Abstract
M-proteins (M-Prts) are major virulence determinants of Group A Streptococcus pyogenes (GAS) that are covalently anchored to the cell wall at their conserved COOH-termini while the NH2-terminal regions extend through the capsule into extracellular space. Functional M-Prts are also secreted and/or released from GAS cells where they exist as helical coiled-coil dimers in solution. Certain GAS strains (Pattern D) uniquely express an M-protein (plasminogen-binding group A streptococcal M-protein; PAM) that directly interacts with human plasminogen (hPg), a process strongly implicated in the virulence of these strains. M-Prt expressed by the emm gene is employed to serotype over 250 known strains of GAS, ~20 of which are hitherto found to express PAMs. We have developed a modular structural model of the PAM dimer that describes the roles of different domains of this protein in various functions. While the helical COOH-terminal domains of PAM are essential for dimerization in solution, regions of its NH2-terminal domains also exhibit a weak potential to dimerize. We find that temperature controls the open (unwound) or closed (wound) states of the functional NH2-terminal domains of PAM. As temperature increases, α-helices are dramatically reduced, which concomitantly destabilizes the helical coiled-coil PAM dimers. PAMs with two a-repeats within the variable NH2-terminal A-domain (class I/III) bind to hPg tightly, but natural PAM isolates with a single a-repeat in this domain (class II) display dramatic changes in hPg binding with temperature. We conclude that coexistence of two a-repeats in PAM is critical to achieve optimal binding to hPg, especially in its monomeric form, at the biologically relevant temperature.
Keywords: Plasminogen acquisition, PAM dimerization, α-helix, dimer dissociation, single a-repeat, coiled coils
Graphical Abstract
1. Introduction
Group A Streptococcus pyogenes (GAS) is a Gram-positive β-hemolytic pathogen that infects ~700 M humans annually. While the majority of these infections are mild and treatable, e.g., pharyngitis and impetigo, >18 M of these cases become severe and ~1 M progress to the highly invasive stage, e.g., necrotizing fasciitis and toxic shock syndrome, resulting in ~550,000 deaths per year (Carapetis et al., 2005). A critical virulence determinant of GAS is its characteristic strain-specific and antigenically-variable fibrous M-protein (M-Prt) that is expressed from the bacterial emm gene. The variability of the emm gene and its M-Prt product likely evolved via recombination-driven gene-specific sweeps (Bao et al., 2016), giving rise to the multiplicity of GAS strains. For epidemiological purposes, the 5’-end of the emm gene is used to serotype GAS isolates (~250 distinct variations), and the 3’-end of M-like protein genes within the mga regulon are employed to pattern-type GAS strain (Patterns A-E) (Facklam et al., 1999).
All M-Prts contain distinct domains. The extracellular regions consist of an NH2-terminal hypervariable region (HVR), followed by A-, B-, C-, and D-domains, and a COOH-terminal cell wall peptidoglycan-located Pro/Gly-rich module. Lastly, a sortase A recognition site and a single pass-through membrane spanning region containing a short cytoplasmic tail make up the emm gene product (Fischetti et al., 1988). Processing at the COOH-terminal sortase A site anchors PAM to the cell wall, after which the fibrous rod-like protein projects through the outer capsule into the extracellular medium, wherein the majority of the protein is available for interactions with the host (Fischetti, 1989; Phillips et al., 1981b; Qiu et al., 2018; Sanderson-Smith et al., 2008). Variations in M-Prts consist of differing numbers and sequences of short homologous repeat sequences within the major domains (Fischetti, 1989; Smeesters et al., 2010).
In the case of skin-tropic Pattern D strains of GAS, the relevant M-Prt is a direct host human plasminogen (hPg) receptor (plasminogen-binding group A streptococcal M-protein; PAM) which recruits the fibrinolytic zymogen, hPg, to the GAS cell surface (Berge and Sjobring, 1993; McArthur et al., 2008; McKay et al., 2003). The hPg bound to its GAS receptor is activated by GAS-secreted streptokinase (SK) (Schick and Castellino, 1974; Zhang et al., 2012), thereby generating a surface bound serine protease, plasmin (hPm), which initially degrades the fibrin mesh that encapsulates the GAS cells as part of the innate immune response to infection, and subsequently disrupts host extracellular matrix proteins and tight cell junctions (Plow et al., 1995; Sumitomo et al., 2013l; Sumitomo et al., 2016; Walker et al., 2014). In this manner, more virulent forms of GAS are able to gain entry into deep tissues of the host (Fulde et al., 2013).
We have previously cloned and expressed the extracellular regions of PAMs from serologically distinct Pattern D GAS isolates (Qiu et al., 2018). As with other M-Prts, PAMs exist in solution as coiled-coil dimers (Cedervall et al., 1997; McNamara et al., 2008; Stewart et al., 2016), and we have constructed a structural model that depicts the manner in which PAMs form non-ideal dimers in solution (Qiu et al., 2018). The exposed NH2-terminal A-domain is responsible for specific binding of PAM to the lysine binding site of the ~80-residue kringle 2 (K2hPg) domain of the multi-modular hPg (Castellino and Ploplis, 2003).
Since M-Prts possess the characteristics to form helical coiled-coils, especially in solution (Glinton et al., 2017; Qiu et al., 2018), it is assumed that dimers are present on the cell surface (Cedervall et al., 1997; Phillips et al., 1981a). However, it is most likely that PAM is translocated through the narrow sortase A secretion (SecA) channel in the cell membrane in an unfolded monomeric state, where the protein is anchored to the pentaglycine residues of the mother and daughter septa of newly forming cell wall and not to previously formed cell wall sites (Raz et al., 2012). Under these circumstances, it is unlikely that the suboptimal spatial proximity of individual anchored PAM monomers favors the type of uniform coiled-coil dimers on cells that are seen in solution. Understanding the solution structure-function relationships of M-Prts is essential since soluble M-Prts are released during infection (Berge and Björck, 1995) where they possess a variety of functions, such as induction of T-cell activation and subsequent inflammation (Påhlman et al., 2008). Thus, it is necessary to explore the significance of both monomers and dimers in the structure and function of PAMs. In this study, we also delve into the relationships between the PAM secondary structure and hPg-binding properties through use of recombinant PAM modules. In particular, we investigate whether substantial amounts of PAM monomers that are generated under relevant conditions cause significant changes in binding to hPg. We finally integrate all biophysical results in order to provide a comprehensive picture of the bacteria-host interactions as related to the virulence-determining fibrinolytic system.
2. Materials and Methods
2.1. Bacterial strains and cloning constructs
The GAS isolates (and the M-Prt serotypes) used in this study, specifically, AP53 (emm53), NS88.2 (emm98.1), NS223 (emm91), NS455 (emm52), SS1448 (emm86.2), SS1572 (emm223), and SS1574 (emm224), have been previously described (Qiu et al., 2018). Genomic DNAs of each strain were extracted for the cloning of PAM (Ward and Leigh, 2002). The pam coding sequences were amplified from each isolate by the polymerase chain reaction (PCR) and then ligated into the vector, pET-28a (EMD Biosciences), without the N-terminal signal peptides and C-terminal LPXTG cell wall anchor regions. A His6-tag was engineered into the reverse primer for each full-length PAM for further facile purification by Ni+-based affinity chromatography. The same cloning strategy above was applied to PAMAP53_short, PAMAP53_medium, and PAMAP53_long (Zhang et al., 2012).
To construct plasmids for expression of shortened peptides, including AGL55NS88.2, KTI55SS1448, VEK75AP53, and VEK75AP53_RH1/AA, an engineered vector, pET-15b (Novagen, Gibbstown, NJ), was employed. This modified plasmid sequentially contains an ATG initiation codon, a His6-tag, a GB1 domain, a 9-residue linker, a thrombin cleavage site (LVPR↓GS), the peptide coding sequence, and a translation stop codon. Therefore, after thrombin cleavage, all truncated peptides in this study possessed the dipeptide GS at their NH2-termini. Tyr in the sequences was used for concentration determinations at 280 nm (Bhattacharya et al., 2014). The class designation, residue ranges, and a-repeat sequences of all materials used in this study are summarized in Table 1.
Table 1.
Summary of recombinant PAM proteins and peptides
| Protein/Peptide | Class | Range | Sequence of A-repeat(s)a |
|---|---|---|---|
| PAMSS1574 | I | 42-360 | DAELQRLKNERHE∥EAELERLKSERHEHDKK |
| PAMAP53 | 42-392 | DAELQRLKNERH∥EAELERLKSERHDHDKK | |
| PAMAP53_short | 42-175 | ||
| PAMAP53_medium | 42-207 | ||
| PAMAP53_long | 42-338 | ||
| VEK75AP53 | 97-171 | ||
| VEK75AP53_RH1/AA | 97-171 | DAELQRLKNEAAE∥EAELERLKSERHDHDKK | |
| PAMNS88.2 | II | 42-385 | DAELKRLNEERHDHDKR |
| AGL55NS88.2 | 95-149 | ||
| PAMSS1448 | 42-375 | DNVELERLKNERHDHDE | |
| KTI55SS1448 | 85-139 | ||
| PAMNS223 | III | 42-399 | EVELERLKNERHDHDE∥EAELNRLREERHDHDKK |
| PAMNS455 | 42-401 | EVALERLKNERHVHDE∥EVELERLKNERHDHDKK | |
| PAMSS1572 | 42-399 | AAELERLKNERHVHDE∥EAELNRLKNERHDHDKK |
In the case of Class I/III PAMs or derivative peptides, a1- and a2-repeats are separated by the symbol “∥”, and RH-motifs are labeled in bold letters.
2.2. Protein expression and purification
Escherichia coli BL21/DE3 cells (New England BioLabs) were transformed with different recombinant expression plasmids (pET-28a/pET-15b). The procedure for protein expression has been previously described (Yuan et al., 2017). In order to increase the yields, terrific broth medium (TB; 12 g tryptone/24 g yeast extract/4 mL glycerol/2.3 g KH2PO4/12.5 g K2HPO4 per liter) was applied instead of conventional Luria-Bertani (LB) medium. A 10 mL overnight culture of transformed E. coli BL21/DE3 cells was inoculated into 1 L TB medium, supplemented with 40 μg/ml kanamycin for full-length and truncated PAMs or 100 μg/ml ampicillin for shortened peptides. The entire culture was incubated at 37° C until an OD600 nm ~0.6 was reached. Protein expression was then induced by the addition of 0.4 mM isopropyl-1-thio-β-D-galactopyranoside (IPTG), and the cells were grown overnight (~16 hr) at 25° C. The cells were next collected at 8,000 rpm in a Sorvall RC-5B refrigerated superspeed centrifuge and resuspended in 40 mL lysis buffer (50 mM Tris/300 mM NaCl/10 mM imidazole, pH 8.0) with 500 μg/ml lysozyme (Ameresco, Solon, OH) and 1 mM phenylmethylsulfonyl fluoride (PMSF). After a 20 min sonication period, the culture was centrifuged at 10,000 rpm for 30 min. The resultant supernatant was then separated from cell pellets and stored at 4° C.
Ni-NTA agarose (MCLAB, S. San Francisco, CA) was used to specifically capture the His6-tag of the protein in the purification step. For full-length and truncated PAMs, the packed column was washed with a solution of 50 mM Tris/300 mM NaCl/40 mM imidazole, pH 8.0, and the proteins were eluted with a buffer containing 50 mM Tris/300 mM NaCl/250 mM imidazole, pH 8.0. This final eluted protein contained the corresponding PAM. For purification of short peptides, viz. AGL55ns88.2, KTI55SS1448, VEK75AP53, and VEK75AP53_RH1/AA, the concentrations of imidazole in lysis, wash, and elution buffer were 5 mM, 20 mM, and 60 mM, respectively. Thrombin (50 μL; 1,000 U; ERL, South Bend, IN) was added to ~10 mL concentrated eluate for cleavage. Cleaved fragments were further separated using Ni-NTA agarose, which specifically bound to His6-GB1-linker-LVPR. Accordingly, the flow-through fraction contained those short PAM-derivative peptides. The protease inhibitor, p-aminobenzamidine, immobilized on agarose (Sigma), was added to the aforementioned fraction to remove thrombin (Yuan et al., 2017) and the resin subsequently removed by centrifugation.
2.3. Protein identification and concentration determination
Identification of all proteins and peptides was conducted by MALDI-TOF mass spectrometry using an Autoflex III (Bruker Daltonics, Bremen, Germany). The extinction coefficients of full-length PAMs, along with PAMAP53_long, were determined by analytical ultracentrifugation. The procedure has been described in detail and specific extinction coefficients were the same as previously described (Babul and Stellwagen, 1969; Qiu et al., 2018). However, since this strategy is only optimally applied to larger proteins, extinction coefficients of AGL55NS88.2, KTI55SS1448, VEK75AP53, VEK75AP53_RH1/AA, PAMAP53_short, and PAMAP53_medium were obtained from ExPASy calculations.
2.4. Circular dichroism (CD) spectroscopy
Far-UV CD wavelength scans were collected on an Aviv model 202 SF Spectrometer (Aviv, Lakewood, NJ) to estimate the helical contents of the proteins and peptides. These data were collected at 4° C, 25° C, and 37° C, from 195 nm to 250 nm, with 1-nm increments. Sodium phosphate (10 mM, pH 7.4) was used as the protein buffer. The spectra represented the average of three scans, and an average buffer reference scan was subtracted from that of each sample. Calculations of the percentage of α-helix for each protein were performed as previously published (Chen et al., 1972; Greenfield, 2006; Qiu et al., 2018). In the temperature scans, the first derivative of the mean residue ellipticities (MRE; Δθ/ΔT) was calculated to determine the slope at every data point, which describes the rate of the secondary structural changes with temperature (Veltri et al., 2017). All data points were plotted using GraphPad Prism 6.0. In the Δθ/ΔT vs T plots, fourth order smoothing was applied in connections of all data points.
2.5. Analytical ultracentrifugation (AUC)
Sedimentation equilibrium experiments were performed at various temperatures (4° C, 25° C, 37° C) on an Optima XL-1 analytical ultracentrifuge (Beckman Coulter, Brea, CA). The proteins/polypeptides were dissolved in 10 mM sodium phosphate (pH 7.4), and diluted to A280nm ~0.15. The buffer density was 1 g/mL. The types of cell centerpieces and cell housings have been described previously (Bhattacharya et al., 2014; Qiu et al., 2018).
In order to compare quaternary structures of PAMs at different temperatures, optimal rotor speeds were used first at 25° C, wherein the status of monomers or dimers could be determined. Then experiments were conducted at 4° C and 37° C to assess weight-average molecular mass changes. Each protein solution was rotated at the speeds of 18,000 rpm and 20,000 rpm for all full-length PAMs; 28,000 rpm and 32,000 rpm for PAMAP53_short; 24,000 rpm and 28,000 rpm for PAMAP53_medium; and 15,000 rpm and 18,000 rpm for PAMAP53_long. The monochromator scanned at 280 nm every hour. When the three most recent scans overlapped, we considered that equilibrium was reached, at which point a different speed was applied. Generally, ~20 hr was needed to reach equilibrium at the initial lowest speed. The partial specific volume of each protein/peptide was determined from SEDNTERP (Qiu et al., 2018).
Weight-average molecular masses were calculated from a non-linear fitting model using Optima XL-A/XL-I data analysis software (Beckman Coulter). To examine the fraction of monomers and dimers in a sample, the model “species analysis” was employed in SEDPHAT (Vistica et al., 2004). The molecular mass, partial specific volume, buffer density, and molar extinction coefficient were entered in the parameter control panel. AUC assays were conducted in triplicate for each protein or peptide at each rotor speed.
2.6. Surface plasmon resonance (SPR)
Binding kinetics of hPg were measured from SPR in real-time using a BIAcore X100 Biosensor system (GE Healthcare). HBS-EP (10 mM Hepes/0.15 M NaCl/3 mM EDTA/ 0.005% polysorbate-20, pH 7.4) was employed as the running buffer at a flow rate of 30 μL/min. An amine coupling kit (GE Healthcare) was used in the immobilization step. hPg, diluted to 30 μg/mL in 10 mM sodium acetate, pH 4.5, was injected into flow cell 2 of a CM5 sensor chip. Flow cell 1 was prepared by the same method, but without immobilizing ligands on the chip (Bhattacharya et al., 2014; Chandrahas et al., 2015; Glinton et al., 2017). Ligand immobilizations and binding experiments were performed at various temperatures (15° C, 25° C, 37° C). Due to the low efficiency in ligand immobilization at 4° C, ligands were applied at 15° C to monitor binding activities of PAMs to hPg at a low temperature. Ethanolamine (1 M, pH 8.5) was injected immediately after ligand immobilization to block non-bound sites on the CM5 chip.
Different concentrations of analytes in HBS-EP buffer were injected over the hPg-bound CM5 chip surface for an association time of 120 sec and a dissociation time of 360 sec (Qiu et al., 2018; Yuan et al., 2017). The gold surface was regenerated using 10 mM glycine, pH 1.5. All binding data collected in flow cell 2 were subtracted from those obtained in the reference cell. Sensorgrams were analyzed using BIA evaluation software 2.0.1 (GE Healthcare). Equilibrium dissociation constants (Kd) were calculated from the dissociation (koff) rates divided by association rates (kon). In cases where kinetic assays showed very fast kinetic rates, affinity analysis was applied to determine the Kd. For this experiment, the RUs of the last points in the association stage were plotted against different concentrations of ligand. Nonlinear fitting was employed for both kinetics and affinity methods. Binding curves are presented in GraphPad Prism 6.0.
3. Results
3.1. The hypervariable region (HVR) and A-domain of PAM show a propensity to dimerize at low temperatures
Based on the amino acid sequences of the hPg-binding A-domain of PAM, we have classified PAMs from various isolates of Pattern D strains as Class I, where two hPg binding a-repeats (a1a2) constitute the A-domain, Class II, with only a single a-repeat, viz., a2, and Class III, where VHD or a similar tripeptide is present at the COOH-terminus of the a1-repeat, between a1 and a2 (Qiu et al., 2018) (Table 1). These classifications of PAM in Pattern D strains as studied here and those in databases appear to be consistent across all Pattern D strains.
A previous study shows that the highly homologous c-repeats in the C-domain are the strongest determinants of PAM dimerization (Qiu et al., 2018). It was also found that aliphatic hydrophobic residues within the NH2-terminal HVR and A-domain occupy some of the a and d heptad sequence positions as required for helical coiled-coil formation (Qiu et al., 2018). This finding suggests that NH2-terminal regions may also contribute to PAM dimerization, but the extent to which these regions participate in dimer stabilization is currently unknown. Since α-helices are necessary for coiled-coil dimerization, we first compared the α-helical contents of three truncated N-terminal peptides containing different modular regions of PAMAP53 (modules in parentheses), viz., PAMAP53_short (HVR-a1a2-B), PAMAP53_medium (HVR-a12-B-c1), and PAMAP53_long (HVR-a1a2-B-c1c2c3-D), at various temperatures. Compared to previous data determined at 25° C (Qiu et al., 2018), the present results show that the α-helical contents of these peptides vary with the temperature and are greatly enhanced at 4° C (Fig. 1A, B; Table 2), as is also the case with full-length PAMs (a typical example is shown in Fig. 1C). At 4° C, α-helices account for ~75 - 85% of the secondary structures in PAMAP53_short, PAMAP53_medium and PAMAP53_long (Fig. 1A, B; Table 2). Corresponding to this increase in α-helical content, AUC experiments show that ~30% of PAMAP53_short becomes dimeric at 4° C, although this peptide does not contain any c-repeats (Table 3). In PAMAP53_medium, the COOH-terminus is extended through one (c1) of the three c-repeats of the C domain, and dimerization is increased from 10% at 25° C to 50% at 4° C (Table 3). These results suggest that the HVR-A-domain at the NH2-terminus of PAM has a higher helical content at lower temperature, and concomitantly has the capacity to partially dimerize, albeit weakly. Lastly, PAMAP53_long has a higher α-helical content than PAMAP53, likely because of the Pro/Gly helical destabilizing domain present at the COOH-terminus of PAMAP53, but removed in PAMAP53_long.
Fig. 1. CD spectra of PAM fragments at different temperatures.
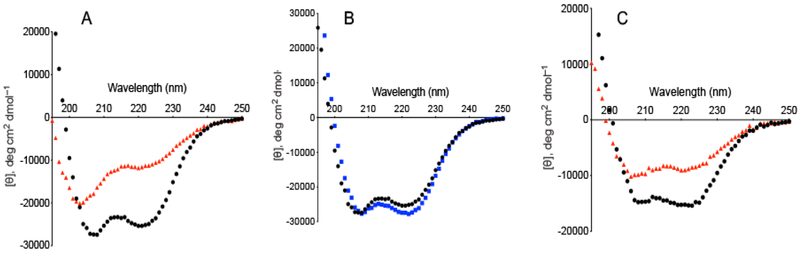
The mean residue ellipticities (MRE; [θ]) of each protein or polypeptide are illustrated as a function of wavelength from 195 nm - 250 nm at the specified temperatures. The results are displayed as data points that represent the average of triplicate scans at a given temperature. The standard error very small at <5% on average for each curve. Panel A represents plots of [θ] vs wavelength for PAMAP53_short at 4° C (black) and 37° C (red). Panel B compares the CD spectra of PAMAP53_short (black) and PAMAP53_medium at 4° C (blue). Panel C illustrates changes in [θ] of full-length PAMNS88.2 (residues 42-385) vs wavelength at 4° C (black) and 37° C (red). Full length PAMs comprise amino acids immediately downstream of the signal peptide up to, but not including, the sortase A recognition peptide (LPXTG), and hence contain the HVR-A-B-C-D-Pro/Gly domains.
Table 2.
α-Helical contents of truncated PAMAP53 peptides and naturally occurring PAMs
| Protein | % α-helices | θ222/θ208 | ||||
|---|---|---|---|---|---|---|
| 4° C | 25° Ca | 37° C | 4° C | 25° C | 37° C | |
| PAMAP53_short | 75 | 39 | 30 | 0.92 | 0.74 | 0.69 |
| PAMAP53_medium | 84 | 32 | 25 | 1.00 | 0.73 | 0.70 |
| PAMAP53-long | 84 | 49 | 28 | 1.08 | 0.96 | 0.86 |
| PAMAP53 | 33 | 27 | 20 | 1.03 | 0.95 | 0.92 |
| PAMNS88.2 | 43 | 33 | 21 | 1.04 | 0.94 | 0.89 |
| PAMNS223 | 75 | 61 | 40 | 1.02 | 0.95 | 0.87 |
| PAMNS455 | 96 | 72 | 42 | 1.05 | 0.95 | 0.82 |
| PAMSS1448 | 52 | 33 | 22 | 1.04 | 0.92 | 0.87 |
| PAMSS1572 | 79 | 59 | 44 | 1.03 | 0.95 | 0.91 |
| PAMSS1574 | 61 | 54 | 29 | 1.05 | 1.01 | 0.89 |
The percentage of α-helix in each PAM at 25° C was published previously (Qiu et al., 2018) and relisted here for comparison purposes.
Table 3.
Weight-average molecular masses of truncated PAMAP53 proteins
| Proteina | T (° C) | Molecular mass (Da)b | % Dimerc |
|---|---|---|---|
| PAMAP53_shortd | 4 | 23,300 ± 600 | 29 ± 4 |
| 25 | 15,200 ± 1,000 | 0 | |
| PAMAP53_mediumd | 4 | 33,600 ± 800 | 48 ± 8 |
| 25 | 22,700 ± 500 | 11 ± 2 | |
| PAMAP53_longe | 25 | 70,600 ± 3,300 | 87 ± 9 |
| 37 | 69,300 ± 2,300 | 75 ± 2 |
Molecular mass of the protein linear sequence from MALDI-TOF: PAMAP53_short, 16,917 Da; PAMAP53_medium, 20,348 Da; PAMAP53_long, 34,937 Da.
Weight-average molecular masses of proteins in solution from AUC experiments at the specified temperatures. Data at 25° C were published previously (Qiu et al., 2018) and are listed here for comparison. Data from the top, middle, and bottom channels of the AUC cell were collected and presented as mean ± S.D.
The portion of dimeric species in solution was calculated using SEDPHAT. Data from the top, middle, and bottom channels were collected and are presented as mean ± S.D.
Not determined at 37° C since minimal dimeric forms for PAMAP53_medium were already present at 25° C.
Not determined at 4° C since maximal dimeric forms for PAMAP53_long were already present at 25° C.
3.3. Dimeric PAM dissociates accompanying the loss of α-helices at high temperature
Although it has been verified that different domains of full-length PAM exhibit dimerization potential at 4° C (this study) or 25° C (Qiu et al., 2018), it is important to examine its secondary structural composition at 37° C, the physiological temperature for GAS. Compared to the values at 4° C, the α-helix contents of all truncated and full-length PAMs significantly decrease at 25° C, and decrease even further at 37° C (Table 2). Meanwhile, the MRE ratios at 222:208 nm (θ222/θ208) also decrease from ~1.0 at 4° C to 0.9-1.0 at 25° C, then to below 0.9 at 37° C (Table 2). It is known that a θ222/θ208>1 implies that coiled-coils predominate the protein secondary structures, and that ratios <0.9 correspond to isolated α-helices (McNamara et al., 2008). Thus, increased temperatures, up to 37° C, destabilize the α-helices in PAMs and may dissociate coiled-coil dimers into monomers.
In order to further correlate the α-helical content of PAM with its ability to dimerize, we next performed AUC experiments for molecular mass determinations at 37° C and further analyzed the data to calculate the portion of dimeric structures in the protein population. Unlike highly dimerized status at 25° C, most full-length PAMs, along with PAMAP53_long (HVR-a1a2-B-C-D domains), exhibit apparent weight-average molecular masses between monomeric and dimeric states (Tables 3 and 4), suggesting that PAMs exist as mixtures of monomers and dimers at 37° C. Interestingly, dimeric portions diverged dramatically among different PAMs. PAMNS223, PAMNS455, and PAMSS1574 only maintained ~20% - 30% dimers at 37° C. However, in the cases of Class II PAMs (PAMNS88.2 and PAMSS1448), along with PAMSS1572, ~50-60% dimers remained at 37° C. Of note, PAMAP53 is almost fully dimeric at this higher temperature (Table 4). Thus, the stability of the coiled-coil dimeric structures differ among PAMs, likely due to the stabilizing or destabilizing amino acid changes at the N-terminus of the proteins, their most variable sequence regions.
Table 4.
Weight-average molecular masses of naturally occurring PAMsa
| Proteinb | T (° C) | Molecular mass (Da)c | % Dimerd |
|---|---|---|---|
| PAMAP53 | 25 | 87,900 ± 1,000 | 100 |
| 37 | 84,700 ± 4,500 | 97.5 ± 2.4 | |
| PAMNS88.2 | 25 | 81,900 ± 300 | 91.4 ± 3.3 |
| 37 | 69,000 ± 800 | 55.5 ± 2.1 | |
| PAMNS223 | 25 | 83,700 ± 1,100 | 94.5 ± 2.8 |
| 37 | 57,500 ± 2,500 | 33.7 ± 6.3 | |
| PAMNS455 | 25 | 83,400 ± 1,200 | 86.8 ± 3.3 |
| 37 | 52,200 ± 2,600 | 20.4 ± 5.2 | |
| PAMSS1448 | 25 | 77,700 ± 400 | 86.7 ± 4.1 |
| 37 | 69,800 ± 2,100 | 56.4 ± 2.5 | |
| PAMSS1572 | 25 | 83,200 ± 900 | 95.0 ± 5.0 |
| 37 | 74,900 ± 2,000 | 75.3 ± 4.1 | |
| PAMSS1574 | 25 | 78,800 ± 1,200 | 100 |
| 37 | 55,700 ± 2,600 | 33.5 ± 7.4 |
Experiments were not performed at 4° C since maximal dimeric forms were present at 25° C.
Molecular masses of the protein linear sequence from MALDI-TOF: PAMAP53, 41,083 Da; PAMNS88.2, 40,020 Da; PAMNS223, 41,760 Da; PAMNS455, 42,055 Da; PAMSS1448, 38,644 Da; PAMSS1572, 41,635 Da; PAMSS1574, 37,401 Da.
Weight-average molecular mass of proteins in solution from AUC experiments at the specified temperatures. Data at 25° C were published previously (Qiu et al., 2018) and are listed here for comparison. Data from the top, middle, and bottom channels were collected and presented as mean ± S.D.
The portion of observable dimeric species in solution was calculated using SEDPHAT. Data from the top, middle, and bottom channels were collected and presented as mean ± S.D.
3.3. The temperature effect is more significant for PAMs with a high α-helical content
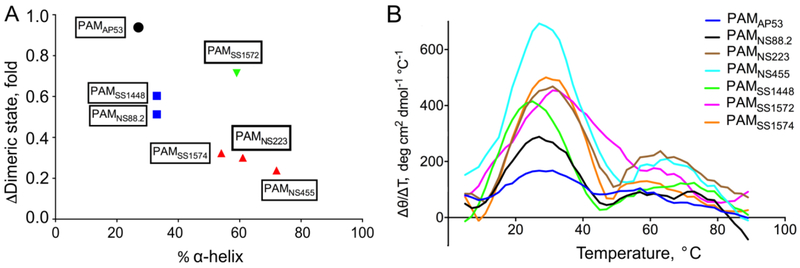
For most full-length PAMs, the fraction of α-helices and tolerance to temperature changes are correlated (Fig. 2A). More rigid, structured dimeric PAMs are affected more by temperature changes, and dissociate to monomers more easily. PAMNS223, PAMNS455, and PAMSS1574 have high percentages of α-helices (50% - 70%) at 25° C, and 90% of these proteins exist as dimers at this temperature. However, at 37° C, only 20%-30% of these PAMs remain as dimers (Table 4). Conversely, although α-helices account for <30% of secondary structures of PAMAP53, this protein demonstrates a higher resistance to temperature changes, and retains its dimeric state at 37° C. The Class II PAMs, PAMNS88.2 and PAMSS1448, exhibit intermediate α-helical contents and relative proportions of dimeric structures (Fig. 2A).
Fig. 2. Correlation between the fraction of α-helices at 25° C and tolerance to temperature changes.
(A) For each full-length PAM, the x-axis represents the α-helix content at 25° C, and the y-axis represents the ratio of the dimeric content at 37° C/25° C, represented as the fold change between these two temperatures. All data points are labeled as the corresponding PAM, and categorized into 4 clusters in this plot, shown as different colors. (B) The first derivative of the mean residue ellipticity [θ] in the CD temperature scan is plotted against temperature at each point. Curves of different PAMs are shown in distinct colors.
The structure-temperature correlation is also observed from the CD temperature scans. We calculated the first derivatives of mean residue ellipticity, Δθ/ΔT, for each data point, and plotted this function against the temperature (Fig. 2B). The value of Δθ/ΔT reflects the slope at each point in the original CD temperature scans. The three highest peaks in Figure 2B correspond to PAMNS455, PAMSS1574, and PAMNS223, indicating that the dimeric states of these three PAMs are the most sensitive to temperature changes. Also, consistent with AUC data at 25° C and 37° C, PAMAP53 shows the lowest peak in Figure 2B, and thus dimeric PAMAP53 displays the strongest tolerance to temperature change. Further, in this plot, PAMNS88.2 and PAMSS1448 show intermediate sensitivity to temperature changes, ascribed to moderate Δθ/ΔT values.
A notable exception to this trend exists in the case of PAMSS1572, which has a high α-helical content at 25° C and a high dimeric content at 37° C. At 37° C, there still is >70% PAMSS1572 in its dimeric state (Fig. 2A; Table 4). The Δθ/ΔT vs T plot of PAMSS1572 depicts a relatively high but broader peak than other PAM proteins (Fig. 2B), indicating that PAMSS1572 responds to temperature variations more vigorously than PAMAP53, but less than PAMNS223, PAMNS455 and PAMSS1574. This unusual exception is likely ascribed to amino acid substitutions in c-repeats of the C-domain. We have found that these dimerization-determining domains are highly conserved in amino acid sequence, especially the c1-repeat. However, PAMSS1572 is distinct from other PAMs in that it encompasses significantly more substitutions in the c2- and c3-repeats (5 and 7 substitutions, respectively), compared to the prototype PAMAP53. Additionally, the D-domain of different PAMs also contains some Leu residues at positions a and d in heptad registers, making it possible for this region of the protein to contribute to dimerization. Whereas PAMSS1572 nonetheless contains 6 substitutions in its D-domain (Qiu et al., 2018), most of these natural mutations occur between apolar/polar and charged residues. This suggests that, besides almost identical hydrophobic niches, two chains in PAMSS1572 may intertwine tighter as a result of the increase in electrostatic attractions or decrease in repulsions, caused by these types of substitutions.
3.4. Physiological temperature attenuates the tight hPg-binding patterns of PAMNS88.2 and PAMSS1448
We have shown previously that at 25° C dimeric PAM proteins interact with hPg at Kd values <50 nM, regardless of their structural classification (Qiu et al., 2018). However, when dimer dissociation occurs upon an increase of temperature to 37° C, the PAM-hPg interaction might also be affected. Thus, we performed SPR analyses at 37° C to examine binding constants of all full-length PAMs to hPg. Kinetic analyses of the binding data showed a small impact on the Kd for hPg binding to ClassI/III PAMs, that contain both a1 and a2 repeats within the A-domain, as shown by the example of PAMNS223 (Fig. 3A). In these cases, while the Kd values of hPg to Class I/III PAMs were larger at 37° C than at 25° C, they were still tightly bound to hPg at a Kd of <50 nM (Table 5). However, the Class II PAMs (PAMNS88.2 and PAMSS1448), which contain only the a2-repeat, displayed much higher dissociation rates than the Class I/III PAMs, as shown by the example of PAMSS1448 (Fig. 3B). Due to the very fast dissociation of these PAMs from hPg, binding constants could not be reliably obtained from kinetic analyses. Therefore, we used steady state analysis to estimate the Kd values for these interactions (Fig. 3C). The Kd values for the PAM-hPg interactions, of 5,700 nM and 7,000 nM, for PAMNS88.2 and PAMSS1448, respectively, were much higher than those for the hPg binding to Class I/III PAMs (Table 5). Overall, when 50% of the dimers dissociated at 37° C, binding of PAMNS88.2 and PAMSS1448 to hPg became much weaker. However, binding behaviors of Class I/III PAMs to hPg were affected to a lesser extent, even when only 20% dimers remain at this high temperature.
Fig. 3. Binding assays of PAMs to hPg at 37° C.
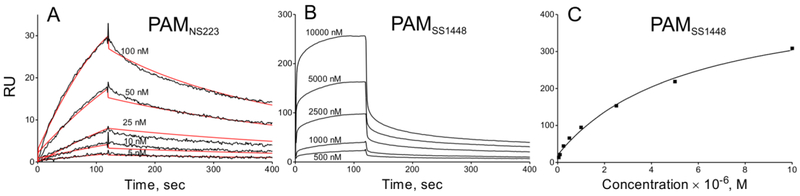
hPg was immobilized on a CM-5 chip and titrated with the indicated full-length PAM at the concentrations specified in each panel. (A) For Class I and Class III PAMs, with two a-repeats in the A-domain, kinetic binding analyses are as shown for the example of PAMNS223 (Class III). Here, SPR response units (RU) were plotted against time and the corresponding Kd values, obtained from koff/kon, are provided in Table 5. The experimental data are shown in black lines and fitted curves are shown in red lines. (B) For Class II PAMs, with only a single a-repeat, very high koff values were observed and were not reliable for calculation, as shown for the example of PAMSS1448. Here a SPR-based steady state approach was utilized, wherein the highest value of the RU at each concentration was plotted against the PAMSS1448 concentration, as in (C), and the Kd value was obtained from the calculated concentration midpoint of the titration. The corresponding Kd values are provided in Table 5.
Table 5.
Binding affinities of naturally occurring PAMs to hPg
| Proteins | 37° C | 25° C | ||
|---|---|---|---|---|
| kon (×104 Ms−1) | koff (×10−3 s−1) | Kda (nM) | Kdb (nM) | |
| Class I PAMs: complete a1a2-repeats | ||||
| PAMAP53 | 21.0 ± 3.3 | 4.3 ± 0.3 | 21 ± 2.0 | 2.8 ± 0.9 |
| PAMSS1574 | 56.2 ± 7.9 | 7.5 ± 0.6 | 14 ± 1.0 | 1.4 ± 0.4 |
| Class II PAMs: lacking a1-repeat | ||||
| PAMNS88.2 | n.d.c | 5,700 ± 240c | 1.5 ± 0.4 | |
| PAMSS1448 | n.d.c | 7,000 ± 990c | 30 ± 6 | |
| Class III PAMs: complete a1a2-repeats (+ VHD/DHD between the a1- and a2-repeats) | ||||
| PAMNS233 | 25 ± 8 | 5.6 ± 1.2 | 28 ± 10 | 2.9 ± 1.3 |
| PAMNS455 | 34 ± 13 | 1.4 ± 0.2 | 6.0 ± 2.3 | 0.3 ± 0.1 |
| PAMSS1572 | 17 ± 6 | 5.5 ± 0.7 | 40 ± 7 | 8.0 ± 3.9 |
Dissociation constants (Kd) were calculated from koff/kon. All data were collected from triplicate runs and are presented as the mean ± S.D.
Dissociation constants (Kd) measured at 25° C were published previously (Qiu et al., 2018) and are listed here for reference.
n.d., not determined. The very fast off rates for hPg to the Class II PAMs did not allow accurate calculations to be made by kinetic analyses of binding. Therefore, the steady state method was used to calculate the Kd values at 37° C.
3.5. Complete a1-and a2-repeats of PAM are needed for tight binding of hPg
It is known that two RH dipeptides in the a-repeats of the A-domain, one each in a1 and a2, are critical for the tight interaction between PAMAP53 and hPg (Sanderson-Smith et al., 2006). The peptides, AGL55NS88.2, KTI55SS1448, and VEK75AP53, are truncated A-domain-containing peptides from the different classes of PAMs (Table 1), and they are all monomeric at 25° C (Qiu et al., 2018). AGL55NS88.2 and KTI55SS1448 only contain the a2-repeat linked to the NH2-terminus of the B-domain, but VEK75AP53, truncated from PAMAP53, contains both a1- and a2-repeats. To test whether complete a1-and a2-repeats are essential for very tight binding of hPg, SPR experiments were carried out to obtain hPg-binding affinities of these PAM-derived peptides at three temperatures. VEK75AP53 interacted with hPg at a Kd of ~1 nM at 25° C, (Fig. 4A; Table 6). In contrast, as with the behavior of their parent PAMs at 37° C, AGL55NS88.2 and KTI55SS1448, displayed much weaker binding to hPg. Here, again, steady state affinity analyses (Fig. 4B, C) were applied to determine Kd values because of the very high koff rates of these peptides truncated from Class II PAMs. From the data obtained, Kd values for AGL55NS88.2 and KTI55SS1448 to hPg were ~830 nM and 1500 nM, at 25° C, respectively, and much higher than that for VEK75AP53 (Table 6).
Fig. 4. Binding assays of PAM-derived peptides to hPg at 25° C.
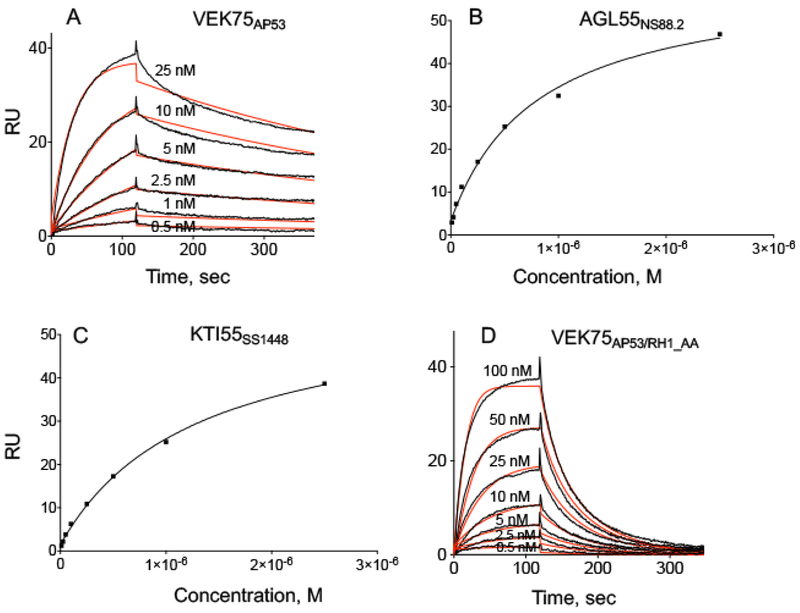
hPg was immobilized on a CM-5 chip, and peptide concentrations used in the binding assay titrations are indicated in Panels (A)-(D). (A) VEK75AP53; (B) AGL55NS88.2, (C) KTI55SS1448, and (D) VEK75AP53_RH1/AA. Kinetic analyses were used for (A) and (D), and steady state analyses for (B) and (C). In kinetic analyses, the experimental data are shown in black lines and fitted curves are shown in red lines. The corresponding binding constants for these curves are provided in Table 6.
Table 6.
Binding affinities of PAM-derived peptides to hPg
| Peptides | T (° C) | Kd (kinetics)a (nM) |
Kd (steady state)b (nM) |
|---|---|---|---|
| AGL55NS88.2 | 15 | 160 ± 5 | 350 ± 2 |
| 25 | 630 ± 11 | 830 ± 45 | |
| 37 | n.d.c | 7,100 ± 500 | |
| KTI55SS1448 | 15 | 360 ± 13 | 540 ± 22 |
| 25 | 2,100 ± 160 | 1,500 ± 110 | |
| 37 | n.d.c | 27,000 ± 640 | |
| VEK75AP53 | 15 | n.d.d | n.d.d |
| 25 | 0.7 ± 0.2 | n.d.d | |
| 37 | 15 ± 7 | 45 ± 18 | |
| VEK75AP53_RH1/AA | 15 | 26 ± 1 | n.d.e |
| 25 | 50 ± 5 | 82 ± 12 | |
| 37 | n.d.c | 890 ± 77 |
Dissociation constants (Kd) were calculated from koff/kon. All data were collected from triplicate runs and are presented as the mean ± S.D.
Dissociation constants (Kd) were calculated from steady state plots of the peak value in the association stage vs concentration of peptides. All data were collected from triplicate runs and are presented as the mean ± S.D.
koff rates were too fast for accurate kinetic assessment.
Binding was too tight to be accurately measured.
Optimal measurements were made by kinetic binding methods.
Because AGL55NS88.2 and KTI55SS1448 only contain a2-repeats with a single RH-motif (RH2), we mutated the first RH-motif (RH1) of VEK75AP53 in the a1-repeat to alanine (R17H18/A17A18). This variant, VEK75AP53_RH1/AA, only maintaining RH2 as in AGL55NS88.2 and KTI55SS1448, shows much tighter binding to hPg (Kd of ~50 nM) at 25° C (Fig. 4D; Table 6). However, the Kd values of VEK75AP53_RH1/AA to hPg are ~50-fold higher than that of VEK75AP53.
The same binding assays of all three truncated PAMAP53 proteins also supported the finding that tight binding to hPg can be attained with the existence of complete a1a2-repeats. PAMAP53_long has been proven to be a dimer at 25° C, and interacted with hPg as tightly as full-length PAMAP53 (Table 7) at this same temperature. Although PAMAP53_medium and PAMAP53_short did not form dimers at 25° C, they nonetheless showed hPg-binding affinities of ~2 nM (Table 7). Again, three peptides maintain tight hPg-binding with Kd of 10–40 nM at 37° C, further suggesting that two intact a-repeats are crucial for monomeric PAM-derived peptides in solution to capture hPg at a nM-magnitude.
Table 7.
Binding affinities of truncated PAMAP53 peptides to hPg
| Proteins | T(° C) | kon (×105 Ms−1) | koff (×10−4s−1) | Kda (nM) |
|---|---|---|---|---|
| PAMAP53_short | 25 | 4.0 ± 0.7 | 7.7 ± 1.6 | 1.9 ± 0.2 |
| 37 | 4.1 ± 0.1 | 43 ± 2 | 11 ± 1 | |
| PAMAP53_medium | 25 | 2.3 ± 0.1 | 5.4 ± 0.9 | 2.4 ± 0.4 |
| 37 | 6.5 ± 0.3 | 103 ± 17 | 16 ± 3 | |
| PAMAP53_long | 25 | 0.9 ± 0.4 | 1.3 ± 0.1 | 2.0 ± 0.7 |
| 37 | 1.2 ± 0.1 | 43 ± 2 | 37 ± 2 |
Dissociation constants (Kd) were calculated from koff/kon. All data were collected from triplicate runs and are presented as the mean ± S.D for each kinetic value.
3.6. α-helical contents in AGL55NS88.2 and KTI55SS1448 affect the hPg-binding affinity
NMR solution structures of AGL55NS88.2 and KTI55SS1448 demonstrated that their a2-repeats contain helices, although the RH binding sites were shown to break the local helix (Qiu et al., 2018). We hypothesized that gain or loss of α-helical content in the a-repeats may impact the hPg-binding patterns. Since we know from the current study that the α-helical content of PAMs greatly depend on temperature, we determined the Kd values for hPg-binding to PAM-derived peptides at different temperatures. For AGL55NS88.2 and KTI55SS1448, each with a single a-repeat, both kinetic and steady state analyses show a substantial decrease in the binding affinity as the temperature is elevated from 15° C to 37° C (Fig. 5A, B, D, E; Table 6). The Kd values at a given temperature were consistent regardless of whether kinetic or steady state analyses were employed to obtain the data. At 37° C, dissociation rates were again too rapid for accurate calculations of the rate constants and the resultant Kd values. Thus, only steady state binding analyses was employed at this higher temperature.
Fig. 5. Binding assays of PAM-derived peptides to hPg at 15° C and 37° C.
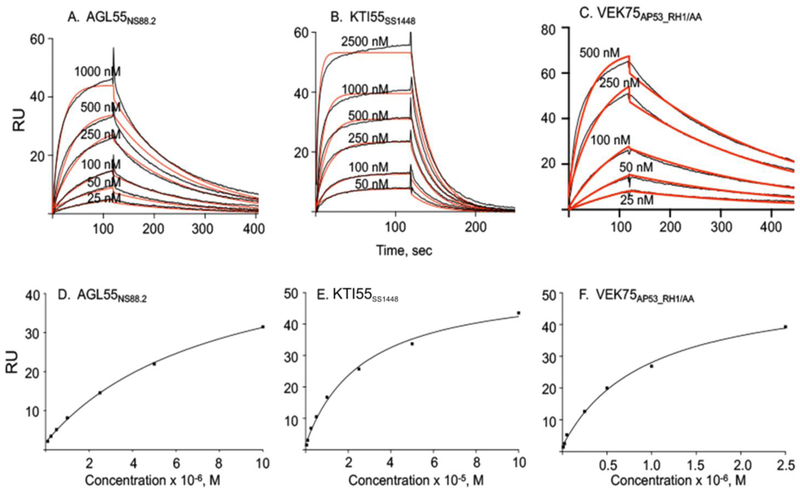
hPg was immobilized on a CM-5 chip and peptide concentrations used for the titrations are indicated in each panel. Kinetic analyses were performed at 15° C for: (A) AGL55NS88.2, (B) KTI55SS1448, (C) VEK75AP53_RHI/AA. Steady state analyses were performed at 37° C for (D) AGL55NS88.2, (E) KTI55SS1448, (F) VEK75AP53_RH1/AA. In kinetic analyses, the experimental data are shown in black lines and fitted curves are shown in red lines. The corresponding binding constants from both kinetic and affinity analyses are provided in Table 6.
VEK75AP53 contains both the a1 and a2 hPg binding sites. Very tight binding is observed for VEK75AP53 to hPg at 25° C, with a Kd value of <1 nM from kinetic analysis (Table 6). The Kd values at 4° C were not determined for the VEK75AP53/hPg complex since the binding would only become stronger and preclude reliable analyses. When the hPg binding site in a1 was inactivated in VEK75AP53 by mutagenesis, the resultant VEK75AP53_RH/AA, which now resembled AGL55NS88.2 and KTI55SS1448 in terms of hPg binding sites, demonstrated an intermediate binding affinity to hPg between VEK75AP53 and AGL55NS88.2/KTI55SS1448 (Fig. 5C, F; Table 6) at various temperatures. Thus, maintenance of both a1 and a2 hPg binding domains is required for optimal strong receptor-type binding of hPg to PAM, even though one of RH-motifs is replaced.
4. Discussion
4.1. Dimeric PAMs switch between open and closed conformations at different temperatures
Some members of the M-like protein family, e.g., protein H, as well as M1-Prt itself, in other strains, have been shown to contain a lower level of α-helices at 37° C than at 25° C (Akerström et al., 1992; Cedervall et al., 1997; Nilson et al., 1995). This loss of their secondary structures, as with the PAMs in our study, causes the subsequent dissociation of dimers into monomers. Meanwhile, it has been found that GAS surface protein H binds to IgG more weakly at 37° C than at 25° C, as is also the case of binding of human fibrinogen to another M-Prt, M1 (Nilson et al., 1995). Such phenomena can be attributed to the fact that unstructured monomers fail to maintain α-helices in the plasma protein binding domain, e.g., B-repeats in M1-Prt, which are required to retain side-chain residues in an optimal orientation for binding. However, the binding of these M and M-like proteins to corresponding plasma proteins exhibit allosteric features (Cedervall et al., 1995). For example, when human serum albumin (HSA) was bound to the C-domain of M1-Prt, fibrinogen binding at the B-domain was enhanced. It appears that HSA binding to the C-domain facilitates maintenance of adequate α-helices for binding of fibrinogen to the B-domain.
It has been determined that the c-repeats within the C-domain are the primary contributors to PAM dimerization. However, we have found that the HVRs from different PAMs retain common structural features, one of which is the scattered hydrophobic residues found at positions a and d in heptad registers (Qiu et al., 2018). AUC data for PAMAP53_short at 4° C provides evidence that Leu, Ile, and Val residues at positions a and d in HVR heptads have the potential to form short coiled-coil dimers. A further example, which also demonstrates the role of the HVR in PAM dimerization, originates from a previous report on the properties of a different truncated PAMAP53 variant, VEK64AP53 (residues 83-145) (Bhattacharya et al., 2014). This 64-amino acid residue peptide contains 20-amino acid residues (83-102) from the COOH-terminal region of the HVR, followed by the a1a2-repeats, and 13 residues of the NH2-terminus of the B-domain. From AUC equilibrium assays, the VEK64AP53 molecular mass of 15,100 Da clearly shows that it is dimeric at 20° C (Bhattacharya et al., 2014). This proves that hydrophobic residues in the HVR are able to form a dimer under certain conditions. However, the longer analogues of VEK64AP53, i.e., VEK75AP53 and PAMAP53_short, are monomers (Qiu et al., 2018). As demonstrated from previous NMR studies, most of the B-domain (D145-Q171) in PAMAP53 is a long flexible loop (Qiu et al., 2018). Additionally, the NH2-terminus of the HVR is unstructured. Thus, when the N- and C-termini of VEK64AP53 are extended to contain the entire HVR and/or B-domain, the dimerization potential of the HVR is not sufficiently strong to counterbalance the destablizing factors, i.e., mobile loops from the A-domain, the NH2-terminus of the HVR, and the COOH-terminus of the B-domain in PAMAP53. Thus, coiled-coil dimers are destabilized by these flexible regions, and the truncated peptides primarily exist as monomers, as seen from the AUC data of VEK75AP53 and PAMAP53_short.
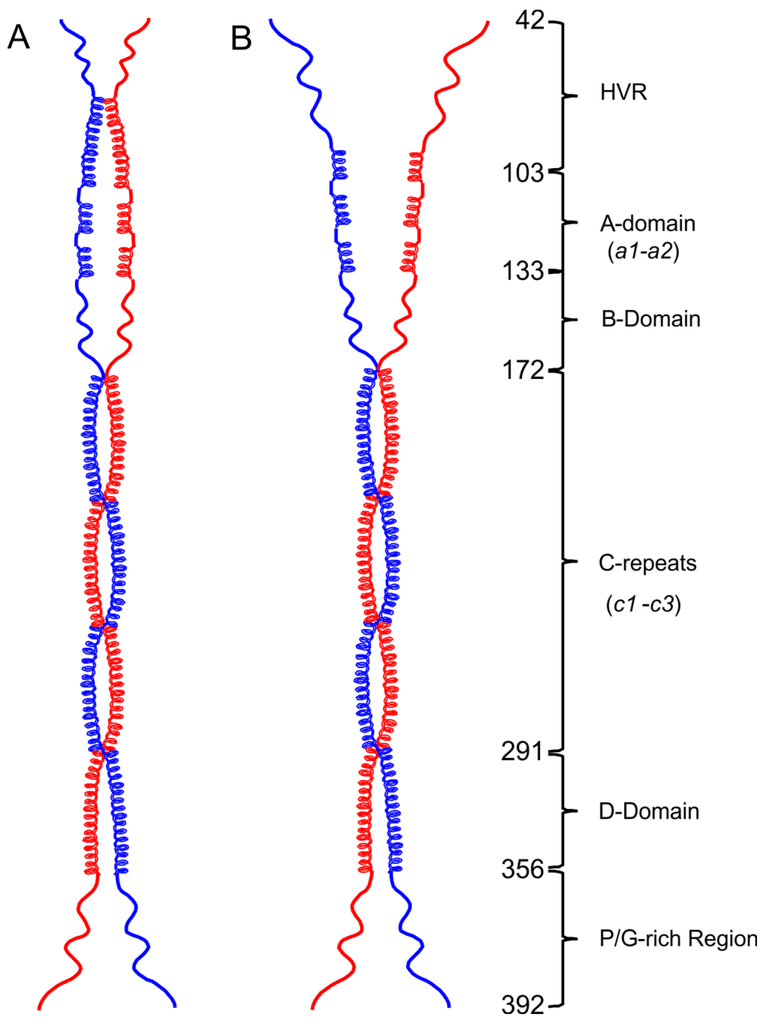
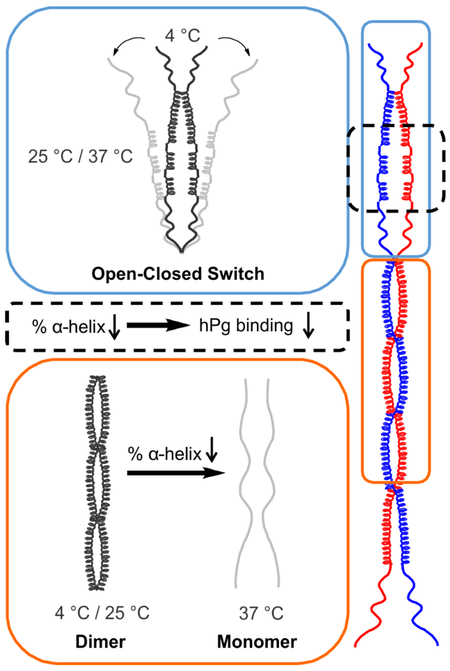
Nearly 30% of PAMAP53_short structures are dimers at 4° C, while at 25° C no dimeric species are observed in solution. Meanwhile, the percentage of α-helix decreases by ~40% from 4° C to 25° C. Higher α-helical contents would be necessary to form helical dimeric coiled-coils. When the α-helical fraction significantly decreases, PAM fails to form these types of dimers. At 25° C or 37° C, a significant loss of α-helices results in dimer dissociation at the N-terminal domains, i.e., the HVR-A-B domains. Thus, the PAM structural model proposed has distinct conformations at the NH2-terminus at different temperatures, i.e., closed at 4° C (Fig. 6A), but unwound and open at 25° C/37° C (Fig. 6B). We conclude that in solution PAMs are composed of structured dimeric portions as shown, along with less structured monomeric fractions at both 25° C and 37° C. Since monomeric species of PAM possess such low α-helical fractions, especially at 37° C, the proteins of such states fail to form helical coiled-coil dimers.
Fig. 6. Open and closed conformations of the PAM structural model.
Dimeric models were drawn to scale in ChemDraw Professional 16.0 based upon the domain organization of PAMAP53. (A) At 4° C, the HVR with some α-helix content dimerizes and forms a closed pattern at the NH2-terminus. (B) At 25° C or 37° C, loss of α-helices within the HVR regions occur, eventually resulting in an open status of NH2-terminal domains, viz., HVR-A-B domains. The numbers of the first residue in each domain are listed on the illustration.
4.2. The a1-repeat protects the secondary structure of the a2-repeat
Previous NMR studies from our group (Wang et al., 2010a; Wang et al., 2010b) have dissected both the structure and dynamics of the complex of K2hpg and VEK30AP53, a 30-residue peptide that contains a short C-terminal region of 6-residues of the HVR (V97-A102) followed by the entire a1-repeat and partial a2-repeat. Numerous interactions make up the binding interface observed between these two moieties. Aromatic residues constitute a hydrophobic groove traversing the interface of the complex, which accommodates residues with hydrophobic side chains in VEK30AP53. The major specific ligand-receptor side-chain interactions are composed of R69 in the COOH-terminus of K2hPg, which serves as a cationic locus for E16 and E20 in VEK30AP53, and D54 and E56 of K2hPg which interact with R17 and H18 (RH1) in VEK30AP53 (Rios-Steiner et al., 2001; Sanderson-Smith et al., 2006). Due to the lack of the RH2-motif in the VEK30AP53, interactions between the a2-repeat-containing region and K2hPg are absent (Wang et al., 2010a; Wang et al., 2010b). However, considering the high homology between a1- and a2-repeats in PAMAP53, the residues in each tandem repeat involved in its binding to K2hPg are likely similar (Yuan et al., 2017).
Our strategy of performing SPR experiments consists of immobilizing hPg on the chip and injecting various concentrations of PAMs or peptides. Therefore, considering the PAM monomer-dimer equilibrium, the Kd values observed using this strategy reflects an average hPg-binding affinity of monomeric and dimeric states in solution. At 25° C, each full-length PAM exists as a dimer, and thus shows small differences in binding to hPg. However, at 37° C, when less structured monomers predominate in most PAMs, the Class II PAMs devoid of the a1-repeat and Class I/III PAMs containing complete a1a2-repeats, diverge in binding affinities to hPg. Specifically, monomeric PAMNS88.2 and PAMSS1448 exhibit lower association rates and consequently bind to hPg weaker than other monomeric PAMs. Accordingly, although either the a1- or a2-repeat is able to interact with one K2hPg domain in a monomeric species, the coexistence of a1- and a2-repeats allow optimal nM-magnitude hPg binding.
This finding is demonstrated more clearly with PAM-derived peptides. PAMAP53_short, VEK75AP53 (Class I), and VKK38NS455 (Class III) are monomeric at 25° C, but these peptides bind tightly to hPg. NMR solution structures of VEK75AP53 and VKK38NS455 demonstrate significant α-helices in both of the a1- and a2-repeats (Qiu et al., 2018; Yuan et al., 2017). Further, VEK75AP53 also shows an α-helix at the NH2-terminus of the B-domain (E133-A144 in the PAMAP53 sequence) (Qiu et al., 2018). In this manner, the secondary structure in the a2-repeat is protected by a-helices in the a1-repeat and B-domain. But the α-helix of the a1-repeat may be less stable due to directly connecting to unstructured HVR, specifically at temperatures above 25° C. Thus, coexistence of two tandem repeats stabilizes the secondary structure of a2-repeat, allowing tighter binding to hPg.
In AGL55NSS8.2 and KTI55SS1448 (Class II), the a2-repeat directly connects to their mobile HVRs and the helix is less stable. Loss of α-helices in the a2-repeat becomes more obvious when the temperature is raised to 37° C. At this temperature, the single RH-motif retains its specificity for hPg, but other residues in the a2-repeat, including those that are charged and apolar, are not ideally oriented for binding. A structural distortion of the a2-repeat weakens its interactions with hPg. Likewise, ~50% of dimeric PAMNS88.2 and PAMSS1448 dissociates into monomers at 37° C accompanying large reductions in α-helices. As a result, these two PAMs show slower binding (lower kon values) to hPg.
Differences in hPg-binding between AGL55NS88.2/KTI55SS1448 and VEK75AP53_RH1/AA further proved that loss of the a1-repeat has disparate impacts on the binding to hPg, compared to the mutation of RH1-motif. These three peptides only contain a single RH2-binding motif, but VEK75AP53_RH1/AA exhibits a Kd ~100 times lower than AGL55NS88.2/KTI55SS1448 at all temperatures examined. Although the RH1 of VEK75AP53_RH1/AA is replaced by two alanine residues, preservation of the helical a1-repeat is still able to stabilize the α-helix in the downstream a2-repeat and this peptide achieves tight binding.
Although residuals of data points in the SPR experiments in each fitted curve are very small throughout the entire binding process, we note that there are still some deviations between fitted curves and raw data in either association or dissociation (Fig. 3A, 4A, 4D, 5A-C). These discrepancies, while minoir, may nonetheless arise from non-specific contacts between hPg and PAM. It is universally accepted that K2hPg and PAM a-repeat(s) are the specific binding domains in each moiety of the complex. However, due to the absence of a hPg-PAM complex structure from X-ray crystallography, it is unclear whether other domains in hPg and/or PAM have some minor interactions. Considering the size of these two proteins (hPg, ~90 kDa; PAM, ~40kDa), non-specific, but weak, contacts possibly exist, leading to a non-ideal 1:1 binding and resultant deviations. For Class I/III PAMs or derivative peptides, another factor that may cause these deviations is diversity in the hPg-binding ability between a1- and a2-repeat (Fig. 3A, 4A, 4D). Despite high homology of a1- and a2-repeat, a few amino acid substitutions still exist in these tandem repeats. Further, a1-repeat is, as discussed, less structured than a2-repeat, which makes orientations of relevant residues unfavorable for binding. Thus, the hPg-binding capacities of a1- and a2-repeat are different. The consequent average hPg-binding affinity is an intermediate value between that of a1- and a2-repeat. This would be another possibility that makes the 1:1 binding somewhat non-ideal. Nevertheless, given that hPg-binding patterns between Class II and Class I/III PAMs are diverged, these minor deviations in SPR fitted curves do not influence the validity of our conclusions or the overall message of this communication, but provide an avenue to interpret the potential complexity behind the hPg-PAM binding.
At this time, GAS strains expressing PAM with two a-repeats are predominant in the population, compared to those expressing PAM with only one a-repeat. Based on this study, it can be explained as the a2-repeat can be maintained in a robust α-helix when the a1-repeat exists. However, it would be redundant for the bacteria to synthesize three or even more a-repeats at the expense of energy and materials, given that two a-repeats in PAM are sufficient for tightly binding to hPg. Thus, even if a GAS strain expressing more than two a-repeats evolved at a certain time point, such cells likely would be unfavorable during natural selection.
5. Conclusions
In this study, we have advanced understanding of PAM secondary and quaternary structures at a biologically relevant temperature of 37° C. We also have established a more refined model that describes the conformational changes of PAMs at different temperatures, and the consequence of these conformations to biologically relevant hPg binding. At 37° C, in spite of dissociation of dimeric PAMs into monomers, PAMs containing intact a1a2-repeats still show nM-scale tight binding to hPg. Contrarily, dimer dissociation has more significant impact on the binding of hPg to Class II PAMs that only encompass one a-repeat in the sequence. This considerable difference corroborates the value of the coexistence of two tandem a-repeats in the A-domain of a PAM protein.
Highlights.
PAMs specifically expressed by Pattern D GAS strains are non-ideal coiled-coil dimers, of which the secondary structure is greatly affected by temperature changes.
Loss of α-helices at 37° C results in the dissociation of most PAM dimers.
Low temperature (4° C) induces an increase in α-helical contents, and thus a closed and partial dimeric conformation at the NH2-terminal region of PAMs.
Dimerization facilitates Class II PAMs to maintain a robust α-helix in their A-domain, leading to a tight hPg binding site.
In the cases of monomeric PAM-derived peptides, only one a-repeat in the A-domain is insufficient to achieve tight binding to hPg.
6.
Funding
This work was supported by National Institutes of Health Grant HL013423.
Footnotes
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Akerström B, Lindah G, Björck L, Lindqvist A, 1992. Protein Arp and protein H from group A streptococci. Ig binding and dimerization are regulated by temperature. J. Immunol 148, 3238–3243. [PubMed] [Google Scholar]
- Babul J, Stellwagen E, 1969. Measurement of protein concentration with interference optics. Anal. Biochem 28, 216–221. [DOI] [PubMed] [Google Scholar]
- Bao YJ, Shapiro BJ, Lee SW, Ploplis VA, Castellino FJ, 2016. Phenotypic differentiation of Streptococcus pyogenes populations is induced by recombination-driven gene-specific sweeps. Sci. Rep 6, 36644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berge A, Sjobring U, 1993. PAM, a novel plasminogen-binding protein from Streptococcus pyogenes. J. Biol. Chem 268, 25417–25424. [PubMed] [Google Scholar]
- Berge A, Björck L, 1995. Streptococcal cysteine proteinase releases biologically active fragments of streptococcal surface proteins. J. Biol. Chem 270, 9862–9867. [DOI] [PubMed] [Google Scholar]
- Bhattacharya S, Liang Z, Quek AJ, Ploplis VA, Law R, Castellino FJ, 2014. Dimerization is not a determining factor for functional high affinity human plasminogen binding by the group A streptococcal virulence factor PAM and is mediated by specific residues within the PAM a1a2 domain. J. Biol. Chem 289, 21684–21693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carapetis JR, Steer AC, Mulholland EK, Weber M, 2005. The global burden of group A streptococcal diseases. Lancet Infect. Dis 5, 685–694. [DOI] [PubMed] [Google Scholar]
- Castellino FJ, Ploplis VA, 2003. Human plasminogen: structure, activation, and function Plasminogen structure, activation, and regulation Kluwer Academic/Plenum Publishers, 3–17. [Google Scholar]
- Cedervall T, Johansson MU, Akerström B, 1997. Coiled-coil structure of group A streptococcal M proteins. Different temperature stability of class A and C proteins by hydrophobic-nonhydrophobic amino acid substitutions at heptad positions a and d. Biochemistry 36, 4987–4994. [DOI] [PubMed] [Google Scholar]
- Cedervall T, Akesson P, Stenberg L, Herrmann A, Akerstrom B, 1995. Allosteric and temperature effects on the plasma protein binding by streptococcal M protein family members. Scand. J. Immunol 42, 433–441. [DOI] [PubMed] [Google Scholar]
- Chandrahas V, Glinton K, Liang Z, Donahue DL, Ploplis VA, Castellino FJ, 2015. Direct host plasminogen binding to bacterial surface M-protein in Pattern D strains of Streptococcus pyogenes is required for activation by its natural coinherited SK2b protein. J. Biol. Chem 290, 18833–18842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YH, Yang JT, Martinez HM, 1972. Determination of the secondary structures of proteins by circular dichroism and optical rotatory dispersion. Biochemistry 11, 4120–4131. [DOI] [PubMed] [Google Scholar]
- Facklam R, Beall B, Efstratiou A, Fischetti V, Johnson D, Kaplan E, Kriz P, Lovgren M, Martin D, Schwartz B, Totolian A, Bessen D, Hollingshead S, Rubin F, Scott J, T. G, 1999. emm typing and validation of provisional M types for group A streptococci. Emerg. Infect. Dis 5, 247–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischetti VA, 1989. Streptococcal M protein: molecular design and biological behavior. Clin. Microbiol. Rev 2, 285–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischetti VA, Parry DA, Trus BL, Hollingshead SK, Scott JR, Manjula BN, 1988. Conformational characteristics of the complete sequence of group A streptococcal M6 protein. Proteins 3, 60–69. [DOI] [PubMed] [Google Scholar]
- Fulde M, Steinert M, Bergmann S, 2013. Interaction of streptococcal plasminogen binding proteins with the host fibrinolytic system. Front. Cell. Infect. Microbiol 3, 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glinton K, Beck J, Liang Z, Qiu C, Lee SW, Ploplis VA, Castellino FJ, 2017. Variable region in streptococcal M-proteins provides stable binding with host fibrinogen for plasminogen-mediated bacterial invasion. J. Biol. Chem 292, 6775–6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenfield NJ, 2006. Using circular dichroism spectra to estimate protein secondary structure. Nat. Protoc 1, 2876–2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McArthur JD, McKay FC, Ramachandran V, Shyam P, Cork AJ, Sanderson-Smith ML, Cole JN, Ringdahl U, Sjobring U, Ranson M, Walker MJ, 2008. Allelic variants of streptokinase from Streptococcus pyogenes display functional differences in plasminogen activation. FASEB J. 22, 3146–3153. [DOI] [PubMed] [Google Scholar]
- McKay FC, McArthur JD, Sanderson-Smith ML, Gardam S, Currie BJ, Sriprakash KS, Fagan PK, Towers RJ, Batzloff MR, Chhatwal GS, Ranson M, Walker MJ, 2003. Plasminogen binding by Group A streptococcal isolates from a region of hyperendemicity for streptococcal skin infection and a highincidence of invasive Iinfection. Infect. Immunol. 72, 364–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara C, Zinkernagel AS, Macheboeuf P, Cunningham MW, Nizet V, Ghosh P, 2008. Coiled-coil irregularities and instabilities in group A Streptococcus M1 are required for virulence. Science 319, 1405–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilson BH, Frick IM, Akesson P, Forsen S, Bjorck L, Akerstrom B, Wikstrom M, 1995. Structure and stability of protein H and the M1 protein from Streptococcus pyogenes. Implications for other surface proteins of gram-positive bacteria. Biochemistry 34, 13688–13698. [DOI] [PubMed] [Google Scholar]
- Påhlman L, Olin A, Darenberg J, Mörgelin M, Kotb M, Herwald H, Norrby-Teglund A, 2008. Soluble M1 protein of Streptococcus pyogenes triggers potent T cell activation. Cell. Microbiol 10, 404–414. [DOI] [PubMed] [Google Scholar]
- Phillips GN, Flicker PF, Cohen C, Manjula BN, Fischetti VA, 1981a. Streptococcal M protein: alpha-helical coiled-coil structure and arrangement on the cell surface. Proc. Natl. Acad. Sci. USA 78, 4689–4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips GN Jr., Flicker PF, Cohen C, Manjula BN, Fischetti VA, 1981b. Streptococcal M protein: alpha-helical coiled-coil structure and arrangement on the cell surface. Proc Natl Acad Sci U S A 78, 4689–4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plow EF, Herren T, Redlitz A, Miles LA, Hooverplow JL, 1995. The cell biology of the plasminogen system. FASEB J. 9, 939–945. [DOI] [PubMed] [Google Scholar]
- Qiu C, Yuan Y, Zajicek J, Liang Z, Balsara RD, Brito-Robionson T, Lee SW, Ploplis VA, Castellino FJ, 2018. Contributions of different modules of the plasminogen-binding Streptococcus pyogenes M-protein that mediate its functional dimerization. J. Struct. Biol 204, 151–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raz A, Talay SR, Fischetti VA, 2012. Cellular aspects of the distinct M protein and SfbI anchoring pathways in Streptococcus pyogenes. Mol. Microbiol 84, 631–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rios-Steiner JL, Schenone M, Mochalkin I, Tulinsky A, Castellino FJ, 2001. Structure and binding determinants of the recombinant kringle-2 domain of human plasminogen to an internal peptide from a group A Streptococcal surface protein. J. Mol. Biol 308, 705–719. [DOI] [PubMed] [Google Scholar]
- Sanderson-Smith ML, Walker MJ, Ranson M, 2006. The maintenance of high affinity plasminogen binding by group A streptococcal plasminogen-binding M-like protein is mediated by arginine and histidine residues within the a1 and a2 repeat domains. J. Biol. Chem 281, 25965–29571. [DOI] [PubMed] [Google Scholar]
- Sanderson-Smith ML, Dinkla K, Cole JN, Cork AJ, Maamary PG, McArthur JD, Chhatwal GS, Walker MJ, 2008. M protein-mediated plasminogen binding is essential for the virulence of an invasive Streptococcus pyogenes isolate. FASEB J. 22, 2715–2722. [DOI] [PubMed] [Google Scholar]
- Schick LA, Castellino FJ, 1974. Direct evidence for the generation of an active site in the plasminogen moiety of the streptokinase-human plasminogen activator complex. Biochem Biophys Res Commun 57, 47–54. [DOI] [PubMed] [Google Scholar]
- Smeesters PR, McMillan DJ, Sriprakash KS, 2010. The streptococcal M protein: a highly versatile molecule. Trends Microbiol 18, 275–282. [DOI] [PubMed] [Google Scholar]
- Stewart CM, Buffalo CZ, Valderrama JA, Henningham A, Cole JN, Nizet V, Ghosh P, 2016. Coiled-coil destabilizing residues in the group A Streptococcus M1 protein are required for functional interaction. Proc. Natl. Acad. Sci. USA 113, 9515–9520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumitomo T, Nakata M, Higashino M, Terao Y, Kawabata S, 2013. Group A streptococcal cysteine protease cleaves epithelial junctions and contributes to bacterial translocation. J. Biol. Chem 288, 13317–13324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumitomo T, Nakata M, Higashino M, Yamaguchi M, Kawabata S, 2016. Group A Streptococcus exploits human plasminogen for bacterial translocation across epithelial barrier via tricellular tight junctions. Sci Rep 7, 20069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veltri T, de Oliveira GAP, Bienkiewicz EA, Palhano FL, Marques MA, Moraes AH, Silva JL, Sorenson MM, Pinto JR, 2017. Amide hydrogens reveal a temperature-dependent structural transition that enhances site-II Ca(2+)-binding affinity in a C-domain mutant of cardiac troponin C. Sci. Rep 7, 691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vistica J, Dam J, Balbo A, Yikilmaz E, Mariuzza RA, Rouault TA, Schuck P, 2004. Sedimentation equilibrium analysis of protein interactions with global implicit mass conservation constraints and systematic noise decomposition. Anal. Biochem 326, 234–256. [DOI] [PubMed] [Google Scholar]
- Walker MJ, Barnett TC, McArthur JD, Cole JN, Gillen CM, Henningham A, Sriprakash KS, Sanderson-Smith ML, Nizet V, 2014. Disease manifestations and pathogenic mechanisms of Group A Streptococcus. Clin. Microbiol. Rev 27, 264–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Prorok M, Castellino FJ, 2010a. NMR backbone dynamics of VEK-30 bound to the human plasminogen kringle 2 domain. Biophys. J. 99, 302–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Zajicek J, Geiger JH, Prorok M, Castellino FJ, 2010b. Solution structure of the complex of VEK-30 and plasminogen kringle 2. J. Struct. Biol 169, 349–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward PN, Leigh JA, 2002. Characterization of PauB, a novel broad-spectrum plasminogen activator from Streptococcus uberis. J. Bacteriol 184, 119–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y, Zajicek J, Qiu C, Chandrahas V, Lee SW, Ploplis VA, Castellino FJ, 2017. Conformationally organized lysine isosteres in Streptococcus pyogenes M protein mediate direct high-affinity binding to human plasminogen. J. Biol. Chem 292, 15016–15027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liang Z, Hsieh HT, Ploplis VA, Castellino FJ, 2012. Characterization of streptokinases from group A Streptococci reveals a strong functional relationship that supports the coinheritance of plasminogen-binding M protein and cluster 2b streptokinase. J. Biol. Chem 287, 42093–42103. [DOI] [PMC free article] [PubMed] [Google Scholar]