Abstract
Background
Estimation of glomerular filtration rate (GFR) using estimated glomerular filtration rate creatinine (eGFRcr) is central to clinical practice but has limitations. We tested the hypothesis that serum metabolomic profiling can identify novel markers that in combination can provide more accurate GFR estimates.
Methods
We performed a cross-sectional study of 200 African American Study of Kidney Disease and Hypertension (AASK) and 265 Multi-Ethnic Study of Atherosclerosis (MESA) participants with measured GFR (mGFR). Untargeted gas chromatography/dual mass spectrometry– and liquid chromatography/dual mass spectrometry–based quantification was followed by the development of targeted assays for 15 metabolites. On the log scale, GFR was estimated from single- and multiple-metabolite panels and compared with eGFR using the Chronic Kidney Disease Epidemiology equations with creatinine and/or cystatin C using established metrics, including the proportion of errors >30% of mGFR (1-P30), before and after bias correction.
Results
Of untargeted metabolites in the AASK and MESA, 283 of 780 (36%) and 387 of 1447 (27%), respectively, were significantly correlated (P ≤ 0.001) with mGFR. A targeted metabolite panel eGFR developed in the AASK and validated in the MESA was more accurate (1-P30 3.7 and 1.9%, respectively) than eGFRcr [11.2 and 18.5%, respectively (P < 0.001 for both)] and estimating GFR using cystatin C (eGFRcys) [10.6% (P = 0.02) and 9.1% (P < 0.05), respectively] but was not consistently better than eGFR using both creatinine and cystatin C [3.7% (P > 0.05) and 9.1% (P < 0.05), respectively]. A panel excluding creatinine and demographics still performed well [1-P30 6.4% (P = 0.11) and 3.4% (P < 0.001) in the AASK and MESA] versus eGFRcr.
Conclusions
Multimetabolite panels can enable accurate GFR estimation. Metabolomic equations, preferably excluding creatinine and demographic characteristics, should be tested for robustness and generalizability as a potential confirmatory test when eGFRcr is unreliable.
Keywords: creatinine, estimating equations, GFR, kidney function, metabolomics
INTRODUCTION
Estimated glomerular filtration rate (eGFR) from the serum concentration of the metabolite creatinine (eGFRcr) is widely used and central to the detection, staging and management of chronic kidney disease (CKD) [1]. eGFRcr equations require demographic characteristics (age, sex and race) as surrogates for creatinine generation by muscle and diet [2]. The Chronic Kidney Disease Epidemiology Collaboration (CKD-EPI) equation for eGFRcr includes coefficients for age, sex and race and is recommended for routine clinical use in white and black populations in North America, Europe and Australia, with modifications as necessary for use in racial and ethnic groups from other geographic regions [3, 4]. Like all eGFRcr equations, it is biased in clinical conditions with alterations in non-GFR determinants of serum creatinine and has limited precision even in populations without known alterations [errors >30% from measured GFR (mGFR) (1-P30) in 10–20% of estimates] [5, 6]. Of particular importance is that chronic illness is often accompanied by a loss of muscle mass, which blunts the increase in serum creatinine concentration at lower GFRs and reduces the sensitivity of eGFRcr for detecting decreased GFR. Thus, while the advantages of creatinine as a filtration marker have led to widespread use for more than a century, it is clear that further improvements in GFR estimation will likely require new filtration markers.
Serum concentrations of cystatin C and other low-molecular weight proteins are less influenced than creatinine by muscle mass and diet. Estimating GFR using cystatin C (eGFRcys) rather than creatinine can reduce bias due to these factors. In a community-based sample of frail older participants, decreased eGFR had a prevalence of 77% based on eGFRcys compared with 45% based on eGFRcr, showing biases can be large, but a gold standard is needed to know which measure is biased [7]. Importantly, precision is improved only when cystatin C is added to creatinine (eGFRcr-cys), not when it replaces creatinine (i.e. eGFRcys) [8]. Although eGFRcys and eGFRcr-cys are recommended as confirmatory tests for decreased eGFRcr [1, 6], estimates without creatinine and demographics are needed.
Metabolomics has promise to be transformative in the field of GFR estimation since it allows for rapid screening of thousands of metabolites (molecular weight ∼50–1500 Da), which are often cleared primarily by glomerular filtration, similar to creatinine. Novel metabolites that are strongly correlated with GFR could serve as filtration markers with or in place of creatinine. Previous metabolomic studies used eGFRcr rather than mGFR as the gold standard, leaving doubt as to whether metabolites estimate GFR or non-GFR determinants of serum creatinine [9–14]. Finally, the Metabolon platform has the advantage of already identifying hundreds of the metabolites, facilitating development of targeted mass spectrometry assays that are both accurate and easily included in multiplex panels. Inclusion of multiple markers should make estimates more robust to factors influencing only one metabolite. Ideally such panels would provide highly accurate, robust estimates of GFR. Exclusion of creatinine would make panels more suitable to use when the validity of creatinine is questionable (e.g. muscle wasting). Exclusion of demographics is desirable since defining race is problematic and the relationship of age and sex to metabolism may vary across different settings (e.g. diet, geography and illness).
This article aims to test central concepts in a pathway toward developing a more accurate and generalizable GFR estimate based on metabolomics. We hypothesized that multiple metabolites with correlations to mGFR that are better than or equal to creatinine can be discovered and validated, targeted assays can be developed and multiplexed with high precision and validity and an initial panel of metabolites can allow for GFR estimation without creatinine or demographics that is at least as accurate as eGFRcr and eGFRcys using the CKD-EPI equation.
MATERIALS AND METHODS
The design of this proof-of-concept study was cross-sectional, with discovery using untargeted assays in one cohort and validation using untargeted assays in a second cohort and repeat testing in both cohorts using targeted assays for a subset of promising metabolites (see Supplementary material for additional details).
Participants
Two populations were chosen to be complementary, with different GFR ranges, methods of GFR measurement and racial composition, to develop metabolite associations with mGFR that are likely to be generalizable. African American Study of Kidney (AASK) participants with consistent mGFR at the 48-month follow-up visit were selected as the discovery study population. Multi-Ethnic Study of Atherosclerosis (MESA) participants with mGFR were selected as the validation study population (n = 265).
Laboratory methods for index tests
Untargeted and targeted metabolomic profiling was performed at Metabolon (Durham, NC, USA). Untargeted gas chromatography/dual mass spectrometry– and liquid chromatography/dual mass spectrometry–based metabolomics semiquantification followed published methods, including updates to the platform finalized during the course of this study, detailed in the Supplementary material [15, 16]. For the 15 most promising metabolites with available pure standards identified in the AASK and previous studies (prior to collection of MESA data), Metabolon developed targeted assays for absolute quantification using negative and positive ionization ultra-performance liquid chromatography–tandem mass spectrometry (UPLC–MS/MS) methods performed on an Agilent 1290 UPLC system coupled to a Sciex QTrap5500 mass spectrometer. The metabolites were also required to have reliable assays and be good candidates for multiplexing. Targeted assays had coefficients of variation typically <5% in a serum quality control sample during qualification of the targeted assays and continued to perform similarly during sample analysis. For consistency with the approach of developing a panel assayed by mass spectrometry we used the targeted mass spectrometry assays for creatinine, which showed excellent agreement with the original clinical chemistry assays. Cystatin C was assayed on serum frozen at −70°C in the MESA on the Roche Cobas using Gentian assays, which was traceable to International Federation for Clinical Chemists (IFCC) Working Group for the Standardization of Serum Cystatin C and the Institute for Reference Materials and Measurements certified reference materials [17] and in the AASK F48 using a particle-enhanced immunonephelometric assay (N Latex Cystatin C, Dade Behring, IL, USA) at the Cleveland Clinic Foundation (CCF) laboratory (calibrated to IFCC values using published methods; 1.12 × (0.105 + 0.848 × CCF cystatin C) [8].
Laboratory methods for reference tests
GFR was measured by urinary clearance of iothalamate in the AASK [18] and plasma clearance of iohexol in the MESA [17] using established procedures detailed previously. In the AASK, mGFR was computed as the weighted mean of four clearance periods of 25–35 min duration. In the MESA, mGFR was computed from samples of 10 to 300 min. mGFR was indexed per 1.73 m2 of body surface area (BSA) in both studies. BSA was calculated using the DuBois and DuBois formula (0.20247 × height in meters0.725 × weight in kilograms0.425).
Data analytical methods
Metabolite levels were log transformed and metabolites were ranked by correlation with log mGFR and compared across the AASK and MESA. Metabolites for targeted assays were selected among those strongly correlated with mGFR with feasible, reliable, targeted assays and with additional metabolites selected if they added independent predictive value in stepwise regression or were viewed as promising in other ongoing studies.
Targeted metabolite assays were tested for linearity, interference and precision. Linear regression using backward stepwise selection was used to develop eGFR equations with the metabolites, with and without creatinine, age, sex and race (P-enter = 0.01 and P-exit = 0.05). The performance of eGFR equations in estimating mGFR was quantified using several established metrics [5]. Root mean square error (RMSE) estimates the residual of the observed from predicted values. Used on the log scale, it approximates the standard deviation (SD) of the percent error in estimation. In addition, we used the interquartile range (IQR) of the mean difference between mGFR and eGFR to quantify precision. The frequency of large errors was quantified by 1-P30 and 1-P20, quantifying the percentage of errors >30% and 20% of mGFR, respectively, reflecting the desire to achieve lower error rates at the more accurate 20% standard. The CKD-EPI 2009 creatinine equation, which models creatinine as a linear spline with a sex-specific knot, was used as the reference equation since it is the established clinical standard [1]. The CKD-EPI 2012 cystatin C and creatinine–cystatin C equations were used as alternative reference equations [8]. Finally, we developed an equation in the AASK and evaluated its performance in the MESA. This analysis excluded 12 individuals in the AASK with missing cystatin C data to allow comparison with established equations including cystatin C as well as creatinine. Since equations developed in other populations could have nonzero bias, we also compared equations after calibration to zero bias (on the log scale).
All statistical tests accounted for the paired design whereby a reference and comparison eGFR are calculated for each mGFR. To calculate P-values, we used a signed-rank test for RMSE, McNemar’s chi-squared for 1-P30 and 1-P20 and bootstrap methods for SD and IQR. Sensitivity analyses calculated area under the curve in the context of an receiver operating curve (AUC) for detecting mGFR <60 mL/min/1.73 m2, recognizing this analysis is limited by dependence on the distribution of mGFRs in the study as well as concordance in predicting mGFR category and additional measures of precision (SD and IQR).
RESULTS
Study populations
The AASK participants were African American with a mean mGFR of 47 mL/min/1.73 m2, while the MESA participants comprised equal proportions of whites and African Americans with a higher mean mGFR of 73 mL/min/1.73 m2 (Table 1).
Table 1.
Characteristics and concentrations of targeted metabolites among study participants in the AASK and MESA
| Characteristic | AASK (n = 200) |
MESA (n = 265) |
||
|---|---|---|---|---|
| Mean (SD) | 5th–95th percentile | Mean (SD) | 5th–95th percentile | |
| Sex (male), % | 69 | 53 | ||
| Black, % | 100 | 46 | ||
| Diabetes, % | 0 | 12 | ||
| Age (years) | 60 (9) | 44–73 | 71 (9) | 57–85 |
| BMI | 30 (6) | 22–42 | 30 (5) | 23–39 |
| Creatinine (mg/dL) | 1.9 (.9) | 1.1–3.8 | 0.93 (0.35) | 0.6–1.36 |
| Cystatin Ca (mg/L) | 1.8 (0.6) | 1.1–3.1 | 1.0 (0.3) | 0.7–1.7 |
| mGFRb (mL/min/1.73 m2) | 47 (17) | 19–75 | 73 (19) | 44–105 |
| mGFR<60 mL/min/1.73 m2, % | 78 | 24 | ||
| Targeted serum metabolites | ||||
| N-acetylthreonine (μg/mL) | 0.16 (0.08) | 0.08–0.34 | 0.09 (0.04) | 0.06–0.14 |
| Pseudouridine (μg/mL) | 1.7 (1.2) | 0.9–4.8 | 0.95 (0.4) | 0.63–1.48 |
| N-acetylserine (μg/mL) | 0.15 (0.07) | 0.08–0.32 | 0.15 (0.08) | 0.10–0.23 |
| Creatinine (mg/dL) | 1.9 (0.9) | 1.0–3.7 | 0.95 (0.35) | 0.61–1.41 |
| Meso-erythritol (μg/mL) | 1.5 (0.9) | 0.7–3.3 | 0.89 (0.38) | 0.48–1.43 |
| Arabitol (μg/mL) | 0.86 (0.46) | 0.41–1.96 | 0.56 (0.24) | 0.34–0.9 |
| Myo-inositol (μg/mL) | 11 (5) | 6–22 | 6.2 (2.4) | 3.7–10.4 |
| Urea (μg/mL) | 34 (18) | 16–65 | 17 (7) | 10–29 |
| N-acetylalanine (μg/mL) | 0.35 (0.11) | 0.23–0.57 | 0.22 (0.06) | 0.16–0.32 |
| 3-Indoxylsulfate (μg/mL) | 2.2 (2.3) | 0.5–4.4 | 1.1 (0.8) | 0.30–2.3 |
| Phenylacetyl-glutamine (μg/mL) | 1.6 (1.7) | 0.3–3.9 | 1 (0.76) | 0.23–2.11 |
| Tryptophan (μg/mL) | 12 (3) | 7–16 | 8.2 (1.9) | 5.2–11.4 |
| Kynurenine (μg/mL) | 0.54 (0.18) | 0.32–0.86 | 0.39 (0.11) | 0.22–0.6 |
| 3-Methyl-histidine (μg/mL) | 3.7 (3.7) | 0.4–11 | 2 (1.7) | 0.30–5.8 |
| Trans-4-hydroxy-proline (μg/mL) | 3.6 (1.8) | 1.6–7.2 | 1.9 (1.1) | 0.90–3.8 |
Cystatin C data are missing for 12 participants in the AASK.
Average of three consistent GFR measurements by design in the AASK; creatinine converted to mg/dL (1 μg/mL = 0.1 mg/dL).
Discovery and validation using untargeted assays
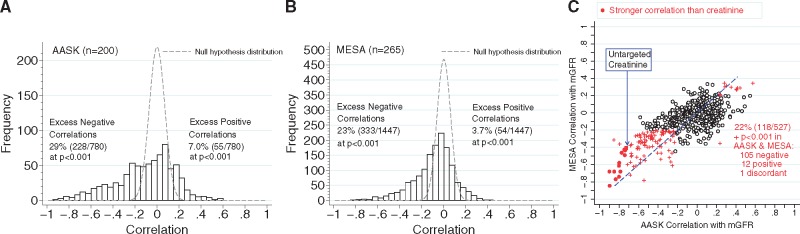
Using untargeted assays, metabolite levels showed a wide distribution of correlations with mGFR in both the AASK and MESA populations. In both populations there was a marked excess of correlations outside the range expected under the null hypothesis, especially for negative correlations, including 29% of the AASK metabolites and 23% of the MESA metabolites (P < 0.001) (Figure 1 and Supplementary data, Table S1). Positive correlations at this level of statistical significance (P < 0.001) were observed in 7.0 and 3.7% of the AASK and MESA populations, far exceeding the 0.1% expected due to chance alone (Figure 1 panels A and B, respectively). Metabolites with stronger correlations in the AASK were more likely to have stronger correlations in the MESA (Figure 1 panel C); of the 283 metabolites with P < 0.001 in the AASK that were also measured in the MESA, 42% had P < 0.001 in the MESA. Among these, the untargeted assay for creatinine had a correlation of −0.73 in the AASK and −0.41 in the MESA (P < 10−11 in each). Eighteen metabolites (13 known and 5 unknown) were more strongly negatively correlated with mGFR in the AASK than the untargeted assay for creatinine, as were 93 metabolites in the MESA (62 known and 31 unknown). Of the 18 in the AASK, 14 were also measured in the MESA, and 12 of the 14 were more negatively correlated than the untargeted assay for creatinine (Supplementary data, Table S1). Among metabolites measured in both the AASK and MESA, tryptophan was the most positively correlated with mGFR in the AASK (r = 0.58, P <10−15) and MESA (r = 0.34, P = 10−8; Supplementary data, Table S1).
FIGURE 1.
Correlations of untargeted metabolite screen with mGFR in the (A) AASK and (B) MESA and (C) a comparison of the correlations for overlapping metabolites in the two studies. In (A) and (B), dashed lines show the null hypothesis expected distribution of correlations. In (C), red indicates correlations significant at P < 0.001 and dots indicate correlations stronger than the untargeted serum creatinine.
Repeated testing using targeted assays
Targeted assays generally improved correlations with mGFR. Of 30 comparisons of targeted and untargeted assays across the two studies, 25 showed correlations with mGFR using the targeted assays whose absolute value was equal or higher than that for the untargeted assay (Supplementary data, Table S2). However, the magnitude of the improvement varied markedly across metabolites. The targeted creatinine assay was much more closely correlated with mGFR than untargeted creatinine in both studies (r =−0.84 and −0.55 versus −0.73 and −0.41). Targeted creatinine was highly correlated with the Jaffe assay in the AASK and the enzymatic creatinine assay in the MESA (r = 0.984 and 0.993, intercepts of −0.04 and 0.02 mg/dL and slopes of 0.989 and 0.996, respectively).
Performance of eGFR based on single metabolites
We first examined the performance of single metabolites in each study separately compared with eGFRcr using the CKD-EPI equation (Table 2). For eGFRcr, 1-P30 was higher in the AASK than in the MESA (11.5% versus 18.5%), but 1-P20 was similar (31.0% and 33.2%), as was RMSE (0.205 and 0.200) (Table 2 top row). Without using demographic characteristics, a number of metabolites performed as well or better in RMSE than creatinine alone (N-acetylthreonine, pseudouridine and N-acetylserine in both studies and meso-erythritol, arabitol, myo-inositol and N-acetylalanine in the MESA only). Creatinine alone performed worse than eGFRcr in both the AASK and MESA. No single metabolite without demographics performed better than eGFRcr in the AASK, while two metabolites performed better than eGFRcr in the MESA (N-acetylthreonine and pseudouridine).
Table 2.
Performance of targeted metabolites in GFR estimation in the AASK and MESA
| Models for estimating mGFR | AASK (n = 200) |
MESA (n = 265) |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Without age and sex |
With age and sex |
Without age and sex |
With age, sex and race |
||||||||||
| RMSE | 1-P30 (%) | 1-P20 (%) | RMSE | 1-P30 (%) | 1-P20 (%) | RMSE | 1-P30 (%) | 1-P20 (%) | RMSE | 1-P30 (%) | 1-P20 (%) | ||
| Reference model | CKD-EPI eGFRcr | 0.205 | 11.5 | 31.0 | 0.200 | 18.5 | 33.2 | ||||||
| Single metabolites | N-acetylthreonine | 0.194 | 13.0 | 24.0 | 0.184 | 11.0 | 25.0 | 0.167*** | 7.9*** | 21.9** | 0.144*** | 4.5*** | 15.1*** |
| Pseudouridine | 0.210 | 12.5 | 29.5 | 0.210 | 12.0 | 30.0 | 0.163*** | 6.0*** | 19.6*** | 0.137*** | 2.6*** | 13.2*** | |
| N-acetylserine | 0.219 | 18.0 | 30.0 | 0.216 | 16.0 | 28.0 | 0.204 | 13.6 | 30.6 | 0.182* | 8.7*** | 23.0*** | |
| Creatinine | 0.226 | 14.5 | 36.0 | 0.190* | 10.5 | 24.5* | 0.238 | 20.8 | 40.8 | 0.149*** | 4.9*** | 15.1*** | |
| Meso-erythritol | 0.228 | 17.5 | 33.5 | 0.228 | 18.5 | 33.5 | 0.211 | 14.0 | 34.0 | 0.190 | 9.8** | 27.2 | |
| Arabitol | 0.241 | 18.5 | 34.5 | 0.239 | 16.5 | 33.0 | 0.216 | 13.2 | 31.7 | 0.185* | 10.2** | 25.7 | |
| Myo-inositol | 0.244 | 20.5 | 38.5 | 0.245 | 21.0 | 36.0 | 0.218 | 14.3 | 32.8 | 0.194 | 9.8** | 31.3 | |
| Urea | 0.261 | 21.5 | 39.5 | 0.258 | 19.5 | 34.0 | 0.245 | 18.5 | 38.5 | 0.210 | 15.1 | 32.1 | |
| N-acetylalanine | 0.267 | 18.5 | 38.5 | 0.263 | 20.0 | 35.5 | 0.192 | 11.3* | 26.0 | 0.169*** | 7.9*** | 16.6*** | |
| 3-Indoxylsulfate | 0.326 | 31.5 | 51.0 | 0.328 | 31.5 | 51.5 | 0.265 | 23.0 | 41.1 | 0.227 | 15.5 | 34.3 | |
| Phenylacetylglutamine | 0.349 | 33.0 | 52.5 | 0.349 | 32.0 | 54.0 | 0.259 | 23.4 | 42.6 | 0.228 | 15.1 | 32.1 | |
| Tryptophan | 0.359 | 36.5 | 58.0 | 0.360 | 35.5 | 57.5 | 0.267 | 21.5 | 41.5 | 0.229 | 17.7 | 30.9 | |
| Kynurenine | 0.359 | 35.0 | 50.0 | 0.360 | 34.5 | 51.0 | 0.258 | 20.4 | 39.6 | 0.217 | 12.8 | 33.6 | |
| 3-Methyl-histidine | 0.380 | 36.0 | 59.5 | 0.380 | 38.0 | 57.5 | 0.273 | 22.6 | 42.3 | 0.230 | 16.2 | 34.0 | |
| Trans-4-hydroxy-proline | 0.394 | 39.5 | 58.5 | 0.396 | 38.0 | 57.0 | 0.282 | 25.7 | 43.8 | 0.237 | 15.5 | 34.3 | |
| Panels | Best by stepwise among metabolites | 0.156** | 3.5** | 17.5** | 0.147*** | 4.5** | 14.5*** | 0.146*** | 3.0*** | 15.8*** | 0.124*** | 1.9*** | 10.6*** |
| Best by stepwise excluding cr | 0.161* | 5.0* | 21.0* | 0.159** | 4.5* | 18.0** | 0.148*** | 3.8*** | 17.4*** | 0.129*** | 2.3*** | 10.8*** | |
Metabolites are ordered by increasing RMSE in the AASK. RMSE is calculated on the log(mGFR) scale.
CKD-EPI equations always include demographics and hence do not have prediction metrics without demographics.
P ≤ 0.05, ** P ≤ 0.01, *** P ≤ 0.001 for improvement compared with CKD-EPI eGFRcr (two-tailed test; worse performance than eGFRcr is less relevant and not footnoted).
Performance of eGFR based on a metabolite panel
We next examined panels of metabolites selected by stepwise regression among the 15 targeted metabolite assays in each study separately. In contrast to the findings of the single metabolites, panels of multiple metabolites showed better performance than eGFRcr (Table 2 bottom section). Panels including metabolites, creatinine and demographic characteristics had by far the lowest RMSE in both the AASK and MESA (0.147 and 0.124, respectively) and the best 1-P30 (4.5 and 1.9%, respectively) and 1-P20 (14.5 and 10.6%, respectively). Panels with only metabolites, excluding creatinine as well as demographic characteristics, were still more accurate (RMSE of 0.161 and 0.148, 1-P30 of 5.0 and 3.8% and 1-P20 of 21.0 and 17.4% in the AASK and MESA, respectively) than eGFRcr; the improvement was statistically significantly better for all metrics with 1-P30 less than half for eGFRcr.
Validation in the MESA of an AASK equation
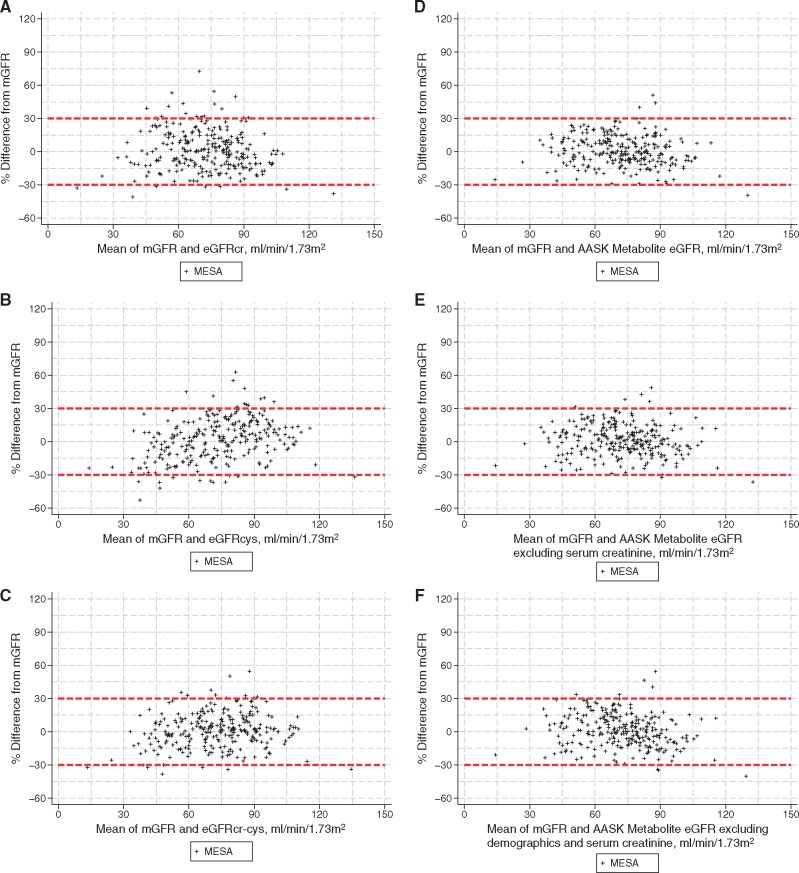
The metabolite equations developed in the AASK (detailed in Supplementary data, Tables S3 and S4) were unbiased and were more accurate than eGFRcr (P < 0.05 for five of six comparisons for both RMSE and 1-P30 metrics) (Table 3). In the MESA, the AASK metabolite equations had comparable accuracy to the equations in the AASK (P < 0.01 for six of six comparisons to eGFRcr) but underestimated mGFR with biases ranging from −1.6 to −3.8 mL/min/1.73 m2. After eGFRcr was recalibrated to remove the bias in the MESA, its accuracy improved (1-P30 decreased from 18.5 to 9.4%), but the AASK metabolite equations were still significantly better. Finally, we used CKD-EPI cystatin equations as alternative reference equations. Similar to prior studies, eGFRcys showed similar performance to eGFRcr in most comparisons while eGFRcr-cys showed better performance than eGFRcr in all comparisons. The AASK metabolite equations were substantially more accurate than eGFRcys in 17 of 18 comparisons, but only 8 of 18 comparisons to eGFRcr-cys in both the AASK and MESA (Table 2). Notably, the AASK metabolite equation excluding creatinine and demographics has numerically higher 1-P30 than eGFRcr-cys, but this difference could have been due to chance (6.4% versus 3.7%; P = 0.30). Categorical analyses for AUC and concordance for mGFR staging categories showed similar results, although power was more limited (Supplementary data, Table S5). Individual estimates for calibrated equations are shown in Figure 2. Subgroup analyses by race and sex in the MESA (Supplementary data, Table S5) suggest improved performance was similar across subgroups, although we observed systematic bias by sex for metabolite equations as was previously observed for eGFRcr and eGFRcr-cys [17].
Table 3.
Discovery in the AASK and validation in the MESA of a metabolite model eGFR compared with using estimated from creatinine, cystatin and demographics
| Equations for estimating mGFR | Development AASK (n=188) |
Validation MESA (n=265) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Development |
Recalibrated to zero bias on log scale |
Validation |
Recalibrated to zero bias on log scale |
||||||||
| RMSE | 1-P30 | Median bias (eGFR- mGFR) | RMSE | 1-P30 | RMSE | 1-P30 | Median bias (eGFR- mGFR) | RMSE | 1-P30 | ||
| Reference equation | CKD-EPI eGFRcr (%) | 0.2071 | 11.2 | 1.024 | 0.206 | 11.7 | 0.200 | 18.5 | 7.186 | 0.176 | 9.4 |
| Alternative reference equations | CKD-EPI eGFRcys (%) | 0.221 | 10.6 | –5.443 | 0.164** | 8.0 | 0.191 | 9.1** | 3.018 | 0.191 | 9.4 |
| CKD-EPI eGFRcr-cys (%) | 0.166*a | 3.7**a | –3.040 | 0.144*** | 5.3** | 0.169* | 9.1*** | 6.369 | 0.155*** | 6.8 | |
| AASK metabolite equation | Including demographics (%) | 0.144***a,b | 3.7***a | –0.640 | 0.144*** | 3.7*** | 0.137***a,b | 1.9***a,b | –1.589 | 0.136***a | 1.5%***a,b |
| Excluding creatinine (%) | 0.153***a | 4.3*a | –0.423 | 0.153*** | 4.3** | 0.152***a,b | 3.0***a,b | –3.858 | 0.139***a | 2.6***a,b | |
| Excluding creatinine and demographics (%) | 0.157*a | 6.4 | –0.137 | 0.157* | 6.4 | 0.157***a | 3.4***a,b | –3.133 | 0.149**a | 3.0**a | |
Equations in the AASK are shown in Supplementary data, Table S4. RMSE is calculated on the log(mGFR) scale.
P-values compared with RMSE and 1-P30 are two-tailed (worse performance than the reference eGFR is less relevant and not footnoted).
P ≤ 0.05, ** P ≤ 0.01, *** P ≤ 0.001 for improvement compared with eGFRcr. aP ≤ 0.05 compared with eGFRcys. bP ≤ 0.05 compared with eGFRcr-cys.
FIGURE 2.
Percent difference between eGFR and mGFR in validation MESA. Panels show estimates calibrated to zero bias for CKD-EPI (A) eGFRcr, (B) eGFRcys, (C) eGFRcr-cys and the AASK metabolite including (D) demographics and excluding (E) creatinine and (F) creatinine and demographics. Supplementary figures show the data without calibration as well as for the AASK and MESA population combined.
DISCUSSION
Our goal in this study was to identify novel glomerular filtration–related markers using metabolomics that are better than or equal to creatinine, the most established metabolite in use for GFR estimation. We found that more than one-quarter of metabolites measured using nontargeted assays were significantly correlated with mGFR. The correlations were mostly negative, consistent across two-study populations and included a dozen metabolites that were more strongly correlated with mGFR than creatinine in both studies. Targeted assays for a subset of the promising metabolites were developed efficiently and increased the strength of correlation with mGFR. Finally, targeted assays for panels of metabolites without creatinine provided more accurate estimation of mGFR than the CKD-EPI eGFRcr equation. Metabolite equations developed in the AASK performed well in the MESA, with low bias and excellent accuracy. These AASK equations were better than eGFRcr and eGFRcys using the CKD-EPI equations, even after recalibration of the CKD-EPI equations to remove bias, but were not consistently significantly better than bias-corrected eGFRcr-cys. Excluding creatinine and demographics still led to a metabolite equation with good accuracy (>30% from mGFR, 1-P30 <6.4%), suggesting a path to eGFR tests that are accurate and independent of creatinine and demographics.
The optimal composition of panels of metabolites and whether and when it may be beneficial to include creatinine, cystatin C or other low molecular weight protein filtration markers (e.g. beta-2 microglobulin or beta-trace protein) for clinical use remains to be determined. However, our results provide proof of the concept that a panel of novel metabolites can accurately estimate mGFR. Metabolite panels that improve on the precision of eGFRcr-cys in a wide range of settings would be useful in clinical settings where precise and accurate GFR estimates are needed for clinical action, possibly providing an alternative to GFR measurement, which although valuable, is cumbersome, not easy to standardize and too often clinically unavailable. Alternatively, metabolite panels with similar performance to that shown here are as accurate as eGFRcr-cys in general population and general CKD settings, would be useful in settings where creatinine or cystatin are unreliable as filtration markers. Studies quantifying the degree of benefit in such settings will be important to guide future use of new estimates such as the one we develop here.
This study is the first to quantify correlations of untargeted metabolomics assays with mGFR rather than eGFR, develop targeted metabolite assays, combine them into a metabolite panel eGFR and validate it in a second population. While untargeted metabolomic assays of ∼1000 compounds, including creatinine and urea, in a single sample are considered semiquantitative, we found that results were highly correlated with mGFR. As hypothesized, these low molecular weight compounds are a promising source for discovering powerful filtration-related markers. The metabolites we focussed on were also found to be strongly correlated with eGFRcr in other studies. Recently, Sekula et al.[9] reported 56 metabolites that replicated as associated with eGFRcr, including 6 metabolites that were consistently strongly (r <−0.50) correlated with eGFRcr (pseudouridine, c-mannosyltryptophan, N-acetylalanine, erythronate, myo-inositol and N-acetylcarnosine). In our study, all except N-acetylcarnosine were very strongly negatively correlated with mGFR. In fact, correlations were even stronger for all of the three we tested using targeted assays. Of interest, tryptophan was positively correlated with mGFR (lower levels at lower levels of mGFR), which is likely due to increased catabolism [19], but contributed to the accuracy of panels for GFR estimation. Thus tryptophan can be considered a GFR marker even though it is not a retained solute.
Using multiple noncorrelated filtration-related markers in a panel reduces the effect of non-GFR determinants of each single marker on the overall panel. For example, the AASK model containing four metabolites without creatinine or demographics has a median coefficient of −0.23 for the metabolites compared with −1.2 for creatinine alone in the CKD-EPI equation (in the high creatinine part of the spline). As such, each coefficient contributes substantially less to the overall GFR estimation than creatinine does in eGFRcr. We hypothesize that panels where no one marker contributes to the overall estimate disproportionately will be more robust across very diverse study populations. In the current panel, the metabolites that we identified are within the amino acid, nucleic acid and carbohydrate metabolic pathways and their serum concentrations may be affected by non-GFR determinants as well as by GFR. Further knowledge of non-GFR determinants of panel metabolites will be helpful to clinical interpretation of corresponding GFR estimates. Genetic non-GFR effects undoubtedly exist but are often small. For example, infrequent variants in ACY1, whose product catalyzes the hydrolysis of acetylated L-amino acids, explain ∼0.5% of the variance in N-acetylthreonine concentrations. This is analogous to common genetic variants that have modest effects on serum creatinine and cystatin C levels but are not included in GFR estimating equations [20, 21]. Theoretical considerations suggest a concern that diet and supplements may influence the serum tryptophan concentration. In particular, this marker will need to be extensively tested for generalizability across a variety of clinical settings before it can be recommended as part of a panel of markers to estimate GFR in a broad population setting.
Our results suggest that studies of metabolite association with subsequent CKD progression or end-stage renal disease incidence are likely to be strongly confounded by baseline GFR, since more than one-quarter of the metabolome is elevated at low GFR. Therefore, it will be important to replicate previously reported associations of metabolites with eGFRcr and risk of CKD progression [10–14, 22] and test their independence of baseline GFR. Improved estimates of GFR such as the ones we presented could allow future studies to distinguish progression markers from filtration markers.
The strengths of the study include validation of the initial discovery and demonstration of the consistency of the results across two complementary populations with mGFR and development of highly precise and accurate targeted assays for metabolites identified in the initial screen. Validation of the global discovery showed the validity of the approach in that 42% of markers with P < 0.001 in the AASK have P < 0.001 in the MESA, much higher than the 0.1% expected due to chance alone. However, it also points out that metabolites that did not validate likely included false positives and the expected overoptimism included in initial discovery prior to replication.
This study also has limitations. The AASK is a study of African Americans with hypertensive kidney disease and as such is a rather homogeneous population with respect to race, geographical location and diet (US based) and cause of kidney disease. We were able to replicate the findings in US Whites and Blacks with and without kidney disease and thus know that these results are not due to Black ethnicity. However, the relative homogeneity of the samples does not allow us to test the generalizability of the findings. Sample handling in the AASK did not follow a standardized protocol and the storage period was many years. As a result, some metabolites may have been missed, but those that were identified are likely to be robust to a range of handling techniques and long-term storage. The identity of some of the most strongly correlated metabolites with mGFR was unknown, limiting our current panel but providing an opportunity for further improvement on this proof of concept. GFR measurement is known to be imprecise, but this inflates the reported GFR estimation errors [18]. Furthermore, iothalamate and iohexol GFR measurement methods differ systematically [23] and standardization of GFR for BSA may not optimally deal with variation in body compositon. Importantly, the performance and practicality of combining metabolites with low molecular weight proteins such as cystatin C was not tested. However, our focus on metabolites measured in a multiplex panel has the potential for economies of scale. Future steps should include evaluation of panels including cystatin C and potentially other low molecular weight proteins. A better understanding of the metabolism of all components of the panel used to estimate GFR will be useful to better predict when they are influenced by non-GFR determinants. The magnitude of such influences on the overall GFR estimate across a range of clinical settings needs to be quantified, particularly in clinical settings where creatinine and cystatin are known to be unreliable.
In summary, the algorithms presented here provide a proof of concept in realizing the potential of translating untargeted metabolomic screening to algorithms. Given the known limitations of serum creatinine and widespread use of GFR estimation, the clinical implications that a panel of metabolites can provide an accurate estimate of GFR with or without serum creatinine or demographics could be substantial if these initial results can be taken through the full diagnostic test development process. Testing in multiple populations should be conducted to confirm the external generalizability of a metabolite panel. Importantly, the final robust algorithm that can be used to estimate GFR would ideally be developed in a more diverse dataset.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank the other investigators, the staff and participants in the MESA for valuable contributions. A full list of participating MESA investigators and institutions can be found at www.mesa-nhlbi.org. This research was also supported by the National Institutes of Health; contracts N01-HC-95159, N01-HC-95160, N01-HC-95161, N01-HC-95162, N01-HC-95163, N01-HC-95164, N01-HC-95165, N01-HC-95166, N01-HC-95167, N01-HC-95168 and N01-HC-95169 from the National Heart, Lung and Blood Institute and grants UL1-TR-000040 and UL1-TR-001079 from the National Center for Research Resources.
FUNDING
Research reported in this publication was supported by the National Institute of Diabetes and Digestive and Kidney Diseases of the National Institutes of Health (NIH; grant R01DK087961). This publication was made possible by the Johns Hopkins Institute for Clinical and Translational Research, which is funded in part by grant UL1 TR 001079 from the National Center for Advancing Translational Sciences, a component of the NIH, and the NIH Roadmap for Medical Research. Its contents are solely the responsibility of the authors and do not necessarily represent the official view of the Johns Hopkins Institute for Clinical and Translational Research, National Center for Advancing Translational Sciences or the NIH. The funders of this study had no role in the study design; collection, analysis, and interpretation of the data; writing of the report or the decision to submit the report for publication.
AUTHORS’ CONTRIBUTIONS
J.C., L.A.I. and A.S.L. contributed to all aspects of the article. Y.S. and J. Chen conducted the analysis with T.G., M.S. and R.P. providing input. T.S. led the MESA data collection with W.S.P. and M.G.S. L.F. and K.G. led and supervised the metabolomic analysis with R.P., who was also involved with the conception and design of the study. All authors assisted with revising the article and approved the final version.
CONFLICT OF INTEREST STATEMENT
Provisional patent: Coresh, Inker and Levey filed 8/15/2014 – ‘Precise estimation of GFR from multiple biomarkers’ (PCT/US2015/044567). Tufts Medical Center, John Hopkins University and Metabolon have a collaboration agreement to develop a product to estimate GFR from a panel of markers. R.P., L.F. and K.G. are employees of Metabolon and as such have affiliations with or financial involvement with Metabolon. Metabolon owns issued and pending patents in the USA and foreign countries, including those based on PCT/US2014/037762, ‘Biomarkers Related to Kidney Function and Methods Using the Same.’ Priority date: 14 May 2013.
The results presented in this article have not been published previously in whole or part except in abstract format.
REFERENCES
- 1. Levin A, Stevens PE, Bilous RW. et al. KDIGO 2012 clinical practice guideline for the evaluation and management of chronic kidney disease. Kidney Int Suppl 2013; 3: 1–150 [DOI] [PubMed] [Google Scholar]
- 2. Stevens LA, Coresh J, Greene T. et al. Assessing kidney function–measured and estimated glomerular filtration rate. N Engl J Med 2006; 354: 2473–2483 [DOI] [PubMed] [Google Scholar]
- 3. Horio M, Imai E, Yasuda Y. et al. Performance of GFR equations in Japanese subjects. Clin Exp Nephrol 2013; 17: 352–358 [DOI] [PubMed] [Google Scholar]
- 4. Jessani S, Levey AS, Bux R. et al. Estimation of GFR in South Asians: a study from the general population in Pakistan. Am J Kidney Dis 2014; 63: 49–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Levey AS, Stevens LA, Schmid CH. et al. A new equation to estimate glomerular filtration rate. Ann Intern Med 2009; 150: 604–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Levey AS, Becker C, Inker LA.. Glomerular filtration rate and albuminuria for detection and staging of acute and chronic kidney disease in adults: a systematic review. JAMA 2015; 313: 837–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ballew SH, Chen Y, Daya NR. et al. Frailty, kidney function, and polypharmacy: the atherosclerosis risk in communities (ARIC) study. Am J Kidney Dis 2017; 69: 228–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Inker LA, Schmid CH, Tighiouart H. et al. Estimating glomerular filtration rate from serum creatinine and cystatin C. N Engl J Med 2012; 367: 20–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sekula P, Goek ON, Quaye L. et al. A metabolome-wide association study of kidney function and disease in the general population. J Am Soc Nephrol 2016; 27: 1175–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Goek ON, Doring A, Gieger C. et al. Serum metabolite concentrations and decreased GFR in the general population. Am J Kidney Dis 2012; 60: 197–206 [DOI] [PubMed] [Google Scholar]
- 11. Goek ON, Prehn C, Sekula P. et al. Metabolites associate with kidney function decline and incident chronic kidney disease in the general population. Nephrol Dial Transplant 2013; 28: 2131–2138 [DOI] [PubMed] [Google Scholar]
- 12. Rhee EP, Clish CB, Ghorbani A. et al. A combined epidemiologic and metabolomic approach improves CKD prediction. J Am Soc Nephrol 2013; 24: 1330–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Niewczas MA, Sirich TL, Mathew AV. et al. Uremic solutes and risk of end-stage renal disease in type 2 diabetes: metabolomic study. Kidney Int 2014; 85: 1214–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yu B, Zheng Y, Nettleton JA. et al. Serum metabolomic profiling and incident CKD among African Americans. Clin J Am Soc Nephrol 2014; 9: 1410–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Evans AM, DeHaven CD, Barrett T. et al. Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Anal Chem 2009; 81: 6656–6667 [DOI] [PubMed] [Google Scholar]
- 16. Evans AM, Bridgewater BR, Liu Q. et al. High resolution mass spectrometry improves data quantity and quality as compared to unit mass resolution mass spectrometry in high-throughput profiling metabolomics. Metabolomics 2014; 4: 132 [Google Scholar]
- 17. Inker LA, Shafi T, Okparavero A. et al. Effects of race and sex on measured GFR: the multi-ethnic study of atherosclerosis. Am J Kidney Dis 2016; 68: 743–751 [DOI] [PubMed] [Google Scholar]
- 18. Kwong YT, Stevens LA, Selvin E. et al. Imprecision of urinary iothalamate clearance as a gold-standard measure of GFR decreases the diagnostic accuracy of kidney function estimating equations. Am J Kidney Dis 2010; 56: 39–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schefold JC, Zeden JP, Fotopoulou C. et al. Increased indoleamine 2, 3-dioxygenase (IDO) activity and elevated serum levels of tryptophan catabolites in patients with chronic kidney disease: a possible link between chronic inflammation and uraemic symptoms. Nephrol Dial Transplant 2009; 24: 1901–1908 [DOI] [PubMed] [Google Scholar]
- 20. Kottgen A, Glazer NL, Dehghan A. et al. Multiple loci associated with indices of renal function and chronic kidney disease. Nat Genet 2009; 41: 712–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. O’Seaghdha CM, Tin A, Yang Q. et al. Association of a cystatin C gene variant with cystatin C levels, CKD, and risk of incident cardiovascular disease and mortality. Am J Kidney Dis 2014; 63: 16–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rhee EP, Clish CB, Pierce KA. et al. Metabolomics of renal venous plasma from individuals with unilateral renal artery stenosis and essential hypertension. J Hypertens 2015; 33: 836–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Seegmiller JC, Burns BE, Schinstock CA. et al. Discordance between iothalamate and iohexol urinary clearances. Am J Kidney Dis 2016; 67: 49–55 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.