Abstract
Type-II single-span membrane proteins, such as CadC or RodZ, lacking a signal-sequence and having a far-downstream hydrophobic segment, require the SecA secretion motor for insertion into the inner membrane of Escherichia coli. Using two chimeric single-span proteins containing a designed hydrophobic segment H, we have determined the requirements for SecA-mediated secretion, the molecular distinction between TM domains and signal peptides, and the propensity for hydrophobic H-segments to remain embedded within the bilayer after targeting. By means of engineered H-segments and a strategically placed SPase I cleavage site, we determined how targeting and stability of the chimeric proteins are affected by the length and hydrophobicity of the H-segment. Very hydrophobic segments (e.g. 16Leu) are stably incorporated into the inner membrane, resulting in a C-terminal anchored membrane protein, while a 24L construct was not targeted to the membrane by SecA and remained in the cytoplasm. However, a construct carrying preMalE at the N-terminus led to SecA targeting to SecYEG via the native signal sequence and stable insertion of the downstream 24L H-segment. We show that the RseP intramembrane protease degrades weakly stable H-segments and is a useful tool for investigating the borderline between stable and unstable TM segments. Using RseP− cells, we find that moderately hydrophobic sequences (e.g. 5Leu+11Ala) are targeted to SecYEG by SecA and inserted, but subsequently drop out of the membrane into the cytoplasm. Therefore, the free energy of transfer from translocon to bilayer is different from the transfer free energy from membrane to water.
Keywords: single-span membrane proteins, protein targeting, membrane protein stability, RseP intramembrane protease, protein secretion
Introduction
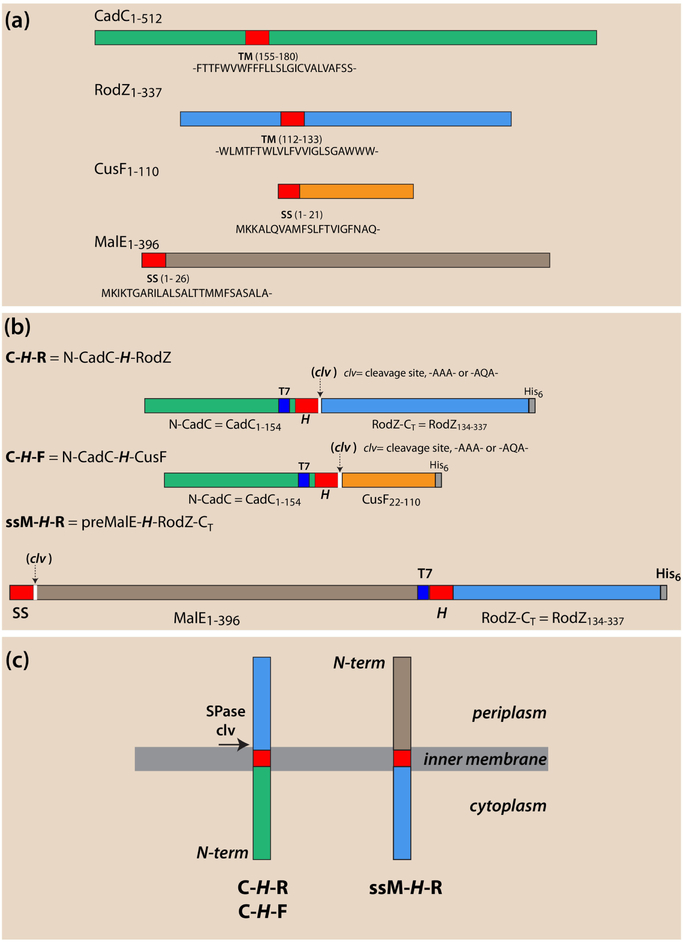
CadC or RodZ (Figure 1a) are unusual Type-II (Nin-Cout) single-span membrane proteins (MPs), because they lack an N-terminal signal-sequence (S-S) and have a far-downstream transmembrane (TM) segment. We have shown that both proteins require the SecA secretion motor for targeting and insertion into the inner membrane of Escherichia coli [1, 2]. CadC activates the cadBA operon during low-pH stress [3–5] while RodZ, plays an important role in the maintenance of the rod shape of Escherichia coli [6, 7]. Targeting and TM insertion are determined solely by hydrophobic segments that are more than 100 residues downstream from the N-terminus (over 150 residues for CadC and 100 residues for RodZ). In contrast, most Type-II MPs have their signal-anchor sequences at, or very close to, the N-terminus.
Figure 1.
Schematic overview of single-span proteins and chimeras used in this study. Locations of transmembrane (TM) segments [H-segments] or signal sequences (ss) are colored red. Location of T7 immuno tags are colored blue, His6 tags gray, and cleavage sites white. CadC and RodZ non-TM domains are colored green and blue, respectively, CusF orange, and MalE brown. (a) CadC or RodZ are unusual Type-II (Nin-Cout) single-span membrane proteins (MPs) that lack an N-terminal signal sequence (ss) and have a far-downstream TM segment. CadC activates the CadBA operon during low-pH stress[3–5] while RodZ, plays an important role in the maintenance of the rod shape of Escherichia coli [6, 7]. CusF, is the periplasmic copper chaperone of the E. coli CusCFBA copper-transporting efflux system[14] and MalE is the well-known maltose binding protein [17]. Both have N-terminal signal sequences (ss) indicated in red. (b) Chimeric proteins composed of foldable fragments of the proteins in panel a. Spase I cleavage sites (clv) are inserted immediately following the H-segment except for the ssM-H-R construct that carries a native cleavable signal sequence. Proteins cleaved by SPase I indicate that the protein has entered the SecA pathway and passed through the SecYEG translocon. (c) Schematic overview of the topology of the proteins in panel b after SecA targeting and SecYEG membrane incorporation.
In an earlier study of the dimerization of the CadC sensor domain [1], which is required for activating the cadBA operon, we developed a tripartite single-span chimera CadC-H-RodZ in which the periplasmic domain of RodZ replaced the periplasmic CadC sensor domain. The two domains were linked by a hydrophobic H-segment of the form GGPG-H-GPGG to serve as the single TM helix. The purpose of the GGPG/GPGG sequences was to isolate the hydrophobic TM domain from the surrounding sequence [8]. We added a signal peptidase cleavage site (clv) [9–12] of the form clv = -AXA- following the H-segment. The inclusion of a T7 tag upstream from H and a His6 tag downstream at the C-terminus (Figures 1b & 1c) allowed us to track the insertion and membrane topology of the chimera using Western blots [13].
The C-H-R chimeras have proven useful for examining SecA-dependent targeting of proteins to the SecYEG translocon for insertion. The presence of a cleavage site allowed the cellular location of the tagged fragments (periplasm, cytoplasm, or membrane) to be determined in order to verify their Nin-Cout topology. We found that targeting by SecA to the SecYEG translocon could be easily judged by whether or not SPase I cleaved the periplasmic domain. For example, SPase I cleavage of a polyleucine construct revealed that the construct was targeted and inserted into the inner membrane, because the CadC fragment was located in the membrane fraction as a C-terminal anchored membrane protein while the RodZ fragment was found in the periplasmic fraction. A polyalanine construct, on the other hand, was not cleaved and was found solely in the cytoplasm, indicating that it was not even recognized by SecA. These results suggested to us a simple means for examining in greater detail the requirements for H-segment recognition by SecA. They further suggested a means of determining H-segment stability in the membrane to answer an important question: Are there H-segments that are sufficiently non-polar to be recognized by SecA and inserted via SecYEG but not sufficiently ‘greasy’ to remain in the membrane after SPase I cleavage? Our results also suggested that we could determine the rules governing recognition of far-downstream H-segments by SecA to answer the question of whether the rules for N-terminal signal sequence recognition apply to far downstream hydrophobic segments.
We present in this paper answers to these questions obtained using the chimeras shown in Figure 1b. The C-H-R construct is the same as used earlier [1]. The C-H-F construct is similar except that the C-terminal fragment is CusF (without its natural signal sequence), which is the periplasmic copper chaperone of the E. coli CusCFBA copper-transporting efflux system [14]. Our experience so far is that the complete mature domain of almost any periplasmic protein is suitable for constructing this class of chimeras. What is important is that the protein form a stably folded domain. Early experiments revealed that fragments of exported proteins that do not form stable folds are rapidly degraded. We show below that C-H-R or C-H-F constructs with very long and hydrophobic H-segments are not recognized by SecA and therefore not targeted to SecYEG for secretion or insertion, consistent with earlier studies of signal peptides by Debra Kendall and her colleagues [15, 16]. We wished to learn whether secretable proteins carrying long and greasy downstream H-segments could nevertheless be inserted by SecYEG via the SecA pathway even if recognition of the substrate did not depend on the H-segment. For this purpose, we created the ssM-H-R construct (Figure 1b and 1c) containing the immature form of the periplasmic maltose binding protein MalE [17] (preMalE) at the N-terminus and RodZ at the C-terminus. We show below that ssM-H-R can be targeted successfully by SecA regardless of the H-segment structure. For example, as revealed by signal sequence cleavage, the H = 16 Ala construct is secreted through SecYEG while the H = 16Leu or 24Leu constructs are partitioned into the membrane by SecYEG.
Results
The fates of long and short H-segments of varying hydrophobicity
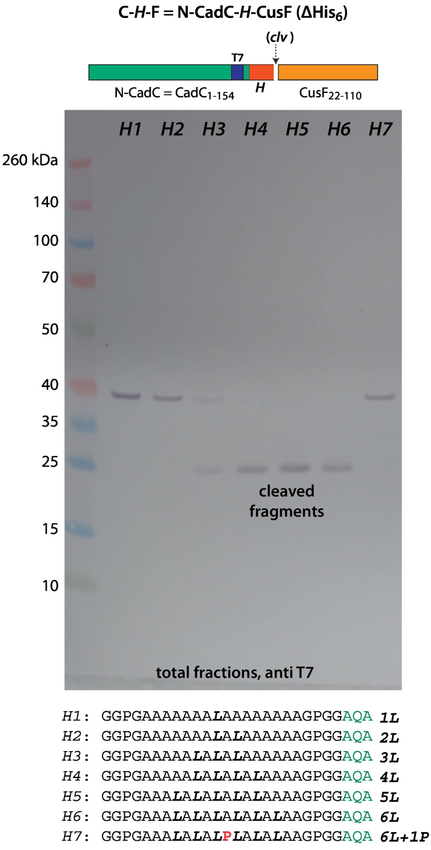
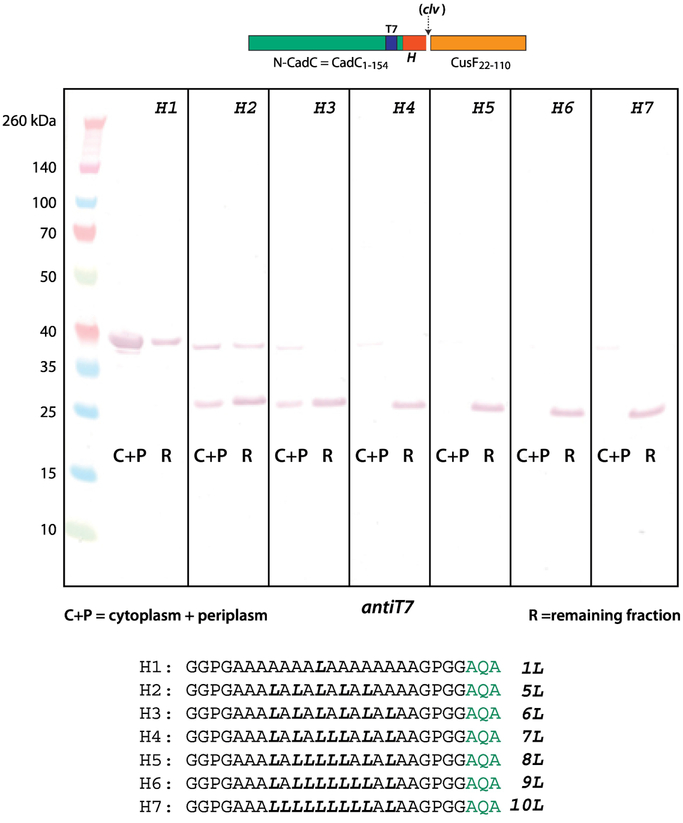
Figure 2 shows an immunoblot (T7 antibody) of C-H-F constructs expressed in E. coli BL21 cells grown in SOC media at 37° C. The H-segment contained 16 residues composed primarily of Ala and Leu ranging A15L1 to A10L6. SecA recognition and SecYEG-guided insertion of the chimera—zcleaved by SPase1—requires at least 4 leucines (lane H4) for complete insertion, but partial insertion occurs with 3 leucines (lane H3). A single Leu-to-Pro substitution at the center of the A10L6 construct to give A10P1L5 prevents recognition of the construct. These results are entirely consistent with early secretion studies of PhoA containing artificial N-terminal signal sequences [18].
Figure 2.
To be recognized by SecA, the 16 residue Ala/Leu H-segment must have 3–4 leucines in order to be recognized by SecA. This immunoblot identifies T7 tags, which are contained within the N-terminal domain carrying the H-segment. The appearance of the lower molecular weight bands indicates cleavage by SPase I and therefore processing of the construct by SecA. The seven H-segments used in the constructs are indicated. The blots show that to be recognized by SecA, the H-segment must contain at least 3 leucines (lane H3), but for complete processing 4 leucines are required (lane H4). A single Leu-to-Pro substitution at the center of the A13L6 construct to give A13P1L5 prevents recognition and secretion of by SecA. The C-H-F constructs were expressed in E. coli BL21 cells grown in SOC media at 37° C (see Materials and Methods).
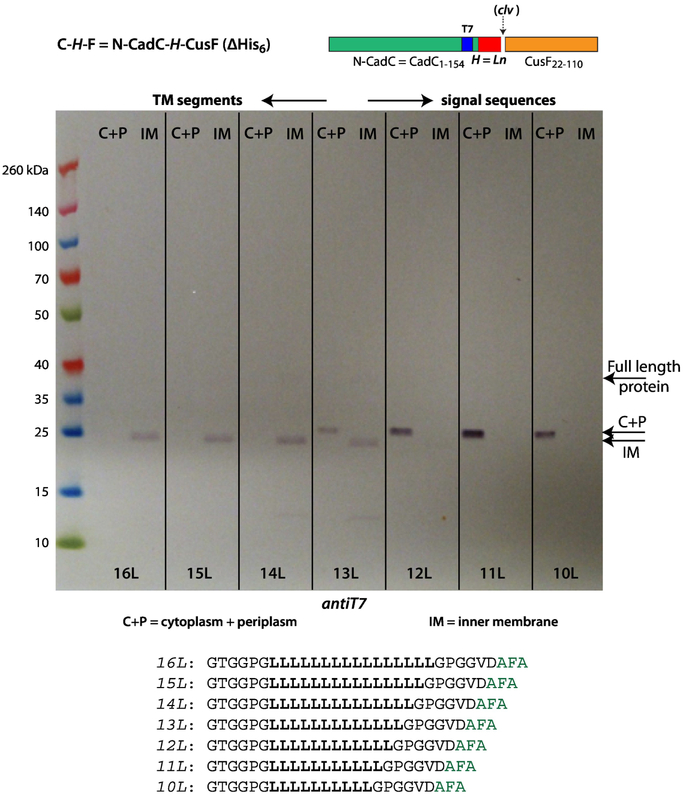
C-H-F constructs such as these could be used for exhaustive studies of signal sequence recognition by SecA, but here we are concerned with the conditions for SecA-based recognition of TM segments. Because polyleucine segments form the most stable stop-transfer sequences [16], we examined the fate of polyleucine H-segments ranging in length from 10 to 16 leucines, using anti-T7 antibodies to identify the location of cleaved fragments produced by SPase I. Figure 3 shows that H-segments shorter than 13L are not seen in the inner membrane fraction; all are located in the soluble fraction. For 13L, the fragments are found equally in the soluble and membrane fractions. All fragments containing 14 or more Leu are found exclusively in the inner membrane fraction. Figure 3 shows that the soluble and the membrane bound fragments differ in size. We hypothesized that the 10L-13L H-segment fragments are further cleaved by RseP [19, 20] and that the remaining RseP cleaved signal sequence drops out of the membrane (see Figure 4). We do not know exactly where RseP cleaves, but it seems that it discriminates between ‘signal sequences’ (10L-13L), which are attacked, and TM segments (14L-16L), which are not attacked. This suggested that RseP could be a useful indicator tool for investigating the borderline between stable or unstable TM segments.
Figure 3.
All of the polyleucine H-segments are recognized and processed by SecA, but full incorporation into the inner membrane (IM) requires 14 leucines. This suggests a translocon/membrane partitioning process occurs as the secreted protein passes through the translocon. As in Figure 2, this immunoblot identifies T7 tags, which are contained within the N-terminal domain carrying the H-segment. The appearance of the lower molecular weight bands indicates cleavage by SPase I and therefore processing of the construct by SecA. In lanes 10L to 12L, the fragments are found only in the soluble (C+P=cytoplasm plus periplasm) fraction. Beginning with 14L, all fragments are found in the IM fraction. Note the fragment molecular weight differences for lanes 14L-16L compared with lanes 10L-12L. The difference is particularly obvious in 13L, which reveals clearly the transition from the cytoplasm to inner membrane. The difference in molecular weight arises from processing by the RseP intramembrane protease (see Figure 4). The C-H-F constructs were expressed in E. coli BL21 cells grown at 37 °C in SOC media augmented with glucose, MgCl2, and MgSO4. See Materials and Methods.
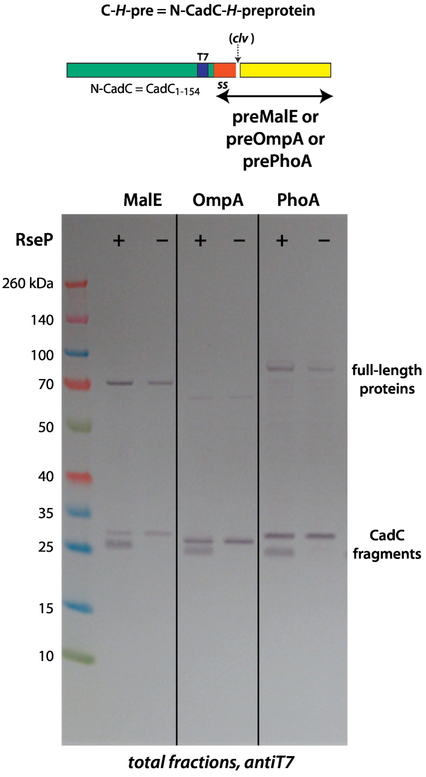
Figure 4.
N-terminal-extended native secreted proteins (MalE, OmpA, and PhoA) can serve as SecA targets. SecA targeted all constructs to the membrane, based upon SPase I cleavage. The cleaved T7-labeled fragments revealed two bands on the immunoblots resulting from cleavage by SPase I (left-hand lanes labeled RseP+). These bands arise from post-cleavage processing by the site-2 intramembrane metalloprotease RseP system [19, 20], as proven by the presence of a single-band in RseP− mutants (right-hand, single bands). These results indicate that less stable TM segments are attacked by RseP. However, RseP processes membrane embedded sequences only if they have been cleaved initially by SPase I. See Materials and Methods for descriptions of the RseP− cells.
To test further the idea that RseP recognizes signal sequences but not TM-like polyleucine segments, we examined the membrane stability of the signal sequences of MalE, OmpA, and PhoA with the CadC cytoplasmic domain as an N-terminal extension (Figure 4), which allowed us to track the signal sequence after SPase I cleavage. As expected from their similarity to the C-H-R construct, SecA targeted all of the constructs to SecYEG as determined by SPase I cleavage. The cleaved T7-labeled fragments revealed two bands on the immunoblots resulting from cleavage by SPase I (left-hand lanes labeled RseP+). These bands must arise from post-cleavage processing by the site-2 intramembrane metalloprotease RseP system [19, 20], as proven by the presence of a single-band in RseP− mutants (right-hand single bands).
Using the RseP− strain and T7-labeling of immunoblots, we looked for the location of the cleaved fragments produced in experiments such as those of Figure 2. We used C-H-R constructs containing 16 residues in the H-segment ranging from A11L5 to A6L10. Figure 5 shows the distribution of the fragments between soluble (periplasm + cytoplasm) and insoluble (membrane) fractions. As indicated by the lack of cleavage product (lane H1), the A15L1 protein is not processed. For the A11L5 construct, on the other hand, the soluble and insoluble fractions contained about equal quantities cleaved fragments. Importantly, as the number of leucines increased, there was a progressive shift of cleaved material in the soluble fraction to the insoluble fraction; virtually all of the fragments were found in the insoluble (membrane) fraction for A8L8 and beyond. We interpreted this to mean that H-segments containing fewer than about 8 leucines are not stably bound to the membrane and consequently “drop out” into the cytoplasm after cleavage. These and the results of Figure 2 suggest that in a 16-residue Leu+Ala H-segment, 4 Leu are sufficient for complete partitioning from translocon (SecYEG) to membrane while about 8 leucines are required to keep the tail-anchored fragment in the membrane.
Figure 5.
In RseP− cells, as the number of leucines in Ala/Leu segments increase from 5 leucines to 10, there is a steady shift of the fragments toward the insoluble fraction from the soluble fraction; the major break point occurred at 7–8 leucines. The RseP− condition prevents further processing of TM segments after SPase I cleavage, which reveals information about the inherent stability of TM segments. The important conclusion from these data is that the hydrophobicity requirements for partitioning a segment from translocon to membrane are different than for partitioning between membrane and the cytoplasm; 4 leucines are sufficient to guarantee translocon-to-bilayer partitioning of the H-segment (Figure 2) whereas 7 or so are required to prevent the CadC-H protein from dropping into the cytoplasm. See Material and Methods for experimental details.
The fates of very long hydrophobic H-segments
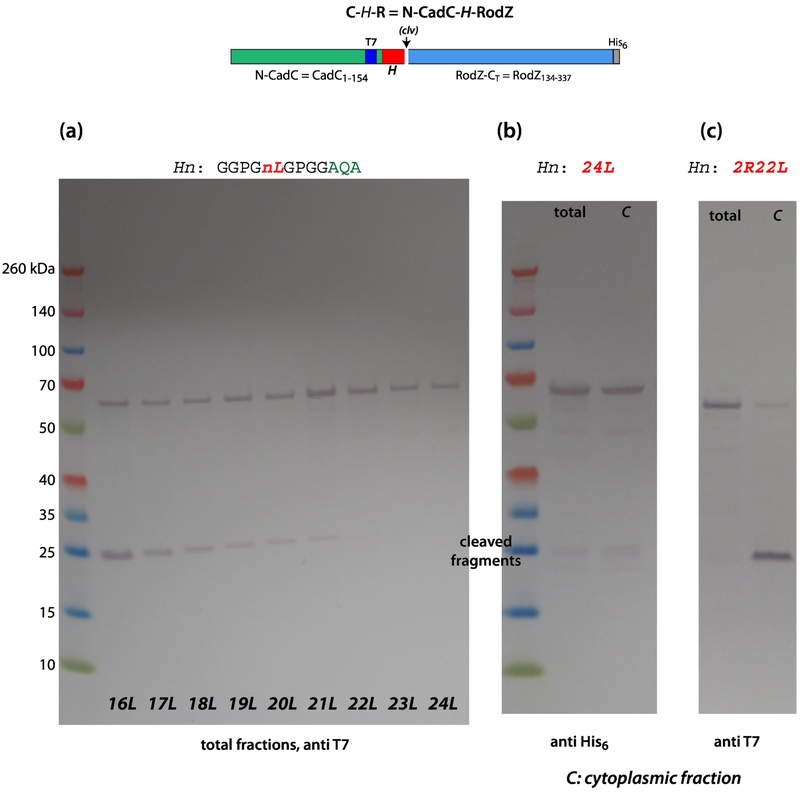
To this point, we have described experiments in which the longest Leu/Ala H-segments were 16 residues. Because it is known from studies of artificial signal sequences that polyleucine segments longer than about 20 residues are ineffective in targeting and secretion PhoA [18], we created C-H-R constructs containing polyleucine H-segments with lengths of 16 to 24 residues (Figure 6a). As expected, a 16L segment was readily targeted to SecYEG, inserted into the membrane, and cleaved by SPaseI. But as the number of leucines in the segments increased, there was steady decline in recognition by SecA as indicated by the diminishing amounts of cleaved RodZ. No cleavage products were apparent for 22 or more leucines, and it appears that SecA begins having difficulty recognizing H-segments longer than 16L. This suggested that SecA could not recognize TM segments longer than 22 residues. Figure 6c reveals, however, that the substitution of arginines for two leucines (positions 10 and 15) can ‘rescue’ the 24L construct; making the sequence less hydrophobic converts it into a SecA target. This implies that the length of the segment is not the critical issue. These very long segments are, of course, unusual and one would not expect to encounter such highly hydrophobic segments in nature. Interestingly, although we expected these constructs to produce insoluble proteins, that was not the case. As shown in Figure 6b (see also Figure S3), the C-24L-RodZ protein is found mostly in the cytoplasmic fraction. This could be because it is misfolded or is stabilized or ‘protected’ in some way by chaperones.
Figure 6.
SecA cannot identify long and highly hydrophobic H-segments. (a) These data show that SecA identification of polyleucine H-segments composed of more than 16 leucines becomes more and more difficult with increasing numbers of leucines, as indicated by the steady decline in the intensity of the cleaved fragments. For segments containing 22 or more residues, the C-H-R constructs were not processed at all. (b) Remarkably, despite the high hydrophobicity of the polyleucine H-segments, the chimeric proteins are found only in the cytoplasmic fraction rather than in inclusion bodies. This might be due to interactions with cytoplasmic chaperones. (c) These data show that replacement of two leucines in the 24Leu construct with two arginine (pos. 10 and 15) transform it to a SecA target (left lane without a cleavage site, right lane with a cleavage site). The occurrence of the cleaved fragment (right lane) indicates SecA targeting. These experiments were carried out using BL21 cells carrying a pET21 vector. Cells were grown in SOC media at 37° C. See Materials and Methods.
Because there is no insertion of the long greasy segments, it is clear that the signal recognition particle (SRP/ffH) co-translational pathway is not being utilized despite the greasiness of the H-segment. This is consistent with our earlier finding [2] that single-span membrane proteins with far downstream TM segments are recognized and inserted via SecA. The data of Figures S1 and S2, carried out using depletion strains under the control of arabinose, confirm the central importance of SecA in the insertion process. In addition, the depletion of SPase I clearly shows that the introduced cleavage site (-AXA-) is exclusively used by this protease. Notice in Figure S2, particularly, that ffH depletion had only minor effects on targeting and secretion, as we observed earlier [2] for the targeting and insertion of RodZ. Although ffH can enhance the SecA pathway for very hydrophobic signal sequences, there is little doubt that SecA is necessary and sufficient for the targeting and secretion of proteins [21]. Because the 24L construct is found only in the cytoplasm, it is extraordinarily unlikely that the SRP pathway is involved.
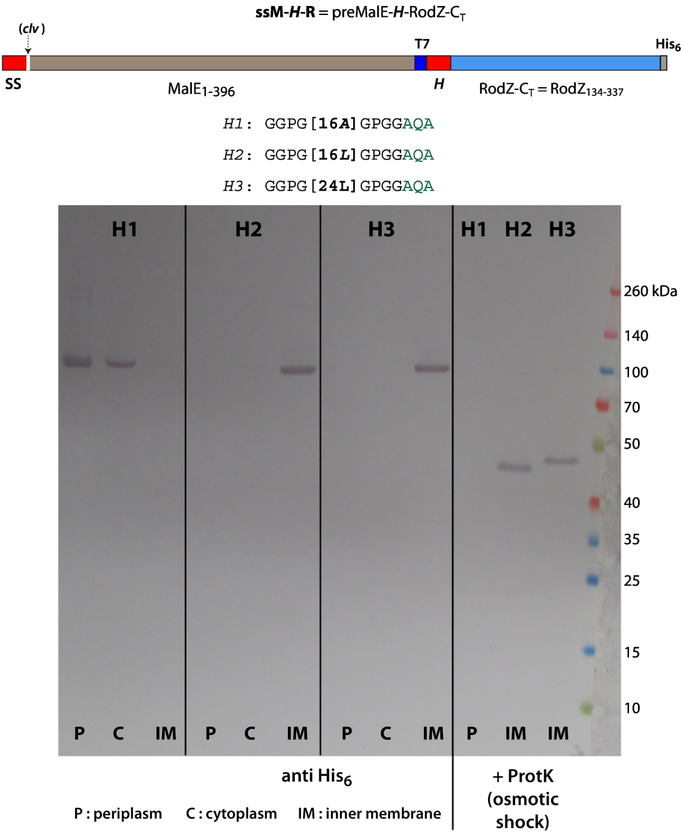
Figure 6 shows rather dramatically that SecA cannot insert C-H-R constructs across the inner membrane if the H-segments are very long and greasy. Two possibilities are that the long segments cannot bind to SecA for recognition or that, even if recognized, SecYEG is incapable of inserting them. The experiment of Figure 6 was designed to eliminate the possibility that SecYEG cannot manage the insertion of a very long H-segment (24L). We hypothesized that a natural secreted protein (ss-MalE) at the N-terminus of the construct would force the construct into the ‘regular’ SecA pathway independent of the late-occurring H-segment. For this purpose, we used the ssM-H-R construct (Figure 1b and 1c) consisting of T7-tagged preMalE and an H-segment followed by the His6-tagged C-terminal sequence of RodZ. Figure 7 shows that the 24L construct is readily inserted into the membrane as is the 16L construct, consistent with earlier work in eukaryotes using an in vitro expression system [22]. The 16A construct, however, passed through SecYEG as a secreted protein. These results are consistent with the idea presented earlier that as the ‘secreted’ protein passes through the translocon, the very greasy H-segment partitions into the membrane from the translocon. Importantly, the ssM-16LR construct contains two SecA targets (the MalE signal sequence and the 16L H-segment). We could not detect any competitor effect, which would have been indicated by the presence of two different topologies. The data show that the first-occurring signal sequence is exclusively recognized.
Figure 7.
These results, obtained using the ssM-H-R, construct show that SecYEG can insert proteins containing very long and greasy TM segments. Despite the presence of a long greasy segment far downstream from the N-terminus, SecA identifies the sequence as a target via the preMalE signal sequence at the N-terminal. If the H-segment is polyalanine, the protein is secreted and found only in the periplasmic and cytoplasmic fractions. For H = 16 and 24 Leu, the protein is found only in the inner membrane fraction, which indicates that SecYEG is capable of incorporating highly hydrophobic segments into the membrane. Osmotic shock followed by proteinase K (ProtK) treatment (right-hand lanes labeled H1, H2, H3) hydrolyzes the periplasmic MalE fragment, leaving the RodZ component as an N-terminal anchored single-span membrane protein. E. coli BL21 cells grown at 37 °C in SOC media augmented with glucose, MgCl2, and MgSO4. See Materials and Methods.
Discussion
We have explored the rules E. coli follows for targeting and secretion/insertion of model Type-II single-span membrane proteins that have a far-downstream hydrophobic segment but lack an N-terminal signal sequence (Figure 1a). The results confirm earlier work on the targeting and membrane insertion this class of single-span membrane proteins into the E. coli inner membrane by the SecA secretion motor [1, 2]. To examine more thoroughly the requirements for SecA recognition and SecYEG-guided membrane insertion, we created several chimeric proteins of the form C-H-X in which C is the cytoplasmic domain of CadC, H is a hydrophobic sequence of the form GGPG-H-GPGG [23], and X is the periplasmic domain of either RodC (R) or CusF (F) (Figure 1b and 1c). We explored initially the H-segment requirements for SecA identification and processing for 16-residue H-segments comprised of leucine and alanine residues (Figure 2). The results showed that at least four Leu leucines are required for membrane partitioning, although partial partitioning occurs with three leucines. This is consistent with earlier studies on SecA recognition of N-terminal signal sequences using PhoA as a model [18]. We conclude that the rules for identification of signal sequences by SecA is independent of location within the proteins. N-terminal extension of the signal sequence has no influence of the secreted C terminus.
Using polyleucine H-segments, we then examined the question of how many leucines are required for an H-segment to be partitioned stably into the membrane rather than being attacked by RseP. Figure 3 shows that complete membrane integration requires 14 leucines with partial integration occuring for 13 leucines. This is the border between TM segments and signal sequences. This finding is consistent with similar results obtained by Jaud et al. [24] using an in vitro eukaryotic system. Those authors established that polyleucine sequences containing 8 or fewer leucines were thermodynamically too costly to insert into the membrane as result of the extreme hydrophobic mismatch between the 40 Å-thick lipid bilayer and an 8-residue helical segment (12 Å length).
Given that SecA is responsible for targeting of our chimeric proteins to SecYEG, we wondered if SecA could process them if the H-R segments were replaced by native secreted proteins. Figure 4 shows that indeed the H-segments could be replaced by the signal sequences of MalE, OmpA, and PhoA, confirming that SecA can identify ‘normal’ N-terminal signal sequences placed downstream from the N-terminus. However, whereas processing of polyleucine H-segments by SPase I resulted in a single species of cleaved product (Figure 3), cleavage of the less hydrophobic native signal sequences resulted in two cleaved species (Figure 4). This is a result of further cleavage on the cytoplasmic membrane surface by the RseP system [19, 20], because RseP− cells produced only a single fragment (Figure 4). Unlike the more polar signal sequences, polyleucine H-segments result in very stable fragments not recognized apparently by RseP. These results provide strong support for the idea that RseP plays a major role in disposing of cleaved signal sequences [20]; cleaved fragments that are not stable in the membrane are cleaved further by RseP and consequently drop into the cytoplasm where they can be hydrolyzed by cytoplasmic enzymes.
Because RseP cleaved less stable TM segments such as N-terminal signal sequences, we examined the fate of our 16-residue Ala/Leu transmembrane segments after SecA insertion into the membrane (Figure 2). Examination of the processing of the Ala/Leu segments containing from 5 to 10 leucines, showed that fragments of segments containing 5 leucines appeared in the both the soluble and insoluble (membrane) fractions whereas segments with 10 leucines were found solely in the insoluble fraction (Figure 5). As the number of leucines was increased, there was a steady shift of the fragments toward the insoluble fraction; the major break point occurred at 7–8 leucines. This is an important result, because it shows that the hydrophobicity requirements for partitioning a segment from translocon to membrane are different than for partitioning between membrane and the cytoplasm; 4 leucines are sufficient to guarantee translocon-to-bilayer partitioning of the H-segment whereas 7 or so are required to prevent the CadC-H protein from dropping out into the cytoplasm. This implies that translocon-to-bilayer partitioning is not equivalent to water-bilayer-partitioning. This is perhaps not surprising in the light of the studies of Capponi et al. [25] who showed using molecular dynamics simulations that water behaves quite anomalously within the translocon. This result also means that proteins such as CadC and RodZ with moderately non-polar H-segments may be stable in the membrane only because of the insolubility of their cytoplasmic and periplasmic domains in the membrane phase.
To establish the criteria for SecA identification of far-downstream TM segments, we determined that it was difficult for SecA to identify polyleucine segments composed of more than 16 leucines; there was a steady decline in the processing of polyleucine segments as the number of leucines increased (Figure 6). For segments of 22 leucines or longer, the C-H-F constructs were not processed at all. This raised the question of whether the failure was due to the inability of SecA to recognize the segment or the inability of SecYEG to incorporate/secrete a highly hydrophobic segment across the membrane. To answer that question, we created the preMalE-H-R construct (Figure 1b and 1c). The data of Figure 7 show that SecA recognized the preMalE signal sequence and initiated secretion across the membrane. For H composed of 16 alanines, the construct was completely secreted. For segments composed of 16 or 24 leucines, however, the greasy segment partitioned into the membrane to form a single-span membrane protein. We conclude that the failure of SecA to target C-H-R constructs with long polyleucine H-segments (24L) was due to failure of SecA to recognize the segments.
Monné et al. [22, 26] examined the consequences of placing helix-breaking residues into very long poly-leucine segments inserted via the signal recognition particle (SRP) pathway using a dog pancreas microsome system. They showed, for example, that the introduction of a single proline or arginine into the middle of a long poly-Leu segment could cause a topology reversal that led to the insertion of the segment as a hairpin rather than a single-TM segment. We never observed such a phenomenon, probably because the SRP pathway allows greater folding flexibility than the SecA pathway. The formation of a hairpin in our system would require either that SecA reverse its direction of transport at some point or that the soluble periplasmic domain pass back across the inner membrane. Both possibilities seem unlikely and were in fact never observed.
Finally, we confirmed that secretion/insertion of our C-H-X is due to the SecA system (Figures S1 and S2), in agreement with Zhou et al. [21] who showed that the SecA pathway is both necessary and sufficient for secretion, although the SRP pathway can enhance the SecA pathway for very hydrophobic sequences.
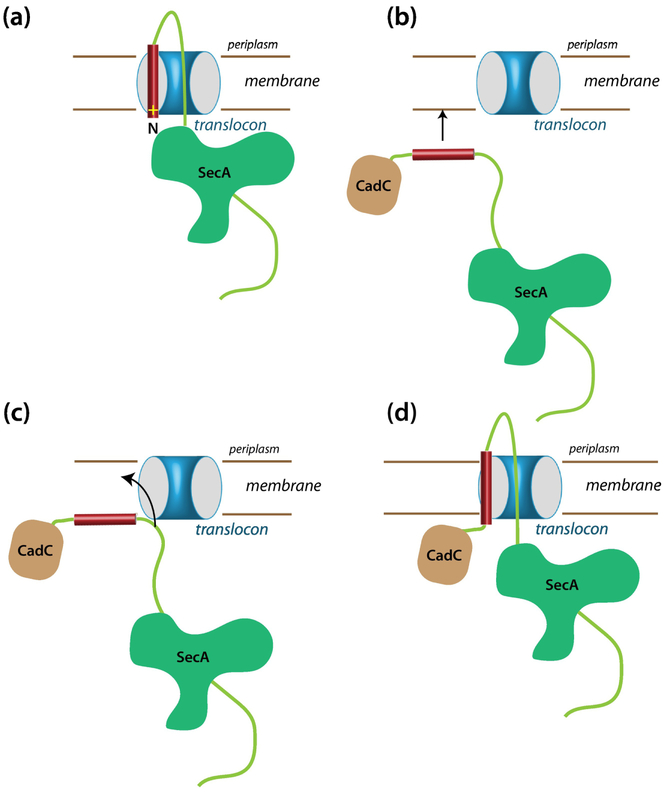
Typically [27], the first step in protein secretion by SecA is assumed to be insertion into the translocon of a hairpin-like structure comprised of the signal sequence and the adjacent mature sequence such that the N-terminus of the signal faces the cytoplasm (Figure 8a). Thinking about the experiments presented here and our earlier work on RodZ and CadC, we wondered about how SecA could manage secretion of proteins carrying a far-downstream transmembrane segment. It is difficult to visualize how SecA could secrete chimeras like ours that have a folded domain at the N-terminus. Consider the C-H-R. The 154-residue CadC cytoplasmic domain must emerge from the ribosome long before the appearance of the H-segment. It seems likely that the domain is folded before the construct is recognized by SecA. We suggest in Figure 8b–d a plausible scheme in which the direct interaction the H-segment with the membrane bilayer plays a dominant role. We suggest that, with the intracellular CadC domain folded, SecA binds to the H-segment and transports it in some uncertain manner to the vicinity of a SecYEG translocon. If, as seems likely, the H-segment has a higher affinity for the membrane bilayer than for SecA, then the segment will spontaneously transfer to the membrane interface. We suggest that because the free energy of the peptide is likely higher in the surface-bound state than in a transmembrane state [28, 29], the segment should spontaneously partition across the membrane carrying its C-terminal domain through the translocon. Thus, we suggest, the secreted protein threads the translocon. This scenario helps explain the presence of positive charges at the N-terminus of signal sequences, which are often dispensable and are required mostly for short less-hydrophobic segments [30, 31]. The positive charge interacting with the negatively charged membrane may anchor the N-terminus of the sequence at the interface to assure the correct topology.
Figure 8.
How can a single-span Type II membrane protein with a far-downstream TM segment be inserted into the membrane via the SecYEG translocon? (a) A typical scheme for the insertion into the translocon of a secreted protein carrying an N-terminal signal sequence. Topologically, this scheme seems unlikely to work for proteins with a far downstream TM domain, because the N-terminal domain is likely already folded when the TM domain emerges from the ribosome. We suggest instead the scheme shown in panels b, c, and d. (b) The folded N-terminal domain problem can be avoided if SecA transports the TM segment to the membrane, bringing it into close proximity of SecYEG. (c) If the affinity of the TM segment for the membrane is higher than for SecA, the segment will bind to the membrane. The nearby translocon would provide a pathway across the membrane for the much more polar part of the chain. For a very hydrophobic segment with a low hydrophobic moment [38, 39], a transmembrane configuration likely has a lower free than a surface-bound state. (d) As the TM segment partitions across the membrane, we suggest that this pulls the more polar C-terminal part of the chain into the translocon so that SecA can secrete the remainder of the chain across the membrane. This scheme works just as well for secreted proteins carrying an N-terminal signal sequence.
Materials and Methods
Bacterial strains, plasmids, and materials
All constructs were amplified from chromosomal DNA (E. coli K12). We used the restriction sites NdeI and XhoI for gene insertion into the pET21 vector (T7 promoter/lac operator, NOVAGEN). We inserted two additional unique restriction sites (KpnI and BamHI) to the cadC gene to exchange the H-segment using cassette cloning or overlap extension. All constructs were confirmed by sequencing. BL21(DE3) (F− ompT gal dcm lon hsdSB(rB− mB−) λ(DE3 [lacI lacUV5-T7 gene 1 ind1 sam7 nin5]) cells were used to express the various CadC constructs, which all carried an internal T7-tag and a C-terminal His6-tag for Western blot detection [13]. For SPase I depletion studies, we used E. coli strain FTL85 in which lepB is under the control of AraC [32]. For SecA depletion studies, we used E. coli strain EO527 in which secA is under the control of AraC. For Ffh depletion studies, we used E. coli strain WAM121 in which ffh is under the control of AraC. All depletion strains were received from Ross E. Dalbey at the Ohio State University who obtained them from Tracy Palmer (FTL85) and Tom Rapoport (EO527), respectively. For RseP studies we used AD1811 (ΔrseA) and AD2328 (ΔrseA, ΔrseP) cells kindly provided by Prof. Yoshinori Akiyama at Kyoto University.
Growth Conditions
Various CadC-based proteins were expressed from an IPTG-inducible and T7 polymerase-dependent system (pET-vector). We used a standard expression strategy: 1h expression in BL21(DE3) cells (presence of T7 polymerase). Protein expression in Figures 2, 3, 6, 7, S1, and S2 was done using BL21(DE3) cells containing the gene for T7-polymerase (CadC protein is regulated by the T7-promoter and the lac-operator). This leads to high protein expression levels even in the presence of small amounts IPTG inducer (10 – 20 μM) and short expression time (1 h). The experiments were done at pH 7 in Luria-Bertani (LB) medium or super optimal broth with catabolite repression (SOC) full media using glucose for repression [33].
SPase I, SecA, and Ffh Depletion Protocols
Depletion experiments. C-H-R constructs with clv = AQA (modified pET-vector, T7-RNA-polymerase independent system using a T5 promoter sequence, which is recognized by the wt E. coli RNA-polymerase) were transformed in depletion cells. Overnight cultures were grown in SOC media in the presence of 0.02 % arabinose (non-depletion condition). A 400 μl inoculum from the culture was added to 10 ml fresh SOC media with or without 0.02 % arabinose. After 2 h (OD600 ~ 0.6) protein expression was induced by adding 10 μM IPTG. After 0.5 h of protein expression, cells were pelleted and analyzed. (Figures S1 and S2)
RseP deletion experiments
C-H-R constructs with clv = AQA (modified pET-vector, T7-RNA-polymerase independent system using a T5 promoter sequence which is recognized by the wt E. coli RNA-polymerase.) were transformed in deletion cells AD1811 (ΔrseA) (positive control RseP plus condition) or AD2328 (ΔrseA, ΔrseP) (RseP minus condition; cells are only viable when rseA is deleted in addition). A 400 μl inoculum from an over-night culture was added to 10 ml fresh SOC media. After 1 h (OD600 ~ 0.6) protein expression was induced by adding 10 – 20 μM IPTG. After 1 h of protein expression, cells were pelleted and analyzed. Figure 4 and 5.
Cell Fractionation
Cell fractionation was performed by cell lysis using freeze-thaw and DNaseI treatment [33]. The bacterial cells were harvested, centrifuged, and the pellet resuspended in Lysis-Equilibration-Wash buffer (LEW buffer: 50 mM NaH2PO4, 300 mM NaCl, pH 8.0) containing DNaseI enzyme, DNaseI buffer, lysozyme, and phenylmethanesulfonyl fluoride (PMSF). Thereafter, the cell pellet was subjected to 10 cycles of freeze (liquid Nitrogen) and thaw (at 37°C water bath) followed by incubation at 37°C for 10 minutes. The cell suspension was centrifuged at 13,000xg for 15 minutes at 4°C, and the supernatant containing the soluble and periplasmic proteins (called the C/P fraction) was transferred to a new tube. The pellet was either washed once with 100 mM ice-cold Na2CO3 to remove membrane-adherent proteins[34] (CW fraction) or directly resuspend in LEW+1.5% CHAPS to solubilize membrane proteins. The suspension was centrifuged at 13,000xg at 4°C for 15 minutes. The supernatant contains the inner membrane (IM) fraction.
Periplasmic fraction
Cells were grown to mid-logarithmic phase and harvested by centrifugation. Osmotic shock was performed by a method adapted from Neu and Heppel [35] and Thorstenson et al. [36] as follows: Cell pellets were resuspended in 100μl osmotic shock buffer (0.5 M sucrose, 0.2 M Tris, 0.5 mM EDTA) and incubated on ice for 15 min, followed by the addition of 400 μl of 5 mM MgSO4. The cells were incubated on ice for an additional 30 min, followed by pelleting at 13,000xg at 4°C for 15 minutes. The supernatant (periplasmic fraction) and the pellet were mixed separately with SDS sample buffer and analyzed by SDS-PAGE [37].
Protease treatment studies
Cells were grown to mid-logarithmic phase and harvested by centrifugation. Cell pellets were resuspended in 100μl osmotic shock buffer (0.5 M sucrose, 0.2 M Tris, 0.5 mM EDTA) and incubated on ice for 15 min. Then, 400 μl of 5 mM MgSO4 containing prot K (80 ng) was added and the cells incubated on ice for an additional 30 min, followed by pelleting at 13,000xg at 4°C for 15 minutes. The supernatant was discarded, the pellet resuspended in SDS sample buffer, and analyzed by SDS-PAGE [37].
Western Blotting
The pellet was resuspended in SDS sample buffer and analyzed by SDS-PAGE (4–20 %)[37] and then Western blotted using iBLOT from Invitrogen® (Invitrogen Corp., Carlsbad, CA), which guarantees complete protein transfer that is necessary under low-expression conditions. The protein was detected by a T7-tag alkaline phosphatase-conjugated antibody from Novagen® (Novagen (EMD) Biosciences, Madison, WI) or by a His6-tag antibody from Roche® (Hoffman La Roche, Basel, Switzerland)
Supplementary Material
Highlights.
Single-span membrane proteins with far downstream TM segments use the SecA pathway
SecA recognizes signal sequences even if not at the protein’s N-terminus
Moderately hydrophobic sequences drop out of the membrane into the cytoplasm
Translocon-to-bilayer transfer ΔGs of TM segments differ from membrane-to-water ΔGs
Bilayer interface is crucial for SecA to the thread sequences through the translocon
Acknowledgements
The work presented in this paper was supported by NIH grant GM74637. We are happy to acknowledge the excellent technical support of Dr. Gargi Dasgupta.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Lindner E, White SH. Topology, dimerization, and stability of the single-span membrane protein CadC. J Mol Biol. 2014;426:2942–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Rawat S, Zhu L, Lindner E, Dalbey R, White SH. SecA drives transmembrane insertion of RodZ, an unusual single-span membrane protein. J Mol Biol. 2014;427:1023–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Watson N, Dunyak DS, Rosey EL, Slonczewski JL, Olson ER. Identification of elements involved in transcriptional regulation of the Escherichia coli cad operon by external pH. J Bacteriol. 1992;174:530–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Küper C, Jung K. CadC-mediated activation of the cadBA promoter in Escherichia coli. J Mol Microbiol Biotechnol. 2005;10:26–39. [DOI] [PubMed] [Google Scholar]
- [5].Haneburger I, Eichinger A, Skerra A, Jung K. New insights into the signaling mechanism of the pH-responsive, membrane-integrated transcriptional activator CadC of Escherichia coli. J Biol Chem. 2011;286:10681–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].van den Ent F, Johnson CM, Persons L, de Boer P, Löwe J. Bacterial actin MreB assembles in complex with cell shape protein RodZ. EMBO J. 2010;29:1081–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Alyahya SA, Alexander R, Costa T, Henriques AO, Emonet T, Jacob-Wagner C. RodZ, a component of the bacterial core morphogenic apparatus. Proc Natl Acad Sci USA. 2009;106:1239–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Hessa T, Kim H, Bihlmaier K, Lundin C, Boekel J, Andersson H, et al. Recognition of transmembrane helices by the endoplasmic reticulum translocon. Nature. 2005;433:377–81. [DOI] [PubMed] [Google Scholar]
- [9].Zwizinski C, Wickner W. Purification and characterization of leader (signal) peptidase from Escherichia coli. J Biol Chem. 1980;255:7973–7. [PubMed] [Google Scholar]
- [10].Date T, Wickner W. Isolation of the Escherichia coli leader peptidase gene and effects of leader peptidase overproduction in vivo. Proc Natl Acad Sci USA. 1981;78:6106–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Paetzel M, Dalbey RE, Strynadka NCJ. The structure and mechanism of bacterial type 1 signal peptidases: a novel antibiotic target. Pharmacol Ther. 2000;87:27. [DOI] [PubMed] [Google Scholar]
- [12].von Heijne G Signal sequences: The limits of variation. J Mol Biol. 1985;184:99–105. [DOI] [PubMed] [Google Scholar]
- [13].Burnette WR. “Western blotting”: Electrophoretic transfer of proteins from sodium dodecyl sulfate-p0lyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiondinated protein A. Anal Biochem. 1981;112:195–203. [DOI] [PubMed] [Google Scholar]
- [14].Franke S, Grass G, Rensing C, Nies DH. Molecular analysis of the copper-transporting efflux system CusCFBA of Escherichia coli. J Bacteriol. 2003;185:3804–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Chen H, Kendall DA. Competition between functional signal peptides demonstrates variation in affinity for the secretion pathway. J Bacteriol. 1996;178:6658–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Chen H, Kendall DA. Artificial transmembrane segments. J Biol Chem. 1995;270:14115–22. [DOI] [PubMed] [Google Scholar]
- [17].Bordignon E, Grote M, Schneider E. The maltose ATP-binding cassette transporter in the 21st century - towards a structural dynamic perspective on its mode of action. Mol Microbiol. 2010;77:1354–66. [DOI] [PubMed] [Google Scholar]
- [18].Chou MM, Kendall DA. Polymeric sequences reveal a functional interrelationship between hydrophobicity and length of signal peptides. J Biol Chem. 1990;265:2873–80. [PubMed] [Google Scholar]
- [19].Feng L, Yan H, Wu Z, Yan N, Wang Z, Jeffrey PD, et al. Structure of a site-2 protease family intramembrane metalloprotease. Science. 2007;318:1608–12. [DOI] [PubMed] [Google Scholar]
- [20].Saito A, Hizukuri Y, Matsuo E- I, Chiba S, Mori H, Nishimura O, et al. Post-liberation cleavage of signal peptides is catalyzed by the site-3 (S2P) in bacteria. Proc Natl Acad Sci USA. 2011;108:13740–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Zhou Y, Ueda T, Müller M. Signal recognition particle and SecA cooperate during export of secretory proteins with highly hydrophobic signal sequences. PLoS One. 2014;9:e92994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Monné M, Hermansson M, von Heijne G. A turn propensity scale for transmembrane helices. J Mol Biol. 1999;288:141–5. [DOI] [PubMed] [Google Scholar]
- [23].Hessa T, Meindl-Beinker NM, Bernsel A, Kim H, Sato Y, Lerch-Bader M, et al. The molecular code for transmembrane-helix recognition by the Sec61 translocon. Nature. 2007;450:1026–30. [DOI] [PubMed] [Google Scholar]
- [24].Jaud S, Fernández-Vidal M, Nillson I, Meindl-Beinker NM, Hübner NC, Tobias DJ, et al. Insertion of short transmembrane helices by the Sec61 translocon. Proc Natl Acad Sci USA. 2009;106:11588–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Capponi S, Heyden M, Bondar A- N, Tobias DJ, White SH. Anomalous behavior of water inside the SecY translocon. Proc Natl Acad Sci USA. 2015;112:9016–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Monné M, Nilsson IM, Elofsson A, von Heijne G. Turns in transmembrane helices: Determination of the minimal length of a “helical hairpin” and derivation of a fine-grained turn propensity scale. J Mol Biol. 1999;293:807–14. [DOI] [PubMed] [Google Scholar]
- [27].Rapoport TA, Li L, Park E. Structural and mechanistic insights into protein translocation. Annu Rev Cell Dev Biol. 2017;33:369–90. [DOI] [PubMed] [Google Scholar]
- [28].Ulmschneider MB, Ulmschneider JP, Schiller N, Wallace BA, von Heijne G, White SH. Spontaneous transmembrane helix insertion thermodynamically mimics translocon-guided insertion. Nature Communications. 2014;5:4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Gumbart JC, Ulmschneider MB, Hazel A, White SH, Ulmschneider JP. Computed free energies of peptide insertion in bilayers are independent of computational method. J Membr Biol. 2018;251:345–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Hikita C, Mizushima S. The requirement of a positive charge at the amino terminus can be compensated for by a longer central hydrophobic stretch in the functioning of signal peptides. J Biol Chem. 1992;267:12375–9. [PubMed] [Google Scholar]
- [31].Izard JW, Rusch SL, Kendall DA. The amino-terminal charge and core region hydrophobicity interdependently contribute to the function of signal sequences. J Biol Chem. 1996;271:21579–82. [DOI] [PubMed] [Google Scholar]
- [32].Lüke I, Handford JI, Palmer T, Sargent F. Proteolytic processing of Escherichia coli twin-argine signal peptides by LepB. Arch Microbiol. 2009;191:919–25. [DOI] [PubMed] [Google Scholar]
- [33].Green MR, Sambrook J. Molecular Cloning. A Laboratory Manual. 4 ed. Cold Spring Harbor: Cold Spring Harbor Press; 2012. [Google Scholar]
- [34].Fujiki Y, Hubbard AL, Fowler S, Lazarow PB. Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. J Cell Biol. 1982;93:97–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Neu HC, Heppel LA. The release of enzymes from Escherichia coli by osmotic shock and during the formation of spheroplasts. J Biol Chem. 1965;240:3685–92. [PubMed] [Google Scholar]
- [36].Thorstenson YR, Zhang Y, Olson PS, Mascarenhas D. Lederless polypeptides efficiently extracted from whole cells by osmotic shock. J Bacteriol. 1997;179:5333–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–5. [DOI] [PubMed] [Google Scholar]
- [38].Eisenberg D, Weiss RM, Terwilliger TC. The helical hydrophobic moment: A measure of the amphiphilicity of a helix. Nature. 1982;299:371–4. [DOI] [PubMed] [Google Scholar]
- [39].Eisenberg D, Weiss RM, Terwilliger TC, Wilcox W. Hydrophobic moments and protein structure. Faraday SympChemSoc. 1982;17:109–20. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.