Abstract
Enumeration of tumor-infiltrating lymphocytes (TILs) in H&E stained tissue sections has demonstrated limited value in predicting immune responses to cancer immunotherapy, likely reflecting the diversity of cell types and immune activation states among tumor infiltrates. Multiparametric flow cytometry enables robust phenotypic and functional analysis to distinguish suppression from activation, but tissue dissociation eliminates spatial context. Multiplex methods for immunohistochemistry (IHC) are emerging, but these interrogate only a single tissue section at a time. Here, we report transparent tissue tomography (T3) as a tool for three-dimensional (3D) imaging cytometry in the complex architecture of the tumor microenvironment, demonstrating multiplexed immunofluorescent analysis in core needle biopsies. Using T3 imaging, image processing and machine learning to map CD3+CD8+ cytotoxic T cells (CTLs) in whole core needle biopsies from Her2+ murine mammary tumors and human head and neck surgical specimens revealed marked inhomogeneity within single needle cores, confirmed by serial section IHC. Applying T3 imaging cytometry, we discovered a strong spatial correlation between CD3+CD8+ CTLs and microvasculature in the EGFR+ parenchyma, revealing significant differences among head and neck cancer patients. These results show that T3 offers simple and rapid access to three-dimensional and quantitative maps of the tumor microenvironment and immune infiltrate, offering a new diagnostic tool for personalized cancer immunotherapy.
Introduction
Core needle biopsy is a minimally invasive, image-guided procedure commonly used to obtain tumor tissue for diagnostic, prognostic, and predictive tests in the clinical setting [1–4]. For tumor immune profiling, a core needle biopsy is typically formalin fixed and paraffin embedded (FFPE), thin sections are cut, and these are stained for morphology by hematoxylin and eosin (H&E) and immune markers by chromogenic immunohistochemistry (IHC) [5]. Analyzing samples by IHC typically takes a few days and requires considerable technical expertise, but allows specific types of tumor infiltrating lymphocyte such as CD8+ cytotoxic T cells (CTLs) or CD4+ FoxP3+ suppressor T cells to be counted and localized with respect to tumor cells [6, 7]. Recent progress in multiplexed fluorescent IHC has enabled simultaneous analysis of multiple immune cell types in a single tissue section, though this additional information comes at the cost of an additional day or two for staining and analysis [8, 9]. Nonetheless, even if fully characterized, individual tissue sections cannot adequately sample the complex three-dimensional (3D) architecture of the tumor microenvironment, which is characterized by a heterogeneous distribution of diverse cell types and a disordered microvasculature [10]. There remains a pressing need for a rapid sample-to-answer technology for multiplex 3D imaging at cellular resolution that is able to fully characterize the tumor immune microenvironment in core needle biopsies.
Driven by demand for new methods to characterize the brain connectome, diverse approaches to tissue clearing, antigen detection and 3D scanning have been introduced recently [11, 12], several of which have also demonstrated potential as tools to map the tumor microenvironment [13–20]. At the same time, these pioneering efforts have revealed fundamental limitations of current approaches including tissue distortion, antigen loss, uneven immunostaining, limited imaging depth, low sample throughput and need for dedicated equipment [21]. Particularly relevant, current tumor staining, clearing and 3D imaging methods remain poorly matched to the demands of the clinical environment, where speed is at a premium and simple, robust methods are required.
To address these challenges, we have adapted Transparent Tissue Tomography (T3) [22] to enable a fast, simple and clinically relevant core needle biopsy imaging method. Here, whole core needle biopsies are lightly fixed, immunostained with fluorescently labeled primary antibodies, optically cleared, and imaged by confocal microscopy. Open source image processing and machine learning tools are applied to determine the 3D distribution of multiple antigens and identify cell types throughout the whole core needle biopsy, characterizing the tumor microenvironment at microscopic, mesoscopic and macroscopic scales. Mapping up to six antigens in a core at cellular resolution can be completed in 3 days. Moreover, as T3 is a non-destructive process, the core can still be processed by FFPE and analyzed by IHC.
Here, we apply this new method to map CTLs in whole core needle biopsies obtained from spontaneous murine mammary tumors and human head and neck cancer surgical specimens. Our data confirm the heterogeneous distribution of CTLs in the tumor microenvironment and reveal that an element of this pattern is defined by the tumor microvasculature. Our work establishes T3 as a novel tool for high resolution, quantitative 3D analysis of immune infiltrates in core needle biopsies of patient tumors.
Materials and methods
Mouse core needle biopsy
Transgenic BALB/c males carrying the mutated rat Her2/neu oncogene driven by the MMTV promoter were bred with wild type females [23]. Genotypes of offspring were determined by PCR of tail snips. BALB-NeuT female mice developed spontaneous mammary carcinoma in each mammary gland between 5 and 33 weeks of age. All mice were maintained under specific pathogen-free conditions in accordance with the animal experimental guidelines set by the Institutional Animal Care and Use Committee. The study has been approved by the Institutional Animal Care and Use Committee of the University of Chicago and all experiments conformed to the relevant regulatory standards. Tumors were collected from mammary glands of BALB-NeuT mice, and then frozen in liquid nitrogen (N2) until analyzed. The tumors were thawed in RPMI 1640 media mixed with 10 mg/ml bovine serum albumin (BSA) on ice. Core biopsies were obtained from thawed tumors using an 18 gauge automated core needle biopsy device (MAX-CORE, BRAD Biopsy), fixed with 2% paraformaldehyde in PBS for 5 min at room temperature, and washed with PBS.
Human head & neck core biopsy
De-identified fresh head and neck surgical biopsies (from 7 patients) were obtained from the University of Chicago Medicine with patient consent (Supplemental Table 1). None of the patients had received any local or systemic treatment prior to surgical excision of the tumor masses examined here. The study has been approved by the Institutional Review Board (IRB) of the University of Chicago. Some biopsies were stored by freezing in liquid nitrogen (N2) until analyzed. Three core needle biopsies were collected from each fresh or thawed tumor, and fixed and washed as above.
Agarose well preparation
Short sections of 18 gauge needle stock were laid into 2 cm diameter glass bottom dishes. Hot 2% agarose (LE Quick Dissolve Agarose, GeneMate) in distilled water was poured into each dish, a glass vial cap placed in the dish, and the dish cooled on ice. Cast agarose discs were cut from the wells and the needles removed. A single core was loaded in each well.
Conjugation of primary antibodies with fluorescent dyes or biotin
Anti-rat Her2 antibody was isolated from hybridoma 7.16.4 (ATCC) culture supernatant at the Frank W. Fitch Monoclonal Antibody Facility at the University of Chicago. Other antibodies were purchased from commercial sources. Primary antibodies diluted in phosphate buffered saline pH 8.0 (PBS, Corning) were conjugated to fluorescent dyes as shown in Supplemental Table 2. Reactions were incubated with gentle agitation at 4°C overnight. Unreacted dye was removed by dialysis (MWCO 10K) against PBS pH 7.4 at 4°C for 3 days. Fluorescent antibody solutions were stored at 4°C. For secondary immunohistochemistry examination of human CD3 T cells in head and neck core FFPE sections, we prepared biotin-anti-CD3 antibody by conjugating rabbit anti-human CD3 antibody (DAKO, Code A0452, 0.6 mg/ml) with biotin using Pierce™ antibody biotinylation kit (ThermoFisher).
Immunofluorescence staining
Core needle biopsies were stained with antibody cocktails in staining buffer (SB) composed of RPMI 1640 media with 10 mg/ml BSA and 0.3% (for mouse core biopsies) or 0.1% (for human core biopsies) Triton X-100. For Fig. 2 and Supplemental Fig. 1, 100 μl DyLight488-anti-rat Her2, 50 μl DyLight550-anti-mouse CD3, 80 μl DyLight594-anti-mouse CD8, 40 μl DyLight-633-anti-mouse CD31, 100 μl DyLight-680-anti-mouse F4/80, and 300 μl DyLight-755-anti-mouse ER-TR7 were combined in 1 ml SB. For Supplemental Fig. 3, 100 μl DyLight488-, DyLight550-, DyLight594-, DyLight633-, DyLight680-, or DyLight755-anti-rat Her2 was added in 1 ml SB. For Fig. 4 and Supplemental Fig. 4, 8 and 11, 160 μl DyLight488-anti-human EGFR, 60 μl DyLight550-anti-human CD3, 60 μl DyLight594-anti-human CD8, and 40 μl DyLight-633-anti-human CD31 were combined in 1 ml SB. For Supplemental Fig. 5, 100 μl DyLight488-anti-rat Her2 and 10 μl LDS 751 (10 mg/ml DMSO) were combined in 1 ml SB. Core biopsies were incubated with the antibody cocktails for 18 h at 4°C. After incubation, core biopsies were washed three times for 10 min in PBS pH 7.4 at 4°C, fixed with 2% paraformaldehyde in PBS for 5 min at room temperature, and washed in PBS pH 7.4. We directly evaluated the effects of blocking and staining in RPMI 1640 on our approach. First, we compared 6-plex primary antibody staining of BALB-NeuT tumor tissue with or without pre-incubation in 10% goat serum solution for 3 h. We observed no difference in staining quality for any of the antibodies with or without a blocking step (Supplemental Fig. 1). Next, we compared autofluorescence after mock staining BALB-NeuT tumor tissue by incubation in RPMI 1640 or PBS staining buffer for 18 h. We did not observe any difference in autofluorescence (Supplemental Fig. 2).
Fig. 2.
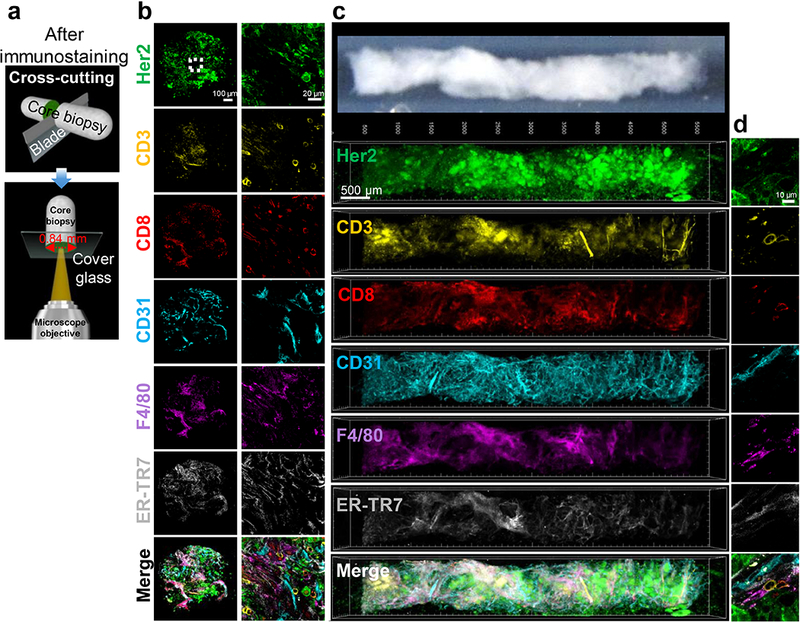
Multiplex fluorescent immunostaining and 3D image reconstruction of whole core needle biopsies using T3. a Diagram shows slicing and imaging of cross-section of cores to assess antibody penetration after multiplex immunostaining. b Cross section images from BALB-NeuT tumor core after immunostaining for Her2 (green), CD3 (yellow), CD8 (red), CD31 (cyan), F4/80 (magenta), and ER-TR7 (gray). Left column shows 10X objective scan of 0.84 mm diameter core. Right column shows 40X objective region-of-interest imaging of the center of the cross section (white dotted square) demonstrating staining of tumor cells, T cells, microvasculature, macrophages, and fibroblasts. Scale bars: 100 μm (left) and 20 μm (right). c Six-plex immunostaining, clearing, scanning and image analysis of a whole BALB-NeuT tumor core (top image) reveals 3D distributions of Her2 tumor cell marker (green), CD3 T cell marker (yellow), CD8 cytotoxic T cell marker (red), CD31 endothelial marker (cyan), F4/80 macrophage marker (magenta), and ER-TR7 fibroblast marker (gray), displaying each channel in pseudocolor individually and in a merged image. Scale bar: 500 μm. d High resolution 2D images of the cell markers in the core. Scale bar: 10 μm.
Fig. 4.
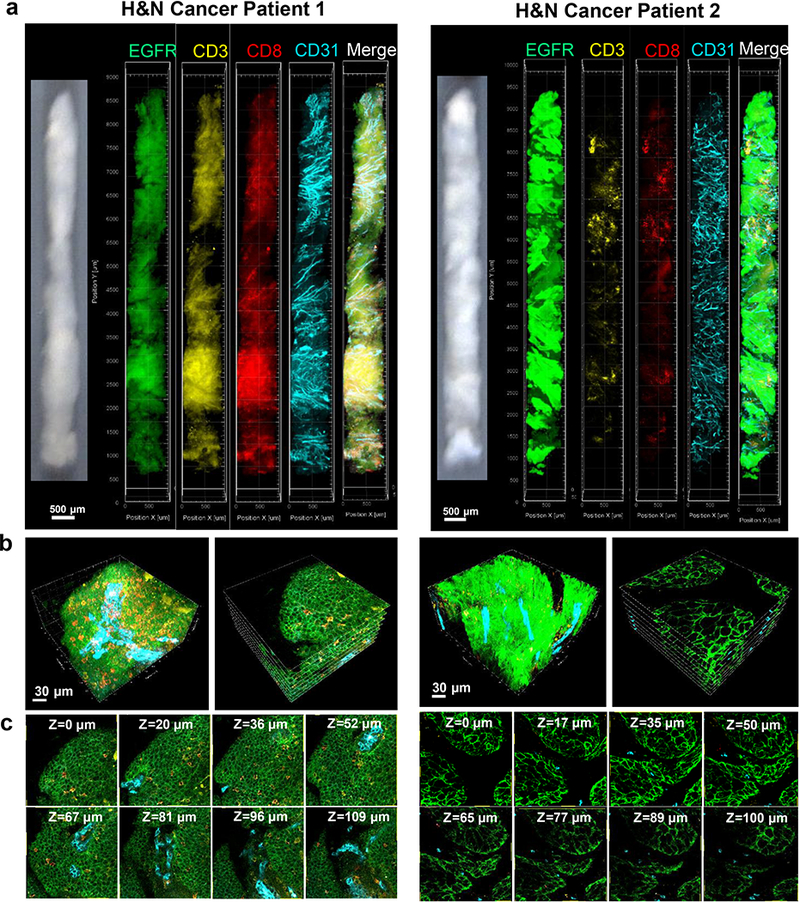
3D visualization and mapping of CTLs in whole head and neck cancer cores. a Cores obtained with an 18 gauge automated device from head and neck cancer samples from patients 1 and 2 (left images) were immunostained for EGFR (green), CD3 (yellow), CD8 (red), and CD31 (cyan), cleared and scanned. 3D rendering of whole cores are shown for each channel and a merged image. Scale bar: 500 μm. b High resolution 3D view (left) and X-Y tomographic visualization (right) of a volume within the rendered core models, showing distributions of CD3+CD8+ CTLs (orange) and CD31+ endothelium (cyan) within the EGFR+ (green) tumor tissue and stroma. Scale bar: 30 μm. c Serial tomographic sections of X-Y planes at different Z-stack depths showing CTLs (orange) and endothelium (cyan) within the EGFR+ (green) tumor tissue.
Optical clearing of core needle biopsies
To prepare D-fructose solutions for optical clearing, 20, 50, and 100% (w/v) solutions of D-fructose were prepared in 10 mM phosphate buffer pH 7.8 to a final volume of 10 ml. 30 μl of α-thioglycerol (Sigma Aldrich) was added to each D-fructose solution. Immunostained core needle biopsies were incubated sequentially in 10 ml of 20, 50, and 100% D-fructose solutions for 30 min, 30 min, and 1 h at 25°C with gentle agitation in 20 ml glass vials, respectively. The immunostained cores and agarose were then mounted between coverslips in 100% (w/v) D-fructose solutions.
Confocal microscopic imaging
We used a Leica TCS SP8 confocal laser scanning microscope, white light laser, Leica HCX PL APO 10X/0.4 NA dry objective (2.2 mm working distance) or Leica HC PL APO 40X/1.25 NA oil objective (0.24 mm working distance), and a SuperZ galvometric scanning stage. Using the 10X objective, one half of an immunostained and cleared core was scanned, the sample flipped over, and the other half scanned. 3D scanning was performed in a defined X/Y/Z (1.82/1.82/7.5 μm) volume with 10 frame averaging and bidirectional scanning using 488 nm excitation and 495–530 nm emission filter for DyLight488, 550 nm and 563–579 nm filters for DyLight550, 594 nm and 603–624 nm filter for DyLight594, 633 nm and 648–662 nm filter for DyLight633, 670 nm and 698–721 nm filter for DyLight680, 670 nm and 749–800 nm filter for DyLight755, and 550 nm excitation and 700–722 nm emission filters for LDS 751 nuclear staining.
Image processing
To evaluate and compensate for fluorescent channel spillover for multiplex immunofluorescence imaging, six BALB-NeuT tumor tissue samples were individually stained with only 7.16.4 anti-rat Her2 antibody conjugated to one of the DyLight dyes used in the 6-plex imaging, 488, 550, 594, 633, 680, or 755, and then imaged with the standard 6-plex capture protocol. To evaluate crosstalk, the mean intensity of fluorescence of a Her2+ region in each capture channel was compared to that of the signal in the optimal channel. Representative data are shown in pseudocolor with the same intensity range in Supplemental Fig. 3a and percent of optimal signal intensity are presented in Supplemental Fig. 3b. In all images for the 755 channel (CH6), laser backscanner contributes 2–15% signal, as is evident in the 488 channel top row rightmost panel. While crosstalk is <10% for most combinations, for DyLight594, 72% of the signal that appeared in the 594 channel was detected in the 633 channel. The second row of 594 channel images demonstrates the effect of the linear compensation by subtracting 72% of the scaled 594 image from the 633 image, thereby reducing the bleed over to <7%.
We developed macros for automated 3D reconstruction of whole core needle biopsy images using open-source plugins in Fiji (http://fiji.sc/Fiji). Using the grid/collection stitching plugin [24], we built macro LIFtile-restitcher to align and stitch 3D mosaics for multi-channel “hyperstack” images (0–999 image tiles). To compensate for depth-related intensity losses, the mean intensity value (of Otsu-thresholded areas; maximum gain limited to 3X) of each optical slice in the core biopsy images was normalized to the mean value of the entire core biopsy for each channel using the macro HPRstackConstantMean, facilitating thresholding and segmentation as used in cell enumeration. We designed the macro biopSEEr to enable automated alignment and registration of the stitched core biopsy images by the trakEM2 plugin (https://imagej.net/TrakEM2) (Detailed instruction in Supplemental Methods). Data overlap is combined and redundancy removed with CombineMatchesMacro. Next, we used macro hyprBKGDfix to clear background outside the stained core biopsy to facilitate 3D volume visualization. Finally, the 3D images were deconvolved using Huygens Pro software v. 4.3 (Scientific Volume Imaging), to correct for refractive index mismatch, and to enhance spatial signals and cell morphology discrimination. We constructed 3D tomographic visualizations and movies of the core biopsy images using Imaris software v. 9.0 (Bitplane).
Data analysis
We developed the macro core biopsy extractor for automated segmentation of EGFR+ lesion and CD31+ vasculature from the EGFR and CD31 channel images. For hyperstack segmentation of CD3+CD8+ cytotoxic T cells from the images, we used macro wekaMacro using the trainable Weka segmentation plugin [25]. The classifier was trained to extract the cells only showing fluorescence signals on the membrane. Tumor vascular features were extracted as follows: Depth-related signal attenuation was normalized to a common mean, and CD31 signal was amplified tenfold where above the mean background per focal slice whereas background pixels were decreased three-fold. Feature edges were enhanced by means of a 3D unsharp mask filter. Vessels were detected by using a “tubeness” extractor (data padded by one image at each end to avoid lack of fitting the end images by the plugin) with sigma set at 3.3 for our data (larger values favor connectedness of nearby structures, but increase feature radii; smaller values can leave the vessel lumen unfilled), and application of a binarization threshold set as the mean (stack) values of triangle and of Otsu methods. Vessel content of each optical slice was measured as the area fraction of the extracted binary map. The binary vessel map was converted to a 3D skeleton and to a 3D Euclidean distance map. The 3D skeleton was used to generate the vessel radial thickness at each voxel by masking the 3D distance map. The 3D skeleton was measured using the “analyze skeleton” plugin to yield values of branching order, segment lengths, tortuosity, etc. Vessel volume was taken as binary area times the slice Z step size. We analyzed the 3D morphological features of the microvasculature throughout three cores each from Patients 1 and 2. To measure distance of CD3+CD8+ cytotoxic T cells from CD31+ tumor vasculature, we applied the 3D distance map plugin to the tumor blood vessels [26]. All other image analysis was conducted with basic analyze functions in Fiji software.
Code availability
Fiji macros (Supplemental Table 3 and Fiji macro script 1–4) can be downloaded at Laboratory Investigation’s website.
Immunohistochemistry
After 3D imaging, cores were incubated sequentially in 10 ml of 50, 20, and 0% D-fructose solutions with gentle agitation in 20 ml glass vials at 25°C for 30 min, 30 min, and 1 h, respectively, fixed with 4% paraformaldehyde in PBS overnight at 4°C, and washed in PBS pH 7.4. Paraffin embedding and serial sectioning at 5 μm thickness were performed by the Human Tissue Resource Center at the University of Chicago. To evaluate CD8+ T cell distribution in a BALB-NeuT tumor core biopsy, 12 sections were selected from a total of 90 serial sections by drawing every 7th section. After deparaffinization and serial rehydration, biopsy sections were treated with citrate buffer pH 6.0 in an epitope retrieval steamer (IHC-Tek, IHCWorld). Rat anti-mouse CD8α antibody (eBioscience, 4SM16, 1:200) was applied overnight at 4°C in a humidity chamber. Following a PBS wash, core biopsy sections were incubated with horseradish peroxidase (HRP) polymer anti-rat IgG reagent (Vector Laboratories, MP-7404) for 30 min at room temperature. Antigen-antibody binding was detected using a peroxidase substrate kit (Vector Laboratories, SK-4105). Human head and neck core biopsy sections also went through the same deparaffinization and epitope retrieval processing with citrate buffer pH 6.0. The sections were incubated with mouse anti-human EGFR antibody-biotin (ThermoFisher Scientific, 111.6, 1:25), mouse anti-human pan-cytokeratin antibody-biotin (Novus Biologicals, C11, 1:25), mouse anti-human CD8α antibody-biotin (ThermoFisher Scientific, AMC908, 1:100), or rabbit anti-human CD3 antibody-biotin (DAKO, Code A0452, 1:25) for 1 h at room temperature. After washing with PBS, core biopsy sections were incubated with horseradish peroxidase (HRP) conjugated streptavidin (Vector Lab., Cat# PK-6100) for 30 min at room temperature. Antigen-antibody binding was detected using a DAB+ substrate chromogen system (DAKO, Cat# K3468). The core biopsy sections were briefly immersed in hematoxylin for counterstaining and mounted with cover glasses. 2D IHC images were obtained using a whole slide scanner (Pannoramic SCAN BF, Perkin Elmer) with 40X objective. We counted CD8+ cells on every seventh section (11 sections from 80 serial sections) of human head and neck cores.
Statistical analysis
Statistical values are expressed as mean ± SEM, and statistical comparisons between groups were made using one-way ANOVA, Student’s t test or K-means cluster analysis as noted. P value of <0.05 was considered statistically significant.
Results
Transparent Tissue Tomography (T3) for core needle biopsy
Using an 18 gauge (0.84 mm ID) automated core needle biopsy device, we obtained tissue cores from fresh or frozen and thawed BALB-NeuT murine mammary tumors and human head and neck cancer surgical masses. Core biopsies were embedded in agarose, lightly fixed, immunostained with multiple fluorescent dye-labeled primary antibodies, optically cleared by incubation in 100 (w/v) % D-fructose, and imaged with a confocal microscope (Fig. 1 and Table 1). Fluorescent primary antibodies were directly labeled by conjugation to amine-reactive, N-hydroxysuccinimide (NHS)-ester activated fluorophores at specific dye:antibody molar ratios (Supplemental Table 2). Typically, four to six fluorescent primary antibodies were combined and applied to the tissue in a cocktail. To evaluate antibody and nuclear staining dye penetration, 0.84 mm diameter cores were stained with fluorescent antibodies and dye to detect cancer cells (Her2 or EGFR for murine and human samples, respectively), cytotoxic T cells (anti-murine or -human CD3 and CD8), vascular endothelium (anti-murine or -human CD31), macrophages (anti-murine F4/80), fibroblasts (anti-murine ER-TR7), and cell nucleus (LDS 751). Imaging cross-sections of cores showed that incubation for 18 h at 4 °C was sufficient for fluorescent antibodies and nuclear staining dye to fully penetrate and stain the tissue (Fig. 2, Supplemental Fig. 4 and Fig. 5). After sandwiching cores between glass coverslips, the tissue was scanned in each color by confocal microscopy with a 10X/0.4 NA air objective at 7.5 μm Z-steps to a depth halfway through the diameter of the core and then the sample flipped over to image the other half (Fig. 1e). Then, using a macro based on the open source image analysis software Fiji [27] and its plugins, the two half cylinder images were fused into one by aligning the CD31+ microvasculature across the interface (Fig. 1f, Supplemental Fig. 6a and Movie 1).
Fig. 1.
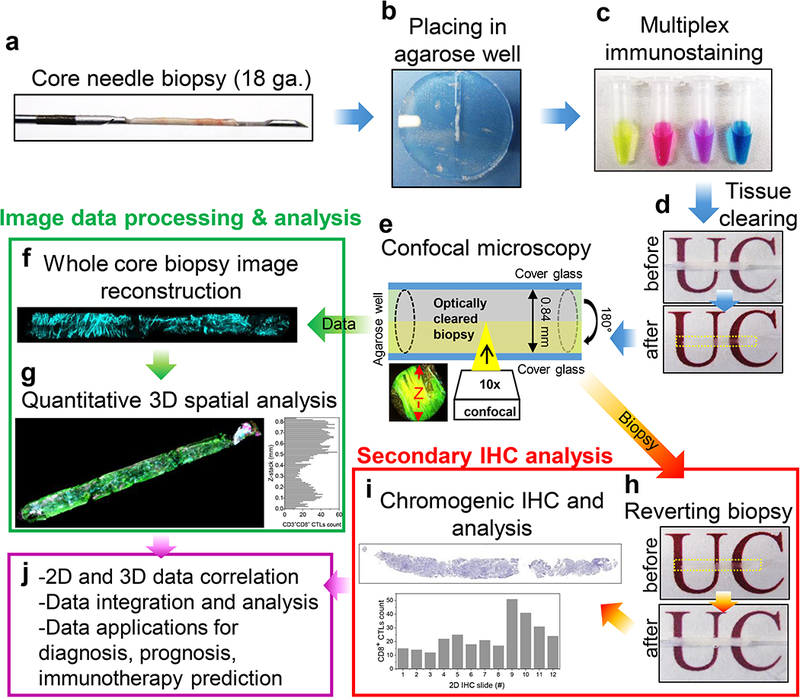
Workflow of Transparent Tissue Tomography (T3) for whole core needle biopsies. a Tissue cylinders (cores) are collected using an automated 18 gauge needle (0.84 mm ID) core biopsy device and b cores are fixed with 2% paraformaldehyde and loaded into agarose wells. c Cores are stained overnight with cocktails of fluorescent primary antibodies. d After washing and fixing, cores are rendered transparent by equilibration with 100% (w/v) D-fructose, mounted between coverslips, and e scanned on one side by confocal microscopy in each fluorescent channel, then flipped and scanned on the other side. f The core is reconstructed by fusing half cylinder images and g spatial analysis of the immune microenvironment is performed with Fiji macros. For secondary conventional IHC analysis, h imaged cores are washed free of D-fructose, fixed, and i submitted for FFPE processing and chromogenic IHC assay. Thereby, j correlated 2D and 3D data can be collected and integrated, providing a starting point for diagnostic, prognostic, and predictive testing for cancer immunotherapy.
Table 1.
Overview of T3 procedure for core needle biopsy and average times required for each step
| PROCESS | AVERAGE TIME | NOTES |
|---|---|---|
| Harvesting core needle biopsy and fixing | 10 min | -Using a core biopsy needle (e.g. 18 gauge) -Fixing in 2% PFA for 5 min at 25°C -Washing in PBS |
| Immunofluorescence staining | 18 h | -Incubating with fluorescent primary antibodies at 4°C -Washing in PBS -Fixing in 2% PFA for 5 min at 25°C |
| Optical clearing | 2 h | -Incubating sequentially with 20% (30 min), 50% (30 min), 100% (1 h) D-fructose solutions (containing 0.3% (v/v) α-thioglycerol) at 25°C |
| Confocal microscopic imaging | 1 h 30 min/biopsy (10X objective, 18 gauge biopsy of 0.5 cm length and 0.84 mm diameter) |
-Using a 10X objective and voxel dimensions of X/Y/Z = 1.82/1.82/7.5 μm -Using 6 excitation and emission filters -Using a 40X objective for high-resolution 3D imaging of area of interest |
| Image processing and analysis | 1 day | -Using commercial and open-source software, and custom macros for image reconstruction, deconvolution, and 3D analysis |
| Reverting biopsy | 2 h | -Incubating sequentially with 50% (30 min), 20% (30 min), 0% (1 h) D-fructose solutions at 25°C |
| FFPE and IHC | ~3–5 days | -Dehydration, paraffin embedding, sectioning (5 μm), IHC staining, light microscopy |
Given uncertainty about depth of focus in thick, refractive index-matched samples as used here, a concern was that scanning at 10X with a 7.5 μm Z-step might fail to capture smaller features such as lymphocytes. Thus, we stained a head and neck cancer tissue core with anti-CD8 antibody, cleared the tissue with fructose, mounted it and then collected data using the 10X objective at 1 μm Z step intervals. Of CD8+ cells detected after image reconstruction using the full data set, 99.3% (1495 of 1505) were also detected using every seventh scan (7 μm Z-step) and 84% using every fourteenth scan (14 μm Z-step). These data suggest that a 7.5 μm Z-step spacing is sufficient to detect lymphocytes with satisfactory sensitivity.
At higher magnification, T3 revealed the 3D distribution of immune infiltrates at cellular resolution (Supplemental Fig. 6b). The reconstructed cores were then subjected to further image processing including channel spillover elimination and multiparameter 3D spatial quantification analysis (Fig. 1g) using Fiji macros (Supplemental Table 3), to map distributions of specific cell types in the immune infiltrate in whole core needle biopsies. After scanning, the cores were washed with PBS to remove the D-fructose and processed by conventional FFPE, thin sectioning and IHC (Fig. 1h, i), allowing direct comparison of 2D and 3D data (Fig. 1j).
3D mapping of immune infiltrates in whole core needle biopsy
To model analysis of immune infiltrate in patient diagnostic biopsies, we examined the distribution of CD3+CD8+ cytotoxic T lymphocytes (CTLs) in whole cores obtained from spontaneous mammary tumors formed in female BALB-NeuT mice expressing rat Her2 [28] and human head and neck cancer surgical mass (Patient #0 in Supplemental Table 1). Without embedding or sectioning, the cores were immunostained for Her2 (or EGFR for human samples), CD3, CD8, and CD31, optically cleared, and imaged (Supplemental Fig. 7 and Fig. 8). High resolution images showed complete staining for each biomarker (Supplemental Fig. 7b and Fig. 8b). While abundance of tumor-infiltrating CTLs has been proposed as a biomarker to predict clinical response to cancer immunotherapies such as immune checkpoint blockade therapy [29–31], the density of inflammatory lymphocytes is often inhomogeneous. Toward examining the feasibility of 3D imaging to evaluate infiltrating CTLs, we used a machine learning approach (Supplemental Fig. 9) to identify and map CD3+CD8+ cytotoxic cells in cores isolated from six different NeuT tumors (Fig. 3a) and from a head and neck cancer mass (Supplemental Fig. 10a). To examine potential advantages of 3D analysis, we counted CTLs on virtual optical sections along the long axis of each mouse or human core biopsy (Fig. 3b, Supplemental Movie 2 and Fig. 10b). Given the cylindrical shape, sections closer to the center will tend to have higher numbers of cells and the data also suggest artifacts may arise during fusion of the two half-cylinder core images, but the data revealed highly non-uniform CTL distributions across the biopsies. To compare this approach to conventional analysis, following T3 imaging, mouse and human cores were washed to remove fructose and subjected to formalin fixation and paraffin embedding (FFPE). Serial thin sections cut along the long axis of the core were examined by immunohistochemistry with a different anti-CD8α antibody clone (Fig. 3c and Supplemental Fig. 10c). Enumeration of the CD8+ cells in multiple sections collected from case #1 core confirmed the inhomogeneous distribution of CTLs determined by T3 analysis (Fig. 3d and Supplemental Fig. 10d).
Fig. 3.
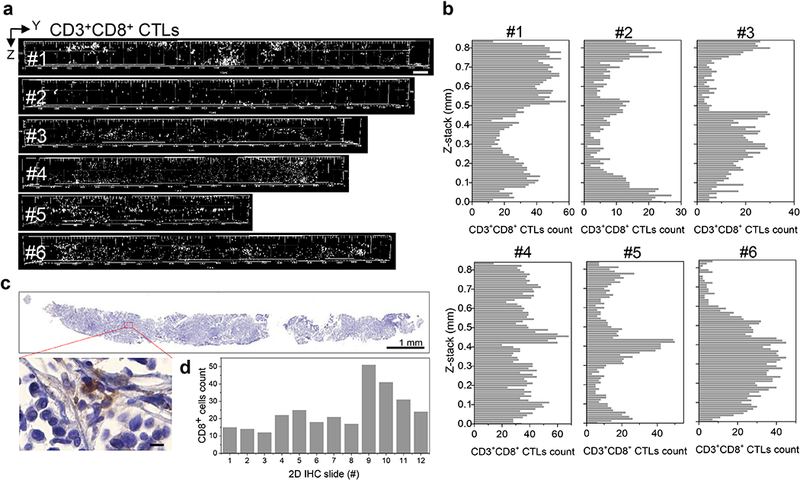
3D mapping and analysis of cytotoxic T lymphocytes in whole mouse core needle biopsy. a 3D spatial mapping of CD3+CD8+ cytotoxic T lymphocytes (CTLs) in cores from six NeuT tumors displays local clustering and inhomogeneity. Scale bar: 500 μm. b Enumeration of CTLs in virtual optical sections along the long axis of each core in a demonstrates heterogeneous distribution of CTLs. c Conventional IHC after T3 confirms non-uniform distribution of CD8+ cells. Core #1 in a was formalin fixed and paraffin embedded, sectioned, immunostained, counterstained, and scanned. Inset displays high-magnification image of CD8+ cells. Scale bar: 1 mm (top) and 10 μm (inset). d Manual counting of CD8+ cells after IHC. Every seventh 5 μm serial section was stained and counted.
3D visualization of immune infiltrates in whole human head and neck core needle biopsy
With the protocol established, we applied T3 to explore the distribution of CD3+CD8+ CTLs in human tumors. Thus, we applied the T3 protocol to sets of three cores cut from each of six tumor specimens excised during surgery to treat head and neck cancer (Supplemental Table 1) and visualized expression of tumor marker EGFR, CTL markers CD3 and CD8, and endothelial marker CD31 (Fig. 4a and Supplemental Fig. 11). At higher magnification, T3 revealed distinct distributions of CTLs and microvasculature in tumor parenchyma versus stroma among the different tumors (Fig. 4b and Supplemental Movie 3). Given the volumetric data on multiple markers, it was also possible to examine virtual sections (Fig. 4c and Supplemental Movie 4), which enabled 2D representations of the data, facilitating evaluation of immune infiltrate distributions and their spatial relationships within the tumor microenvironment. T3 images of Patient 1’s core biopsy displayed high numbers of infiltrating CTLs along with a highly vascularized tumor parenchyma. In contrast, Patient 2’s core biopsy displayed a “cold” immune microenvironment with less vascularization in the tumor parenchyma.
3D quantitative mapping of CTLs with respect to microvasculature
To evaluate the spatial correlation of CTLs and the tumor microvasculature, we defined the parenchyma based on EGFR immunoreactivity and quantified CD3+CD8+ CTLs and CD31+ vasculature in whole cores (Fig. 5). Based on the 3D segmentation, we determined the volume (%) of EGFR+ parenchyma in sets of three cores cut from head and neck tumor tissue excised from six cancer patients (Fig. 5a). Automated machine learning-based segmentation facilitated the extraction and 8 bit mapping of CD3+CD8+ CTLs and CD31+ endothelial vasculature in the 3D core images, enabling the volume (%) measurement of CTLs and microvasculature within whole cores as well as EGFR+ domains (Fig. 5b, c). After normalization to account for the different fractions of tumor tissue among the cores, we detected a strong correlation between CTL infiltration and microvasculature density within the EGFR+ domains using K-means cluster analysis (Fig. 5d). The six head and neck cancer samples fell into three groups based on common image features (P < 0.05 for both X and Y axes), separating Patients 1 and 2 into “hot” and “cold” tumors [32, 33]. High resolution images of cores from Patients 1 and 2 confirmed distinct and correlated patterns of immune infiltrate and microvasculature in stroma and EGFR+ parenchyma (Fig. 5e). 3D spatial distance mapping of the “hot”, inflamed Patient 1 sample revealed that over 54% of CD3+CD8+ CTLs localized at less than 10 μm distance from micro-blood vessels (Fig. 5f). To further examine features of the microvasculature, the T3 images were segmented to reveal the 3D vascular network across whole cores from Patients 1 and 2 (Fig. 6a). We quantitatively analyzed the topological features such as vessel diameter, branch count, branch length, and tortuosity (Fig. 6b, c). Analysis of the core from Patient 1, which showed a higher level of immune infiltrate, revealed a distinctly more branched and tortuous vascular network. Together, these results suggest that the microvasculature may play a role in determining amount and distribution of immune infiltrate in tumors.
Fig. 5.
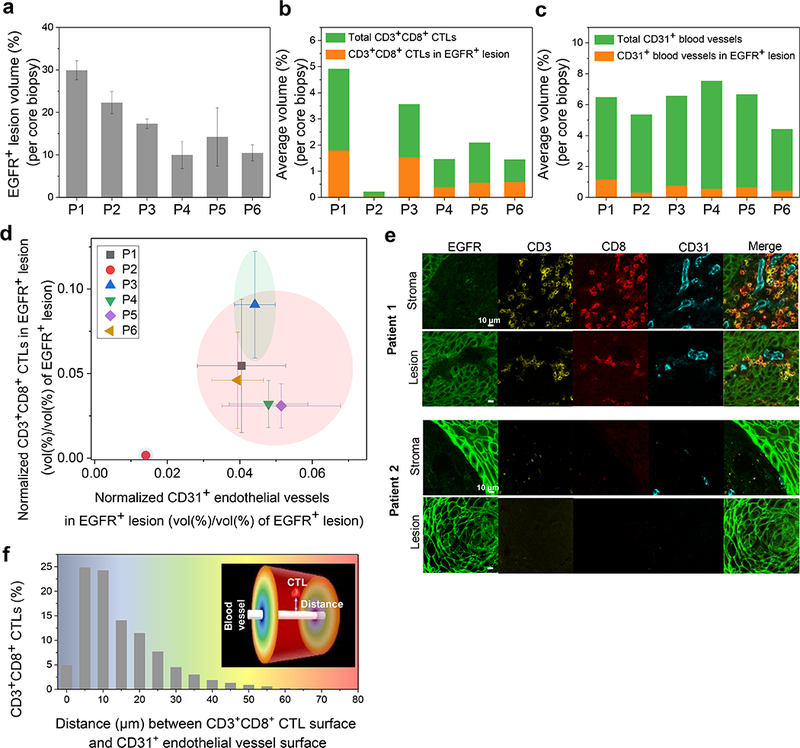
Spatial correlation between CTLs and microvasculature in tumor parenchyma in head and neck cancer. a Relative quantification of EGFR+ parenchyma (%) in sets of three cores each from six excised head and neck cancer patient samples. Data are expressed as mean ± SEM. b Quantification of relative volumes (%, mean ± SEM, n = 3) of CD3+CD8+ CTLs within the EGFR+ parenchyma (orange) and the whole cores (green) for each patient sample. c Quantification of relative volumes (%, mean ± SEM, n = 3) of CD31+ microvasculature within the EGFR+ parenchyma (orange) and the whole cores (green) for each patient. d Plot comparing normalized densities of CD3+CD8+ CTLs and CD31+ microvasculature in EGFR+ parenchyma for each patient (% CD3+CD8+ (b) or % CD31+ (c) within EGFR+ volume/% EGFR+ volume (a), mean ± SEM, n = 3). Three groups (blue, red, and green) were defined using K-means cluster analysis (P<0.05 for both X and Y axes), distinguishing one patient, 1, with a “hot”, inflamed tumor and another patient, 2, with a “cold”, non-inflamed tumor. e High resolution 2D images of CD3+CD8+ CTLs and CD31+ endothelium in EGFR+ parenchyma and stroma in Patients 1 (“hot”) and 2 (“cold”) cores. Scale bar: 10 μm. f 3D distance profiles of CD3+CD8+ CTLs away from CD31+ blood vessels in Patient 1 (“hot”) core. Inset shows a color distance map between blood vessel and CTL surfaces.
Fig. 6.
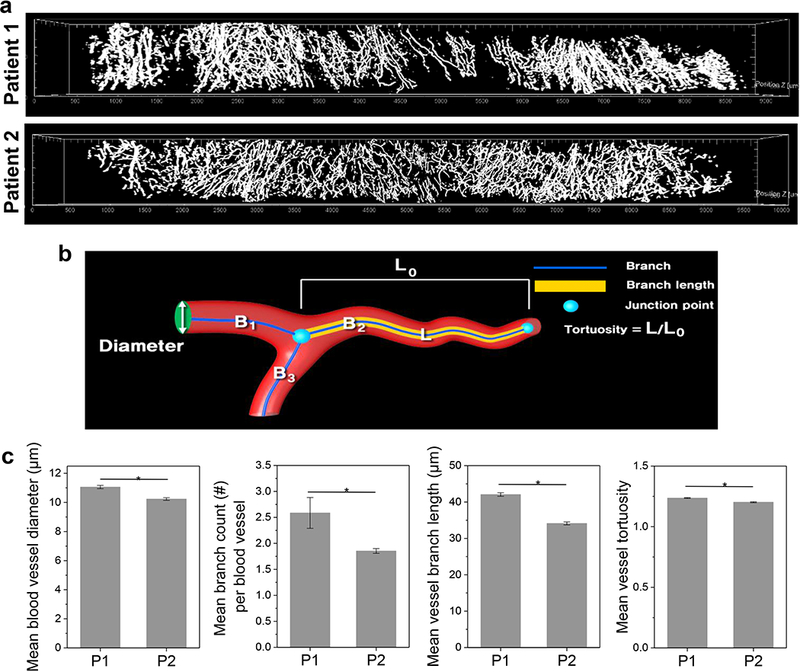
3D topological features of microvasculature in whole head and neck cancer cores. a 3D rendering of segmented CD31+ microvasculature in cores from Patient 1 and 2 samples. b Diagram showing quantitative parameters used to characterize 3D topology of microvasculature. c Quantification of diameter, branch quantity and length, and tortuosity of microvasculature in Patient 1 (“hot”) and 2 (“cold”) cores. The microvasculature of three cores each from Patients 1 and 2 were analyzed (mean ± SEM, *P<0.01).
IHC examination of core needle biopsy following T3
To further examine the potential to combine T3 analysis and conventional immunohistochemistry (IHC), single Patient 1 and 2 biopsies were washed free of D-fructose with PBS and submitted for conventional FFPE processing. Thin sections were examined by hematoxylin and eosin (H&E) staining and chromogenic IHC with biotinylated primary antibodies (Supplemental Fig. 12 and Fig. 13). H&E stained sections showed nests and islands of keratinizing squamous cell carcinoma (SCC). The SCC cells were readily identified by staining with anti-pan-cytokeratin (CK). The tissue was also probed with anti-EGFR, anti-CD8, and anti-CD3 using different antibody clones than those used for T3 imaging. In the pathological scoring for the EGFR IHC slides on a 0–3 scale, the cores from Patient 1 and 2 were scored as 0 (negative) and 2 (positive), respectively. Note that T3 appeared to display higher sensitivity that IHC, having detected EGFR immunoreactivity in both samples although at lower fluorescence intensity for Patient 1 than Patient 2. Consistent with T3 analysis, IHC detected a high density of CD8+ and CD3+ cells infiltrating the parenchyma in Patient 1’s tumor, while CD8+ and CD3+ cells were rare in Patient 2’s sample.
Discussion
The remarkable series of clinical successes and FDA approvals for immune checkpoint blockade antibodies in the last decade may represent only the first phase of a dramatic shift in cancer treatment away from genotoxic agents and toward immunotherapy [34–36]. Antibodies targeting the programmed cell death protein-1/-ligand 1 (PD-1/PD-L1) immune evasion pathway have shown robust clinical responses by reactivating antitumor immunity in a wide range of malignancies [37–39]. Matching the proper drug to each patient is a fundamental challenge for targeted therapies that is particularly problematic for checkpoint blockade [40, 41], where patient-specific molecular tests are lacking [42]. Together with PD-L1 expression in tumors, the abundance of tumor-infiltrating lymphocytes (TILs) and particularly of cytotoxic T lymphocytes (CTLs) have been regarded as key biomarkers to predict response to PD-1/PD-L1 checkpoint blockade [43–45]. As a gold-standard method, tissue samples collected by core needle biopsy are examined for CTLs by chromogenic immunohistochemistry (IHC) [46]. The regular IHC workflow of formaldehyde fixation, embedding in paraffin, thin sectioning, deparaffinization, epitope retrieval, and probing with primary antibodies and secondary enzyme conjugates provides cell-resolution immunolocalization. For practical reasons, IHC analysis is typically performed on a single section, which may poorly represent the heterogeneity of the tumor immune microenvironment. In turn, while IHC is sensitive and specific, simultaneous detection of multiple antigens remains challenging, making assignment of antigens to specific cell types challenging. While multiplexed immunodetection might be combined with tomographic reconstruction from serial sections [47, 48], the strategy would be too slow and cumbersome for the clinical environment, where speed is at a premium and simple, robust methods are required.
Recent progress in tissue optical clearing and light microscopy has led to significant advances in 3D immunofluorescence imaging of tissues. In particular, CLARITY [49] and related methods [50] have provided key tools for establishing connectivity in the central nervous system. Applications to cancer are emerging, but initial studies have revealed challenges including rendering the tumor tissue sufficiently transparent while maintaining structural integrity and protein immunogenicity, obtaining satisfactory penetration of antibodies and conjugates, and performing multiplexed detection with high sensitivity and specificity at cellular resolution. A realistic concern is that tissue clearing and 3D imaging methods that require dedicated equipment such as electrophoresis systems [51] and light-sheet microscopes [15, 16] may be impractical to deploy in the clinical environment. Even then, application of these methods to 3D imaging cytometry of biopsies to assess the immune microenvironment has yet to be described.
To overcome these challenges, we have adapted the Transparent Tissue Tomography (T3) [22] platform to enable fast, simple, and non-destructive 3D imaging cytometry in core needle biopsy tissue and perform semi-automated analysis of TIL types, numbers and distributions. Over little more than a day, whole cores can be stained with directly labeled, fluorescent primary antibodies, optically cleared by equilibration with concentrated D-fructose and scanned using a standard confocal scanning microscope. Image analysis using open-source Fiji macros enables 3D mapping of multiple cellular features including TILs, tumor cells and vasculature. Further, insofar as T3 is non-destructive, the same tissue can be subjected to conventional IHC assay to confirm the 3D analysis and/or assess additional antigens. Given these favorable characteristics, T3 has considerable potential for translation to the clinic.
Using T3 imaging cytometry, we were able to map the distribution of CD3+CD8+ cytotoxic T lymphocytes (CTLs) in whole needle core biopsies, capturing heterogeneity in spatial distribution on multiple scales. Studying a small cohort study of human head and neck cancer tissue samples, we found a strong correlation between tumor infiltrating CTLs and tumor penetrating microvasculature. In particular, a highly branched and tortuous microvasculature appeared associated with capillary penetration into the tumor parenchyma and greater CTL infiltration. Interrogating the 3D morphology of the microvasculature in biopsies may provide complementary information indicating the potential for immunotherapy to drive increased inflammation [52–54].
In summary, we describe Transparent Tissue Tomography (T3) as a new tool for 3D imaging cytometry and spatial analysis of core needle biopsies. Without the need for sectioning or special equipment, intact whole core needle biopsies can be simply and rapidly stained, rendered transparent, and scanned, yielding 3D high resolution maps of multiple immune infiltrate markers, all the while conserving the samples for conventional immunohistochemistry. Examining the potential role of T3 as a tool for clinical diagnostics, we demonstrated non-destructive 3D spatial mapping of immune infiltrates, a signature of anti-tumor immune responses, in whole core biopsies. Thus, we anticipate that T3 can be broadly applied to quantitative multiparameter spatial analysis of patient tumor biopsies, with the potential to improve predictive testing and on-treatment diagnostics for immune responses to checkpoint blockade immunotherapy.
Supplementary Material
Acknowledgments
We thank our colleague Nishant Agrawal for generous access to excised head and neck cancer surgical tissue. This study was supported by NCI R01s CA164492 and CA199663 to SJK and by NIBIB K99 EB022636 and Susan G. Komen Postdoctoral Fellowship PDF15333618 (made possible through funding from American Airlines) to SSL. Imaging, image processing, and data analysis were performed at the University of Chicago Integrated Light Microscopy Facility, supported by NCI P30 CA014599.
Footnotes
Conflict of interest
S.S.-Y. L., V.P.B., and S.J.K. have founded Transnostics LLC to commercialize related technologies.
Supplementary information is available at Laboratory Investigation’s website.
References
- 1.Ben-Yehuda D, Polliack A, Okon E, et al. Image-guided core-needle biopsy in malignant lymphoma: experience with 100 patients that suggests the technique is reliable. J Clin Oncol. 1996;14:2431–2434. [DOI] [PubMed] [Google Scholar]
- 2.Heslin MJ, Lewis JJ, Woodruff JM, et al. Core needle biopsy for diagnosis of extremity soft tissue sarcoma. Ann Surg Oncol. 1997;4:425–431. [DOI] [PubMed] [Google Scholar]
- 3.Rakha EA, Ellis IO. An overview of assessment of prognostic and predictive factors in breast cancer needle core biopsy specimens. J Clin Pathol. 2007;60:1300–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Motamedolshariati M, Memar B, Aliakbaian M, et al. Accuracy of prognostic and predictive markers in core needle breast biopsies compared with excisional specimens. Breast Care. 2014;9:107–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hagemann AR, Cadungog M, Hagemann IS, et al. Tissue-based immune monitoring I. Cancer Biol Ther. 2011;12:357–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tumeh PC, Harview CL, Yearley JH, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515:568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jensen HK, Donskov F, Nordsmark M, et al. Increased intratumoral FOXP3-positive regulatory immune cells during interleukin-2 treatment in metastatic renal cell carcinoma. Clin Cancer Res. 2009;15:1052–1058. [DOI] [PubMed] [Google Scholar]
- 8.Parra ER, Uraoka N, Jiang M, et al. Validation of multiplex immunofluorescence panels using multispectral microscopy for immune-profiling of formalin-fixed and paraffin-embedded human tumor tissues. Sci Rep. 2017;7:13380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsujikawa T, Kumar S, Borkar RN, et al. Quantitative multiplex immunohistochemistry reveals myeloid-inflamed tumor-immune complexity associated with poor prognosis. Cell Rep. 2017;19:203–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Egeblad M, Nakasone ES, Werb Z. Tumors as organs: Complex tissues that interface with the entire organism. Dev Cell. 2010;18:884–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Azaripour A, Lagerweij T, Scharfbillig C, et al. A survey of clearing techniques for 3D imaging of tissues with special reference to connective tissue. Prog Histochem Cyto. 2016;51:9–23. [DOI] [PubMed] [Google Scholar]
- 12.Feuchtinger A, Walch A, Dobosz M. Deep tissue imaging: a review from a preclinical cancer research perspective. Histochem Cell Biol. 2016;146:781–806. [DOI] [PubMed] [Google Scholar]
- 13.Lagerweij T, Dusoswa SA, Negrean A, et al. Optical clearing and fluorescence deep-tissue imaging for 3D quantitative analysis of the brain tumor microenvironment. Angiogenesis. 2017;20:533–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lloyd-Lewis B, Davis FM, Harris OB, et al. Imaging the mammary gland and mammary tumours in 3D: Optical tissue clearing and immunofluorescence methods. Breast Cancer Res. 2016;18:127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tanaka N, Kanatani S, Tomer R, et al. Whole-tissue biopsy phenotyping of three-dimensional tumours reveals patterns of cancer heterogeneity. Nat Biomed Eng. 2017;1:796–806. [DOI] [PubMed] [Google Scholar]
- 16.Glaser AK, Reder NP, Chen Y, et al. Light-sheet microscopy for slide-free non-destructive pathology of large clinical specimens. Nat Biomed Eng. 2017;1:0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cuccarese MF, Dubach JM, Pfirschke C, et al. Heterogeneity of macrophage infiltration and therapeutic response in lung carcinoma revealed by 3D organ imaging. Nat Commun. 2017;8:14293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sindhwani S, Syed AM, Wilhelm S, et al. Three-dimensional optical mapping of nanoparticle distribution in intact tissues. ACS Nano. 2016;10:5468–5478. [DOI] [PubMed] [Google Scholar]
- 19.Nojima S, Susaki EA, Yoshida K, et al. CUBIC pathology: three-dimensional imaging for pathological diagnosis. Sci Rep. 2017;7:9269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li W, Germain RN, Gerner MY. Multiplex, quantitative cellular analysis in large tissue volumes with clearing-enhanced 3D microscopy (Ce3D). Proc Natl Acad Sci USA. 2017;114:E7321–E7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Richardson DS, Lichtman JW. Clarifying tissue clearing. Cell. 2015;162:246–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee SS-Y, Bindokas VP, Kron SJ. Multiplex three-dimensional optical mapping of tumor immune microenvironment. Sci Rep. 2017;7:17031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boggio K, Nicoletti G, Di Carlo E, et al. Interleukin 12–mediated prevention of spontaneous mammary adenocarcinomas in two lines of Her-2/neu transgenic mice. J Exp Med. 1998;188:589–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Preibisch S, Saalfeld S, Tomancak P. Globally optimal stitching of tiled 3D microscopic image acquisitions. Bioinformatics. 2009;25:1463–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hall M, Frank E, Holmes G, et al. The WEKA data mining software: an update. ACM SIGKDD Explor Newsl. 2009;11:10–18. [Google Scholar]
- 26.Ollion J, Cochennec J, Loll F, et al. TANGO: a generic tool for high-throughput 3D image analysis for studying nuclear organization. Bioinformatics. 2013;29:1840–1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schindelin J, Arganda-Carreras I, Frise E, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Conti L, Ruiu R, Barutello G, et al. Microenvironment, oncoantigens, and antitumor vaccination: lessons learned from BALB-neuT mice. BioMed Res Int. 2014;2014:534969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Topalian SL, Sznol M, McDermott DF, et al. Survival, durable tumor remission, and long-term safety in patients with advanced melanoma receiving nivolumab. J Clin Oncol. 2014;32:1020–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Topalian SL, Taube JM, Anders RA, et al. Mechanism-driven biomarkers to guide immune checkpoint blockade in cancer therapy. Nat Rev Cancer. 2016;16:275–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Teng MWL, Ngiow SF, Ribas A, et al. Classifying cancers based on T-cell infiltration and PD-L1. Cancer Res. 2015;75:2139–2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gajewski TF, Schreiber H, Fu Y-X. Innate and adaptive immune cells in the tumor microenvironment. Nature Immunol. 2013;14:1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van der Woude LL, Gorris MAJ, Halilovic A, et al. Migrating into the tumor: A roadmap for T cells. Trends Cancer. 2017;3:797–808. [DOI] [PubMed] [Google Scholar]
- 34.Emens LA, Ascierto PA, Darcy PK, et al. Cancer immunotherapy: Opportunities and challenges in the rapidly evolving clinical landscape. Eur J Cancer. 2017;81:116–129. [DOI] [PubMed] [Google Scholar]
- 35.Lonberg N, Korman AJ. Masterful antibodies: Checkpoint blockade. Cancer Immunol Res. 2017;5:275–281. [DOI] [PubMed] [Google Scholar]
- 36.Moskovitz J, Moy J, Ferris RL. Immunotherapy for head and neck squamous cell carcinoma. Curr Oncol Rep. 2018;20:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tan S, Zhang CWH, Gao GF. Seeing is believing: anti-PD-1/PD-L1 monoclonal antibodies in action for checkpoint blockade tumor immunotherapy. Signal Transduct Target Ther. 2016;1:16029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Balar AV, Weber JS. PD-1 and PD-L1 antibodies in cancer: current status and future directions. Cancer Immunol Immunother. 2017;66:551–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Megan M, Michael C, Clint A. Exploring the rationale for combining ionizing radiation and immune checkpoint blockade in head and neck cancer. Head Neck. 2018;40:1321–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nishino M, Ramaiya NH, Hatabu H, et al. Monitoring immune-checkpoint blockade: response evaluation and biomarker development. Nat Rev Clin Oncol. 2017;14:655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taube JM, Galon J, Sholl LM, et al. Implications of the tumor immune microenvironment for staging and therapeutics. Mod Pathol. 2017;00:1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lesterhuis WJ, Bosco A, Millward MJ, et al. Dynamic versus static biomarkers in cancer immune checkpoint blockade: unravelling complexity. Nat Rev Drug Discov. 2017;16:264. [DOI] [PubMed] [Google Scholar]
- 43.Fridman WH, Zitvogel L, Sautès–Fridman C, et al. The immune contexture in cancer prognosis and treatment. Nat Rev Clin Oncol. 2017;14:717. [DOI] [PubMed] [Google Scholar]
- 44.Cogdill AP, Andrews MC, Wargo JA. Hallmarks of response to immune checkpoint blockade. Br J Cancer. 2017;117:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lovitch SB, Rodig SJ. The role of surgical pathology in guiding cancer immunotherapy. Annu Rev Pathol-Mech. 2016;11:313–341. [DOI] [PubMed] [Google Scholar]
- 46.Wargo JA, Reddy SM, Reuben A, et al. Monitoring immune responses in the tumor microenvironment. Curr Opin Immunol. 2016;41:23–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parfitt GJ, Xie Y, Reid KM, et al. A novel immunofluorescent computed tomography (ICT) method to localise and quantify multiple antigens in large tissue volumes at high resolution. PLOS ONE. 2013;7:e53245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Farahani N, Braun A, Jutt D, et al. Three-dimensional imaging and scanning: Current and future applications for pathology. J Pathol Inform. 2017;8:36–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chung K, Wallace J, Kim S-Y, et al. Structural and molecular interrogation of intact biological systems. Nature. 2013;497:332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tainaka K, Kuno A, Kubota SI, et al. Chemical principles in tissue clearing and staining protocols for whole-body cell profiling. Annu Rev Cell Dev Biol. 2016;32:713–741. [DOI] [PubMed] [Google Scholar]
- 51.Tomer R, Ye L, Hsueh B, et al. Advanced CLARITY for rapid and high-resolution imaging of intact tissues. Nat Protoc. 2014;9:1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tan LY, Martini C, Fridlender ZG, et al. Control of immune cell entry through the tumour vasculature: a missing link in optimising melanoma immunotherapy. Clin Trans Immunol. 2017;6:e134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lanitis E, Irving M, Coukos G. Targeting the tumor vasculature to enhance T cell activity. Curr Opin Immunol. 2015;33:55–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fukumura D, Kloepper J, Amoozgar Z, et al. Enhancing cancer immunotherapy using antiangiogenics: opportunities and challenges. Nat Rev Clin Oncol. 2018;15:325–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


