Abstract
Background
Gastric cancer (GC), the second leading cause of cancer mortality behind lung cancer worldwide, is caused by both genetic and environmental factors. In this study, we evaluated the association between the genetic polymorphisms of methylenetetrahydrofolate reductase (MTHFR), methionine synthesis reductase (MTR), and methyltransferase reductase (MTRR) genes and ischemic stroke risk in Chinese population.
Methods
A case–control study was conducted including 681 patients with GC and 756 healthy controls. Chi‐squared test/Fisher's exact test and genetic model were used to evaluate associations. Odds ratios (ORs) and 95% confidence intervals (CIs) were calculated using unconditional logistic regression.
Results
In the allele model, using the chi‐square test, we found that the rs1532268 in MTRR with a minor allele T was significantly associated with increased risk of GC (OR = 1.24, 95% CI, 1.00–1.53; p = 0.048). In the genetic model analysis, we identified that the single‐nucleotide polymorphism of the rs1801133 in MTHFR could increase the GC risk in the recessive model (OR = 1.31, 95% CI, 1.01–1.70; p = 0.042) and log‐additive model (OR = 1.19, 95% CI, 1.02–1.38; p = 0.025). In MTHFR, a strong linkage of rs2274976 and rs1801133 was detected. The haplotype “GC” in the MTHFR gene was found to prominently increase the risk of GC (OR = 1.26, 95% CI: 1.07–1.47; p = 0.005). Other haplotypes did not display the correlativity.
Conclusion
This study suggested that MTR and MTHFR polymorphisms may contribute to increase the risk of GC.
Keywords: gastric cancer, genetics polymorphisms, MTHFR, MTR, MTRR
1. BACKGROUND
Gastric cancer (GC), one of the most common cancers, is the second leading cause of cancer mortality behind lung cancer worldwide. Particularly in China, an obvious clustering of geographical distribution of GC and a high mortality are estimated (Li et al., 2017; Yang, 2006). Despite remarkable decline in GC mortality is noticed, because of the poor prognosis and limited treatment options, there remains a major challenge in clinical with population growth. Smoking, high salt intake, smoking, and a familial genetic component are also recognized as predisposing factors. Additionally, hereditary genes were assessed to contribute 28% inducement by model fitting (De, Forman, & Plummer, 2003).
Genome‐wide association studies (GWASs), aimed at detecting variants at genomic loci such as single‐nucleotide polymorphisms (SNPs) that are associated with complex traits in populations, indicate new genetics insights about malignancies. Through GWAS, a number of genetic variation at different genes involved in gastric carcinogenesis and prognosis have been identified such as XPG, PLCE1, HFE, ERCC5, EZH2, DOC2, CYP19A1, ALDH2, and CDH1 (González, Sala, & Rokkas, 2013; Xia et al., 2015). Folate, an important constituent of vegetables and fruits, has been proved to decrease the risk of colorectal, pancreatic, and esophageal cancers by cumulative evidence. Besides an inadequate folate intake, polymorphisms in folate metabolism‐related enzyme‐coding genes, which play critical roles in DNA methylation and synthesis, have been previously suggested the correlation with the risk of cancer in various sites, including gastric carcinoma. So far, in terms of GC, SNPs like rs1801133 and rs1801131 in the methylenetetrahydrofolate reductase (MTHFR) (OMIM: 607093) are most extensively studied in. These SNPs may decrease the activity of MTHFR, resulting in increased levels of homocysteine (Kumar et al., 2012). Genetic variations of 5‐methyltetrahydrofolate‐homocysteine methyltransferase reductase (MTRR) (OMIM: 602568) and methionine synthesis reductase (MTR) (OMIM: 156570) genes were recently reported mainly associated to other cancers (breast, colon, prostate, pancreatic) (Ohnami et al., 2008; Shrubsole et al., 2006; Wu, Tang, & An, 2014).
Previous studies suggested there were differences of susceptibility variants in different races. China has one of the highest GC incidences in the world (Rafiei, Mohammadian‐Hafshejani, Towhidi, Makhsosi, & Salehiniya, 2016). For evaluating the correlations between genetic polymorphisms in the folate pathway and the risk of GC in Chinese, herein, we expanded the sample scale and focused on metabolism‐related genes MTHFR, MTR, and MTRR. The investigation was hoped to provide theoretical foundation of the study in potential functional SNPs.
2. MATERIALS AND METHODS
2.1. Ethics statement
This investigation was conducted in accordance with the ethical standards of the Declaration of Helsinki and following national and international guidelines. Additionally, our study has been approved by the ethics committee of the first affiliated Hospital of Xi'an Jiaotong University. All participants were informed both in writing and verbally of the procedures and purpose of our research and signed informed consent documents were obtained from all subjects.
2.2. Research participates
The study groups comprised 681 patients with GC and 756 healthy controls. All the subjects were recruited from Northwestern China at the first affiliated Hospital of Xi'an Jiaotong University. The related clinical information, including residence, age, ethnicity, sex, dietary habits, and previous disease history were collected through a detailed questionnaire. The patients were recently diagnosed with primary GC on the basis of clinical manifestations with further confirmation by endoscopic and histopathological analysis. Cases with other types of cancers or who underwent radiotherapy or chemotherapy were excluded. Unrelated healthy individuals through strict physical examinations in the same hospital were enrolled as controls. Patients had any other form of cancer, gastritis, or gastric ulcers or had blood relatives with GC going back three generations were excluded.
2.3. SNP selection and genotyping
We screened the SNPs of MTHFR, MTR, and MTRR with over 5% minor allele frequency (MAF) and disease relevance in 1,000 genome (http://www.internationalgenome.org/). For this study, we selected seven related SNPs, according to previous studies, rs2274976 and rs1801133 in MTHFR, rs1805087 and rs2853522 in MTR,Rs1801394, rs1532268, and rs162036 in MTRR, for the association analysis (Jae‐Young et al., 2012).
A venous blood sample (5 ml) was collected from each participates and were stored at −80°C. Patients’ blood sample collection was implemented before radiation or chemotherapy. Genomic DNA isolation was executed using a Gold Mag‐Mini Whole Blood Genomic DNA Purification Kit (GoldMag Ltd, Xian, China). Spectrometry (DU530 UV/VIS spectrophotometer, Beckman Instruments, Fullerton, CA) was used to measure the DNA concentration. Agena MassARRAY Assay Design 4.0 software was used to design the multiplexed SNP Mass EXTEND assay and Agena MassARRAY RS1000 was used to perform SNP genotyping according to the standard protocol. Then, Agena Typer 4.0 software was applied to analyze and manage our data (Chen et al., 2017; Gabriel, Ziaugra, & Tabbaa, 2009; Tian et al., 2018).
2.4. Statistical analysis
We used SPSS version 19.0 software (SPSS, Chicago, IL) and Microsoft Excel (Redmond, WA) to analyze all the related data. A value of p < 0.05 was considered as statistically significant. A chi‐squared test was used to evaluate the genotype frequencies of cases and controls. Frequencies of the variants were estimated using the Hardy–Weinberg equilibrium (HWE) (p value calculated by exact test) to compare the expected frequencies of the genotypes in the control groups. PLINK software (http://pngu.mgh.harvard.edu/purcell/plink/) were used to calculate the 95% confidence intervals (CIs) and odds ratios (ORs) by unconditional logistic regression analysis with adjustment for age and gender (Lin et al., 2017), in order to assess the strength for the association of genotypes and their combinations with GC risk in the four models (codominant, dominant, recessive, and log‐additive) (Jin et al., 2016; Lin et al., 2017; Liu et al., 2017). Finally, we measured the linkage disequilibrium (LD) between loci, haplotype construction, and genetic association was calculated by unconditional logistic regression. The Haploview were used to construct haplotype and genetic association at significant polymorphism loci and to estimate the pairwise LD, haplotype software (version4.2) (Barrett, Fry, Maller, & Daly, 2005).
3. RESULTS
3.1. Characteristics of the participants
A case–control study containing 681 GC patients with a mean age of 57.57 ± 10.826 and 756 healthy controls with a mean age of 52.58 ± 8.709 was performed. The general characteristics are listed in Table 1.
Table 1.
General characteristics of the study population
| Variables | Case (%) | Control (%) | Total | p |
|---|---|---|---|---|
| Total | 681 | 756 | ||
| Gender | <0.001 | |||
| Male | 527 (77.4) | 489 (64.7%) | 1,016 | |
| Female | 154 (22.6) | 267 (35.3%) | 421 | |
| Age | <0.001 | |||
| Mean age ± SD | 57.57 ± 10.826 | 52.58 ± 8.709 |
Values of p were calculated from a two‐sided chi‐squared test/Fisher's exact test. Value of p ≤ 0.05 was statistically significant.
3.2. The associations between SNPs and GC risk
Basic information containing position, alleles, MAF distribution, HWE‐p value, ORs, 95% CIs of all the candidate SNPs is shown in Table 2. The minor allele of each SNP was assumed as a risk allele compared to the wild‐type allele and MAF was listed. The HWE‐p values showed none of the seven SNPs had a significant departure from the HWE. OR = 1 indicates that the factor had no effect on the disease; OR >1 means a risk factor; and OR <1 means protective factor. The p values were estimated by the chi‐square test. From the seven SNPs, only rs1532268 on MTRR with a minor allele T was significantly associated with increased risk of GC (OR = 1.24, 95% CI, 1.00–1.53; p = 0.048).
Table 2.
Basic information of candidate SNPs in all the individuals examined in this study
| SNP ID | Gene | Position | Band | Allele (aA/B) | MAF | HWE‐p | OR (95% CI) | p value | |
|---|---|---|---|---|---|---|---|---|---|
| Case | Control | ||||||||
| rs2274976 | MTHFR | 11790870 | 1p36.22 | G/A | 0.059 | 0.065 | 1.000 | 0.90 (0.67−1.22) | 0.507 |
| rs1801133 | MTHFR | 11796321 | 1p36.22 | T/C | 0.476 | 0.451 | 0.340 | 1.11 (0.96−1.28) | 0.173 |
| rs1805087 | MTR | 236885200 | 1q43 | G/A | 0.08 | 0.1 | 1.000 | 0.78 (0.60−1.01) | 0.061 |
| rs2853522 | MTR | 236897756 | 1q43 | A/C | 0.428 | 0.421 | 0.455 | 1.03 (0.89−1.19) | 0.698 |
| rs1801394 | MTRR | 7870860 | 5p15.31 | G/A | 0.268 | 0.284 | 0.858 | 0.92 (0.78−1.09) | 0.333 |
| rs1532268 | MTRR | 7878066 | 5p15.31 | T/C | 0.153 | 0.128 | 0.071 | 1.24 (1.01−1.53) | 0.048* |
| rs162036 | MTRR | 7885846 | 5p15.31 | G/A | 0.163 | 0.174 | 0.377 | 0.92 (0.76−1.12) | 0.422 |
Value of HWE‐p ≤ 0.05 was excluded.
SNP: single nucleotide polymorphism; A: minor allele; B: major allele. MAF: minor allele frequency; HWE: Hardy–Weinberg equilibrium; ORs: odds ratios; 95% CI: 95% confidence interval.
Minor allele.
Value of p ≤ 0.05 indicates statistical significance.
3.3. Associations between genotype frequencies and GC risk
The correlations between polymorphisms and GC susceptibility were analyzed based on four genetic models (codominant, dominant, recessive, and log‐additive) using logistic tests, as shown in Table 3. After adjusting for age and gender, one SNP was found to be conspicuous: rs1801133 in MTHFR increased the GC risk in the recessive model (OR = 1.31, 95% CI, 1.01–1.70; p = 0.042) and log‐additive model (OR = 1.19, 95% CI, 1.02–1.38; p = 0.025). Besides, no polymorphism displayed the association based on statistically significance.
Table 3.
Logistic regression analysis of the association between prominent SNPs with the gastric cancer (adjusted sex and age)
| Gene | SNP ID | Model | Genotype | Control, n (%) | Case, n (%) | Without adjusted | With adjusted | ||
|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | p | OR (95% CI) | p | ||||||
| MTHFR | rs2274976 | Codominant | A/A | 661 (87.3) | 604 (88.6) | 1.00 | 0.740 | 1.00 | 0.990 |
| A/G | 93 (12.3) | 75 (11) | 0.88 (0.64−1.22) | 0.98 (0.70−1.38) | |||||
| G/G | 3 (0.4) | 3 (0.4) | 1.09 (0.22−5.44) | 1.10 (0.21−5.70) | |||||
| Dominant | A/A | 661 (87.3) | 604 (88.6) | 1.00 | 0.470 | 1.00 | 0.940 | ||
| A/G‐G/G | 96 (12.7) | 78 (11.4) | 0.89 (0.65−1.22) | 0.99 (0.71−1.38) | |||||
| Recessive | A/A‐A/G | 754 (99.6) | 679 (99.6) | 1.00 | 0.900 | 1.00 | 0.910 | ||
| G/G | 3 (0.4) | 3 (0.4) | 1.11 (0.22−5.52) | 1.10 (0.21−5.70) | |||||
| Log‐addition | — | — | — | 0.90 (0.67−1.22) | 0.510 | 0.99 (0.72−1.36) | 0.960 | ||
| MTHFR | rs1801133 | Codominant | C/C | 234 (30.9) | 192 (28.3) | 1.00 | 0.430 | 1.00 | 0.076 |
| C/T | 361 (47.8) | 327 (48.2) | 1.10 (0.87−1.41) | 1.14 (0.88−1.47) | |||||
| T/T | 161 (21.3) | 160 (23.6) | 1.21 (0.91−1.62) | 1.42 (1.05−1.93) | |||||
| Dominant | C/C | 234 (30.9) | 192 (28.3) | 1.00 | 0.270 | 1.00 | 0.096 | ||
| C/T‐T/T | 522 (69) | 487 (71.7) | 1.14 (0.91−1.43) | 1.22 (0.96−1.55) | |||||
| Recessive | C/C‐C/T | 595 (78.7) | 519 (76.4) | 1.00 | 0.300 | 1.00 | 0.042* | ||
| T/T | 161 (21.3) | 160 (23.6) | 1.14 (0.89−1.46) | 1.31 (1.01−1.70) | |||||
| Log‐addition | — | — | — | 1.10 (0.95−1.27) | 0.190 | 1.19 (1.02−1.38) | 0.025* | ||
| MTR | rs1805087 | Codominant | A/A | 611 (80.9) | 577 (84.6) | 1.00 | 0.170 | 1.00 | 0.290 |
| A/G | 137 (18.1) | 101 (14.8) | 0.78 (0.59−1.03) | 0.86 (0.64−1.15) | |||||
| G/G | 7 (0.9) | 4 (0.6) | 0.61 (0.18−2.08) | 0.46 (0.13−1.65) | |||||
| Dominant | A/A | 611 (80.9) | 577 (84.6) | 1.00 | 0.065 | 1.00 | 0.210 | ||
| A/G‐G/G | 144 (19.1) | 105 (15.4) | 0.77 (0.59−1.02) | 0.83 (0.62−1.11) | |||||
| Recessive | A/A‐A/G | 748 (99.1) | 678 (99.4) | 1.00 | 0.460 | 1.00 | 0.240 | ||
| G/G | 7 (0.9) | 4 (0.6) | 0.63 (0.18−2.16) | 0.47 (0.13−1.69) | |||||
| Log‐addition | — | — | — | 0.78 (0.60−1.01) | 0.059 | 0.82 (0.63−1.08) | 0.160 | ||
| MTR | rs2853522 | Codominant | C/C | 247 (32.9) | 224 (32.9) | 1.00 | 0.780 | 1.00 | 0.950 |
| A/C | 377 (50.1) | 332 (48.8) | 0.97 (0.77−1.23) | 1.01 (0.79−1.29) | |||||
| A/A | 128 (17) | 125 (18.4) | 1.08 (0.79−1.46) | 1.05 (0.76−1.45) | |||||
| Dominant | C/C | 247 (32.9) | 224 (32.9) | 1.00 | 0.980 | 1.00 | 0.840 | ||
| A/C‐A/A | 505 (67.2) | 457 (67.1) | 1.00 (0.80−1.24) | 1.02 (0.81−1.29) | |||||
| Recessive | C/C‐A/A | 624 (83) | 556 (81.6) | 1.00 | 0.510 | 1.00 | 0.760 | ||
| A/C | 377 (50.1) | 332 (48.8) | 1.10 (0.84−1.44) | ||||||
| Log‐addition | — | — | — | 1.03 (0.88−1.19) | 0.730 | 1.02 (0.88−1.20) | 0.760 | ||
| MTRR | rs1801394 | Codominant | A/A | 388 (51.4) | 366 (53.7) | 1.00 | 0.600 | 1.00 | 0.600 |
| A/G | 305 (40.4) | 266 (39.1) | 0.92 (0.74−1.15) | 0.93 (0.74−1.17) | |||||
| G/G | 62 (8.2) | 49 (7.2) | 0.84 (0.56−1.25) | 0.82 (0.54−1.25) | |||||
| Dominant | A/A | 388 (51.4) | 366 (53.7) | 1.00 | 0.370 | 1.00 | 0.410 | ||
| A/G‐G/G | 367 (48.6) | 315 (46.3) | 0.91 (0.74−1.12) | 0.91 (0.73−1.14) | |||||
| Recessive | A/A‐A/G | 693 (91.8) | 632 (92.8) | 1.00 | 0.470 | 1.00 | 0.410 | ||
| G/G | 62 (8.2) | 49 (7.2) | 0.87 (0.59−1.28) | 0.84 (0.56−1.27) | |||||
| Log‐addition | — | — | — | 0.92 (0.78−1.08) | 0.310 | 0.92 (0.77−1.09) | 0.320 | ||
| MTRR | rs1532268 | Codominant | C/C | 581 (76.8) | 498 (73) | 1.00 | 0.150 | 1.00 | 0.280 |
| C/T | 157 (20.8) | 159 (23.3) | 1.18 (0.92−1.52) | 1.14 (0.88−1.49) | |||||
| T/T | 18 (2.4) | 25 (3.7) | 1.62 (0.87−3.00) | 1.55 (0.81−2.94) | |||||
| Dominant | C/C | 581 (76.8) | 498 (73) | 1.00 | 0.094 | 1.00 | 0.180 | ||
| C/T‐T/T | 175 (23.1) | 184 (27) | 1.23 (0.97−1.56) | 1.19 (0.92−1.52) | |||||
| Recessive | C/C‐C/T | 738 (97.6) | 657 (96.3) | 1.00 | 0.150 | 1.00 | 0.210 | ||
| T/T | 18 (2.4) | 25 (3.7) | 1.56 (0.84−2.89) | 1.50 (0.79−2.84) | |||||
| Log‐addition | — | — | — | 1.22 (0.99−1.49) | 0.058 | 1.18 (0.96−1.46) | 0.120 | ||
| MTRR | rs162036 | Codominant | A/A | 519 (68.7) | 477 (70) | 1.00 | 0.550 | 1.00 | 0.620 |
| A/G | 210 (27.8) | 187 (27.5) | 0.97 (0.77−1.22) | 0.97 (0.76−1.24) | |||||
| G/G | 26 (3.4) | 17 (2.5) | 0.71 (0.38−1.33) | 0.72 (0.37−1.40) | |||||
| Dominant | A/A | 519 (68.7) | 477 (70) | 1.00 | 0.590 | 1.00 | 0.640 | ||
| A/G‐G/G | 236 (31.3) | 204 (30) | 0.94 (0.75−1.18) | 0.95 (0.75−1.20) | |||||
| Recessive | A/A‐A/G | 729 (96.6) | 664 (97.5) | 1.00 | 0.290 | 1.00 | 0.340 | ||
| G/G | 26 (3.4) | 17 (2.5) | 0.72 (0.39−1.33) | 0.73 (0.38−1.40) | |||||
| Log‐addition | — | — | — | 0.92 (0.76−1.12) | 0.420 | 0.93 (0.76−1.14) | 0.480 | ||
Values of p were calculated by unconditional logistic regression adjusted for age and gender.
SNP: single nucleotide polymorphism; ORs: odds ratios; 95% CI: 95% confidence interval.
Value of p ≤ 0.05 indicates statistical significance.
3.4. Associations between haplotype analyses and GC risk
Haplotype associations were detected based on pairwise LD of MTHFR, MTR, and MTRR. D’ value was calculated for the LD of pairwise SNPs, and the adjacent SNPs with pairwise D’ = 0.98 were classified in the same block. LD is indicated by bright red (very strong: LOD ≥2, D’ = 1) and light red (LOD ≥2, D’ < 1). As shown in Figure 1, rs2274976 with rs1801133, and rs1805087 with rs2853522 consisted of one block in MTHFR and MTR, respectively. In MTR, a strong linkage of rs1532268 and rs162036 was detected. As shown in Table 4, we used the chi‐square test and logistic test to analyze the haplotype. Haplotype “GC” in the MTHFR gene was found to prominently increase the risk of GC (OR = 1.26, 95% CI, 1.07–1.47; p = 0.005). Other haplotypes did not display the correlativity.
Figure 1.
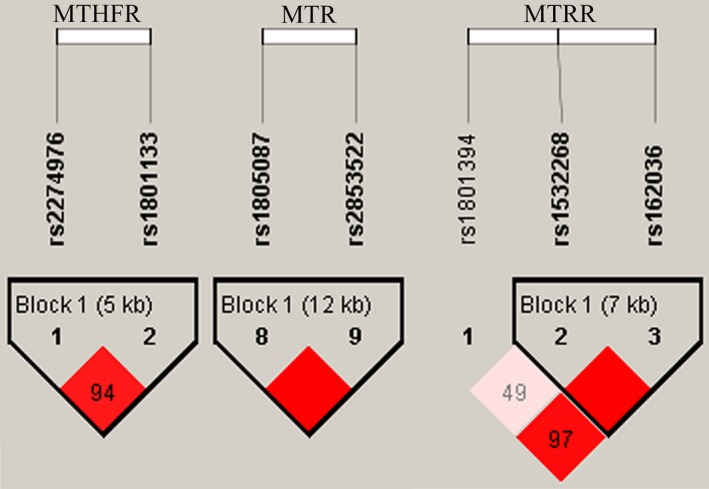
Linkage disequilibrium (LD) plots
Table 4.
Haplotype frequencies and the association with gastric cancer
| SNPs | Haplotype | Freq (case) | Freq (control) | Without adjusted | With adjusted | ||
|---|---|---|---|---|---|---|---|
| OR (95% CI) | p a | OR (95% CI) | P a | ||||
| rs2274976/rs1801133 | AC | 0.517 | 0.545 | 1.00 | — | 1.00 | — |
| GC | 0.424 | 0.390 | 1.16 (1.00–1.35) | 0.057 | 1.26 (1.07–1.47) | 0.005* | |
| GT | 0.052 | 0.062 | 0.84 (0.60–1.18) | 0.320 | 0.94 (0.66–1.33) | 0.710 | |
| rs1805087/rs2853522 | AC | 0.493 | 0.481 | 1.00 | — | 1.00 | — |
| AA | 0.427 | 0.420 | 0.99 (0.85–1.16) | 0.940 | 1.00 (0.85–1.17) | 0.990 | |
| GC | 0.080 | 0.100 | 0.78 (0.60–1.02) | 0.069 | 0.82 (0.62–1.09) | 0.170 | |
| rs1532268/rs162036 | CA | 0.684 | 0.699 | 1.00 | — | 1.00 | — |
| CG | 0.162 | 0.174 | 0.96 (0.78–1.17) | 0.660 | 0.96 (0.78–1.18) | 0.680 | |
| TA | 0.153 | 0.128 | 1.20 (0.98–1.48) | 0.078 | 1.17 (0.94–1.45) | 0.150 | |
SNP: single nucleotide polymorphism; ORs: odds ratios; 95% CI: 95% confidence interval.
Adjusted by sex and age.
Value of p ≤ 0.05 indicates statistical significance.
3.5. Stratification analysis
To further explore the gender influence on the potential GC susceptibility of selected polymorphisms in MTHFR, MTR, and MTRR genes, we performed the same statistical analysis in males and females, separately. In the allele model, stratified by gender, we discovered two SNPs (rs1805087 and rs1532268) were notably correlated with a decreased risk of GC in male, respectively. The rs1805087 in MTR, a protective factor, was associated with a decreased risk of GC (OR = 0.66, 95% CI, 0.48–0.91; p = 0.01), and on the contrary, the rs1532268 in MTRR showed an increased risk to the GC susceptibility (OR = 1.31, 95% CI, 1.02–1.68; p = 0.036) (Table 5 ).
Table 5.
Basic analysis of association between sex and the risk of gastric cancer
| SNP | Allele (Aa/B) | Male | Female | ||||||
|---|---|---|---|---|---|---|---|---|---|
| MAF (case) | MAF (control) | OR (95% CI) | p value | MAF (case) | MAF (control) | OR (95% CI) | p value | ||
| rs2274976 | G/A | 0.053 | 0.056 | 0.94 (0.64−1.38) | 0.758 | 0.081 | 0.082 | 0.98 (0.59−1.64) | 0.950 |
| rs1801133 | T/C | 0.462 | 0.440 | 1.09 (0.92−1.30) | 0.314 | 0.526 | 0.472 | 1.24 (0.94−1.64) | 0.131 |
| rs1805087 | G/A | 0.070 | 0.102 | 0.66 (0.48−0.91) | 0.010* | 0.114 | 0.096 | 1.21 (0.77−1.91) | 0.413 |
| rs2853522 | A/C | 0.422 | 0.419 | 1.01 (0.85−1.21) | 0.885 | 0.448 | 0.424 | 1.10 (0.83−1.46) | 0.503 |
| rs1801394 | G/A | 0.279 | 0.301 | 0.90 (0.74−1.09) | 0.281 | 0.227 | 0.252 | 0.87 (0.63−1.22) | 0.423 |
| rs1532268 | T/C | 0.159 | 0.127 | 1.31 (1.02−1.68) | 0.036* | 0.133 | 0.130 | 1.03 (0.68−1.56) | 0.887 |
| rs162036 | G/A | 0.163 | 0.167 | 0.97 (0.77−1.22) | 0.787 | 0.162 | 0.186 | 0.85 (0.58−1.23) | 0.385 |
Value of HWE‐p ≤ 0.05 was excluded. Value of p was adjusted by age.
SNP: single nucleotide polymorphism; MAF: minor allele frequency; ORs: odds ratios; 95% CI: 95% confidence interval.
Minor allele.
Value of p ≤ 0.05 indicates statistical significance.
In the genetic model (Table 6), no significant association was found in females. However, in male, the rs2274976 in MTHFR was correlated with 1.50‐fold increased risk of GC (OR = 1.50, 95% CI, 1.07–2.10; p = 0.018) in the recessive model, while the rs1805087 in MTR was correlated with 0.65‐fold decreased risk (OR = 0.65, 95% CI, 0.45–0.94; p = 0.021) in the dominant model. Besides, as for haplotype analysis, no remarkable SNPs were related neither in males nor in females.
Table 6.
Analysis of SNPs and gastric cancer risk in males and females based on logistic tests
| SNP ID | Model | Genotype | Male | Female | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Control, n (%) | Case, n (%) | OR (95% CI) | p | Control, n (%) | Case, n (%) | OR (95% CI) | p | |||
| rs1805087 (MTR) | Codominant | A/A | 394 (80.7) | 457 (86.7) | 1.00 | 0.063 | 216 (81.2) | 119 (77.3) | 1.00 | 0.520 |
| A/G | 88 (18.0) | 66 (12.5) | 0.66 (0.45−0.96) | 49 (18.4) | 35 (22.7) | 1.24 (0.76−2.04) | ||||
| G/G | 6 (1.2) | 4 (0.8) | 0.49 (0.12−1.96) | 1 (0.4) | 0 (0) | 0.00 (0.00‐NA) | ||||
| Dominant | A/A | 394 (80.7) | 457 (86.7) | 1.00 | 0.021* | 216 (81.2) | 119 (77.3) | 1.00 | 0.420 | |
| A/G‐G/G | 94 (19.3) | 70 (13.3) | 0.65 (0.45−0.94) | 50 (18.8) | 35 (22.7) | 1.22 (0.75−2.00) | ||||
| Recessive | A/A‐A/G | 482 (98.8) | 523 (99.2) | 1.00 | 0.350 | 265 (99.6) | 154 (100) | 1.00 | 0.450 | |
| G/G | 6 (1.2) | 4 (0.8) | 0.52 (0.13−2.08) | 1 (0.4) | 0 (0) | 0.00 (0.00‐NA) | ||||
| Log‐addition | 0.67 (0.48−0.94) | 0.019* | 1.20 (0.74−1.94) | 0.470 | ||||||
Value of p was adjusted by age.
SNP: single nucleotide polymorphism; ORs: odds ratios; 95% CI: 95% confidence interval.
Value of p ≤ 0.05 indicates statistical significance.
4. DISCUSSION
It is well documented that individual susceptibility to the development of cancer can vary. Despite the fact that the polymorphism in folate pathway was involved in carcinogenesis, the conclusions of case–control analysis were inconsistent among different ethnic groups. We researched the associations between SNPs on folate metabolism‐related genes—MTHFR, MTR, and MTRR—and the risk of GC. In view of the whole collected subjects, rs1532268 and rs1801133 were detected as risk factors. Haplotype GC constructed by polymorphisms rs2274976 and rs1801133 in MTHFR gene was associated with increased GC risk. In addition, by further stratification analysis, gender distribution had differences between the cases and controls. In male population, rs1532268 increased the GC risk and rs1805087 was recognized as a protective factor.
The correlation of a common polymorphism of MTHFR, rs1801133 (C677T), with GC risk has been analyzed repeatedly. In addition, its GC susceptibility was determined positively only in Asian individuals. A subgroup meta‐analysis by Lina Sun et al showed an increased risk in the Asians, but not in North American or European populations (Sun, Liang, Yuan, Jiangqi, & Jiang, 2014). Long Chen et al. identified a statistically significantly elevated risk of GC in Asian MTHFR C677T polymorphism populations. E Zintzaras carried out a 1,584/2,785 cases/controls study and recognized that evidence of the association was mainly in East Asian, and in Caucasians, the value was not significant. Additionally, the "T/T" genotype of rs1801133 was associated with a higher GC risk than the "C/C" genotype, which is similar to that reported in our case‐control studies (Zintzaras, 2006).
MTR encodes enzyme catalyzing the methylation of homocysteine to methionine with simultaneous conversion of 5‐methyl‐THF to tetrahydrofolate (THF), which is essential for DNA synthesis. The present study indicated rs1805087 as a predictor for GC. Evidences have been provided that G allele of MTR A2756G polymorphism was associated with multiple diseases, such as autism, head, and neck cancer (Galbiatti et al., 2010; Haghiri, Mashayekhi, Bidabadi, & Salehi, 2016). The GG genotype was reported to affect the DNA hypomethylation and individuals who carried 2756GG showed a lower frequency of CpG island hypermethylation in tumor suppressor genes (Goode, Potter, Bigler, & Ulrich, 2004; Zhao et al., 2013).
MTRR is a key enzyme in the homocysteine/methionine metabolic pathway. For functional polymorphism in MTRR, rs1801394 was commonly estimated. An interaction between MTRR A66G polymorphism (rs1801394) and colorectal cancer susceptibility was identified (Wang, Li, Wang, He, & Xi, 2017). An association between the A66G gene polymorphism and LC risk in a Turkish population was also been suggested (Aksoy‐Sagirli, Erdenay, Kaytan‐Saglam, & Kizir, 2017). Jae‐Young Yoo et al also reported that rs1801394 was associated with GC risk in Koreans by genotype analysis (Yoo et al., 2012; Yuan et al., 2012). Interestingly, in the current population, no positive association was observed, and the linkage of rs1532268 in MTRR and GC risk was first revealed in the Han Chinese population.
Nevertheless, there are limitations that need to be noticed in the current study. First of all, the inherent selection bias and information bias were inevitable problems, because the subjects of investigation were enrolled from the identical hospital. Second, the number of cases in our study was not large, which cannot preclude false‐negative results and cannot be extrapolated to other populations. So, larger sample size and further confirmation in other ethnic populations are needed for further verification. Despite the limitations mentioned above, the results of our present study provided scientific evidence of genes—MTHFR, MTR and MTRR—with the risk of GC in the future studies.
5. CONCLUSION
To sum up, the results of our study indicate that the rs1532268 and rs1801133 polymorphisms are potential risk factors for the development of GC in the Chinese population. rs1805087 decreased GC risk in Chinese males. For our assessment, no interaction appeared in female population. Published data demonstrated that the worldwide incidence of GC is higher in men than in women. The reason is unclear. Presumably, endogenous ovarian sex hormones may lower GC incidence in women (Duell et al., 2010; Yu, He, & Guo, 2014). Our current research is fundamental, and more functional studies are required to dig deeper.
CONFLICTS OF INTEREST
The authors have no conflicts of interest to disclose.
ACKNOWLEDGMENTS
We are grateful to the patients and control subjects for their participation in this study. We also thank the clinicians and hospital staff who obtained the blood samples and performed data collection for this study.
Wei L, Niu F, Wu J, et al. Association study between genetic polymorphisms in folate metabolism and gastric cancer susceptibility in Chinese Han population: A case–control study. Mol Genet Genomic Med. 2019;7:e633 10.1002/mgg3.633
Funding information
This study was funded by Shaanxi Normal University with funding from Central Universities (No. GK20170306), the Key Laboratory of Se‐enriched Products Development and Quality Control, Ministry of Agriculture/National‐Lacal Joint Engineering Laboratory of Se‐enriched Food Development (No. Se‐2018B03), and the National Natural Science Foundation of China (No. 60171009).
Contributor Information
Tianbo Jin, Email: jtb111111@163.com.
Yifei Wu, Email: wuyifei622@163.com.
REFERENCES
- Aksoy‐Sagirli, P. , Erdenay, A. , Kaytan‐Saglam, E. , & Kizir, A. (2017). Association of three single nucleotide polymorphisms in MTR and MTRR genes with lung cancer in a Turkish population. Genetic Testing and Molecular Biomarkers, 21(7), 428–432. 10.1089/gtmb.2017.0062 [DOI] [PubMed] [Google Scholar]
- Barrett, J. C. , Fry, B. , Maller, J. , & Daly, M. J. (2005). Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics, 21(2), 263–265. 10.1093/bioinformatics/bth457 [DOI] [PubMed] [Google Scholar]
- Chen, J. , Liu, W. , Cao, Y. , Zhang, X. , Guo, Y. , Zhu, Y. , … Wang, G. (2017). MMP‐3 and MMP‐8 single‐nucleotide polymorphisms are related to alcohol‐induced osteonecrosis of the femoral head in Chinese males. Oncotarget, 8(15), 25177 10.18632/oncotarget.15587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De, M. C. , Forman, D. , & Plummer, M. (2003). Gastric cancer: Epidemiology and risk factors. Journal of Surgical Oncology, 56(1), 1. [Google Scholar]
- Duell, E. J. , Travier, N. , Lujan‐Barroso, L. , Boutron‐Ruault, M. C. , Clavel‐Chapelon, F. , Palli, D. , … González, C. A. (2010). Menstrual and reproductive factors, exogenous hormone use, and gastric cancer risk in a cohort of women from the European Prospective Investigation Into Cancer and Nutrition. American Journal of Epidemiology, 172(12), 1384–1393. 10.1093/aje/kwq321 [DOI] [PubMed] [Google Scholar]
- Gabriel, S. , Ziaugra, L. , & Tabbaa, D. (2009). SNP genotyping using the Sequenom MassARRAY iPLEX platform. Current Protocols in Human Genetics, Chapter 2(Unit 2), Unit 2.12 10.1002/0471142905.hg0212s60 [DOI] [PubMed] [Google Scholar]
- Galbiatti, A. L. , Ruiz, M. T. , Biselli‐Chicote, P. M. , Raposo, L. S. , Maniglia, J. V. , Pavarino‐Bertelli, E. C. , & Goloni‐Bertollo, E. M. (2010). 5‐Methyltetrahydrofolate‐homocysteine methyltransferase gene polymorphism (MTR) and risk of head and neck cancer. Brazilian Journal of Medical and Biological Research, 43(5), 445–450. 10.1590/S0100-879X2010007500034 [DOI] [PubMed] [Google Scholar]
- González, C. A. , Sala, N. , & Rokkas, T. G. (2013). Gastric cancer: Epidemiologic aspects. Helicobacter, 18(Supplement s1), 34–38. 10.1111/hel.12082 [DOI] [PubMed] [Google Scholar]
- Goode, E. L. , Potter, J. D. , Bigler, J. , & Ulrich, C. M. (2004). Methionine synthase D919G polymorphism, folate metabolism, and colorectal adenoma risk. Cancer Epidemiology, Biomarkers & Prevention, 13(1), 157–162. 10.1158/1055-9965.EPI-03-0097 [DOI] [PubMed] [Google Scholar]
- Haghiri, R. , Mashayekhi, F. , Bidabadi, E. , & Salehi, Z. (2016). Analysis of methionine synthase (rs1805087) gene polymorphism in autism patients in Northern Iran. Acta Neurobiologiae Experimentalis, 76(4), 318–323. 10.21307/ane-2017-030 [DOI] [PubMed] [Google Scholar]
- Jae‐Young, Y. , Sook‐Young, K. , Jung‐Ah, H. , Hong, S. H. , Aesun, S. , Ju, C. I. , & Yeon‐Su, L. (2012). Association study between folate pathway gene single nucleotide polymorphisms and gastric cancer in Koreans. Genomics & Informatics, 10(3), 184 10.5808/GI.2012.10.3.184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, T. , Li, B. , He, N. , Zhang, Y. , Xia, R. , Kang, L. , … Yuan, D. (2016). CLPTM1L polymorphism as a protective factor for lung cancer: A case‐control study in southern Chinese population. Tumor Biology, 37(8), 1–6. 10.1007/s13277-016-4938-9 [DOI] [PubMed] [Google Scholar]
- Kumar, J. , Das, S. K. , Sharma, P. , Karthikeyan, G. , Ramakrishnan, L. , & Sengupta, S. (2012). Homocysteine levels are associated with MTHFR A1298C polymorphism in Indian population. Journal of Human Genetics, 50(12), 655–663. 10.1007/s10038-005-0313-1 [DOI] [PubMed] [Google Scholar]
- Li, J. , Ma, G. , Zhu, X. , Jin, T. , Wang, J. , & Li, C. (2017). Association analysis of telomere length related gene ACYP2 with the gastric cancer risk in the northwest Chinese Han population. Oncotarget, 8(19), 31144–31152. 10.18632/oncotarget.16097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, S. , Wang, M. , Liu, X. , Zhu, W. , Guo, Y. , Dai, Z. , … Dai, Z. (2017). Association of genetic polymorphisms in MIF with breast cancer risk in Chinese women. Clinical and Experimental Medicine, 17(3), 395–401. 10.1007/s10238-016-0439-9 [DOI] [PubMed] [Google Scholar]
- Liu, D. , Wang, M. , Tian, T. , Wang, X.‐J. , Kang, H.‐F. , Jin, T.‐B. , … Dai, Z.‐J. (2017). Genetic polymorphisms (rs10636 and rs28366003) in metallothionein 2A increase breast cancer risk in Chinese Han population. Aging, 9(2), 547–555. 10.18632/aging.101177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnami, S. , Sato, Y. , Yoshimura, K. , Ohnami, S. , Sakamoto, H. , Aoki, K. , … Yoshida, T. (2008). His595Tyr polymorphism in the methionine synthase reductase (MTRR) gene is associated with pancreatic cancer risk. Gastroenterology, 135(2), 477–488. 10.1053/j.gastro.2008.04.016 [DOI] [PubMed] [Google Scholar]
- Rafiei, E. , Mohammadian‐Hafshejani, A. , Towhidi, F. , Makhsosi, B. R. , & Salehiniya, H. (2016). Lack of any relationship of stomach cancer incidence and mortality with development in Asia. Asian Pacific Journal of Cancer Prevention, 17(8), 3777. [PubMed] [Google Scholar]
- Shrubsole, M. J. , Gao, Y. T. , Cai, Q. , Shu, X. O. , Dai, Q. , Jin, F. , & Zheng, W. (2006). MTR and MTRR polymorphisms, dietary intake, and breast cancer risk. Cancer Epidemiology Biomarkers & Prevention, 15(3), 586–588. 10.1158/1055-9965.EPI-05-0576 [DOI] [PubMed] [Google Scholar]
- Sun, L. , Liang, F. , Yuan, B. , Jiangqi, L. I. , & Jiang, L. (2014). A meta‐analysis on relationship of MTHFR C677T polymorphism and susceptibility to gastric cancer. Cancer Research on Prevention & Treatment, 41 10.3971/j.iSSN.1000-8578.2014.11.015 [DOI] [Google Scholar]
- Tian, T. , Wang, M. , Zheng, Y. , Yang, T. , Zhu, W. , Li, H. , … Dai, Z. (2018). Association of two FOXP3 polymorphisms with breast cancer susceptibility in Chinese Han women. Cancer Management and Research, 10, 867–872. 10.2147/cmar.s158433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, P. , Li, S. , Wang, M. , He, J. , & Xi, S. (2017). Association of MTRR A66G polymorphism with cancer susceptibility: Evidence from 85 studies. Journal of Cancer, 8(2), 266–277. 10.7150/jca.17379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, P. P. , Tang, R. N. , & An, L. (2014). A meta‐analysis of MTRR A66G polymorphism and colorectal cancer susceptibility. Journal of the Balkan Union of Oncology, 20(3), 918. [PubMed] [Google Scholar]
- Xia, P. , Li, B. , Geng, T. , Deng, Z. , Dang, C. , Chang, D. , … Chen, C. (2015). FGFR2 gene polymorphisms are associated with breast cancer risk in the Han Chinese population. American Journal of Cancer Research, 5(5), 1854. [PMC free article] [PubMed] [Google Scholar]
- Yang, L. (2006). Incidence and mortality of gastric cancer in China. World Journal of Gastroenterology, 12(1), 17 10.3748/wjg.v12.i1.17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo, J. Y. , Kim, S. Y. , Hwang, J. A. , Hong, S. H. , Shin, A. , Choi, I. J. , & Lee, Y. S. (2012). Association study between folate pathway gene single nucleotide polymorphisms and gastric cancer in Koreans. Genomics & Informatics, 10(3), 184–193. 10.5808/gi.2012.10.3.184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J. , He, Y. , & Guo, Z. (2014). Age trend of the male to female sex ratio in surgical gastric cancer patients at a single institution. World Journal of Surgical Oncology, 12, 269 10.1186/1477-7819-12-269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, L. J. , Jin, T. B. , Yin, J. K. , Du, X. L. , Wang, Q. , Dong, R. , … Lu, J. G. (2012). Polymorphisms of tumor‐related genes IL‐10, PSCA, MTRR and NOC3L are associated with the risk of gastric cancer in the Chinese Han population. Cancer Epidemiology, 36(6), e366–372. 10.1016/j.canep.2012.05.016 [DOI] [PubMed] [Google Scholar]
- Zhao, Y. , Chen, Z. , Ma, Y. , Xia, Q. , Zhang, F. , Fu, D. , & Wang, X.‐F. (2013). Lack of association between methionine synthase A2756G polymorphism and digestive system cancer risk: Evidence from 39327 subjects. PLoS ONE, 8(4), e61511 10.1371/journal.pone.0061511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zintzaras, E. (2006). Association of methylenetetrahydrofolate reductase (MTHFR) polymorphisms with genetic susceptibility to gastric cancer: A meta‐analysis. Journal of Human Genetics, 51(7), 618–624. 10.1007/s10038-006-0405-6 [DOI] [PubMed] [Google Scholar]


