Abstract
Osteoporosis is a silent systemic disease that causes bone deterioration, and affects over 10 million people in the US alone. This study was undertaken to develop a potential stem cell therapy for osteoporosis. We have isolated and expanded human dental pulp-derived stem cells (DPSCs), characterized them, and confirmed their multipotential differentiation abilities. Stem cells often remain quiescent and require activation to differentiate and function. Herein, we show that ferutinin activates DPSCs by modulating the Wnt/β-catenin signaling pathway and key osteoblast-secreted proteins osteocalcin and collagen 1A1 both mRNA and protein levels. To confirm that ferutinin modulates the Wnt pathway, we inhibited glycogen synthase kinase 3 (GSK3) and found that protein expression patterns were similar to those found in ferutinin-treated DPSCs. To evaluate the role of ferutinin in epigenetic regulation of canonical Wnt signaling, the pathway molecules Wnt3a and Dvl3 were analyzed using chromatin immunoprecipitation (ChIP)-quantitative PCR approaches. We confirmed that active marks of both H3K9 acetylation and H3K4 trimethylation were significantly enhanced in the promoter sites of the WNT3A and DVL3 genes in DPSCs after addition of ferutinin. These data provide evidence that ferutinin activates and promotes osteogenic differentiation of DPSCs, and could be used as an inducer as a potentially effective stem cell therapy for osteoporosis.
Keywords: DPSC, ferutinin, osteogenic differentiation, Wnt/β-catenin, epigenetic regulation
1. Introduction
Osteoporosis is a silent systemic skeletal disease of progressive bone loss that each year causes 9 million fractures worldwide [1, 2] and about 1.5 million fractures in the United States [3]. In the US alone, over 10 million people aged 50 or older suffer from osteoporosis and an additional 43 million have low bone mass that may progress into osteoporosis [4]. The incidence of osteoporosis is roughly twice greater in women than in men [5]. A number of environmental, endocrine, and genetic factors contribute to the development of osteoporosis [6]. Other contributing factors include nutritional deficiencies, smoking, lack of exercise, and the presence of other diseases or medications [7]. Several treatment options for osteoporosis exist currently, including calcium and vitamin D supplements as preventative measures [6], bisphosphonates as the first line of therapy [8], and other hormonal and biologic treatments.
Under normal physiologic conditions, bone undergoes a constant process of remodeling whereby it is simultaneously resorbed and deposited by osteoclasts and osteoblasts, respectively [17]. Osteoporosis results from excessive resorption by osteoclasts coupled with inadequate osteoblast activity that is insufficient to properly restore bone [18], leading to structural alterations and reduction in BMD. Osteoblasts are derived from mesenchymal precursors and osteoclasts are derived from myeloid cells. Dental pulp stem cells (DPSCs) are mesenchymal in nature, retain self-renewal and mutipotential capacity, and are capable of mediating tissue regeneration [19, 20]. These cells have been shown to maintain their “stemness” without changes in either morphology or expression of stem cell markers over time in culture [21]. DPSCs have been used to regenerate lost dental pulp as well as dentin [22]. The potential of DPSCs for bone tissue engineering has been demonstrated both in vitro and in vivo [23, 24]. Osteogenic differentiation has been enhanced by the addition of bone morphogenetic protein (BMP)-2 [25] and by growth on various polymeric and biologic scaffolds [26]. However, it is not well established how DPSC are regulated during osteogenic differentiation.
Wnt signaling plays essential roles in cell proliferation and differentiation during embryogenesis, post-natal development, and tissue homeostasis [27]. Wnt signaling is also important for maintenance and expansion of stem cells [28, 29]. Moreover, it is important in regulating the osteogenic process [30, 31], and its disruption is associated with several bone diseases, including osteoporosis [32].
Ferutinin is a daucane phytoestrogen found in plants of the Ferula genus which binds to both isoforms of the estrogen receptor [33]. Ferutinin has been shown to prevent bone loss in rats [34]. Furthermore, its efficacy in promoting the recovery of bone density following ovariectomy-induced osteoporosis has also been demonstrated [35]. The molecular pathways by which Ferutinin promotes osteogenesis have yet to be elucidated. Furthermore, its potential therapeutic efficacy is currently unknown.
This study was undertaken to develop an effective potential stem cell therapy using human DPSCs for the treatment of osteoporosis. The pharmacological compound ferutinin was tested to activate and promote osteogenesis in DPSCs. Furthermore, molecular pathways and epigenetic mechanisms by which ferutinin promotes DPSC differentiation were evaluated.
2. Materials and Methods
2.1. DPSC isolation and expansion
Human dental pulp derived stem cells (DPSC) were isolated from discarded third molar teeth, which were obtained after surgical extraction from a healthy adolescent donor with prior approval from the Institutional Review Board (IRB) and consent from donor. Teeth were thoroughly (at least 3 times) washed with phosphate buffered saline (PBS) containing 1% Penicillin-Streptomycin-Glutamine (PSG) (Gibco, Thermo Fisher, Waltham, MA). Teeth were cut open to harvest the pulp, which was then minced into approximately 1 mm cubes and plated onto 60 mm cell culture plates and cultured with alpha (α) Modified Eagle Medium (MEM) (Gibco) with 20% FBS (Hyclone, Thermo Fisher, USA) and 1% PSG. Fresh medium was added every third day of culture after removing old medium. Cells that migrated from the pulp tissues and became confluent were collected by dissociation by scraping and were re-cultured as passage 1 and maintained using the same medium. Cell viability was determined using the trypan blue exclusion method. Experiments were performed using cells between 3–7 passages.
2.2. Flow cytometry
Fluorescently labeled antibodies for cell surface markers included CD73, CD90, and CD105 (eBioscience, San Diego, CA), CD133 and CD34 (Miltenyi Biotec, San Diego, CA), IgG, CD31, CD45R, CD14, CD11b, CXCR4, and MHC class II (BD Biosciences, San Jose, CA). Flow cytometry was carried out according to a previously described method [36]. The DPSC aliquots were incubated at 4°C for more than 30 min in 2% FBS containing Hanks’ buffer, each in presence of one of the aforementioned antibodies. After antibody staining, cells were washed twice using 2% FBS containing Hanks’ buffer and fixed in 1% paraformaldehyde. Flowcytometric analysis was performed by using a FACS Calibur analyzer machine (BD Biosciences). Relevant isotype controls were also included for compensation setting. At least 20,000 events were acquired for each sample for analysis using CellQuest Pro software (BD Biosciences).
2.3. Induced differentiation
The multi-differentiation potential of DPSCs was evaluated in vitro using mesenchymal stem cell (MSC) differentiation kits (Millipore, Burlington, MA). Cells (passage 3) were plated in 10 cm cell culture dishes and grown to confluency in α MEM medium with 20% FBS and 1% PSG before induced differentiation.
2.4. Alizarin red S staining
Alizarin red S staining was carried out according to a previously described procedure [37]. DPSCs were cultured in osteogenic induction medium (Millipore) consisting of basic growth medium with 0.1 μM dexamethasone, 0.2 mM ascorbic 2-phosphate, and 10 mM glycerol 2-phosphate for 14 days on a 6-well collagen-coated plate. Medium was replaced every 3rd day. After 14 days of differentiation, cells were subjected to Alizarin red S staining, which stains calcified deposits produced by osteoblast cells and thus provides evidence of osteogenic differentiation. For staining, cells were fixed with ice-cold 70% ethanol for 1 h at room temperature. Ethanol was removed and cells were washed twice with water. Water was aspirated and Alizarin red solution was added; cells were then incubated for 30 minutes in the dye at room temperature. Cells were then washed 4 times with water and left in water to avoid drying. Images were captured under a light microscope at various magnifications.
2.5. Alcian blue staining
Chondrogenic differentiation of hMSCs was accomplished by a modification of the protocol outlined by Johnstone et al. [38]. In brief, aliquots of 250,000 DPSCs suspended in 0.5 mL medium were distributed to 15 mL conical polypropylene centrifuge tubes (VWR, West Chester, PA). The cells were centrifuged for 5 min at 600 g and pelleted at the bottom of the tube, and cultured in serum-free chondrogenic medium (Millipore). Tubes were placed in an incubator with caps loosened to permit gas exchange. The sedimented cells formed a spherical mass at the bottom of the tube within 24 h. Medium was replaced three times per week. Cell pellets were harvested by rinsing in D-PBS followed by fixation for 1 h in 4% formaldehyde in D-PBS, made fresh. Samples were then transferred into 70% ethanol, dehydrated in ethanol and xylene series, and paraffin-embedded. Sections of 5 μm were cut through the center of each pellet. Sections were stained with Alcian blue stain and images were captured with a light microscope.
2.6. Oil Red O staining
DPSCs were cultured in adipogenic induction medium (Millipore) for 14 days. Differentiated cells produced lipid droplets that were subsequently stained using Oil Red O dye. To stain cells, medium was removed and cells were washed 3 times for 5 minutes each with 1X PBS. Cells were fixed in 4% paraformaldehyde for 10 minutes at room temperature. The fixative was aspirated and the cells were washed 3 times for 5 minutes each with 1X PBS, then washed twice with water. The water was aspirated and Oil Red O solution was added; cells were incubated for 50 minutes at room temperature. The Oil Red O solution was removed and cells were washed 3 times with water. Nuclei were stained with hematoxylin solution for 3 minutes. Images were captured under a light microscope at various magnifications.
2.7. Quantitative RT-PCR
Total RNA was isolated from DPSCs treated with ferutinin for 12, 24, and 48 h and from vehicle-treated DPSCs using a TRIzol purification method (Thermo Fisher). Complementary (c) DNA synthesized from the mRNA was used for quantitative PCR using a Bio-Rad CFX96 Real-Time System. Primers for COL1A1, BGLAP (Osteocalcin), LRP6, DVL3, GSK3B, CTNNB1 (β-catenin), and GAPDH were purchased from Integrated DNA Technologies (Coralville, IA). Sequences are presented in Supplemental Table II. Cq measurements were obtained, and data are presented as fold difference of ΔΔCT values corrected with GAPDH expression.
2.8. Western blot
Whole cell lysates were obtained from DPSCs cultured under control conditions or stimulated with ferutinin (10 μg/mL in αMEM) or CHIR-98014 (50 nM) for 12, 24, and 48 h. Protein was quantified by colorimetric assay using the Bradford method (Bio-Rad, Hercules, CA) and the proteins were separated in a polyacrylamide gel. Briefly, a polyacrylamide gel was cast and denatured proteins (20 µg) were loaded and separated through the gel by electrophoresis; a protein ladder was loaded as a marker (Sigma, St. Louis, MO). The proteins were transferred from the gel to a 0.45 µm nitrocellulose membrane (Bio-Rad) at 4 °C. The membrane was blocked for 1 h at room temperature (RT) with a blocking buffer composed of 5% nonfat milk in TBS-Tween-20 (TBST) (Boston BioProducts, Ashland, MA). The membrane was washed and incubated with primary antibody against LRP6, Dvl3, Axin1, Naked1, β-catenin, GAPDH (Cell Signaling, Danvers, MA), and GSK3 (Santa Cruz Biotechnology, Dallas, TX) (1:1000 diluted in a solution of 5% BSA in TBST) for 2 h. The membrane was washed, then incubated in secondary antibody (1:3000 in a solution of 5% milk in TBST) (Cell Signaling). The membrane was then washed, placed in the cassette holder and incubated briefly in chemiluminescent substrate (Sigma). Films were then exposed and developed. Densitometric quantification of bands was performed using ImageJ software (NIH).
2.9. Immunostaining
DPSCs were plated at medium density on glass cover slips in 6 well plates and with 1.5 mL complete medium (α MEM, 20% FBS, 1% PSG). Next day, medium was changed for complete medium containing 10 µg/mL ferutinin or 1 µL/mL DMSO in complete medium as a vehicle-treated control. 24 h later, medium was aspirated and cells were washed with PBS, fixed with 4% paraformaldehyde, and permeabilized with 1% Triton-X 100 at room temperature (RT). After washing with PBS, cells were blocked in 5% FBS in PBS overnight at 4 °C. Next day, they were washed with wash buffer (PBS with 0.5% FBS and 0.5% TBST) and incubated in primary antibodies (α-Osteocalcin and α-Collagen1A1; diluted 1:100 in wash buffer) for 2 h at RT; two samples were incubated in secondary antibody alone to serve as negative controls. Samples were washed with wash buffer and were incubated with fluorophore conjugated secondary antibody (Alexa Fluor 488 α-Rabbit and TX Red α-Goat as necessitated by the primary antibodies used; diluted 1:2000 in wash buffer) for 1 h at RT. Cover slips were mounted upon glass microscopy slides with ProLong Gold Antifade Mountant with DAPI (Invitrogen, Carlsbad, CA). Upon drying, fluorescence microscopy was performed and images were obtained.
2.10. Chromatin immunoprecipitation (ChIP) and quantitative PCR
ChIP analysis was performed using Imprint® Chromatin Immunoprecipitation Kit (Sigma) according to previously described methods [39]. Briefly, after chromatin cross-linking with 1% formaldehyde and DNA shearing, chromatin-protein complexes were immunoprecipitated from DPSCs with or without stimulation of ferutinin for 24 h, with antibodies against H3K9Ac (Millipore Sigma), H3K4me3 (Millipore Sigma). Antibody against goat IgG (Abcam, Cambridge, UK) was used as a negative control. Quantitative PCR analysis was performed with the primers described (in Supplementary Table-I and Fig. 1) using SYBR green PCR master mix (Thermo Fisher) and a real-time PCR machine (Bio-Rad CFX96 Real-Time System). Values obtained from the ChIP assay were normalized to the background obtained from the precipitation with a non-specific antibody. Percent (%) of input was analyzed by following standard formula. Each experiment was performed in triplicate at least three times.
Figure 1. DPSC morphology during expansion.
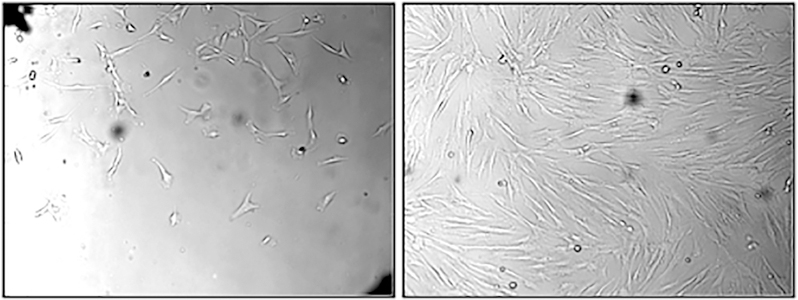
Dental pulp tissues were plated to expand stem cells. Left panel shows early growth of stem cells and right panel shows stem cells after confluence.
3. Results
3.1. DPSC isolation, expansion and characterization
DPSCs were isolated from the pulp of donor human third molar teeth and were expanded in plentiful numbers in vitro. These cells display a fibroblast-like morphology during expansion. Captured images were shown during early stage of expansion (Fig. 1, left panel), and after confluence of the culture (Fig. 1, right panel). The phenotype of DPSCs is elucidated by flow cytometry. Flowcytometric analysis revealed that the expanded DPSCs are a population of homogenous cells (Fig. 2). These cells express all progenitor stromal stem cell biomarkers; 99.97% were positive for CD90, 99.94% were positive for CD105, and 99.96% were positive for CD73. They were negative for hematopoietic stem cell markers CD133 (2.03%) and CD34 (3.19%). They were negative for platelet endothelial cell adhesion molecule CD31 (2.01%). They were negative for CD45R (5.67%), a marker present on B cells and other antigen presenting cells. They were negative for CD14 (2.47%), a monocytic marker. They were negative for CD11b (2.38%), a dendritic cell marker. They were negative for chemokine receptor CXCR4 (3.14%). They were also negative for the antigen presenting molecule MHC class II (1.48%).
Figure 2. DPSC phenotype.
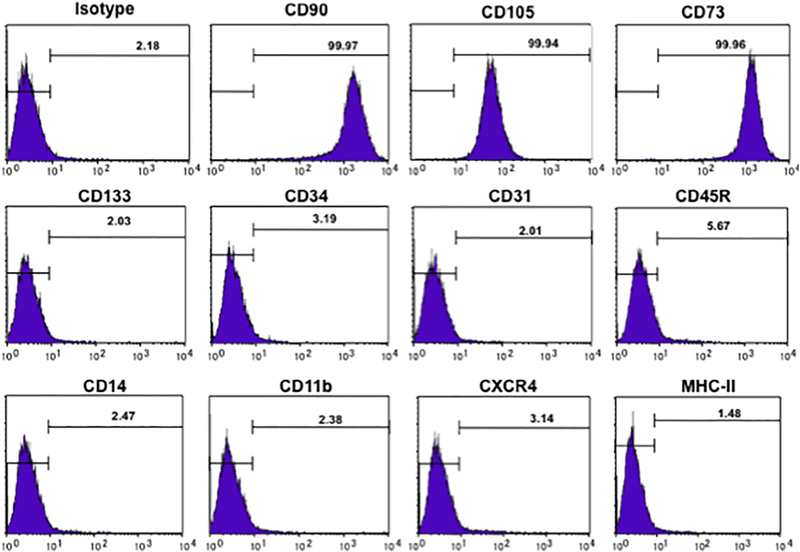
Flowcytometric analysis reveals that expanded cells represent a homogeneous population characterized by expression of CD90, CD105, and CD73. They do not express CD133 and CD34 (hematopoietic stem cell markers), CD31 (endothelial progenitor cell marker), CD45R (B cell marker), CD14 (monocyte marker), CD11b (dendritic cell marker), CXCR4 (chemokine receptor), or MHC class II (antigen presentation protein).
3.2. Differentiation potential of DPSCs
To investigate whether expanded DPSCs retain their multipotential capabilities, we induced them along the osteogenic, chondrogenic and adipogenic lineages in vitro for 14 days. Osteogenic differentiation was detected by using Alizarin red S staining, which stains calcium deposited by differentiated cells. Images at various magnifications were shown (Fig. 3, upper panels). Red staining indicates that DPSCs have differentiated to osteoblastic cells, which deposit calcium extracellularly. Chondrogenic differentiation was detected by Alcian blue staining that stains acidic polysaccharides found in cartilage. Following induced differentiation, DPSCs were found to possess chondrogenic properties as demonstrated by positive Alcian blue staining (Fig. 3, middle panels).
Figure 3. Expanded DPSCs maintain multipotential differentiation abilities.
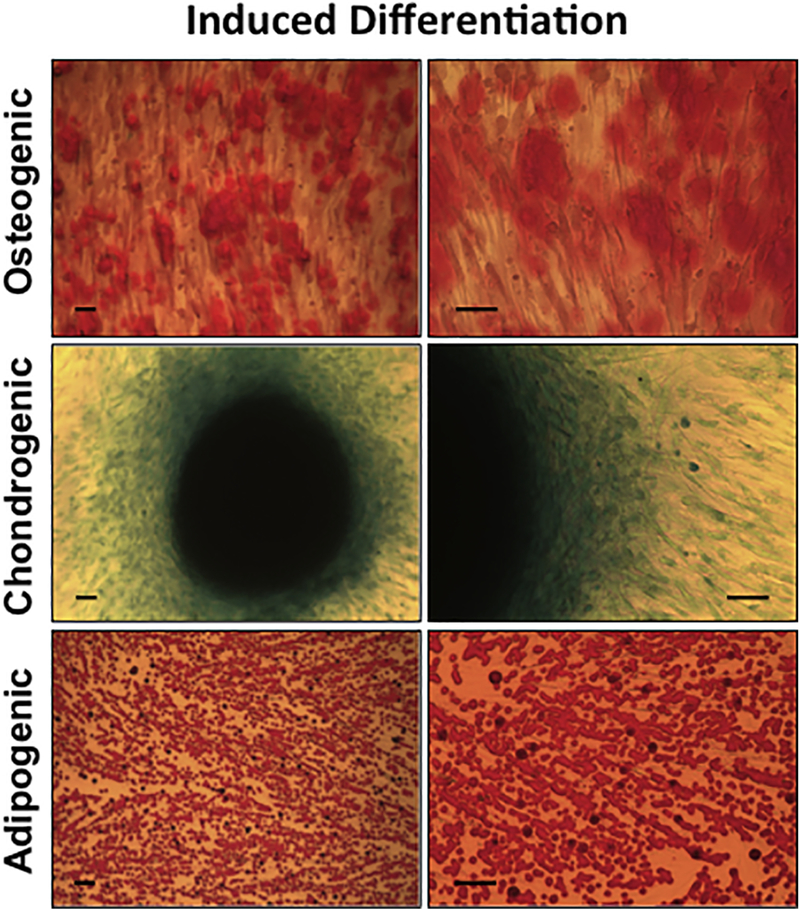
DPSCs were differentiated towards the osteogenic, chondrogenic, and adipogenic lineages. (A) Micrographs at various magnifications of differentiated DPSCs following alizarin red staining. (B) Chondrogenic differentiation visualized by microscopy after Alcian blue staining. (C) Micrographs of adipogenic differentiation shown after oil red o staining.
Adipogenic differentiation was detected by Oil Red O dye, which stains neutral lipids in cells. After induced differentiation, lipid droplets within cells were stained red, while cell nuclei were stained black by hematoxylin. Abundant oil droplets were observed, indicating that DPSCs are capable of differentiation along the adipogenic lineage (Fig. 3, lower panels).
3.3. Effect of ferutinin on DPSC activation and differentiation
To further elucidate the role of ferutinin in DPSC activation and differentiation, expression of osteoblast specific genes (COL1A1 and BGLAP), and key Wnt pathway genes (LRP6, DVL3, GSK3B, CTNNB1), and GAPDH as an internal control were evaluated by quantitative RT-PCR. Quantitative RT-PCR data revealed that all tested genes were elevated upon stimulation with Ferutinin at various degrees during the time course study (Fig. 4A). The effects of ferutinin on expression of Wnt/β-catenin pathway molecules were evaluated by western blot analysis (Fig. 4B). Increased expression of LRP6 was observed at 12 and 24 h of stimulation. Dvl3 expression increased at 24 h of stimulation. Wnt3a expression was markedly increased at 12 and 24 h of stimulation and was completely attenuated by the 48 h time point. Wnt5a/b expression did not substantially change over the course of the experiment. Expression of Naked1 increased at 12 and 24 h of stimulation, before returning to levels akin to the control condition by 48 h. Axin was very minimally expressed throughout, but was observed at slightly higher levels at the 24 h time point. GSK3 expression was attenuated at 12, 24, and 48 h of stimulation. Correspondingly, expression of β-catenin increased at 12 and 24 h of stimulation before returning to control levels by 48 h. GAPDH expression was assessed as a control to ensure equal loading (quantified data is presented in Supplemental Fig. 2). Data at the gene expression level corroborate protein-level observations. A notable exception is the observation of significantly increased mRNA expression of GSK3B at the 24 and 48 h time points following ferutinin stimulation, while at the same time points a marked decrease is observed at the protein level. This discrepancy may be due to post-translational modification of GSK3 or other regulatory mechanisms [32]; however, this stands to be evaluated in greater detail in the future.
Figure 4.
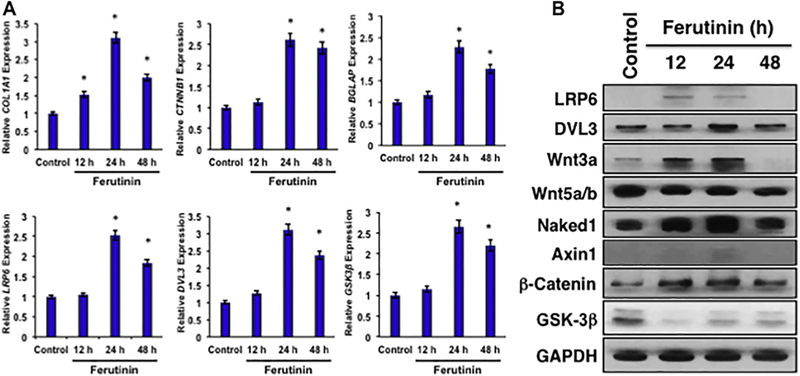
A. Ferutinin modulates mRNA expression of osteoblast specific and key Wnt pathway genes. RT-PCR was carried out to evaluate gene expression of COL1A1, BGLAP, LRP6, DVL3, GSK3B, and CTNNB1. Expression is shown as fold difference ± SEM derived from calculated ΔΔCT values. Statistical significance, p<0.05 was shown (*), compared to vehicle-treated controls. B. Ferutinin modulates Wnt/ β -catenin signaling pathway molecules in DPSCs. Various Wnt/β-catenin signaling pathway proteins were evaluated in DPSCs after stimulation with ferutinin (10 µg/mL) for 12, 24, and 48 h using western blot methods. GAPDH was used as an internal loading control. Band density is presented as protein expression relative to GAPDH ± SEM.
3.4. Effect of ferutinin on osteogenic markers
Expression of key matrix proteins secreted by osteoblasts was evaluated by immunostaining techniques in DPSCs stimulated with ferutinin (10 µg/mL) for 24 h. Immunocytochemical evaluation revealed that the level of expression of collagen 1A1 was far greater in ferutinin-treated cells compared to cells treated with the vehicle (Fig. 5A). In addition, expression of osteocalcin was also increased in stimulated cells compared to vehicle treated cells (Fig. 5B). Samples were visualized and photographed using a Leica DMi8 microscope and Leica Application Suite X software.
Figure 5. DPSCs treated with ferutinin express osteogenic molecules.
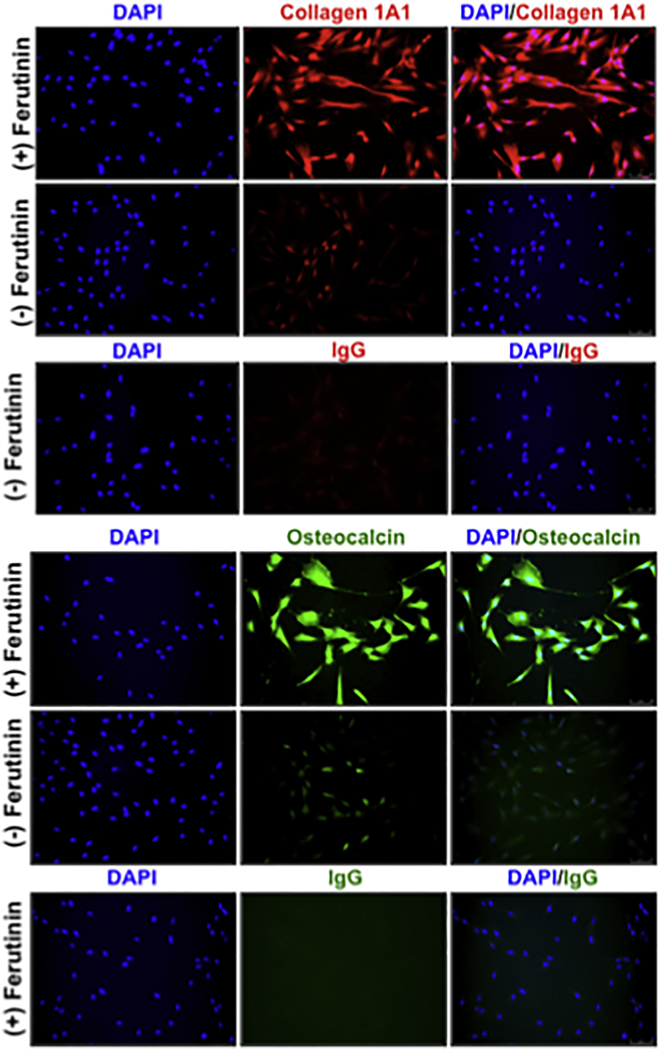
Immunostaining was performed to visualize expression levels of (A) collagen 1A1, and (B) osteocalcin in ferutinin-treated DPSCs compared to vehicle-treated cells. DAPI was used for nuclear staining.
3.5. Effect of GSK3 inhibitor on Wnt/β-Catenin signaling pathway molecules in DPSCs
The effect of CHIR-98014, a pharmacological compound that inhibits GSK3 was tested on Wnt/β-Catenin signaling pathway molecules of DPSCs at various time points (Fig. 6). In untreated DPSCs, LRP6 is expressed faintly; after 24 h of stimulation, however, it was substantially upregulated. Dvl3 was minimally expressed in untreated cells and was increased over time. Wnt3a was significantly upregulated over the course of differentiation. Wnt5a/b expression was upregulated following 12 and 24 h of stimulation before attenuating by 48 h. Naked1 was upregulated at all time points after stimulation. Axin1 was most strongly expressed at 24 h following stimulation. Decreased expression of GSK3 was observed in stimulated cells. Expression of β-catenin was increased over all three time points. GAPDH served as a loading control and minimal changes were observed (quantified data is presented in Supplemental Fig. 3). These results indicate that inhibition of GSK3 pathway causes modulation of Wnt pathway molecules similar to that observed when DPSCs are stimulated with ferutinin.
Figure 6. GSK3 inhibition modulates Wnt/β-catenin signaling pathway molecules in DPSCs.
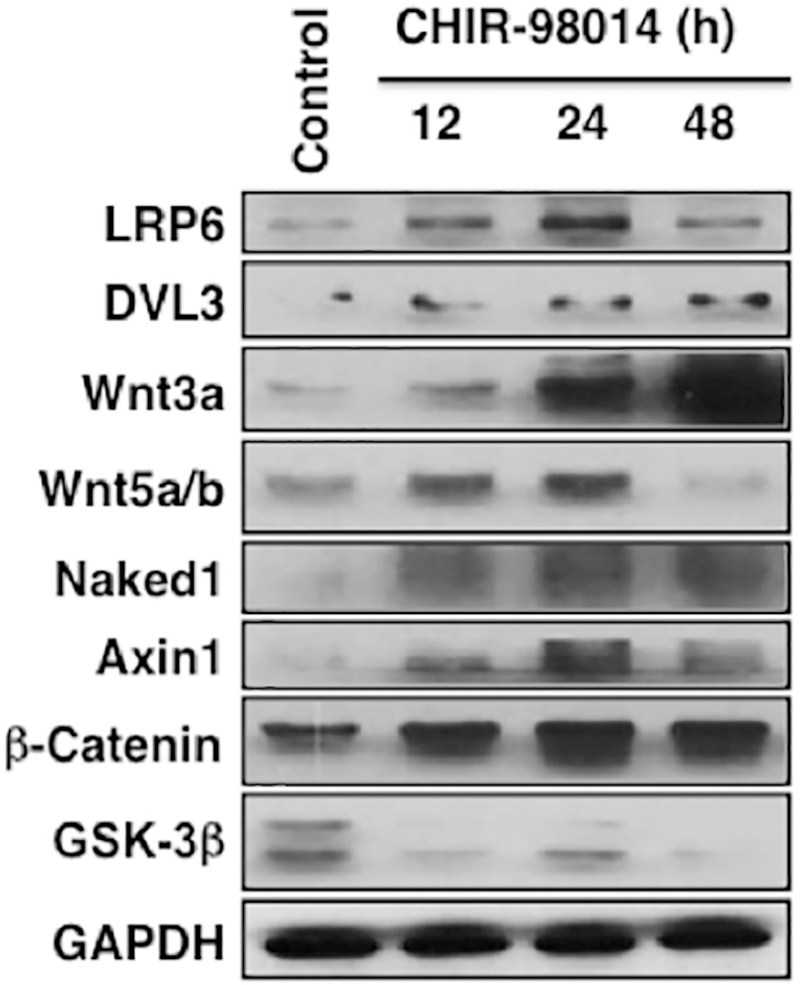
Various Wnt/β-catenin signaling pathway molecules in DPSCs were evaluated after stimulation with a GSK3 inhibitor molecule CHIR-98014 for 12, 24 and 48 h. Western blot results are shown.
3.6. Epigenetic evaluation of Wnt3a and DVL3 molecules upon ferutinin stimulation
To evaluate the role of ferutinin in epigenetic regulation of canonical Wnt signaling in DPSCs, the pathway molecules Wnt3a and Dvl3 were analyzed by chromatin immunoprecipitation (ChIP) quantitative PCR approaches with and without stimulation by ferutinin. ChIP analysis revealed that the active marks of both histone 3 lysine 9 (H3K9) acetylation and histone 3 lysine 4 (H3K4) trimethylation were significantly enhanced in the promoter sites of the WNT3A and DVL3 genes in DPSCs after stimulation with ferutinin (Fig. 7).
Figure 7. Ferutinin regulates Wnt3a and Dvl3 genes epigenetically.
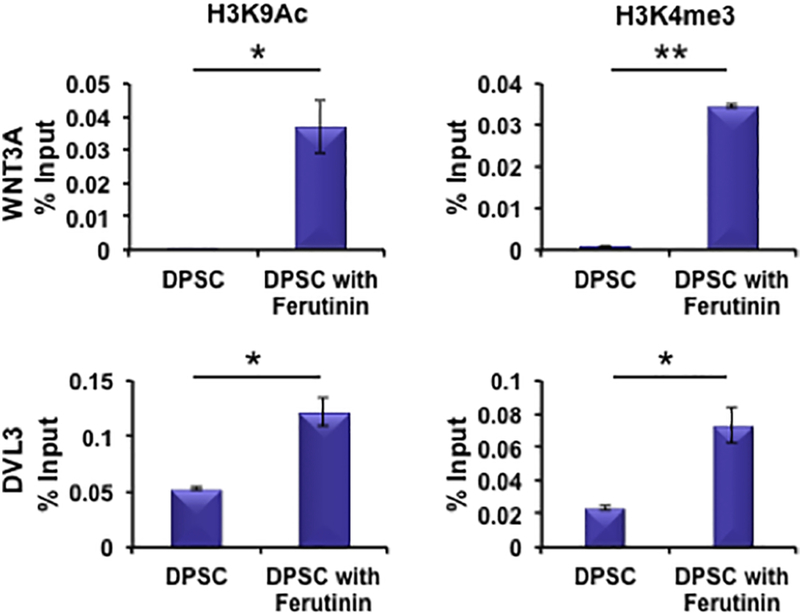
Wnt signaling pathway molecules Wnt3a and Dvl3 were analyzed using chromatin immunoprecipitation (ChIP) quantitative PCR methods to assess epigenetic regulation in DPSCs after stimulation with ferutinin for 24 h. Promoter site active marks of Wnt3a and Dvl3 genes for both H3K9 acetylation and H3K4 trimethylation were evaluated and shown graphically. (* indicates p < 0.05, ** indicates p < 001).
4. Discussion
Osteoblasts are derived from mesenchymal precursor cells and are the key mediators of bone regeneration in health and disease states. Human dental pulp tissues from the wisdom teeth contain clonogenic, highly proliferative stem cells capable of tissue regeneration [19]. These DPSCs show fibroblast-like morphology and express markers similar to those of human bone marrow stromal cells (BMSC). DPSCs have also been shown to self-renew in vivo after in vitro expansion [20]. These cells have been shown to maintain their “stemness” without changes to morphology or expression of stem cell markers over time in culture [21]. DPSCs have been used to regenerate lost dental pulp as well as dentin by differentiation into odontoblasts [22]. DPSCs show promise for bone tissue engineering, as their capacity for osteogenic differentiation has been demonstrated both in vitro and in vivo [23, 24]. However, the molecular mechanism by which DPSCs differentiate has yet to be defined. In this study, we have demonstrated that DPSCs can be cultured successfully in vitro (Fig. 1) while maintaining their MSC-like multilineage differentiation capacity along the osteogenic, chondrogenic and adipogenic lineages (Fig. 3). They possess specifically mesenchymal properties, as they express the cell surface markers CD73, CD90, and CD105, and do not express markers of hematopoietic lineage cells (CD34, CD133, CD11b, CD14, CD31, CD45R, CXCR4, MHC class II) (Fig. 2). Because they do not express MHC class II, they are considered to possess low immunogenicity and thus be suited for transplantation with low risk of rejection. Although they possess multilineage differentiation potential, our goal is to direct them towards the osteogenic lineage for the regeneration of bone. For that purpose, we have investigated several compounds and found a phytoestrogen, ferutinin, which could be a potential molecule of interest.
Phytoestrogens are plant-derived polyphenols that share structural similarity to the endogenous estrogen 17β-estradiol. Many such compounds have been shown to share the osteoprotective effects of estrogens without possessing their carcinogenic side effects [40]. Ferutinin is a daucane phytoestrogen found in plants of the Ferula genus and has affinity for both isoforms of the estrogen receptor; it is therefore speculated that the compound may be useful as a selective estrogen receptor modulator [41]. The effects of ferutinin on bone metabolism have previously been evaluated in ovariectomized rats with known protective effects on bone density [34]. Furthermore, it has been shown to aid in recovery of bone density in animal models of osteoporosis [35]. However, the mechanisms by which ferutinin promotes bone density have not yet been evaluated. We found that ferutinin promotes osteogenic differentiation by modulating the various molecules of the Wnt/β-catenin signaling pathway, which critically regulates osteogenesis (Fig. 4).
Wnt signaling plays essential roles in embryogenesis, post-natal development, and tissue homeostasis as a regulator of cell proliferation and differentiation [27]. In the absence of the Wnt ligand, the transcription factor β-catenin is held within the destruction complex, composed of Axin, adenomatous polyposis coli protein (APC), and GSK3. GSK3 phosphorylates β-catenin, marking it for ubiquitination and proteasomal degradation [42]. When Wnt binds to the Frizzled/LRP receptor complex on the cell surface, the destruction complex is inhibited. Hypophosphorylated β-catenin is released from its interaction with destruction complex proteins and subsequently accumulates in the cytoplasm and enters the nucleus, where it acts to regulate transcription [43]. Moreover, its importance as a pathway involved in regulating osteogenic differentiation of mesenchymal stem cells has been demonstrated [30, 31]. Wnt signaling is central to bone modeling and especially to osteoblast function; its disruption is associated with several bone diseases including osteoporosis [32]. Our observations of the effect of ferutinin on molecules of the Wnt/β-catenin signaling pathway are in accord with the canonical mechanism of this pathway’s activation.
Observing Wnt pathway dynamics alone is insufficient to state that cells are undergoing osteoblastic differentiation, as this pathway is involved in myriad cell growth, proliferation, and differentiation processes. Expression of key osteogenic markers was thus assessed to confirm DPSC differentiation toward the osteogenic lineage. Collagen 1A1 is expressed in osteoblasts from an early stage of differentiation and is a key component of the organic bone matrix [44, 45]; we observed its upregulation in DPSCs as early as 24 h after stimulation with ferutinin (Fig. 5A). Osteocalcin is an osteoblast-specific protein with important metabolic functions in bone [46]; we observed that it was markedly upregulated in ferutinin-stimulated DPSCs (Fig. 5B). Expression of these key proteins indicates that ferutinin promotes osteoblastic differentiation of DPSCs.
Recent reports of DPSC activation and subsequent tooth repair by GSK3 antagonists piqued our interest in the possibility that these cells can be stimulated with such pharmacologic agents to undergo osteogenic differentiation [47]. For this reason, we employed the GSK3 inhibitor CHIR-98014 to explore the role of Wnt signaling in osteoblastogenesis and validate the role of ferutinin as a Wnt pathway modulator (Fig. 6). When CHIR-98014 was administered, protein expression patterns similar to those seen with ferutinin were observed. This leads us to propose that ferutinin may act by a similar mechanism of GSK3 inhibition.
To confirm further, we have performed molecular analysis at the epigenetic level to assess the regulation of the genes that we found were most changed in DPSCs after stimulation with ferutinin. We evaluated histone 3 lysine 9 acetylation (H3K9ac) and histone 3 lysine 4 trimethylation (H3K4me3). Both of these are hallmarks of active promotors and therefore indicate that transcription of the target gene is active [48, 49]. We found that active marks of both H3K9 acetylation and H3K4 trimethylation were significantly enhanced in the promoter sites of the WNT3A and DVL3 genes in DPSCs after stimulation with ferutinin (Fig. 7).
Though the Wnt pathway plays a significant role in osteoblastogenesis, it is one of the several key pathways in this process. The BMP2 pathway, for instance, is a critical regulator at various stages of osteoblastic differentiation and maturation [43]. Signaling via estrogen receptors is also of great importance in osteoblastogenesis and bone health [44]. Moreover, these pathways hardly act independently of one another; there is growing evidence of cross-talk among the many pathways involved [45]. The effects of ferutinin on the activity of pathways, such as shown above in DPSCs, represents an ongoing research effort in our laboratory.
5. Conclusion
These data provide evidence that ferutinin activates and promotes osteogenic differentiation of DPSCs canonical Wnt/β-catenin signaling pathway by activating marks of both H3K9 acetylation and H3K4 trimethylation in the promoter regions of WNT3A and DVL3. It could be used as an inducer to modulate DPSCs towards osteogenic differentiation for a potentially effective stem cell therapy for osteoporosis.
Supplementary Material
Highlights.
Isolation and homogeneous expansion of multipotent human dental pulp-derived stem cells (DPSCs).
Ferutinin activates DPSCs via the Wnt/β-catenin signaling pathway, and induces osteocalcin and collagen 1A1 both mRNA and proteins.
GSK3 inhibitor also activates DPSCs through the Wnt/β-catenin signaling pathway.
Ferutinin induces H3K9 acetylation and H3K4 trimethylation in the promoter sites of the WNT3A and DVL3 genes in DPSCs.
Acknowledgments
This work was supported in part by National Institutes of Health grants, R01AR068279 (NIAMS), STTR 1R41EY024217 (NEI) and STTR 1R41AG057242 (NIA). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cosman F, de Beur SJ, LeBoff MS, Lewiecki EM, Tanner B, Randall S, Lindsay R, F. National Osteoporosis, Clinician’s Guide to Prevention and Treatment of Osteoporosis, Osteoporos Int 25(10) (2014) 2359–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnell O, Kanis JA, An estimate of the worldwide prevalence and disability associated with osteoporotic fractures, Osteoporos Int 17(12) (2006) 1726–33. [DOI] [PubMed] [Google Scholar]
- 3.Bone Health and Osteoporosis: A Report of the Surgeon General, Rockville (MD: ), 2004. [PubMed] [Google Scholar]
- 4.Wright NC, Looker AC, Saag KG, Curtis JR, Delzell ES, Randall S, Dawson-Hughes B, The recent prevalence of osteoporosis and low bone mass in the United States based on bone mineral density at the femoral neck or lumbar spine, J Bone Miner Res 29(11) (2014) 2520–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cummings SR, Melton LJ, Epidemiology and outcomes of osteoporotic fractures, Lancet 359(9319) (2002) 1761–7. [DOI] [PubMed] [Google Scholar]
- 6.Sozen T, Ozisik L, Basaran NC, An overview and management of osteoporosis, Eur J Rheumatol 4(1) (2017) 46–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Camacho PM, Petak SM, Binkley N, Clarke BL, Harris ST, Hurley DL, Kleerekoper M, Lewiecki EM, Miller PD, Narula HS, Pessah-Pollack R, Tangpricha V, Wimalawansa SJ, Watts NB, American Association of Clinical Endocrinologists and American College of Endocrinology Clinical Practice Guidelines for the Diagnosis and Treatment of Postmenopausal Osteoporosis - 2016, Endocr Pract 22(Suppl 4) (2016) 1–42. [DOI] [PubMed] [Google Scholar]
- 8.Watts NB, Bilezikian JP, Camacho PM, Greenspan SL, Harris ST, Hodgson SF, Kleerekoper M, Luckey MM, McClung MR, Pollack RP, Petak SM, American Association of Clinical Endocrinologists Medical Guidelines for Clinical Practice for the diagnosis and treatment of postmenopausal osteoporosis: executive summary of recommendations, Endocr Pract 16(6) (2010) 1016–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kennel KA, Drake MT, Adverse effects of bisphosphonates: implications for osteoporosis management, Mayo Clin Proc 84(7) (2009) 632–7; quiz 638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Black DM, Delmas PD, Eastell R, Reid IR, Boonen S, Cauley JA, Cosman F, Lakatos P, Leung PC, Man Z, Mautalen C, Mesenbrink P, Hu H, Caminis J, Tong K, Rosario-Jansen T, Krasnow J, Hue TF, Sellmeyer D, Eriksen EF, Cummings SR, Trial HPF, Once-yearly zoledronic acid for treatment of postmenopausal osteoporosis, N Engl J Med 356(18) (2007) 1809–22. [DOI] [PubMed] [Google Scholar]
- 11.Marx RE, Sawatari Y, Fortin M, Broumand V, Bisphosphonate-induced exposed bone (osteonecrosis/osteopetrosis) of the jaws: risk factors, recognition, prevention, and treatment, J Oral Maxillofac Surg 63(11) (2005) 1567–75. [DOI] [PubMed] [Google Scholar]
- 12.Gennari L, Merlotti D, Nuti R, Selective estrogen receptor modulator (SERM) for the treatment of osteoporosis in postmenopausal women: focus on lasofoxifene, Clin Interv Aging 5 (2010) 19–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lindberg MK, Moverare S, Skrtic S, Gao H, Dahlman-Wright K, Gustafsson JA, Ohlsson C, Estrogen receptor (ER)-beta reduces ERalpha-regulated gene transcription, supporting a “ying yang” relationship between ERalpha and ERbeta in mice, Mol Endocrinol 17(2) (2003) 203–8. [DOI] [PubMed] [Google Scholar]
- 14.Love RR, Mazess RB, Barden HS, Epstein S, Newcomb PA, Jordan VC, Carbone PP, DeMets DL, Effects of tamoxifen on bone mineral density in postmenopausal women with breast cancer, N Engl J Med 326(13) (1992) 852–6. [DOI] [PubMed] [Google Scholar]
- 15.Bord S, Horner A, Beavan S, Compston J, Estrogen receptors alpha and beta are differentially expressed in developing human bone, J Clin Endocrinol Metab 86(5) (2001) 2309–14. [DOI] [PubMed] [Google Scholar]
- 16.Wensel TM, Iranikhah MM, Wilborn TW, Effects of denosumab on bone mineral density and bone turnover in postmenopausal women, Pharmacotherapy 31(5) (2011) 510–23. [DOI] [PubMed] [Google Scholar]
- 17.Kular J, Tickner J, Chim SM, Xu J, An overview of the regulation of bone remodelling at the cellular level, Clin Biochem 45(12) (2012) 863–73. [DOI] [PubMed] [Google Scholar]
- 18.Raisz LG, Pathogenesis of osteoporosis: concepts, conflicts, and prospects, J Clin Invest 115(12) (2005) 3318–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gronthos S, Mankani M, Brahim J, Robey PG, Shi S, Postnatal human dental pulp stem cells (DPSCs) in vitro and in vivo, P Natl Acad Sci USA 97(25) (2000) 13625–13630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gronthos S, Brahim J, Li W, Fisher LW, Cherman N, Boyde A, DenBesten P, Robey PG, Shi S, Stem cell properties of human dental pulp stem cells, J Dent Res 81(8) (2002) 531–5. [DOI] [PubMed] [Google Scholar]
- 21.Lizier NF, Kerkis A, Gomes CM, Hebling J, Oliveira CF, Caplan AI, Kerkis I, Scaling-up of dental pulp stem cells isolated from multiple niches, PLoS One 7(6) (2012) e39885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iohara K, Nakashima M, Ito M, Ishikawa M, Nakasima A, Akamine A, Dentin regeneration by dental pulp stem cell therapy with recombinant human bone morphogenetic protein 2, J Dent Res 83(8) (2004) 590–5. [DOI] [PubMed] [Google Scholar]
- 23.Lindroos B, Maenpaa K, Ylikomi T, Oja H, Suuronen R, Miettinen S, Characterisation of human dental stem cells and buccal mucosa fibroblasts, Biochem Biophys Res Commun 368(2) (2008) 329–35. [DOI] [PubMed] [Google Scholar]
- 24.d’Aquino R, Graziano A, Sampaolesi M, Laino G, Pirozzi G, De Rosa A, Papaccio G, Human postnatal dental pulp cells co-differentiate into osteoblasts and endotheliocytes: a pivotal synergy leading to adult bone tissue formation, Cell Death Differ 14(6) (2007) 1162–71. [DOI] [PubMed] [Google Scholar]
- 25.Yang X, Walboomers XF, van den Beucken JJ, Bian Z, Fan M, Jansen JA, Hard tissue formation of STRO-1-selected rat dental pulp stem cells in vivo, Tissue Eng Part A 15(2) (2009) 367–75. [DOI] [PubMed] [Google Scholar]
- 26.Akkouch A, Zhang Z, Rouabhia M, Engineering bone tissue using human dental pulp stem cells and an osteogenic collagen-hydroxyapatite-poly (L-lactide-co-epsilon-caprolactone) scaffold, J Biomater Appl 28(6) (2014) 922–36. [DOI] [PubMed] [Google Scholar]
- 27.Logan CY, Nusse R, The Wnt signaling pathway in development and disease, Annu Rev Cell Dev Biol 20 (2004) 781–810. [DOI] [PubMed] [Google Scholar]
- 28.Clevers H, Nusse R, Wnt/beta-catenin signaling and disease, Cell 149(6) (2012) 1192–205. [DOI] [PubMed] [Google Scholar]
- 29.Gat U, DasGupta R, Degenstein L, Fuchs E, De Novo hair follicle morphogenesis and hair tumors in mice expressing a truncated beta-catenin in skin, Cell 95(5) (1998) 605–14. [DOI] [PubMed] [Google Scholar]
- 30.Cawthorn WP, Bree AJ, Yao Y, Du B, Hemati N, Martinez-Santibanez G, MacDougald OA, Wnt6, Wnt10a and Wnt10b inhibit adipogenesis and stimulate osteoblastogenesis through a beta-catenin-dependent mechanism, Bone 50(2) (2012) 477–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim JH, Liu X, Wang J, Chen X, Zhang H, Kim SH, Cui J, Li R, Zhang W, Kong Y, Zhang J, Shui W, Lamplot J, Rogers MR, Zhao C, Wang N, Rajan P, Tomal J, Statz J, Wu N, Luu HH, Haydon RC, He TC, Wnt signaling in bone formation and its therapeutic potential for bone diseases, Ther Adv Musculoskelet Dis 5(1) (2013) 13–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Y, Li Y-P, Paulson C, Shao J-Z, Zhang X, Wu M, Chen W, Wnt and Wnt signaling pathway in bone development and disease, Front Biosci (2014) 379–407. [DOI] [PMC free article] [PubMed]
- 33.Appendino G, Spagliardi P, Cravotto G, Pocock V, Milligan S, Daucane phytoestrogens: a structure-activity study, J Nat Prod 65(11) (2002) 1612–5. [DOI] [PubMed] [Google Scholar]
- 34.Palumbo C, Ferretti M, Bertoni L, Cavani F, Resca E, Casolari B, Carnevale G, Zavatti M, Montanari C, Benelli A, Zanoli P, Influence of ferutinin on bone metabolism in ovariectomized rats. I: role in preventing osteoporosis, J Bone Miner Metab 27(5) (2009) 538–45. [DOI] [PubMed] [Google Scholar]
- 35.Ferretti M, Bertoni L, Cavani F, Zavatti M, Resca E, Carnevale G, Benelli A, Zanoli P, Palumbo C, Influence of ferutinin on bone metabolism in ovariectomized rats. II: role in recovering osteoporosis, J Anat 217(1) (2010) 48–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Joseph M, Das M, Kanji S, Lu J, Aggarwal R, Chakroborty D, Sarkar C, Yu H, Mao HQ, Basu S, Pompili VJ, Das H, Retention of stemness and vasculogenic potential of human umbilical cord blood stem cells after repeated expansions on PES-nanofiber matrices, Biomaterials 35(30) (2014) 8566–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aggarwal R, Lu J, Kanji S, Joseph M, Das M, Noble GJ, McMichael BK, Agarwal S, Hart RT, Sun Z, Lee BS, Rosol TJ, Jackson R, Mao HQ, Pompili VJ, Das H, Human umbilical cord blood-derived CD34+ cells reverse osteoporosis in NOD/SCID mice by altering osteoblastic and osteoclastic activities, PLoS One 7(6) (2012) e39365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnstone B, Hering TM, Caplan AI, Goldberg VM, Yoo JU, In vitro chondrogenesis of bone marrow-derived mesenchymal progenitor cells, Exp Cell Res 238(1) (1998) 265–72. [DOI] [PubMed] [Google Scholar]
- 39.Sengupta D, Deb M, Rath SK, Kar S, Parbin S, Pradhan N, Patra SK, DNA methylation and not H3K4 trimethylation dictates the expression status of miR-152 gene which inhibits migration of breast cancer cells via DNMT1/CDH1 loop, Exp Cell Res 346(2) (2016) 176–87. [DOI] [PubMed] [Google Scholar]
- 40.Rice S, Whitehead SA, Phytoestrogens and breast cancer--promoters or protectors?, Endocr Relat Cancer 13(4) (2006) 995–1015. [DOI] [PubMed] [Google Scholar]
- 41.Ikeda K, Arao Y, Otsuka H, Nomoto S, Horiguchi H, Kato S, Kayama F, Terpenoids found in the umbelliferae family act as agonists/antagonists for ER(alpha) and ERbeta: differential transcription activity between ferutinine-liganded ER(alpha) and ERbeta, Biochem Biophys Res Commun 291(2) (2002) 354–60. [DOI] [PubMed] [Google Scholar]
- 42.Staal FJ, Clevers HC, WNT signalling and haematopoiesis: a WNT-WNT situation, Nat Rev Immunol 5(1) (2005) 21–30. [DOI] [PubMed] [Google Scholar]
- 43.Komiya Y, Habas R, Wnt signal transduction pathways, Organogenesis 4(2) (2008) 68–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Komori T, Regulation of bone development and extracellular matrix protein genes by RUNX2, Cell Tissue Res 339(1) (2010) 189–95. [DOI] [PubMed] [Google Scholar]
- 45.Young MF, Bone matrix proteins: their function, regulation, and relationship to osteoporosis, Osteoporos Int 14 Suppl 3 (2003) S35–42. [DOI] [PubMed] [Google Scholar]
- 46.Wei J, Karsenty G, An overview of the metabolic functions of osteocalcin, Rev Endocr Metab Disord 16(2) (2015) 93–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Neves VC, Babb R, Chandrasekaran D, Sharpe PT, Promotion of natural tooth repair by small molecule GSK3 antagonists, Sci Rep 7 (2017) 39654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Karmodiya K, Krebs AR, Oulad-Abdelghani M, Kimura H, Tora L, H3K9 and H3K14 acetylation co-occur at many gene regulatory elements, while H3K14ac marks a subset of inactive inducible promoters in mouse embryonic stem cells, BMC Genomics 13 (2012) 424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Soares LM, He PC, Chun Y, Suh H, Kim T, Buratowski S, Determinants of Histone H3K4 Methylation Patterns, Mol Cell 68(4) (2017) 773–785 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


