Abstract
Interactions between mutations play a central role in shaping the fitness landscape, but a clear picture of intragenic epistasis has yet to emerge. To further reveal the prevalence and patterns of intragenic epistasis, we present a survey of epistatic interactions between sequential mutations in TEM-1 β-lactamase. We measured the fitness effect of ~12,000 pairs of consecutive amino acid substitutions and used our previous study of the fitness effects of single amino acid substitutions to calculate epistasis for over 8,000 mutation pairs. Since sequential mutations are prone to physically interact, we postulated that our study would be surveying specific epistasis instead of nonspecific epistasis. We found widespread negative epistasis, especially in beta-strands, and a high frequency of negative sign epistasis among individually beneficial mutations. Negative epistasis (52%) occurred 7.6 times as frequently as positive epistasis (6.8%). Buried residues experienced more negative epistasis that surface exposed residues. However, TEM-1 exhibited a couple of hotspots for positive epistasis, most notably L221/R222 at which many combinations of mutations positively interacted. This study is the first to systematically examine pairwise epistasis throughout an entire protein performing its native function in its native host.
Keywords: Fitness landscapes, epistasis, antibiotic resistance protein, protein evolution
Introduction
Understanding the fitness effects of mutations is fundamental to the study of molecular evolution. Mutations can have different effects depending on the genetic background in which they occur. For example, a mutation that is beneficial in one context may become deleterious in another, limiting mutational trajectories or yielding evolutionary dead-ends. This interaction between two or more mutations, called epistasis, plays a central role in evolution. Epistasis affects speciation [1, 2], the benefits of recombination and sex [3], genetic robustness [4, 5], and the predictability of evolution [6].
Genetic interactions can manifest in various ways. When two or more mutations interact such that their combined effect is more beneficial than predicted from their individual effects, it is termed positive epistasis. Alternatively, negative epistasis occurs when the combined effect is more deleterious than predicted. The magnitude of epistasis can have important consequences for the dynamics of evolution by affecting the curvature of the fitness landscape [7]. Sign epistasis occurs when a mutation’s effect changes from deleterious to beneficial by the presence of an additional mutation. The opposite is termed negative sign epistasis. A particular case of sign epistasis is reciprocal sign epistasis, in which two or more individual mutations are deleterious individually, but their combined effect is beneficial. This type of epistasis is particularly consequential in shaping the topography of the fitness landscape, causing local ruggedness and rendering certain peaks inaccessible [8].
Despite its theoretical importance in evolution, epistasis is understudied empirically and its contribution to evolution is not well understood. Empirical studies have aimed to elucidate aspects of epistasis in various ways. For example, examination of the fitness effects of multiple mutations from ancestral sequences of Hsp90 [9], and computational prediction epistatic effects from large fitness datasets for naturally-occurring mutations HIV-1 protease and reverse transcriptase [10] provide evidence that epistasis shapes natural protein evolution. As a complementary approach, the advent of deep sequencing technology has provided the ability to explicitly quantifying the functional or fitness effects of two or more mutations within a gene. Studies of intragenic epistasis have found it to be widespread [11–13] or rare [14, 15], mutational interactions to be typically strong [16] or weak [17], and sign epistasis to occur at a wide range of frequencies [13, 18]. The lack of consensus reflects the variety of molecules studied, differences in function or fitness measurements, modes of analysis, and fundamental limitations of multi-mutant studies. Recent studies of epistatic interactions in RNA molecules, which are attractive due to their typically shorter gene lengths and fewer possible combinations of mutations, reveal a predominance of negative epistasis [19]. While it is possible to characterize nearly all combinations of two point mutations in a small RNA molecule, capturing the full landscape of every pair of amino acid substitutions in an average size protein is currently beyond our limits. Intragenic epistasis studies of proteins necessarily compensate by looking at combinations of a small subset of mutations, focusing on a small region, or surveying a small fraction of the possible pairs.
Many studies have focused on combinations of a small set of mutations, or random mutations in the background of a few “anchor mutations”. For example, a study by Schenk et al. [18] looked exclusively at combinations of beneficial mutations, quantifying epistasis in sets of four single mutations that had a known “large effect” or “small effect” on improving antibiotic resistance. They found significant negative epistasis in both landscapes and pervasive negative sign epistasis, especially among large effect mutations. Parera and Martinez [16] tested epistasis by introducing a known deleterious amino acid substitution into various backgrounds of a protease and measuring catalytic efficiency compared to wildtype. Significant epistasis was observed in 50 of the 56 backgrounds tested. A study by Bank at el. [11] analyzed more than 1,000 double mutants comprised of 7 point mutation backgrounds of neutral to slightly deleterious effect and found common negative epistasis (46%) and rare positive epistasis. While these studies show important patterns in epistasis among a few known mutations, or among random mutations in the background of a few anchor mutations, they may be limited in their ability to capture larger epistatic trends. We previously reported epistatic landscapes along an evolutionary pathway [20] wherein ~12,500 single amino acid mutants were analyzed in the background of the mutations that make up an adaptive pathway from TEM-1 to TEM-15 β-lactamase, which contains the E104K and G238S mutations. The anchor mutation in each landscape (E104K or G238S) was found to be a determining factor in the patterns of epistasis observed. For instance, while epistasis with E104K was rare (8%), it was observed for 53% of mutant pairs with G238S. This suggests that the use of anchor mutations to capture general trends in epistasis may bias the conclusions.
Studies that looked at random pairs of mutations often focused their scope to a small domain within a protein. Often the domain has been excised from its native protein, necessitating the characterization of interactions affecting a biophysical property, such as binding, in a non-native context. For instance, Araya et al. calculated epistasis for ~5000 variants in a 34-amino acid WW binding domain using phage display [17]. They found epistasis to be rare, with values small in magnitude, and no population tendency toward positive or negative epistasis. In a 2014 study, Olson et al. quantified the effects of all double mutations between all positions in the IgG- binding domain of protein G (GB1), using in vitro mRNA display [14]. They reported notable instances of both positive and negative epistasis, as well as sign epistasis, but overall observed that epistasis was rare. Likewise, Melamed et al. [15] analyzed double mutants within a 90 amino acid RNA recognition motif in a poly(A)-binding protein and found that only 3.6% exhibited negative epistasis and 1.0% exhibited positive epistasis. They also found that pairs of mutations zero to five residues apart along the primary sequence exhibited a significantly higher frequency of both positive and negative epistasis than pairs further apart. Bank et al. [11] examined epistasis among all possible combinations of 13 amino acid mutations at 6 sites in the heat shock protein, Hsp90. They found a prevalent pattern of negative epistasis and ruggedness in their local landscape, concluding that predicting fitness landscapes from the effects of individual mutations is made exceedingly difficult by genetic interactions. These studies on small domains are instrumental in revealing local epistatic interactions involved in a particular biophysical property, but are limited in capturing epistatic effects involving the entire protein in its native, biological context [13].
Few studies have examined interactions between random pairs of mutations throughout an entire protein. A 2016 study of the fitness landscape of the green fluorescent protein defined fitness as the level of fluorescence in E. coli [21]. The authors sampled ~2% of all possible pairs of mutations, representing 30% of pairs of positions in the protein, and found that less than 5% exhibited epistasis. They observed pairs exhibiting epistasis to be located at sites across the gene, but slightly closer together than random. They found that pairs containing weak-effect mutations exhibited epistasis more often than pairs containing strong effect mutations, and suggest that the combined effect of weak mutations exhausts a stability threshold. Finally, they observed both strong and weak epistasis more prevalently among pairs of two buried sites, compared to pairs containing at least one solvent exposed site. Overall, they conclude that pairwise epistasis is more common at sites important to function.
Existing studies lack a survey of pairwise intragenic epistasis of a protein performing its native function in its native host in which the mutations are not limited to a particular domain or involve a small set of anchor mutations. Here, we examine pairwise epistasis throughout TEM-1 β-lactamase, a 286 amino acid antibiotic resistance protein native to E. coli. Informed by the observation that epistasis is more prevalent in pairs close together in primary sequence [15], we asked a specific question: how does epistasis present in pairs of consecutive amino acid substitutions throughout the protein? Previously, we quantified the fitness effect of nearly all (95.6%) possible single amino acid substitutions in TEM-1 [22]. We use this data set to compare individual effects of mutations to the fitness effects of over 8,000 sequential double mutants. We mapped the resulting epistasis values on sequence and structure and present overall trends in epistasis between sequential mutations in TEM-1.
Results
Fitness landscape of sequential double mutants
TEM-1 is a convenient model for the study of gene/protein evolution, as it confers an easily identifiable and quantifiable phenotype – resistance to penicillin antibiotics, such as ampicillin (Amp). Although growth competition experiments in the presence of Amp can be used to measure enrichment of various alleles as a proxy for fitness, the values obtain depend on the concentration of Amp used [23]. In addition, the relative growth rate of cells with different alleles will change over time as the Amp in the culture is degraded, so the fitness values obtained are not precise relative growth rate comparisons. In addition, growth competition experiments have low resolution of low fitness alleles. As an alternative, minimum inhibitory concentration (MIC) assays can be used as a proxy for fitness, quantifying the ability of the allele to confer resistance to the antibiotic [24, 25], but MIC assays are not high throughput. Here, we use our previously described synthetic biology approach to quantify Amp resistance in a MIC-like fashion as a proxy for fitness [22, 26]. This method overcomes the limitations of growth competition experiments and standard MIC assays, as the fitness measures are ampicillin concentration independent and low fitness values are as precisely measured as high fitness values. Our fitness values measure the level of ampicillin resistance conferred by the gene and are predictive of fitness values measured by growth competition experiments in the presence of a range of ampicillin concentrations (Supplementary Fig. S1).
We created a library of ~30,000 sequential double mutants in TEM-1 using inverse PCR using abutting, degenerate primers in which the 5’-end of one primer had the sequence (NNN)2 [27]. We created separate libraries for each third of the gene to be compatible with the read length of the Illumina MiSeq 2×300 deep sequencing platform. Due to the nature of the genetic code, a consequence of using NNN to encode the mutations is that our libraries were biased towards certain amino acid substitutions (e.g. mutation to serine will occur six times as often as mutation to tryptophan) (Supplementary Fig. S2). However, mutations occurring naturally are biased in a similar way based on the genetic code (i.e. mutation to serine occurs more frequently than mutation to tryptophan). In addition, our library is not biased towards those amino acid substitutions that would most likely occur in TEM-1, which would be those occurring by single base substitution in the TEM-1 codons. We focused on studying the epistasis between sequential amino acid substitutions in the TEM-1 protein, not on epistatic interactions that would likely occur by single base substitutions in the TEM-1 gene.
We plated transformed SN0301 E. coli cells with the double mutant library on plates containing tetracycline and 13 different Amp concentrations ranging from 0.25 μg/ml to 1024 μg/ml. Whereas Amp prevents growth if the Amp concentration is too high relative to the amount of Amp resistance conferred, tetracycline prevents growth if the concentration of Amp is too low relative to the amount of Amp resistance conferred. As a result, a particular allele will confer growth only in a narrow range of Amp concentrations – a behavior that results from the band-pass synthetic gene circuit in SNO301 cells (see Firnberg et al. [22] for a detailed explanation). We recovered the resulting sublibraries from the plates, PCR-amplified the appropriate third of the gene with Illumina MiSeq compatible barcodes, and deep sequenced the amplicons to determine how often each allele appeared on each plate.
Sequencing reads of alleles containing synonymous codons were grouped together to gain statistical power. This comes at the expense of examining potential differences in fitness between alleles with synonymous codons. This is most relevant at the beginning of the gene, where synonymous mutations can cause differences in translations rates due to differences in RNA structure[22, 28, 29]. Although we have evidence that synonymous mutations in the first ten codons of TEM-1 can have fitness effects [22], these effects are the exception and not the rule. In addition, fitness values for individual codon substitutions in TEM-1 (as opposed to combining data for synonymous codons for each amino acid substitution) usually have a high uncertainty due to a low number of counts in Firnberg et al., an uncertainty that would make detection of epistatic effects difficult. A result of our grouping of synonymous codons is the potential for a slight overrepresentation of the extent of epistasis, particularly at the beginning of the signal sequence. Supplementary Data S1 tabulates all sequencing counts. The reported fitness (w) is the calculated Amp concentration at which the mutant allele appeared most frequently relative to the same value calculated for wildtype allele (tabulated in Supplementary Data S2). We calculated fitness values only for double amino acid mutants with 20 or more sequencing counts to focus on those fitness measurements with lower uncertainty (see Materials and Methods for a more detailed explanation).
We next applied an adjustment to these fitness measurements to account for potential experimental differences between the two sets of fitness measurements. Our epistasis calculations rely on consistent fitness measurements between our previous fitness measurements of single mutants [22] and the measurements of double mutants presented here. Thus, we took measures to ensure that the fitness values were consistent between the two experiments. We hypothesized that small differences in plating, incubation temperature, or other experimental factors may affect a cell’s propensity to form a colony on each plate, perhaps resulting in a slight shift higher or lower in the Amp concentrations that favor growth. Such phenomena would result in systematic shifts in fitness values between the two experiments, which could be different for different ranges of fitness values.
To examine this possibility, we compared single mutant fitness values measured in each experiment. Our double mutant library creation technique also produced alleles containing an amino acid substitution next to a synonymous mutation. We assumed that all observed synonymous mutations were neutral, consistent with our previous observations that the vast majority of synonymous mutations in TEM-1 are neutral [22]. We compared the fitness values for the 1,470 such alleles in our experiment with the corresponding single mutant fitness values from Firnberg et al. We observed small offsets in fitness values that were different for different fitness value ranges. For example, fitness values less than ~0.125 were ~30% higher in the double mutant data set than the single mutant data set, whereas fitness values nearer to the wildtype value had a much smaller offset. Based on this observation, we adjusted the double mutant fitness measurements set to account for these differences. These adjustments are provided as Supplementary Data S4. We judge this cross-experiment normalization procedure to be the most justifiable way to compare the two sets of data. However, we also analyzed the data without the fitness value adjustments, and the overall trends presented in this study remained the same.
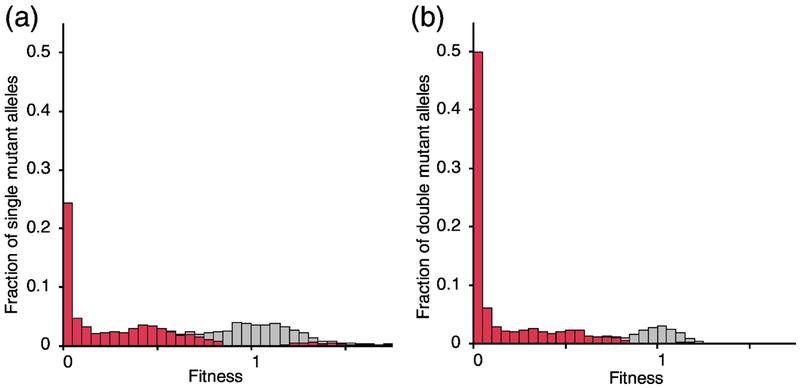
We obtained fitness values for 12,374 alleles of unique double mutant pairs, with an average of 30 pairs per position. This number represents 12.0% (12,374/102,855) of all possible consecutive double mutants. The distribution of fitness values of the double-mutants shows a shift toward lower fitness values (Fig. 1b), compared to the distribution of fitness values of the single mutants (Fig. 1a). Only 89 double mutations resulted in fitness values significantly higher than wildtype. Nearly half (49.9%) of double mutations resulted in a near-complete loss of function (W<0.05). This shift toward low fitness is expected and in agreement with other mutation accumulation studies [15, 21], including one on TEM-1 [5].
Fig. 1. Distribution of mutational fitness effects of single and double mutants IN TEM-1.
(a) Distribution of 5460 single mutant fitness values from Firnberg et al. [22]. (b) Distribution of 12,374 sequential double mutant fitness values. The single mutant distribution has a very small number of fitness values >1.8 that are not shown. Bars are stacked to show total fractions. Fitness values are normalized to that of wildtype TEM-1 beta-lactamase. Fitness values that are significantly different from 1.0 are indicated in red. Supplementary Data S2 tabulates all fitness values of double mutants.
Epistatic landscape of sequential double mutants
We define pairwise epistasis as occurring when the product of the fitness values of two individual mutations differs from the fitness of the combined pair. Epistasis (ε) between mutation A with fitness WA and mutation B with fitness WB is calculated as:
| (1) |
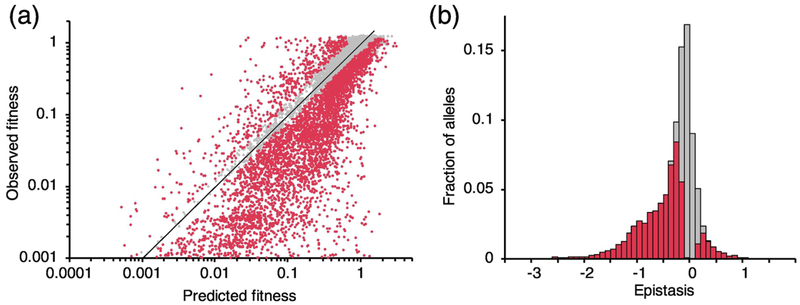
where Wo is the fitness of wildtype TEM-1 and WAB is the fitness of the double mutant. We calculated epistasis values for 8.1% (8,302/102,885) of all possible pairs of sequential amino acid substitutions. For our epistasis analysis, we exclude pairs containing mutations with individual fitness values less than 0.02 to avoid the lower limit in fitness measurements causing high epistasis values by artifact. Epistasis measurements were determined to be significantly different than zero if they were greater or less than 0 by twice the error estimate in the epistasis measurement (see Methods). Over half (58%) of all double mutants analyzed exhibited significant epistasis (Fig. 2b). The high prevalence of epistasis compared to most other studies is consistent with the previous observation that epistasis is more common in sequential mutations than in non-sequential mutations [15], although we do not have a corresponding measure of the frequency of epistasis among random mutations in TEM-1 to which we can directly compare. It may also reflect differences in the prevalence of epistasis with regard to fitness (here the ability of the allele to confer Amp resistance to live cells), compared to epistasis with regard to a less complex biophysical property, as hypothesized by Sackman and Rokyta [13]. The distribution of epistasis values was skewed toward negative values, with a mean epistasis of −0.32 and a median of −0.18, indicating that the combined fitness effect of two mutations is often more deleterious than predicted in the absence of epistasis. Negative epistasis (51%) occurred 7.5 times as frequently as positive epistasis (6.8%). This pervasive negative epistasis is consistent with a TEM-1 mutation accumulation study that concluded its fitness landscape is characterized by negative epistasis [5].
Fig. 2. Distribution of epistasis values among sequential mutations in TEM-1.
(a) Observed fitness versus predicted fitness for 8,302 double mutant alleles. Predicted fitness assumes no epistasis. (b) The distribution of epistasis values among 8,302 double mutant alleles. Bars are stacked to show total fractions. Significant epistasis values are indicated in red. Supplementary Data S2 tabulates all epistasis values.
A comparison of observed fitness values (WAB) to predicted fitness in the absence of epistasis (WAWB) clearly shows the prevalence of negative epistasis (Fig. 2a). We found that the product of single mutant fitness values (i.e. the predicted fitness in the absence of epistasis) predicted double mutant fitness values with a Pearson’s R2 of 0.71. This is within the range of the correlations found in other epistasis studies, which had R2 values ranging from 0.67 [17] to 0.76 [15].
Relationship between epistasis and protein sequence/structure
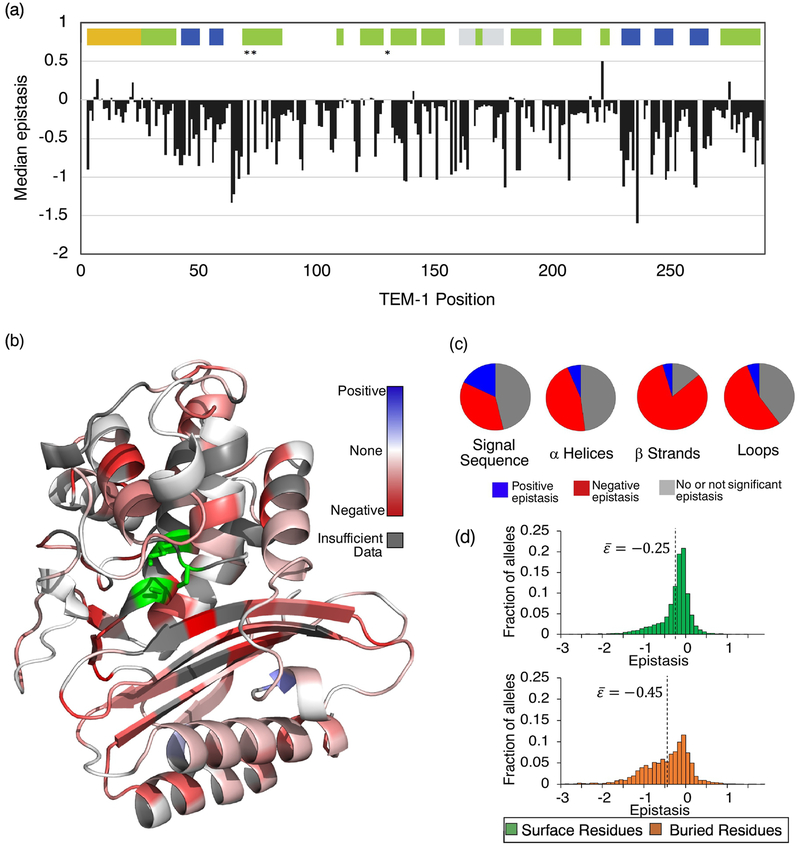
Examining epistasis among sequential double mutant pairs allowed us to map median epistasis at each position and look at trends within secondary structures (Fig. 3). Although negative epistasis dominates, there were 19 pairs of positions with positive median epistasis values, indicating hot spots for synergistic potential (Fig. 3a). Interestingly, we note a particularly high median epistasis at positional pair 221–222. This median was calculated from a total of 21 observations. With the exception of one pair, the double mutants at this position were combinations of deleterious single mutations (median fitness of 0.052). Residues L221 and R222 make up the first two amino acids of a four-residue helical element (helix 10). Positive epistasis, indicating a higher than expected fitness between individually deleterious mutations at this positional pair suggests hotspot for compensatory interactions, possibly buffering structural disruptions in the helix.
Fig. 3. The relationship between protein sequence, structure, and epistasis in TEM-1.
(a) Median epistasis values across the TEM-1 primary sequence. Median values were calculated only for position pairs with 5 or more epistasis values. Median epistasis for a mutation pair is plotted at the first position of that pair. Colored bars indicate regions that code for the signal sequence (yellow), alpha helices (green), beta strands (blue), and the omega loop (grey). Asterisks indicate the location of important catalytic residues. (b) Median epistasis values mapped onto the TEM-1 structure. Active site residues are indicated in green. (c) Frequency of positive epistasis (blue), negative epistasis (red), and no or not significant epistasis (grey) in the signal sequence and secondary structure elements. Data are categorized by the structural identity of the first mutation. Distribution of fitness values for these categories is provided as Supplementary Fig. S3. (d) Epistasis distributions for buried and surface residues. The median value of the distribution is indicated.
Positive epistasis occurred three times more frequently in the signal sequence (17.8%) than in the mature protein (5.82%) (P<0.0001, Fisher’s exact test) (Fig. 3c), although our reported frequency of both positive and negative epistasis in the signal sequence may be slightly inflated due to the potential for fitness effects of synonymous mutations in the first 10 codons (as discussed above). The signal sequence is a 23 amino acid peptide that directs export of the protein to the periplasmic space of E. coli. The signal sequence is removed in the periplasm and is not part of the mature protein. However, mutations within the signal sequence can change protein abundance and therefore affect fitness. Over half (52%) of the occurrences of positive epistasis in the signal sequence were between one beneficial and one deleterious mutation, with the remaining 48% being between mutations that are deleterious individually. Positive epistasis in this region suggests detrimental mutations are easily partially compensated by mutations at adjoining positions.
In the mature protein, negative epistasis occurred most often in beta-strands (Fig. 3c), indicating that two sequential mutations within these structures is often more detrimental than the combination of their individual effects. A majority (68%) of mutations occurring in beta-strands were individually deleterious. The side chains of sequential mutations are not expected to physically interact because sequential amino acids point in different directions in a beta strand. Rather, deleterious mutations probably cause backbone shifts that affect the fitness effects of sequential mutations in non-additive ways – an interaction through the backbone. As deleterious mutations in beta-strands likely result from packing problems and decreases in stability, these findings suggest that the threshold robustness to additional deleterious mutations [5] is more often exhausted in beta-strands, presumably because the complexity of the structure has more constraints on the amino acids at each position.
We also examined epistasis among surface residues versus buried residues. We define surface residues as those with >20% solvent accessibility, and buried residues as those with <20% solvent accessibility. On average, buried residue pairs exhibited lower epistasis values than surface residue pairs (P <0.0001, by Student’s t-test), suggesting that multiple mutations at internally oriented residues are more likely to interact antagonistically (Fig. 3d). Epistasis values for buried residues also had a broader distribution of values than epistasis values for solvent accessible residues (P<0.0001 by Brown–Forsythe test). We observed no obvious pattern in epistasis between different pairs of amino acids; however, we note that the lowest two median epistasis values occurred between pairs of two cysteines and pairs of two aspartic acids (Supplementary Fig. S4). We found no correlation between epistasis and the distance from the active site (Supplementary Fig. S5).
The influence of fitness effect sign and size on epistasis.
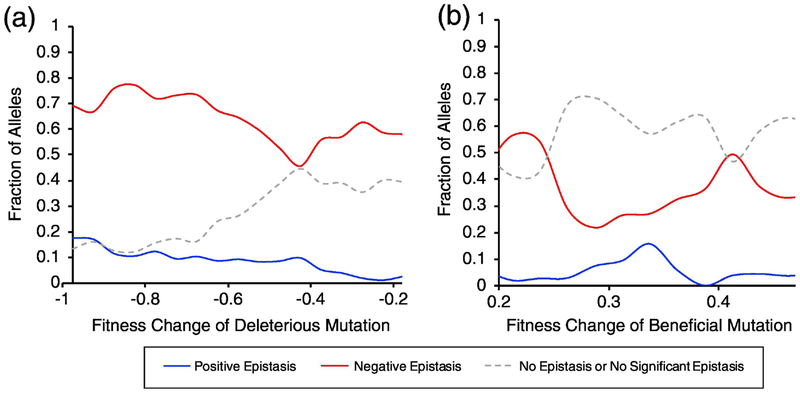
Previous studies have noted differences in epistasis among individually beneficial versus deleterious mutations [11, 18]. Additionally, Pumir et al. [30] posited that the effect size of the mutation may influence its epistatic effect in the context of another mutation. To probe this further, we examined epistasis versus the effect size of the individual mutations contained in the pair. We define a mutation as deleterious if its fitness is more than two times its error below wildtype fitness and beneficial if its fitness is more than two times its error above wildtype fitness. In general, we observed epistasis more frequently in pairs containing at least one deleterious mutation than in pairs containing at least one beneficial mutation (Fig. 4). Epistasis was especially prevalent among large effect deleterious mutations (W<0.1), with nearly 90% of all pairs containing a large effect deleterious mutation exhibiting either positive or negative epistasis (Fig. 4a). In particular, pairs containing large effect deleterious mutations have a higher frequency of positive epistasis than pairs containing small effect deleterious mutations, suggesting that the fitness cost of highly deleterious mutations can be somewhat dampened by the presence of an additional mutation. Our inability to observe any meaningful trends in epistasis for pairs containing at least one beneficial mutation (Fig. 4b) may be due to the small number of mutations with statistically significant beneficial effects (Fig. 1a).
Fig. 4. The effect of size and nature of the mutational effect on the frequency of positive and negative epistasis.
Frequency of epistasis among mutation pairs with (a) at least one deleterious mutation and (b) at least one beneficial mutation. The deleterious or beneficial mutation must have a statistically significant effect on fitness, but the other mutation in the pair may be deleterious, beneficial, or neutral. Boxcar smoothing was applied to the data to improve visualization of trends.
Sign epistasis
We also examined sign epistasis for 11,679 double mutant alleles for which we had corresponding single mutant fitness values. Sign epistasis is solely determined by the sign of fitness measurements (beneficial or deleterious). Unlike magnitude epistasis, it is not calculated from the product or ratio of two fitness values. Therefore, we included pairs containing single mutants with W<0.02 in the analysis of sign epistasis. By definition, positive sign epistasis can occur only for pairs containing at least one deleterious mutation and negative sign epistasis can occur only for pairs containing at least one beneficial mutation. We observe positive sign epistasis in only 13 out of 9673 pairs containing a deleterious mutation. The low frequency of positive sign epistasis indicates a scarcity of paths to climb above wildtype fitness in a single step from a deleterious mutation. Negative sign epistasis is much more prevalent, occurring in 55.4% of pairs containing a beneficial mutation. This indicates a moderately rugged landscape for sequential double mutants that is dominated by fitness valleys. We examined the relationship between negative sign epistasis and individual mutation effect size, but found the frequency to be >50% across all effect sizes. Thus, for beneficial mutations, the magnitude of the fitness effect does not predict the likelihood of surrounding fitness valleys. We found no cases of reciprocal sign epistasis, suggesting that many peaks may be accessible on the TEM-1 fitness landscape through accumulation of one mutation at a time.
Discussion
The picture of epistasis in protein evolution is still emerging. Our study examines pairwise intragenic epistasis in TEM-1 beta-lactamase in the context of it performing its native function (antibiotic resistance) in its native host (E. coli). Although TEM-1 is native to E. coli, it differs from most E. coli genes because it is found on plasmids (instead of the chromosome), which can be transferred among different bacteria. Whether this difference would impact the extent of epistasis is unknown. We contend that fitness and epistasis measurements are best performed in their native host when the goal is to understand the landscapes that shape the natural evolution of proteins. Studies that extract proteins from their native environment will miss native fitness and epistatic effects arising from the interaction of the protein and the cell and may be colored by non-native fitness and epistatic effects arising from non-native interactions. For example, mutations may promote misfolding [31] or misinteractions [32] that have deleterious effects on cell growth (i.e. fitness), and such effects will be environment dependent.
We specifically examined pairwise epistasis between sequential amino acid substitutions across the entire length of the primary sequence. Our intent was to study the inherent susceptibility of the TEM-1 protein to epistatic interactions between sequential amino acid substitutions, not to study epistatic interactions between the mutations that are most likely to occur in the TEM-1 gene (i.e. those achieved with a single bp substitution). The results of our study should be viewed with these limitations in mind. We postulated that consecutive double mutants represent a subset of possible mutational pairs that are more likely to exhibit epistatic effects due to spatial proximity and direct physical link in the backbone. As such, these epistatic effects are likely examples of specific epistasis as opposed to nonspecific epistasis.
Epistatic interactions can be classified as specific or nonspecific [33]. Specific epistasis results from direct physical interactions, and as such, these mutations result in nonadditivity on the level of biophysical properties, such as stability, activity, or binding. To the extent that these properties determine organismal fitness, the nonadditivity of biophysical properties explains epistasis on the fitness level. In contrast, nonspecific epistasis results from a nonlinear dependence of fitness on the biophysical properties themselves. With nonspecific epistasis, mutations may act additively on the level of the protein, but epistasis exists on the level of organismal fitness. Mutations that exhibit nonspecific epistasis often do so with a relatively large number of mutations. For example, the M182T mutation in TEM-1 stabilizes the folded state and is a global suppressor mutation [34]. The presence of M182T reduces the deleterious fitness effect of many mutations throughout the entire protein [24] – mutations that are presumably destabilizing. This nonspecific, positive epistasis manifests not from nonadditive effects of the two mutations on stability, but from the nonlinear mapping of stability to the probability of the folded state [5] and thus the cellular abundance of the protein (a biological property that effects fitness). This results in proteins having a stability robustness threshold [5]. Nonspecific epistasis may represent a significant fraction of all intragenic epistatic effects, as Dasmeh et al. estimate that 30–40% of epistasis can be attributed to protein folding stability [35]. A protein’s interaction with the cells protein quality control machinery (chaperones and proteases), and a mutation’s effect on those interactions will also shape fitness/epistatic effects and the stability threshold [36].
The relative contributions of specific and nonspecific epistasis to protein evolution is an important open question [37]. One challenge in addressing this question is the difficulty in attributing a measured epistatic effect as specific or nonspecific in a high-throughput manner. By studying sequential mutations – mutations that are highly likely to interact due to their proximity – we postulate that we are predominantly measuring specific epistasis. Here, we take a broad definition of “physically interact” to include mutations that interact through movement of the peptide backbone (as might well occur in sequential mutations). For instance, when a position is mutated in the interior of the protein to a larger amino acid, the protein structure must compensate. One way it may do this is through adjusting the relative position of the peptide backbone that includes the mutated amino acid. This adjustment would be prone to affect, in nonadditive ways, the fitness effects of mutations at adjacent positions. In this manner, we believe that many of the epistatic effects in this study are likely to be specific. We contrast this study with our previous study on epistasis in TEM-1 involving the G238S mutation [20]. The G238S mutation exhibited negative epistasis with 58% of other mutations throughout TEM-1 and decreases stability about 2 kcal/mol [38]. Most epistasis with G238S is likely nonspecific epistasis and manifests from G238S’s deleterious effect on stability. However, negative epistasis involving G238S and some other mutations may well be specific in nature. We note that a protein’s stability threshold can be exhausted by specific negative epistasis and nonspecific negative epistasis.
We find widespread negative epistasis between sequential mutations in TEM-1 evaluated in its native environment, though hotspots for positive epistasis existed. This high frequency of epistasis contrasts with the typically low frequency of epistasis found in studies that focus on epistasis on the level of biophysical properties measured in non-native environments [14–17, 21]. Our study does not address the reasons for the higher frequency of epistasis. The higher frequency is likely some combination of measuring fitness on the level of the cell (instead of the protein level) [39][33, 40], measuring fitness of the protein in its native environment, and measuring fitness only for sequential mutations. We can say that measuring fitness effects of mutations in a protein in its native environment and at the level of the cell should better provide fitness and epistatic landscapes that reflect those that constrain and shape protein evolution. Our study lends support to the emerging picture of pervasive negative epistasis among mutations studied in their native context, the threshold robustness hypothesis, and the relationship between solvent accessibility and epistasis. Our findings lend support to the hypothesis that epistasis may be pervasive with regard to biological fitness despite underlying additive mutational effects on biophysical properties such as stability.
Materials and Methods
Library Creation
The TEM-1 gene was expressed on pSkunk3, a 4.36 kb plasmid containing spectinomycin resistance and the p15 origin of replication, under the IPTG-inducible tac promotor in E. coli. We used inverse PCR with primers (IDT) designed to create every possible sequential double mutant in TEM-1, using NNN-NNN degenerate nucleotide oligos and a compatible reverse primer designed for each position. PCR products were visualized using gel electrophoresis, to confirm the creation of a linearized plasmid product at each of the 286 positions. We pooled the PCR products, isolated the ~4 kb band from an agarose electrophoresis gel, phosphorylated the DNA at 37°C (NEB T4 PNK), and ligated it overnight at 16°C. NEB 5-alpha F’ lacIq E. coli were transformed with the ligation product and plated on LB-agar plates containing 50 μg/ml spectinomycin and 2% glucose (w/v). At least 500,000 transformants were obtained for each third.
We recovered each library from the plate in LB media and isolated the plasmid library. We transformed electrocompetent SNO301 E. coli cells with each library and plated on LB-agar plates containing 50 μg/ml spectinomycin, 50 μg/ml chloramphenicol, and 2% glucose. At least 80,000 transformants were obtained from each third. We recovered each library from the plate in LB media and made glycerol stocks. The library sizes were greater than the number of sequences we could analyze by deep sequencing. Thus, we prepared a smaller sublibraries of each library by plating ~10,000 CFU from each library on LB-agar plates with 50 μg/ml spectinomycin, 50 μg/ml chloramphenicol, and 2% glucose (i.e. permissive growth conditions), recovering those cells, and creating final frozen sublibrary stocks for selection.
Selection and Sequencing
High-throughput selection for resistance to ampicillin (Amp) was performed using a band-pass genetic circuit, described previously [22]. Briefly, E. coli SNO301 cells containing the double mutant library were plated on LB-agar plates containing 20 μg/ml tetracycline and 13 different Amp concentrations, ranging from 0.25 μg/ml to 1024 μg/ml, in 2-fold increments. Plates were incubated for 21 hours at 37°C. The library was plated in triplicate on each Amp concentration and the CFUs from each plate were counted to determine the frequency of colonies appearing on each plate. Based on these counts, a proportional amount of barcoded PCR amplicon from each plate was deep sequenced. Amplicons were prepared by recovering the cells from each selection plate, isolating the plasmid DNA, and performing PCR with appropriate primers as described previously [20, 22]. Barcodes to identify each plate and adapters compatible with Illumina MiSeq platform were added in this PCR step. Amplicons were pooled and sequenced using Illumina MiSeq with 300 base pair, paired-end reads.
Data Analysis
The de-multiplexed MiSeq reads were analyzed using custom MATLAB scripts. Paired-end reads were trimmed and concatenated to yield full length reads. Each read was then aligned to TEM-1 using a Smith-Waterman algorithm with a gap opening penalty of 100. Reads with an alignment score lower than 300 were filtered out and only reads containing two sequential codon substitutions were used for analysis. Fitness was calculated for each unique double amino acid mutant based on the counts from each plate (Amp concentration). Synonymous codons were grouped together and total counts were used to calculate the single amino acid fitness. First, counts were adjusted based on the number of sequencing reads obtained from each plate relative to the CFUs observed on that plate, as described previously [20]. Detailed description of the fitness calculation can be found in our previous studies [20, 22], which we followed with a few minor differences. In this study, we excluded alleles with fewer than 20 counts and alleles with a maximum single plate count less than 1/3 the total count. We exclude alleles with fewer than 20 counts in order to focus on fitness measurements that had smaller uncertainty. We excluded alleles with a maximum single plate count less than 1/3 the total count to eliminate alleles for which the count distribution made the correct fitness ambiguous. For example, a small number of alleles had two clusters of counts (we hypothesize that this arises from some plasmids with the indicated mutation having an additional, spontaneous mutation outside the sequencing range) and a small number of alleles had a low level of counts on many plates without a clear cluster of counts (we hypothesize that this arises when an allele is absent or present at low frequency in the library and the position is prone to sequencing errors). For each allele (i) that passed these criteria, the plate with the highest adjusted counts and the four plates on either side (i.e. two plates with higher Amp and two plates with lower Amp) were used to calculate an unnormalized fitness value, representing the midpoint resistance to Amp:
| (2) |
where ci,p is the adjusted count of allele i on plate p, and ap is the Amp concentration on plate p (in μg/ml). The reported fitness values are normalized to wildtype TEM-1:
| (3) |
Wildtype fitness was calculated in the same way (i.e. using adjusted sequencing counts) and verified separately by separately plating cells expressing wildtype TEM-1 in triplicate during the bandpass selection step. Both colony counts of the wildtype plates and wildtype sequencing counts revealed a midpoint Amp resistance of ~185 μg/ml (186.1μg/ml, 184.8 μg/ml, and 182.3μg/ml for each of the thirds, and 187.4 μg/ml for the colony counts). Wildtype sequencing counts and colony counts are provided in Supplementary Data S3.
We adjusted the fitness measurements based on a comparison between fitness values for 1,470 single amino acid substitutions containing a synonymous wild type mutation and the corresponding single amino acid fitness values from Firnberg et al. [22]. We calculated a ratio of the two fitness values across different fitness value ranges. Based on the offset of this value from 1, we determined adjustment factors for each range of fitness values, which ranged from 0.52 to 0.97. We multiplied the calculated double mutant fitness values by these adjustment factors and used these cross-experiment normalized fitness values for all subsequent analysis, which is presented in this study. We also analyzed the data without the fitness value adjustments, and the overall trends presented in this study remained the same.
Error in fitness was estimated via Eqs 4 and 5, using our previously determined correlation between sequencing counts (ni) and the standard deviation of the difference in fitness between synonymous alleles [20, 22].
| (4) |
where ei, the upper-level estimate of the fraction error in fitness, is given by:
| (5) |
Fitness values were determined to be significantly different than 1 if they were greater or less than 1 by twice the error estimate.
Epistasis was calculated using Eq. 1. To determine epistasis values that were significantly different than 0, upper and lower limits were calculated using Eqs 6 and 7:
| (6) |
| (7) |
Epistasis values were determined to be significantly positive or significantly negative based on Eq 8 and 9, respectively:
| (8) |
| (9) |
Sign epistasis was determined based on fitness measurements of the individual mutations and double mutant pair. Positive sign epistasis was defined as occurring when at least one of the mutants was individually deleterious (less than twice the error below 1), and the double mutant was beneficial (greater than twice the error above 1). Likewise, negative sign epistasis was defined as occurring when at least one of the mutants was individually beneficial, and the double mutant was deleterious. Reciprocal sign epistasis required both mutants to be individually deleterious, while the double mutant was beneficial. Negative reciprocal sign epistasis was the inverse.
Supplementary Material
Highlights.
Epistasis plays a key role in shaping protein fitness landscapes.
Quantified effect of 12,000 consecutive mutants in TEM-1 β-lactamase.
Provided a structural map of intragenic epistasis involving consecutive mutations.
Widespread negative epistasis especially in beta-strands and buried residues.
First systematic epistasis survey throughout a protein in native context.
Study contrasts epistasis on biophysical properties level vs. biological context.
Acknowledgements
This research was supported by the National Science Foundation (DEB-1353143, CBET-1402101, and MCB-1817646 to M.O.) and by the National Institutes of Health under a Ruth L. Kirschstein National Research Service Award (F31GM101941) to C.E.G.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Gavrilets S, Fitness landscapes and the origin of species. Princeton University Press, Princeton, N.J. (2004). [Google Scholar]
- [2].Dettman JR, Sirjusingh C, Kohn LM, Anderson JB, Incipient speciation by divergent adaptation and antagonistic epistasis in yeast, Nature 447 (2007) 585–588. [DOI] [PubMed] [Google Scholar]
- [3].de Visser JAGM, Elena SF, The evolution of sex: Empirical insights into the roles of epistasis and drift, Nature Reviews Genetics 8 (2007) 139. [DOI] [PubMed] [Google Scholar]
- [4].Wagner A, Robustness and evolvability in living systems. Princeton University Press, (2005). [Google Scholar]
- [5].Bershtein S, Segal M, Bekerman R, Tokuriki N, Tawfik DS, Robustness-epistasis link shapes the fitness landscape of a randomly drifting protein, Nature 444 (2006) 929–932. [DOI] [PubMed] [Google Scholar]
- [6].de Visser JAGM, Krug J, Empirical fitness landscapes and the predictability of evolution, Nature Reviews Genetics 15 (2014) 480. [DOI] [PubMed] [Google Scholar]
- [7].Ivan GS, Martijn FS, Jasper F, Joachim K, d. Visser JAGM, Quantitative analyses of empirical fitness landscapes, Journal of Statistical Mechanics: Theory and Experiment 2013 (2013) P01005. [Google Scholar]
- [8].Weinreich DM, Watson RA, Chao L, Perspective: Sign epistasis and genetic constraint on evolutionary trajectories, Evolution 59 (2005) 1165–1174. [PubMed] [Google Scholar]
- [9].Starr TN, Flynn JM, Mishra P, Bolon DNA, Thornton JW, Pervasive contingency and entrenchment in a billion years of hsp90 evolution, Proceedings of the National Academy of Sciences of the United States of America 115 (2018) 4453–4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Hinkley T, Martins J, Chappey C, Haddad M, Stawiski E, Whitcomb JM, Petropoulos CJ, Bonhoeffer S, A systems analysis of mutational effects in hiv-1 protease and reverse transcriptase, Nature Genetics 43 (2011) 487. [DOI] [PubMed] [Google Scholar]
- [11].Bank C, Hietpas RT, Jensen JD, Bolon DNA, A systematic survey of an intragenic epistatic landscape, Molecular Biology and Evolution 32 (2015) 229–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Bank C, Matuszewski S, Hietpas RT, Jensen JD, On the (un)predictability of a large intragenic fitness landscape, Proceedings of the National Academy of Sciences 113 (2016) 14085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Sackman AM, Rokyta DR, Additive phenotypes underlie epistasis of fitness effects, Genetics 208 (2018) 339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Olson CA, Wu NC, Sun R, A comprehensive biophysical description of pairwise epistasis throughout an entire protein domain, Current biology : CB 24 (2014) 2643–2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Melamed D, Young DL, Gamble CE, Miller CR, Fields S, Deep mutational scanning of an rrm domain of the saccharomyces cerevisiae poly(a)-binding protein, RNA 19 (2013) 1537–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Parera M, Martinez MA, Strong epistatic interactions within a single protein, Molecular Biology and Evolution 31 (2014) 1546–1553. [DOI] [PubMed] [Google Scholar]
- [17].Araya CL, Fowler DM, Chen W, Muniez I, Kelly JW, Fields S, A fundamental protein property, thermodynamic stability, revealed solely from large-scale measurements of protein function, Proceedings of the National Academy of Sciences of the United States of America 109 (2012) 16858–16863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Schenk MF, Szendro IG, Salverda ML, Krug J, de Visser JA, Patterns of epistasis between beneficial mutations in an antibiotic resistance gene, Mol Biol Evol 30 (2013) 1779–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Bendixsen DP, Ostman B, Hayden EJ, Negative epistasis in experimental rna fitness landscapes, J Mol Evol 85 (2017) 159–168. [DOI] [PubMed] [Google Scholar]
- [20].Steinberg B, Ostermeier M, Shifting fitness and epistatic landscapes reflect tradeoffs along an evolutionary pathway, J Mol Biol 428 (2016) 2730–2743. [DOI] [PubMed] [Google Scholar]
- [21].Sarkisyan KS, Bolotin DA, Meer MV, Usmanova DR, Mishin AS, Sharonov GV, Ivankov DN, Bozhanova NG, Baranov MS, Soylemez O, Bogatyreva NS, Vlasov PK, Egorov ES, Logacheva MD, Kondrashov AS, Chudakov DM, Putintseva EV, Mamedov IZ, Tawfik DS, Lukyanov KA, Kondrashov FA, Local fitness landscape of the green fluorescent protein, Nature 533 (2016) 397–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Firnberg E, Labonte JW, Gray JJ, Ostermeier M, A comprehensive, high-resolution map of a gene’s fitness landscape, Mol Biol Evol 31 (2014) 1581–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Stiffler Michael A., Hekstra Doeke R., Ranganathan R, Evolvability as a function of purifying selection in tem-1 β-lactamase, Cell 160 (2015) 882–892. [DOI] [PubMed] [Google Scholar]
- [24].Jacquier H, Birgy A, Le Nagard H, Mechulam Y, Schmitt E, Glodt J, Bercot B, Petit E, Poulain J, Barnaud G, Gros P-A, Tenaillon O, Capturing the mutational landscape of the beta-lactamase tem-1, Proceedings of the National Academy of Sciences 110 (2013) 13067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Schenk MF, Szendro IG, Salverda MLM, Krug J, de Visser JAGM, Patterns of epistasis between beneficial mutations in an antibiotic resistance gene, Molecular Biology and Evolution 30 (2013) 1779–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Sohka T, Heins RA, Phelan RM, Greisler JM, Townsend CA, Ostermeier M, An externally tunable bacterial band-pass filter, Proceedings of the National Academy of Sciences 106 (2009) 10135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Ochman H, Gerber AS, Hartl DL, Genetic applications of an inverse polymerase chain reaction, Genetics 120 (1988) 621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Goodman DB, Church GM, Kosuri S, Causes and effects of n-terminal codon bias in bacterial genes, Science 342 (2013) 475. [DOI] [PubMed] [Google Scholar]
- [29].Bentele K, Saffert P, Rauscher R, Ignatova Z, Blüthgen N, Efficient translation initiation dictates codon usage at gene start, Molecular systems biology 9 (2013) 675–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Pumir A, Shraiman B, Epistasis in a model of molecular signal transduction, PLoS Computational Biology 7 (2011) e1001134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Geiler-Samerotte KA, Dion MF, Budnik BA, Wang SM, Hartl DL, Drummond DA, Misfolded proteins impose a dosage-dependent fitness cost and trigger a cytosolic unfolded protein response in yeast, Proceedings of the National Academy of Sciences of the United States of America 108 (2011) 680–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Yang J-R, Liao B-Y, Zhuang S-M, Zhang J, Protein misinteraction avoidance causes highly expressed proteins to evolve slowly, Proceedings of the National Academy of Sciences of the United States of America 109 (2012) E831–E840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Starr TN, Thornton JW, Epistasis in protein evolution, Protein Science : A Publication of the Protein Society 25 (2016) 1204–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Huang W, Palzkill T, A natural polymorphism in beta-lactamase is a global suppressor, Proceedings of the National Academy of Sciences of the United States of America 94 (1997) 8801–8806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Dasmeh P, Serohijos AWR, Estimating the contribution of folding stability to nonspecific epistasis in protein evolution, Proteins: Structure, Function, and Bioinformatics 0 (2018). [DOI] [PubMed] [Google Scholar]
- [36].Bershtein S, Mu W, Serohijos AWR, Zhou J, Shakhnovich EI, Protein quality control acts on folding intermediates to shape the effects of mutations on organismal fitness, Molecular cell 49 (2013) 133–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Harms MJ, Thornton JW, Evolutionary biochemistry: Revealing the historical and physical causes of protein properties, Nature reviews Genetics 14 (2013) 559–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Wang X, Minasov G, Shoichet BK, Evolution of an antibiotic resistance enzyme constrained by stability and activity trade-offs, Journal of Molecular Biology 320 (2002) 85–95. [DOI] [PubMed] [Google Scholar]
- [39].Kaltenbach M, Tokuriki N, Dynamics and constraints of enzyme evolution, Journal of Experimental Zoology Part B: Molecular and Developmental Evolution 322 (2014) 468–487. [DOI] [PubMed] [Google Scholar]
- [40].Klesmith JR, Bacik J-P, Wrenbeck EE, Michalczyk R, Whitehead TA, Tradeoffs between enzyme fitness and solubility illuminated by deep mutational scanning, Proceedings of the National Academy of Sciences of the United States of America 114 (2017) 2265–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.