Abstract
Leafy head formation in Chinese cabbage (B. rapa ssp. pekinensis cv. Bre) results from leaf curvature, which is under the tight control of genes involved in the adaxial-abaxial patterning during leaf development. The transcriptional coactivator ANGUSTIFOLIA3 (AN3) binds to the SWI/SNF chromatin remodeling complexes formed around ATPases such as BRAHMA (BRM) in order to regulate transcription in various aspects of leaf development such as cell proliferation, leaf primordia expansion, and leaf adaxial/abaxial patterning in Arabidopsis. However, its regulatory function in Chinese cabbage remains poorly understood. Here, we analyzed the expression patterns of the Chinese cabbage AN3 gene (BrAN3) before and after leafy head formation, and produced BrAN3 gene silencing plants by using the turnip yellow mosaic virus (TYMV)-derived vector in order to explore its potential function in leafy head formation in Chinese cabbage. We found that BrAN3 had distinct expression patterns in the leaves of Chinese cabbage at the rosette and heading stages. We also found silencing of BrAN3 stimulated leafy head formation at the early stage. Transcriptome analysis indicated that silencing of BrAN3 modulated the hormone signaling pathways of auxin, ethylene, GA, JA, ABA, BR, CK, and SA in Chinese cabbage. Our study offers unique insights into the function of BrAN3 in leafy head formation in Chinese cabbage.
Keywords: AN3, BRM, leafy head formation, VIGS, transcriptome analysis, Chinese cabbage
Introduction
Heading Chinese cabbage (Brassica rapa ssp. pekinensis cv. Bre; 2n = 2× = 20) is one of the most important horticultural crops in China and, to a lesser extent, an oilseed crop (Zhao et al., 2005). Leafy head formation undergoes four developmental stages, i.e., the seedling, rosette, folding, and heading stages (He et al., 2000; Yu et al., 2000; Ke, 2010; Wang et al., 2014b). It forms uniform leafy heads with extremely incurved leaves surrounding the shoot apexes after the rosette stage. The leafy heading trait has been selected for several Brassica species including Chinese cabbage and cabbage (B. oleracea; 2n = 2× = 18) during crop domestication and breeding (Cheng et al., 2014). Well-formed leafy heads function normally for photosynthesis and serve as storage organs for essential minimal nutrients, fibers, and vitamins, while poorly-developed heads significantly decrease the crop’s commercial values (Yu et al., 2000, 2013; Cheng et al., 2016). Leafy head formation is affected by many factors such as low temperature, low light intensity, uneven auxin distribution, and low carbohydrate/nitrogen ratio (Ito, 1957). Plants have to respond to these factors properly for leaf bending and folding (i.e., leaf curvature), which results from the differential cell division and enlargement in leaf regions, and which are under the tight control of genes involved in the adaxial-abaxial patterning during leaf development (Mao et al., 2014).
Recent efforts had been taken to study the molecular mechanisms controlling leafy head formation in Chinese cabbage by using genetic mapping (Zhao et al., 2005; Ge et al., 2011; Yu et al., 2013; Zhang et al., 2013; Zhang X. et al., 2016), transcriptome profiling (Wang et al., 2012; Zhang C.W. et al., 2016; Gu et al., 2017; Li et al., 2018), proteomic analysis (Zhang C.W. et al., 2016), and miRNA expression profiling (Wang et al., 2013). Moreover, the effects of a few individual genes on leafy head formation had been studied in Chinese cabbage. These genes include the Agrobacterium genes Aux1 and Aux2 (He et al., 2000), the Chinese cabbage genes BcpLH (Yu et al., 2000), TCP (Mao et al., 2014), BrpSPL9 (Wang et al., 2014b), and auxin transport genes BrLAX, BrPIN, and BrPGP (Gao et al., 2017). Most of these genes are involved in the adaxial-abaxial patterning during leaf development in Chinese cabbage.
The Arabidopsis ANGUSTIFOLIA3/GRF-INTERACTIN FACTOR1 (AtAN3/AtGIF1), an important transcriptional activator of the GIF family, is also involved in the determination of leaf adaxial-abaxial patterning and growth (Horiguchi et al., 2011). Loss-of-function mutant of AtAN3 exhibited smaller and narrower leaves due to a decrease in cell number (Kim and Kende, 2004; Horiguchi et al., 2005), while ectopic overexpression of AtAN3 resulted in enlarged leaf size (Horiguchi et al., 2005; Lee et al., 2009). AtAN3 binds to the SWI/SNF chromatin remodeling complex formed around ATPases such as BRAHMA (BRM). By using the energy from ATP hydrolysis, the Arabidopsis AN3-SWI/SNF-BRM complex regulates gene expression by changing the interactions between histone octamers and the DNA for the access of transcription factors (Clapier and Cairns, 2009). The brm mutant had small spiral-shaped leaves with downward curling edges (Hurtado et al., 2006; Vercruyssen et al., 2014). The AN3-SWI/SNF-BRM complex also interacts with GROWTH REGULATING FACTOR (GRF) proteins, a class of plant-specific transcription activators in Arabidopsis (Kim and Kende, 2004; Liu et al., 2009; Debernardi et al., 2014). Ectopic overexpression of the GRFs increased leaf size in Arabidopsis due to enhanced cell proliferation (Kim et al., 2003; Horiguchi et al., 2005; Liu et al., 2009; Debernardi et al., 2014; Wang et al., 2014a). However, the regulatory function of AN3 in Chinese cabbage remains poorly understood.
In the present study, we explored the expression patterns of the Chinese cabbage AN3 gene (i.e., BrAN3) in different leaf locations of Chinese cabbage at the rosette and heading stages. We generated BrAN3-silencing Chinese cabbage plants by using the turnip yellow mosaic virus (TYMV)-derived vector, and examined the effect of BrAN3 silencing on the stimulation of leafy head formation. We also conducted transcriptome analysis of the BrAN3-silencing leaves and identified the differentially expressed genes (DEGs) caused by BrAN3 silencing. All of the results provide insights into the function of the BrAN3 gene in leafy head formation in Chinese cabbage.
Materials and Methods
Sequence Alignment and Phylogenetic Analysis
The protein sequences of the Arabidopsis AtAN3 and AtBRM genes were used individually as the query sequences to BlastP against the Brassica database1 in order to obtain their homologous sequences in B. rapa. The protein sequences of the Arabidopsis AtAN3 and AtBRM genes were also used individually as the query sequences to BlastP against Phytozome2 in order to obtain their homologous sequences in B. oleracea, Zea mays, and Oryza sativa. The genomic DNA sequences and cDNA sequences of these homologous sequences in these plant species were also obtained from Phytozome. All of the homologous protein sequences of each gene in different species were aligned together using CluastalX 2.1 (Larkin et al., 2007), and subjected to the phylogentic analysis using the MEGA5 program3 and the neighbor-joining method with 1,000 bootstraps (Tamura et al., 2013).
Plant Material and Growth Conditions
Inbred line seeds of Chinese cabbage cv. Bre were germinated in soil in a tray with holes. Three weeks old seedlings were transferred to pots containing a mixture of nutrient soil and vermiculite (3:1) and grown in a growth chamber at 22°C with a 16/8 h light/dark photocycle and 50% relative humidity.
Gene Cloning and Plasmid Construction
Total RNA was extracted from the leaves of Chinese cabbage for gene cloning using an RNA extraction kit (TaKaRa; Dalian, Liaoning, China). DNase I treatment and first-strand cDNA synthesis were performed sequentially using the PrimeScript® 1st Strand cDNA Synthesis Kit (TaKaRa; Dalian, Liaoning, China). One microgram of total RNA was reverse-transcribed with the oligo(dT) primer according to the manufacturer’s protocol. The cDNA fragments of BrAN3 and BrBRM were PCR amplified individually using the first-strand cDNA as the templates and the gene-specific primers (Supplementary Table S1). The PCR products were purified using an AxyPrep DNA Gel Extraction Kit (Axygen Biosciences; Union City, CA, United States) and then cloned into the pMD19-T vector (TaKaRa; Dalian, Liaoning, China), followed by Sanger sequencing. A gene-specific fragment of 40 nt in length was selected for each of the two genes to create an in-frame stop codon (TAA, TGA, or TAG) on the second, third or fourth amino acid position on each fragment due to frame shift. The gene-specific fragment was selected to target all the homologous sequences of each gene in the Chinese cabbage genome. Each fragment and its reverse complementary sequence (Supplementary Table S1) were synthesized from TaKaRa (Dalian, Liaoning, China) and used to form a palindromic oligonucleotide dimer after self-hybridization, which was cloned into the plasmid pTY-S with the help of SnaBI (Pflieger et al., 2008). The resulting plasmids were named as pTY-BrAN3 and pTY-BrBRM, respectively.
Real-Time RT-PCR
Total RNA was extracted from the apical location, the lateral 1∼3 locations, and the base location of individual leaves and the whole leaves of Chinese cabbage (Gao et al., 2017) at both the rosette and heading stages, respectively, for the analysis of expression profiles of BrAN3 on different locations of Chinese cabbage leaves. As for the analysis of the effects of virus-induced gene silencing, total RNA was extracted individually from the Chinese cabbage leaves inoculated with each virus-induced gene silencing vector, the negative control vector, and positive control vector. The purity and integrity of the RNA were analyzed using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies; Wilmington, DE, United States) and verified by gel electrophoresis. The real-time RT-PCR was conducted using the SYBR Green PrimeScript Plus RT-PCR Kit (TaKaRa; Dalian, Liaoning, China) on an ABI PrismR 7900HT (Applied Biosystems; Carlsbad, CA, United States) according to the manufacturer’s instructions. Gene-specific primers were designed outside of the 40 nt long fragments for both BrAN3 and BrBRM genes as well as the BrAN3 downstream genes ARABIDOPSIS RESPONSE REGULATOR4 (ARR4) and CYTOKININ RESPONSE FACTOR2 (CRF2) (Vercruyssen et al., 2014) using the Beacon Designer 7.9 (Premier Biosoft International, Palo Alto, CA, United States) (Supplementary Table S1). These primers were designed to target all the homologous sequences of each gene in the Chinese cabbage genome. The amplification procedure was as follows: pre-denaturation at 94°C for 10 s, followed by 40 cycles of 94°C for 30 s and 60°C for 30 s, and finally a melting curve was performed (95°C for 15 s and at increments of 0.5°C from 60 to 95°C). Three biological and three technical replicates were carried out for each sample. The 2-ΔΔCT method (Schmittgen and Livak, 2008) was used for relative transcript quantification normalized by the internal control gene BrAct (Bra028615) as described (Dheda et al., 2004). The quality cutoff of the real-time RT-PCR efficiency was set at >95% with R2 > 0.99 for the standard curve.
Particle Bombardment-Mediated Delivery of the TYMV-Derived Vectors
The plasmids pTY-BrAN3 and pTY-BrBRM, the empty plasmid pTY-S and the positive control plasmid pTY-BrPDS (Yu et al., 2018) were transformed into the 6 weeks old Chinese cabbage seedlings for virus-induced gene silencing (VIGS) by particle bombardment as described in Yu et al. (2018). Leaves were harvested from three individual plants before and after head formation and kept in liquid nitrogen for RNA extraction (see above).
Library Preparation for Transcriptome Sequencing
Total RNA was extracted individually from the leaves of two Chinese cabbage plants inoculated with each virus-induced gene silencing vector. The purity and integrity of the RNA was assessed using the RNA Nano 6000 Assay Kit of the Agilent Bioanalyzer 2100 system (Agilent Technologies; Palo Alto, CA, United States) and the NanoPhotometer® spectrophotometer (IMPLEN; Westlake Village, CA, United States), while the concentration was measured using the Qubit® RNA Assay Kit in a Qubit® 2.0 Flurometer (Life Technologies; Carlsbad, CA, United States). Three μg RNA per sample was used for RNA sample preparations. Sequencing libraries were generated using the NEB Next® UltraTM RNA Library Prep Kit for Illumina® (NEB; Beverley, MA, United States) following the manufacturer’s recommendations and index codes were added to attribute sequences to each sample. The library quality was assessed on the Agilent Bioanalyzer 2100 system (Agilent Technologies; Palo Alto, CA, United States).
Clustering and Reads Alignment to the Reference Genome
The clustering of the index-coded samples was performed on a cBot Cluster Generation System using TruSeq PE Cluster Kit v3-cBot-HS (Illumina; Hayward, CA, United States) according to the manufacturer’s instructions. Then, the libraries were sequenced on an Illumina Hiseq 2500 platform and 150 bp paired-end reads were generated. Reference genome and gene model annotation files were downloaded from Brassica database4. Index of the reference genome4 was built and paired-end clean reads were aligned to the reference genome using HISAT2 (Kim et al., 2015).
The expression level was calculated as fragments per kilobase of exon model per million mapped (FPKM) values and gene expression pattern using Clusterv3.05 and the heat map of hierarchical clustering was established using TreeView v.3.06 based on the log2-converted FPKM values. Bubble Charts were performed using the OmicShare tools, a free online platform for data analysis7.
GO Enrichment and KEGG Pathway Analysis of Differentially Expressed Genes
Differentially expressed genes (DEGs) were defined if the False Discovery Rate is smaller than 0.01 and the Fold Change equals to or is larger than 2. Gene Ontology (GO) enrichment analysis of the DEGs was implemented by the GOseq R packages based on the Wallenius non-central hyper-geometric distribution (Young et al., 2010), which allows the adjustment for gene length bias in DEGs. KOBAS software was used to test the statistical enrichment of DEGs in KEGG pathways (Mao et al., 2005).
Statistical Analysis
Statistical analyses was performed using analysis of variance (ANOVA; SAS 9.4 for Windows; SAS Institute, Cary, NC; p<0.05).
Results
Identification of the BrAN3 and BrBRM Genes in Chinese Cabbage
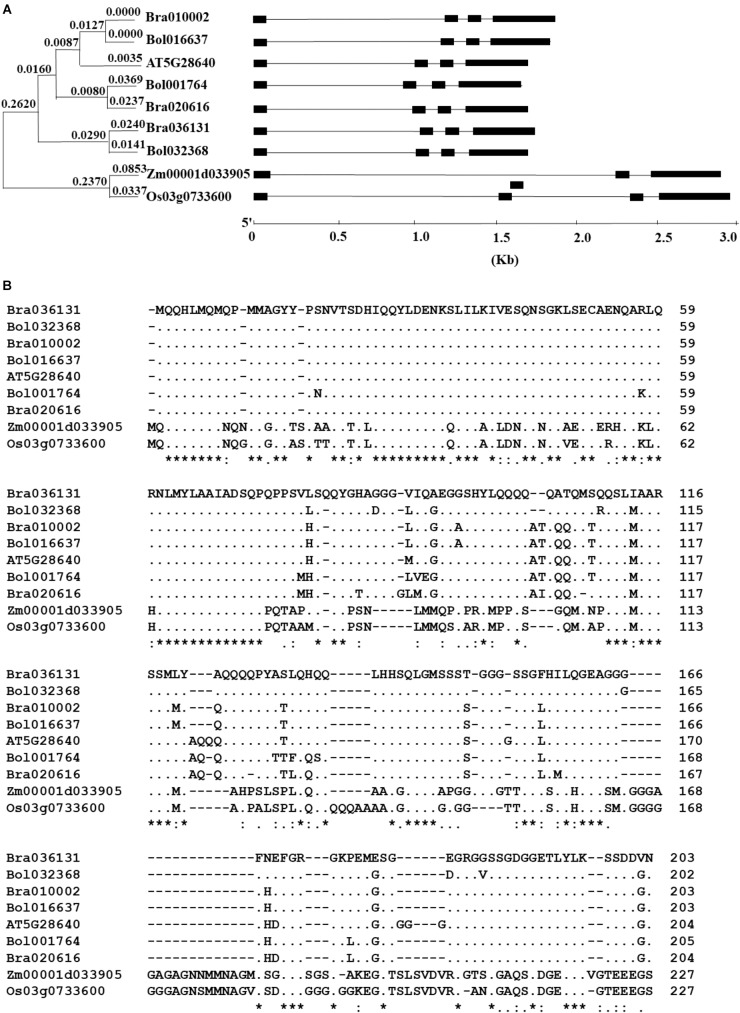
The BlastP search for the homologous sequences of the AtAN3 gene in the Brassica database and Phytozome returned three homologous genes in both B. rapa and B. oleracea, and one homologous gene in both Z. mays and O. sativa (Figure 1). Phylogenetic analysis revealed that the homologous genes in B. rapa and B. oleracea formed three monophyletic groups, indicating an origin of gene duplication (Figure 1A). The solo Arabidopsis homolog grouped with one of the three groups. The comparison between the cDNA sequences with the genomic DNA sequences revealed that all of these homologous genes have similar exon/intron structures (Figure 1B). Each homologous gene consists of four exons and three introns at relatively conserved positions. When using the protein sequence of the AtBRM gene as the query, we found two homologs in both B. rapa and B. oleracea, and one homolog in both Z. mays and O. sativa (Supplementary Figure S1). The homologous genes in B. rapa and B. oleracea formed two monophyletic groups, indicating a single gene duplication event (Supplementary Figure S1). All of these homologous genes have similar exon/intron structures (Supplementary Figure S1). The existence of the same numbers of homologous genes of both AtAN3 and AtBRM in the two Brassica genomes is consistent to the finding that parallel selection of homologous genes for leaf heading in the two species (Cheng et al., 2016).
FIGURE 1.
Comparison of the homologs of the AtAN3 protein in Brassica rapa, B. oleracea, Zea mays, and Oryza sativa. (A) Phylogenetic relationship and gene structures of the homologs of the AtAN3 protein in those species. The unrooted phylogenetic tree was generated by the neighbor-joining method using the MEGA5 program with 1,000 bootstraps. Lines represent the introns while solid boxes represent exons. (B) Comparison of the deduced amino acid sequences of the homologs of the AtAN3 protein in those species. Dashes represent gaps, dots represent the identical amino acid residues, and the numbers represent the length of the amino acid residues.
Distinct Expression Patterns of BrAN3 in the Leaves of Chinese Cabbage at the Rosette and Heading Stages
To explore the potential roles of the BrAN3 gene in head formation in Chinese cabbage, we conducted real-time RT-PCR analysis of the relative transcription levels of BrAN3 in the Chinese cabbage leaves at both the rosette and heading stages. We found that BrAN3 had a constantly stable relative expression levels in different locations of the leaves as well as the whole leaf at the rosette stage except in the leaf apical tip where BrAN3 expression levels doubled (Figure 2). However, BrAN3 exhibited highly variable but significantly decreased relative expression levels in different locations of the leaves of Chinese cabbage at the heading stage (Figure 2). Their relative expression peaked in the leaf apical tip, followed by the leaf base, and the lateral 2 and 3 locations. The least relative expression was observed in the lateral 1 location. The significant variations on the relative expression in different locations of the Chinese cabbage leaf at the heading stage indicate a potential role in heading formation since the lateral parts affect the leaf curvature and leaf head formation than the mid-vein part (i.e., the lateral 1 location) does.
FIGURE 2.
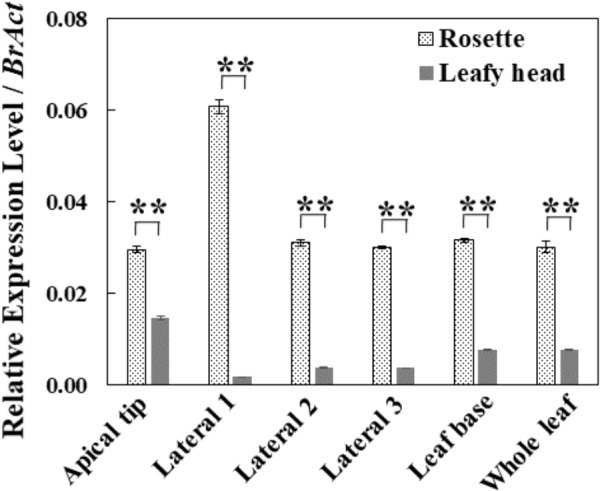
Relative expression levels of BrAN3 in different leaf positions and the whole leaf of Chinese cabbage at the rosette and heading stages measured by real-time RT-PCR. Total RNA was extracted from the apical location, the lateral 1∼3 locations, and the base location of individual leaves and the whole leaves of Chinese cabbage (Gao et al., 2017) at both the rosette and heading stages, respectively. BrAN3-specific primers were designed to target all the three homologous sequences of BrAN3 in Chinese cabbage. Relative expression levels were calculated using the standard curve method with BrAct (Bra028615; Dheda et al., 2004) as the internal control gene. Bars represent the means of three replicates ± standard errors (vertical bars). Asterisks indicate a significant difference between the two stages at p ≤ 0.05 as calculated by t-test.
The Effects of VIGS-Mediated Knockdown of BrAN3 and BrBRM Individually in Head Formation in Chinese Cabbage
To further explore the potential roles of the BrAN3 and its genetically interacting BrBRM gene in head formation in Chinese cabbage, VIGS was employed to knock down the expression of BrAN3 and BrBRM individually in Chinese cabbage. At 15 days post inoculation (DPI), all the TYMV-derived vector-inoculated plants displayed typical viral symptoms when compared with the untreated plants. These include plant stunting, leaf distortion, and mosaic symptoms. At 60 DPI, the untreated and the empty vector pTY-S-inoculated plants showed a normal increase in leaf size, while the positive control vector pTY-BrPDS-inoculated plants exhibited a photobleaching phenotype as expected (Figure 3A; Yu et al., 2018). However, the pTY-BrAN3- and pTY-BrBRM-inoculated plants produced curled leaves and formed smaller leafy heads at the rosette age (Figure 3A), indicating an important role for both genes in head formation at the early stage.
FIGURE 3.
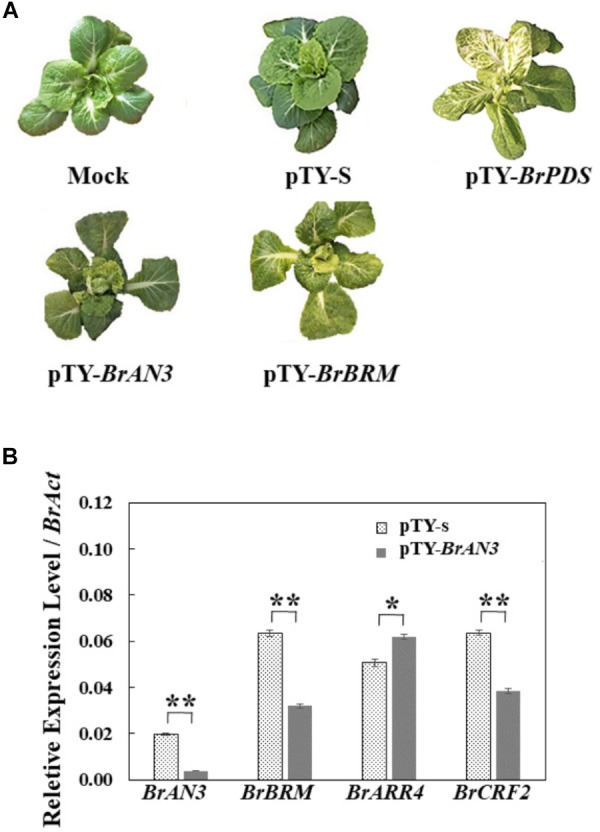
Effects of gene silencing of BrAN3 and BrBRM in Chinese cabbage. (A) Representative images of the silencing plants by using the TYMV-derived vector pTY-S at 60 DPI. A gene-specific fragment of 40 nt in length was selected for each of the two genes to create an in-frame stop codon (TAA, TGA, or TAG) on the second, third or fourth amino acid position on each fragment due to frame shift. The gene-specific fragment was selected to target all the homologous sequences of each gene in the Chinese cabbage genome. Six weeks old Chinese cabbage plants were inoculated with each gene silencing vector. The mock treatment and the empty vector pTY-S were used as the negative control, while the pTY-BrPDS was used as the positive control. Leaf head formation was observed for pTY-BrAN3 and pTY-BrBRM-inoculated plants. (B) Relative expression levels of BrAN3, BrBRM, and two of BrAN3 downstream genes BrARR4 and BrCRF2 in the BrAN3-silencing plants measured by Real-time RT-PCR. RNA was extracted from the BrAN3-silencing leaves with the empty vector pTY-S-inoculated leaves as the control at 60 DPI. Relative expression levels were calculated using the standard curve method with BrAct (Bra028615; Dheda et al., 2004) as the internal control gene. Bars represent the means of three replicates ± standard errors (vertical bars). Asterisks indicate a significant difference between the two stages at p ≤ 0.05 as calculated by t-test.
To confirm the VIGS effect of BrAN3 in the BrAN3-silencing Chinese cabbage, real-time RT-PCR was performed to detect the relative expression levels of BrAN3 in pTY-BrAN3-inoculated plants, respectively. When compared to its relative expression levels on the empty vector pTY-S-inoculated plants, the relative expression levels of BrAN3 were significantly decreased on the pTY-BrAN3-inoculated plants (Figure 3B). In addition, we found that the relative expression levels of BrBRM significantly decreased in the BrAN3-silencing plants (Figure 3B), indicating BrAN3 positively regulates the expression of BrBRM.
The Involvement of the Cytokinin Signaling Pathway in Head Formation in Chinese Cabbage
Since AtAN3 plays an important role in the regulation of cytokinin signaling to stimulate leaf cell proliferation and adaxial-abaxial patterning, it represses and induces the expression of its downstream genes AtARR4 and AtCRF2, respectively, which are cytokinin response regulators (Rashotte et al., 2006; Werner and Schmuelling, 2009; Holst et al., 2011; Vercruyssen et al., 2014). Thus, real-time RT-PCR was also conducted to detect the relative expression levels of BrARR4 and BrCRF2 in the BrAN3-silencing plants. As expected, the relative expression levels of BrARR4 and BrCRF2 significantly increased and decreased, respectively, in the BrAN3-silencing plants when compared to the empty vector pTY-S-inoculated plants (Figure 3B).
RNA-Seq Analysis and Differentially Expressed Gene (DEG) Identification in the BrAN3-Silencing Chinese Cabbage
To further characterize the molecular mechanisms of BrAN3 in leafy head formation, RNA-Seq was performed on the BrAN3-silencing plants with the empty vector pTY-S-inoculated plants being used as the control. About 20,992,641 and 19,553,742 clean reads obtained from the BrAN3-silencing plants and the control plants, respectively, were mapped against the Brassica reference sequence, indicating equal match ratios on the reference genome for both groups of data. When compared to the control plants, a total of 1692 DEGs were detected in the BrAN3-silencing plants based on the transcript abundances; among which, 800 DEGs were significantly up-regulated and 892 DEGs were significantly down-regulated (Figure 4).
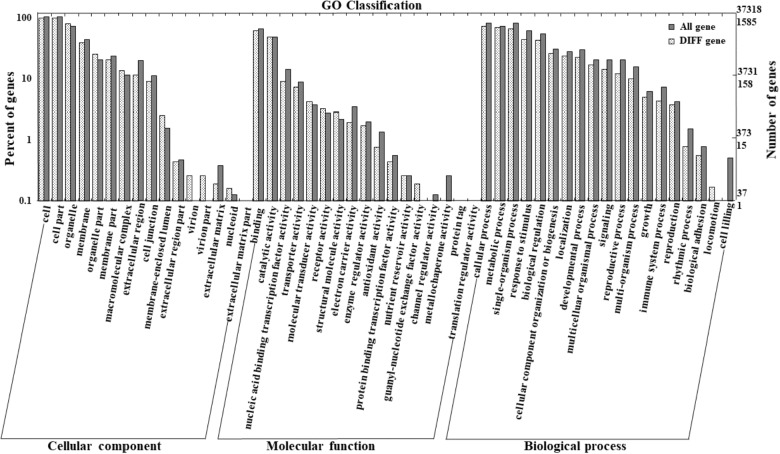
FIGURE 4.
Functional Classification of the DEGs by Gene Ontology (GO) Enrichment Analysis. RNA was extracted from the BrAN3-silencing leaves with the empty vector pTY-S-inoculated leaves as the control at 60 days post inoculation (DPI), and used for library construction and RNA-Seq analysis. GO enrichment analysis of the DEGs was implemented by the GOseq R packages based on the Wallenius non-central hyper-geometric distribution (Young et al., 2010), which allows the adjustment for gene length bias in DEGs. It assigned all the genes and the DEGs to the three GO categories: biological processes, cell components, and molecular functions.
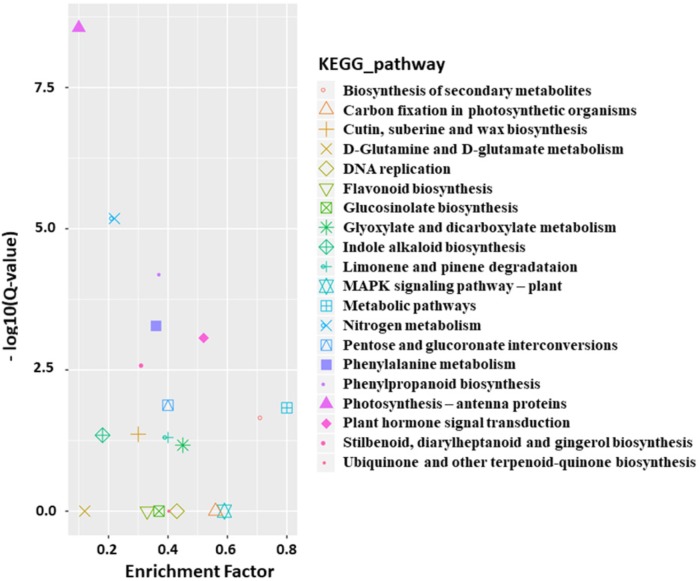
Functional Classification of DEGs by Gene Ontology (GO) Enrichment Analysis and KEGG Pathway Analysis
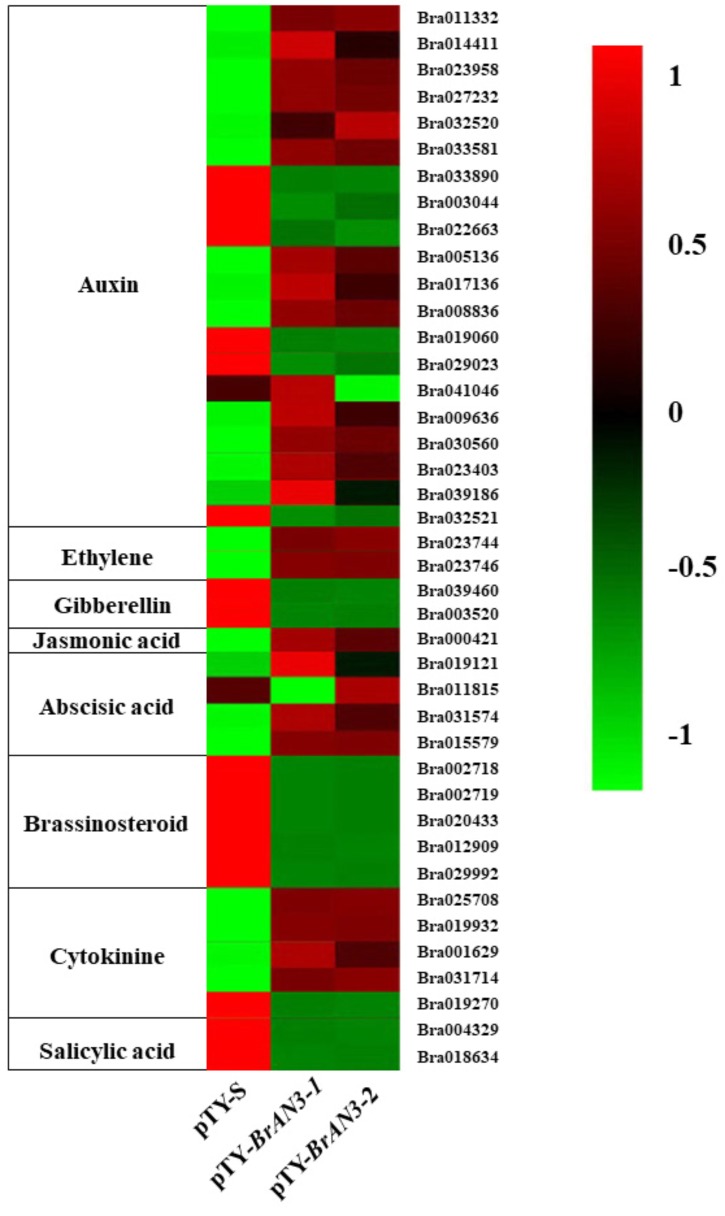
Functional classification of the identified DEGs using the GO term enrichment analysis assigned all the genes and the DEGs to the three GO categories: biological processes, cell components, and molecular functions (Figure 4). Overall, DEGs was more enriched in biological processes category than that in the other two categories as well as that in all genes background. Specifically, the over-represented (p < 0.05) GO terms of biological processes category included cellular process, metabolic process, single-organism process, response to stimulus, biological regulation, developmental process, etc., indicating the silencing of BrAN3 may affect the developmental and biological processes. Furthermore, the KEGG pathway analysis of the identified DEGs showed that the DEGs involving in the pathways of photosynthesis-antenna proteins, nitrogen metabolism, phenylpropanoid biosynthesis, phenylalanine metabolism, and plant hormone signal transduction were significantly enriched in the BrAN3-silencing plants (Figure 5). We found there existed 41 DEGs in the plant hormone signal transduction term with 23 and 16 DEGs being significantly up-regulated and down-regulated, respectively (Figure 6). The remaining 2 DEGs showed controversial results, i.e., a significant increase in one BrAN3-silencing plant and a significant decrease in the other BrAN3-silencing plant (Figure 6). Specifically, the DEGs involved in the signaling pathways of ethylene (ETH; Bra023744 and Bra023746) and jasmonic acid (JA; Bra000421) were significantly increased in the BrAN3-silencing leaves when compared to the empty vector pTY-S-inovulated leaves (Figure 6). The DEGs involved in the signaling pathways of gibberellin (GA; Bra039460 and Bra003520), brassinosteroid (BR; Bra002718, Bra002719, Bra020433, Bra012909, and Bra029992), and salicylic acid (SA; Bra004329 and Bra018634) were significantly decreased in BrAN3-silencing leaves in comparison to the empty vector-inoculated leaves (Figure 6). However, the DEGs involved in the signaling pathways of auxin (AUX), abscisic acid (ABA), and cytokinin (CK) showed opposite expression patterns. For example, 13 DEGs involved in the AUX signaling pathway (Bra011332, Bra014411, Bra023958, Bra027232, Bra032520, Bra033581, Bra005136, Bra017136, Bra008836, Bra009636, Bra030560, Bra023403, and Bra039186), 3 DEGs involved in the ABA signaling pathway (Bra019121, Bra031574, and Bra015579), and 4 DEGs involved in the CK signaling pathway (Bra025708, Bra019932, Bra001629, and Bra031714) were significantly increased in the BrAN3-silencing leaves (Figure 6). And 6 and 1 DEGs in the signaling pathways of AUX (Bra033890, Bra003044, Bra022663, Bra019060, Bra029023, and Bra032521) and CK [Bra019270 (BrCRF2)] showed a significantly decrease in the BrAN3-silencing plants (Figure 6). In addition, 1 DEG involved in each of the auxin (Bra041046) and ABA (Bra011815) signaling pathways exhibited controversial expression patterns when compared to the empty vector-inoculated leaves (Figure 6).
FIGURE 5.
Over-represented KEGG terms of the differentially expressed genes (DEGs). KOBAS software was used to test the statistical enrichment of DEGs in KEGG pathways (Mao et al., 2005).
FIGURE 6.
Heatmap showing the differential expression patterns of the 41 differentially expressed genes (DEGs).
To validate the accuracy of the RNA-Seq data, we selected 9 out of the 41 DEGs from the plant hormone signal transduction term for real-time RT-PCR analysis. When compared to the empty vector pTY-S-inoculated leaves, we found that 7 (Bra011332, Bra014411, Bra023958, Bra027232, Bra005136, Bra017136, and Bra025708) out of the 9 DEGs were significantly increased while the other 2 DEGs (Bra003044 and Bra022663) were significantly decreased in the BrAN3-silencing plants (Figure 7). We also found the same expression patterns of these 9 DEGs were observed in the RNA-Seq data when calculated as the FPKM (Figure 7). Thus, the real-time RT-PCR and RNA-Seq data correlated very closely (Figure 7).
FIGURE 7.
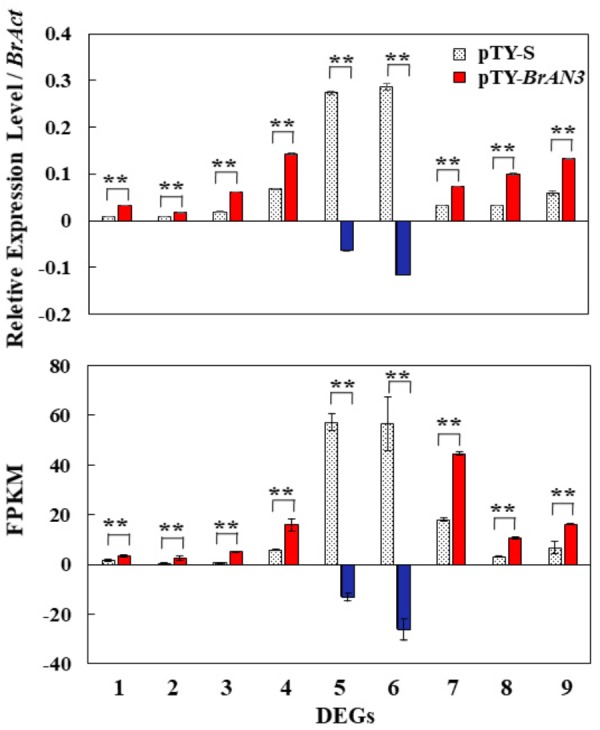
Validation of the expression of 9 out of the 41 differential expression genes (DEGs) by real-time RT-PCR. Gene-specific primers were designed to target all the homologous sequences of each gene in Chinese cabbage. Relative expression levels were calculated using the standard curve method with BrAct (Bra028615; Dheda et al., 2004) as the internal control gene. The FPKM expression levels were calculated as fragments per kilobase of exon model per million mapped (FPKM) values. Bars represent the means of three replicates ± standard errors (vertical bars). Genes: 1, Bra011332; 2, Bra014411; 3, Bra023958; 4, Bra027232; 5, Bra003044; 6, Bra022663; 7, Bra005136; 8, Bra017136; 9, Bra025708. Asterisks indicate a significant difference between pTY-S- and pTY-BrAN3-inoculated plants at p ≤ 0.01 as calculated by t-test.
Discussion
Leafy head formation in Chinese cabbage is a tightly controlled process. Many proteins such as the SWI/SNF chromatin remodeling protein complex, ATPases, and transcription activators could play important roles in head formation. This is the first time to functionally confirm that the BrAN3 gene could induce head formation in Chinese cabbage. Its distinct expression patterns in different leaf locations and at different head developmental stages are consistent with its function in head formation. Similar expression patterns had also been observed for the BrTCP gene (Mao et al., 2014) and the BrLAX, BrPIN, BrPGP genes (Gao et al., 2017), which exhibited higher expression levels in the leaf apical tip and at the rosette stage than in the leaf base and at the heading stage in Chinese cabbage. However, the BcpLH (Yu et al., 2000), BrGRF (Wang et al., 2014a), and BrpSPL9 (Wang et al., 2014b) genes showed a gradual increase in transcript abundance from the rosette stage to the heading stage, implying different functions.
Gene knockout/knockdown methods such as the CRISPR-Cas9 system, RNA interference, T-DNA insertion, transposons, and some chemical reagent-induced mutations suffer from the limitations of off-target effects, functional redundancy, embryonic lethality, and multi-insertions (Pflieger et al., 2008). To the contrary, VIGS is a rapid and cost-effective RNA-mediated reverse genetics technology, and does not rely on the acquisition of mutants or transgenic plants. It can be easily applied in many species to study gene function either individually or on a large scale. pTY-S is a newly developed VIGS vector, which contains the entire genome of TYMV, and the 35S CAMV promoter and terminator (Pflieger et al., 2008; Huang et al., 2018a,b; Muntha et al., 2018). It possesses a strong infection ability, produces liable and robust VIGS effects that lasts throughout the plant life (Pflieger et al., 2008), and has been successfully applied in Chinese cabbage (Yu et al., 2018). In the present study, the expression of BrAN3 and BrBRM genes had been effectively knocked down by each silencing vector, and the formation of leafy heads had been observed in the inoculated Chinese cabbage plants at the early stage (Figure 3A). The results demonstrated the function of the BrAN3 and BrBRM genes in leafy head formation in Chinese cabbage. Since the efficiency of VIGS is highly dependent on the virus-host affinity, and the infected TYMV eventually took over and killed the host plants at the later stage of head formation, it should be noted that the effect of knockdown of the BrAN3 and BrBRM genes on leafy head formation could be better examined in stable transgenic China cabbage in the near future.
Hormones such as auxin and gibberellins play important roles in the adaxial-abaxial patterning during leaf development in Chinese cabbage (He et al., 2000; Yu et al., 2000; Wang et al., 2014a; Gao et al., 2017). The identification of 41 DEGs from the plant hormone signal transduction term in the BrAN3-silencing leaf by RNA-Seq further demonstrated that plant hormones are key players in leafy head formation. In the present study, we found that cytokinin plays an important role in leafy head formation in Chinese cabbage. The significant increase and decrease in the expression of the BrARR4 and BrCRF2 genes in the BrAN3-silencing plants (Figure 3B) indicate that BrAN3 is involved in the cytokinin signaling pathway since both BrARR4 and BrCRF2 genes are cytokinin response regulators (Rashotte et al., 2006; Werner and Schmuelling, 2009; Holst et al., 2011; Vercruyssen et al., 2014).
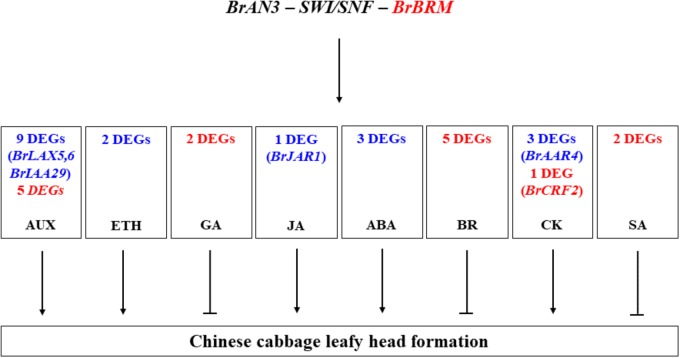
Moreover, our RNA-Seq results revealed that the BrAN3 protein functions to induce the GA, BR, and SA signaling pathways, which inhibit leafy head formation in Chinese cabbage (Figure 6). The BrAN3 also inhibit the ethylene and JA signaling pathways, which promote leafy head formation in Chinese cabbage (Figure 6). The BrAN3 protein increases and decreases DEGs involved in the auxin, ABA, and CK signaling pathways, indicating the three pathways play both a positive and a negative roles in leafy head formation in Chinese cabbage (Figure 6). The accumulation and uneven distribution of auxin in the head leaves play an important role in Chinese cabbage leafy head formation (He et al., 2000; Gao et al., 2017). This is in accordance with our findings that a total of 20 auxin DEGs were significantly expressed in the BrAN3-silencing plants, including 13 up-regulated and 6 down-regulated DEGs. Among them, 3 DEGs (Bra017136, Bra005136, and Bra 023958, which correspond to BrLAX5, BrLAX6, and BrIAA29) had been demonstrated to be involved in leafy head formation in Chinese cabbage (Wang et al., 2012; Gao et al., 2017). Auxin mediates plant growth by affecting DELLA proteins (Fu and Harberd, 2003), while SWI3C, a core component of Arabidopsis SWI/SNF chromatin-remodeling complexes, also interacts with several DELLA proteins (Sarnowska et al., 2013). DELLA was reported as a repressor of ethylene signaling (Sarnowska et al., 2013), and interacts with JAZ in response of JA signaling (Lor and Olszewski, 2015; Vleesschauwer et al., 2016). Ethylene has been shown to induce petiole unequal growth between the adaxial and abaxial axes in Arabidopsis (Polko et al., 2012), and JAZ represses the JA signaling (Lor and Olszewski, 2015; Vleesschauwer et al., 2016). Accordingly, we found that the ethylene and JA signaling pathways were significantly induced in the BrAN3-silencing plants. The only DEG (Bra000421) which corresponds to the BrJAR1 gene was identified to be a DEG in leafy head formation in Chinese cabbage by transcriptomics and proteomics analyses (Zhang C.W. et al., 2016). In addition, GA signaling was proved to be a positive regulator of the SWI/SNF chromatin remodeling complexes (Rafal et al., 2013; Jose, 2014). BRM regulates primary root elongation by repressing the activity of the ABA signaling pathway (Han et al., 2012). Our findings that the BrAN3-silencing plants inhibited and promoted GA and ABA genes expression, respectively, are consistent with those reports (Han et al., 2012; Rafal et al., 2013; Jose, 2014). Moreover, BRM binds to the promoter of ARR16 for boundary establishment in Arabidopsis cotyledon, which is an inhibitor of CK responses (Efroni et al., 2013). Similarly, we found that 4 and 1 DEGs involved in the CK signaling pathway were up-regulated and down-regulated in the BrAN3-silencing plants, respectively (Figure 6). The potential roles of these hormone signaling pathways in leafy head formation in Chinese cabbage were summarized in Figure 8. Further studies should be conducted to investigate the involvement of each hormone signaling pathway in leafy head formation in Chinese cabbage.
FIGURE 8.
The potential roles of the signaling pathways of auxin (AUX), ethylene (ETH), gibberellin (GA), jasmonic acid (JA), abscisic acid (ABA), brassinosteroid (BR), cytokinine (CK), and salicylic acid (SA) in leafy head formation in Chinese cabbage. Red and blue font colors indicate the significantly induced and decreased DEGs detected by RNA-Seq in the present study. The genes in the parenthesis, which were functionally studied in previous studies, had been identified as the DEGs in the present study. Arrows represent the positive effects while inhibition arrows represents the negative effects.
Conclusion
We identified and characterized the function of the BrAN3 gene in leafy head formation in Chinese cabbage. We also found that DEGs involved in auxin, ethylene, GA, JA, ABA, BR, CK, and SA signaling pathways play either positive or negative roles in leafy head formation in Chinese cabbage. This information could be useful for genetic engineering, gene editing, and molecular breeding of Chinese cabbage to improve heading and thus yield.
Author Contributions
CZ and XH conceived the project. JY and LG conducted the experiments. JY, LG, WL, CZ, DX, XH, JY, and TL performed the data analysis. JY, LG, WL, CZ, XH, and LS wrote the manuscript. All authors read and approved the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Antoine Bouteilly (Centre National dela Recherche Scientifique) for providing the plasmid pTY-S.
Funding. This work was supported by the grants from the National Natural Science Foundation of China (31272172), the Fundamental Research Funds for the Central Universities (KYZ201826), and the Nature Science Foundation of Jiangsu Province (BK20141364).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.00520/full#supplementary-material
References
- Cheng F., Sun R., Hou X., Zheng H., Zhang F., Zhang Y., et al. (2016). Subgenome parallel selection is associated with morphotype diversification and convergent crop domestication in Brassica rapa and B. oleracea. Nat. Genet. 48 1218–1224. 10.1038/ng.3634 [DOI] [PubMed] [Google Scholar]
- Cheng F., Wu J., Wang X. (2014). Genome triplication drove the diversification of Brassica plants. Hort. Res. 1:14024. 10.1038/hortres.2014.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapier C. R., Cairns B. R. (2009). The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 78 273–304. 10.1146/annurev.biochem.77.062706.153223 [DOI] [PubMed] [Google Scholar]
- Debernardi J. M., Mecchia M. A., Vercruyssen L., Smaczniak C., Kaufmann K., Inze D., et al. (2014). Post-transcriptional control of GRF transcription factors by microRNA miR396 and GIF co-activator affects leaf size and longevity. Plant J. 79 413–426. 10.1111/tpj.12567 [DOI] [PubMed] [Google Scholar]
- Dheda K., Huggett J. F., Bustin S. A., Johnson M. A., Rook G., Zumla A. (2004). Validation of housekeeping genes for normalizing RNA expression in real-time PCR. BioTechniques 37 112–119. 10.2144/04371rr03 [DOI] [PubMed] [Google Scholar]
- Efroni I., Han S., Kim H., Wu M., Steiner E., Birnbaum K., et al. (2013). Regulation of leaf maturation by chromatin-mediated modulation of cytokinin responses. Dev. Cell 24 438–445. 10.1016/j.devcel.2013.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X., Harberd N. (2003). Auxin promotes Arabidopsis root growth by modulating gibberellin response. Nature 421 740–743. 10.1038/nature01387 [DOI] [PubMed] [Google Scholar]
- Gao L., Lyu S., Tang J., Zhou D., Bonnema G., Xiao D., et al. (2017). Genome-wide analysis of auxin transport genes identifies the hormone responsive patterns associated with leafy head formation in Chinese cabbage. Sci. Rep. 7:42229. 10.1038/srep42229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Y., Ramchiary N., Wang T., Liang C., Wang N., Wang Z., et al. (2011). Mapping quantitative trait loci for leaf and heading-related traits in Chinese cabbage (Brassica rapa L. ssp. pekinesis). Hort. Environ. Biotechnol. 52 494–501. 10.1007/s13580-011-0031-x [DOI] [Google Scholar]
- Gu A., Meng C., Chen Y., Wei L., Dong H., Lu Y., et al. (2017). Coupling Seq-BSA and RNA-Seq analyses reveal the molecular pathway and genes associated with heading type in Chinese cabbage. Front. Genet. 8:176. 10.3389/fgene.2017.00176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S., Sang Y., Rodrigues A., Wu M., Rodriguez P., Wagnera D. (2012). The SWI2/SNF2 chromatin remodeling ATPase BRAHMA represses abscisic acid responses in the absence of the stress stimulus in Arabidopsis. Plant Cell 24 4892–4906. 10.1105/tpc.112.105114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y. K., Xue W., Sun Y., Yu X., Liu P. (2000). Leafy head formation of the progenies of transgenic plants of Chinese cabbage with exogenous auxin genes. Cell Res. 10 151–160. 10.1038/sj.cr.7290044 [DOI] [PubMed] [Google Scholar]
- Holst K., Schmuelling T., Werner T. (2011). Enhanced cytokinin degradation in leaf primordia of transgenic Arabidopsis plants reduces leaf size and shoot organ primordia formation. J. Plant Physiol. 168 1328–1334. 10.1016/j.jplph.2011.03.003 [DOI] [PubMed] [Google Scholar]
- Horiguchi G., Kim G.-T., Tsukaya H. (2005). The transcription factor AtGRF5 and the transcription coactivator AN3 regulate cell proliferation in leaf primordia of Arabidopsis thaliana. Plant J. 43 68–78. 10.1111/j.1365-313x.2005.02429.x [DOI] [PubMed] [Google Scholar]
- Horiguchi G., Nakayama H., Ishikawa N., Kubo M., Demura T., Fukuda H., et al. (2011). ANGUSTIFOLIA 3 plays roles in adaxial/abaxial patterning and growth in leaf morphogenesis. Plant Cell Physiol. 52 112–124. 10.1093/pcp/pcq178 [DOI] [PubMed] [Google Scholar]
- Huang F., Liu T., Hou X. (2018a). Isolation and functional characterization of a floral repressor, BcMAF1, from Pak-choi (Brassica rapa ssp. chinensis). Front. Plant Sci. 9:290. 10.3389/fpls.2018.00290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F., Liu T., Wang J., Hou X. (2018b). Isolation and functional characterization of a floral repressor, BcFLC2, from Pak-choi (Brassica rapa ssp. chinensis). Planta 248 423–435. 10.1007/s00425-018-2891-0 [DOI] [PubMed] [Google Scholar]
- Hurtado L., Farrona S., Reyes J. C. (2006). The putative SWI/SNF complex subunit BRAHMA activates flower homeotic genes in Arabidopsis thaliana. Plant Mol. Biol. 62 291–304. 10.1007/s11103-006-9021-2 [DOI] [PubMed] [Google Scholar]
- Ito H. (1957). Effect of temperature and photoperiod on head formation of leafy head of Chinese cabbage. J. Hort. Assoc. Jpn. 26:154 10.2503/jjshs.26.154 [DOI] [Google Scholar]
- Jose C. R. (2014). The many faces of plant SWI/SNF complex. Mol. Plant 7 454–458. 10.1093/mp/sst147 [DOI] [PubMed] [Google Scholar]
- Ke G. (2010). Chinese Cabbage Breeding. Beijing: China Agricult. Press. 10.1093/mp/sst147 [DOI] [Google Scholar]
- Kim D., Langmead B., Salzberg S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12 357–360. 10.1038/nmeth.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. H., Choi D., Kende H. (2003). The AtGRF family of putative transcription factors is involved in leaf and cotyledon growth in Arabidopsis. Plant J. 36 94–104. 10.1046/j.1365-313x.2003.01862.x [DOI] [PubMed] [Google Scholar]
- Kim J. H., Kende H. (2004). A transcriptional coactivator, AtGIF1, is involved in regulating leaf growth and morphology in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 101 13374–13379. 10.1073/pnas.0405450101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23 2947–2948. 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- Lee B. H., Ko J.-H., Lee S., Lee Y., Pak J.-H., Kim J. H. (2009). The Arabidopsis GRF-INTERACTING FACTOR gene family performs an overlapping function in determining organ size as well as multiple developmental properties. Plant Physiol. 151 655–668. 10.1104/pp.109.141838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Fan Y., Jiao Y., Wu J., Zhang Z., Yu X., et al. (2018). Transcriptome profiling of yellow leafy head development during the heading stage in Chinese cabbage (Brassica rapa subsp. pekinesis). Physiol. Plant 165 800–813. 10.1111/ppl.12784 [DOI] [PubMed] [Google Scholar]
- Liu D., Song Y., Chen Z., Yu D. (2009). Ectopic expression of miR396 suppresses GRF target gene expression and alters leaf growth in Arabidopsis. Physiol. Plant 136 223–236. 10.1111/j.1399-3054.2009.01229.x [DOI] [PubMed] [Google Scholar]
- Lor V. S., Olszewski N. (2015). GA signalling and cross-talk with other signaling pathways. Essays Biochem. 58 49–60. 10.1042/bse0580049 [DOI] [PubMed] [Google Scholar]
- Mao X., Cai T., Olyarchuk J. G., Wei L. (2005). Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 21 3787–3793. 10.1093/bioinformatics/bti430 [DOI] [PubMed] [Google Scholar]
- Mao Y., Wu F., Yu X., Bai J., Zhong W., He Y. (2014). microRNA319a-targeted Brassica rapa ssp. Pekinensis TCP genes modulate head shape in Chinese cabbage by differential cell division arrest in leaf regions. Plant Physiol. 164 710–720. 10.1104/pp.113.228007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muntha S. T., Zhang L., Zhou Y., Zhao X., Hu Z., Yang J., et al. (2018). Phytochrome A signal transduction 1 and CONSTANS-LIKE 13 coordinately orchestrate shoot branching and flowering in leafy Brassica juncea. Plant Biotechnol. J. 10.1111/pbi.13057 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pflieger S., Blanchet S., Camborde L., Drugeon G., Rousseau A., Noizet M., et al. (2008). Efficient virus-induced gene silencing in Arabidopsis using a ‘one-step’ TYMV-derived vector. Plant J. 56 678–690. 10.1111/j.1365-313X.2008.03620.x [DOI] [PubMed] [Google Scholar]
- Polko J., van Zanten M., van Rooij J., Marée A., Voesenek L., Peeters A., et al. (2012). Ethylene-induced differential petiole growth in Arabidopsis thaliana involves local microtubule reorientation and cell expansion. New Phytol. 193 339–348. 10.1111/j.1469-8137.2011.03920.x [DOI] [PubMed] [Google Scholar]
- Rafal A., Daniel B., Tomasz J., Elzbieta S., Anna T., Takayuki T., et al. (2013). BRAHMA ATPase of the SWI/SNF chromatin remodeling complex acts as a positive regulator of gibberellin-mediated responses in Arabidopsis. PLoS One 3:e58588. 10.1371/journal.pone.0058588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashotte A. M., Mason M. G., Hutchison C. E., Ferreira F. J., Schaller G. E., Kieber J. J. (2006). A subset of Arabidopsis AP2 transcription factors mediates cytokinin responses in concert with a two-component pathway. Proc. Natl. Acad. Sci. U.S.A. 103 11081–11085. 10.1073/pnas.0602038103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarnowska E. A., Rolicka A. T., Bucior E., Cwiek P., Tohge T., Fernie A. R., et al. (2013). DELLA-interacting SWI3C core subunit of switch/sucrose nonfermenting chromatin remodeling complex modulates gibberellin responses and hormonal cross talk in Arabidopsis. Plant Physiol. 163 305–317. 10.1104/pp.113.223933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen T. D., Livak K. J. (2008). Analyzing real-time PCR data by the comparative C-T method. Nat. Protoc. 3 1101–1108. 10.1038/nprot.2008.73 [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vercruyssen L., Verkest A., Gonzalez N., Heyndrickx K. S., Eeckhout D., Han S.-K., et al. (2014). ANGUSTIFOLIA3 binds to SWI/SNF chromatin remodeling complexes to regulate transcription during Arabidopsis leaf development. Plant Cell 26 210–229. 10.1105/tpc.113.115907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vleesschauwer V., Seifi H., Filipe O., Haeck A., Huu S., Demeestere K., et al. (2016). The DELLA protein SLR1 integrates and amplifies salicylic acid- and jasmonic acid-dependent innate immunity in rice. Plant Physiol. 170 1831–1847. 10.1104/pp.15.01515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Li H., Zhang Y., Li J., Li L., Liu L., et al. (2013). microRNA expression analysis of rosette and folding leaves in Chinese cabbage using high-throughput Solexa sequencing. Gene 532 222–229. 10.1016/j.gene.2013.09.039 [DOI] [PubMed] [Google Scholar]
- Wang F., Li L., Li H., Liu L., Zhang Y., Gao J., et al. (2012). Transcriptome analysis of rosette and folding leaves in Chinese cabbage using high-throughput RNA sequencing. Genomics 99 299–307. 10.1016/j.ygeno.2012.02.005 [DOI] [PubMed] [Google Scholar]
- Wang F., Qiu N., Ding Q., Li J., Zhang Y., Li H., et al. (2014a). Genome-wide identification and analysis of the growth-regulating factor family in Chinese cabbage (Brassica rapa ssp. pekinensis). BMC Genomics 15:807. 10.1186/1471-2164-15-807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Wu F., Bai J., He Y. (2014b). BrpSPL9 (Brassica rapa ssp. pekinensis SPL9) controls the earliness of heading time in Chinese cabbage. Plant Biotechnol. J. 12 312–321. 10.1111/pbi.12138 [DOI] [PubMed] [Google Scholar]
- Werner T., Schmuelling T. (2009). Cytokinin action in plant development. Curr. Opin. Plant Biol. 12 527–538. 10.1016/j.pbi.2009.07.002 [DOI] [PubMed] [Google Scholar]
- Young M. D., Wakefield M. J., Smyth G. K., Oshlack A. (2010). Gene ontology analysis for RNA-Seq: accounting for selection bias. Genome Biol. 11:R14. 10.1186/gb-2010-11-2-r14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Yang X. D., Wang Q., Cao L. W., Yang Y., Xiao D., et al. (2018). Efficient virus-induced gene silencing in Brassica rapa using a turnip yellow mosaic virus vector. Biol. Plantarum 62 826–834. 10.1007/s10535-018-0803-6 [DOI] [Google Scholar]
- Yu X., Peng J., Feng X., Yang S., Zheng Z., Tang X., et al. (2000). Cloning and structural and expressional characterization of BcpLH gene preferentially expressed in folding leaf of Chinese cabbage. Sci. China C Life Sci. 43 321–329. 10.1007/BF02879292 [DOI] [PubMed] [Google Scholar]
- Yu X., Wang H., Zhong W., Bai J., Liu P., He Y. (2013). QTL mapping of leafy heads by genome resequencing in the RIL population of Brassica rapa. PLoS One 8:e76059. 10.1371/journal.pone.0076059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Li H., Zhang M., Hui M., Wang Q., Li L., et al. (2013). Fine mapping and identification of candidate Br-or gene controlling orange head of Chinese cabbage (Brassica rapa ssp. pekinensis). Mol. Breed. 32 799–805. 10.4238/gmr.15028370 [DOI] [PubMed] [Google Scholar]
- Zhang X., Su Y., Liu Y., Fang Z., Zhuang M., Zhang Y., et al. (2016). Genetic analysis and QTL mapping of traits related to head shape in cabbage (Brassica rapa ssp. pekinensis). Sci. Hort. 207 82–88. 10.1016/j.scienta.2016.05.015 [DOI] [Google Scholar]
- Zhang C. W., Wei Y. P., Xiao D., Gao L. W., Lyu S. W., Hou X. L., et al. (2016). Transcriptomic and proteomic analyses provide new insights into the regulation mechanism of low-temperature-induced leafy head formation in Chinese cabbage. J. Proteomics 144 1–10. 10.1016/j.jprot.2016.05.022 [DOI] [PubMed] [Google Scholar]
- Zhao J. J., Wang X. W., Deng B., Lou P., Wu J., Sun R. F., et al. (2005). Genetic relationships within Brassica rapa as inferred from AFLP fingerprints. Theor. Appl. Genet. 110 1301–1314. 10.1007/s00122-005-1967-y [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.