2H1) antibodies were acquired from Cell Signaling Technologies; CD63 (poly) was acquired from R&D Systems; and EpCAM (G8.8) was acquired from eBioscience. CD40 and IDH1-R132H antibodies were conjugated to Pacific Blue; TSG101, VAMP-3, and EGFRvIII antibodies were conjugated to AF488; IDH1 and CD63 were conjugated to AF555; integrin beta 1 and Alix antibodies were conjugated to AF594; CD45 and CD41 antibodies were conjugated to AF647; and EpCAM, CD81, and Arf6 antibodies were conjugated to AF680 utilizing Antibody Labeling Kits (Thermo Fisher Scientific) per manufacturer’s instructions.
Single EV Analysis Protocol
Experiments were performed on a BX-63 Upright Automated Fluorescent Microscope (Olympus) with a 100x oil objective using Metamorph Software. Biotinylated EV in 1x PBS were captured on a streptavidin-coated coverslip for 30 min at room temperature. The coverslip was then washed 3 times using 0.2% bovine serum albumin (BSA) in PBS and then blocked for 30 min with this BSA solution. Next, fluorescently labeled antibodies were incubated on the coverslip for 1.5 hours followed by 3 washes of 5 min with 0.2% BSA in PBS. Coverslips were then mounted on slides using ProLong Gold Antifade Mountant (Thermo Fisher Scientific) overnight. Slides were sealed and fluorescent images were taken.
Image Processing
Image analysis was performed using ImageJ. We used the Cell Tracker CM-DiI imaging channel to create masks at EV locations and isolate the signal of interest in all channels. Imaging conditions (ie, objective, exposure times, camera settings, sample preparation, illumination) were fixed across all experiments. Instances of spectral bleed-through among adjacent imaging channels were eliminated using spectral un-mixing plugins in ImageJ. At each mask position, we obtained average pixel intensities for each respective molecular target.
Data Analysis
Data were analyzed in a similar fashion as done in flow cytometry. Single parameter histograms were first obtained by measuring the signal distribution for each marker as compared with controls where the primary antibody was omitted or was nonsense immunoglobulin G. A gating threshold was then determined and each data channel was normalized to this value. Vesicles with signal above the threshold were then counted as positive and the percentages of total EV positive for a given marker was determined for each channel. Heat maps were built with each fluorescence channel normalized to its maximum thresholded value.
Results
Microvesicles Are Abundant
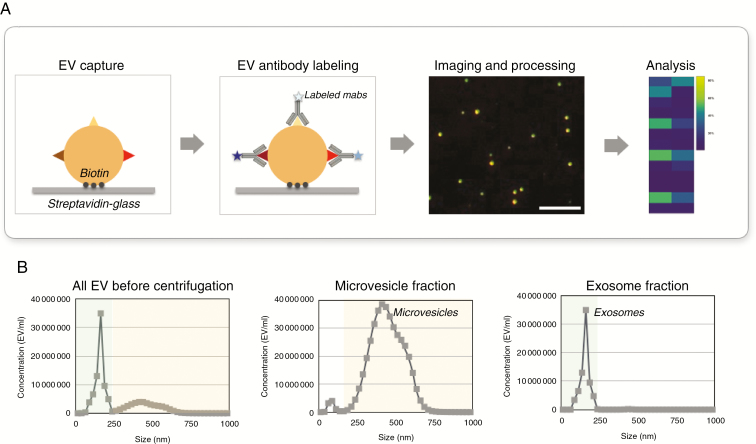
Our study sought to characterize vesicles utilizing a single vesicle analytical strategy, outlined in Fig. 1. We biotinylated vesicles obtained by differential ultracentifugation and captured them on streptavidin-coated glass slides to immobilize vesicles. Following this capture vesicles were stained with fluorochrome labeled antibodies and imaged, and the data then analyzed. This generic principle was applied to vesicles obtained by 10000 g centrifugation and a subsequent 100000 g centrifugation. Since it is not possible to determine the precise ontogeny of vesicles based on ultracentrifugation alone, we have adopted the nomenclature of “large (MV-like)” and “small (EX-like)” vesicles. We were primarily interested in the large MV-like fraction, since (i) MV are more abundant than EX, (ii) MV contain most of the mRNAs that can report on mutational status and gene amplification,32 (iii) much less is known about MV protein makeup, and (iv) MV-like fractions contain larger vesicles that afford easier analysis and isolation procedures. Since some prior studies had looked at single EX in cell lines, we also directly compared MV-like fraction and EX-like fraction makeup released by cell lines.
Fig. 1.
Overview of single vesicle analysis. (A) Overview of the procedural steps for single vesicle analysis. EV are biotinylated and captured on streptavidin-coated glass coverslips and stained by fluorescent antibodies. Coverslips are imaged by microscopy and multidimensional data are analyzed. Scale bar = 10 µm (B) Representative Nanosight Tracking Analysis (NTA) size distribution of vesicles from GBM8 media. NTA analyses are shown for samples before and after separation.
As can be seen from Fig. 1, the different vesicle fractions were abundant following their respective enrichment procedures. In general we obtained ~109 vesicles/mL of harvested media and used ~10 µL of that for analysis. Supplementary Fig. 1 shows a representative example of the typical MV-like fraction obtained from the plasma of GB patients. In this particular example, there were about 2000 vesicles per field of view (100x objective). The target-to-background ratio following staining was ~110:1 (Supplementary Fig. 2) once the different antibodies had been optimized (Supplementary Table 1).
Unique Patterns of Biomarker Expressions in Microvesicles
Following the above optimization procedures we next analyzed vesicles from 2 different cell lines—primary human GBM8 and mouse glioma GL261. GBM8 is a patient-derived xenografted GB cell line that had been directly propagated following resection under anchorage-independent sphere culture conditions and then maintained by serial intracranial engraftment.33 GBM8 cells exhibit primitive neuroectodermal characteristics and proliferate as loose neurosphere aggregates in vitro. Inoculation of GBM8 cells into the brain of immunosuppressed NSG (NOD-scid IL2Rgammanull) mice leads to a diffusely invasive tumor. In contrast, GL261 is a mouse glioma cell line that can be grown in immunocompetent C57/BL6 mice.34 Another reason to analyze EV from cell lines first was to focus on tumor cell derived vesicles prior to embarking on clinical samples where EV in plasma represent a combination of both normal host and tumor cells.
We assessed single EV compositions, and a representative example of this from GBM8 culture is demonstrated in Fig. 2A. Focusing on MV-like vesicles, in this particular example about 400 individual vesicles were examined in a single field of view. Of interest, putative MV marker integrin beta 1 was present on only half of the isolated MV-like vesicles. Furthermore, the putative MV marker CD40 was present on even lower numbers of MV-like vesicles or even absent. Conversely, classic markers that are often used in the literature to characterize EX in mixed EV populations were also positive in this MV-like fraction.5 In particular, tetraspanins (CD9, CD63, CD81) were found at varying levels in the large MV-like fraction. Finally GB markers (EGFR, EGFRvIII, EpCAM) were present at various concentrations. We calculated the proportion of individual MV-like vesicles that were positive for a given marker in GBM8 (Fig. 2B). The positive markers ranked as follows (Fig. 2B, Table 1): integrin beta 1 (52% of MV were positive), CD81 (49%), IDH1 wildtype (48%), TSG101 (43%), EGFR (32%), CD9 (26%), CD63 (13%), EGFRvIII (12%), CD40 (9%), Alix (6%), EpCAM (6%), Arf6 (5%), VAMP-3 (5%), IDH1-R132H (negative). To determine whether the above MV observations were unique to the GBM8 model, we also investigated GL261 vesicles. As can be seen from Fig. 3C, a similar picture emerged. The most abundant MV markers (number of vesicles above the dashed red line Fig. 3A, C) were integrin beta 1, CD81, and EGFR.
Fig. 2.
Marker expression in microvesicles obtained from GBM8. (A) Images of MV-like vesicles labeled for generic EV markers (CD81, TSG101, CD63, Alix, integrin beta 1, CD9, CD40, Arf6, VAMP-3) and glioblastoma markers (EGFR, EGFRvIII, EpCAM, IDH1-R132H) were quantified per single vesicle. Heat map of data is organized by high expression to low expression. Note the heterogeneity profile of each individual MV-like vesicle. (B) Waterfall plot displaying the percent of the total MV-like vesicles that are positive for a given marker. IDH1-R132H was not detected in any of the vesicles.
Table 1.
Summary of marker expression in MV-like fractions obtained from different specimens
| Vesicle Type | Protein | Microvesicles Obtained from | ||
|---|---|---|---|---|
| GBM8 Cells | GL261 Cells | Human Plasma | ||
| All vesicles | CD9 | 0.26 | 0.02 | 0.12 |
| CD63 | 0.13 | 0.02 | 0.09 | |
| CD81 | 0.49 | 0.67 | 0.14 | |
| TSG101 | 0.43 | 0.15 | 0.11 | |
| Alix | 0.06 | 0.1 | 0.41 | |
| CD40 | 0.09 | 0.1 | 0.1 | |
| Arf6 | 0.05 | 0.12 | 0.29 | |
| VAMP-3 | 0.05 | 0.04 | 0.09 | |
| IDH1WT | 0.48 | 0.22 | 0.03 | |
| Integrin beta 1 | 0.52 | 0.73 | 0.26 | |
| Tumor vesicles | EGFR | 0.32 | 0.11 | See Fig. 4 |
| EGFRvIII | 0.12 | 0.07 | ||
| IDH1-R132H | 0 | NA | ||
| EpCAM | 0.06 | 0.09 | ||
The table summarizes the fractions of MV-like vesicles that are positive for a given protein across cell lines and human samples (n = 10). For example, the putative ubiquitous MV biomarker integrin beta 1 was present in only 52% of GBM8. In GL261 cells it was present in 73% of vesicles but in primary human samples only in 26% of MV-like vesicles. In human samples, this fraction was lower (26%). IDH1-R132H was not detected in any of the samples and all human tumors were IDH wildtype by sequencing. WT = wildtype; NA = no mouse antibody available.
Fig. 3.
Comparison of biomarker expression in MV-like and EX-like fractions in GBM8 (A, B) and GL261 (C, D). (A) GBM8 MV-like vesicles were analyzed for different proteins and data are plotted for individual vesicles. A signal intensity above 1 (red line) denotes “positivity” of a given marker and values below 1 indicate negativity. Note the heterogeneous distribution for any given marker. (B) Comparative analysis of MV-like and EX-like fractions obtained from GBM8. Shown are the percentages of vesicles that are positive for a given protein marker. The heat map on the right represents a ratio of MV over EX: red = higher in MV; blue = higher in EX; yellow = similar expression in both subfractions. (C) Similar analysis as in (A) but for GL261 MV. (D) Similar analysis as in (B) but for GL261.
We next addressed the question of whether MV patterns of the analyzed proteins would be different in concurrently isolated single EX. For this purpose, we analyzed GBM8 (Fig. 3B) and Gli26 (Fig. 3D) vesicles and simply classified them as either positive or negative for a given protein, using the same metric as in Fig. 3A, C, that is, larger than the cutoff (1). Data were expressed as percent of vesicles positive for a given marker and as the MV/EX ratio, that is, whether a marker was preferentially expressed at higher levels in the MV-like fraction compared with the EX-like fraction. As can be seen from Fig. 3, Arf6 and CD9 were expressed at a higher fraction in the EX-like fraction, but most other proteins studied were expressed at similar levels in both EX-like and MV-like fractions. The exception was in expression levels of GB markers (EGFR, EGFRvIII, and IDH1), where different patterns of MV/EX were observed in Gli261 compared with GBM8. It should be pointed out that these numbers represent ratios of ratios and that all MV-like and EX-like fractions contained vesicles positive for these tumor markers.
Microvesicles in Human Plasma Samples Are Heterogeneous
We next analyzed large MV-like vesicles obtained from plasma of GB patients (n = 8) and age-matched normal controls (n = 2). In a first set of experiments, we analyzed total MV-like vesicle concentrations in plasma samples. As can be seen in Fig. 4A, MV-like vesicles were fairly abundant in all patients, with concentrations ranging from 8.1 × 107 to 2.1 × 109 MV/mL. As expected, there was overlap in concentrations between GB patients and controls, so that this metric is unlikely to be of diagnostic utility. We thus set out to determine individual biomarker expression in the MV-like fraction. To facilitate the differentiation between host and putative tumor cell MV-like vesicles we used a cocktail of antibodies to account for the majority of host cell MV. This cocktail contained antibodies against CD45 (leukocytes), CD31 (endothelial), CD41/CD42b (platelet), and CD235a (red blood cell), all with the same fluorochrome. While this staining strategy is simple and akin to a “negative selection,” it still cannot unequivocally define GB MV-like vesicles. To enable the latter, we also analyzed tumor markers (EGFR, EGFRvIII, EpCAM, and IDH1-R132H). This study was focused on characterization of the most common molecular markers associated with clinical GB and for which good antibodies are commercially available (EGFR amplification/mutation and IDH1 mutation). Additionally, EpCAM overexpression has been suggested to be predictive of malignancy,35 proliferation, and prognosis in GB and was thus included for exploratory reasons.
Fig. 4.
Global marker expression in human GB plasma samples. (A) Total MV-like vesicle numbers in the plasma from each patient were determined via nanoparticle tracking analysis (NTA). (B) The amount of TMV was determined by first accounting for host cell MV (using a cocktail of CD31, CD41, CD42β, CD45, and CD235a) then staining remaining vesicles for tumor markers (EGFR, EGFRvIII, EpCAM, and IDH1-R132H). TMV were present and detectable in all GB patients analyzed. (C) The relative contribution of TMV to the total MV-like vesicle pool was variable among GB patients. (D) The heterogeneity of EGFR, EGFRvIII, and EpCAM expression (IDH1-R132H was not detected in any of the EV observed) among the subpopulation of TMV for each patient was assessed.
Fig. 4B–D shows representative tumor-derived MV-like vesicle (TMV) analyses. The absolute TMV concentrations ranged considerably from 3.2 × 105 to 7.6 × 108 in the 8 GB patients (Fig. 4B) and there was no clear pattern in patients who were treatment naive or who had received standard-of-care treatment. The majority of plasma MV-like vesicles in these samples originated from host cells, on average making up ~90% of the MV-like vesicles. The exception was a single patient (G1) in whom TMV were more abundant.
The makeup of the TMV in each of the 8 patients is summarized in Fig. 4D. In these samples, EGFR, EGFRvIII, and EpCAM were positive in a fraction of vesicles. EGFR overexpression was commonly observed. The main source of TMV in G1, the patient with the highest TMV concentration, was due to EGFRvIII vesicles. None of the age-matched control samples stained positive for any of the 3 tumor markers. Furthermore, vesicle analysis was conducted blinded to the IDH1 mutation status of the patients, and patients were not selected with specific IDH1 mutation specifications. In the small cohort of patients, IDH1-R132H mutations were not identified by either sequencing or by the Single EV Analysis (SEA) protocol. Table 1 summarizes the fraction of marker positive MV-like vesicles across all cell lines and human samples for comparison. The plasma MV analysis was largely concordant with pathological analysis of tumor tissue (Table 2).
Table 2.
Patient demographics and clinical characteristics
| Pathology Tumor Tissue Dx | Plasma MV Analysis | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| # | Sex | Age | Treatment Prior to Blood Draw | Diagnosis | EGFR | EGFRvIII | IDH1-R132H | EGFR | EGFRvIII | IDH1-R132H |
| G1 | M | 49 | None | GB mesench | Amp | NA | WT | Pos | Pos | Neg |
| G2 | F | 75 | None | GB proneural | Polysomy Chr7 | NA | WT | Pos | Neg | Neg |
| G3 | M | 73 | None | GB proneural | Neg | Neg | WT | Neg | Neg | Neg |
| G4 | F | 63 | None | GB proneural | Amp | Neg | WT | Pos | Neg | Neg |
| G5 | M | 55 | Surgery + XRT + TMZ | GB mesench | Amp | Neg | WT | Pos | Neg | Neg |
| G6 | M | 61 | Surgery + XRT + TMZ | GB mesench | Amp | Neg | WT | Pos | Neg | Neg |
| G7 | M | 73 | Surgery + XRT + TMZ | GB NA | Amp | NA | WT | Pos | Neg | Neg |
| G8 | F | 54 | Surgery + XRT + TMZ | GB mesench | Neg | Neg | WT | Neg | Neg | Neg |
| H1 | M | 57 | None | Normal | NA | NA | NA | NA | NA | NA |
| H2 | M | 50 | None | Normal | NA | NA | NA | NA | NA | NA |
XRT = external beam radiation therapy; TMZ = temozolomide; WT = wildtype. The plasma MV analysis was largely concordant with pathological analysis of tumor tissue, although some specimens were small biopsies only. NA = not available.
Discussion
Extracellular vesicles are shed by tumor cells but little is known about their individual molecular phenotypes and heterogeneity. While exosomes have received considerable attention, much less is known about larger MV. Microvesicles are a common type of EV and are produced by direct membrane shedding from many host and tumor cells. These vesicles measure 100–1000 nm in diameter and reflect the plasma membrane protein composition and orientation of their respective host cells. In addition, they contain intracellular proteins, mRNAs, miRNAs, and many noncoding small RNAs.32 Changes in vesicle number and composition have been observed in cancer and a variety of other diseases.36,37 These changes can potentially be used as biomarkers, but several questions and clinical challenges remain. For example: what are the host cell MV (HMV) numbers and variations in normal humans, what are the number and composition of TMV, and how does one best differentiate HMV and TMV in clinical samples?
The current study was designed to shed light on some of the above questions. In addition—and as a necessary step to answer the above questions—we were interested in single vesicle analyses to provide unequivocal answers. Finally, we asked the question whether MV analysis would be complementary, additive to, or exclusive from EX analysis. This question is of importance as many current clinical assays first isolate EX-like vesicles but discard larger MV-like fractions. In other words: is such a separation warranted or could diagnostics be performed on the larger pool of all EV, irrespective of their origin? To address this question we compared biomarker expression in both MV-like and EX-like fractions from well-established model lines. We found that (i) MV-like vesicles contained biomarkers once thought of as typical of EX fractions, (ii) EX-like vesicles also contain MV markers, and (iii) there was considerable heterogeneity in marker expression across MV-like and EX-like fractions. These results question the current paradigm of isolating “specific” exosome fractions for diagnostic purposes and argue for more comprehensive analysis of all EV fractions, ideally with single vesicle resolution.
Our technique applied to patient samples showed that MV-like vesicles were abundant in normal patients (~7.1 × 108 MV/mL) and on average were slightly more numerous in GB patients (~1.5 × 109 MV/mL) but with considerable heterogeneity. The use of a host cell cocktail (containing antibodies against CD45, CD31, CD41/CD42b, and CD235a to account for MV from leukocytes, endothelial cells, platelets, and red blood cells, respectively) allowed us to broadly separate host and tumor MV-like vesicles in a given sample in a time-efficient manner. We found that the molecular makeup of MV-like vesicles in patient samples was surprising. We found that only a relatively small fraction of MV-like vesicles were positive for presumed “ubiquitous” MV markers such as integrin beta 1 (20–27% of vesicles), VAMP-3 (9–10% of vesicles), and Arf6 (28–31% of vesicles). Similar findings were also true for tetraspanins (CD63+ in 7–9% of MV, CD9+ in 6–13% of MV, CD81+ in 8–15% of MV) and other MV markers. The observation that EV are heterogeneous is not entirely unexpected given heterogeneous/mosaic marker expression on parental tumor cells.2,38–41 What is less established, however, is that even large MV-like vesicles from homogeneous cell lines show heterogeneity and occasionally lack expression of defining markers, such as shown here. As expected, tumor markers (eg, EpCAM, EGFR, EGFRvIII, IDH1-R132H) were also heterogeneous across samples from different patients (Supplementary Fig. 3). These results are not entirely surprising and likely reflect the heterogeneity of GB makeup as well as the varying proportion of TMV in plasma in different GB patients.
While the current study was preliminary in nature and focused on developing and validating new technology and then applying it to thousands of vesicles per sample, future studies are warranted to study larger patient cohorts and different cancer types. Similar single vesicle analyses are being performed utilizing nanoflow methods42; however, the SEA method has several advantages, including higher signal-to-noise ratios and the ability to interrogate vesicles in more than 2–3 channels. The implications of the current study are clear: (i) TMV are heterogeneous in their makeup and only a small fraction harbor tumor-specific mutated or overexpressed proteins; (ii) single vesicle analysis is likely necessary to capture these “rare” events and thus detect early forms of cancers; and (iii) both host-cell and tumor-cell specific staining protocols can be used to analyze large numbers of EV in short periods of time.
Funding
This study was funded in part by P01CA069246 (EAC, XOB, RW), R01CA204019 (RW), and R21CA205322 (HL).
Conflict of interest statement
None declared.
Authorship statement.
Study design, data acquisition: KF, AJ, KY, CV
Data analysis. KF, AJ, JG
Patient sample collection. PP
Data interpretation. KF, AJ, EAC, XB, HL, RW
Drafted manuscript. KF, RW
Approved final manuscript. KF, AJ, JG, CV, KY, PP, EAC, XB, HL, RW
Supplementary Material
References
- 1. Gould SJ, Raposo G. As we wait: coping with an imperfect nomenclature for extracellular vesicles. J Extracell Vesicles. 2013;2:20389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Graner MW, Alzate O, Dechkovskaia AM, et al. Proteomic and immunologic analyses of brain tumor exosomes. FASEB J. 2009;23(5):1541–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kowal J, Tkach M, Théry C. Biogenesis and secretion of exosomes. Curr Opin Cell Biol. 2014;29:116–125. [DOI] [PubMed] [Google Scholar]
- 4. Kowal J, Arras G, Colombo M, et al. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc Natl Acad Sci U S A. 2016;113(8):E968–E977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yáñez-Mó M, Siljander PR, Andreu Z, et al. Biological properties of extracellular vesicles and their physiological functions. J Extracell Vesicles. 2015;4:27066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. D’Asti E, Chennakrishnaiah S, Lee TH, Rak J. Extracellular vesicles in brain tumor progression. Cell Mol Neurobiol. 2016;36(3):383–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pucci F, Garris C, Lai CP, et al. SCS macrophages suppress melanoma by restricting tumor-derived vesicle-B cell interactions. Science. 2016;352(6282):242–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ricklefs FL, Alayo Q, Krenzlin H, et al. Immune evasion mediated by PD-L1 on glioblastoma-derived extracellular vesicles. Sci Adv. 2018;4(3):eaar2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tkach M, Théry C. Communication by extracellular vesicles: where we are and where we need to go. Cell. 2016;164(6):1226–1232. [DOI] [PubMed] [Google Scholar]
- 10. Ai X, Hu M, Wang Z, et al. Recent advances of membrane-cloaked nanoplatforms for biomedical applications. Bioconjug Chem. 2018;29(4):838–851. [DOI] [PubMed] [Google Scholar]
- 11. Armstrong JP, Holme MN, Stevens MM. Re-engineering extracellular vesicles as smart nanoscale therapeutics. ACS Nano. 2017;11(1):69–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang X, Zhang H, Yang H, et al. Cell-derived exosomes as promising carriers for drug delivery and targeted therapy. Curr Cancer Drug Targets. 2018;18(4):347–354. [DOI] [PubMed] [Google Scholar]
- 13. Yang KS, Im H, Hong S, et al. Multiparametric plasma EV profiling facilitates diagnosis of pancreatic malignancy. Sci Transl Med. 2017;9(391):pii: eaal3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Meehan B, Rak J, Di Vizio D. Oncosomes—large and small: what are they, where they came from?J Extracell Vesicles. 2016;5:33109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Théry C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol. 2006;Chapter 3:Unit 3.22. [DOI] [PubMed] [Google Scholar]
- 16. van der Pol E, Sturk A, van Leeuwen T, Nieuwland R, Coumans F; ISTH-SSC-VB Working group Standardization of extracellular vesicle measurements by flow cytometry through vesicle diameter approximation. J Thromb Haemost. 2018;16(6):1236–1245. [DOI] [PubMed] [Google Scholar]
- 17. Akers JC, Gonda D, Kim R, Carter BS, Chen CC. Biogenesis of extracellular vesicles (EV): exosomes, microvesicles, retrovirus-like vesicles, and apoptotic bodies. J Neurooncol. 2013;113(1):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Muralidharan-Chari V, Clancy J, Plou C, et al. ARF6-regulated shedding of tumor cell-derived plasma membrane microvesicles. Curr Biol. 2009;19(22):1875–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Théry C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol. 2009;9(8):581–593. [DOI] [PubMed] [Google Scholar]
- 20. Théry C. Cancer: diagnosis by extracellular vesicles. Nature. 2015;523(7559):161–162. [DOI] [PubMed] [Google Scholar]
- 21. Shao H, Chung J, Balaj L, et al. Protein typing of circulating microvesicles allows real-time monitoring of glioblastoma therapy. Nat Med. 2012;18(12):1835–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shao H, Im H, Castro CM, Breakefield X, Weissleder R, Lee H. New technologies for analysis of extracellular vesicles. Chem Rev. 2018;118(4):1917–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Im H, Shao H, Park YI, et al. Label-free detection and molecular profiling of exosomes with a nano-plasmonic sensor. Nat Biotechnol. 2014;32(5):490–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Simpson RJ, Lim JW, Moritz RL, Mathivanan S. Exosomes: proteomic insights and diagnostic potential. Expert Rev Proteomics. 2009;6(3):267–283. [DOI] [PubMed] [Google Scholar]
- 25. Arakelyan A, Ivanova O, Vasilieva E, Grivel JC, Margolis L. Antigenic composition of single nano-sized extracellular blood vesicles. Nanomedicine. 2015;11(3):489–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kibria G, Ramos EK, Lee KE, et al. A rapid, automated surface protein profiling of single circulating exosomes in human blood. Sci Rep. 2016;6:36502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lee K, Fraser K, Ghaddar B, et al. Multiplexed profiling of single extracellular vesicles. ACS Nano. 2018;12(1):494–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Libregts SFWM, Arkesteijn GJA, Németh A, Nolte-‘t Hoen ENM, Wauben MHM. Flow cytometric analysis of extracellular vesicle subsets in plasma: impact of swarm by particles of non-interest. J Thromb Haemost. 2018;16(7):1423–1436. [DOI] [PubMed] [Google Scholar]
- 29. Smith ZJ, Lee C, Rojalin T, et al. Single exosome study reveals subpopulations distributed among cell lines with variability related to membrane content. J Extracell Vesicles. 2015;4:28533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lötvall J, Hill AF, Hochberg F, et al. Minimal experimental requirements for definition of extracellular vesicles and their functions: a position statement from the International Society for Extracellular Vesicles. J Extracell Vesicles. 2014;3:26913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Uhlen M, Bandrowski A, Carr S, et al. A proposal for validation of antibodies. Nat Methods. 2016;13(10):823–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wei Z, Batagov AO, Schinelli S, et al. Coding and noncoding landscape of extracellular RNA released by human glioma stem cells. Nat Commun. 2017;8(1):1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wakimoto H, Mohapatra G, Kanai R, et al. Maintenance of primary tumor phenotype and genotype in glioblastoma stem cells. Neuro Oncol. 2012;14(2):132–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Szatmári T, Lumniczky K, Désaknai S, et al. Detailed characterization of the mouse glioma 261 tumor model for experimental glioblastoma therapy. Cancer Sci. 2006;97(6):546–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chen X, Ma WY, Xu SC, et al. The overexpression of epithelial cell adhesion molecule (EpCAM) in glioma. J Neurooncol. 2014;119(1):39–47. [DOI] [PubMed] [Google Scholar]
- 36. Minciacchi VR, Freeman MR, Di Vizio D. Extracellular vesicles in cancer: exosomes, microvesicles and the emerging role of large oncosomes. Semin Cell Dev Biol. 2015;40:41–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Skog J, Würdinger T, van Rijn S, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10(12):1470–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Al-Nedawi K, Meehan B, Micallef J, et al. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat Cell Biol. 2008;10(5):619–624. [DOI] [PubMed] [Google Scholar]
- 39. Higginbotham JN, Zhang Q, Jeppesen DK, et al. Identification and characterization of EGF receptor in individual exosomes by fluorescence-activated vesicle sorting. J Extracell Vesicles. 2016;5:29254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Montermini L, Meehan B, Garnier D, et al. Inhibition of oncogenic epidermal growth factor receptor kinase triggers release of exosome-like extracellular vesicles and impacts their phosphoprotein and DNA content. J Biol Chem. 2015;290(40):24534–24546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nolan JP, Duggan E. Analysis of individual extracellular vesicles by flow cytometry. Methods Mol Biol. 2018;1678:79–92. [DOI] [PubMed] [Google Scholar]
- 42. Choi D, Montermini L, Kim DK, Meehan B, Roth FP, Rak J. The impact of oncogenic EGFRvIII on the proteome of extracellular vesicles released from glioblastoma cells. Mol Cell Proteomics. 2018;17(10):1948–1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.