Abstract
Coat color genetics successfully adapted and applied to different animal species, which provides a good demonstration of the concept of comparative genetics. In this study, we sequenced 945 bp fragments of melanocortin 1 receptor (MC1R) gene, 421 bp fragments of exon 1 of tyrosinase (TYR) gene and 266 bp fragments of exon 3 of agouti signaling protein (ASIP) gene for 250 individuals with five plumage color patterns. We detected a total of three SNPs (T398A, T637C, and G920C) in MC1R and built six haplotypes (H1–H6) based on the three SNPs. H5 and H6 haplotypes were mainly concentrated in white and grey chicken. And diplotypes H2H3 occurred in white feather and black-speckle feather with the same frequency. Moreover, a total of three SNPs (C47G, T120C, and T172C) in TYR were found and built six haplotypes (P1–P6) based on the three SNPs. Among them, haplotype P2, P3 and P6 were not occurred in black chicken, the diplotypes P1P6 and P4P6 were only distributed in white, gray and black-speckled feather. We only detected one SNP (T168C) in ASIP gene and found that genotype TT was advantage genotype in the different plumage color groups of chickens. Collectively, our study suggested an association between plumage color and genetic variation of MC1R, TYR and ASIP in chicken.
Keywords: Chicken; MC1R, TYR and ASIP genes; SNP; Plumage Color
Introduction
With the development of society and the increase of population, the number of chickens is increasing and the distribution area is also growing. The chicken’s plumage is gradually enriched by continuous natural selection and artificial selection. People have different preference for different color of chicken feathers. Thus, studying the relationship between feather phenotype and genotype paves the way for appropriate products that meet consumers’ demands (Pang 2001).
The plumage color of birds is mainly related to the pigment distribution or proportion of eumelanin and pheomelanin (Giuseppe 1980). The relative ratio of eumelanin and pheomelanin is regulated by melanocortin receptor-1 (MC1R) and its antagonist agouti protein (Schiöth et al. 2003). MC1R encodes a seven-transmembrane domain G-protein-coupled receptor, which expressed primarily in melanocytes of developing feathers and hair (Mundy 2005). The process of eumelanin synthesis is triggered by the binding of α-melanocyte stimulating hormone (α-MSH) to MC1R (Scherer and Kumar 2010).Then, it will lead to an increase in intracellular cAMP which activates tyrosinase and catalyzes the first step of melanogenesis (Mountjoy et al. 1992).
Agouti, now known as Agouti signaling protein (ASIP), is a ligand to the MC1R, and plays an important role in coat colour (Schütz 2015). ASIP acts as an antagonist of MC1R by nullifying the action of a-melanocyte-stimulating hormone (a-MSH). Loss-of-function of MC1R will result in yellow pigment (pheomelanin), whereas gain-of-function of MC1R or loss-of-function of ASIP seems to result in the production of black pigment—eumelanin (Barsh 1996). In some animals, members of the defensins, a protein family previously implicated in immunity, have also been found to involved in melanocortin pathway by binding competitively to the MC1R (Candille et al. 2007; Zhang et al. 2019).
The tyrosinase, encoded by tyrosinase (TYR) gene, is known to be the rate-limiting enzyme affecting the production of melanin pigment. The functions includes oxidizing tyrosine to dihydroxyphenylalanine (DOPA) and determines which type of melanin (eumelanin or phaeomelanin) could be synthesized (Ito 2000). Researcher found that the C locus is the structural site for tyrosinase in fowl, and TYR gene was cloned from chickens for the first time on this basis (Mochii et al. 1992).
In previous studies, it has been confirmed that mutations of the MC1R, TYR and ASIP genes were associated with melanin trait variation or skin cancer and other diseases in a number of mammalian species, such as human, mouse, cattle, horse, fox, pig, sheep, and dog (Oetting 2010; Robbins et al. 1993; Klungland et al. 1995; Schmutz et al. 2004; Yokoyama et al. 1990; Lu et al. 1994; Våge et al. 1999; Zdarsky et al. 1990). In birds, studies on molecular mechanism of melanin deposition are relatively rare at present. The aim of the current study is to detect SNPs in MC1R, TYR and ASIP genes and explore their phenotypes association with plumage color in chicken.
Materials and methods
Samples
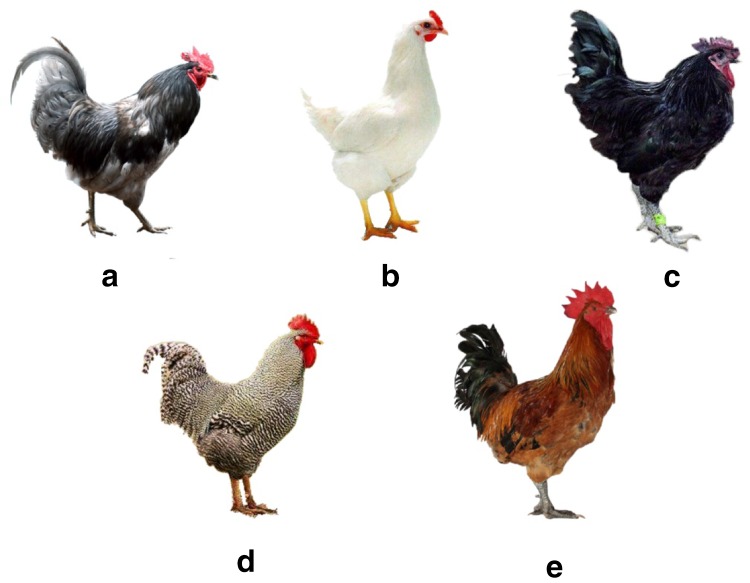
Blood samples were collected from 250 chickens with five plumage color phenotypes, including 50 Jiuyuan chickens (pure black feather), 50 Guanyuan gray chickens (completely gray plumage), 50 Lohmann Laying Hens (pure white plumage), 50 barred Plymouth Rock chicken (horizontal spot feather), and 50 HS1 chickens (black-speckled feather) (Fig. 1). The chickens came from the poultry breeding farm of Sichuan Agricultural University. The protocol was approved by the Committee on the Care and Use of Laboratory Animals of the State-Level Animal Experimental Teaching Demonstration Center of Sichuan Agricultural University. All samples were refrigerated in drikold before stored at − 20 °C.
Fig. 1.
Completely gray plumage (a), pure white plumage (b), pure black feather (c), horizontal spot feather (d) and black-speckled feather (e)
DNA isolation and PCR amplification
Total genomic DNA was extracted from chicken blood using the TIAN amp Genomic DNA Kit in accordance with the manufacturer’s instructions. DNA was eluted in a final volume of 200 μl using AE buffer and then stored at − 20 °C. The primers were designed by Oligo 6.0 and Primer 5.0 software based on the conserved sequence of MC1R gene (Gene ID: 427562), exon 1 of TYR gene (Gene ID: 373971), and exon 3 of ASIP gene (Gene ID: 419147) in Gallus gallus (Table 1).
Table 1.
Primers of PCR in chicken MC1R, TYR and ASIP genes
| Gene | Sequence (5′–3′) | Annealing temperature ( °C) | Fragment (bp) |
|---|---|---|---|
| MC1R | F:GTAGGTGCTGCAGTTGTGCT | 59.4 | 1187 |
| R:CTTTATTTGGGAGCGCGAGG | 59.4 | ||
| TYR | F:TCACATGGATTGGGCTGTGG | 57.5 | 667 |
| R:ACTCTGAGCCTTCCAGTGTTA | 57.5 | ||
| ASIP | F:GAATTGTGGTTTGCCGCATTG | 59 | 451 |
| R:AACTGCTGGATGTGACAGAAT | 59 |
PCR (25 μl) consists of 12.5 μl 2 × Taq Master Mix, 1 μl forward primer (10μM), 1μl reverse primer (10 μM), 2 μl DNA and 8.5 μl ddH2O. PCR cycles included 95 °C for 5 min; 35 cycles of 95 °C for 30 s, annealing temperature for 30 s, and 72 °C for 90 s; and a final extension at 12 °C for 10 min ending with incubation at 4 °C. PCR products were checked on 1% agarose gel and sequenced by direct sequencing in TSINGKE Biological Technology Corporation (Beijing, China).
Sequence analysis
The general linear model (GLM) procedure of SAS 6.12 (Statistical Analysis Systems Institute Inc. Cary, NC, USA) was built to test associations between the genotype and feather color; significant associations were declared when p < 0.05. The mixed model is as follows:
where Y is the dependent variable, μ is the population mean, G is the genotype value, S is the fixed effects of sex, B is the fixed effects of breed, F is the family effect, and eijkf is the random error.
The identified SNPs in these genes were tested for Hardy–Weinberg equilibrium, when p > 0.05 indicated the genetic balance of population gene (Wigginton et al. 2005). The linkage disequilibria D’ and r2 value of the SNPs were estimated by Haploview (Barrett et al. 2005). Significance of the least squares means was tested with the Duncan’s Multiple Range test. The polymorphism information content (PIC) was established (PIC > 0.5 is high polymorphism, 0.25 < PIC < 0.5 is the intermediate polymorphism, and PIC < 0.25 is low polymorphism) (Elston 2005).
Haplotypes were constructed based on each SNP of these genes in all experimental animals using the PHASE program v. 2.0. The function of this program is to reconstruct haplotypes from the population data. The genetic status of the subjects was expressed as the combination of two haplotypes. The SAS 6.12 (Statistical Analysis Systems Institute Inc., Cary, NC, USA) was used to analyze the associations between the haplotypes and feather color. Significant associations were declared when p < 0.05.
Results
Sequence polymorphism in the MC1R, TYR and ASIP genes
In this study, we obtained 945 bp fragments of MC1R gene, 421 bp fragments of exon 1 of TYR gene and 266 bp fragments of exon 3 of ASIP gene. Three SNPs (T398A, T637C, and G920C) were detected from MC1R gene; all of them were leading to amino acid substitution (Leu133Gln, Cys213Arg and Arg307Pro). Three SNPs (C47G, T120C, and T172C) were detected from TYR gene: two of them were leading to amino acid substitution (Pro16Arg and Phe58Leu), and only one synonymous SNP (T168C) was detected from ASIP gene.
Allele and genotype frequency of the mutated loci
The results of the allele and genotype frequency of the 7 SNPs in population are shown in Tables 2, 3 and 4. The Chi-squared test was used to compare the allele frequencies in the MC1R, TYR and ASIP genes between the different plumage color groups. The genotype distribution of all mutations in five chicken populations of different plumage color reached significant level (p < 0.05). In the different plumage color groups of chickens, SNP1 (T398A) and SNP2 (T637C) showed a homozygous genotype TT for advantage genotype, while SNP3 (G920C) showed GG genotype for advantage. We analyzed the relationship between different plumage color pattern and the genotypes of SNP1 and SNP2; the results showed that TT genotype was the advantageous genotype in black, gray, black-speckle and horizontal spot chicken in both sites, while TA and TC genotypes was predominated in white feather chicken in SNP1 and SNP2, respectively. For SNP3 loci, GG genotype was the advantage genotype in all groups.
Table 2.
The genotype distribution and allele frequencies of MC1R gene SNPs locus
| SNPs | Genotype | Phenotype class | Total | Frequency/% | χ2 value | ||||
|---|---|---|---|---|---|---|---|---|---|
| Black | White | Gray | Black-speckle | Horizontal spot | |||||
| SNP1 (T398A) | TT | 50 (0.2) | 15 (0.06) | 47 (0.188) | 50 (0.2) | 24 (0.096) | 186 | 74.4 |
χ2 = 6.56 p < 0.05 |
| TA | 0 (0) | 26 (0.104) | 3 (0.012) | 0 (0) | 19 (0.076) | 48 | 19.2 | ||
| AA | 0 (0) | 9 (0.036) | 0 (0.00) | 0 (0) | 7 (0.028) | 16 | 6.4 | ||
| Total | 50 | 50 | 50 | 50 | 50 | 250 | 100 | ||
| T | 100 | 56 | 97 | 100 | 67 | 420 | 84.0 | ||
| A | 0 | 44 | 3 | 0 | 33 | 80 | 16.0 | ||
| SNP2 (T637C) | TT | 46 (0.184) | 8 (0.032) | 32 (0.128) | 43 (0.172) | 22 (0.088) | 151 | 60.4 |
χ2 = 7.25 p < 0.05 |
| TC | 4 (0.016) | 29 (0.116) | 14 (0.056) | 6 (0.024) | 18 (0.072) | 71 | 28.4 | ||
| CC | 0 (0.00) | 13 (0.052) | 4 (0.016) | 1 (0.004) | 10 (0.04) | 28 | 11.2 | ||
| Total | 50 | 50 | 50 | 50 | 50 | 250 | 100 | ||
| T | 96 | 45 | 78 | 92 | 62 | 373 | 74.6 | ||
| C | 4 | 55 | 22 | 8 | 38 | 127 | 25.4 | ||
| SNP3 (G920C) | GG | 50 (0.2) | 31 (0.124) | 39 (0.156) | 41 (0.164) | 41 (0.164) | 202 | 80.8 |
χ2 = 6.03 p < 0.05 |
| GC | 0 (0) | 12 (0.048) | 8 (0.032) | 9 (0.036) | 9 (0.036) | 38 | 15.2 | ||
| CC | 0 (0) | 7 (0.028) | 3 (0.012) | 0 (0) | 0 (0.08) | 10 | 4 | ||
| Total | 50 | 50 | 50 | 50 | 50 | 250 | 100 | ||
| G | 100 | 74 | 86 | 91 | 91 | 442 | 88.4 | ||
| C | 0 | 26 | 14 | 9 | 9 | 58 | 11.6 | ||
Table 3.
The genotype distribution and allele frequencies of TYR gene SNPs locus
| SNPs | Genotype | Phenotype class | Total | Frequency/% | χ2 value | ||||
|---|---|---|---|---|---|---|---|---|---|
| Black | White | Gray | Black-speckle | Horizontal spot | |||||
| SNP4 (C47G) | CC | 44 (0.176) | 45 (0.18) | 41 (0.164) | 39 (0.156) | 50 (0.2) | 210 | 84.0 |
χ2 = 4.15 p < 0.05 |
| CG | 5 (0.02) | 5 (0.02) | 6 (0.024) | 9 (0.036) | 0 (0) | 25 | 10.0 | ||
| GG | 1 (0.004) | 9 (0.036) | 3 (0.012) | 2 (0.008) | 0 (0) | 15 | 6.00 | ||
| Total | 50 | 50 | 50 | 50 | 50 | 250 | 100 | ||
| C | 93 | 77 | 88 | 87 | 100 | 445 | 89.0 | ||
| G | 7 | 23 | 12 | 13 | 0 | 55 | 11.0 | ||
| SNP5 (T120C) | TT | 50 (0.2) | 45 (0.18) | 45 (0.18) | 34 (0.136) | 48 (0.192) | 222 | 88.8 |
χ2 = 6.88 p < 0.05 |
| TC | 0 (0) | 5 (0.02) | 5 (0.02) | 11 (0.044) | 2 (0.008) | 23 | 9.2 | ||
| CC | 0 (0) | 0 (0) | 0 (0) | 5 (0.02) | 0 (0) | 5 | 2.0 | ||
| Total | 50 | 50 | 50 | 50 | 50 | 250 | 100 | ||
| T | 100 | 95 | 95 | 79 | 98 | 467 | 93.4 | ||
| C | 0 | 5 | 5 | 21 | 2 | 33 | 6.6 | ||
| SNP6 (T172C) | TT | 35 (0.14) | 45 (0.18) | 35 (0.14) | 30 (0.12) | 0 (0) | 145 | 58.0 |
χ2 = 6.71 p < 0.05 |
| TC | 11 (0.044) | 5 (0.02) | 8 (0.032) | 15 (0.06) | 27 (0) | 66 | 26.4 | ||
| CC | 4 (0.016) | 0 (0) | 7 (0.028) | 5 (0.02) | 23 (0) | 39 | 15.6 | ||
| Total | 50 | 50 | 50 | 50 | 50 | 250 | 100 | ||
| T | 81 | 95 | 78 | 75 | 27 | 356 | 71.2 | ||
| C | 19 | 5 | 22 | 25 | 73 | 144 | 28.8 | ||
Table 4.
The genotype distribution and allele frequencies of ASIP gene SNP locus
| SNPs | Genotype | Phenotype class | Total | Frequency/% | χ2 value | ||||
|---|---|---|---|---|---|---|---|---|---|
| Black | White | Gray | Black-speckle | Horizontal spot | |||||
| SNP7 (T168C) | TT | 44 (0.176) | 50 (0.2) | 43 (0.172) | 44 (0.176) | 46 (0.184) | 227 | 90.8 |
χ2 = 5.96 p < 0.05 |
| TC | 6 (0.024) | 0 (0) | 7 (0.028) | 6 (0.024) | 4 (0.016) | 23 | 9.2 | ||
| CC | 0 (0.00) | 0 (0) | 0 (0.00) | 0 (0.00) | 0 (0.00) | 0 | 0.00 | ||
| Total | 50 | 50 | 50 | 50 | 50 | 250 | 100 | ||
| T | 94 | 100 | 93 | 94 | 98 | 477 | 95.4 | ||
| C | 6 | 0 | 7 | 6 | 4 | 23 | 4.6 | ||
In addition, SNP4 (C47G) showed a homozygous genotype CC for advantage genotype in all groups, while SNP5 (T120C) and SNP6 (T172C) showed TT genotype for advantage. However, for SNP6 loci, there was no TT genotype in horizontal spot chicken, and TC genotype was predominated. Homozygous genotype TT was the advantage genotype of SNP7 (T168C) in all groups, while there was only advantage genotype in white chicken.
Haplotype analysis
Six haplotypes (H1–H6) were obtained based on the three SNPs (T398A, T637C, and G920C) of the MC1R gene (Table 5); six haplotypes (P1–P6) of the TYR gene are shown in Table 6 and the corresponding diplotypes are displayed in Tables 7 and 8, separately. The results showed that haplotypes H1 and H3 were found in all kinds of feather colors with a high frequency, and speculated haplotype H1 was the advantage haplotype. The H5 and H6 haplotypes were mainly concentrated in white and gray chicken, but not found in black chicken. We guessed that two haplotypes may partially inhibit the production of melanin. Haplotype P1 was found in all kinds of feather colors with a high frequency, and speculated that it was the advantage haplotype. Haplotypes P2, P3 and P6 were not occurred in black chicken.
Table 5.
Distribution of the MC1R gene haplotypes
| Haplotype | Number | Frequency | Phenotype class | ||||
|---|---|---|---|---|---|---|---|
| Black | White | Gray | Black-speckle | Horizontal spot | |||
| H1 (TTG) | 156 | 0.624 | 26 | 32 | 28 | 31 | 39 |
| H2 (TCC) | 21 | 0.084 | 0 | 6 | 5 | 3 | 7 |
| H3 (TCG) | 35 | 0.140 | 6 | 10 | 9 | 5 | 5 |
| H4 (ATG) | 17 | 0.068 | 0 | 7 | 3 | 2 | 5 |
| H5 (ATC) | 12 | 0.048 | 0 | 6 | 2 | 0 | 4 |
| H6 (ACC) | 9 | 0.036 | 0 | 5 | 2 | 0 | 2 |
| Total | 250 | 1.0000 | 32 | 66 | 49 | 41 | 62 |
Table 6.
Distribution of the TYR gene haplotypes
| Haplotype | Number | Frequency | Phenotype class | ||||
|---|---|---|---|---|---|---|---|
| Black | White | Gray | Black-speckle | Horizontal spot | |||
| P1 (CTT) | 127 | 0.508 | 26 | 28 | 16 | 23 | 34 |
| P2 (CCT) | 34 | 0.136 | 0 | 6 | 9 | 8 | 11 |
| P3 (CCC) | 26 | 0.104 | 0 | 4 | 8 | 6 | 8 |
| P4 (GTT) | 30 | 0.120 | 7 | 9 | 8 | 6 | 0 |
| P5 (GTC) | 21 | 0.084 | 6 | 8 | 1 | 6 | 0 |
| P6 (GCC) | 12 | 0.048 | 0 | 3 | 4 | 5 | 0 |
| Total | 250 | 1.0000 | 39 | 58 | 46 | 54 | 53 |
Table 7.
Distribution of the MC1R gene diplotypes
| Diplotype | Number | Frequency | Phenotype class | ||||
|---|---|---|---|---|---|---|---|
| Black | White | Gray | Black-speckle | Horizontal spot | |||
| H1H1 | 148 | 0.592 | 29 | 31 | 22 | 30 | 36 |
| H1H2 | 31 | 0.124 | 0 | 7 | 12 | 9 | 3 |
| H1H5 | 26 | 0.104 | 0 | 10 | 8 | 4 | 4 |
| H2H3 | 19 | 0.076 | 0 | 5 | 7 | 5 | 2 |
| H3H4 | 16 | 0.064 | 0 | 4 | 3 | 2 | 7 |
| H3H6 | 10 | 0.040 | 0 | 4 | 3 | 0 | 3 |
| Total | 250 | 1.0000 | 29 | 61 | 55 | 50 | 55 |
Table 8.
Distribution of the TYR gene diplotypes
| Diplotype | Number | Frequency | Phenotype class | ||||
|---|---|---|---|---|---|---|---|
| Black | White | Gray | Black-speckle | Horizontal spot | |||
| P1P1 | 121 | 0.484 | 27 | 21 | 26 | 31 | 16 |
| P1P2 | 41 | 0.164 | 0 | 5 | 14 | 7 | 15 |
| P1P6 | 16 | 0.064 | 0 | 7 | 3 | 6 | 0 |
| P2P3 | 29 | 0.116 | 0 | 8 | 11 | 4 | 6 |
| P4P5 | 26 | 0.104 | 10 | 8 | 3 | 5 | 0 |
| P4P6 | 17 | 0.068 | 0 | 6 | 8 | 3 | 0 |
| Total | 250 | 1.0000 | 37 | 55 | 65 | 56 | 37 |
Furthermore, the H1H1 combination was distributed in all groups with the largest frequency. Black chicken only had a haplotype combination of H1H1. Particularly, diplotypes H2H3 was occurred in white feather and black-speckled feather with the same frequency. Diplotype P1P1 was distributed in all groups with the largest proportion, and diplotype P1P6 with the smallest proportion. The diplotypes P1P6 and P4P6 were only distributed in white, gray and black-speckled feather.
Discussion
The plumage color of poultry has been widely used as a morphological marker for genetic selection and also considered as an important economic trait that caters to consumer preference. A functional melanocortin-1-receptor is fundamental for eumelanin expression. Gain-of-function mutations MC1R or loss-of-function mutations in the MC1R antagonist ASIP are believed to enhance this process, resulting in black coat color phenotypes. In our previous study, we found that there was significant association between the MC1R genetic variation with plumage colors of geese and pigeon (Huang et al. 2014; Ran et al. 2016).
In the past fewer years, lots of MC1R, TYR and ASIP genes mutation and the feather color difference relations research could form the perspective of the molecular to expound melanin synthesis regulative process, and for birds and mammals the molecular mechanism on the formation of melanin provides a solid theoretical basis. In this study, we identified five missense and two synonymous mutations in 250 samples of three genes. The SNP1, SNP2, SNP3, SNP4 and SNP6 lead to a change in amino acids. In genetics, a missense mutation, a type of nonsynonymous substitution, could result in the changes of protein. When the amino acid loci 156 of MC1R protein mutated into proline, chicken plumage color occurs as albino (grey, white, black-speckle and horizontal spot), which is similar to Guernsey’s research (Guernsey et al. 2013). In addition, although the two SNPs did not cause amino acid change, association analysis showed that they were significantly associated with chicken plumage colors (p < 0.05). Previous studies have shown that different synonymous degenerate codon on these loci would affect protein translation efficiency and structural conformation, which lead to phenotypic changes finally (Shastry 2009; Kurland 1991; Kimchi-Sarfaty et al. 2007). Anthony’s study hypothesized that when frequent codons are changed to rare codons in a cluster of infrequently used codons, the timing of cotranslational folding is affected and may result in altered function (Anthony and Skach 2002). For the sample size which is relatively limited, further more studies should be carried forward on the relationship between loci and plumage color traits in chicken.
For the purpose of investigating the possible function of the mutations, we analyzed the association between MC1R and TYR genotypes with plumage color trait. Haplotype analysis provided a practical solution to resolve these problems. Haplotypes were constructed with the three SNPs and were used to analyze the association of haplotypes with plumage color traits. We found that haplotype H1 was distributed in all kinds of feather colors with a high frequency but the H5 and H6 haplotypes were mainly concentrated in white and gray chicken. We also found that haplotype P2, P3 and P6 were not occurred in black chicken. And we could obtain that haplotypes and diplotypes were significantly associated with plumage colors. These results demonstrated that haplotypes and diplotypes of the MC1R and TYR genes bear the characteristic of regional distribution and were associated with plumage color in chicken, although the plumage color control is a very complex trait.
Conclusion
On the basis of our results in this study, we speculated that there were significant associations between plumage colors and genetic variants of the MC1R, TYR and ASIP genes in chicken. However, as a complex trait, plumage color is determined by a complex pathway system and multiple interactive patterns; further studies would be helpful to confirm this conclusion.
Acknowledgements
Research was supported by the China Agriculture Research System (Grant No. CARS-41-G04); the National Science and Technology support planning project (Grant No. 2015BAD03B03); the Province Key Technologies R & D Program of Livestock and Poultry Breeding Programs of Sichuan Province (Grant Nos. 2016NYZ0025 & 2016NYZ0043 & 2016NZ0104).
Compliance with ethical standards
Conflict of interest
All authors have no declared conflict of interest.
Ethical approval
All procedures carried out in this experiment were reviewed and approved by the Institutional Animal Care and Use Committee of Sichuan Agricultural University, China.
Contributor Information
Yi-Ping Liu, Email: liuyp578@yahoo.com.
Xiao-song Jiang, Email: xsjiang@sasa.cn.
References
- Anthony V, Skach W. Molecular mechanism of Pglycoprotein assembly into cellular membranes. Curr Protein Pept Sci. 2002;3(5):485–501. doi: 10.2174/1389203023380503. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Barsh GS. The genetics of pigmentation: from fancy genes to complex traits. Trends Genet. 1996;12(2):299–305. doi: 10.1016/0168-9525(96)10031-7. [DOI] [PubMed] [Google Scholar]
- Candille I, Kaelin CB, Cattanach BM, Yu B, Thompson DA, Nix MA, Kerns JA, Schmutz SM, Millhauser GL, Barsh GS. A β-defensin mutation causes black coat color in domestic dogs. Science. 2007;318(5855):1418–1423. doi: 10.1126/science.1147880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elston RC (2005) Polymorphism information content. In: Armitage P, Colton T (eds) Encyclopedia of biostatistics. Wiley
- Giuseppe P. Recent advances in the chemistry of melanogenesis in mammals. J Investig Dermatol. 1980;75(1):122–127. doi: 10.1111/1523-1747.ep12521344. [DOI] [PubMed] [Google Scholar]
- Guernsey MW, Ritscher L, Miller MA, et al. A Val85Met mutation in melanocortin-1 receptor is associated with reductions in eumelanic pigmentation and cell surface expression in domestic rock pigeons (Columba livia) PLoS One. 2013;8(8):e74475. doi: 10.1371/journal.pone.0074475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Zhou B, He DQ, et al. Sequence variation of melanocortin 1 receptor (MC1R) gene and association with plumage color in domestic geese. Jpn Poult Sci Assoc. 2014;35(1):270–274. doi: 10.2141/jpsa.0130066. [DOI] [Google Scholar]
- Ito S. Chemical analysis of melanins and its application to the study of the regulation of melanogenesis. Pigment Cell Res. 2000;13(s8):103–109. doi: 10.1034/j.1600-0749.13.s8.19.x. [DOI] [PubMed] [Google Scholar]
- Kimchi-Sarfaty C, Oh JM, Kim IW, et al. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315(5811):525–528. doi: 10.1126/science.1135308. [DOI] [PubMed] [Google Scholar]
- Klungland H, Våge DI, Gomez-Raya L, et al. The role of melanocyte-stimulating hormone (MSH) receptor in bovine coat color determination. Mamm Genome. 1995;6(9):636. doi: 10.1007/BF00352371. [DOI] [PubMed] [Google Scholar]
- Kurland CG. Codon bias and gene expression. FEBS Lett. 1991;285(2):165–169. doi: 10.1016/0014-5793(91)80797-7. [DOI] [PubMed] [Google Scholar]
- Lu D, Willard D, Patel IR, et al. Agouti protein is an antagonist of the melanocyte-stimulating-hormone receptor. Nature. 1994;371(6500):799–802. doi: 10.1038/371799a0. [DOI] [PubMed] [Google Scholar]
- Mochii A, Iio H, Yamamoto H, Takeuchi T, Eguchi G. Isolation and characterization of a chicken tyrosinase cDNA. Pigment Cell Res. 1992;5(4):162–167. doi: 10.1111/j.1600-0749.1992.tb00454.x. [DOI] [PubMed] [Google Scholar]
- Mountjoy KG, Robbins LS, Mortrud MT, et al. The cloning of a family of genes that encode the melanocortin receptors. Science. 1992;257(5074):1248–1251. doi: 10.1126/science.1325670. [DOI] [PubMed] [Google Scholar]
- Mundy NI. A window on the genetics of evolution: MC1R and plumage colouration in birds. Proc R Soc B Biol Sci. 2005;272(1573):1633–1640. doi: 10.1098/rspb.2005.3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oetting WS. The tyrosinase gene and oculocutaneous albinism type 1 (OCA1): a model for understanding the molecular biology of melanin formation. Pigment Cell Res. 2010;13(5):320–325. doi: 10.1034/j.1600-0749.2000.130503.x. [DOI] [PubMed] [Google Scholar]
- Pang Y. The design and analysis for genetics experiment concerning sex-linked gene plumage color in quail. Lab Anim Sci Adm. 2001;4(1):15–18. [Google Scholar]
- Ran JS, You XY, Jin J, et al. The relationship between MC1R mutation and plumage color variation in pigeons. Biomed Res Int. 2016;3:1–6. doi: 10.1155/2016/3059756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins LS, Nadeau JH, Johnson KR, et al. Pigmentation phenotypes of variant extension locus alleles result from point mutations that alter MSH receptor function. Cell. 1993;72(6):827–834. doi: 10.1016/0092-8674(93)90572-8. [DOI] [PubMed] [Google Scholar]
- Scherer D, Kumar R. Genetics of pigmentation in skin cancer—a review. Mutat Res. 2010;705(2):141–153. doi: 10.1016/j.mrrev.2010.06.002. [DOI] [PubMed] [Google Scholar]
- Schiöth HB, Raudsepp T, Ringholm A, et al. Remarkable synteny conservation of melanocortin receptors in chicken, human, and other vertebrates. Genomics. 2003;81(5):504–509. doi: 10.1016/S0888-7543(03)00028-4. [DOI] [PubMed] [Google Scholar]
- Schmutz SM, Berryere TG, Ciobanu DC, et al. A form of albinism in cattle is caused by a tyrosinase frameshift mutation. Mamm Genome. 2004;15(1):62–67. doi: 10.1007/s00335-002-2249-5. [DOI] [PubMed] [Google Scholar]
- Schütz K. Melanocortin 1-receptor (MC1R) mutations are associated with plumage colour in chicken. Anim Genet. 2015;34(4):241–248. doi: 10.1046/j.1365-2052.2003.00991.x. [DOI] [PubMed] [Google Scholar]
- Shastry BS. SNPs: impact on gene function and phenotype. Methods Mol Biol. 2009;578:3–22. doi: 10.1007/978-1-60327-411-1_1. [DOI] [PubMed] [Google Scholar]
- Våge DI, Klungland H, Lu D, et al. Molecular and pharmacological characterization of dominant black coat color in sheep. Mamm Genome. 1999;10(1):39–43. doi: 10.1007/s003359900939. [DOI] [PubMed] [Google Scholar]
- Wigginton JE, Cutler DJ, Abecasis GR. A note on exact tests of Hardy–Weinberg equilibrium. Am J Hum Genet. 2005;76(5):887–893. doi: 10.1086/429864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama T, Silversides DW, Waymire KG, et al. Conserved cysteine to serine mutation in tyrosinase is responsible for the classical albino mutation in laboratory mice. Nucleic Acids Res. 1990;18(24):7293–7298. doi: 10.1093/nar/18.24.7293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdarsky E, Favor J, Jackson IJ. The molecular basis of brown, an old mouse mutation, and of an induced revertant to wild type. Genetics. 1990;126(2):443–449. doi: 10.1093/genetics/126.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Chen D, Yu L, Wei Y, Li J, Zhou C. Genome-wide analysis of the ovodefensin gene family: monophyletic origin, independent gene duplication and presence of different selection patterns. Infect Genet Evol. 2019;68:265–272. doi: 10.1016/j.meegid.2019.01.001. [DOI] [PubMed] [Google Scholar]