Abstract
Excessive fat intake is associated with changes in gut microbiota composition. In the present study, we focused on the secretory immunoglobulin A (SIgA) coating of gut microbiota as a mucosal immune response affecting the gut microbiota following a high-fat diet (HFD). The level of SIgA coating of gut microbiota was evaluated in normal-fat diet (NFD)- and HFD-fed mice. HFD significantly decreased the level of SIgA coating the gut microbiota compared with NFD. Of note, substitution of HFD with NFD resulted in a complete recovery of the level of SIgA coating. These findings suggest that dietary fat influences the SIgA coating of the gut microbiota. Furthermore, we analyzed the composition of the gut microbiota and the concentration of cecal short-chain fatty acids. HFD feeding changed the gut microbiota composition at the phylum and family levels. Pearson correlation analysis between the level of SIgA coating of gut microbiota and the relative abundance of gut microbiota showed that the relative abundances of Clostridiaceae, Mogibacteriaceae, Turicibacteraceae, and Bifidobacteriaceae were negatively correlated with the level of SIgA coating of gut microbiota. Conversely, the relative abundances of Desulfovibrionaceae, S24-7, and Lactobacillaceae were positively correlated with the level of SIgA coating. The concentrations of cecal acetate and butyrate were lower in HFD-fed mice and positively correlated with the level of SIgA coating of gut microbiota. Our observations suggest that a decrease in the level of SIgA coating of the gut microbiota through a HFD might relate to HFD-induced changes in microbial composition and microbial metabolites production.
Keywords: secretory immunoglobulin A, high-fat diet, commensal gut microbiota
INTRODUCTION
Excessive consumption of dietary fat alters the composition of gut microbiota. High-fat diet (HFD) consumption increases the ratio of Firmicutes:Bacteroidetes, the dominant phyla in the human and mouse gut [1]. Family-level changes in gut microbiota have also been reported, including increases in Enterobacteriaceae, Enterococcaceae, and Bifidobacteriaceae, as well as decreases in Lactobacillaceae and Prevotellaceae [2,3,4]. In addition, HFD consumption has been associated with decreased microbial diversity [5]. Several factors, such as bile acid, dietary fat, and short-chain fatty acids (SCFAs), may induce shifts in microbial composition as a result of HFD feeding. Firstly, elevated fat consumption triggers increased bile acid synthesis, a process required in lipid digestion and absorption. Unabsorbed bile acids are hydrolyzed into secondary bile acids by gut microbial bile salt hydrolase [6]. The antimicrobial nature of the resultant secondary bile acids has been suggested to favor the growth of bile-tolerant microbiota [7]. Secondly, movement of unabsorbed dietary fat into the distal intestine after HFD consumption has been shown to cause an increase in the Firmicutes:Bacteroidetes ratio due to the bacteriostatic properties of saturated fatty acids [8, 9]. Further, prolonged consumption of a diet rich in saturated fats is associated with ER stress-mediated reduction in colonic mucin production, resulting in changes in gut microbiota composition [10]. These HFD-related changes in gut microbiota result in lower SCFA production in the intestinal lumen [11]. The subsequent increase in luminal pH inhibits the growth of pH-sensitive bacteria, further modulating the gut microbiota composition [12]. In addition to these factors, we hypothesized that the secretory immunoglobulin A (SIgA) coating of gut microbiota is related to changes in microbial composition upon HFD consumption. This is because SIgA plays an important role in maintaining a stable gut microbial composition [13]. SIgA is the predominant antibody isotype secreted into the intestinal lumen [14]. SIgA specifically coats gut microbiota [15] and suppresses the overgrowth of gut microbiota [16, 17]. While it is evident that the SIgA coating of gut microbiota modulates the gut microbial composition, the relationship between the SIgA coating of gut microbiota and HFD-induced changes in gut microbiota remains unclear. To investigate this relationship, we evaluated the level of IgA coating of gut microbiota and the gut microbial composition in NFD- and HFD-fed mice and explored the correlation between them.
MATERIALS AND METHODS
Experimental design
The experimental protocol was approved by the Animal Care and Use Committee of Okayama University, Japan (approval no. OKU-2016305). Thirty male BALB/c mice (9 weeks old) were allocated into groups based on body weight and acclimated for a week. We conducted two experiments which differed in dietary treatment and dissection period.
In experiment 1, 20 mice were allocated into 2 groups and allowed free access to water and to either a normal-fat diet (NFD) or HFD. The diet composition can be found in Table 1. Fecal samples were collected at the start of the experiment and at weeks 6 and 12. Before dissection at week 12, body weight was measured. The mice were then euthanized by exsanguination via cardiac puncture under pentobarbital anesthesia. The cecal content and colonic tissue were collected.
Table 1. Composition of experimental diets.
| NFD | HFD | |
|---|---|---|
| Ingredients (g/kg diet) | ||
| Maize starch1 | 465.692 | 290.692 |
| α-Maize starch1 | 155 | - |
| Casein1 | 140 | 140 |
| Sucrose2 | 100 | 100 |
| Cellulose1 | 50 | 50 |
| Soybean oil3 | 40 | 70 |
| Lard4 | - | 300 |
| AIN-93 Mineral mix1 | 35 | 35 |
| AIN- 93 Vitamin mix1 | 10 | 10 |
| L-cystine3 | 1.8 | 1.8 |
| Choline bitartrate5 | 2.5 | 2.5 |
| Tert-butylhydroquinone6 | 0.008 | 0.008 |
| Total energy (kcal/g diet) | 3.8 | 5.5 |
NFD: normal fat diet; HFD: high fat diet.
1Purchased from CLEA Japan, Japan.
2Purchased from Nippon Beet Sugar, Japan.
3Purchased from Nacalai Tesque, Inc, Japan.
4Purchased from Yukijirushi, Japan.
5Purchased from Tokyo Chemical Industry, Japan.
6Purchased from Wako Pure Chemical Industries, Japan.
In experiment 2, 10 mice were divided into 2 groups; one group was fed NFD for 18 weeks, and the other group was fed HFD for the first 12 weeks and then switched to NFD for the following 6 weeks. At week 18, fecal samples were collected.
Flow cytometry analysis of IgA-coated bacteria
One fecal pellet was suspended in 500 µl PBS and sedimented by centrifugation (100 × g for 20 min). The supernatant was then centrifuged at 9,000 × g for 10 min. After centrifugation, the supernatant was collected for fecal IgA ELISA. The resultant bacterial pellet was washed twice with PBS. The bacterial cells were then fixed with 4% paraformaldehyde (Wako) overnight at 4°C. After washing twice with PBS, the bacterial cells were stained with FITC-labelled anti-mouse IgA (BD Pharmingen) for 30 min. After washing twice with PBS, the bacterial cells were stained with propidium iodide (PI; Sigma Aldrich) solution (4 µg/ml). The bacterial suspension was analyzed by a Gallios flow cytometer (Beckman Coulter). The gating strategy used to identify SIgA-coated and non-coated bacteria is shown in Fig. 1. FCS Express V3 (De Novo Software) was used to calculate the average FITC intensity emitted by a single SIgA-coated bacterium. The average FITC intensity emitted by a single SIgA-coated bacterium was defined as the average level of SIgA coating per fecal bacterium.
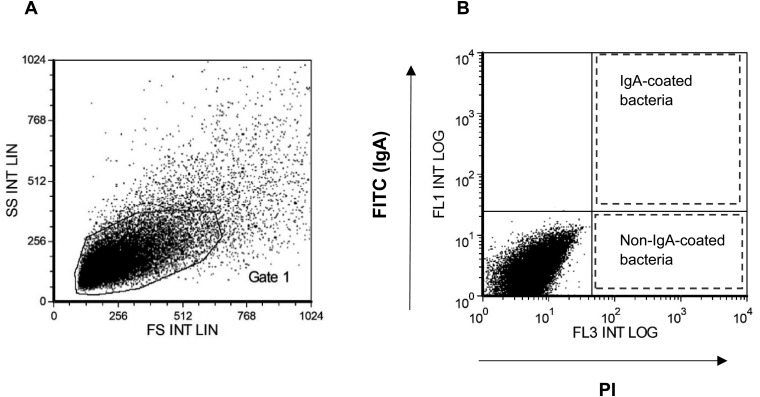
Fig. 1.
Gating based on an unstained bacterial pellet was used to identify bacteria from mouse feces. (A) The microbial fraction was first identified by forward scatter (FS) and side scatter (SS) properties, as shown in gate 1. (B) Quadrants on an FL1 vs. FL3 dot plot using gate 1 were used to identify bacteria. IgA-coated bacteria were identified as PI- and FITC-positive populations; non-IgA-coated bacteria were identified as PI-positive and FITC-negative populations.
Quantification of fecal IgA concentration
The concentration of fecal IgA in the aforementioned fecal supernatant was measured using a Mouse IgA ELISA Quantitation Set (Bethyl) according to the manufacturer’s instructions. The absorbance of each well was read at 450 nm using a microplate reader (Bio-Rad).
Western blot of IgA bound to fecal bacteria
The samples containing fecal bacteria were prepared as described above. The bacterial pellet was lysed with 15 µl RIPA lysis buffer. Mouse serum IgA (Bethyl) was also lysed and used as standard for the quantification of IgA. The bacterial lysate was mixed with 4 × Laemmli buffer and separated on 10% SDS-PAGE. The separated proteins were transferred onto a PVDF membrane using the Power Blotter System (Thermo Fisher Scientific). After blocking with 1% skim milk buffer, the membrane was stained with anti-mouse IgA antibody (Bethyl) and HRP-conjugated anti-goat IgG antibody (R&D Systems) as primary and secondary antibodies, respectively, and visualized using Chemi-Lumi One Super substrates (Nacalai). Imaging and quantification of IgA coating fecal bacteria were performed using a ChemiDoc XRS+ system (Bio-Rad). The amount of IgA coating fecal bacteria was quantified by measuring the band intensity and comparing it to reference bands of serum IgA.
Analysis of gene expression
Total RNA was extracted from colonic samples using ISOGEN II according to the manufacturer’s protocol (Nippon Gene). Random primers were then used to reverse transcribe the RNA (Takara). qPCR was performed using an AriaMx Real-Time PCR System (Agilent Technologies). Sample quantification cycle (Cq) values were normalized using the housekeeping gene GAPDH. Fold changes were then calculated relative to the average NFD group value using the ∆∆Cq method. The primer pair sequences used were as follows: 5′-TCAAGAAGGTGGTGAAGCAG-3′, forward primer, and 5′-AAGGTGGAAGAGTGGGAGTTG-3, reverse primer, for mouse GAPDH; 5′-TGCACAGCTTTCTTCTGCAC-3′, forward primer, and 5′-TGCCAGCCTCACATGTACTC-3′, reverse primer, for mouse IgA [18]; and 5′-GCTGACGAGTGGTTGGTGAATG-3′, forward primer, and 5′-GATGAGGTGGCAGACAGGAGAC-3, reverse primer, for mouse Muc2 [19].
Illumina MiSeq sequencing
Bacterial DNA was extracted from fecal samples by using a QIAamp Stool Mini Kit (Qiagen) according to the manufacturer’s instructions, with additional freeze, thaw, and bead-beating steps. Next-generation sequencing analysis was performed as previously described [20]. All bacterial species obtained from fecal samples were classified, and the proportion of different phyla and families was computed using QIIME (version 1.9.1). The alpha diversity (Chao1 and Shannon indexes) of species-level microbial taxa was computed for rarefied operational taxonomic units (OTUs) (5,000 reads) using the Primer version 7 with the Permanova+ add-on software (Primer-E, Plymouth Marine Laboratory, Plymouth, UK).
SCFAs in the cecum
Determination of cecal SCFAs was conducted using a gas chromatograph (GC-14a, Shimadzu) fitted with a glass capillary column coated with nitroterephthalic acid modified polyethylene glycol (TC- FFAP, GL Sciences). Cecum samples were weighed, homogenized, and then deproteinized using 10% trichloroacetic acid. The samples were then centrifuged before injection into the TC-FFAP column. The column oven temperature was programmed at 80°C for the first 2 minutes and then increased to 200°C at a rate of 10°C per minute [21].
Statistical analysis
Data are expressed as means ± SEM. All statistical analysis was conducted using GraphPad Prism version 7.00 for Windows (GraphPad Software, USA). Values obtained from experiments 1 and 2 were compared between diet groups using Student’s t-test at each time point. Correlations between the average level of SIgA coating per fecal bacterium and fecal IgA concentration, body weight, colonic IgA mRNA expression, relative abundance of gut microbiota, and cecal SCFA concentration were analyzed using Pearson correlation coefficients. p<0.05 was considered statistically significant in all experiments.
RESULTS
HFD feeding reduces SIgA coating of gut microbiota
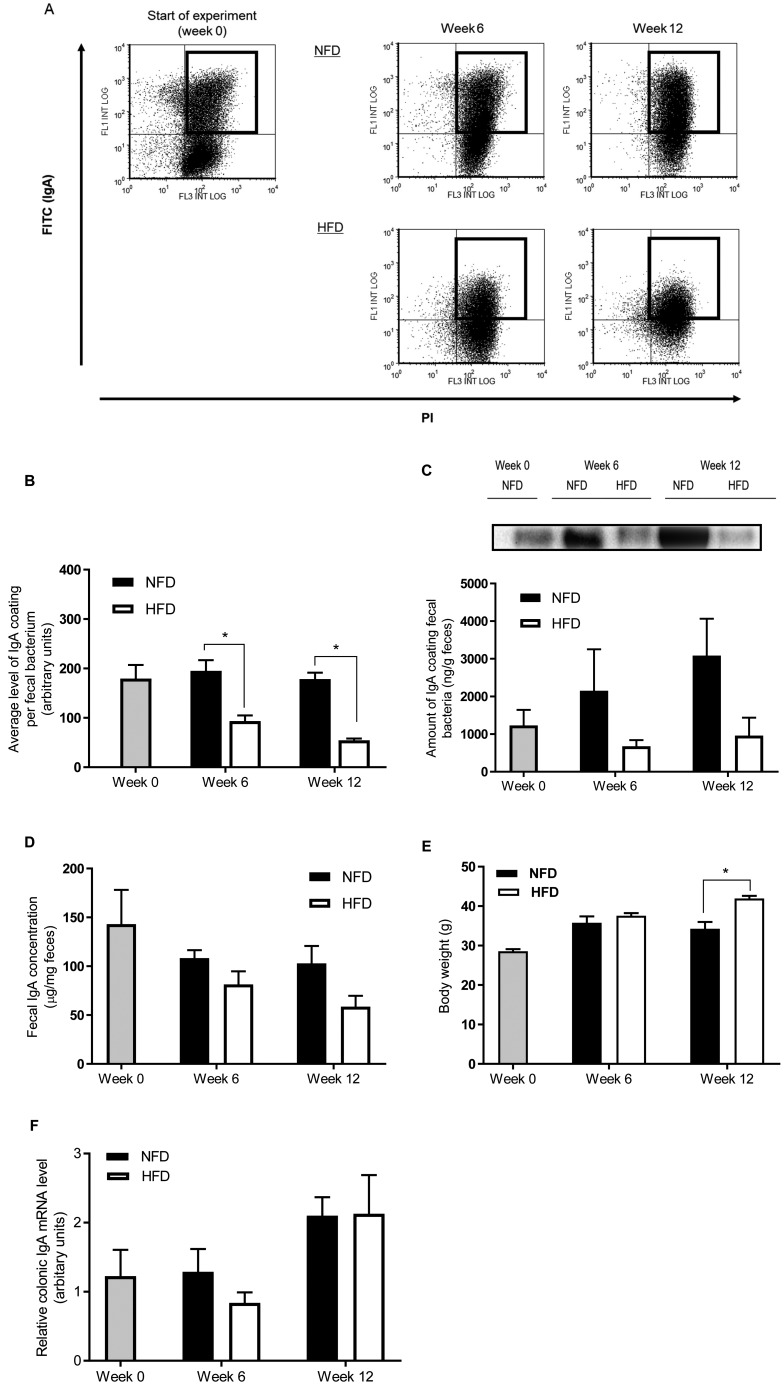
In experiment 1, we detected by flow cytometry SIgA-coated bacteria in the feces of mice fed either NFD or HFD for 6 and 12 weeks. Representative results of the flow cytometry for the detection of SIgA-coated bacteria are shown in Fig. 2A. SIgA-coated bacteria were recognized as both FITC- and PI-positive populations and were also gated. The average level of SIgA coating per fecal bacterium was significantly lower in HFD-fed mice at weeks 6 and 12 than in NFD-fed mice (Fig. 2B). Although no significant differences between the diet groups were detected, the amount of SIgA coating of fecal bacteria measured by western blotting was also lower in HFD-fed mice compared with NFD-fed mice at week 6 and week 12 (Fig. 2C). Fecal IgA concentration tended to be lower in HFD-fed mice than in NFD-fed mice at weeks 6 and 12 (Fig. 2D). Body weight was significantly higher in HFD-fed mice than in NFD-fed mice at week 12 (Fig. 2E). The gene expression of IgA in the colon did not differ between diet groups at week 6 and week 12 (Fig. 2F).
Fig. 2.
HFD consumption decreases the level of SIgA coating of gut microbiota (experiment 1). (A) Representative results of flow cytometry for the detection of SIgA-coated bacteria in feces at week 0 and after 6 and 12 weeks of NFD or HFD feeding. Gated population represents SIgA-coated bacteria. (B) Average level of SIgA coating per fecal bacterium analyzed by flow cytometry. Average FITC intensity of SIgA-coated bacteria measured by flow cytometry and defined as the average level of SIgA coating per fecal bacterium. (C) Amount of IgA coating fecal bacteria analyzed by western blotting. Fecal bacteria were subjected to western blotting using an anti-IgA antibody. A representative blot is shown above the graph. The amount of IgA coating fecal bacteria was quantified by reference to band intensity of a reference serum IgA. (D) Fecal IgA concentration. (E) Body weight. (F) Colonic mRNA expression of IgA was determined by qPCR. Values are given as means ± SEM (n=5 per group). *p<0.05 for HFD vs. NFD at week 6 and week 12 by unpaired Student’s t-test.
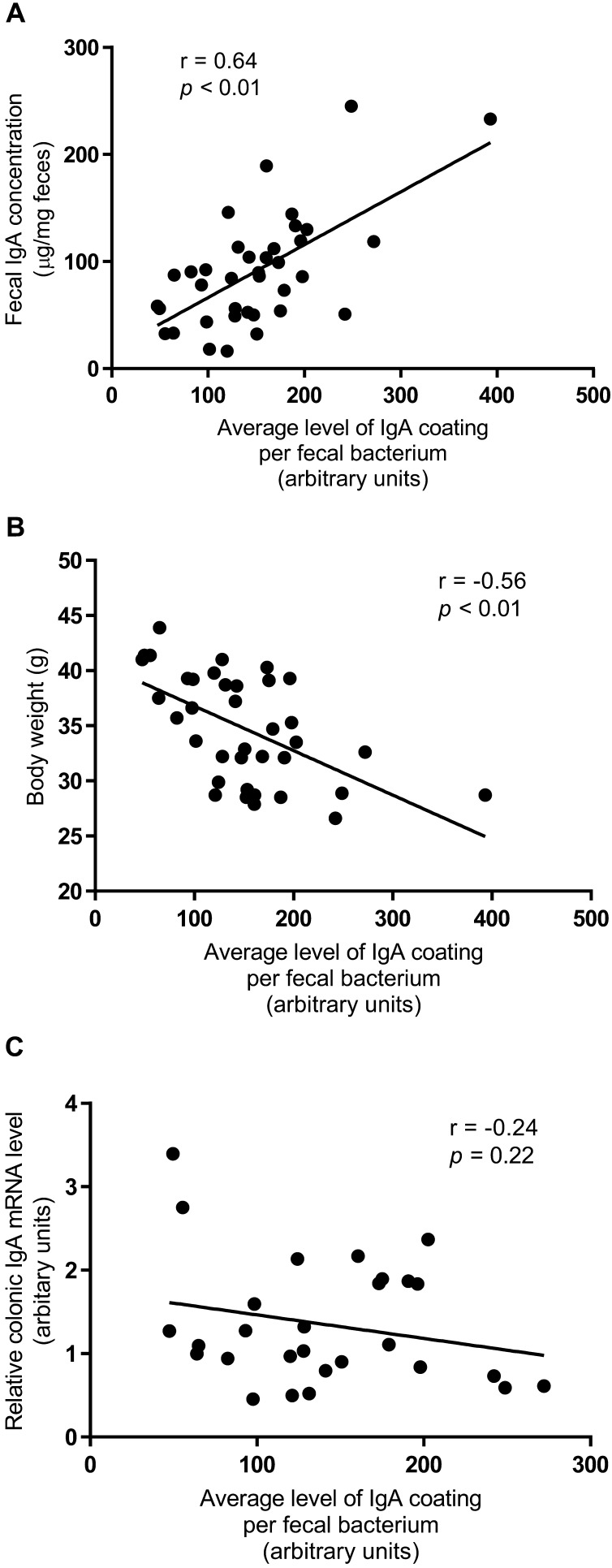
Level of SIgA coating of gut microbiota is associated with fecal IgA concentration and body weight
The fecal IgA concentration positively correlated with the average level of SIgA coating per fecal bacterium (r=0.64, p<0.01) (Fig. 3A). Body weight negatively correlated with the average level of SIgA coating per fecal bacterium (r=−0.56, p<0.01) (Fig. 3B). There was no correlation between the gene expression of IgA in the colon and the average level of SIgA coating per fecal bacterium (r=−0.24, p=0.22) (Fig. 3C).
Fig. 3.
Level of SIgA coating of gut microbiota is associated with fecal IgA concentration and body weight. Pearson correlation between (A) fecal IgA concentration, (B) body weight, and (C) relative expression of colonic IgA and the average level of SIgA coating per fecal bacterium. Each dot represents measurements of a single mouse. The correlation coefficient (r), the corresponding p value, and the linear regression line are shown. Values are given as means ± SEM (n=5 per group).
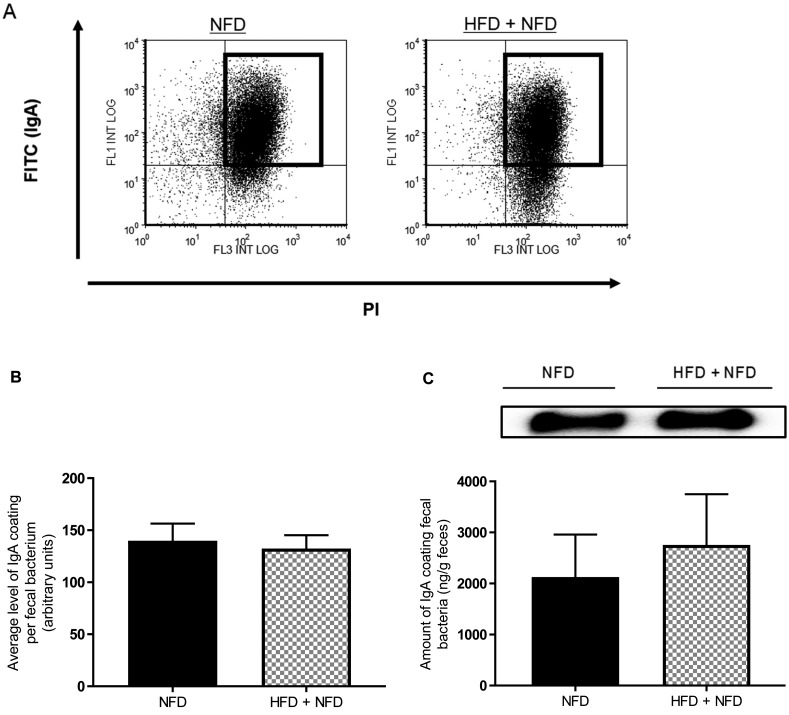
HFD-induced reduction of SIgA coating of gut microbiota is reversed by NFD feeding
In experiment 2, we detected SIgA-coated bacteria in the feces of mice fed NFD for 18 weeks (NFD) or fed HFD for the first 12 weeks and then NFD for the subsequent 6 weeks (HFD + NFD). Representative results of flow cytometry for the detection of SIgA-coated bacteria are shown in Fig. 4A. There were no significant differences in the average level of SIgA coating per fecal bacterium between NFD and HFD + NFD (Fig. 4B). The level of SIgA coating fecal bacteria measured by western blotting was also not significantly different between the diet groups (Fig. 4C).
Fig. 4.
Reduction in the level of SIgA coating of gut microbiota induced by HFD consumption is reversed by NFD consumption (experiment 2). (A) Representative results of flow cytometry for the detection of SIgA-coated bacteria in mice fed NFD for 18 weeks (NFD) and in mice fed HFD for the first 12 weeks and then NFD for the following 6 weeks (NFD+HFD). (B) Average level of SIgA coating per fecal bacterium analyzed by flow cytometry. (C) Amount of IgA coating fecal bacteria analyzed by western blotting. Values are given as means ± SEM (n=5 per group).
HFD-induced changes in gut microbiota and cecal SCFA concentrations are associated with the level of SIgA coating of gut microbiota
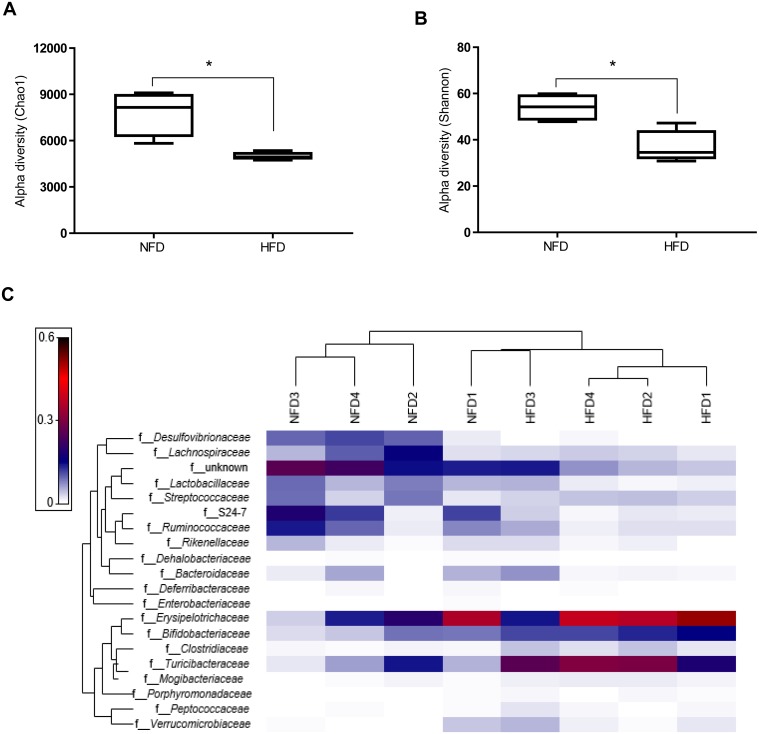
The relative abundances of fecal microbial taxa at the phylum and family levels and of cecal SCFAs in mice fed NFD or HFD for 12 weeks are shown in Table 2. HFD feeding elevated the relative abundance of Firmicutes (p=0.06) and reduced that of Bacteroidetes (p=0.09). The ratio of Firmicutes:Bacteroidetes was not significantly different between NFD-fed (20.1 ± 16.7) and HFD-fed mice (34.3 ± 12.7). The relative abundance of Actinobacteria was significantly higher, while that of Proteobacteria was significantly lower in HFD-fed mice, compared with NFD-fed mice. Further, HFD-fed mice showed significantly higher relative abundances of Bifidobacteriaceae, Clostridiaceae, Mogibacteriaceae and Turicibacteraceae compared with NFD-fed mice. Conversely, Lactobacillaceae, S24-7, and Desulfovibrionaceae were less abundant in HFD-fed mice. To determine the effect of HFD on microbial diversity, the Shannon index and the Chao1 index were computed. HFD-fed mice exhibited a significantly lower diversity at the species level based on observed richness (Chao1) and Shannon’s diversity index (Fig. 5A and 5B). A heat map of the 20 most abundant microbiota at the family level was generated to examine the individual microbial differences at 12 weeks. The heat map showed that the samples were separated into two different clusters based on diet, with the exception of one individual in the NFD group (Fig. 5C). HFD-feeding decreased the concentrations of cecal acetate, propionate, and butyrate; however, the differences were not significant between the diet groups.
Table 2. Relative abundances (%) of fecal microbial taxa at the phylum and family levels1 and cecal SCFAs in mice fed on NFD and HFD for 12 weeks (experiment 1).
| NFD | HFD | Correlation with average level of IgA coating per fecal bacterium |
||||
|---|---|---|---|---|---|---|
| r | p value | |||||
| Relative abundance (%) | ||||||
| Firmicutes | 69.98 ± 3.79 | 80.43 ± 2.54 | –0.65 | 0.08 | ||
| Clostridiaceae | 0.49 ± 0.10 | 2.59 ± 0.58* | –0.79 | 0.02¶ | ||
| Lactobacillaceae | 6.19 ± 1.13 | 1.81 ± 0.90* | 0.62 | 0.1 | ||
| Mogibacteriaceae | 0.32 ± 0.12 | 0.97 ± 0.12* | –0.78 | 0.02¶ | ||
| Turicibacteraceae | 6.26 ± 2.57 | 25.52 ± 2.68* | –0.85 | 0.01¶ | ||
| Bacteroidetes | 15.15 ± 4.84 | 4.60 ± 2.38 | 0.59 | 0.12 | ||
| S24-7 | 10.42 ± 3.57 | 1.43 ± 0.49* | 0.65 | 0.08 | ||
| Actinobacteria | 5.45 ± 1.55 | 12.35 ± 0.97* | –0.77 | 0.03¶ | ||
| Bifidobacteriaceae | 5.36 ± 1.57 | 12.24 ± 0.95* | –0.77 | 0.03¶ | ||
| Proteobacteria | 7.60 ± 2.11 | 0.18 ± 0.08* | 0.73 | 0.04¶ | ||
| Desulfovibrionaceae | 7.48 ± 2.15 | 0.14 ± 0.08* | 0.72 | 0.04¶ | ||
| SCFA (µmol/g) | ||||||
| Acetate | 3.11 ± 0.78 | 1.81 ± 0.32 | 0.76 | 0.05¶ | ||
| Propionate | 0.40 ± 0.08 | 0.21 ± 0.01 | 0.68 | 0.09 | ||
| Butyrate | 0.40 ± 0.11 | 0.22 ± 0.01 | 0.78 | 0.04¶ | ||
1Results include only family-level microbial taxa that were significantly different between diet groups. Values are given as means ± SEM (n=5 per group). *Different from NFD (p<0.05). ¶Significant correlation (p<0.05).
Fig. 5.
HFD consumption influences fecal microbial composition of mice fed NFD or HFD for 12 weeks. Alpha diversity of species-level microbial taxa using (A) Chao1 and (B) Shannon indexes for rarefied OTUs (5000 reads). (C) Heat map representing the 20 most abundant families in NFD- and HFD-fed mice. A Euclidean distance metric was used to group individuals into clusters (n=4 per diet group). Values are given as means ± SEM. *Different from NFD (p<0.05).
The relative abundance of Firmicutes tended to be negatively correlated with the average level of SIgA coating per fecal bacterium (Table 2). The relative abundances of Clostridiaceae, Mogibacteriaceae, Turicibaceteraceae and Bifidobacteriaceae were also negatively correlated with average level of SIgA coating per fecal bacterium. The relative abundances of Lactobacillaceae, S24-7, and Desulfovibrionaceae were positively correlated with average level of SIgA coating per fecal bacterium.
The concentrations of cecal acetate and butyrate were positively correlated with the average level of SIgA coating per fecal bacterium.
DISCUSSION
In the present study, we investigated the relationship between SIgA coating of gut microbiota and HFD-induced changes in gut microbiota. Our study demonstrates that the level of SIgA coating fecal bacteria greatly decreased in HFD-fed mice compared with NFD-fed mice, which suggests that SIgA coating of gut microbiota may be suppressed by HFD feeding. Furthermore, we observed that the suppression of SIgA coating of gut microbiota induced by HFD is completely reversed by substitution of HFD with NFD. It was reconfirmed that excessive fat intake is a major cause of suppression of SIgA coating of gut microbiota. Although the exact underlying mechanism remains unclear, we showed the possibility that fat content in diet is one of the determinant factors modulating the adaptive mucosal immune response associated with SIgA against gut microbiota.
There are two possible causes for the decrease of SIgA coating of gut microbiota induced by HFD: one is the fecal IgA concentration, and the other is SIgA specificity against gut microbiota. Although no significant difference was found, the fecal IgA concentration was decreased by HFD ingestion. Furthermore, it was positively correlated with the average level of SIgA coating per fecal bacterium. This implies that the decreased SIgA secretion induced by HFD feeding might decrease the level of SIgA coating the gut microbiota. However, a previous study demonstrated that the fecal IgA concentration is not a determinant factor for the amount of SIgA coating gut microbiota, because only less than 1% of fecal IgA is used for the coating of gut microbiota [22]. Therefore, this possibility should be further examined in a future study.
SIgA specificity against luminal antigens is partly regulated by dendritic cells and T cells present in the Peyer’s patches [23]. James et al. reported that HFD feeding reduces the ability of dendritic cells to induce T cell expansion, which plays a critical role in the differentiation of antigen-specific IgA plasmablasts [24]. Together with these studies, our results suggest that HFD consumption might suppress the differentiation of IgA plasmablasts specific for the gut microbiota, resulting in a decrease of SIgA coating of gut microbiota.
In line with previous studies, we observed that HFD feeding induced changes in the microbial composition at the phylum level [1, 5]. HFD-fed mice had a higher abundance of Firmicutes and a lower abundance of Bacteroidetes. At the family level, the relative abundances of Clostridiaceae and Bifidobacteriaceae were significantly increased in HFD-fed mice compared with NFD-fed mice, while those of Lactobacillaceae and S24-7 were significantly decreased as in previous studies [2, 3]. In the present study, we found a clear correlation between the level of SIgA coating gut microbiota and the relative abundance of gut microbiota in mice fed NFD or HFD for 12 weeks. There was a tendency of negative correlation between the relative abundance of Firmicutes and the level of SIgA coating gut microbiota. Furthermore, we observed a significant negative correlation between the relative abundances of Clostridiaceae, Mogibacteriaceae, Turicibacteraceae, and Bifidobacteriaceae and the level of SIgA coating gut microbiota. Peterson et al. demonstrated that the growth of Bacteroides thetaiotaomicron, a dominant gut bacterium, is suppressed in IgA-deficient mice transplanted with hybridoma cells secreting IgA that specifically binds to B. thetaiotaomicron, compared with IgA-deficient mice without hybridoma cells [25]. Furthermore, Wei et al. reported that both aerobic and anaerobic gut microbiota grow excessively in AIDG23S mice, whose intestinal IgA has low specificity against gut microbiota compared with wild-type mice [26]. These reports suggest that SIgA coating of gut microbiota plays a crucial role in suppressing gut microbiota coated by SIgA. Therefore, the negative correlation between the level of SIgA coating of gut microbiota and the relative abundances of the phylum Firmicutes and the families Clostridiaceae, Mogibacteriaceae, Turicibacteraceae, and Bifidobacteriaceae indicates that the overgrowth of these microbial groups might have occurred due to a decrease in the SIgA coating against these microbial groups upon HFD feeding. As opposed to these microbial groups, a positive correlation between the level of SIgA coating of gut microbiota and relative abundance was observed for S24-7, a major family of Bacteroidales, and for Lactobacillaceae. A report showed that Bacteroides and Lactobacillus, major genera of Bacteroidales and Lactobacillaceae, resist the SIgA coating in NFD-fed mice [27]. This report and our observations suggest that S24-7 and Lactobacillaceae might show reduced levels of SIgA coating and thus be less affected by the ability of SIgA to suppress the growth of gut microbiota. Consequently, the relative abundance of these microbial families might be higher in NFD-fed mice. However, it is unclear from our study why the relative abundances of these microbial families decrease when the level of SIgA coating gut microbiota is decreased by HFD feeding. There is the possibility that HFD feeding promotes the overgrowth of other bacteria that depress the growth of these microbial families.
Our study also showed that HFD-fed mice have a significantly lower microbial diversity compared with NFD-fed mice, which is consistent with previous reports [28]. Using AIDG23S mice, Wei and colleagues demonstrated that a decrease of SIgA specificity against gut microbiota results in low microbial diversity [26]. So, a decrease of SIgA coating of gut microbiota induced by HFD feeding might be related to reduced microbial diversity in HFD-fed mice.
Acetate, propionate, and butyrate are major SCFAs produced by intestinal fermentation of dietary fibers. These SCFA ameliorate HFD-induced obesity and insulin resistance to a similar extent when given as a dietary supplement [29]. In the present study, the concentration of SCFAs was decreased in HFD-fed mice, which is in line with previous reports [30]. In fact, in the present study, SCFA-producing gut microbiota, such as Ruminococcaceae and S24-7 [31, 32], were decreased in HFD-fed mice compared with NFD-fed mice. There is a possibility that the reduction in SCFA producers may be due to reduction in available substrate owing to the reduced starch content in the HFD. Further research needs to be conducted to clarify this.
Interestingly, there was a significant positive correlation between the concentrations of cecal acetate and butyrate and the level of SIgA coating of gut microbiota. Kim et al. reported that oral administration of an SCFA mixture containing acetate, propionate, and butyrate increases the ratio of SIgA-coated bacteria to total intestinal bacteria, suggesting that SCFAs can promote SIgA coating of gut microbiota [33]. Therefore, our observations suggest that reduction in the concentration of SCFAs in the gut induced by HFD consumption might be linked to a decrease of SIgA coating of gut microbiota.
In conclusion, our study clearly showed that excessive dietary-fat intake decreases the level of SIgA coating of gut microbiota. The reduced levels of SIgA coating gut microbiota after HFD consumption might be related to HFD-induced changes in microbial composition and microbial metabolites production.
Conflict of interest
The authors have no conflicts of interest to disclose.
Acknowledgments
We would like to express our appreciation to the Assisted Reproductive Technology Center and Department of Genomics and Proteomics (Okayama University) for technical support.
References
- 1.Hildebrandt MA, Hoffmann C, Sherrill-Mix SA, Keilbaugh SA, Hamady M, Chen YYU, Knight R, Ahima RS, Bushman F, Wu GD. 2009. High-fat diet determines the composition of the murine gut microbiome independently of obesity. Gastroenterology 137: 1716–24.e1, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. 2009. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med 1: 6ra14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cani PD, Bibiloni R, Knauf C, Waget A, Neyrinck AM, Delzenne NM, Burcelin R. 2008. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes 57: 1470–1481. [DOI] [PubMed] [Google Scholar]
- 4.Mujico JR, Baccan GC, Gheorghe A, Díaz LE, Marcos A. 2013. Changes in gut microbiota due to supplemented fatty acids in diet-induced obese mice. Br J Nutr 110: 711–720. [DOI] [PubMed] [Google Scholar]
- 5.Turnbaugh PJ, Bäckhed F, Fulton L, Gordon JI. 2008. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe 3: 213–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ridlon JM, Kang DJ, Hylemon PB. 2006. Bile salt biotransformations by human intestinal bacteria. J Lipid Res 47: 241–259. [DOI] [PubMed] [Google Scholar]
- 7.Kurdi P, Kawanishi K, Mizutani K, Yokota A. 2006. Mechanism of growth inhibition by free bile acids in lactobacilli and bifidobacteria. J Bacteriol 188: 1979–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Wit N, Derrien M, Bosch-Vermeulen H, Oosterink E, Keshtkar S, Duval C, de Vogel-van den Bosch J, Kleerebezem M, Müller M, van der Meer R. 2012. Saturated fat stimulates obesity and hepatic steatosis and affects gut microbiota composition by an enhanced overflow of dietary fat to the distal intestine. Am J Physiol Gastrointest Liver Physiol 303: G589–G599. [DOI] [PubMed] [Google Scholar]
- 9.Kabara JJ, Swieczkowski DM, Conley AJ, Truant JP. 1972. Fatty acids and derivatives as antimicrobial agents. Antimicrob Agents Chemother 2: 23–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gulhane M, Murray L, Lourie R, Tong H, Sheng YH, Wang R, Kang A, Schreiber V, Wong KY, Magor G, Denman S, Begun J, Florin TH, Perkins A, Cuív PÓ, McGuckin MA, Hasnain SZ. 2016. High fat diets induce colonic epithelial cell stress and inflammation that is reversed by IL-22. Sci Rep 6: 28990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murphy EF, Cotter PD, Healy S, Marques TM, O’Sullivan O, Fouhy F, Clarke SF, O’Toole PW, Quigley EM, Stanton C, Ross PR, O’Doherty RM, Shanahan F. 2010. Composition and energy harvesting capacity of the gut microbiota: relationship to diet, obesity and time in mouse models. Gut 59: 1635–1642. [DOI] [PubMed] [Google Scholar]
- 12.Duncan SH, Louis P, Thomson JM, Flint HJ. 2009. The role of pH in determining the species composition of the human colonic microbiota. Environ Microbiol 11: 2112–2122. [DOI] [PubMed] [Google Scholar]
- 13.Pabst O. 2012. New concepts in the generation and functions of IgA. Nat Rev Immunol 12: 821–832. [DOI] [PubMed] [Google Scholar]
- 14.Cerutti A, Chen K, Chorny A. 2011. Immunoglobulin responses at the mucosal interface. Annu Rev Immunol 29: 273–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Palm NW, de Zoete MR, Cullen TW, Barry NA, Stefanowski J, Hao L, Degnan PH, Hu J, Peter I, Zhang W, Ruggiero E, Cho JH, Goodman AL, Flavell RA. 2014. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell 158: 1000–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suzuki K, Meek B, Doi Y, Muramatsu M, Chiba T, Honjo T, Fagarasan S. 2004. Aberrant expansion of segmented filamentous bacteria in IgA-deficient gut. Proc Natl Acad Sci USA 101: 1981–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mirpuri J, Raetz M, Sturge CR, Wilhelm CL, Benson A, Savani RC, Hooper LV, Yarovinsky F. 2014. Proteobacteria-specific IgA regulates maturation of the intestinal microbiota. Gut Microbes 5: 28–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nishi K, Muranaka A, Nishimoto S, Kadota A, Sugahara T. 2012. Immunostimulatory effect of β-cryptoxanthin in vitro and in vivo. J Funct Foods 4: 618–625. [Google Scholar]
- 19.Hoebler C, Gaudier E, De Coppet P, Rival M, Cherbut C. 2006. MUC genes are differently expressed during onset and maintenance of inflammation in dextran sodium sulfate-treated mice. Dig Dis Sci 51: 381–389. [DOI] [PubMed] [Google Scholar]
- 20.Tang MT, Han H, Yu Z, Tsuruta T, Nishino N. 2017. Variability, stability, and resilience of fecal microbiota in dairy cows fed whole crop corn silage. Appl Microbiol Biotechnol 101: 6355–6364. [DOI] [PubMed] [Google Scholar]
- 21.Li Y, Nishino N. 2011. Monitoring the bacterial community of maize silage stored in a bunker silo inoculated with Enterococcus faecium, Lactobacillus plantarum and Lactobacillus buchneri. J Appl Microbiol 110: 1561–1570. [DOI] [PubMed] [Google Scholar]
- 22.Tsuruta T, Inoue R, Nojima I, Tsukahara T, Hara H, Yajima T. 2009. The amount of secreted IgA may not determine the secretory IgA coating ratio of gastrointestinal bacteria. FEMS Immunol Med Microbiol 56: 185–189. [DOI] [PubMed] [Google Scholar]
- 23.Iwasaki A, Kelsall BL. 2000. Localization of distinct Peyer’s patch dendritic cell subsets and their recruitment by chemokines macrophage inflammatory protein (MIP)-3alpha, MIP-3beta, and secondary lymphoid organ chemokine. J Exp Med 191: 1381–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.James BR, Tomanek-Chalkley A, Askeland EJ, Kucaba T, Griffith TS, Norian LA. 2012. Diet-induced obesity alters dendritic cell function in the presence and absence of tumor growth. J Immunol 189: 1311–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peterson DA, McNulty NP, Guruge JL, Gordon JI. 2007. IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell Host Microbe 2: 328–339. [DOI] [PubMed] [Google Scholar]
- 26.Wei M, Shinkura R, Doi Y, Maruya M, Fagarasan S, Honjo T. 2011. Mice carrying a knock-in mutation of Aicda resulting in a defect in somatic hypermutation have impaired gut homeostasis and compromised mucosal defense. Nat Immunol 12: 264–270. [DOI] [PubMed] [Google Scholar]
- 27.Tsuruta T, Inoue R, Iwanaga T, Hara H, Yajima T. 2010. Development of a method for the identification of S-IgA-coated bacterial composition in mouse and human feces. Biosci Biotechnol Biochem 74: 968–973. [DOI] [PubMed] [Google Scholar]
- 28.Qiao Y, Sun J, Xie Z, Shi Y, Le G. 2014. Propensity to high-fat diet-induced obesity in mice is associated with the indigenous opportunistic bacteria on the interior of Peyer’s patches. J Clin Biochem Nutr 55: 120–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin HV, Frassetto A, Kowalik EJ, Jr, Nawrocki AR, Lu MM, Kosinski JR, Hubert JA, Szeto D, Yao X, Forrest G, Marsh DJ. 2012. Butyrate and propionate protect against diet-induced obesity and regulate gut hormones via free fatty acid receptor 3-independent mechanisms. PLoS One 7: e35240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berger K, Falck P, Linninge C, Nilsson U, Axling U, Grey C, Stålbrand H, Nordberg Karlsson E, Nyman M, Holm C, Adlercreutz P. 2014. Cereal byproducts have prebiotic potential in mice fed a high-fat diet. J Agric Food Chem 62: 8169–8178. [DOI] [PubMed] [Google Scholar]
- 31.Louis P, Flint HJ. 2017. Formation of propionate and butyrate by the human colonic microbiota. Environ Microbiol 19: 29–41. [DOI] [PubMed] [Google Scholar]
- 32.Evans CC, LePard KJ, Kwak JW, Stancukas MC, Laskowski S, Dougherty J, Moulton L, Glawe A, Wang Y, Leone V, Antonopoulos DA, Smith D, Chang EB, Ciancio MJ. 2014. Exercise prevents weight gain and alters the gut microbiota in a mouse model of high fat diet-induced obesity. PLoS One 9: e92193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim M, Qie Y, Park J, Kim CH. 2016. Gut microbial metabolites fuel host antibody responses. Cell Host Microbe 20: 202–214. [DOI] [PMC free article] [PubMed] [Google Scholar]