Abstract
Purpose: Bladder cancer (BC) is the most common urinary cancer among men with a high rate of deaths despite the improved medical technology and treatment. Recent evidence demonstrated that Mex-3 RNA-Binding Family Member C (MEX3C) plays various roles in different biological activities, but its molecular mechanisms underlying the pathogenesis of BC remain unclear yet. The aim of this research was to explore the expression patterns of MEX3C and its biological functions in human BC.
Materials and methods: The Cancer Genome Atlas (TCGA) and Oncomine databases were jointly used to analyze the expression of MEX3C in BC and its correlation with the clinicopathological features, while real-time PCR and immunohistochemistry analysis were used to verify the predicted results. Wound-healing assay, Matrigel invasion assay, BODIPY staining and Western blot analysis were used in a cell model to assess the effect of MEX3C on the lipid metabolism, invasion and migration of BC and its mechanisms.
Results: MEX3C was highly expressed in BC tissues and cells compared with their normal counterparts, and its expression was positively correlated with the clinicopathological features, especially the invasiveness phenotype. Overexpression of MEX3C accumulated lipid droplets and promoted cell adhesion, invasion and migration. We further demonstrated that MEX3C regulated lipid metabolism and promoted tumor development and progression through activation of JNK signaling and upregulating the JNK downstream protein levels of sterol regulatory element-binding proteins-1, fatty acid synthase and acetyl-CoA carboxylase-1.
Conclusion: Here we identified MEX3C as a new oncogene to promote bladder tumorigenesis by regulating lipid metabolism through Mitogen-activated protein kinase/c-Jun N-terminal kinase (MAPK/JNK) pathway. These findings suggest a new role of MEX3C in promoting BC tumorigenesis and provide a novel biomarker or molecular target for diagnosis or treating BC.
Keywords: MEX3C, lipid metabolism, tumorigenesis, JNK pathway, bladder cancer
Introduction
Bladder cancer (BC) is one of the top-ten frequently occurring malignant cancers and the 9th most common cause of death in the world.1 About 80,000 BC patients were diagnosed in the United States with a second incidence rate ranked behind only prostate cancer in 2017.2 Approximately 75% of BC are nonmuscle invasive at initial diagnosis, 14–45% of which will progress to invasive BC after primary surgery.3 The exact molecular mechanism of metastasis and invasion is still largely unknown, although it is the main cause of death or unfavorable treatment for BC patients.
MEX3C (Mex-3 RNA-Binding Family Member C, also known as RKHD2) encodes a protein with two K homology (KH) RNA-binding domains and a C-terminal RING-finger domain and plays various roles in different biological activities, including immune responses,4 RNA molecule transferring,5 translational repression,6 cancer chromosomal instability,7 energy balance and adiposity,8,9 and postnatal growth.10 Our previous study found that miR-451 acted as a tumor suppressor in BC and inhibit its metastasis and proliferation by regulating EMT,11 and bioinformatics analysis in our preliminary experiment showed that MEX3C was a critical target gene of miR-451.Therefore, it is imperative to explore the expression patterns of MEX3C and its biological functions in human BC.
In this study, we found an increased expression of MEX3C in BC tissues and cells and positive correlations between its expression and BC clinicopathological features. Overexpression of MEX3C accumulated lipid droplets and promoted cell intercellular adhesion, invasion, and migration. We further identified that the overexpression of MEX3C was associated with activated JNK signaling through upregulating the downstream protein levels of sterol regulatory element-binding protein-1 (SREBP1),fatty acid synthase (FASN), and acetyl-CoA carboxylase-1 (ACC1).
Materials and methods
Bioinformatics analysis identified the role of MEX3C in BC
To investigate MEX3C’s role in BC, we analyzed MEX3C gene expression in two large datasets from The Cancer Genome Atlas (TCGA) and Oncomine databases (https://www.oncomine.org/). Mining MEX3C gene chip database was used to analyze the expression of MEX3C in BC tissues and its relationship with the type of BC and the prognosis of patients by Oncomine database. Gene set enrichment analysis (GSEA) was applied to determine sets of genes from signaling pathways that showed statistically significant differences between higher and lower MEX3C expression groups.
Tissue specimens and cell lines
Human BC samples (60 cases) with the matched normal tissues (10 cases) as controls were obtained from patients who underwent radical cystectomy at the Jiangxi Provincial People’s Hospital Affiliated to Nanchang University from January 2012 to December 2016. This study and the written informed consents obtained from all patients were approved by the Ethics Committee and the Institutional Review Board of the Jiangxi Provincial People’s Hospital and conducted in compliance with the Declaration of Helsinki. The histopathology of tissue samples was determined and confirmed by two pathologists according to the criteria of the WHO and the Nevin stage system. Human BC cell lines T24, HT1376, TCCSUP, and immortalized human bladder cell SV-HUC-1 were purchased from China Academia Sinica Cell Repository (Shanghai, China), T24 was cultured in RPMI 1640 supplemented with 10% FBS(GIBCO, USA),while HT1376 and TCCSUP were cultured in DMEM (GIBCO, Grand Island, NE, USA), and SV-HUC-1 was cultured in F12K (GIBCO, USA) and all were incubated in incubator at 37°C with a 5% CO2 atmosphere. T24 was treated with 10 μM SP600125 (Selleck Chemicals, Houston, TX, USA) for 1 hr to inhibit the JNK pathway.
RNA extraction and quantitative real-time PCR (qrt-PCR) analysis
Total RNA was isolated from cells of different groups using Trizol Reagents (Invitrogen, Carlsbad, CA, USA). The RNA were used as templates for the synthesis of cDNA using PrimeScripts®RT (Takara Biotech, Dalian, China). A 20 μL volume that contained 1 μL cDNA, 0.5 μM forward and reverse primers, and SYBR Green PCR Master Mix (Takara Biotech, Dalian, China) were subjected to PCR reaction using a Real-time PCR instrument BioRad CFX96 (BioRad, USA). Primer pairs were 5ʹATGAGGTTATTGCTGCCCTAG3’ (forward) and 5ʹGTGAATTTGGATTGCCTGAGTA’3’ (reverse) for MEX3C, and 5ʹTGACTTCAACAGCGACACCCA3’ (forward) and 5ʹCACCCTGTTGCTGTAGCCAAA3’ (reverse) for GAPDH. The amplification cycle was set as below: 2 mins at 50°C; 10 mins at 95°C; 40 cycles of 15 s at 95°C and 60 s at 60°C. Compared with control, the fold changes of mRNA levels were calculated via the ΔΔCt method. GAPDH was used as an internal reference for normalization.
Immunohistochemistry (IHC) analysis
IHC staining was used following the standard protocols. Paraffin-embedded tissue specimens were cut into 4-μm sections and deparaffinized. slides were incubated with anti-MEX3C (ab79041, Abcam, USA, 1:100 dilutions), FASN antibody (ab22759, Abcam, MA, USA, 1:100 dilutions), ACC1 antibody (21923–1-AP, Proteintech, Wu Han, China, 1:100) and SREBF1 (SERBP1) antibody (14088–1-AP, Proteintech, China, 1:100) overnight at 4°C, then incubated with a biotin-labeled secondary antibody (1:100 dilutions) for 40 mins at 37°C. Sections were incubated with 3,3′-diaminobenzidine (DAB), counterstained with hematoxylin, and visualized using light microscopy. Positive and negative controls were performed for each run of IHC. The staining signals were recorded by Olympus light, Tokyo, Japan microscope, and the staining index was measured and calculated by Olympus FV10-ASW software.
Plasmids and lentiviral infection
In order to increase the expression of MEX3C gene, target cells were cultured with a good growth status, andT24 cells with stable expression of MEX3C were established by infection with a lentivirus carrying a MEX3C cDNA-encoding expression vector or an empty vector (GV492, Shanghai Genechem) according to the manufacturer’s instructions. Lentivirus was packaged into T24 cells using virus titers and promoter as soon as cell density reached 30%. The lentivirus carried a copy of GFP and infection efficiency was assessed based on the numbers of Green Fluorescent Protein (GFP)-expressing cells using fluorescence microscopy.
Wound-healing assay
T24 cells were grown as monolayers in triplicates in 6-well plates until 80% confluence. Cell wound was generated by an artificial scratch after culturing in RPMI 1640 without FBS for 12 hrs, and then the cells were washed thrice with PBS to remove the suspended cells and cultured sequentially without FBS for avoiding the influence of cell proliferation. Cell migration was photographed at 0, 8, 24 hrs after scratching, and the width of the wound was measured.
Matrigel invasion assay
Invasion assays were performed in chambers with a 6.5-mm insert in 24-well plates (Corning company, Cat No: 3422, Corning, NY, USA). The polycarbonate membrane was precoated with 12 μL Matrigel Matrix (BD Biosciences, Cat No: 356234, Bedford, MA, USA), and reconstituted at 37°C for 24 hrs to gel. Cells (5x104 per well) were added to the upper chamber in 200 μL serum-free RPMI 1640, while the lower chamber was filled with 600 μL of medium containing 30% FBS. After 24 hrs of incubation, the cells were then fixed with 3.7% paraformaldehyde (PFA) and stained with 1% crystal violet. Five visual fields were selected randomly under the inverted microscope and cell numbers were counted.
BODIPY staining and detection of triglyceride level
BODIPY 493/503 (Molecular Probes, Carlsbad, CA, USA) was diluted in PBS at a concentration of 1 mg/mL. Following fixation with 4% PFA for 10 mins and DAPI (Millipore, County Cork, Ireland) was used to stain the nucleus. Lipid droplets were stained with Bodipy®493/503 (1:500, B&P Biotech, 131083–16-4, Hangzhou, China) following manufacturer’s instruction. Digital images obtained with an NIKON fluorescent microscope Eclipse CI in at least four randomly chosen areas were measured by fluorescent image processing using Image Pro Plus 6.0. The cells were lysed on ice with pyrolytic solution for 30 mins, then heated in water bath at 70°C for 10 mins, and centrifuged for 5 mins at 2,000 r/min. The 10 μL supernatant was added to the 190 μL working fluid, and then the triglyceride content was detected by a microplate reader.
Western blot analysis
Western blotting analyses were performed using standard protocols. The protein samples were resolved by SDS-PAGE and transferred onto Polyvinylidene Fluoride (PVDF) membranes (Basel, Switzerland, Roche). The membranes were blocked with Phosphate Buffered Solution (PBST) in 10% low-fat dry milk solution at room temperature for 1 hr, then incubated at 4°C overnight with rabbit polyclonal MEX3C antibody (1:2,000, Cat NO: ab79041, Abcam Co. Ltd.), FASN antibody (1:5,000, Cat NO: ab22759, Abcam Co. Ltd.), JNK antibody (1:5,000, Cat NO: ab179461, Abcam Co. Ltd.), p-JNK antibody (1:3,000, Cat NO: ab124956, Abcam Co. Ltd.), ACC1 antibody (1:1,000, Cat NO: 21923–1-AP, Proteintech Co. Ltd.), SREBP1 antibody (1:1,000, Cat NO: 14088–1-AP, Proteintech Co. Ltd.) and mouse anti-β-actin antibodies (1:2,000, Cat NO: T0022, Affinity). Horseradish Peroxidase (HRP)-conjugated goat anti-rabbit IgG (1:5,000, cw0103S, Kangweishiji Biotechnology Co. Ltd., Beijing, China) and goat anti-mouse IgG (1:,5000, cw0102S, Kangweishiji Biotechnology Co. Ltd) antibodies were used as secondary antibodies to detect the proteins. The signal was detected using ECL reagent (cw0048S, Kangweishiji Biotechnology Co. Ltd.). The relative quantities of protein band were determined with Image J software (Barcelona, Spain) by analyzing the sum density of each protein band image. The quantity of β-actin was used for internal control. The density value of each sample was normalized to its β-actin density value to get its relative quantity value.
Statistical analysis
All the above experiments were independently repeated at least three times. The results were given as mean ± SD and analyzed using SPSS (19.0 version software, IBM, USA). Statistical analyses to determine significance were tested with the two-tailed Student’s t-test and χ2 test. Statistical significance between two groups was calculated by unpaired Student’s t-test, and p-value <0.05 is considered significant changes.
Results
MEX3C is highly expressed in BC and closely related to its clinicopathological features
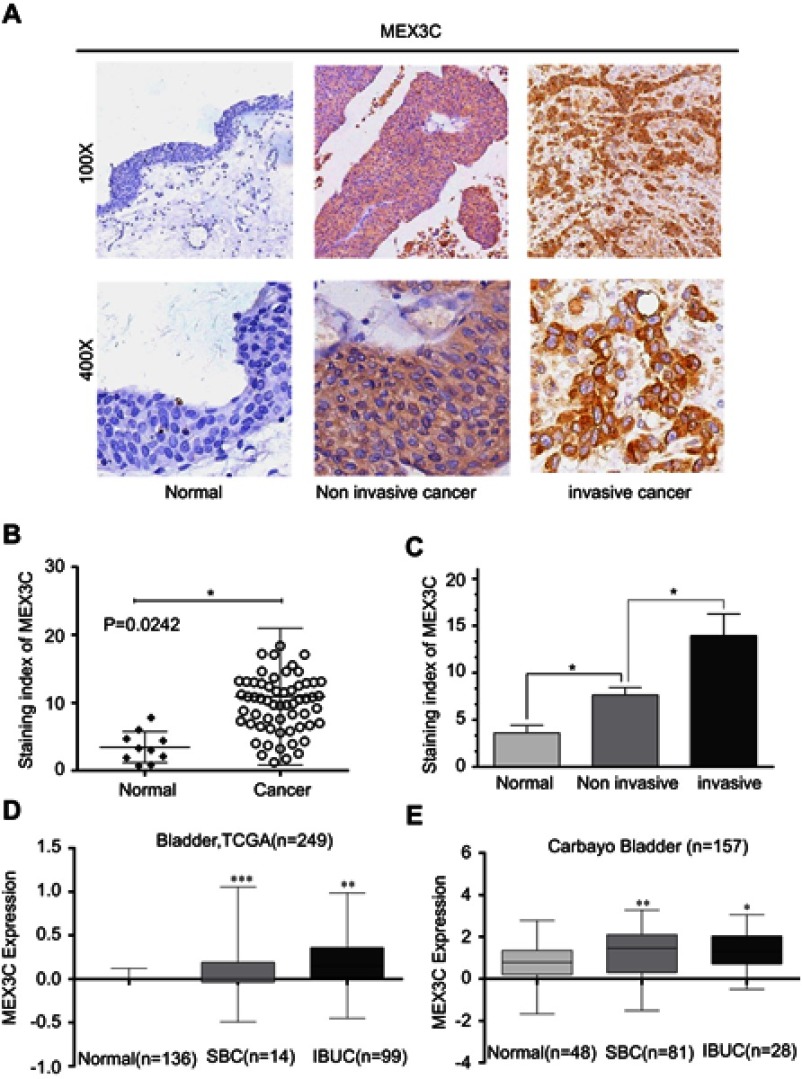
To examine the expression of MEX3C in BC tissues, we used streptavidin-peroxidase (SP) immunohistochemical assay to stain 60 cases of BC and 10 cases of paired normal tissue samples. Our IHC staining analyses showed that MEX3C was highly expressed in BC compared with the normal bladder tissues (Figure 1A and B). Moreover, higher expression of MEX3C was associated with the higher histological grade and more aggressive (invasive) clinical stage in 60 BC patients (P<0.01), indicating that the expression level of MEX3C was positively correlated with clinical stage and pathological grade (Figure 1C). In consistent with the tissue IHC results, analyses of TCGA RNA Seq data and Oncomine database also showed that MEX3C was upregulated in BC tissues (n=249, n=157) compared with normal control tissues, and MEX3C expression was higher in invasive BC tissues than that in superficial cancer tissues (Figure 1D and E).
Figure 1.
MEX3C expression was significantly increased in BC. (A) Representative images of MEX3C immunostaining in human BC tissues and para-tumor tissues. (B and C) Immunostaining index of MEX3C in normal bladder tissue, noninvasive and invasive BC tissues. (D and E) MEX3C expression in normal bladder tissue (normal), noninvasive (SBC) and IBUC, respectively, from Oncomine database. *p<0.05, **p<0.01, ***p<0.001 vs the normal.
Abbreviations: MEX3C, Mex-3 RNA- Binding Family Member C; BC, bladder cancer; IBUC, invasive bladder cancer; TCGA, The Cancer Genome Atlas.
Overexpression of MEX3C promotes adhesion, migration, and invasion of BC cells
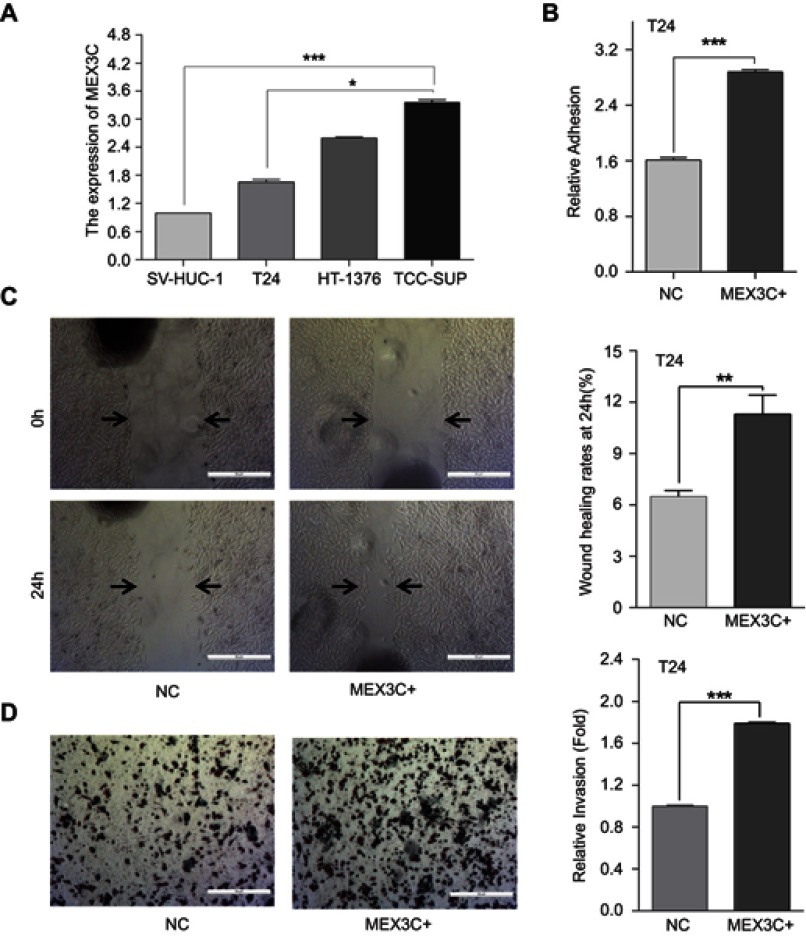
To investigate the cellular functions of MEX3C in BC, we used qRT-PCR assay to first measure the mRNA level of MEX3C in different BC cells. As shown in Figure 2A, MEX3C mRNA level was significantly upregulated in all three BC cells compared to the immortalized human bladder cell SV-HUC-1. Meanwhile, qRT-PCR and western blot were performed to evaluate the expression level of MEX3C mRNA and protein in T24 cells transfected with MEX3C lentiviral plasmid. As shown in Figure 3A and B, MEX3C was stably overexpressed in T24 cells after infection. Overexpression of MEX3C promoted the adhesion of T24 cells (Figure 2B) and further enhanced the migration and invasion of T24 cells by the wound-healing and matrigel transwell assays (Figure 2C and D). These findings suggest that MEX3C may function as an oncogene to promote the adhesion, invasion, and migration of BC cells, which is closely associated with cancer metastasis and the malignant behavior.
Figure 2.
Overexpression of MEX3C increases cell adhesion, migration, and invasion of bladder cancer cells. (A) MEX3C mRNA levels as determined by a quantitative reverse-transcription polymerase chain reaction (qRT-qPCR) in bladder cancer cells T24, HT1376, TCCSUP, and immortalized human bladder cell SV-HUC-1. (B) Normalized adhesive of T24 cells stably transfected with overexpression of MEX3C or control (NC). (C) Images and normalized wound healing of T24 cells stably transfected with overexpression of MEX3C or control (NC) after 24 hrs. (D) Images and normalized invasion of T24 cells following overexpression of MEX3C or NC expression. All experiments were performed in triplicate, and data are presented as the mean ± SE of the mean, n=3. *p<0.05, **p<0.01, ***p<0.001 vs the NC. Abbreviations: MEX3C, Mex-3 RNA- Binding Family Member C.
Figure 3.
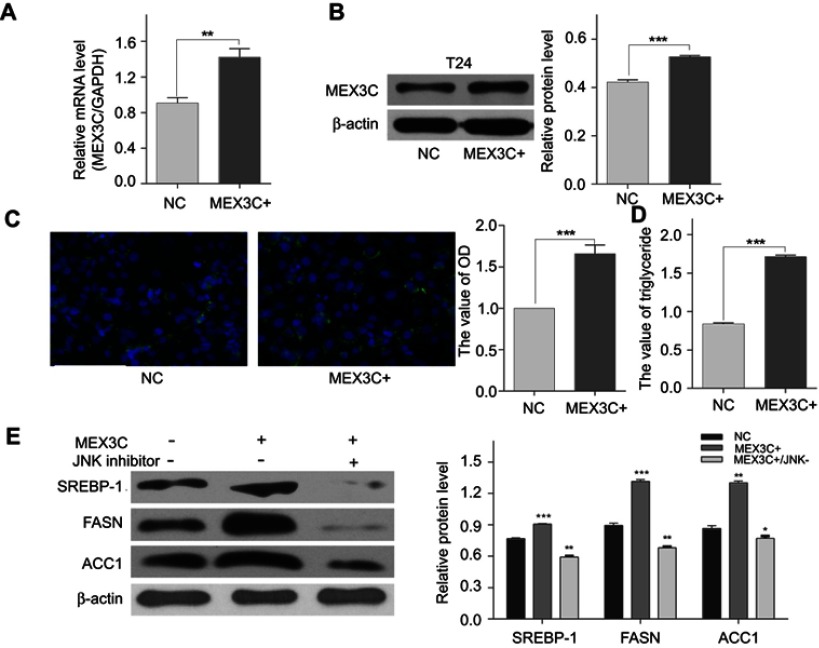
MEX3C accumulated lipid droplets and triglycerides by upregulating FASN, ACC1, and SREBP1 protein.(A) MEX3C mRNA levels as determined by a qRT-qPCR in T24 cells following stable transfection with control (NC) or MEX3C-18357 plasmid. The relative mRNA expression was normalized to GAPDH expression. * vs the NC. (B) The level of the MEX3C protein in T24 cells following MEX3C-18357 was analyzed by western blotting. (C) BODIPY assay analysis of lipid droplet staining in T24 cells with stable MEX3C overexpression relative to control cells. (D) The level of triglycerides in T24 cells. (E) Expression levels of SREBP1, FASN, and ACC1 protein were analyzed by western blotting in T24 cell, and β-actin was used as an internal control (mean ± SEM, **P<0.01, *** P<0.001, t-test).
Abbreviations: MEX3C, Mex-3 RNA- Binding Family Member C; FASN, fatty acid synthase; SREBP1, sterol regulatory element-binding protein-1;ACC1,acetyl-CoA carboxylase-1;qRT-PCR,quantitative reverse-transcription polymerase chain reaction.
MEX3C participates in the metabolic process of fatty acid in BC
To determine whether MEX3C affects fatty acid metabolism in BC cells, we examined the level of intracellular lipid and triglyceride droplets in MEX3C overexpressing T24 cells. As shown in Figure 3C and D, overexpression of MEX3C promoted the deposition of intracellular lipid droplets and triglycerides in T24 cells (p=0.0004, p<0.0001). As SREBP1, FASN, and ACC1 are key enzymes in the fatty acid synthesis pathway, we then examined whether the protein levels of SREBP1, FASN, and ACC1 are regulated by MEX3C overexpression in T24 cells. As shown in Figure 3E, the expression of SREBP1, FASN, and ACC1 was upregulated in MEX3C-overexpressing T24 cells compared to their control cells (p<0.05, p<0.05, p<0.05). Thus, these results may suggest that MEX3C upregulates FASN, ACC1, and SREBP1 to increase the fatty acid synthesis in BC.
MEX3C expression is positively correlated with SERBP1, ACC1, FASN in BC samples
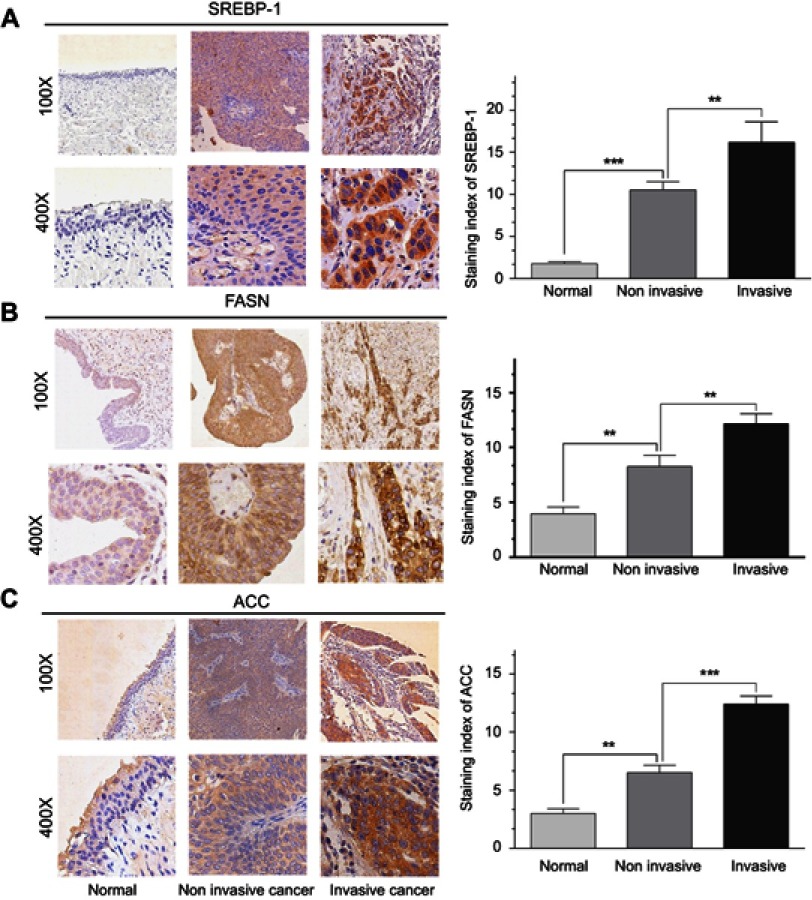
Previous studies have shown that FASN, ACC1, and SREBP1, all play a significant role in tumor progression, especially in the lipid metabolism.12 Therefore, we speculated that MEX3C might be associated with FASN, ACC1, and SREBP1 in the development of BC. We then used IHC staining to examine the expression of FASN, ACC1, and SREBP1 in MEX3C-positive tissues of noninvasive and invasive BC and normal bladder tissues, respectively. As expected, all three proteins showed the strongest nuclear and cytoplasm staining (brown) in the invasive BC, and the weakest staining in normal bladder tissue (Figure 4A–C).
Figure 4.
(A,B) and (C) showFASN, ACC1, and SREBP1 expression was significantly increased in bladder cancer, respectively. Left: Representative images of FASN, ACC1, and SREBP1 in normal bladder tissue, noninvasive, and invasive bladder cancer tissues determined by IHC, respectively (magnification: 100×, 400×). Right: Immunostaining index of FASN, ACC1, and SREBP1 in normal bladder tissue, noninvasive, and invasive bladder cancer tissues.
Abbreviations: FASN, fatty acid synthase; SREBP1, sterol regulatory element-binding protein-1; ACC1, acetyl-CoA carboxylase-1.
Activation of MEX3C is associated with the JNK/SREBP1/ACC&FASN pathway
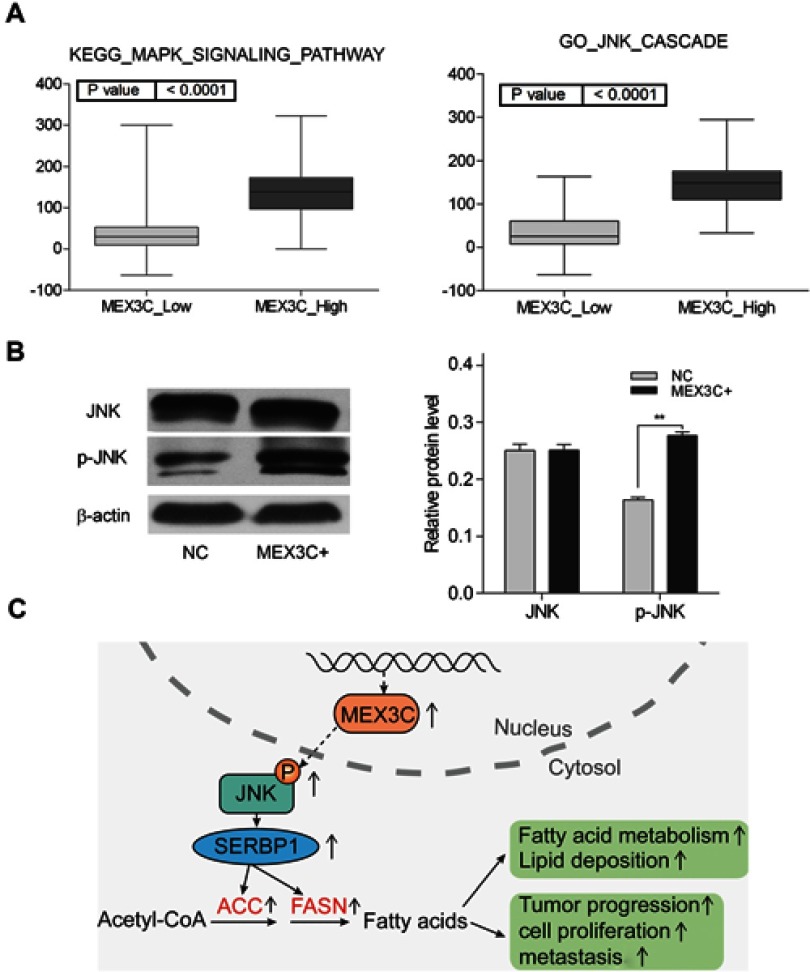
To investigate the specific signaling pathways that may mediate the effects of MEX3C on lipid metabolism, we performed a GSEA based on the TCGA BC databases. We found that higher MEX3C expression was positively correlated with the MAPK/JNK signaling pathway (Figure 5A). To validate the bioinformatics results, we performed western blot analysis of the MAPK/JNK pathways in T24 cells. We found that overexpression of MEX3C evidently increased the p-JNK protein levels, but the protein levels of JNK still unchanged (Figure 5B). Then, we performed experiment using JNK inhibitor (SP600125,) to examine the effects of MEX3C overexpression on the JNK/SREBP1/ACC&FASN pathway by Western blotting. We found that JNK inhibitor can diminish the ability of MEX3C on upregulating the level of SREBP1/ACC&FASN protein (Figure 3E). These findings suggested that MEX3C may play an important role in regulating JNK/SREBP1/ACC&FASN pathway, leading to BC tumorigenesis (Figure 5C).
Figure 5.
MEX3C activated JNK pathway.(A) GSEA indicates that high expression of MEX3C is associated with the MAPK/JNK signaling pathway in the TCGA bladder cancer databases. (B) Western blot showed that overexpression of MEX3C promoted p-JNK protein level, while total JNK protein level remained unchanged compared with negative control (mean ± SEM, **, p<0.01, t test). (C) A model depicting MEX3C,s regulation of fatty acid metabolism in bladder cancer. Overexpression of MEX3C increased the protein levels of SREBP-1,FASN and ACC1, and FASN, which activated by MAPK/JNK signaling, leading to tumor progression.
Abbreviations: MEX3C, Mex-3 RNA- Binding Family Member C; GSEA, gene set enrichment analysis; TCGA, The Cancer Genome Atlas; FASN, fatty acid synthase; SREBP1, sterol regulatory element-binding protein-1; ACC1, acetyl-CoA carboxylase-1.
Discussion
MEX3C, also known as RKHD2, is a member of a family of Mex-3 protein. The MEX3C protein contains two KH RNA-binding domains and a C-terminal RING-finger domain, allowing it to bind RNA and making it become an ubiquitin E3 ligase.13 Mex-3 protein family contains four protein members, MEX3A, MEX3B, MEX3C, and MEX3D, which encode genes on human chromosomes 1, 15, 18, and 19, respectively. MEX3A has been shown to be significantly overexpressed in bladder tumor tissues compared to the adjacent normal tissues, and its expression level in the papillary type of BC was higher than that in nonpapillary type.14 Barriga et al identified MEX3A as a biomarker to represent slowly dividing Lgr5 + intestinal stem cells.15 In addition, Krepischi et al found that MEX3A overexpressed in Wilms tumor and was associated with tumor recurrence.16 Oda et al reported that MEX3B was critical for cellular stress responses as it can modulate DNA damage stress-induced apoptosis.17 Other research group provided evidence that MEX3B mediates immune escape from cancer immunotherapy by destabilizing its downstream gene HLA-A.18 These findings strongly suggest the critical role of Mex-3 protein family in tumorigenesis and progression. However, few is known about the role of another important member, MEX3C, in cancer despite its wide expression in many tissues. As Burrell et al showed that MEX3C, which is located in chromosome 18q and often lost in CIN (+) colorectal cancer (CRC), can function as a new cancer chromosomal instability (CIN) suppressor gene to regulate DNA replication stress and chromosome stability and segregation.7 Other research suggested that MEX3C plays various roles in different biological activities, including immune responses,4 RNA molecule transferring,5 translational repression,6 energy balance and adiposity,8,9 and postnatal growth.10 How ever, there is so far little information about MEX3C and its relationship with human BC. Our previous study found that miR-451 acted as a tumor suppressor in BC,11 and bioinformatics analysis in our preliminary experiment showed that MEX3C was a critical target gene of miR-451. Thus, it is interesting to investigate its biological functions and the underlying mechanisms for BC tumorigenesis.
Our studies confirmed the hypothesis that MEX3C is a potential oncogene to promote BC tumorigenesis. Microarray data gathered from the TCGA RNA Seq database and Oncomine database showed that MEX3C was upregulated in BC tissues compared with normal control tissues, as well as higher expression of MEX3C in invasive BC tissues than superficial cancer tissues. Furthermore, MEX3C levels were associated with the higher histological grade and clinical stage. We validated these bioinformatics analyses by SP staining in bladder tissues and qRT-PCR assays in BC cell lines to show that MEX3C expression was increased in BC tissues and cell lines. Finally, through bioinformatics analysis, we found that MEX3C was closely associated with fatty acid metabolism and MAPK/JNK signaling pathway. We validated that MEX3C overexpression does accumulate lipid and triglyceride droplet in BC cells and promoted cell adhesion, invasion, and migration. Some research reported that SREBP1, FASN, and ACC1 all play a significant role in tumor progression, especially in the lipid metabolism,12,19 and JNK is the critical regulating factor in the signaling pathway.19 We then demonstrated that MEX3C overexpression promoted tumor development and progression through activation of JNK signaling and upregulating the downstream protein levels of SREBP1, FASN, and ACC1. The ability of MEX3C on up-regulating the level of SREBP1/ACC&FASN protein can be diminished by the JNK inhibitor. Taken together, our studies strongly suggest MEX3C acts as a potential oncogene, instead of tumor suppressor like in CRC, to promote BC tumorigenesis by increasing activated JNK pathway-mediated fatty acid synthesis and providing a new biomarker or a molecular target for its clinical application.
Acknowledgment
This work was supported by the project of Youth Science Foundation of Jiangxi Science and Technology Office to Haichao Chao (No. 20171BAB215015).
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387 [DOI] [PubMed] [Google Scholar]
- 3.Kaufman DS, Shipley WU, Feldman AS. Bladder cancer. Lancet. 2009;374:239–249. doi: 10.1016/S0140-6736(09)61069-2 [DOI] [PubMed] [Google Scholar]
- 4.Okamoto M, Kouwaki T, Fukushima Y, et al. Regulation of RIG-I Activation by K63-Linked Polyubiquitination. Front Immunol. 2018;8:1942. doi: 10.3389/fimmu.2017.01960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu P, Li H, Li N, et al. MEX3C interacts with adaptor-related protein complex 2 and involves in miR-451a exosomal sorting. PLoS One. 2017;12:e0185992. doi: 10.1371/journal.pone.0185992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang L, Wang C, Li F, et al. The human RNA-binding protein and E3 ligase MEX3C binds the MEX-3-recognition element (MRE) motif with high affinity. J Biol Chem. 2017;292:16221–16234. doi: 10.1074/jbc.M117.797746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burrell RA, McClelland SE, Endesfelder D, et al. Replication stress links structural and numerical cancer chromosomal instability. Nature. 2013;494:492–496. doi: 10.1038/nature11935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Han C, Jiao Y, Zhao Q, Lu B. Mex3c mutation reduces adiposity partially through increasing physical activity. J Endocrinol. 2014;221:457–468. doi: 10.1530/JOE-14-0071 [DOI] [PubMed] [Google Scholar]
- 9.Jiao Y, George SK, Zhao Q, et al. Mex3c mutation reduces adiposity and increases energy expenditure. Mol Cell Biol. 2012;32:4350–4362. doi: 10.1128/MCB.00452-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiao Y, Bishop CE, Lu B. Mex3c regulates insulin-like growth factor 1 (IGF1) expression and promotes postnatal growth. Mol Biol Cell. 2012;23:1404–1413. doi: 10.1091/mbc.E11-11-0960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zeng T, Peng L, Chao C, et al. miR-451 inhibits invasion and proliferation of bladder cancer by regulating EMT. Int J Clin Exp Pathol. 2014;7:7653–7662. [PMC free article] [PubMed] [Google Scholar]
- 12.Chakraborty PK, Xiong X, Mustafi SB, et al. Role of cystathionine beta synthase in lipid metabolism in ovarian cancer. Oncotarget. 2015;6:37367–37384. doi: 10.18632/oncotarget.5424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buchet-Poyau K, Courchet J, Le HH, et al. Identification and characterization of human Mex-3 proteins, a novel family of evolutionarily conserved RNA-binding proteins differentially localized to processing bodies. Nucleic Acids Research. 2007;35:1289–1300. doi: 10.1093/nar/gkm016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi JW, Huang Y. Mex3a expression and survival analysis of bladder urothelial carcinoma. Oncotarget. 2017;8:54764–54774. doi: 10.18632/oncotarget.18399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barriga FM, Montagni E, Mana M, et al. Mex3a marks a slowly dividing subpopulation of Lgr5+intestinal stem cells. Cell Stem Cell. 2017;20:801–816. doi: 10.1016/j.stem.2017.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krepischi AC, Maschietto M, Ferreira EN, et al. Genomic imbalances pinpoint potential oncogenes and tumor suppressors in Wilms tumors. Mol Cytogenet. 2016;9:20. doi: 10.1186/s13039-016-0227-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oda T, Yamazumi Y, Hiroko T, et al. Mex-3B induces apoptosis by inhibiting miR-92a access to the Bim-3ʹUTR. Oncogene. 2018;37:5233–5247. doi: 10.1038/s41388-018-0336-7 [DOI] [PubMed] [Google Scholar]
- 18.Huang L, Malu S, McKenzie JA, et al. The RNA-binding protein MEX3B mediates resistance to cancer immunotherapy by downregulating HLA-A expression. Clin Cancer Res. 2018;4:3366–3376. doi: 10.1158/1078-0432.CCR-17-2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee JS, Sul JY, Park JB, et al. Fatty acid synthase inhibition by amentoflavone suppresses HER2/neu (erbB2) oncogene in SKBR3 human breast cancer cells. Phytother Res;2013. 713–720. doi: 10.1002/ptr.4778 [DOI] [PubMed] [Google Scholar]