Abstract
Introduction
We sought to determine if our previously validated proteomic profile for detecting Alzheimer's disease would detect Parkinson's disease (PD) and distinguish PD from other neurodegenerative diseases.
Methods
Plasma samples were assayed from 150 patients of the Harvard Biomarkers Study (PD, n = 50; other neurodegenerative diseases, n = 50; healthy controls, n = 50) using electrochemiluminescence and Simoa platforms.
Results
The first step proteomic profile distinguished neurodegenerative diseases from controls with a diagnostic accuracy of 0.94. The second step profile distinguished PD cases from other neurodegenerative diseases with a diagnostic accuracy of 0.98. The proteomic profile differed in step 1 versus step 2, suggesting that a multistep proteomic profile algorithm to detecting and distinguishing between neurodegenerative diseases may be optimal.
Discussion
These data provide evidence of the potential use of a multitiered blood-based proteomic screening method for detecting individuals with neurodegenerative disease and then distinguishing PD from other neurodegenerative diseases.
Keywords: Parkinson's disease, Precision medicine, Proteomics, Blood biomarkers, Diagnostic accuracy
1. Background
Parkinson's disease (PD) is the second most common neurodegenerative disease affecting over 1% of people aged 65 years and older in the United States [1]. The cost of PD to our society was reported to be $23 billion annually in the United States in 2005 [2]. Considering the estimated 15% growth in the elderly US population during the last decade, these costs can be expected to increase dramatically as the population ages. Neuropathologically, PD is a progressive disorder of unknown cause affecting multiple neurotransmitter systems. Common nonmotor features of the disease include autonomic failure, urinary incontinence, hallucinations, and dementia [3]. Although a number of treatments have been developed that improve the “dopaminergic deficit,” no treatment has been demonstrated to slow the neuronal degeneration of the substantia nigra neurons. Novel therapeutic approaches are needed with new disease-modifying therapies currently being examined that may ultimately improve patient outcomes.
A major impediment to treatment developments and clinical trials for neurodegenerative diseases is the lack of a sensitive, easily obtained biomarker of disease presence [4], [5], [6], [7], [8]. The “cornerstone” to the development of novel disease-modifying therapies in PD is the identification and validation of biomarkers of disease presence and progression [9]. Over the last several decades, the search for biomarkers that have diagnostic and prognostic use in neurodegenerative diseases has grown exponentially [5], [10], [11] with most work focusing on neuroimaging and cerebrospinal (CSF) methods [5], [10], [11], [12], [13], [14] and increasingly clinical-genetic algorithms [15], [16]. In fact, amyloid-beta (Aβ) positron emission tomography (PET) scanning tracers and CSF assays have been approved by the Food and Drug Administration for use in the diagnostic process for Alzheimer's disease (AD), and dopamine transporter single photon emission computed tomography [17] has been established for PD. Recent work suggests CSF markers may also have use in the differential diagnosis of neurodegenerative diseases [18]. Although advance imaging and CSF methods have tremendous potential as biomarkers of PD and other neurodegenerative diseases, invasiveness, accessibility, and cost barriers preclude these from being used as initial detection procedures [6], [7], [19], [20]. Therefore, it has been proposed that blood-based methods require additional investigation [21], [22], [23] and may serve as first step in a multitier detection process [6], [19] similar to the models used in cancer [24].
There has been a surge in the search for blood-based biomarkers for PD [25], [26], [27]. Blood-based biomarkers have the potential to serve as the initial step in the neurodiagnostic process used in large-scale screening, in primary care settings [19], as well as screening into novel clinical trials, the latter of which will result in substantial cost savings to the overall trial itself. As is the case with all initial screening tests, the goal of the first step is to screen out those patients who should not undergo more expensive and invasive confirmatory diagnostic procedures [19]. This is the same model used by cancer biomarkers that have received both regulatory and reimbursement approval [24]. Our work on blood-based biomarkers of AD has consistently shown that a multimarker approach identifying biomarker profiles of disease presence can yield excellent results [28], [29], [30], and our initial work suggested that this same proteomic profile could distinguish AD from PD [31], as well as accurately detect neurodegenerative disease [32]. Therefore, we hypothesize that our blood-based biomarker profile approach may serve to provide a cost- and time-effective means for establishing a rapidly scalable multitiered neurodiagnostic process [19], [32] for detecting neurodegenerative disease, including PD. With this initial screening approach, appropriate referrals can be made for subsequent specialty examinations and confirmatory diagnostic biomarkers (imaging, CSF), following the multistage models used for diagnosing cancer [24].
Here, we test the hypothesis that our previously validated proteomic profile for detecting AD [31], [32] would be successful in (1) detecting neurodegenerative diseases (PD and other neurodegenerative diseases vs. controls) and (2) discriminating PD from other neurodegenerative disease. This study was conducted by examination of plasma samples from the Harvard Biomarker Study (HBS).
2. Methods
2.1. Subjects
The study sample included 150 patients from the HBS (PD n = 50; other neurodegenerative diseases n = 50, controls n = 50). The other neurodegenerative diseases category included AD (n = 12), frontotemporal dementia (n = 25), progressive supranuclear palsy (n = 7), and corticobasal degeneration (n = 6) (Table 1). HBS is a longitudinal, case-control study that tracks clinical phenotypes and linked biospecimens of individuals with neurodegenerative diseases and controls without neurologic disease. High-quality biosamples and high-resolution clinical phenotypes are longitudinally tracked over time. HBS was designed for the primary goal of developing biomarkers that track disease progression and allow go/no go decisions in phase II clinical trials. The HBS specifically fosters research across neurodegenerative diseases, such as the proof-of-concept study described in this study. HBS has been published extensively [15], [33], [34], [35], [36], [37], [38], [39], [40].
Table 1.
Descriptive characteristics of the sample
| Characteristic | PD | Neurodegenerative controls | Healthy controls |
|---|---|---|---|
| Total, N | 50 | 50 | 50 |
| Male/female, N | 25/25 | 25/25 | 25/25 |
| UPDRS | 49.6 ± 23.9 | - | - |
| Age | 72.4 ± 9.4 | 72.64 ± 10.3 | 69.08 ± 9.7 |
| MMSE | 26.5 ± 3.7 | 20.4 ± 6.7 | 29.2 ± 1.6 |
| PD medications | 36 (72%) | 0 (0%) | 0 (0%) |
Abbreviations: MMSE, Mini-Mental State Examination; PD, Parkinson's disease; UPDRS, unified Parkinson's disease rating scale.
2.2. Proteomics
Plasma samples were assayed using two technological platforms. The proteomic assays were conducted using two automated systems. The electrochemiluminescence (ECL) assays from our previously validated AD blood screen were captured via the multiplex platform QuickPlex from Meso Scale Discovery per our previously published methods [31], [32], with assay preparation performed via automation using the Hamilton Robotics StarPlus system. We recently reported the analytic performance of each of these markers for >1300 samples across multiple cohorts and diagnoses (normal cognition, mild cognitive impairment, AD) [32]. The assays are reliable, and our experience with these assays show excellent spiked recovery, dilution linearity, coefficient of variation (CV), and detection limits. Interassay and intraassay variability has been excellent. A total of 250 μl of plasma was used to assay the following markers: fatty acid–binding protein (FABP), β2-microglobulin, pancreatic polypeptide, C-reactive protein, intercellular adhesion molecule 1, thrombopoietin, α2-macroglobulin, eotaxin 3, tumor necrosis factor α, tenascin C, interleukin (IL)-5, IL6, IL7, IL10, IL18, I309, factor VII, vascular cell adhesion molecule 1, thymus and activation regulated chemokine, and serum amyloid A. With automation, the average CV for these assays on >1000 samples in our laboratory has been excellent with nearly all having CVs < 10% and 62% having CVs < 5%. Given the recent surge in the literature examining ultrasensitive blood-based markers of neuropathological markers in neurodegenerative diseases, here the Simoa assays for Aβ40, Aβ42, tau, α-synuclein, and neurofilament light polypeptide were conducted using the automated HD-1 analyzer from Quanterix. The performance of the assays in our laboratory from >1000 samples has been excellent with all CVs ≤ 5%.
2.3. Proteomic profile
In our prior work, we have generated and cross-validated an AD proteomic profile across platforms [28], [31], cohorts [28], [30], [32], [41], [42], species (human, mouse) [31], tissue (brain, serum, plasma) [31], and ethnicities (non-Hispanic white, Mexican American) [28], [43]. In our preliminary work, we found that this same proteomic profile could discriminate PD from AD [31]. In that work, we found that the relative importance of the proteins varied between PD and AD, but the overall algorithm was still highly accurate in detecting both diseases. In a subsequent study, we found that our proteomic profile was highly accurate in detecting neurodegenerative diseases [32]. Therefore, here we sought to cross-validate and expand on that work by demonstrating the accuracy of our proteomic profile approach for detecting neurodegenerative diseases and discriminating PD from other neurodegenerative diseases.
2.4. Statistical analysis
Statistical analyses were conducted using R (V 3.3.3) statistical software [44] and SPSS 24 (IBM). Diagnostic accuracy was calculated via receiver operating characteristic (ROC) curves. First, support vector machine (SVM) analyses were used to discriminate controls from neurodegenerative disease (i.e., PD/Other), with resulting diagnostic accuracy statistics generated (step 1). Then, SVM analysis was restricted only to PD versus other neurodegenerative diseases (step 2). SVM analyses were conducted with internal 5-fold cross-validation. In our prior work, we have found that the overall proteomic profile varies between different neurodegenerative diseases [31]. Therefore, our two-step approach was used to capitalize on these differences to increase accuracy and also to allow for the overall algorithm to be more robust and avoid multilevel analyses simultaneously. The latter reduces risk for error and sample overidentification.
3. Results
Descriptive statistics of the sample are provided in Table 1. The average age of the sample was 71.37 years (standard deviation [SD] = 9.9). There were even numbers of males and females across all three groups. An analysis of variance showed there were no significant age differences among the PD group, the healthy control group, and the other neurodegenerative disorders group (F(2, 147) = 2.04, P = .13). There were significant group differences in Mini-Mental State Examination score among the three groups (F(2, 118) = 39.9, P = .<.001). Tukey's honestly significant difference post hoc analysis revealed that participants with PD (M = 26.5, SD = 3.7) scored significantly lower than healthy controls (M = 29.2, SD = 1.6) but higher than those with other neurodegenerative diseases (M = 20.4, SD = 6.7).
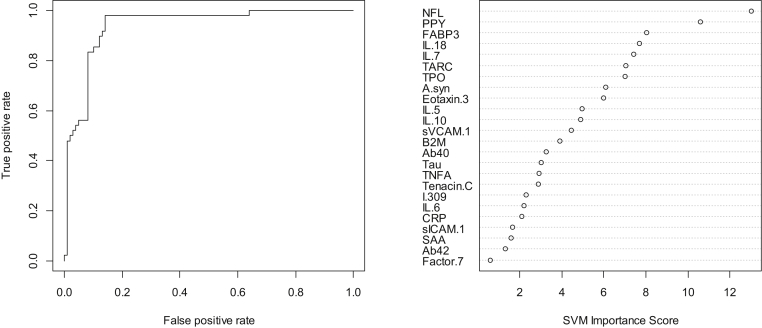
In step 1, our SVM-based proteomic profile was highly accurate in detecting neurodegenerative disease (PD and other) as compared to normal controls. The overall area under the receiver operating characteristic curve (AUC) was 0.94 with an observed sensitivity (SN) of 0.92 and specificity (SP) of 0.65. Table 2 shows all the correct and incorrect predictions while the variable importance plot and ROC curve are presented in Fig. 1. Inclusion of demographic factors did not significantly change the AUC.
Table 2.
Accuracy of step 1 in detecting neurodegenerative diseases
| Predicted | SVM model |
|
|---|---|---|
| PD/AD/FTD/others | NC | |
| PD/AD/FTD/others | 92 | 17 |
| NC | 8 | 31 |
| Sensitivity | 92.0% | |
| Specificity | 64.6% | |
| AUC | 0.94 | |
Abbreviations: AD, Alzheimer's disease; AUC, area under the receiver operating characteristic curve; FTD, frontotemporal dementia; NC, normal control; PD, Parkinson's disease; SVM, support vector machine.
Fig. 1.
ROC curve and variable importance plot for proteomic profile for detecting neurodegenerative disease. Abbreviations: ROC, receiver operating characteristic; IL, interleukin.
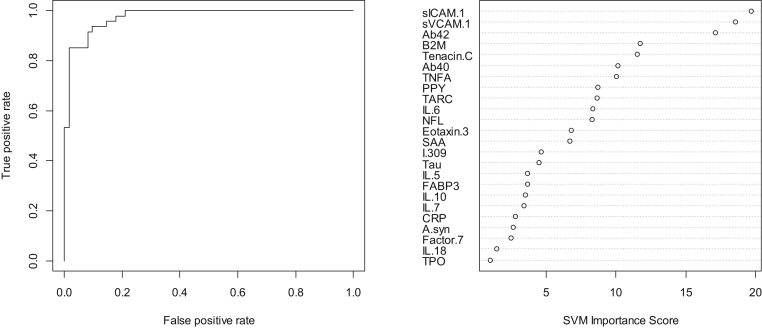
In the step 2, the overall SVM-proteomic profile also showed excellent accuracy at distinguishing PD from other neurodegenerative diseases. In this model, the AUC = 0.98, SN = 0.94, and SP = .89. Table 3 shows all classifications (correct and incorrect) while the variable importance plot and ROC curve are presented in Fig. 2. Inclusion of demographic factors did not significantly change the AUC.
Table 3.
Classification accuracy for proteomic profile for distinguishing PD from other neurodegenerative diseases
| Predicted | SVM model |
|
|---|---|---|
| PD | AD/FTD/others | |
| PD | 44 | 7 |
| AD/FTD/others | 3 | 55 |
| Sensitivity | 93.6% | |
| Specificity | 88.7% | |
| AUC | 0.98 | |
Abbreviations: AD, Alzheimer's disease; AUC, area under the receiver operating characteristic curve; FTD, frontotemporal dementia; PD, Parkinson's disease; SVM, support vector machine.
Fig. 2.
ROC curve and variable importance plot for proteomic profile for distinguishing PD from other neurodegenerative diseases. Abbreviations: ROC, receiver operating characteristic; IL, interleukin; PD, Parkinson's disease.
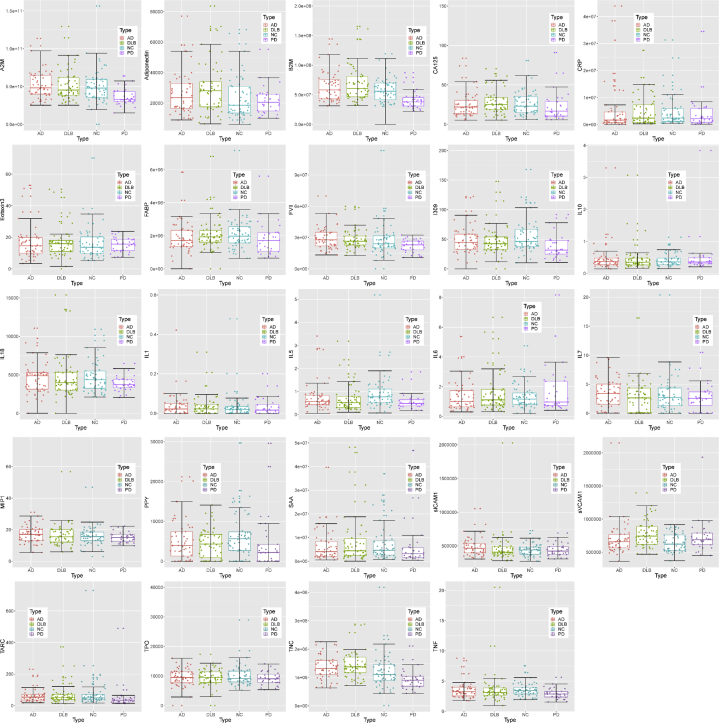
When reviewing the variable importance plots (Figs. 1 and 2), the overall profiles for discriminating PD/other neurodegenerative diseases from controls were different than the profile for discriminating PD from other neurodegenerative diseases, as was the case in our prior work. The top 10 markers for discriminating neurodegenerative diseases from controls were as follows: neurofilament light polypeptide, pancreatic polypeptide, FABP3, IL18, IL7, thymus and activation regulated chemokine, thrombopoietin, α-synuclein, Eotaxin 3, and IL5. However, the top 10 variables for discriminating PD from other neurodegenerative diseases were intercellular adhesion molecule 1, vascular cell adhesion molecule 1, Aβ42, B2M, Tenascin C, Aβ40, tumor necrosis factor alpha, pancreatic polypeptide, thymus and activation regulated chemokine, and IL6. Fig. 3 provides box plots by protein across the three diagnostic groups for all proteins.
Fig. 3.
Box plots for the top 10 variables. Abbreviations: AD, Alzheimer's disease; PD, Parkinson's disease; DLB, dementia with Lewy bodies; IL, interleukin.
Given our prior work looking at our proteomic profile in AD, we conducted preliminary analyses with (1) only our AD proteomic algorithm, (2) only the Simoa assays, and (3) all markers combined for discriminating PD from AD as well as PD from controls in this sample. For PD versus AD, the Simoa assays alone yielded an excellent SN of 1.0 but only an SP of 0.25. However, our standard ECL proteomic profile (described earlier) yielded a superior balance of SN (also 1.0) and SP (0.75). When the Simoa assays were combined with our standard ECL proteomic panel, there was a modest increase in SP to 0.80. When distinguishing PD from controls, the Simoa assays yielded an SN = 0.74 and SP = .83. Our standard ECL profile yielded an improved SN = 0.92 and SP = .90. The combined algorithm with our ECL and Simoa assays resulted in an increase of SP to 0.94. These results are very preliminary given the size of the sample.
4. Discussion
In the present study, we cross-validated in an independent cohort the findings that our previously validated proteomic profile for AD can also (1) detect neurodegenerative diseases and (2) discriminate PD from other neurodegenerative diseases. In detecting neurodegenerative disease versus controls, the current AUC was 0.94 with an observed SN of 0.92 and SP of 0.65. When distinguishing PD from other neurodegenerative diseases, the overall accuracy improved to an AUC = 0.98, SN = 0.94, and SP = .89.
While our proteomic profile has previously been validated for detecting AD and neurodegenerative diseases combined, there is also significant literature to implicate many of these markers in PD. For example, multiple inflammatory markers such as tumor necrosis factor alpha, C-reactive protein, and IL6 have previously been linked with PD [45], [46], and inflammation has been shown to improve after exercise interventions in persons with PD [47], [48]. Mollenhauer et al. [49] found FABP to be differentially expressed in PD and dementia with Lewy bodies compared with controls, and FABP was among the top 10 markers in discriminating PD from AD in our prior work [31]. A meta-analysis of 877 PD cases and 1296 controls found polymorphisms associated with alpha-2-macroglobulin (r669 in particular) are associated with the risk for PD [50]. Recent work demonstrated that the VCAM1 gene was one of 7 novel genes that displayed significant changes in PD [51] while Tan et al. [52] also recently showed that intercellular adhesion molecule 1 was a hub that participated in the pathogenesis of PD. Scherzer et al [40] found differential expression of PD gene α-synuclein (SNCA) in PD, and low SNCA transcript abundance predicted cognitive decline longitudinally in PD [40]. Therefore, there is substantial extant of literature to support the underlying rationale for these markers being altered PD.
It is important to put these SN and SP estimates into perspective relative to the specific context of use. All first-line screening tools are designed to rule out disease, not rule in disease given the population base rates of disease presence. Therefore, assuming a 20% neurodegenerative disease base rate in the population of those aged 65 years and older, the SN = 0.92 and SP = .64 would yield a negative predictive power of 0.97 with a positive predictive power of 0.39 using Bayesian statistics for appropriate calculations. This means that a trial would be accurate in saying that a specific patient should not undergo a lumbar puncture, PET scan, or additional clinical evaluations 97% of the time, thereby allowing large-scale screening at substantially reduced cost. Our group has previously provided the same sorts of calculations for AD clinical trials [32].
This work also provides novel data when putting the newly designed ultrasensitive assays of amyloid, tau, α-synuclein, and neurofilament light polypeptide in context with other proteomic markers. In our prior work, our refined algorithm has been highly accurate in detecting both AD and PD. Here, we cross-validate the accuracy of the approach for detecting PD in an independent cohort (HBS). However, we also demonstrate that adding these new markers may increase the accuracy. On the other hand, these new markers were not very accurate at detecting PD or distinguishing PD from AD alone. The SN of 1.0 obtained by both approaches is likely an artifact of sample size and will not hold in larger samples. The current team is assaying additional PD samples to (1) cross-validate the current findings in independent samples/cohorts and (2) working to build a larger database for combined analyses across cohorts for a clinically relevant estimate of the overall accuracy of these algorithms and markers. If cross-validated, this approach should be applied prospectively within the specific population reflective of the intended context of use as the current group is actively doing with our AD blood screen.
There are limitations to the present study. First, the sample size is relatively small, and the results are proof of concept and must be validated in independent cohorts and larger sample sizes. Instead of splitting the sample into training and test samples, internal 5-fold cross-validation was conducted. However, the results strongly support the justification for such validation studies, which are being carried out by the current team. Second, addition analyses are needed to determine the impact of preanalytic conditions on the assay performance as we have previously pointed out in the AD space [53]. Interestingly, our group recently assayed an independent cohort of PD and dementia with Lewy bodies with preanalytic protocols different from HBS and found comparable diagnostic accuracy [54]. Additional variables such as fasting duration, storage time, medication status, and so forth should be examined in future studies. Third, the reliability of the findings over time should also be tested. The usability of any blood test is reliant on the accuracy of the test over time. Therefore, longitudinal application of the blood test to the same samples over time is warranted. Finally, the current analyses do not compare the blood proteomics to CSF-defined or PET scan–defined pathology. It is certainly possible that the blood proteomic profiles are detecting underlying amyloid-, tau-, or α-synuclein-related pathologies, but that has not yet been tested. It is also possible that the algorithms, specifically of the underlying targeted pathology (e.g., amyloid vs. tau), would be different. This should also be tested. If validated, the next step will be to determine the scalability of the methods to meet the population needs at the primary care office level.
Overall, the current findings are strongly supportive of follow-up application of the current proteomic profiles to larger biorepository samples, such as the full HBS cohort. The current team is working toward that goal. Ultimately, the goal is to provide clinicians and companies with a rapidly scalable tool (or tools) that can streamline and increase access (while cost containing) to novel clinical trials to improve patient outcomes.
Research in Context.
-
1.
Systematic Review: Literature was identified and reviewed using PubMed. Several articles described the importance of rapid and cost-effective biomarkers for neurodegenerative diseases, including Parkinson's disease (PD). However, no such blood-based biomarkers currently exist as a first step in a multitiered neurodiagnostic process.
-
2.
Interpretation: Our findings show that a blood-based biomarker profile can detect neurodegenerative disease (PD/other) and distinguish PD from other neurodegenerative diseases.
-
3.
Future Directions: This article provides support for the notion that a blood-based biomarker profile can accurately detect PD and even distinguish PD from other neurodegenerative diseases. Future work will be conducted to expand the sample size, particularly among other neurodegenerative diseases. A blood-based biomarker profile for PD would be of tremendous use for screening into novel therapeutic trials.
Acknowledgments
The authors thank all study participants, their families, and friends for their support and participation and our study coordinators. C.R.S. was supported in part by the American Parkinson Disease Association and NIH grants U01NS082157, U01NS095736, U01NS100603, and R01AG057331. HBS was supported by the APDA Advanced Center for Parkinson's Disease Research, Harvard NeuroDiscovery Center, NINDS U01NS082157, U01NS100603, and the Massachusetts Alzheimer's Disease Research Center NIA P50AG005134. S.E.O. was supported in part by grants from the National Institute on Aging of the National Institutes of Health under award numbers R01AG054073, R01AG051848, R01AG058252, and R01AG058537. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The authors thank the following studies and investigators:
Harvard Biomarkers Study:
Co-Directors of the Harvard NeuroDiscovery Center: Clemens R. Scherzer, Bradley T. Hyman, Charles G. Jennings; investigators and study coordinators at the Harvard NeuroDiscovery Center: Yuliya I. Kuras, Daly Franco, Frank Zhu, Karbi Choudhury; investigators and study coordinators at the Brigham and Women's Hospital: Lewis R. Sudarsky, Michael T. Hayes, Chizoba C. Umeh, Reisa Sperling; investigators and study coordinators at the Massachusetts General Hospital: John H. Growdon, Michael A. Schwarzschild, Albert Y. Hung, Alice W. Flaherty, Deborah Blacker, Anne-Marie Wills, U. Shivraj Sohur, Vivek K. Unni, Nicte I. Mejia, Anand Viswanathan, Stephen N. Gomperts, Vikram Khurana, Mark W. Albers, Maria Allora-Palli, Alireza Atri, David Hsu, Alexandra Kimball, Scott McGinnis, Nutan Sharma, John Becker, Randy Buckner, Thomas Byrne, Maura Copeland, Bradford Dickerson, Matthew Frosch, Theresa Gomez-Isla, Steven Greenberg, James Gusella, Julius Hedden, Elizabeth Hedley-Whyte, Keith Johnson, Raymond Kelleher, Aaron Koenig, Maria Marquis-Sayagues, Gad Marshall, Sergi Martinez-Ramirez, Donald McLaren, Olivia Okereke, Elena Ratti, Christopher William, Koene Van Dij, Shuko Takeda, Anat Stemmer-Rachaminov, Jessica Kloppenburg, Catherine Munro, Rachel Schmid, Sarah Wigman, Sara Wlodarcsyk; investigators and study coordinators at the University of Ottawa: Michael G. Schlossmacher; Scientific Advisory Board member of the Massachusetts General Hospital: John H. Growdon; Scientific Advisory Board members of the Brigham and Women's Hospital: Dennis J. Selkoe and Reisa Sperling; Scientific Advisory Board member of the Harvard School of Public Health: Alberto Ascherio; Data Coordinator at the Harvard NeuroDiscovery Center: Thomas Yi; Data Coordinators at the Massachusetts General Hospital: Joseph J. Locascio and Haining Li; Biobank Management Staff at the Harvard NeuroDiscovery Center: Gabriel Stalberg and Zhixiang Liao.
Footnotes
Potential Conflicts of Interest: C.R.S. has collaborated with Pfizer, OPKO, Proteome Sciences, Sanofi, and Berg Health; has consulted for Sanofi; has served as advisor to the Michael J. Fox Foundation, NIH, Department of Defense; is on the scientific advisory board of the American Parkinson Disease Association; has received funding from the NIH, the US Department of Defense, the Harvard NeuroDiscovery Center, the Michael J. Fox Foundation, and American Parkinson Disease Association. S.E.O. has multiple patents pending on precision medicine for neurodegenerative diseases; is the founding scientist for Cx Precision Medicine; and has served as a consultant to Roche Diagnostics. S.E.O. has received funding from the National Institute on Aging, Michael J. Fox Foundation, Alzheimer's Association, has multiple patents pending on precision medicine for neurodegenerative diseases; is the founding scientist for Cx Precision Medicine and has served as a consultant to Roche Diagnostics.
References
- 1.Wright W.A., Evanoff B.A., Lian M., Criswell S.R., Racette B.A. Geographic and ethnic variation in Parkinson disease: a population-based study of US Medicare beneficiaries. Neuroepidemiology. 2010;34:143–151. doi: 10.1159/000275491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huse D.M., Schulman K., Orisni L., Castelli-Haley J., Kennedy S., Lenhardt G. Burden of illness in Parkinson's disease. Mov Disord. 2005;20:1449–1454. doi: 10.1002/mds.20609. [DOI] [PubMed] [Google Scholar]
- 3.Hely M.A., Reid W.G., Adena M.A., Halliday G.M., Morris J.G. The Sydney multicenter study of Parkinson's disease: the inevitability of dementia at 20 years. Mov Disord. 2008;23:837–844. doi: 10.1002/mds.21956. [DOI] [PubMed] [Google Scholar]
- 4.Henriksen K., O'Bryant S.E., Hampel H., Trojanowski J.Q., Montine T.J., Jeromin A. The future of blood-based biomarkers for Alzheimer's disease. Alzheimers Dement. 2014;10:115–131. doi: 10.1016/j.jalz.2013.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thal L.J., Kantarci K., Reiman E.M., Klunk W.E., Weiner M.W., Zetterberg H. The role of biomarkers in clinical trials for Alzheimer disease. Alzheimer Dis Assoc Disord. 2006;20:6–15. doi: 10.1097/01.wad.0000191420.61260.a8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schneider P., Hampel H., Buerger K. Biological marker candidates of alzheimer's disease in blood, plasma, and serum. CNS Neurosci Ther. 2009;15:358–374. doi: 10.1111/j.1755-5949.2009.00104.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eller M., Williams D.R. α-Synuclein in Parkinson disease and other neurodegenerative disorders. Clin Chem Lab Med. 2011;49:403–408. doi: 10.1515/CCLM.2011.077. [DOI] [PubMed] [Google Scholar]
- 8.Sinha N., Firbank M., O'Brien J.T. Biomarkers in dementia with Lewy bodies: A review. Int J Geriatr Psychiatry. 2012;27:443–453. doi: 10.1002/gps.2749. [DOI] [PubMed] [Google Scholar]
- 9.Gerlach M., Maetzler W., Brioch K., Hampel H., Rems L., Reum T. Biomarker candidates of neurodegeneration in Parkinson's disease for the evaluation of disease-modifying therapeutics. J Neural Transm. 2012;119:39–52. doi: 10.1007/s00702-011-0682-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anonymous. Consensus report of the Working Group on: “Molecular and Biochemical Markers of Alzheimer's Disease”. The Ronald and Nancy Reagan Research Institute of the Alzheimer's Association and the National Institute on Aging Working Group. Neurobiol Aging. 1998;19:109–116. [see comment][erratum appears in Neurobiol Aging 1998 May-Jun;19(3):285] [PubMed] [Google Scholar]
- 11.O'Bryant S.E., Hobson V., Hall J.R., Warring S.C., Chan W., Massman P. Brain-derived neurotrophic factor levels in alzheimer's disease. J Alzheimers Dis. 2009;17:337–341. doi: 10.3233/JAD-2009-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Graff-Radford J., Boeve B.F., Pedraza O., Verman T.J., Przybelski S., Lesnick T.C. Imaging and acetylcholinesterase inhibitor response in dementia with Lewy bodies. Brain. 2012;135:2470–2477. doi: 10.1093/brain/aws173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Colloby S.J., Firbank M.J., Pakrasi S., Lloyd J.J., Driver I., McKeith I.G. A comparison of 99mTc-exametazime and 123I-FP-CIT SPECT imaging in the differential diagnosis of Alzheimer's disease and dementia with Lewy bodies. Int Psychogeriatr. 2008;20:1124–1140. doi: 10.1017/S1041610208007709. [DOI] [PubMed] [Google Scholar]
- 14.McKeith I., O'Brien J., Walker Z., Tatsch K., Booij J., Darcourt J. Sensitivity and specificity of dopamine transporter imaging with 123I-FP-CIT SPECT in dementia with Lewy bodies: a phase III, multicentre study. Lancet Neurol. 2007;6:305–313. doi: 10.1016/S1474-4422(07)70057-1. [DOI] [PubMed] [Google Scholar]
- 15.Liu G., Boot B., Locascio J.J., Jansen I.E., Winder-Rhodes S., Eberly S. Specifically neuropathic Gaucher's mutations accelerate cognitive decline in Parkinson's. Ann Neurol. 2016;80:674–685. doi: 10.1002/ana.24781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu G., Locascio J.J., Corvol J.C., Boot B., Liao Z., Page K. Prediction of cognition in Parkinson's disease with a clinical-genetic score: a longitudinal analysis of nine cohorts. Lancet Neurol. 2017;16:620–629. doi: 10.1016/S1474-4422(17)30122-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park E., Hwang Y.M., Lee C.N., Kim S., Oh S.Y., Kim Y.C. Differential Diagnosis of Patients with Inconclusive Parkinsonian Features Using [18F]FP-CIT PET/CT. Nucl Med Mol Imaging. 2014;48:106–113. doi: 10.1007/s13139-013-0253-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaerst L., Kuhlmann A., Wedekind D., Stoeck K., Lange P., Zerr I. Using cerebrospinal fluid marker profiles in clinical diagnosis of dementia with lewy bodies, Parkinson's disease, and Alzheimer's disease. J Alzheimers Dis. 2014;38:63–73. doi: 10.3233/JAD-130995. [DOI] [PubMed] [Google Scholar]
- 19.O'Bryant S.E., Mielke M.M., Rissman R.A., Lista S., Vanderstichele H., Zetterberg H. Blood-based biomarkers in Alzheimer disease: Current state of the science and a novel collaborative paradigm for advancing from discovery to clinic. Alzheimers Dement. 2017;13:45–58. doi: 10.1016/j.jalz.2016.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henchcliffe C., Dodel R., Beal M.F. Biomarkers of Parkinson's disease and Dementia with Lewy bodies. Prog Neurobiol. 2011;95:601–613. doi: 10.1016/j.pneurobio.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 21.Shtilbans A., Henchcliffe C. Biomarkers in Parkinson's disease: An update. Curr Opin Neurol. 2012;25:460–465. doi: 10.1097/WCO.0b013e3283550c0d. [DOI] [PubMed] [Google Scholar]
- 22.Henriksen K., O'Bryant S.E., Hampel H., Trojanowski J.Q., Montine T.J., Jeromin A. The future of blood-based biomarkers for Alzheimer's disease. Alzheimers Demen. 2014;10:115–131. doi: 10.1016/j.jalz.2013.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hennecke G., Scherzer C.R. RNA biomarkers of Parkinson's disease: developing tools for novel therapies. Biomark Med. 2008;2:41–53. doi: 10.2217/17520363.2.1.41. [DOI] [PubMed] [Google Scholar]
- 24.Gold L.S., Klein G., Carr L., Kessler L., Sullivan S.D. The emergence of diagnostic imaging technologies in breast cancer: Discovery, regulatory approval, reimbursement, and adoption in clinical guidelines. Cancer Imaging. 2012;12:13–24. doi: 10.1102/1470-7330.2012.0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hansson O., Janelidze S., Hall S., Magdalinou N., Lees A.J., Andreasson U. Blood-based NfL: A biomarker for differential diagnosis of parkinsonian disorder. Neurology. 2017;88:930–937. doi: 10.1212/WNL.0000000000003680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Green H., Zhang X., Tiklova K., Volakakis N., Brodin L., Berg L. Alterations of p11 in brain tissue and peripheral blood leukocytes in Parkinson's disease. Proc Natl Acad Sci U S A. 2017;114:2735–2740. doi: 10.1073/pnas.1621218114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chatterjee P., Roy D. Comparative analysis of RNA-Seq data from brain and blood samples of Parkinson's disease. Biochem Biophys Res Commun. 2017 doi: 10.1016/j.bbrc.2017.01.121. [DOI] [PubMed] [Google Scholar]
- 28.O'Bryant S.E., Xiao G., Barber R., Reisch J., Doody R., Fairchild T. A serum protein-based algorithm for the detection of Alzheimer disease. Arch Neurol. 2010;67:1077–1081. doi: 10.1001/archneurol.2010.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Bryant S.E., Xiao G., Barber R., Reisch J., Hall J., Cullum C.M. A blood-based algorithm for the detection of Alzheimer's disease. Demen Geriatr Cogn Disord. 2011;32:55–62. doi: 10.1159/000330750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O'Bryant S.E., Xiao G., Barber R., Huebinger R., Wilhelmsen K., Edwards M. A blood-based screening tool for Alzheimer's disease that spans serum and plasma: findings from TARC and ADNI. PLoS One. 2011;6:e28092. doi: 10.1371/journal.pone.0028092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O'Bryant S.E., Xiao G., Zhang F., Edwards M., German D.C., Yin X. Validation of a serum screen for alzheimer's disease across assay platforms, species, and tissues. J Alzheimers Dis. 2014;42:1325–1335. doi: 10.3233/JAD-141041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O'Bryant S.E., Edwards M., Johnson L., Hall J., Villarreal A.E., Britton G.B. A blood screening test for Alzheimer's disease. Alzheimers Dement (Amst) 2016;3:83–90. doi: 10.1016/j.dadm.2016.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cho H.H., Cahill C.M., Vanderburg C.R., Scherzer C.R., Wang B., Huang X. Selective translational control of the Alzheimer amyloid precursor protein transcript by iron regulatory protein-1. J Biol Chem. 2010;285:31217–31232. doi: 10.1074/jbc.M110.149161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ding H., Sarokhan A.K., Roderick S.S., Bakshi R., Maher N.E., Ashourian P. Association of SNCA with Parkinson: replication in the Harvard NeuroDiscovery Center Biomarker Study. Mov Disord. 2011;26:2283–2286. doi: 10.1002/mds.23934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hakimi M., Selvanantham T., Swinton E., Padmore R.F., Tong Y., Kabbach G. Parkinson's disease-linked LRRK2 is expressed in circulating and tissue immune cells and upregulated following recognition of microbial structures. J Neural Transm. 2011;118:795–808. doi: 10.1007/s00702-011-0653-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu Y., Chopra V., Chopra R., Locascio J.J., Liao Z., Ding H. Transcriptional modulator H2A histone family, member Y (H2AFY) marks Huntington disease activity in man and mouse. Proc Natl Acad Sci U S A. 2011;108:17141–17146. doi: 10.1073/pnas.1104409108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scherzer C.R., Eklund A.C., Morse L.J., Liao Z., Locascio J.J., Fefer D. Molecular markers of early Parkinson’s disease based on gene expression in blood. Proc Natl Acad Sci U S A. 2007;104:955–960. doi: 10.1073/pnas.0610204104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scherzer C.R., Grass J.A., Liao Z., Pepivani I., Zheng B., Eklund A.C. GATA transcription factors directly regulate the Parkinson's disease-linked gene alpha-synuclein. Proc Natl Acad Sci U S A. 2008;105:10907–10912. doi: 10.1073/pnas.0802437105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Bitterswikj M., Gulati S., Smoot E., Jaffa M., Maher N., Hyman B.T. Anti-superoxide dismutase antibodies are associated with survival in patients with sporadic amyotrophic lateral sclerosis. Amyotroph Lateral Scler. 2011;12:430–438. doi: 10.3109/17482968.2011.585163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Locascio J.J., Eberly S., Liao Z., Liu G., Hoesing A.N., Duong K. Association between alpha-synuclein blood transcripts and early, neuroimaging-supported Parkinson's disease. Brain. 2015;138:2659–2671. doi: 10.1093/brain/awv202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Villarreal A.E., O'Bryant S.E., Edwards M., Grajales S., Britton G.B., Panama Aging Research Initiative Serum-based protein profiles of Alzheimer's disease and mild cognitive impairmetn in elderly Hispanics. Neurodegener Dis Manag. 2016;6:203–213. doi: 10.2217/nmt-2015-0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Edwards M., Hall J., Williams B., Johnson L.A., O'Bryant S.E. Molecular markers of amnestic mild cognitive impairment among Mexican Americans. J Alzheimers Dis. 2016;49:221–228. doi: 10.3233/JAD-150553. [DOI] [PubMed] [Google Scholar]
- 43.O'Bryant S.E., Xiao G., Edwards M., Devous M., Gupta V.B., Martins R. Biomarkers of Alzheimer's disease among Mexican Americans. J Alzheimer's Dis. 2013;34:841–849. doi: 10.3233/JAD-122074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.R_Development_Core_Team R: A language and environment for statistical computing. 2009. Accessed October 2018. http://www.R-project.org 2009. Available from.
- 45.Kim S.W., Kang H.J., Bae K.Y., Shin I.S., Hong Y.J., Ahn Y.K. Interactions between pro-inflammatory cytokines and statins on depression in patients with acute coronary syndrome. Prog Neuropsychopharmacol Biol Psychiatry. 2018;80:250–254. doi: 10.1016/j.pnpbp.2017.07.003. [DOI] [PubMed] [Google Scholar]
- 46.Hall S., Janelidze S., Surova Y., Widner H., Zetterberg H., Hansson O. Cerebrospinal fluid concentrations of inflammatory markers in Parkinson's disease and atypical parkinsonian disorders. Sci Rep. 2018;8:13276. doi: 10.1038/s41598-018-31517-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Landers M.R., Navalta J.W., Murtishaw A.S., Kinney J.W., Pirio Richardson S. A high-intensity exercise boot camp for persons with Parkinson disease: a phase II, pragmatic, randomized clinical trial of feasibility, safety, signal of efficacy, and disease mechanisms. J Neurol Phys Ther. 2019;43:12–25. doi: 10.1097/NPT.0000000000000249. [DOI] [PubMed] [Google Scholar]
- 48.Al-Jarrah M.D., Erekat N.S. Treadmill exercise training could attenuate the upregulation of Interleukin-1beta and tumor necrosis factor alpha in the skeletal muscle of mouse model of chronic/progressive Parkinson disease. NeuroRehabilitation. 2018;43:501–507. doi: 10.3233/NRE-182492. [DOI] [PubMed] [Google Scholar]
- 49.Mollenhauer B., Steinacker P., Bahn E., Bibl M., Brechlin P., Schlossmacher M.G. Serum heart-type fatty acid-binding protein and cerebrospinal fluid tau: marker candidates for dementia with Lewy bodies. Neurodegener Dis. 2007;4:366–375. doi: 10.1159/000105157. [DOI] [PubMed] [Google Scholar]
- 50.Guo X., Tang P., Li X., Chong L., Zhang X., Li R. Association between two alpha-2-macroglobulin gene polymorphisms and Parkinson's disease: a meta-analysis. Int J Neurosci. 2016;126:193–198. doi: 10.3109/00207454.2014.996641. [DOI] [PubMed] [Google Scholar]
- 51.George G., Valiya Parambath S., Lokappa S.B., Varkey J. Construction of Parkinson's disease marker-based weighted protein-protein interaction network for prioritization of co-expressed genes. Gene. 2019;697:67–77. doi: 10.1016/j.gene.2019.02.026. [DOI] [PubMed] [Google Scholar]
- 52.Tan C., Liu X., Chen J. Microarray analysis of the molecular mechanism involved in Parkinson's disease. Parkinsons Dis. 2018;2018:1590465. doi: 10.1155/2018/1590465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.O'Bryant SE G.V., Henriksen K., Edwards M., Jeromin A., Lista S., Bazenet C. Guidelines for the standardization fo preanalytic variables for blood-based biomarker studies in Alzheimer's disease. Alzheimer's Demen. 2015;11:549–560. doi: 10.1016/j.jalz.2014.08.099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.O'Bryant SE F.T., Zhang F., Pedraza O., Wszolek Z.K., Commo T., Julovich D. A proteomic signature for dementia with Lewy bodies. Alzheimers Dement (Amst) 2019;11:270–276. doi: 10.1016/j.dadm.2019.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]