Abstract
In the present review, we focus on the phenomenon of chromothripsis, a new type of complex chromosomal rearrangements. We discuss the challenges of chromothripsis detection and its distinction from other chromoanagenesis events. Along with already known causes and mechanisms, we introduce aberrant epigenetic regulation as a possible pathway to chromothripsis. We address the issue of chromothripsis characteristics in cancers and benign tumours, as well as chromothripsis inheritance in cases of its occurrence in germ cells, zygotes and early embryos. Summarising the presented data on different phenotypic effect of chromothripsis, we assume that its consequences are most likely determined not by the chromosome shattering and reassembly themselves, but by the genome regions involved in the rearrangement.
Keywords: chromothripsis, complex chromosomal rearrangements, epigenetics, cancer, benign tumour, chromosome pulverisation, constitutional chromothripsis
Introduction
Complex chromosomal rearrangements have been found since introduction of cytogenetic techniques. At present, due to development of new molecular-cytogenetic and molecular methods, the nature of CCRs became apparent making possible their classification.
The first documented CCR case was a translocation affecting three chromosomes in a child with mental retardation and associated dysmorphic features (Nuzzo et al., 1968). In 1970, a team of Lund University researchers discovered another translocation involving three, or possibly, four chromosomes and characterised it as a “complex translocation” and “complex rearrangement” (Fredga and Hall, 1970). Subsequently, complex chromosomal translocations were given the definition that currently extends to the term “CCRs”: complex chromosomal translocations involve more than a reciprocal exchange of segments between two chromosomes resulting in multiple derivative chromosomes (Pai et al., 1980). As molecular genetic techniques gained popularity, our understanding of the nature and origins of structural chromosomal abnormalities increased. As a result, the initial definition of CCRs is frequently updated in terms of the number of breakpoints and number of involved chromosomes. At present, CCRs are understood to be structural chromosomal abnormalities that arise as a result of three or more breakpoints in one or more chromosomes, with the exception of inter- and intrachromosomal insertions (Madan, 2013; McGowan-Jordan et al., 2016).
In January 2011, Stephens et al. (2011) published a paper on CCRs in chronic lymphocytic leukaemia. Using paired-end DNA sequencing, they revealed 42 rearrangements affecting chromosome 4 and several rearrangements affecting chromosomes 1, 12, and 15 in the tumour cells of one patient. The detected rearrangements were characterised not only by numerous breakpoints in a relatively short genome region but also by multiple deletions in the almost complete absence of duplications. Subsequently, when studying similar genome alterations in a small-cell lung cancer cell line (SCLC-21H), the authors observed the formation of double minutes from fragments of derivative chromosome 8 (Stephens et al., 2011). Notably, the rearranged chromosomes and the double minutes comprised material from only one of the homologous chromosomes, the other remaining intact. The authors suggested the term “chromothripsis” to describe this phenomenon (from the Greek “chromos” – “chromosome” – and “thripsis” – “shattering” into small fragments) (Stephens et al., 2011).
Importantly, apart from chromothripsis, over the last 7 years two more CCR types have been described: chromoanasynthesis and chromoplexy. The three types of aberrations are covered by the umbrella term “chromoanagenesis” (from the Greek “anagenesis” – “rebirth”), which indicates a structural chromosome reorganisation (Holland and Cleveland, 2012). It is believed, however, that chromothripsis differs from other chromoanagenesis phenomena by the mechanisms of its occurrence and the nature of genetic alterations (Poot, 2018).
Chromothripsis and Other Types of Chromoanagenesis
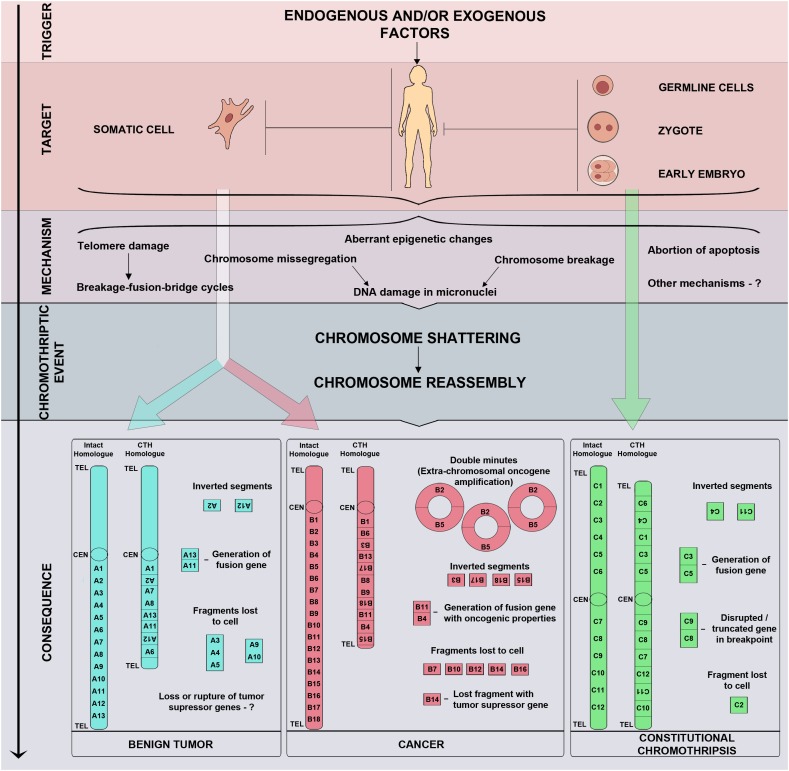
The results of whole genome sequencing, followed by mapping reads against a reference genome, lead us to believe that chromothripsis is based on the process of chromosome shattering triggered by double-strand DNA breaks (Stephens et al., 2011). The repair of double-strand breaks in a cell may occur through either a homologous recombination or NHEJ (reviewed in Ceccaldi et al., 2016). NHEJ is believed to be the primary repair mechanism in chromothripsis cases (Stephens et al., 2011). Once the DNA has been repaired through NHEJ, the reassembled chromosome may have errors in the order and orientation of segments. Fragments that do not ligate together with a centromere may be lost during subsequent cell divisions resulting in deletions (Figure 1; MacKinnon and Campbell, 2013). When double-strand breaks occur in two or more chromosomes, chromosome fragments may fuse, forming derivative chromosomes.
FIGURE 1.
Triggers, mechanisms, and consequences of chromothripsis. Chromothripsis may arise in any cell, including somatic cells, germline cells, zygotes, and blastomeres of preimplantation embryos, thus, determining the fate of an affected organ or the whole organism. Chromothripsis is induced by exogenous and/or endogenous factors which trigger chromosome shattering and sequential reassembly of fragments through micronuclei formation, breakage-fusion-bridge cycles, aberrant epigenetic regulation, abortive apoptosis, and other yet unknown mechanisms.
In theory, such CCRs may result from either chromosome pulverisation or sequential, independent rearrangements. The Monte-Carlo simulation method, which includes repeated random sampling and is traditionally used in stochastic process research, has established that the chromosome pulverisation model, which implies an absence of duplications, more accurately matches the genome alterations observed in chromothripsis. These data have given rise to an assumption that chromothripsis is the result of a single catastrophic event (Stephens et al., 2011).
The discovery of chromothripsis in the tumour cells of patients with chronic lymphocytic leukaemia was followed by a description of constitutional chromosomal rearrangements that are comparable with chromothripsis by number of breakpoints and breakpoint clustering but have different copy-number profiles. Microarray results have revealed that the karyotype of 17 patients with various developmental problems featured not only deletions but also multiple duplications and triplications, which could not have arisen as a result of NHEJ (Liu et al., 2011). This enabled the authors to hypothesise that such copy number alterations may result from replication and repair errors caused by DNA microhomology (MMBIR, microhomology-mediated break-induced replication; MMIR, microhomology/microsatellite-induced replication) (Payen et al., 2008; Hastings et al., 2009). Since chromothripsis does not fully reflect the characteristics of the observed genome alterations, the authors suggested replacing the term “chromothripsis” with “chromoanasynthesis,” which stands for chromosome reconstitution or chromosome reassortment (Liu et al., 2011).
In their review article on the hypothetical mechanisms and consequences of chromoanagenesis, Holland and Cleveland (2012) contrast the terms “chromoanasynthesis” and “chromothripsis.” According to the authors, chromoanasynthesis and chromothripsis are two independent phenomena with different underlying mechanisms. However, multiple chromosomal aberrations, which are observed in both, are most likely the result of a single catastrophic event, and not a successive series of rearrangements.
By contrast, chromoplexy, the third example of chromoanagenesis, is the result of an accumulation of chromosome rearrangements. The term “chromoplexy” (from the Greek “pleko” – “to weave”) was introduced in 2013 to indicate complex rearrangements of prostate cancer genomes (Baca et al., 2013). To analyse the results of whole-genome sequencing and microarray-based comparative genomic hybridisation (aCGH), the researchers developed the ChainFinder algorithm, which identifies the chained rearrangements that resulted in the CCRs. They demonstrated that, in the majority of samples (50 out of 57), multiple deletions and translocations occurred successively, which is uncharacteristic of either chromothripsis or chromoanasynthesis. Chromoplexy is also characterised by fewer breakpoints and a larger number of rearranged chromosomes (up to eight) compared to chromothripsis (Baca et al., 2013). Importantly, the breakpoints are presumably localised in open chromatin regions (Berger et al., 2011). Therefore, the high transcription level of certain loci may serve as a chromoplexy trigger.
Recent study showed a novel potential mechanism of chromoanagenesis: DNA polymerase 𝜃-dependent alternative homologous end joining (Masset et al., 2016). Thus, chromoanagenesis may be induced by a variety of mechanisms that lead to CCRs. In contrast to chromoplexy and chromoanasynthesis, chromothripsis is characterised by a larger number of breakpoints and a random order and orientation of chromosome segments after reassembly. Chromothripsis features a high frequency of deletions in the almost complete absence of duplications in localised genome regions. However, chromothripsis identification among the multitude of CCRs is challenged by a lack of distinct limitations on the number of breakpoints and other features. The authors suggest six criteria to distinguish chromothripsis from other CCRs (Korbel and Campbell, 2013):
-
simple 1.
Clustering of breakpoints;
-
simple 2.
Oscillation of copy number states between one and two which is consistent with mono- or disomy;
-
simple 3.
A prevalence of regions with interspersed loss and retention of heterozygosity;
-
simple 4.
A prevalence of rearrangements affecting a single haplotype, i.e., one of two homologous chromosomes;
-
simple 5.
Randomness of DNA fragment joins and order, and;
-
simple 6.
Ability to “walk” the derivative chromosome by joining breakpoints.
The authors used statistical algorithms to justify some of the criteria, but they did not report the minimal number of breakpoints, and admitted the possibility of partial tri- and tetrasomies (Korbel and Campbell, 2013).
Initially, CCRs with over 50 breakpoints were classified as chromothripsis (Stephens et al., 2011). However, this criterion was not always fulfiled in subsequent works. In a number of cases, rearrangements with 20 (Molenaar et al., 2012), 10 (Northcott et al., 2012; Rausch et al., 2012), or fewer (Chiang et al., 2012) breakpoints were treated as chromothripsis. Kinsella et al. (2014) drew attention to this issue in 2014. Using statistical simulation, they demonstrated that chromothripsis-like rearrangements may result from sequential rearrangement. Importantly, these results do not debunk the traditional hypothesis of the origins of chromothripsis but only emphasise the need for further research.
Methods of Chromothripsis Detection
It has been possible to describe the features of chromothripsis due to mate-pair sequencing and paired-end sequencing. These methods work for structural variant detection and CCRs, as well as genome assembly and de novo sequencing (Miller et al., 2010). In the case of a CCR, mate-pair and paired-end sequencing with subsequent verification by Sanger sequencing not only determine the precise localisation of breakpoints, but also gains data on nucleic acid sequences at breakpoint junctions (Gao and Smith, 2017). In spite of their high cost and challenging methodology, mate-pair sequencing and paired-end sequencing are widely used in chromothripsis studies.
Another efficient method of detecting and studying chromothripsis is microarray-based comparative genomic hybridisation (array CGH, aCGH), which is frequently referred to as “virtual karyotyping” or “chromosomal microarray analysis.” Copy number analysis allows detection of deletions, duplications, and other aberrations as well as identification of their precise genome localisation and size. The resolution of this method is sufficient to detect submicroscopic aberrations. For higher resolution and information capacity, aCGH is combined with a single nucleotide polymorphism (SNP) array (Keren, 2014). As a method, aCGH is not without considerable limitations: it cannot detect balanced structural chromosomal aberrations or determine the order and orientation of derivative chromosome segments (Balajee and Hande, 2018).
For detection and localisation of a specific DNA or RNA sequence on a chromosome or in a cell, fluorescence in situ hybridisation (FISH) is frequently used. In chromothripsis studies, various FISH techniques are used, each of them addressing specific aspects in the identification of the derivative chromosome structure. SKY and multicolour FISH (M-FISH), with the use of whole chromosome probes conjugated with different fluorochromes, enables identification of chromosomes involved in a rearrangement. The multicolour-banding FISH technique (MCB-FISH) is a segment-specific variant of chromosome banding that allows one to determine the structure of an aberrant chromosome (Balajee and Hande, 2018). To map breakpoints on the chromosomes, locus-specific probes with known cytogenetic localisation may be used for FISH. A combination of SKY and in situ hybridisation with fluorescent locus-specific probes is used to determine the precise structure not only of derivative chromosomes but also of double minutes (Stephens et al., 2011).
In patients with hereditary diseases, chromothripsis may be detected by a conventional karyotyping of metaphases from peripheral lymphocytes. This technique allows identification of numerical and structural chromosomal abnormalities including translocations and inversions, which are frequently observed in CCR cases. However, the complex nature of CCRs makes their interpretation by conventional karyotyping alone difficult. Therefore, to precisely determine the structure of rearrangements in chromothripsis, it is necessary to use a complex approach that includes classical chromosome banding, visualisation of the aberrations on metaphase chromosomes by FISH and molecular genetic techniques.
Causes and Mechanisms of Chromothripsis
The first assumptions regarding the mechanisms of chromothripsis were made by Stephens et al. (2011). The authors argue that DNA junction sequences and their localisation in the genome attests to chromosome pulverisation during mitosis at the stage of their highest condensation, not at the interphase stage. Today, several presumed causes of chromothripsis are listed (Meyerson and Pellman, 2011; Forment et al., 2012; Jones and Jallepalli, 2012; Maher and Wilson, 2012).
DNA Damage in Micronuclei
The most accepted hypothesis of chromothripsis occurrence is chromosome pulverisation in micronuclei. Chromosomes and their acentric fragments that lag during segregation in mitosis may be incorporated in a nuclear envelope outside of the main nucleus, which leads to the formation of micronuclei (Leibowitz et al., 2015). Certain features of the micronuclear envelope facilitate the access of cytoplasmic nucleases to the DNA (Géraud et al., 1989; Terradas et al., 2016). Micronuclei are characterised by abnormalities in chromatin condensation, which may lead to chromosome breaks (Terzoudi et al., 2015; Zhang et al., 2015). Experimental studies have shown the possibility of chromosome fragmentation and the formation of double minutes in micronuclei (Crasta et al., 2012; Hatch and Hetzer, 2015; Terradas et al., 2016). Using SKY, the authors determined that the majority of metaphases from cells with micronuclei feature multiple small fragments from one or two chromosomes (Crasta et al., 2012). The experiment on chromosome Y centromere inactivation also shed light on certain details of chromothripsis in micronuclei (Ly et al., 2017). The missegregated chromosome Y was included in a micronucleus and fragmented as a result of premature chromatin condensation. After the DNA breaks were repaired through NHEJ, the re-ligated chromosome Y showed typical characteristics of chromothripsis. It has been established that chromothripsis in micronuclei results from chromosome missegregation, their fragmentation, and the repair of breaks that occur during three cell cycles (Ly et al., 2017).
Aborted Programmed Cell Death
The abortion of apoptosis is regarded as one of the causes of chromothripsis (Tubio and Estivill, 2011; Tang et al., 2012). The first data on the association of chromothripsis with mutations of TP53, which encodes p53 protein – the key apoptosis regulator – were obtained in 2012. Chromothripsis was detected in Sonic Hedgehog (SHH) medulloblastoma cells in a patient with hereditary Li-Fraumeni syndrome (a germline mutation of TP53) (Rausch et al., 2012). In acute and chronic lymphocytic leukaemia, TP53 mutations may co-occur with chromothripsis in tumour cells (Pei et al., 2012).
In 2015, the occurrence of chromothripsis in TP53-/- cells after doxorubicin treatment on a cell-based model system was confirmed (Mardin et al., 2015). Observing a higher frequency of chromothripsis in hyperploid medulloblastomas, as compared to diploid ones, the authors established an association between cell hyperploidisation and chromothripsis. In this regard, it has been suggested that hyperploidisation may serve as a risk factor for chromothripsis (Mardin et al., 2015).
Telomere Shortening and Formation of Dicentric Chromosomes
Highly localised rearrangements in chromothripsis can also be explained by breakage-fusion-bridge cycles in dicentric chromosomes, which arise from DNA damage or telomere fusion caused, in turn, by telomere shortening or loss (Stephens et al., 2011; Sorzano et al., 2013). When dicentric chromosomes segregate during mitosis, chromatin bridges are formed and undergo subsequent rupturing (McClintock, 1939). Having induced the formation of an envelope with an aberrant structure, the chromatin bridge is destroyed by cytoplasmic 3′-exonuclease TREX1 (Maciejowski et al., 2015; Maciejowski and de Lange, 2017). This may result not only in multiple losses and inversions of chromosome segments but also in the formation of double minutes. Breakage-fusion-bridge cycles may co-occur with fragment amplification, as demonstrated on regions of chromosome 21 (iAMP21) in a dicentric chromosome formed as a result of a Robertsonian translocation of chromosomes 15 and 21 (Li et al., 2014). The risk of iAMP21 acute lymphoblastic leukaemia in carriers of rob (15;21) is assessed to be ∼2700 times higher than in the population (Li et al., 2014). The presence of breakage-fusion-bridge cycles in cells with chromothripsis has also been demonstrated in studies of cancer genome alterations (Nones et al., 2014; Maciejowski et al., 2015; Ernst et al., 2016). These cycles, however, may be a part of neochromosome evolution and therefore, considering that neochromosomes arise through chromothripsis, may be the consequence, not the cause, of the phenomenon (Garsed et al., 2014).
Chromosome Pulverisation Caused by Exogenous Factors
Chromosome pulverisation is an extreme example of DNA fragmentation. Multiple double-strand breaks in the DNA may result from exposure to a range of DNA-damaging agents including drugs, therapeutic or environmental ionising radiation, oxidative stress and virus infections.
Despite the initial suggestion that ionising radiation may induce chromothripsis, experimental proof was not obtained until several years later. In their experiments, Morishita et al. (2016) used a focused vertical microbeam system designed to irradiate a spot within the nuclei – the Single Particle Irradiation system to Cell (SPICE) – on oral squamous-cell carcinoma cells. The authors then established irradiated monoclonal sublines from them and analysed genome abnormalities using SKY and SNP array. One of the 46 monoclonal sublines showed chromothripsis-like complex chromosomal alterations with 14 breakpoints. The involvement of 10 chromosomes in the rearrangement is explained by the exposure of the interphase nuclei to a powerful particle beam. The authors presume that cell irradiation during mitosis may induce chromosome missegregation and, as a result, lead to micronuclei formation (Morishita et al., 2016).
Another potential cause of chromothripsis is chromosome pulverisation in viral infections. A connexion has been established between chromosome pulverisation and fragmentation and infection of cell cultures with measles, herpes zoster, herpes simplex, and adenovirus types 4, 12, and 18 (Benyesh-Melnick et al., 1964; Nichols et al., 1965; O’Neill and Miles, 1970; Peat and Stanley, 1986). In addition, herpes simplex may induce cell polyploidisation, which is also a risk factor for chromothripsis (Chenet-Monte et al., 1986; Mardin et al., 2015). Tumour cells infected with the Epstein-Barr virus have an increased level of both transmissible and unstable chromosomal abnormalities (dicentric chromosomes, chromatid fragments, ring chromosomes, double minutes, satellite associations of acrocentric chromosomes, and chromatin breaks) (Kamranvar et al., 2007). However, only one of the studies (Schütze et al., 2016) confirms the association of chromothripsis with viral infections. In human foreskin keratinocytes culture infected with human papillomavirus, chromothripsis-like complex chromosomal alterations within chromosome 8 occurred after passage 30, were detected at passage 40, and resulted in a gain of MYC. Concurrently, immortalisation of the cell line in vitro with non-transformed phenotype was observed (Schütze et al., 2016).
While the listed causes of chromothripsis appear to be the most likely, it is necessary to consider other possible contributing factors such as mutations in DNA repair genes or abnormal chromatin condensation.
Aberrant Epigenetic Patterns as a Cause of Chromosome Damage
Chromothripsis is characterised by a high frequency of deletions, translocations and inversions (Stephens et al., 2011). These chromosomal aberrations result from multiple double-strand breaks (DSBs) possibly occurring during M or G1 phase. DSBs are most probably repaired by error-prone NHEJ or microhomology-mediated end joining (MMEJ) mechanisms (Jones and Jallepalli, 2012). In most cases, very short or no microhomology in the chromothripsis breakpoint junctions can be found (Stephens et al., 2011; Chiang et al., 2012; Kloosterman et al., 2012; Malhotra et al., 2013; Weckselblatt et al., 2015; Aristidou et al., 2018; Slamova et al., 2018). However, in a few cases of CC, DSBs were found in high-copy repeats (Nazaryan et al., 2014; Nazaryan-Petersen et al., 2016).
The chromatin conformation is of importance for occurrence of spontaneous DSBs. The transition from closed to open chromatin, which is necessary for transcription, makes DNA vulnerable to damage (Kuo, 1981; Falk et al., 2008; Meschini et al., 2015). Chromatin looping facilitates DNA cleavage by nucleases, including endogenous ones originating from transposable elements (Maniotis et al., 2005). In the study on the events involved in the occurrence of stably segregating CC, DNA cleavage by catalytically active L1-endonuclease and translocations between distally located DNA regions were explained by Alu-mediated chromatin looping (Nazaryan-Petersen et al., 2016). Enhanced activation of transposable elements is associated with a response to environmental change and as well as with syndromes caused by MeCP2 (methyl-CpG binding protein 2; involved in transcription regulation) and ATM (ataxia telangiectasia, mutated; involved in DNA repair machinery) mutations (Bundo et al., 2014).
A key role in the regulation of chromatin structure belongs to epigenetic mechanisms: DNA methylation, histone variants, and non-coding RNAs (Geiman and Robertson, 2002; Li, 2014). Both tumorigenesis and cell differentiation including embryonic and germline cells are characterised by extensive epigenetic changes (Yamaguchi et al., 2013; Efimova et al., 2015, 2017, 2018; Avgustinova and Benitah, 2016; Atlasi and Stunnenberg, 2017; Pendina et al., 2018). Epigenetic machinery provides fast response to environmental change through gene-specific and/or genome-wide alterations of DNA methylation with subsequent change in expression patterns of genes coding proteins and regulatory RNAs (Wang et al., 2017; West, 2017). Abnormal DNA methylation also may compromise genome integrity. In vivo increase of chromosome aberrations has been documented in tissues with reduced global DNA methylation caused by ionising radiation (Lee et al., 2015), oxidative stress (Tunc and Tremellen, 2009), or deregulated DNMTs (Gaudet et al., 2003). In blood cells of ICF patients having DNMT3a mutation, hypomethylation of 1q, 9q, 16q heterochromatin regions is associated with abnormal chromatin looping, telomeric associations, anaphase bridges, lagging chromosomes, chromosome breakage and micronuclei formation (Gisselsson et al., 2005). In addition, hypomethylation of pericentromeric heterochromatin may trouble kinetochore orientation and spindle attachment, resulting in chromosome missegregation and micronuclei formation (Luzhna et al., 2013). Thus, aberrant DNA methylation contributes to abnormal chromatin compaction and, as a consequence, to DNA damage.
The involvement of epigenetic mechanisms in the pathway between damaging agents and genome integrity has been established in the studies of the radiation-induced bystander effect. The bystander effect is a phenomenon whereby irradiated cells communicate damage to non-irradiated nearby bystander cells, thus destabilising their genome and contributing to carcinogenesis (Koturbash et al., 2007). In rodents, localised X-ray exposure modifies expression of DNA methyltransferases and 5-methylcytosine-binding protein MeCP2 genes leading to global hypomethylation both in irradiated and non-irradiated tissues in vivo (Koturbash et al., 2006, 2007; Tamminga et al., 2008). DNA damage in non-irradiated bystander tissues is associated with induction of apoptosis (Koturbash et al., 2008; Kovalchuk et al., 2010; Cordelli et al., 2012). Recent advances in bystander effect aetiology assumed that communication between irradiated and non-irradiated cells involves numerous microRNAs (Xu et al., 2015; Yuan et al., 2016; Cai et al., 2017). In addition to microRNAs, cell-free chromatin released from radiation-induced dying cells is involved in extensive chromosome instability of bystander cells (Kirolikar et al., 2018).
Summarising the abovementioned issues, it could be assumed that activation of the cellular mechanisms involved in the chromothripsis formation by exogenous and/or endogenous insult is epigenetically mediated. However, lack of experimental evidence directly linking disruption of epigenetic regulation to the initiation of chromothripsis substantiates further studies in this field.
Chromothripsis and Neoplasia
In 2015, ChromothripsisDB1 database was created (Yang et al., 2016) to categorise cases of chromothripsis in human and model organisms by disease, research method, and criteria that enabled the authors to classify the observed chromosomal abnormalities as chromothripsis. As of March 2018, the database counted 500 chromothripsis cases across 46 cancers. The authors of ChromothripsisDB update it on a regular basis and standardise the information on all the rearrangements that are treated as chromothripsis (Cai, 2018). At present, ChromothripsisDB is the most informative source of information for accessing and comparing the results of chromothripsis studies.
Chromothripsis in Cancers
Chromothripsis is typical for 2–3% of cancer types (Stephens et al., 2011). As of today, chromothripsis has been observed in blood cancers, central nervous system cancers, soft tissue tumours, and carcinomas (Rode et al., 2016).
The frequency of chromothripsis varies across tumour entities (Table 1). Chromothripsis occurs most frequently in bone cancers – osteosarcoma and chordoma (Stephens et al., 2011). It is associated with advanced stages of the disease and poor clinical outcomes (Forment et al., 2012). At times, chromothripsis is coupled with additional mutations in tumour cells, for instance, IDH mutations (Cohen et al., 2015). In addition, the occurrence of chromothripsis in cancers is considerably higher in patients with inherited genetic disorders that are linked to cell-cycle and DNA repair gene mutations: Li-Fraumeni and Louis-Bar syndromes (Rausch et al., 2012; Ratnaparkhe et al., 2017). The risk of chromothripsis also varies across different genome regions: chromosomes 17, 8, 12, and 11 are the most likely to be involved in such rearrangements. As it appears, the highest frequency of chromothripsis in chromosome 17 is predetermined by the presence of the TP53 gene in its short arm (Cai et al., 2014).
Table 1.
Types of cancer with the highest occurrence of chromothripsis.
| References | Cancer type | Cases with chromothripsis/ total number of cases | Chromothripsis frequency |
|---|---|---|---|
| Rausch et al., 2012 | SHH medulloblastoma with mut TP53 | 10/10 | 100% |
| Rausch et al., 2012 | SHH medulloblastoma with wt TP53 | 0/22 | 0% |
| Northcott et al., 2012; Rausch et al., 2012 | Medulloblastoma, all subgroups | 13/98; 139/1087 | 13% |
| Li et al., 2014 | Acute lymphoblastic leukaemia with iAMP21 | 8/9 | 89% |
| Morrison et al., 2014 | Invasive bladder carcinoma | 81/150 | 60% |
| Zemanova et al., 2014 | Myelodysplastic syndrome with CCR | 77/157 | 49% |
| Rausch et al., 2012 | Acute myeloid leukaemia with mut TP53 | 8/17 | 47% |
| Rausch et al., 2012 | Acute myeloid leukaemia with wt TP53 | 1/91 | 1% |
| Przybytkowski et al., 2014 | High-risk breast cancer | 12/29 | 41% |
| Malhotra et al., 2013 | Grade IV glioma (glioblastoma) | 7/18 | 39% |
| Cohen et al., 2015 | Grade IV glioma with mut IDH | 9/24 | 37% |
| Cohen et al., 2015 | Grade II–III glioma | 5/45 | 11% |
| Malhotra et al., 2013 | Lung adenocarcinoma | 2/6 | 33% |
| Stephens et al., 2011 | Osteosarcoma | 3/9 | 33% |
| Nones et al., 2014 | Esophageal adenocarcinoma | 40/123 | 32% |
iAMP, amplification of a chromosome 21 region; IDH, isocitrate dehydrogenase; SHH, Sonic Hedgehog.
Chromothripsis in Benign Tumours
Chromothripsis does not occur exclusively in malignant tumours; cases of chromothripsis have been observed in benign tumours as well. The year 2013 brought the first descriptions of chromothripsis in uterine leiomyoma (also called uterine fibroid) cells – a benign tumour of the uterine myometrium, which is characterised by a high frequency of chromosomal abnormalities. By various estimates, chromothripsis occurs in 13–42% of uterine fibroids (Mehine et al., 2013; Holzmann et al., 2014; Mehine et al., 2014).
Unlike malignant tumours, chromothripsis in uterine fibroid cells is characterised by fewer breakpoints (20 or more) and a larger number of affected chromosomes (up to four) (Figure 1). Such aberrations are normally observed in uterine fibroids without fibroid-specific MED12 (mediator complex subunit 12) and FH (fumarate hydratase) mutations. They do not feature TP53 mutations or histological signs of malignancy (Mehine et al., 2013; Holzmann et al., 2014; Mehine et al., 2014; Pendina et al., 2017). Furthermore, chromothripsis with large deletions (from 43 to 13,647 kbp) has been observed in non-cultured sample of uterine fibroid which demonstrated normal karyotype in culture conditions (Holzmann et al., 2014). This could be associated with a lower proliferative potential of tumour cells with chromothripsis in vitro. However, a case of unbalanced chromothripsis has been observed in both the cultured and non-cultured fibroid cells (Pendina et al., 2017). It is likely that the ability of fibroid cells with chromothripsis to proliferate in vitro is determined not so much by the size of deletions and number of breaks as by the genomic loci involved in rearrangement. It should be noted, however, that the absence of malignisation signs in fibroids with chromothripsis by no means implies that their growth and malignant potential does not require thorough study.
Constitutional Chromothripsis as a Consequence of Genome Damage in Germ Cells and Preimplantation Embryos
Chromothripsis may also be a constitutional karyotype abnormality caused by chromosome damage in germline cells or preimplantation embryos. Cases of CC are extremely rare and usually coincide with congenital malformations or reproductive failure in the patient (Table 2; Kloosterman et al., 2011;de Pagter et al., 2015). In the virtually complete absence of any genetic imbalance, CC may co-occur with breakage of multiple genes or changes in their expression (Table 2; van Heesch et al., 2014; de Pagter et al., 2015; Bertelsen et al., 2016; Middelkamp et al., 2017). CC may include structural chromosomal abnormalities associated with genetic disorders (Table 2; Fontana et al., 2014; Genesio et al., 2015; Kurtas et al., 2018). In this case, the patient displays symptoms of an inherited disease. However, certain non-specific phenotypical features complicate the diagnosis and prognosis of the clinical outcome of the CC (Table 2).
Table 2.
Clinical outcomes of constitutional chromothripsis.
| References | Chromosome regions involved in chromothripsis | Chromothripsis detection method | Imbalance (size, copy number alterations) | Affected genes | Phenotype of a carrier(s) |
|---|---|---|---|---|---|
| Bertelsen et al., 2016; Nazaryan-Petersen et al., 2016 | 3q22.3-q23 5q23.1 |
Conventional cytogenetics, mate-pair sequencing | Four deletions (2–110 kb) | Truncated genes: PPP2R3A, CLDN18, A4GNT, DBR1, HSD17B4, ATR Fusion genes: CLDN18-HSD17B4, HSD17B4-DBR1 Deleted genes: DZIP1L |
No apparent association with a disorder |
| Anderson et al., 2016 | 13q33.1-q33.3 Xp11.22-p21.3 Xq21.31-q22.1 |
Conventional cytogenetics, FISH, aCGH | 10 deletions (327 kb – 8 Mb): a total 4.4 Mb of chr. 13 material and 28.1 Mb of chr. X material | Deleted genes: Chr. 13 – ERCC5, SLC10A2 Chr. X – IL1RAPL1, DMD, GK, NROB1, CYBB, OTC, RPGR, TSPAN7, XK, ATP6AP2, BCOR, CASK, CFP, KDM6A, MAOA, NDP, NYX, RBM10, RP2, SYN1, UBA1, USP9X, ZNF81, BMP15, CACNA1F, CLCN5, FOXP3, HSD17B10, IQSEC2, KDM5C, PHF8, FGD1, HUWE1, HSD17B10, DIAPH2, SRPX2 |
Developmental delay and dysmorphism |
| Weckselblatt et al., 2015 | 1q21 4q31 7p14.3 15q22 |
Conventional cytogenetics, FISH, targeted sequencing | 530-kb deletion of chr. 1 material; 4,2-Mb duplication of chr. 7 material | No disrupted genes by the breakpoints | Developmental delay, autism, intellectual disability, and/or congenital anomalies |
| 3q25-q26 8q23 9p22-p24 11p14.1 3q21.1 |
Conventional cytogenetics, FISH, WGS | Mb-sized deletions of chr. 8 and 9 material; a total of 99 bp deleted of other chromosomes material | Disrupted genes by the breakpoints: PTPRD, SH3GL2 | Developmental delay, autism, intellectual disability, and/or congenital anomalies | |
| 2q32-qter 3q13 7q21.11-q22.1 10q21.3 11q14.1 |
Conventional cytogenetics, FISH, WGS | 800-kb deletion of chr. 7 material, 2.2-Mb deletion of chr. 11 material; in addition, there are 55 total bp deleted at breakpoint junctions on other chromosomes | Disrupted genes by the breakpoints: GRM3, KPNA1, DLG2, CACNA2D1, GULP1, COL5A2, KCNH7, PCLO, TRRAP | Developmental delay, autism, intellectual disability, and/or congenital anomalies | |
| Nazaryan et al., 2014 | 2p16.1-p22.1 5p14.2-p15.2 7p21.3-q31.1 |
Conventional cytogenetics, FISH, mate-pair sequencing | No copy number alterations | Truncated genes: CDH12, DGKB, FOXP2 | Global developmental and psychomotor delay, severe speech disorder |
| Gamba et al., 2015 | 1p36.33-p35.3 | Conventional cytogenetics, aCGH | Five deletions: 0.83, 0.94, 1.4, 1.7, 3.7 Mb 1 duplication: 5.9 Mb | No data | Multiple congenital malformations presenting some features overlapping the 1p36 deletion phenotype |
| Gu et al., 2013 | 5p13.3-p15.33 7p22 7q32 11q23 21q21 |
Conventional cytogenetics, FISH, aCGH | No copy number alterations | No data | Phenotypically normal |
| 5p13.3-p15.33 11q23 |
Conventional cytogenetics, FISH, aCGH | Three deletions: 2.89, 0.56, and 3.21 Mb | Deleted genes: LOC340094, ADAMTS16, KIAA0947, FLJ33360, MED10, UBE2QL1, LOC255167, NSUN2, SRD5A1, PAPD7, MIR4278 | Phenotypically normal | |
| 5p13.3-5p15.33 | Conventional cytogenetics, FISH, aCGH | ∼26.22-Mb deletion | No data | Developmental delay, dysmorphic and autistic features | |
| Kloosterman et al., 2011 | 1p32.3 4q24 10q21.1 |
Conventional cytogenetics, SNP array, mate-pair sequencing | Small deletions and duplications (<50 bp) | Disrupted gene: PCDH15 | Severe psychomotor retardation, speech delay, hypertelorism and kyphoscoliosis |
| Slamova et al., 2018 | 1q23-q25 6q15-q24 14q13? 18p11.2-p11.3 18q11.2 |
Conventional cytogenetics, FISH, aCGH, mate-pair sequencing | Two deletions: 0.7 and 2.5 Mb | Deleted genes: DNM3, PIGC, C1ORF105, SUCO, NMBR, VTA1, ADGRG6, HIVEP2, AIG1, ADAT2, PEX3, FUCA2, PHACTR2, LTV1, ZC2HC1B, PLAGL1, SF3B5, STX11, UTRN, PAX9 Disrupted genes by the breakpoints: FILIP1, PHIP, HMGN3, AK097143, GAREM |
Developmental and growth delay |
| Wang et al., 2015 | 19p13.13-p13.2 19p12 19q12 19q13.11-q13.12 |
Conventional cytogenetics, FISH, aCGH | Four duplications: 4.3, 0.98, 1.12, and 5.13 Mb | No data | Subtle dysmorphic features |
| Macera et al., 2015 | 3p24.3 5q14 7q35 9p23 18p11.31 18q21.31 |
Conventional cytogenetics, FISH, SNP array, NGS | No loss or gain of chromosomal material at any of the breakpoints | Disrupted genes by the breakpoints: CNTN6, TBC1D5, CNTNAP2, PTPRD, L3MBTL4, LOC1001304840, WDR7 | Bilateral ventriculomegaly (13 and 15 mm), colpocephaly, with partial agenesis of the corpus callosum, and an absent left kidney and small right kidney |
| Kurtas et al., 2018 | 22q13.1-q13.3 | Conventional cytogenetics, FISH, aCGH, WGS, WES | Two duplications: 2.4 Mb, 148 kb 1 deletion: 8.4 Mb | Disrupted genes by the breakpoints: EP300, NFAM1, MYO18B, GTPBP1 | Phelan-McDermid syndrome |
| Genesio et al., 2015 | 9p21-q31 | Conventional cytogenetics, FISH, aCGH | Two deletions: 176.56 kb, 7.44 Mb | Deleted genes: RORB, TRPM6, NMRK1, OSTF1, GNAQ, and the critical region of the 9q21.13 deletion syndrome | Platelet disorder and thyroid dysfunction in addition to the classical neurobehavioral phenotype of the 9q21.13 microdeletion syndrome |
| Del Rey et al., 2016 | 2q34-q37.3 | Conventional cytogenetics, FISH, HR-CGH, MLPA | Deletion: 2.58 Mb duplication of 2q34q37.2 | Deleted genes: K1F1A, PASK, HDLBP, FARP2 | Multiple congenital disorders and intellectual disability |
| Fontana et al., 2014 | 1q41 1q43 9p24.3 21q22.12 |
Conventional cytogenetics, aCGH | Four deletions: 5.23, 1.33, 0.15871, and 0.826 Mb | Deleted genes: SMYD2, PTPN14, CENPF, KCNK2, KCTD3, USH2A, ESRRG, SPATA17, RRP15, TGFB2, CHRM3, KANK1, RCAN1, CLIC6, RUNX1 | Loeys–Dietz syndrome, type 4; borderline mental impairment |
| Kurtas et al., 2019 | 3q22.3-q26.2 | Conventional cytogenetics, FISH, aCGH, paired-end sequencing | Deletion: 6.8 kb | Disrupted genes by the breakpoints: ROPN1B, NAALADL2, TF | Healthy |
| 3q22.3-q26.2 | Conventional cytogenetics, FISH, aCGH, paired-end sequencing | Duplication: 10 Mb deletion: 5 Mb | Disrupted genes by the breakpoints: ROPN1B, NAALADL2, TF | Multiple phenotypic abnormalities and psychomotor delay | |
| chr. 6 14q31.3 |
Conventional cytogenetics, FISH, aCGH, paired-end sequencing | Two deletions: 5.3 and 3.7 kb | Disrupted genes by the breakpoints: OPRM, RNGTT | Healthy | |
| chr. 6 14q31.3 |
Conventional cytogenetics, FISH, aCGH, paired-end sequencing | Deletion: 1 Mb | Disrupted genes by the breakpoints: OPRM, RNGTT | Healthy | |
| 15q15.1 6p21.3-p25.1 6q14.2 6q21-q22.31 7q32.3 |
Conventional cytogenetics, FISH, SNP-CGH array, paired-end sequencing | Deletion: 6 kb | Disrupted genes by the breakpoints: CASC5, RPF2, CHCHD3, CLVS2 | Healthy | |
| 15q15.1 6p21.3-p25.1 6q14.2 6q21-q22.31 7q32.3 |
Conventional cytogenetics, FISH, SNP-CGH array, paired-end sequencing | Four deletions up to 100 bp 6-bp microduplication | Disrupted genes by the breakpoints: CASC5, RPF2, CHCHD3, CLVS2. One parental breakpoint junction is absent | Developmental and speech delay, dysmorphic features |
aCGH, array comparative genomic hybridisation; FISH, fluorescent in situ hybridisation; HR-CGH, high resolution comparative genomic hybridisation; MLPA, multiplex ligation-dependent probe amplification; NGS, next generation sequencing; SNP array, single nucleotide polymorphism array; WGS, whole genome sequencing; WES, whole exome sequencing.
Constitutional chromothripsis carriers may transmit the rearrangement to the offspring either stably or with de novo events (Gu et al., 2013; Weckselblatt et al., 2015; Bertelsen et al., 2016; Nazaryan-Petersen et al., 2016; Kurtas et al., 2019). Whereas the majority of de novo CC cases result from chromosomal aberrations arising from male gametogenesis (Pellestor et al., 2014), chromothripsis is inherited primarily from the mother (Table 3). To all appearances, it is determined by differences in DNA repair capacity and specific features of spermatogenesis and oogenesis.
Table 3.
Chromothripsis inheritance.
| References | Maternal inheritance, cases | Paternal inheritance, cases | De novo chromothripsis, cases |
|---|---|---|---|
| Kloosterman et al., 2011 | – | – | 1 (pat) |
| Kloosterman et al., 2012 | 1 | – | 7 (4/7 – pat; 3/7 – n/d) |
| Gu et al., 2013 | 1 | – | – |
| Nazaryan et al., 2014 | – | – | 1 |
| Fontana et al., 2014 | – | – | 1 |
| Wang et al., 2015 | – | – | 1 |
| Genesio et al., 2015 | – | – | 1 |
| de Pagter et al., 2015 | 3 | – | – |
| Gamba et al., 2015 | – | – | 1 |
| Weckselblatt et al., 2015 | 1 | 1 | 1 (pat) |
| Del Rey et al., 2016 | – | – | 1 |
| Anderson et al., 2016 | – | – | 1 |
| Bertelsen et al., 2016 | 3 | 1 | – |
| Collins et al., 2017 | – | – | 2 |
| Kurtas et al., 2018 | – | – | 1 |
| Kurtas et al., 2019 | 2 | 1 | – |
| Total: | 10 | 3 | 19 |
De novo chromothripsis cases include information on the parental origin of the rearranged chromosomes (if known). pat, paternal chromosomes; n/d, no data about derivative chromosome origin.
Chromothripsis may arise during mitotic and meiotic divisions of spermatogenic cells as well as during spermiogenesis (round spermatid differentiation in spermatogonia) (Pellestor and Gatinois, 2018). Considering that spermatogonia undergo a succession of mitotic divisions, the replication stress may lead to errors during mitosis. Meiotic recombination may also feature double-strand break repair errors (Pellestor and Gatinois, 2018). The DNA breaks in spermatids that occur at the stage of histone-to-protamine transition during spermiogenesis can only be repaired through NHEJ because of the haploid chromosome number in cells at this stage (Gunes et al., 2015). In rodent male germ cells, scaffold/matrix-attached and differentially packaged chromatin regions are highly sensitive to endogenous nucleases, and, thus, to damage (Arpanahi et al., 2009; Grégoire et al., 2013). Accumulation of DNA strand breaks may be also caused by the epigenetically mediated bystander effect in non-irradiated whole testis tissue (Tamminga et al., 2008). This phenomenon is also involved in the production of delayed DNA damage in mouse elongated spermatids due to upregulation of proapoptotic genes 21–33 days later after spermatogonia exposure to X-rays (Cordelli et al., 2012). However, the apoptotic elimination of spermatogenic cells with DNA damage may be aborted before completion (the so-called abortive apoptosis or anoikis), allowing such cells to continue to differentiate and participate in fertilisation (Tubio and Estivill, 2011; Tang et al., 2012). In addition, there are some evidence of aberrant DNA methylation and tissue-specific accumulation of chromosome aberrations in unexposed progeny of cranially irradiated rodents (Koturbash et al., 2006; Tamminga et al., 2008). These data indicate an epigenetic link between DNA damaging agents and occurrence of chromosome aberrations both in unexposed parental germline and offspring’s somatic cells.
In contrast to male germ cells, oocytes may repair breaks through both homologous recombination and NHEJ (Marchetti et al., 2007). Consequently, chromothripsis during oogenesis appears to be less likely than during spermatogenesis. Aberrations in chromosome segregation and premature chromatid separation may cause chromosomal rearrangements during female gametogenesis (Pellestor and Gatinois, 2018). In addition, the DNA repair capacity of an oocyte is the crucial factor of zygote viability, because the repair of maternal and paternal chromosome damage after fertilisation and prior to embryo genome activation occurs through DNA repair factors accumulated in the oocyte cytoplasm.
De novo CC may also be induced by DNA damage during early embryogenesis. Preimplantation embryos typically demonstrate micronuclei formation, blastomere fragmentation, and abnormal mitosis at the cleavage stage (Chavez et al., 2012). This could be a consequence of imperfect repair in germ cells or DNA damage in embryo. In addition, asynchronous pronuclear development and resulting under-replication of the paternal DNA may induce chromosome pulverisation in a zygote (Eichenlaub-Ritter et al., 1995).
Importantly, CCRs are hardly ever detected during conventional karyotyping of chorion cells in a miscarriage, which is conducted starting from 4 to 5 weeks of gestation (i.e., after embryo implantation) (Pendina et al., 2014; Massalska et al., 2017; Soler et al., 2017; Pylyp et al., 2018). As of today, the literature describes only one case of CC in an embryo with multiple malformations (Macera et al., 2015). Apparently, most embryos with CCRs, including chromothripsis, are eliminated at the implantation stage. Despite the wide use of preimplantation genetic testing, the actual frequency and the specific mechanisms of chromothripsis occurrence in gametes and embryos at early stages of development are yet to be determined.
Constitutional chromothripsis is generally characterised by fewer chromosome breaks and almost complete absence of deletions in comparison with malignant tumours (Figure 1; Kloosterman and Cuppen, 2013). A number of studies treat CCR cases with duplications of chromosome regions as chromothripsis (Table 2; Gamba et al., 2015; Wang et al., 2015; Del Rey et al., 2016; Kurtas et al., 2018). It is yet to be established, however, whether such genetic abnormalities in patients are cases of true chromothripsis or variations of other CCRs.
Concluding Remarks
As is the case with any recently discovered phenomenon, the concept of chromothripsis is ambiguous. In our opinion, the most comprehensive definition of chromothripsis has been suggested by Ly and Cleveland: “Chromothripsis is a catastrophic event in which one or a few chromosomes are shattered and stitched back together in random order, producing a derivative chromosome with complex rearrangements within a few cell cycles” (Ly and Cleveland, 2017). Considering that chromothripsis is a highly complex genomic aberration, its reliable detection necessitates the use of a comprehensive approach, combining molecular genetic, molecular cytogenetic, and cytogenetic methods.
Chromothripsis was first detected in chronic lymphocytic leukaemia. To date, it is most frequently found in cancers, even though there are registered cases of chromothripsis both in benign tumours and as constitutional chromosomal abnormality. Both somatic and CC feature multiple rearrangements of one or more chromosomes with a random order and orientation of reassembled fragments, as well as alteration of regions with loss and retention of heterozygosity. However, these aberrations are less pronounced in CC, which normally has fewer breaks and shorter chromosome regions with copy number alterations or a complete absence of such.
The causes and mechanisms underlying chromothripsis remain a subject for discussion. The most probable are telomere damage, exposure to ionising radiation, and viral infections. Along with these already known causes and mechanisms, we suggest aberrant epigenetic regulation as a possible pathway to chromothripsis. The above-mentioned factors may directly destruct chromosomes or activate cell mechanisms associated with chromothripsis. To clearly understand chromothripsis mechanisms, it is necessary to develop models of chromosome pulverisation in micronuclei, reversible apoptosis, and dicentric chromosome breaks.
As of today, it is not clear whether somatic chromothripsis is the cause of tumours or a consequence of pathological processes in tumour cells. Considering that cases of chromothripsis are observed in both malignant and benign tumours, as well as in the karyotype of healthy individuals, it cannot be unambiguously associated with poor clinical outcomes. Apparently, what matters most for neoplasia pathogenesis and a chromothripsis carrier’s phenotype are the genome regions involved in the rearrangement, their localisation, and the size of deleted or amplified fragments – not the presence of chromothripsis itself.
Regardless of the fact that chromothripsis was discovered over 7 years ago, we are still facing challenges in its differentiation from other multiple chromosomal rearrangements and in the understanding of its causes, mechanisms, and consequences – all of which requires further in-depth research.
Author Contributions
AK, AP, OE, OC, TK, and VB contributed to the conception, writing, and checking of the manuscript for important intellectual content.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank Ksenia O. Khudadyan and Anastasiia V. Petrovskaia-Kaminskaia for helpful advice during preparation of the manuscript.
Abbreviations
- aCGH
array comparative genomic hybridisation
- CC
constitutional chromothripsis
- CCR
complex chromosomal rearrangement
- DSB
DNA double-strand break
- FISH
fluorescent in situ hybridisation
- NHEJ
non-homologous end joining
- SKY
spectral karyotyping
- SNP array
single nucleotide polymorphism array
Funding. The section “Constitutional Chromothripsis as a Consequence of Genome Damage in Germ Cells and Preimplantation Embryos” was prepared by AK, AP, and OE in the framework of the project funded by the Russian Science Foundation (Grant Number 18-75-10046). The other article sections were prepared by AK, OC, TK, and VB within the framework of the Basic Research Programme funded by the Ministry of Science and Higher Education of the Russian Federation in 2019–2021 (Programme Number AAAA-A19-119021290033-1). AK is a grantee of RF President Scholarship.
References
- Anderson S. E., Kamath A., Pilz D. T., Morgan S. M. (2016). A rare example of germ-line chromothripsis resulting in large genomic imbalance. Clin. Dysmorphol. 25 58–62. 10.1097/MCD.0000000000000113 [DOI] [PubMed] [Google Scholar]
- Aristidou C., Theodosiou A., Bak M., Mehrjouy M. M., Constantinou E., Alexandrou A., et al. (2018). Position effect, cryptic complexity, and direct gene disruption as disease mechanisms in de novo apparently balanced translocation cases. PLoS One 13:e0205298. 10.1371/journal.pone.0205298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arpanahi A., Brinkworth M., Iles D., Krawetz S. A., Paradowska A., Platts A. E., et al. (2009). Endonuclease-sensitive regions of human spermatozoal chromatin are highly enriched in promoter and CTCF binding sequences. Genome Res. 19 1338–1349. 10.1101/gr.094953.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atlasi Y., Stunnenberg H. G. (2017). The interplay of epigenetic marks during stem cell differentiation and development. Nat. Rev. Genet. 18 643–658. 10.1038/nrg.2017.57 [DOI] [PubMed] [Google Scholar]
- Avgustinova A., Benitah S. A. (2016). The epigenetics of tumour initiation: cancer stem cells and their chromatin. Curr. Opin. Genet. Dev. 36 8–15. 10.1016/j.gde.2016.01.003 [DOI] [PubMed] [Google Scholar]
- Baca S. C., Prandi D., Lawrence M. S., Mosquera J. M., Romanel A., Drier Y., et al. (2013). Punctuated evolution of prostate cancer genomes. Cell 153 666–677. 10.1016/j.cell.2013.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balajee A. S., Hande M. P. (2018). History and evolution of cytogenetic techniques: current and future applications in basic and clinical research. Mutat. Res. 836 3–12. 10.1016/j.mrgentox.2018.08.008 [DOI] [PubMed] [Google Scholar]
- Benyesh-Melnick M., Stich H. F., Rapp F., Hsu T. C. (1964). Viruses and mammalian chromosomes. 3. Effect of herpes zoster virus on human embryonal lung cultures. Proc. Soc. Exp. Biol. Med. 117 546–549. 10.3181/00379727-117-29633 [DOI] [PubMed] [Google Scholar]
- Berger M. F., Lawrence M. S., Demichelis F., Drier Y., Cibulskis K., Sivachenko A. Y., et al. (2011). The genomic complexity of primary human prostate cancer. Nature 470 214–220. 10.1038/nature09744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertelsen B., Nazaryan-Petersen L., Sun W., Mehrjouy M. M., Xie G., Chen W., et al. (2016). A germline chromothripsis event stably segregating in 11 individuals through three generations. Genet. Med. 18 494–500. 10.1038/gim.2015.112 [DOI] [PubMed] [Google Scholar]
- Bundo M., Toyoshima M., Okada Y., Akamatsu W., Ueda J., Nemoto-Miyauchi T., et al. (2014). Increased L1 retrotransposition in the neuronal genome in schizophrenia. Neuron 81 306–313. 10.1016/j.neuron.2013.10.053 [DOI] [PubMed] [Google Scholar]
- Cai H. (2018). Chromothripsis DB: a curated database for the documentation, visualization, and mining of chromothripsis data. Methods Mol. Biol. 1769 279–289. 10.1007/978-1-4939-7780-2_18 [DOI] [PubMed] [Google Scholar]
- Cai H., Kumar N., Bagheri H. C., von Mering C., Robinson M. D., Baudis M. (2014). Chromothripsis-like patterns are recurring but heterogeneously distributed features in a survey of 22,347 cancer genome screens. BMC Genomics 15:82. 10.1186/1471-2164-15-82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai S., Shi G. S., Cheng H. Y., Zeng Y. N., Li G., Zhang M., et al. (2017). Exosomal miR-7 mediates bystander autophagy in lung after focal brain irradiation in mice. Int. J. Biol. Sci. 13 1287–1296. 10.7150/ijbs.18890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccaldi R., Rondinelli B., D’Andrea A. D. (2016). Repair pathway choices and consequences at the double-strand break. Trends Cell Biol. 26 52–64. 10.1016/j.tcb.2015.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez S. L., Loewke K. E., Han J., Moussavi F., Colls P., Munne S., et al. (2012). Dynamic blastomere behaviour reflects human embryo ploidy by the four-cell stage. Nat. Commun. 3:1251. 10.1038/ncomms2249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenet-Monte C., Mohammad F., Celluzzi C. M., Schaffer P. A., Farber F. E. (1986). Herpes simplex virus gene products involved in the induction of chromosomal aberrations. Virus Res. 6 245–260. 10.1016/0168-1702(86)90073-0 [DOI] [PubMed] [Google Scholar]
- Chiang C., Jacobsen J. C., Ernst C., Hanscom C., Heilbut A., Blumenthal I., et al. (2012). Complex reorganization and predominant non-homologous repair following chromosomal breakage in karyotypically balanced germline rearrangements and transgenic integration. Nat. Genet. 44:S1. 10.1038/ng.2202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen A., Sato M., Aldape K., Mason C. C., Alfaro-Munoz K., Heathcock L., et al. (2015). DNA copy number analysis of Grade II-III and Grade IV gliomas reveals differences in molecular ontogeny including chromothripsis associated with IDH mutation status. Acta Neuropathol. Commun. 3:34. 10.1186/s40478-015-0213-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins R. L., Brand H., Redin C. E., Hanscom C., Antolik C., Stone M. R., et al. (2017). Defining the diverse spectrum of inversions, complex structural variation, and chromothripsis in the morbid human genome. Genome Biol. 18:36. 10.1186/s13059-017-1158-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordelli E., Eleuteri P., Grollino M. G., Benassi B., Blandino G., Bartoleschi C., et al. (2012). Direct and delayed X-ray-induced DNA damage in male mouse germ cells. Environ. Mol. Mutagen. 53 429–439. 10.1002/em.21703 [DOI] [PubMed] [Google Scholar]
- Crasta K., Ganem N. J., Dagher R., Lantermann A. B., Ivanova E. V., Pan Y., et al. (2012). DNA breaks and chromosome pulverization from errors in mitosis. Nature 482 53–58. 10.1038/nature10802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Pagter M. S., van Roosmalen M. J., Baas A. F., Renkens I., Duran K. J., van Binsbergen E., et al. (2015). Chromothripsis in healthy individuals affects multiple protein-coding genes and can result in severe congenital abnormalities in offspring. Am. J. Hum. Genet. 96 651–656. 10.1016/j.ajhg.2015.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Rey J., Santos M., González-Meneses A., Milà M., Fuster C. (2016). Heterogeneity of a constitutional complex chromosomal rearrangement in 2q. Cytogenet. Genome Res. 148 156–164. 10.1159/000445859 [DOI] [PubMed] [Google Scholar]
- Efimova O. A., Pendina A. A., Krapivin M. I., Kopat V. V., Tikhonov A. V., Petrovskaia-Kaminskaia A. V., et al. (2018). Inter-cell and inter-chromosome variability of 5-hydroxymethylcytosine patterns in noncultured human embryonic and extraembryonic cells. Cytogenet. Genome Res. 156 150–157. 10.1159/000493906 [DOI] [PubMed] [Google Scholar]
- Efimova O. A., Pendina A. A., Tikhonov A. V., Fedorova I. D., Krapivin M. I., Chiryaeva O. G., et al. (2015). Chromosome hydroxymethylation patterns in human zygotes and cleavage-stage embryos. Reproduction 149 223–233. 10.1530/REP-14-0343 [DOI] [PubMed] [Google Scholar]
- Efimova O. A., Pendina A. A., Tikhonov A. V., Parfenyev S. E., Mekina I. D., Komarova E. M., et al. (2017). Genome-wide 5-hydroxymethylcytosine patterns in human spermatogenesis are associated with semen quality. Oncotarget 8 88294–88307. 10.18632/oncotarget.18331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichenlaub-Ritter U., Schmiady H., Kentenich H., Soewarto D. (1995). Recurrent failure in polar body formation and premature chromosome condensation in oocytes from a human patient: indicators of asynchrony in nuclear and cytoplasmic maturation. Hum. Reprod. 10 2343–2349. 10.1093/oxfordjournals.humrep.a136297 [DOI] [PubMed] [Google Scholar]
- Ernst A., Jones D. T., Maass K. K., Rode A., Deeg K. I., Jebaraj B. M., et al. (2016). Telomere dysfunction and chromothripsis. Int. J. Cancer 138 2905–2914. 10.1002/ijc.30033 [DOI] [PubMed] [Google Scholar]
- Falk M., Lukásová E., Kozubek S. (2008). Chromatin structure influences the sensitivity of DNA to gamma-radiation. Biochim. Biophys. Acta 1783 2398–2414. 10.1016/j.bbamcr.2008.07.010 [DOI] [PubMed] [Google Scholar]
- Fontana P., Genesio R., Casertano A., Cappuccio G., Mormile A., Nitsch L., et al. (2014). Loeys-Dietz syndrome type 4, caused by chromothripsis, involving the TGFB2 gene. Gene 538 69–73. 10.1016/j.gene.2014.01.017 [DOI] [PubMed] [Google Scholar]
- Forment J. V., Kaidi A., Jackson S. P. (2012). Chromothripsis and cancer: causes and consequences of chromosome shattering. Nat. Rev. Cancer 12 663–670. 10.1038/nrc3352 [DOI] [PubMed] [Google Scholar]
- Fredga K., Hall B. (1970). A complex familial translocation involving chromosomes 5, 9 and 13. Cytogenetics 9 294–306. 10.1159/000130099 [DOI] [PubMed] [Google Scholar]
- Gamba B. F., Richieri-Costa A., Costa S., Rosenberg C., Ribeiro-Bicudo L. A. (2015). Chromothripsis with at least 12 breaks at 1p36.33-p35.3 in a boy with multiple congenital anomalies. Mol. Genet. Genomics 290 2213–2216. 10.1007/s00438-015-1072-0 [DOI] [PubMed] [Google Scholar]
- Gao G., Smith D. I. (2017). How can mate-pair sequencing be utilized for cancer patients? Expert Rev. Mol. Diagn. 17 1–3. 10.1080/14737159.2017.1259569 [DOI] [PubMed] [Google Scholar]
- Garsed D. W., Marshall O. J., Corbin V. D., Hsu A., Di Stefano L., Schröder J., et al. (2014). The architecture and evolution of cancer neochromosomes. Cancer Cell 26 653–667. 10.1016/j.ccell.2014.09.010 [DOI] [PubMed] [Google Scholar]
- Gaudet F., Hodgson J. G., Eden A., Jackson-Grusby L., Dausman J., Gray J. W., et al. (2003). Induction of tumors in mice by genomic hypomethylation. Science 300 489–492. 10.1126/science.1083558 [DOI] [PubMed] [Google Scholar]
- Geiman T. M., Robertson K. D. (2002). Chromatin remodeling, histone modifications, and DNA methylation-how does it all fit together? J. Cell Biochem. 87 117–125. 10.1002/jcb.10286 [DOI] [PubMed] [Google Scholar]
- Genesio R., Fontana P., Mormile A., Casertano A., Falco M., Conti A., et al. (2015). Constitutional chromothripsis involving the critical region of 9q21.13 microdeletion syndrome. Mol. Cytogenet. 8:96. 10.1186/s13039-015-0199-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Géraud G., Laquerrière F., Masson C., Arnoult J., Labidi B., Hernandez-Verdun D. (1989). Three-dimensional organization of micronuclei induced by colchicine in PtK1 cells. Exp. Cell Res. 181 27–39. 10.1016/0014-4827(89)90179-1 [DOI] [PubMed] [Google Scholar]
- Gisselsson D., Shao C., Tuck-Muller C. M., Sogorovic S., Pålsson E., Smeets D., et al. (2005). Interphase chromosomal abnormalities and mitotic missegregation of hypomethylated sequences in ICF syndrome cells. Chromosoma 114 118–126. 10.1007/s00412-005-0343-7 [DOI] [PubMed] [Google Scholar]
- Grégoire M. C., Massonneau J., Simard O., Gouraud A., Brazeau M. A., Arguin M., et al. (2013). Male-driven de novo mutations in haploid germ cells. Mol. Hum. Reprod. 19 495–499. 10.1093/molehr/gat022 [DOI] [PubMed] [Google Scholar]
- Gu H., Jiang J. H., Li J. Y., Zhang Y. N., Dong X. S., Huang Y. Y., et al. (2013). A familial Cri-du-Chat/5p deletion syndrome resulted from rare maternal complex chromosomal rearrangements (CCRs) and/or possible chromosome 5p chromothripsis. PLoS One 8:e76985. 10.1371/journal.pone.0076985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunes S., Al-Sadaan M., Agarwal A. (2015). Spermatogenesis, DNA damage and DNA repair mechanisms in male infertility. Reprod. Biomed. Online 31 309–319. 10.1016/j.rbmo.2015.06.010 [DOI] [PubMed] [Google Scholar]
- Hastings P. J., Ira G., Lupski J. R. (2009). A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet. 5:e1000327. 10.1371/journal.pgen.1000327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch E. M., Hetzer M. W. (2015). Linking micronuclei to chromosome fragmentation. Cell 161 1502–1504. 10.1016/j.cell.2015.06.005 [DOI] [PubMed] [Google Scholar]
- Holland A. J., Cleveland D. W. (2012). Chromoanagenesis and cancer: mechanisms and consequences of localized, complex chromosomal rearrangements. Nat. Med. 18 1630–1638. 10.1038/nm.2988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzmann C., Markowski D. N., Koczan D., Küpker W., Helmke B. M., Bullerdiek J. (2014). Cytogenetically normal uterine leiomyomas without MED12-mutations - a source to identify unknown mechanisms of the development of uterine smooth muscle tumors. Mol. Cytogenet. 7:88. 10.1186/s13039-014-0088-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M. J., Jallepalli P. V. (2012). Chromothripsis: chromosomes in crisis. Dev. Cell 23 908–917. 10.1016/j.devcel.2012.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamranvar S. A., Gruhne B., Szeles A., Masucci M. G. (2007). Epstein-Barr virus promotes genomic instability in Burkitt’s lymphoma. Oncogene 26 5115–5123. 10.1038/sj.onc.1210324 [DOI] [PubMed] [Google Scholar]
- Keren B. (2014). The advantages of SNP arrays over CGH arrays. Mol. Cytogenet. 7(Suppl. 1):I31. 10.1186/1755-8166-7-S1-I31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinsella M., Patel A., Bafna V. (2014). The elusive evidence for chromothripsis. Nucleic Acids Res. 42 8231–8242. 10.1093/nar/gku525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirolikar S., Prasannan P., Raghuram G. V., Pancholi N., Saha T., Tidke P., et al. (2018). Prevention of radiation-induced bystander effects by agents that inactivate cell-free chromatin released from irradiated dying cells. Cell Death Dis. 9:1142. 10.1038/s41419-018-1181-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloosterman W. P., Cuppen E. (2013). Chromothripsis in congenital disorders and cancer: similarities and differences. Curr. Opin. Cell Biol. 25 341–348. 10.1016/j.ceb.2013.02.008 [DOI] [PubMed] [Google Scholar]
- Kloosterman W. P., Guryev V., van Roosmalen M., Duran K. J., de Bruijn E., Bakker S. C., et al. (2011). Chromothripsis as a mechanism driving complex de novo structural rearrangements in the germline. Hum. Mol. Genet. 20 1916–1924. 10.1093/hmg/ddr073 [DOI] [PubMed] [Google Scholar]
- Kloosterman W. P., Tavakoli-Yaraki M., van Roosmalen M. J., van Binsbergen E., Renkens I., Duran K., et al. (2012). Constitutional chromothripsis rearrangements involve clustered double-stranded DNA breaks and nonhomologous repair mechanisms. Cell Rep. 1 648–655. 10.1016/j.celrep.2012.05.009 [DOI] [PubMed] [Google Scholar]
- Korbel J. O., Campbell P. J. (2013). Criteria for inference of chromothripsis in cancer genomes. Cell 152 1226–1236. 10.1016/j.cell.2013.02.023 [DOI] [PubMed] [Google Scholar]
- Koturbash I., Boyko A., Rodriguez-Juarez R., McDonald R. J., Tryndyak V. P., Kovalchuk I., et al. (2007). Role of epigenetic effectors in maintenance of the long-term persistent bystander effect in spleen in vivo. Carcinogenesis 28 1831–1838. 10.1093/carcin/bgm053 [DOI] [PubMed] [Google Scholar]
- Koturbash I., Loree J., Kutanzi K., Koganow C., Pogribny I., Kovalchuk O. (2008). In vivo bystander effect: cranial X-irradiation leads to elevated DNA damage, altered cellular proliferation and apoptosis, and increased p53 levels in shielded spleen. Int. J. Radiat. Oncol. Biol. Phys. 70 554–562. 10.1016/j.ijrobp.2007.09.039 [DOI] [PubMed] [Google Scholar]
- Koturbash I., Rugo R. E., Hendricks C. A., Loree J., Thibault B., Kutanzi K., et al. (2006). Irradiation induces DNA damage and modulates epigenetic effectors in distant bystander tissue in vivo. Oncogene 25 4267–4275. 10.1038/sj.onc.1209467 [DOI] [PubMed] [Google Scholar]
- Kovalchuk O., Zemp F. J., Filkowski J. N., Altamirano A. M., Dickey J. S., Jenkins-Baker G., et al. (2010). microRNAome changes in bystander three-dimensional human tissue models suggest priming of apoptotic pathways. Carcinogenesis 31 1882–1888. 10.1093/carcin/bgq119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo M. T. (1981). Preferential damage of active chromatin by bleomycin. Cancer Res. 41 2439–2443. [PubMed] [Google Scholar]
- Kurtas N., Arrigoni F., Errichiello E., Zucca C., Maghini C., D’Angelo M. G., et al. (2018). Chromothripsis and ring chromosome 22: a paradigm of genomic complexity in the Phelan-McDermid syndrome (22q13 deletion syndrome). J. Med. Genet. 55 269–277. 10.1136/jmedgenet-2017-105125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtas N. E., Xumerle L., Giussani U., Pansa A., Cardarelli L., Bertini V., et al. (2019). Insertional translocation involving an additional nonchromothriptic chromosome in constitutional chromothripsis: rule or exception? Mol. Genet. Genomic Med. 7:e00496. 10.1002/mgg3.496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Kim Y. J., Choi Y. J., Lee J. W., Lee S., Cho Y. H., et al. (2015). Radiation-induced changes in DNA methylation and their relationship to chromosome aberrations in nuclear power plant workers. Int. J. Radiat. Biol. 91 142–149. 10.3109/09553002.2015.969847 [DOI] [PubMed] [Google Scholar]
- Leibowitz M. L., Zhang C. Z., Pellman D. (2015). Chromothripsis: a new mechanism for rapid karyotype evolution. Annu. Rev. Genet. 49 183–211. 10.1146/annurev-genet-120213-092228 [DOI] [PubMed] [Google Scholar]
- Li L. C. (2014). Chromatin remodeling by the small RNA machinery in mammalian cells. Epigenetics 9 45–52. 10.4161/epi.26830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Schwab C., Ryan S., Papaemmanuil E., Robinson H. M., Jacobs P., et al. (2014). Constitutional and somatic rearrangement of chromosome 21 in acute lymphoblastic leukaemia. Nature 508 98–102. 10.1038/nature13115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P., Erez A., Nagamani S. C., Dhar S. U., Kołodziejska K. E., Dharmadhikari A. V., et al. (2011). Chromosome catastrophes involve replication mechanisms generating complex genomic rearrangements. Cell 146 889–903. 10.1016/j.cell.2011.07.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luzhna L., Kathiria P., Kovalchuk O. (2013). Micronuclei in genotoxicity assessment: from genetics to epigenetics and beyond. Front. Genet. 4:131. 10.3389/fgene.2013.00131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ly P., Cleveland D. W. (2017). Rebuilding chromosomes after catastrophe: emerging mechanisms of chromothripsis. Trends Cell Biol. 27 917–930. 10.1016/j.tcb.2017.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ly P., Teitz L. S., Kim D. H., Shoshani O., Skaletsky H., Fachinetti D., et al. (2017). Selective Y centromere inactivation triggers chromosome shattering in micronuclei and repair by non-homologous end joining. Nat. Cell Biol. 19 68–75. 10.1038/ncb3450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macera M. J., Sobrino A., Levy B., Jobanputra V., Aggarwal V., Mills A., et al. (2015). Prenatal diagnosis of chromothripsis, with nine breaks characterized by karyotyping, FISH, microarray and whole-genome sequencing. Prenat. Diagn. 35 299–301. 10.1002/pd.4456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciejowski J., de Lange T. (2017). Telomeres in cancer: tumour suppression and genome instability. Nat. Rev. Mol. Cell Biol. 18 175–186. 10.1038/nrm.2016.171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciejowski J., Li Y., Bosco N., Campbell P. J., de Lange T. (2015). Chromothripsis and kataegis induced by telomere crisis. Cell 163 1641–1654. 10.1016/j.cell.2015.11.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKinnon R. N., Campbell L. J. (2013). Chromothripsis under the microscope: a cytogenetic perspective of two cases of AML with catastrophic chromosome rearrangement. Cancer Genet. 206 238–251. 10.1016/j.cancergen.2013.05.021 [DOI] [PubMed] [Google Scholar]
- Madan K. (2013). What is a complex chromosome rearrangement? Am. J. Med. Genet. A 161A 1181–1184. 10.1002/ajmg.a.35834 [DOI] [PubMed] [Google Scholar]
- Maher C. A., Wilson R. K. (2012). Chromothripsis and human disease: piecing together the shattering process. Cell 148 29–32. 10.1016/j.cell.2012.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra A., Lindberg M., Faust G. G., Leibowitz M. L., Clark R. A., Layer R. M., et al. (2013). Breakpoint profiling of 64 cancer genomes reveals numerous complex rearrangements spawned by homology-independent mechanisms. Genome Res. 23 762–776. 10.1101/gr.143677.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniotis A. J., Valyi-Nagy K., Karavitis J., Moses J., Boddipali V., Wang Y., et al. (2005). Chromatin organization measured by AluI restriction enzyme changes with malignancy and is regulated by the extracellular matrix and the cytoskeleton. Am. J. Pathol. 166 1187–1203. 10.1016/S0002-9440(10)62338-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetti F., Essers J., Kanaar R., Wyrobek A. J. (2007). Disruption of maternal DNA repair increases sperm-derived chromosomal aberrations. Proc. Natl. Acad. Sci. U.S.A. 104 17725–17729. 10.1073/pnas.0705257104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardin B. R., Drainas A. P., Waszak S. M., Weischenfeldt J., Isokane M., Stütz A. M., et al. (2015). A cell-based model system links chromothripsis with hyperploidy. Mol. Syst. Biol. 11:828. 10.15252/msb.20156505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massalska D., Zimowski J. G., Bijok J., Pawelec M., Czubak-Barlik M., Jakiel G., et al. (2017). First trimester pregnancy loss: clinical implications of genetic testing. J. Obstet. Gynaecol. Res. 43 23–29. 10.1111/jog.13179 [DOI] [PubMed] [Google Scholar]
- Masset H., Hestand M. S., Van Esch H., Kleinfinger P., Plaisancié J., Afenjar A., et al. (2016). A distinct class of chromoanagenesis events characterized by focal copy number gains. Hum. Mutat. 37 661–668. 10.1002/humu.22984 [DOI] [PubMed] [Google Scholar]
- McClintock B. (1939). The behavior in successive nuclear divisions of a chromosome broken at meiosis. Proc. Natl. Acad. Sci. U.S.A. 25 405–416. 10.1073/pnas.25.8.405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGowan-Jordan J., Simons A., Schmid M. (2016). An International System for Human Cytogenomic Nomenclature. Basel: S. Karger, 139. [Google Scholar]
- Mehine M., Kaasinen E., Mäkinen N., Katainen R., Kämpjärvi K., Pitkänen E., et al. (2013). Characterization of uterine leiomyomas by whole-genome sequencing. N. Engl. J. Med. 369 43–53. 10.1056/NEJMoa1302736 [DOI] [PubMed] [Google Scholar]
- Mehine M., Mäkinen N., Heinonen H. R., Aaltonen L. A., Vahteristo P. (2014). Genomics of uterine leiomyomas: insights from high-throughput sequencing. Fertil. Steril. 102 621–629. 10.1016/j.fertnstert.2014.06.050 [DOI] [PubMed] [Google Scholar]
- Meschini R., Morucci E., Berni A., Lopez-Martinez W., Palitti F. (2015). Role of chromatin structure modulation by the histone deacetylase inhibitor trichostatin A on the radio-sensitivity of ataxia telangiectasia. Mutat. Res. 777 52–59. 10.1016/j.mrfmmm.2015.04.009 [DOI] [PubMed] [Google Scholar]
- Meyerson M., Pellman D. (2011). Cancer genomes evolve by pulverizing single chromosomes. Cell 144 9–10. 10.1016/j.cell.2010.12.025 [DOI] [PubMed] [Google Scholar]
- Middelkamp S., van Heesch S., Braat A. K., de Ligt J., van Iterson M., Simonis M., et al. (2017). Molecular dissection of germline chromothripsis in a developmental context using patient-derived iPS cells. Genome Med. 9:9. 10.1186/s13073-017-0399-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. R., Koren S., Sutton G. (2010). Assembly algorithms for next-generation sequencing data. Genomics 95 315–327. 10.1016/j.ygeno.2010.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molenaar J. J., Koster J., Zwijnenburg D. A., van Sluis P., Valentijn L. J., van der Ploeg I., et al. (2012). Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature 483 589–593. 10.1038/nature10910 [DOI] [PubMed] [Google Scholar]
- Morishita M., Muramatsu T., Suto Y., Hirai M., Konishi T., Hayashi S., et al. (2016). Chromothripsis-like chromosomal rearrangements induced by ionizing radiation using proton microbeam irradiation system. Oncotarget 7 10182–10192. 10.18632/oncotarget.7186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison C. D., Liu P., Woloszynska-Read A., Zhang J., Luo W., Qin M., et al. (2014). Whole-genome sequencing identifies genomic heterogeneity at a nucleotide and chromosomal level in bladder cancer. Proc. Natl. Acad. Sci. U.S.A. 111 672–681. 10.1073/pnas.1313580111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazaryan L., Stefanou E. G., Hansen C., Kosyakova N., Bak M., Sharkey F. H., et al. (2014). The strength of combined cytogenetic and mate-pair sequencing techniques illustrated by a germline chromothripsis rearrangement involving FOXP2. Eur. J. Hum. Genet. 22 338–343. 10.1038/ejhg.2013.147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazaryan-Petersen L., Bertelsen B., Bak M., Jønson L., Tommerup N., Hancks D. C., et al. (2016). Germline chromothripsis driven by L1-mediated retrotransposition and Alu/Alu homologous recombination. Hum. Mutat. 37 385–395. 10.1002/humu.22953 [DOI] [PubMed] [Google Scholar]
- Nichols W. W., Levan A., Aula P., Norrby E. (1965). Chromosome damage associated with the measles virus in vitro. Hereditas 54 101–118. 10.1111/j.1601-5223.1965.tb02008.x [DOI] [PubMed] [Google Scholar]
- Nones K., Waddell N., Wayte N., Patch A. M., Bailey P., Newell F., et al. (2014). Genomic catastrophes frequently arise in esophageal adenocarcinoma and drive tumorigenesis. Nat. Commun. 5:5224. 10.1038/ncomms6224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northcott P. A., Shih D. J., Peacock J., Garzia L., Morrissy A. S., Zichner T., et al. (2012). Subgroup-specific structural variation across 1,000 medulloblastoma genomes. Nature 488 49–56. 10.1038/nature11327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuzzo F., Marini A., Baglioni C., Ford C. E., De Carli L., Piceni Sereni L. (1968). A case of multiple chromosomal rearrangements with persistence of foetal haemoglobin. Cytogenetics 7 169–182. 10.1159/000129981 [DOI] [PubMed] [Google Scholar]
- O’Neill F. J., Miles C. P. (1970). Virus-induced chromosome pulverization in human diploid cells. Proc. Soc. Exp. Biol. Med. 134 825–830. 10.3181/00379727-134-34892 [DOI] [PubMed] [Google Scholar]
- Pai G. S., Thomas G. H., Mahoney W., Migeon B. R. (1980). Complex chromosome rearrangements. Report of a new case and literature review. Clin. Genet. 18 436–444. 10.1111/j.1399-0004.1980.tb01790.x [DOI] [PubMed] [Google Scholar]
- Payen C., Koszul R., Dujon B., Fischer G. (2008). Segmental duplications arise from Pol32-dependent repair of broken forks through two alternative replication-based mechanisms. PLoS Genet. 4:e1000175. 10.1371/journal.pgen.1000175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peat D. S., Stanley M. A. (1986). Chromosome damage induced by herpes simplex virus type 1 in early infection. J. Gen. Virol. 67 2273–2277. 10.1099/0022-1317-67-10-2273 [DOI] [PubMed] [Google Scholar]
- Pei J., Jhanwar S. C., Testa J. R. (2012). Chromothripsis in a case of TP53-deficient chronic lymphocytic leukemia. Leuk. Res. Rep. 1 4–6. 10.1016/j.lrr.2012.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellestor F., Gatinois V. (2018). Potential role of chromothripsis in the genesis of complex chromosomal rearrangements in human gametes and preimplantation embryo. Methods Mol. Biol. 1769 35–41. 10.1007/978-1-4939-7780-2_3 [DOI] [PubMed] [Google Scholar]
- Pellestor F., Gatinois V., Puechberty J., Geneviève D., Lefort G. (2014). Chromothripsis: potential origin in gametogenesis and preimplantation cell divisions. A review. Fertil. Steril. 102 1785–1796. 10.1016/j.fertnstert.2014.09.006 [DOI] [PubMed] [Google Scholar]
- Pendina A. A., Efimova O. A., Chiryaeva O. G., Tikhonov A. V., Petrova L. I., Dudkina V. S., et al. (2014). A comparative cytogenetic study of miscarriages after IVF and natural conception in women aged under and over 35 years. J. Assist. Reprod. Genet. 31 149–155. 10.1007/s10815-013-0148-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendina A. A., Efimova O. A., Krapivin M. I., Mekina I. D., Tikhonov A. V., Koltsova A. S., et al. (2018). Genomic distribution of 5-formylcytosine and 5-carboxylcytosine in human preimplantation embryos. Mol. Reprod. Dev. 85 893–895. 10.1002/mrd.23074 [DOI] [PubMed] [Google Scholar]
- Pendina A. A., Koltsova A. S., Efimova O. A., Malysheva O. V., Osinovskaya N. S., Sultanov I. Y., et al. (2017). Case of chromothripsis in a large solitary non-recurrent uterine leiomyoma. Eur. J. Obstet. Gynecol. Reprod. Biol. 219 134–136. 10.1016/j.ejogrb.2017.10.028 [DOI] [PubMed] [Google Scholar]
- Poot M. (2018). Genes, proteins, and biological pathways preventing chromothripsis. Methods Mol. Biol. 1769 231–251. 10.1007/978-1-4939-7780-2_15 [DOI] [PubMed] [Google Scholar]
- Przybytkowski E., Lenkiewicz E., Barrett M. T., Klein K., Nabavi S., Greenwood C. M., et al. (2014). Chromosome-breakage genomic instability and chromothripsis in breast cancer. BMC Genomics 15:579. 10.1186/1471-2164-15-579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pylyp L. Y., Spynenko L. O., Verhoglyad N. V., Mishenko A. O., Mykytenko D. O., Zukin V. D. (2018). Chromosomal abnormalities in products of conception of first-trimester miscarriages detected by conventional cytogenetic analysis: a review of 1000 cases. J. Assist. Reprod. Genet. 35 265–271. 10.1007/s10815-017-1069-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratnaparkhe M., Hlevnjak M., Kolb T., Jauch A., Maass K. K., Devens K., et al. (2017). Genomic profiling of Acute lymphoblastic leukemia in ataxia telangiectasia patients reveals tight link between ATM mutations and chromothripsis. Leukemia 31 2048–2056. 10.1038/leu.2017.55 [DOI] [PubMed] [Google Scholar]
- Rausch T., Jones D. T., Zapatka M., Stütz A. M., Zichner T., Weischenfeldt J., et al. (2012). Genome sequencing of pediatric medulloblastoma links catastrophic DNA rearrangements with TP53 mutations. Cell 148 59–71. 10.1016/j.cell.2011.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rode A., Maass K. K., Willmund K. V., Lichter P., Ernst A. (2016). Chromothripsis in cancer cells: an update. Int. J. Cancer 138 2322–2333. 10.1002/ijc.29888 [DOI] [PubMed] [Google Scholar]
- Schütze D. M., Krijgsman O., Snijders P. J., Ylstra B., Weischenfeldt J., Mardin B. R., et al. (2016). Immortalization capacity of HPV types is inversely related to chromosomal instability. Oncotarget 7 37608–37621. 10.18632/oncotarget.8058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slamova Z., Nazaryan-Petersen L., Mehrjouy M. M., Drabova J., Hancarova M., Marikova T., et al. (2018). Very short DNA segments can be detected and handled by the repair machinery during germline chromothriptic chromosome reassembly. Hum. Mutat. 39 709–716. 10.1002/humu.23408 [DOI] [PubMed] [Google Scholar]
- Soler A., Morales C., Mademont-Soler I., Margarit E., Borrell A., Borobio V., et al. (2017). Overview of chromosome abnormalities in first trimester miscarriages: a series of 1,011 consecutive chorionic villi sample karyotypes. Cytogenet. Genome Res. 152 81–89. 10.1159/000477707 [DOI] [PubMed] [Google Scholar]
- Sorzano C. O., Pascual-Montano A., Sánchez de Diego A., Martínez-A C., van Wely K. H. (2013). Chromothripsis: breakage-fusion-bridge over and over again. Cell Cycle 12 2016–2023. 10.4161/cc.25266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens P. J., Greenman C. D., Fu B., Yang F., Bignell G. R., Mudie L. J., et al. (2011). Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell 144 27–40. 10.1016/j.cell.2010.11.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamminga J., Koturbash I., Baker M., Kutanzi K., Kathiria P., Pogribny I. P., et al. (2008). Paternal cranial irradiation induces distant bystander DNA damage in the germline and leads to epigenetic alterations in the offspring. Cell Cycle 7 1238–1245. 10.4161/cc.7.9.5806 [DOI] [PubMed] [Google Scholar]
- Tang H. L., Tang H. M., Mak K. H., Hu S., Wang S. S., Wong K. M., et al. (2012). Cell survival, DNA damage, and oncogenic transformation after a transient and reversible apoptotic response. Mol. Biol. Cell 23 2240–2252. 10.1091/mbc.E11-11-0926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terradas M., Martín M., Genescà A. (2016). Impaired nuclear functions in micronuclei results in genome instability and chromothripsis. Arch. Toxicol. 90 2657–2667. 10.1007/s00204-016-1818-4 [DOI] [PubMed] [Google Scholar]
- Terzoudi G. I., Karakosta M., Pantelias A., Hatzi V. I., Karachristou I., Pantelias G. (2015). Stress induced by premature chromatin condensation triggers chromosome shattering and chromothripsis at DNA sites still replicating in micronuclei or multinucleate cells when primary nuclei enter mitosis. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 793 185–198. 10.1016/j.mrgentox.2015.07.014 [DOI] [PubMed] [Google Scholar]
- Tubio J. M., Estivill X. (2011). Cancer: when catastrophe strikes a cell. Nature 470 476–477. 10.1038/470476a [DOI] [PubMed] [Google Scholar]
- Tunc O., Tremellen K. (2009). Oxidative DNA damage impairs global sperm DNA methylation in infertile men. J. Assist. Reprod. Genet. 26 537–544. 10.1007/s10815-009-9346-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heesch S., Simonis M., van Roosmalen M. J., Pillalamarri V., Brand H., Kuijk E. W., et al. (2014). Genomic and functional overlap between somatic and germline chromosomal rearrangements. Cell Rep. 9 2001–2010. 10.1016/j.celrep.2014.11.022 [DOI] [PubMed] [Google Scholar]
- Wang J. C., Fisker T., Sahoo T. (2015). Constitutional chromothripsis involving chromosome 19 in a child with subtle dysmorphic features. Am. J. Med. Genet. A 167A 910–913. 10.1002/ajmg.a.36962 [DOI] [PubMed] [Google Scholar]
- Wang S., Wu W., Claret F. X. (2017). Mutual regulation of microRNAs and DNA methylation in human cancers. Epigenetics 12 187–197. 10.1080/15592294.2016.1273308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weckselblatt B., Hermetz K. E., Rudd M. K. (2015). Unbalanced translocations arise from diverse mutational mechanisms including chromothripsis. Genome Res. 25 937–947. 10.1101/gr.191247.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West M. J. (2017). Chromatin reorganisation in Epstein-Barr virus-infected cells and its role in cancer development. Curr. Opin. Virol. 26 149–155. 10.1016/j.coviro.2017.08.004 [DOI] [PubMed] [Google Scholar]
- Xu S., Wang J., Ding N., Hu W., Zhang X., Wang B., et al. (2015). Exosome-mediated microRNA transfer plays a role in radiation-induced bystander effect. RNA Biol. 12 1355–1363. 10.1080/15476286.2015.1100795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S., Hong K., Liu R., Inoue A., Shen L., Zhang K., et al. (2013). Dynamics of 5-methylcytosine and 5-hydroxymethylcytosine during germ cell reprogramming. Cell Res. 23 329–339. 10.1038/cr.2013.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Deng G., Cai H. (2016). Chromothripsis DB: a curated database of chromothripsis. Bioinformatics 32 1433–1435. 10.1093/bioinformatics/btv757 [DOI] [PubMed] [Google Scholar]
- Yuan D., Xu J., Wang J., Pan Y., Fu J., Bai Y., et al. (2016). Extracellular miR-1246 promotes lung cancer cell proliferation and enhances radioresistance by directly targeting DR5. Oncotarget 7 32707–32722. 10.18632/oncotarget.9017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemanova Z., Michalova K., Buryova H., Brezinova J., Kostylkova K., Bystricka D., et al. (2014). Involvement of deleted chromosome 5 in complex chromosomal aberrations in newly diagnosed myelodysplastic syndromes (MDS) is correlated with extremely adverse prognosis. Leuk. Res. 38 537–544. 10.1016/j.leukres.2014.01.012 [DOI] [PubMed] [Google Scholar]
- Zhang C. Z., Spektor A., Cornils H., Francis J. M., Jackson E. K., Liu S., et al. (2015). Chromothripsis from DNA damage in micronuclei. Nature 522 179–184. 10.1038/nature14493 [DOI] [PMC free article] [PubMed] [Google Scholar]