Abstract
Recombinant rabies viral vectors have proven useful for applications including retrograde targeting of projection neurons and monosynaptic tracing, but their cytotoxicity has limited their use to short-term experiments. Here we introduce a new class of double-deletion-mutant rabies viral vectors that leave transduced cells alive and healthy indefinitely. Deletion of the viral polymerase gene abolishes cytotoxicity and reduces transgene expression to trace levels but leaves vectors still able to retrogradely infect projection neurons and express recombinases, allowing downstream expression of other transgene products such as fluorophores and calcium indicators. The morphology of retrogradely targeted cells appears unperturbed at one year postinjection. Whole-cell patch-clamp recordings show no physiological abnormalities at eight weeks. Longitudinal two-photon structural and functional imaging in vivo, tracking thousands of individual neurons for up to four months, shows that transduced neurons do not die and retain stable visual response properties even at the longest time points imaged.
Introduction
Since their introduction to neuroscience in 20071, recombinant rabies viral vectors have come to occupy a unique role in the field. Their most important and widespread use is in monosynaptic tracing, which allows identification and study of neurons directly presynaptic to single neurons or targeted cell types2–12. Another important application is retrograde infection of projection neurons, to express transgene products such as fluorophores1,9,13, opsins14–19, or activity indicators20–22 selectively within groups of neurons targeted on the basis of their axonal projections. Both of these applications have yielded insights into the organization of neural circuits in a wide variety of regions, ranging from cerebral cortex to spinal cord.
These vectors, however, come with the major downside of causing nontrivial cytotoxicity. All rabies viral (RV) vectors used to date for retrograde targeting and monosynaptic tracing (which we refer to below as “first-generation” or “ΔG” rabies viral vectors) have a single gene, “G” (NCBI symbol: RABVgp4), encoding the viral envelope glycoprotein, deleted from their genomes, with the other four viral genes left intact1 (Fig. 1a). Because the glycoprotein is only required for the final steps of the viral life cycle, this deletion prevents the virus from spreading beyond initially infected cells, but it does not prevent the virus from completing the entirety of its replication cycle up to that point. This is a double-edged sword. On the one hand, it results in rapid and high-level expression of transgenes, which can be extremely useful for such applications as imaging of fine cellular processes or optogenetic manipulation of neuronal activity1,14,15,18,23. On the other hand, it also results in fairly rapid cytotoxicity, with death of many or most infected cells occurring on a timescale of ~2 weeks1. This of course prevents the vectors from being used for long-term imaging or optogenetic experiments.
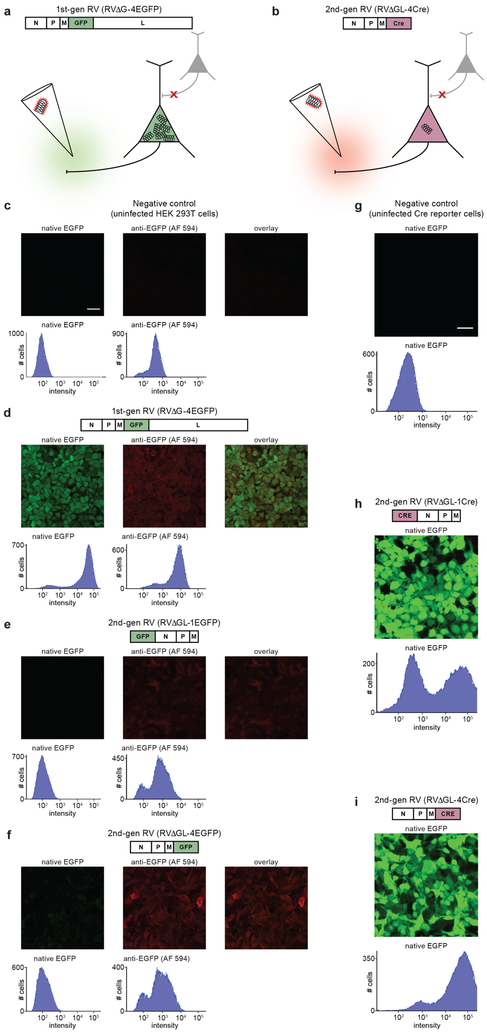
Figure 1. Deletion of the rabies viral polymerase gene reduces gene expression to trace levels but leaves recombinase-encoding vectors still able to cause reporter expression in cell culture.
(a–b) Strategy for eliminating the toxicity of rabies viral vectors. (a) In first-generation vectors, only the G gene, encoding the envelope glycoprotein, is deleted. This prevents the virus from spreading beyond directly infected cells but still allows the virus to replicate within them, resulting in rapid, high transgene expression as well as severe toxicity on the timescale of 1–2 weeks. (b) In the second-generation RV vectors introduced here, the L gene, encoding the viral polymerase, has also been deleted to reduce expression from the viral genome to trace levels. Because transgene expression should therefore also be reduced, the gene for a recombinase, such as Cre or Flp, is used in order to allow the expected very low expression levels to cause subsequent expression of a reporter at experimentally useful levels. (c–i) Results in cell culture: images and flow cytometric histograms show EGFP fluorescence levels in cells infected by first- and second-generation RV vectors. Left-hand images and histograms in (c–f) show native EGFP fluorescence; middle images and right-hand histograms in (c–f) show anti-GFP immunolabeling with AlexaFluor 594 secondary antibody; right-hand images show overlay of left-hand and middle images. X-axes of histograms are log scale with arbitrary units. (c) Negative control: HEK 293T cells not infected with any virus do not express EGFP, as indicated by the single peak at a very low fluorescence level. (d) A first-generation RV vector encoding EGFP causes infected HEK cells to fluoresce brightly, as shown by the peak far to the right of the histogram (the leftmost peak represents uninfected cells in the same well). (e–f) Second-generation RV vectors encoding EGFP cause almost undetectable EGFP expression in infected cells, as shown by histograms that resemble that of the uninfected cells shown in (c); immunostaining for EGFP shows that virus is nonetheless present and producing EGFP at low levels. This is true whether the EGFP gene is placed in the most highly-expressing locus at the start of the viral genome (e) or in the “usual” transgene locus of the deleted G gene (f). (g–i) We then constructed second-generation RV vectors encoding Cre recombinase and tested them in reporter cells that express EGFP when Cre is expressed. (g) Negative control: uninfected reporter cells express very little EGFP. (h–i) Reporter cells infected with second-generation RV vectors encoding Cre express lots of EGFP. This is true whether the Cre gene is inserted at the start of the RV genome (h) or into the G locus (i). Experiments in this figure were performed twice with similar results each time. Scale bar in (c): 33 μm, applies to (c–f); scale bar in (g): 33 μm, applies to (g–i).
Efforts to reduce the toxicity of RV vectors have been made previously. It has recently been reported, for example, that first-generation rabies viral vectors from a different parent strain (CVS N2c, rather than SAD B19) allow infected neurons to survive longer24. However, no claim has been made, and no evidence suggests, that the use of CVS N2c or any other strain completely eliminates the toxicity of first-generation vectors. It has also recently been reported that addition of a proteasome-targeting destabilization domain to the viral nucleoprotein removes viral toxicity25. However, because individual infected neurons were not tracked over time in these studies, it is difficult to assess whether cytotoxicity was completely abolished.
Here we take a quite different approach toward eliminating toxicity (Fig. 1b). We introduce a new class of rabies viral vector, from the genome of which we have deleted a second and arguably much more fundamental gene. Known as L (for “large protein” gene, P being already taken as the name of the viral phosphoprotein gene) (NCBI symbol: RABVgp5), this second deleted gene encodes the viral polymerase, which is required both for transcription of viral genes and for replication of the viral genome26. We hypothesized that deletion of the polymerase gene would reduce expression of viral genes and transgenes to miniscule levels, resulting in a vector that would cause no detectable cytotoxicity whatsoever. We further hypothesized that, if such vectors encoded recombinases, the tiny remnant transcription levels (resulting from the activity of the few starter copies of the polymerase that are packaged in each viral particle27) would nevertheless be sufficient to achieve site-specific recombination in genomic reporter alleles in reporter mouse lines.
We show that these new (“second-generation” or “ΔGL”) viral vectors can be used to express both Cre and Flp recombinases at levels sufficient to effect reporter gene expression in widely available reporter mice and in wild-type animals when used in combination with Cre-dependent adeno-associated viral (AAV) vectors, that the new vectors retain the first generation’s characteristic ability to retrogradely infect projection neurons, and that targeted neurons appear morphologically normal up to a year postinjection. Using whole-cell recording in slices containing retrogradely targeted basolateral amygdala neurons, we show that transduced neurons exhibit normal membrane properties at two months postinjection. We use longitudinal two-photon structural and functional imaging in vivo in mouse primary visual cortex to show that targeted neurons exhibit no signs of toxicity, and that responses to visual stimuli remain stable for at least four months, the longest time point in our dataset. Finally, we find that the new rabies viral vector class has broader tropism for corticocortical projections than two other viral vector species commonly used for retrogradely targeting projection neurons.
This new class of vectors is immediately useful as a means of retrogradely targeting projection neurons to express Cre or other recombinases with no apparent toxic effects, allowing for the systematic selection of different classes of neurons for nonperturbative long-term anatomical or physiological study. This work also lays the foundation for the construction of a future second-generation monosynaptic tracing system that will leave transsynaptically transduced neurons alive and healthy indefinitely.
Results
We began by testing our hypothesis that L deletion would reduce vector expression to trace levels. Because genes in the rabies viral genome are expressed at levels that monotonically decrease with their positional order in the genome26,28,29, we made and tested two variants of ΔGL rabies virus encoding enhanced green fluorescent protein (EGFP)30. In the first, RVΔGL-1EGFP, the EGFP gene was inserted in the highest-expressing locus, at the beginning of the genome before the remaining viral genes (note that the numbers in the vectors’ names refer to the position of the transgene with respect to those of the other genes in the viral genome). In the second, RVΔGL-4EGFP, the EGFP gene was inserted in the lowest-expressing locus, at the end of the viral genome. Histograms of EGFP fluorescence in HEK 293T cells show that, whereas first-generation vectors encoding EGFP cause very bright fluorescence in infected cells (Fig. 1d; cf. uninfected negative control in Fig. 1c), second-generation vectors encoding EGFP (Fig. 1e,f) cause cells to express so little EGFP as to be nearly indistinguishable from uninfected controls, although anti-GFP immunostaining (right-hand histograms in Fig. 1c–e) confirms that EGFP is indeed present at low levels. This is true whether the EGFP gene is inserted in the highest-expressing (Fig. 1e) or lowest-expressing (Fig. 1f) locus in the vector genome, indicating that L deletion indeed reduces gene expression to levels far below those of first-generation vectors, confirming our expectations.
However, these results raised the possibility that the L deletion could reduce transgene expression so much as to render the new vectors useless for neurobiological applications. Our next step was therefore to test whether second-generation rabies viral vectors encoding a recombinase, as opposed to a fluorophore, are capable of expression at levels sufficient to activate reporter gene expression when used in the presence of a suitable recombinase-dependent reporter construct. We constructed two ΔGL vectors encoding Cre recombinase31, the first (RVΔGL-1Cre) with the Cre gene inserted into the highest-expressing locus and the second (RVΔGL-4Cre) with it inserted into the lowest-expressing locus. We tested the viruses on a reporter cell line that expresses EGFP following Cre recombination, and the results were encouraging. Whereas uninfected control cells are not fluorescent (Fig. 1g), cells infected with either of the ΔGL vectors turned brightly fluorescent (Fig. 1h,i), indicating successful recombination of the reporter cells’ EGFP expression cassette by viral expression of Cre. Because even RVΔGL-4Cre, the virus with the Cre gene in the lowest-expressing locus, was able to recombine the reporter cassettes, we used this version for the subsequent in vivo assays and refer to it simply as “RVΔGL-Cre” below.
For our tests of the new class of vectors in vivo, our primary questions were: 1) Are ΔGL vectors encoding recombinases capable of causing reporter expression in brains? 2) Are the new vectors retrogradely infectious like the first-generation ones, so that projection neurons can be selectively targeted? 3) Do the new vectors leave infected neurons alive and apparently healthy, and if so for how long?
Our in vivo experiments gave positive results on all three counts. To test the versatility of the new ΔGL vector class, we constructed two versions: the Cre-encoding vector RVΔGL-Cre described above, and RVΔGL-Flpo encoding Flp recombinase32,33. We injected these vectors into the somatosensory thalamus in mice of two reporter strains expressing the red fluorophore tdTomato34 after reporter cassette recombination by the appropriate recombinase (see Methods). Note that all vectors described in this paper are coated with the native rabies virus envelope protein, which confers on first-generation rabies viral vectors the ability to retrogradely infect large numbers of neurons that project to the injection site1.
The result of these thalamic injections was bright fluorescence in hundreds of corticothalamic neurons in layer 6 of overlying somatosensory cortex for both RVΔGL-Cre (Fig. 2a,c,e) and RVΔGL-Flpo (Fig. 2b,d,f). Even with short survival times (Fig. 2a, RVΔGL-Cre after 7 days; Fig. 2b, RVΔGL-Flpo after 14 days), abundant cortical neurons were labeled. This indicated two important facts: first, that RVΔGL-Cre and RVΔGL-Flpo are quite capable of activating reporter gene expression in common reporter mice and, second, that second-generation rabies viral vectors efficiently and retrogradely infect projection neurons, just as do their first-generation counterparts1. Also, the fact that the efficacy of ΔGL vectors is not restricted to use with a single recombinase indicates that they will likely be useful for a variety of intersectional and simultaneous targeting approaches35,36.
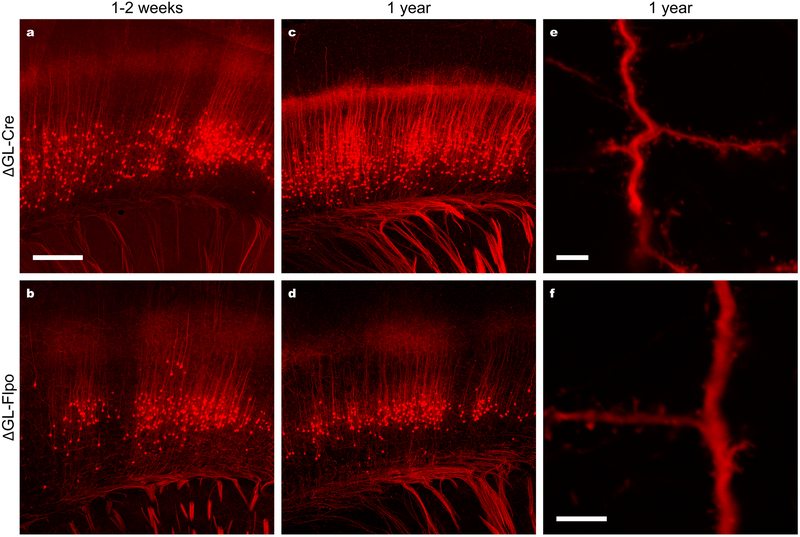
Figure 2. Second-generation rabies viral vectors encoding Cre and Flp retrogradely infect projection neurons, activate reporter expression, and cause no apparent morphological abnormalities even up to a year after injection.
(a–b) Cortical neurons retrogradely labeled by second-generation RV vectors encoding either Cre (a; 7d survival) or Flp (b; 14 d survival) recombinase injected into somatosensory thalamus of tdTomato reporter mice. (c–d) Cortical neurons retrogradely labeled by second-generation RV vectors encoding Cre (c) or Flp recombinase (d) injected into the thalamus of a Flp reporter mouse one year before perfusion. (e–f) High-magnification images of dendrites and other fine processes of cortical neurons from the same sections imaged for (c) and (e), showing apparently normal morphology with a complete absence of decomposition, blebbing, or other abnormalities. These results indicate, first, that second-generation RV vectors are capable of expressing recombinases at levels sufficient to activate downstream transgene expression in readily available reporter mice; second, that second-generation RV vectors are capable of efficient retrograde infection of projection neurons; third, that the new vectors can be used to express different recombinases, allowing a variety of potential intersectional applications; fourth, that the new vectors do not appear to cause cytotoxicity even on time scales comparable to the lifetime of the animal. Experiments in this figure were performed in 2–3 animals each condition with similar results in all cases per condition. Scale bar in (a): 250 μm, applies to (a–b); scale bars in (e,f): 5 μm.
We also investigated whether the second-generation rabies viral vectors are compatible with recombinase-dependent AAV vectors, so that their use would not be restricted to applications in reporter mice. We found that this approach works well in both mice and rats, as shown in Supplementary Figs. S1 and S2. This indicates that the vectors can be used in wild-type animals, in multiple species, for targeting expression of fluorophores and opsins to projection neurons selected on the basis of the locations of both their somata and axonal arborizations.
While these results demonstrate that the new second-generation rabies viral vectors retain the ability of first-generation ones to retrogradely infect projection neurons and that they are capable of causing recombinase-dependent expression of reporter genes, it remained to be seen to what degree deletion of L achieved the main desired effect, namely the elimination of the cytotoxicity that characterizes previous rabies viral vectors. For a preliminary, qualitative answer to the question of preservation of the health of infected neurons, we simply conducted a series of thalamic injections of RVΔGL-Cre in Ai14 mice and RVΔGL-Flpo in Ai65F mice, sacrificing them after increasing lengths of time. Even after leaving mice for an entire year after injection, brightly fluorescent cortical neurons were found in large numbers throughout layer 6 of somatosensory cortex (Fig. 2c,d). At higher magnification (Fig. 2e,f) dendritic spines and other fine processes were clearly intact and showed no blebbing or otherwise altered structures indicative of toxicity.
Although it appeared from these results that the second-generation rabies viral vectors do not kill neurons, there remained the possibility that only some of the labeled neurons survived over long periods, with others having died off. For a more rigorous examination, we conducted longitudinal two-photon imaging in vivo of labeled neurons in visual cortex to monitor individual neurons over the course of months. This stringent test allowed us to see how many of the labeled neurons survived for long periods.
For these longitudinal imaging experiments, we compared the effects of the second-generation virus RVΔGL-Cre to those of two first-generation (ΔG) control viruses. The first control virus, RVΔG-tdTomato, contains the gene for tdTomato in its genome, while the second control virus, RVΔG-Cre, encodes Cre in its genome. We injected each virus into primary visual cortex of mice (Ai14 tdTomato reporter mice for the Cre-encoding viruses, wild-type C57BL/6J for RVΔG-tdTomato), then imaged the same fields of view (FOVs) with the same labeled neurons over multiple imaging sessions. This allowed us to track the fate of individual neurons for up to four months, in order to obtain precise counts of the numbers of neurons that persisted as a function of time after infection (Fig. 3).
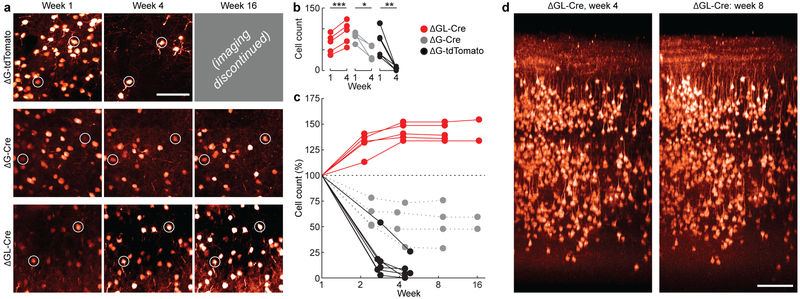
Figure 3. Longitudinal two-photon structural imaging shows that cortical neurons transduced with second-generation rabies viral vectors remain alive and structurally normal for at least 4 months.
(a) Example two-photon fields of view (FOVs) transduced with first-generation RV encoding tdTomato (top row), first-generation RV encoding Cre in a tdTomato reporter mouse (middle row), and second-generation RV encoding Cre in a tdTomato reporter mouse (bottom row), at 1 week (left column), 4 weeks (middle column), and 16 weeks (right column) after injection. Images within each column are of the same FOV imaged at the different time points. Two cells surviving across all time points for each animal have been circled as fiducial markers. As seen in the top row, first-generation RV encoding tdTomato kills almost all infected cells within 4 weeks of infection, in agreement with earlier work with similar vectors. Imaging was therefore not continued past 4 weeks for these mice. As seen in the middle row, first-generation RV encoding Cre (as opposed to a fluorophore) unexpectedly kills only about half of infected neurons over the first few weeks. As seen in the bottom row, none of the neurons visibly labeled by the second-generation RV disappear over time. (b) Population data for all structural FOVs in the study, comparing absolute cell counts between weeks 1 and 4. There is a significant increase in labeled ΔGL-Cre neurons (red, 5 FOVs; two-sided paired t(4) = 11.3, ***p = 3.5×10–4) and significant die-off in both ΔG-Cre (gray, 4 FOVs; paired t(3) = 4.47, *p = 0.021) and ΔG-tdTomato conditions (black, 6 FOVs; paired t(5) = 4.73, **p = 5.2×10–3). (c) Percentage visible labeled cells over time, relative to the number visible at 1 week postinjection, for each of the three viruses; connected sets of dots represent counts obtained from the same FOV within the same brain at the different time points. Cells labeled by first-generation RV encoding tdTomato (black) have almost entirely disappeared by four weeks postinjection. Cells labeled by first-generation RV encoding Cre (gray) have in many cases disappeared by four weeks, but approximately half remain and survive even to eight weeks. Cells labeled by second-generation RV encoding Cre (red) do not die at any point: the numbers of visibly labeled cells increase up to four weeks postinfection (presumably as previously subthreshold tdTomato levels accumulate to the level of detectability) and then remain constant for as long as imaging is continued and presumably for the lifetime of the animal. (d) Example renderings of the same volume of cortex labeled by second-generation RV encoding Cre at two different imaging time points, four weeks and eight weeks. Every single neuron visible at four weeks is still present at eight weeks. This was consistent across the 5 ΔGL-Cre cases. Scale bars: 100 μm.
As expected, most neurons infected by the first-generation control vector RVΔG-tdTomato that were visible one week after injection were not seen after 4 weeks (Fig. 3a, top row; Fig. 3b,c, black traces; volume rendering in Supplementary Video S1), consistent with previous reports of the rapid toxicity of first-generation rabies viral vectors1. Imaging for the mice injected with this control virus was not continued after week 4 because there were very few neurons left to image, and most that remained showed dysmorphic processes and somata (Supplementary Fig. S3a–f).
We obtained more surprising results with our other first-generation control virus, which encoded Cre instead of tdTomato. This virus killed a large fraction of infected neurons, but by no means all of them (Fig. 3a, middle row; volume rendering in Supplementary Video S2). Approximately 50% of cells infected with the ΔG-Cre survived out to four months postinfection (Fig. 3b,c, gray traces). This result, that simply encoding Cre instead of tdTomato makes for a much less toxic first-generation rabies viral vector, is consistent with a previous report that some neurons infected with a replication-competent rabies virus encoding Cre can survive infection, albeit with altered gene expression37. Nevertheless, even this less-toxic ΔG vector killed neurons over the first few weeks of imaging, and, as will be seen from the physiological results below (Figs. 4,5), we found signs that at least some of the surviving neurons had abnormal membrane and visual response properties.
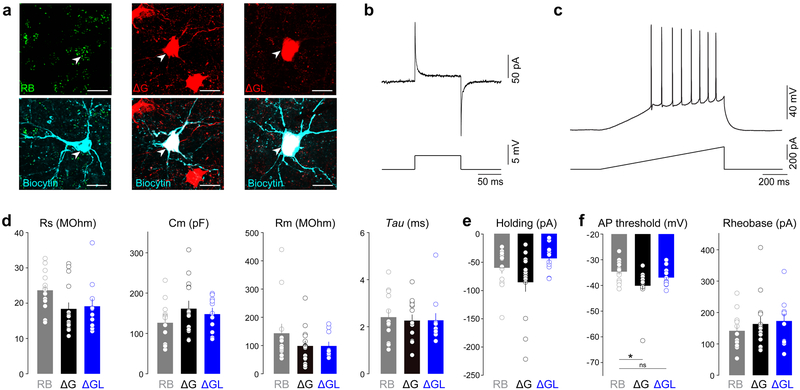
Figure 4. Membrane properties of neurons transduced with second-generation rabies viral vectors remain normal for at least eight weeks.
(a) Confocal images of basolateral amygdala neurons projecting to the nucleus accumbens retrogradely labeled with either retrobeads (RB, left panels), first generation rabies virus expressing Cre (ΔG, middle panels), or second generation of rabies virus expressing Cre (ΔGL, right panels), imaged after targeted whole-cell recording. (b) Representative traces of a seal test response in voltage-clamp, and (c) a ramp test to induce action potential firing in current-clamp mode. (d) The series resistance (Rs), capacitance (Cm), membrane resistance (Rm), decay time constant (Tau), and (e) holding current at −70 mV were not different between the three experimental groups (one-way ANOVA, F(2,36)=2.51, p=0.096 (Rs); F(2,36)=1.35, p=0.27 (Cm); F(2,36)=1.38, p=0.26 (Rm); F(2,36)=0.126, p=0.88 (Tau) – n=13(RB), 15(ΔG), and 11(ΔGL); one-way ANOVA, F(2,35)=3.14, p=0.056 (holding current) – n=13(RB), 14(ΔG), and 11(ΔGL)). (f) The action potential (AP) threshold was significantly more negative for the surviving cells transduced with the first generation RV compared to cells containing RB while cells transduced with the second generation RV did not show a difference compared to RB containing cells (one-way ANOVA, F(2,36)=4.29, *p=0.021, RB vs ΔG **p<0.05, RB vs ΔGL p>0.05; n=13(RB), 15(ΔG), and 11 (ΔGL)). The minimal current necessary to induce firing (rheobase) was not significantly different between the three groups (one-way ANOVA, F(2,36)=0.531, p=0.59; n=13(RB), 15(ΔG), and 11(ΔGL)). All bar graphs display mean ± s.e.m. Experiments in this figure were performed in 6 animals with similar results in all cases.
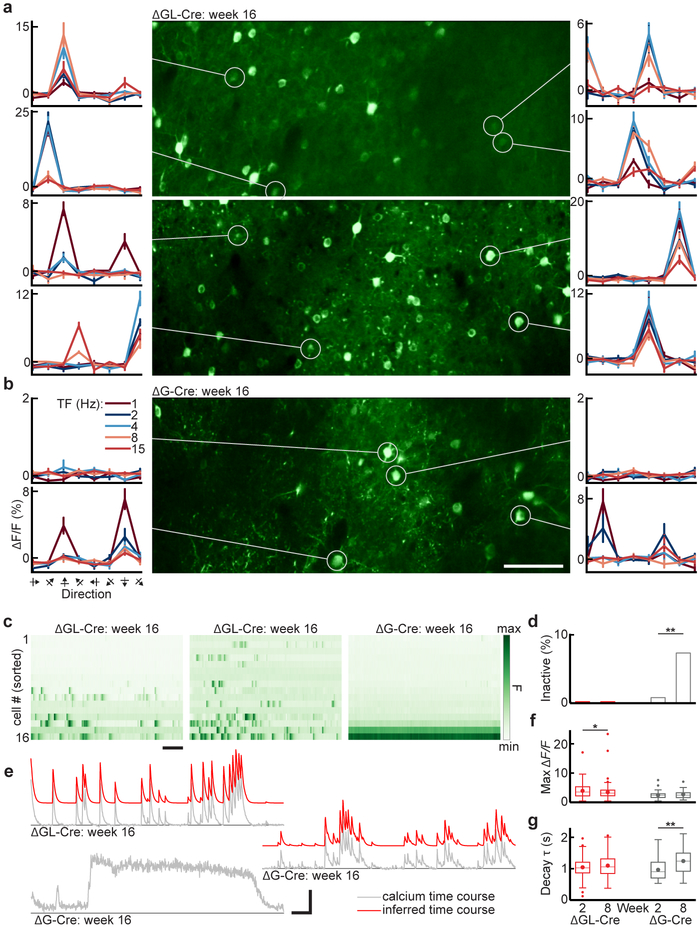
Figure 5. In vivo two-photon calcium imaging using GCaMP6f shows that cortical neurons transduced with second-generation rabies viral vectors retain stable visual response properties for at least 4 months.
(a) Calcium imaging from layer 2/3 of primary visual cortex (V1) in two Cre-dependent GCaMP6f reporter mice, 16 weeks after injection of second-generation virus RVΔGL-Cre. The FOVs (center) are maximum intensity projections of the imaging time series. The curves to the right and left of each FOV show example direction tuning curves of single cells (circled), obtained with drifting gratings presented at 8 directions of motion and 5 temporal frequencies (TF), repeated 15 times (mean ΔF/F ± s.e.m.). Labels on leftmost plots in (b) apply to all plots in (a,b). Tuning curves were consistently responsive with clear direction preferences, as expected in V1. All cells bright enough to be segmented from the max projection showed at least spontaneous activity; bright-inactive cells were never seen in the RVΔGL-Cre cases. (b) Calcium imaging from layer 2/3 of V1, 16 weeks after injection of second-generation virus RVΔGL-Cre. While there are cells that show consistent direction preferences (lower curves) or spontaneous activity, there also appear bright cells with no activity of any kind (upper curves). Scale bar: 100 μm. (c) Single-cell fluorescence time courses for the 16 cells with highest mean luminance in each FOV in (a,b) (normalized within FOV, sorted top to bottom from least to most bright), showing the first 150 seconds of visual stimulation. The first and second panels (second generation, RVΔGL-Cre) show robust spontaneous and evoked activity. The third panel (first generation, RVΔG-Cre) explicitly shows the pathology described in (b), with the brightest cells being completely inactive, consistent with previous reports that cells with the highest baseline GCaMP fluorescence and filled-in appearance (no nuclear exclusion) often show altered or reduced activity39,50. Scale bar: 20 s. (d) Proportions of bright-inactive cells across the RVΔGL-Cre (3 mice, 9 FOVs) and RVΔG-Cre (2 mice, 6 FOVs) populations, for weeks 2 and 8 (horizontal axis labels shared with (f,g)). Cells in the upper 50% of mean fluorescence for each FOV are shown, since poorly labeled GCaMP6f cells with no activity are often too dim to unambiguously segment, and the altered response properties associated with unhealthy RVΔG-Cre-labeled cells are observed in the brightest cells. None of the RVΔGL-Cre cells from week 2 (0 of 502 cells) or week 8 (0 of 717) were inactive, measured over the full 30 minutes of visual stimulation. Inactive cells were noticed as early as week 2 in the RVΔG-Cre condition (1 of 142, 0.7%) and increased significantly by week 8 (10 of 139, 7.2%; two-sample z-score proportion test, z=2.81, p = **5.2×10−3). (e) Example calcium time courses from week 16 (top left: RVΔGL-Cre; middle right and bottom left: RVΔG-Cre). Raw time courses (gray) of active cells can generally be approximated by an autoregressive (AR) process of order 1 (see Methods; inferred trace offset, in red). The bottom RVΔG-Cre time course comes from an aberrant cell with a slow, sustained increase in brightness that is not well fit by the AR model, and very sparse activity otherwise. This is distinct from the bright-inactive cells shown in (c) but may be part of the same progression toward inactivity or cell death. The two RVΔG-Cre examples come from the same FOV, within a hundred microns of each other, suggesting that the prolonged changes in fluorescence cannot be accounted for by z-motion artifacts. There were 13 such sustained-activity cells out of 46 (28.3 %) segmented in the AR analysis of week-8 RVΔG-Cre cells. Vertical scale bar, ΔF/F: 3.0 (1.2, 0.3) for top (middle, bottom) traces. Horizontal scale bar: 20 s. (f) Peak ΔF/F across the RVΔG-Cre and RVΔGL-Cre population, for weeks 2 and 8. The mean (± s.e.m) of RVΔGL-Cre cells decreased significantly from week 2 (3.95 ± 0.17, n=191) to week 8 (3.47 ± 0.14, n=247; two-sided unpaired t(436)=2.25, *p =0.025) as baseline fluorescence gradually increased to final levels, but did not change significantly for the more rapidly expressing RVΔG-Cre cells between week 2 (2.46 ±0.20, n=44) and week 8 (2.71 ± 0.24, n=33; t(75)=0.81, p=0.42). (g) Decay time constant (τ) of calcium transients across the RVΔG-Cre and RVΔGL-Cre population, for weeks 2 and 8. The mean τ of RVΔGL-Cre cells did not increase significantly from week 2 (1.07 ± 0.02 s, n=191) to week 8 (1.12 ± 0.02 s, n= 247; two-sided unpaired t(436)=1.72, p=0.086). For RVΔG-Cre cells, τ showed a pronounced increase from week 2 (0.99 ± 0.05 s, n=44) to week 8 (1.27 ± 0.08 s, n=33; t(75)=3.31, **p=0.0014). Sustained-activity cells were excluded from the analysis, so the large increase in τ in RVΔG-Cre cells likely underestimates the extent of changes in the RVΔG-Cre population. Data are represented as box plots showing mean (large points), median (horizontal line), 25th-75th percentile (boxes, IQR), 1.5xIQR above or below 75th or 25th percentiles (whiskers), and outliers beyond this range (small points).
In contrast to the complete or partial lethality of the first-generation control vectors, the second-generation vector RVΔGL-Cre did not kill neurons at all (Fig. 3a, bottom row; Fig. 3b,c, red traces; volume rendering in Supplementary Video S3). The numbers of detected neurons actually increased over the first several imaging sessions and remained constant thereafter, presumably reflecting the gradual buildup of tdTomato fluorescence above detection threshold (note the relative dimness of the cells in the week 1 image compared to that in subsequent images). After the population of labeled cells stabilized, every cell in large imaged volumes (Fig. 3d and Supplementary Video S4) could be found at later time points, and we saw no cell death in any imaged FOV across the longitudinal study.
While it was clear at this point that the second-generation vector was not killing neurons, it remained to be seen whether neurons transduced with the second-generation vector, and those remnant neurons that survived infection with its first-generation (RVΔG-Cre) counterpart, were physiologically normal. We therefore injected first-generation (RVΔG-Cre) and second-generation (RVΔGL-Cre) vectors into the nucleus accumbens of Ai14 (tdTomato) reporter mice, along with fluorescent retrobeads in the contralateral nucleus accumbens of each mouse, then conducted whole-cell recordings in slice from retrogradely labeled basolateral amygdala neurons at 8 weeks postinjection (Fig. 4). We found no significant difference in any measured membrane property between the neurons labeled with the second-generation vector versus the retrobead-labeled controls after 8 weeks of expression (Fig. 4d–f). In contrast, neurons that survived labeling with the first-generation vector had significantly more negative action potential thresholds than retrobead-labeled neurons (Fig. 4f). These data indicated that second-generation vector RVΔGL-Cre did not produce detectable changes in the membrane properties of neurons, whereas the first-generation Cre-encoding control vector RVΔG-Cre, even though it leaves some infected neurons alive, affects surviving cells’ function.
To determine whether neurons transduced with the second-generation vector retain normal circuit response properties long-term, and to determine the degree to which those cells which survive transduction with the first-generation vector encoding Cre do as well, we conducted longitudinal in vivo two-photon calcium imaging of visual responses using GCaMP6f38–40, again with repeated imaging of the same FOVs. We attempted to conduct long-term imaging of neurons infected with first-generation virus encoding GCaMP6f directly (RVΔG-4GCaMP6f) but found no responsive neurons beyond the two-week time point when using this virus (see Supplementary Fig. S3g–i). We therefore injected either first-generation (RVΔG-Cre) or second-generation (RVΔGL-Cre) virus into the visual cortex of GCaMP6f reporter mice and imaged the visual response properties of transduced neurons to drifting gratings presented to the awake animal at time points up to four months.
At 16 weeks after injection, we found that neurons transduced with the second-generation vector had completely normal response properties. Those that responded to drifting gratings were well tuned to specific orientations (Fig. 5a), most showed spontaneous activity, and there were no bright-inactive neurons in any FOV or time point (Fig. 5c–e and Supplementary Video S5). Tuning preferences of highly responsive individual neurons were stable over sessions spanning the entire four months of imaging (Supplementary Fig. S4). By contrast, neurons surviving infection with the first-generation vector RVΔG-Cre often had abnormal response properties. Many RVΔG-Cre-infected neurons displayed the bright-inactive phenotype (Fig. 5b,c; population data in Fig. 5d) of very high fluorescence levels with no calcium transients or any indication of spiking activity during visual stimulation. Some showed blebbing. Further (Fig. 5e, bottom trace), some cells showed occasional calcium transients followed by long, sustained increases in brightness that were distinct from the bright-inactive phenotype (see Supplementary Video S6 for examples of these pathologies). None of these aberrant physiological profiles were observed in any neurons labeled with the second-generation vector. As in the structural imaging experiments with tdTomato, neurons transduced with second-generation vector showed a gradual buildup of GCaMP6f, with the dimmer baseline fluorescence (F) reflected in higher signal amplitude ΔF/F at the earliest time points (Fig. 5f). Excluding cells with gross pathologies like bright-inactivity and sustained activity from the analysis, the population of neurons infected with the first-generation vector also showed a tendency toward more ‘sluggish’ calcium dynamics, with a large mean increase (29.1%) in decay time constant (τ) of calcium transients between the 2-week and 8-week imaging sessions (Fig. 5g). By contrast, neurons labeled with the second-generation vector showed stable time constants with no significant change in τ. All of these results indicate that the neurons labeled with the second-generation vector retain stable response properties at 8 and 16 weeks postinjection, whereas neurons infected with the first-generation vector, even when they survive for long durations, show significant changes and dysfunction over time.
As a final characterization, we conducted a direct comparison in vivo of the retrograde transduction ability of our second-generation rabies virus to two other viral species used for this application: canine adenovirus type 2 (CAV2)41 and the newly developed rAAV2-retro42. We injected each of the three viruses in the anterior cingulate cortex of Ai75 reporter mice, in which nuclear-localized tdTomato is expressed following Cre recombination. The results are shown in Fig. 6. While all three viruses retrogradely labeled many cells in primary visual cortex, CAV2-Cre exhibited a pronounced preference for layer 5 cells, whereas rAAV2-retro labeled many cells in upper and lower layers but very few in layer 5. In contrast, RVΔGL-Cre labeled many cells in all cortical areas. Injections of the three vectors in other cortical areas showed similar results (Supplementary Fig. S5). These data are consistent with previous reports of both CAV2-Cre43 and rAAV2-retro42 having strong tropism for particular cell types over others (in projections other than the ones examined here) and suggest that tropism may be less of an issue with second-generation RV than with these other two vector species.
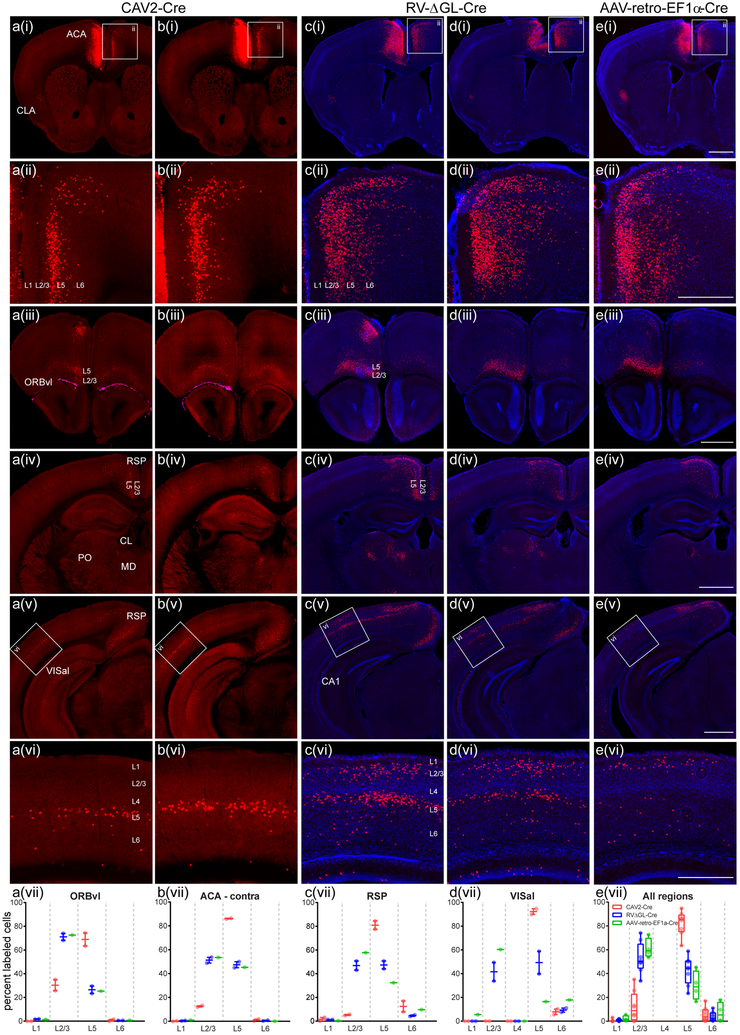
Figure 6. Second-generation rabies viral vectors retrogradely infect more types of corticocortical cells than either canine adenoviral or rAAV2-retro vectors.
CAV2-Cre (a,b), RVΔGL-Cre (c,d), or rAAV2-retro-EF1a-Cre (e) was injected into anterior cingulate area (ACA) of Ai75 mice so that cells across the brain which provide input to ACA were labeled via Cre-induced nuclear tdTomato expression. The approximate center of the injection site for each experiment is shown in (a(i)–e(i)) (left side of images). Most brain regions containing retrogradely labeled cells from ACA were consistently identified across all three types of virus experiments, with at least one notable exception (CA1 contained labeled cells in RVΔGL-Cre, (c(v),d(v)). However, within the same areas, specifically in the cortex, we observed differences suggesting tropism or other biases in the specific types of neurons labeled. For example, labeled input cells in the contralateral ACA (boxes in row i indicate panels enlarged in row ii) were seen predominantly in layer 5 (L5) for CAV2-Cre (a(ii),b(ii)), but are located more uniformly across layers with RVΔGL-Cre (c(ii),d(ii)) and AAV-retro-EF1a-Cre (e(ii)). Similar differences between Cre+ input cells across layers were observed in most cortical areas, including ventrolateral orbital cortex (ORBvl, a(iii)-e(iii)), retrosplenial cortex (RSP, a(iv)-e(iv) and a(v)-e(v)), and anterolateral visual cortex (VISal, a(v)-e(v), a(vi)-e(vi)). Quantification of the number of labeled cells per layer indicated that, in all cases, CAV2-Cre labeling is biased toward L5 neurons, whereas RV-ΔGL-Cre results in input cells labeled across all layers providing long-range inter-areal inputs (L2/3, L5, L6; a(vii)-e(vii)). In contrast to RVΔGL-Cre and CAV2-Cre in VISal and RSP, AAV-retro-EF1a-Cre appeared to be biased towards L2/3, with only sparse labeling in L5 (c(vii) and d(vii)). Cre+ input cells were also observed in subcortical structures, including several thalamic nuclei (e.g. mediodorsal nucleus (MD), posterior complex (PO), and the central lateral nucleus (CL), seen in (iv), consistent with known connectivity of ACA. The more prominent labeling in RVΔGL-Cre and AAV-retro-EF1a-Cre cases in the thalamus (c(iv)-e(iv)) may be due to true differences in efficiency between viruses, or in the amount of viral uptake for each experiment; further experiments are necessary to explore this observation. Images in (a,b) were acquired by serial 2-photon tomography. Images in (c–e) were acquired by epifluorescence microscopy and sections counterstained with DAPI (blue). Graphs in (a(vii)-d(vii)) show the mean and full range of values (min to max, n=2 independent experiments for CAV2-Cre and RVΔGL-Cre;n=1 experiment for AAV-retro-EF1a-Cre). Boxplots in (e(vii)) indicate the median and full range (min to max) in all four regions from (a(vii)-d(vii)) (n=5 independent experiments). Scale bars: rows i,iii-v = 1 mm; rows ii,vi = 500 μm.
Discussion
Here we have introduced a new class of rabies viral vectors that do not cause any detectable structural or functional abnormalities in transduced neurons, in stark contrast to their first-generation counterparts. We have shown that these second-generation rabies viral vectors encoding either Cre or Flp recombinase also are effective at causing reporter gene expression both in cell culture and in vivo in readily available reporter mouse lines and in wild-type animals with reporter AAVs, that they retain first-generation vectors’ ability to efficiently retrogradely transduce projection neurons far from the injection site, with wide tropism, and that these retrogradely transduced neurons appear morphologically and physiologically normal even at the longest time points examined. We expect that this new class of vectors will be a useful new tool for neuroscientists investigating neurons selected on the basis of their extrinsic projections.
Requirements for second-generation monosynaptic tracing
Given that these second-generation RV vectors do not structurally or functionally perturb neurons over any time scale examined and can be used to express recombinases in retrogradely infected projection neurons, an exciting potential application for this new class of vectors would be monosynaptic tracing. Monosynaptic tracing with first-generation RV vectors already allows neurons directly presynaptic to a targeted neuronal population or single neuron to be labeled for short-term anatomical identification3,4,9,36,44, optogenetic manipulation8,45, or activity imaging7,10, the toxicity of first-generation RV vectors confines these experiments to short-term paradigms. A monosynaptic tracing system based on the second-generation RV vectors should allow labeled presynaptic networks to be imaged or optogenetically manipulated for months or even years. First-generation monosynaptic tracing requires coexpression of the single deleted gene (G), while a second-generation monosynaptic tracing system will also require expression of the deleted polymerase gene (L). In some contexts, this is theoretically very simple. Single-cell monosynaptic tracing, for example, currently entails introduction into single neurons, via a patch pipette, a mixture of plasmids encoding G, the receptor TVA (for allowing entry of the EnvA-enveloped RV into the targeted neuron), and a fluorophore to mark the targeted cell4,5,7,10. Second-generation RV would only require inclusion of a fourth plasmid, encoding L. While such direct transfection of single cells is the conceptually simplest implementation of monosynaptic tracing, the much more accessible and widely adopted approach is to label inputs to groups of neurons, using Cre or Flp mouse lines in combination with adeno-associated “helper viruses” that express G, TVA, and a fluorophore in the recombinase-expressing cells. This approach has allowed, for example, whole-brain mapping of inputs to dopaminergic neurons in the midbrain6 and noradrenergic neurons in the locus coeruleus46. Adaptation of the helper virus approach to a second-generation version is in principle also straightforward. However, simply including an L-expressing AAV vector in the mix is problematic, because, at 6 kb in length, L exceeds the packaging capacity of AAV47,48. Alternative approaches currently being pursued include generating L-expressing mouse lines and the splitting of the polymerase into two complementary halves, each within the packaging capacity of AAV. An aspect of RV biology that is likely to be very helpful to these efforts is the fact that the polymerase gene is expressed at very low levels by wild-type rabies virus, much lower than those of the other four viral genes, including G26,49. Achieving sufficient expression levels of L from the complementing systems - whether direct transfection, genomic alleles, or helper viruses - therefore appears unlikely to be a problem.
Online methods
All experimental procedures related to the use of mice were conducted according to NIH guidelines. Experiments performed at the Allen Institute were approved by the Institutional Animal Care and Use Committee (IACUC) of the Allen Institute for Brain Science. Experiments performed at MIT were approved by the MIT Committee for Animal Care (CAC). Animals were housed singly postinjection, and were kept on a normal light/dark cycle for all experiments except awake two-photon imaging.
Cloning
Expression vectors for rabies viral genes and the T7 polymerase were made by cloning the N, P, G, and L genes from pTIT-N, pTIT-P, pTIT-L51, pTIT-G52, and pCAGGS-T753 into pCAG-GFP54 (Addgene #11150) to make pCAG-B19N (Addgene #59924), pCAG-B19P (Addgene #59925), pCAG-B19G (Addgene #59921), pCAG-B19L (Addgene #59922), and pCAG-T7pol (Addgene #59926).
The Flp reporter construct pLV-CAG-F14F15S-Switch was made by cloning into pCSC-SP-PW-GFP55 (Addgene #12337) the CAG promoter56 and a Flp-dependent “FLEX”57 construct consisting of pairs of compatible FRT sites flanking a back-to-back fusion of the reverse-complemented gene for TagRFP-T58 immediately followed by the forward gene for EGFP.
First-generation vector genome plasmids were made by insertion of the tdTomato59 or mammalian codon-optimized Cre60 gene in place of the EGFP gene in pRVΔG-4GFP (a.k.a. cSPBN-4GFP (Addgene #52487) to make pRVΔG-4tdTomato (Addgene #52500), pRVΔG-4Cre (Addgene #98034), and pRVΔG-4GCaMP6f (Addgene #98035) (the “4” in the plasmid/virus names refers to the position of the transgene in the viral genome, following the convention introduced in Wickersham et al. 201061).
Second-generation vector genome plasmids were made by complete deletion of the polymerase gene as well as the G-L intergenic sequence from cSPBN-4GFP and pRVΔG-1EGFP29, with and without replacement of the EGFP gene with the Cre or Flpo genes, by seamless cloning (In-Fusion, Takara), resulting in pRVΔGL-4EGFP (Addgene #98036), pRVΔGL-1EGFP (Addgene #98037), pRVΔGL-1Cre (Addgene #98038), pRVΔGL-4Cre (Addgene #98039), and pRVΔGL-4Flpo (Addgene #98040).
Rabies viral vector production and titering
First-generation rabies viral vectors (RVΔG-EGFP, RVΔG-tdTomato, and RVΔG-Cre) were made as described62. Briefly, HEK 293T cells (ATCC CRL-11268) were transfected with expression vectors for the ribozyme-flanked viral genome (pRVΔG-4EGFP, pRVΔG-4tdTomato, or pRVΔG-4Cre), rabies viral genes (pCAG-B19N, pCAG-B19P, pCAG-B19G, and pCAG-B19L), and the T7 polymerase (pCAG-T7Pol). Supernatants were collected from 4 to 7 days after transfection, filtered, and pooled, then passaged 3–4 times on BHK-B19G2 cells61 at a multiplicity of infection of 2–5. Purification, concentration, and titering were done as described61, with the Cre reporter cell line 293T-Switch263 used for titering RVΔG-Cre.
Production of second-generation rabies viral vectors (RVΔGL-EGFP, RVΔGL-Cre, and RVΔGL-Flpo) proceeded similarly but with the supernatants from the transfection plates passaged not on BHK-B19G cells but on HEK 293T cells transfected with pCAG-B19G and pCAG-B19L. Purification, concentration, and titering were as described61, with RVΔGL-Cre titered on 293T-Switch2 cells and RVΔGL-FlpO titered on HEK 293T cells transfected with the Flp reporter construct pLV-CAG-F14F15S-Switch.
Immunostaining, imaging, and flow cytometric analysis of HEK cells
HEK 293T cells infected with serial dilutions of RVΔG-EGFP and RVΔGL-EGFP as described61 were fixed with 4% paraformaldehyde, permeabilized with 0.05% Triton-X (Sigma) in 5% normal donkey serum (Jackson Immuno, #017–000–121), then incubated with 1:1000 chicken anti-GFP polyclonal primary antibody (Aves Labs, GFP-1020) for one hour followed by washing and incubation with 1:500 AlexaFluor 594-conjugated donkey anti-chicken secondary antibody (Jackson Immuno, #703–585–155). Images were collected on a Zeiss 710 confocal microscope, and flow cytometric analysis was conducted as described61.
Mouse thalamic injections, histology, & confocal imaging
Mouse lines used for thalamic injections were the Cre-dependent tdTomato reporter line Ai1464 (Jackson Laboratory #007908) and the Flp-dependent tdTomato reporter line Ai65F, produced by crossing the Cre- & Flp-dependent tdTomato double reporter line Ai65D65 (Jackson Laboratory 021875) to the Cre deleter line Meox2-Cre66 (Jackson Laboratory 003755), so that only Flp is required for expression of tdTomato.
50–100 nl of viral vector solution were injected into ventral posteromedial nucleus (−1.90 mm AP, 1.57 mm LM, 3.66 mm DV; all stereotaxic coordinates are with respect to bregma) of anesthetized adult mice (Ai14 for RVΔGL-Cre or Ai65F for RVΔGL-Flpo) using a stereotaxic instrument (Stoelting Co.) and custom injection apparatus consisting of a hydraulic manipulator (MO-10, Narishige) with headstage coupled via custom adaptors to a wire plunger advanced through pulled glass capillaries (Wiretrol II, Drummond) back-filled with mineral oil and front-filled with viral vector solution.
Seven to 398 days following virus injection, mice were perfused transcardially with 4% paraformaldehyde in phosphate-buffered saline. Brains were postfixed in 4% paraformaldehyde overnight at 4°C and cut into 50 μm sections on a vibrating microtome (VT-1000S, Leica). Mounted sections were imaged on a LSM 710 confocal microscope (Zeiss).
Rat intersectional injections, histology, & confocal imaging
Adult Long-Evans rats (269–299 g) were injected with both RVΔGL-Cre and a FLEX AAV using the custom injection apparatus described above. For labeling thalamocortical neurons, 100 nl of RVΔGL-Cre was injected into barrel cortex (−1.44 mm AP, +5.40 mm LM, −3.00 mm DV) and 300 nl of AAV8-hSyn-DIO-mCherry (Addgene #50459) or AAV5-Syn-FLEX-rc[ChrimsonR-tdTomato] (Addgene #62723) was injected into ventral posteromedial nucleus (−3.50 mm AP, +3.00 mm LM, −6.00 mm DV) in the same surgery. For labeling corticothalamic neurons, 100 nl of RVΔGL-Cre was injected into ventral posteromedial nucleus, and 300 nl of AAV8-hSyn-DIO-mCherry (Addgene #50459) or AAV5-Syn-FLEX-rc[ChrimsonR-tdTomato] (Addgene #62723) was injected into barrel cortex in the same surgery. 21 days following injection, rats were perfused, brains sectioned, and imaged on a confocal microscope as described above.
Surgical preparation and virus injections for two-photon imaging
Mouse lines used for in vivo two-photon imaging were the Cre-dependent tdTomato reporter line Ai14 for RVΔG-Cre and RVΔGL-Cre structural imaging studies; the Cre-/tTA-dependent GCaMP6f39 reporter line Ai9365 (Jackson Laboratory #024103) crossed with the tTA driver line Camk2a-tTA67 (Jackson Laboratory #007004) for RVΔG-Cre and RVΔGL-Cre functional imaging studies; and C57BL/6 mice (Jackson Laboratory #000664) for all RVΔG-tdTomato and RVΔG-GCaMP6f experiments.
Adult mice (>P45; male and female) were anesthetized under isoflurane (3% induction, 1.25% maintenance, 100% O2) and ketamine/xylazine (70 mg/kg, i.p.). An incision was made in the scalp and headplate implanted with Metabond (Parkell) using aseptic technique. A craniotomy (5 mm diameter) and durotomy were performed over left hemisphere V1 and higher visual areas (ML 3.1, AP 1.3 mm from lambda), and a glass window was cemented over exposed cortex.
After allowing a minimum of 7 days to recover from surgery, we mapped visual area boundaries with widefield intrinsic signal imaging of autofluorescence under 470 nm excitation68,69. The emitted fluorescence was collected by a 4x objective (Olympus, NA 0.28), longpass filtered (500 nm cutoff) and recorded on a PCO.edge CMOS camera (60 Hz frame rate, 2160 × 1920 pixels). The awake, head-fixed animal was shown 7 retinotopic mapping conditions on an LCD screen (60 Hz refresh rate) positioned 25 cm from the contralateral eye. Each condition consisted of a circular patch (30 deg diameter) of square wave drifting grating on a black background (100% grating contrast, 8 directions shown in quick succession, 0.05 cycles/degree, drifting at 4 Hz) for 5 seconds, followed by 15 seconds of black background. The conditions varied only in screen position (2 rows of 3 evenly spaced patches each, spanning the screen, followed by a seventh patch in the center). The stimulus set was repeated 5 times. After downsampling the imaging data to 2 Hz, ΔF/F for each condition was calculated as the difference between fluorescence during the stimulation period (taken as the average signal of the first 4 seconds) and blank (9–20 seconds), divided by fluorescence during the blank, averaged over all repeats. This procedure clearly delineated the boundaries of visual areas V1, LM, PM, and AL.
Using these intrinsic signal images, we targeted all injections to the part of V1 mapped to the center of the LCD screen. The animal was anesthetized under 1.25% isoflurane in 100% O2 and the glass window removed. A replacement window with a pre-drilled 500–750 μm hole was secured over exposed cortex with silicone elastomer (Kwik-Cast, WPI). Virus solutions were titer-matched by diluting with HBSS (diluted titer for all viruses was approximately 5 × 109 infectious units/mL), and Alexa Fluor 594 hydrazide (30 μM final concentration; Thermo Fisher) was added as a contrast agent. Injections were done under two-photon visual guidance using glass micropipettes broken to a tip size of 7.5 μm. After advancing the micropipette through the hole in the window and down to >300 μm depth in cortex, we waited 5 min, then injected for 2.5–3 min at 80–100 mbar pressure, visually monitoring the spread of Alexa Fluor during the entire injection. After waiting 5 more minutes, we repeated the injection procedure. The temporary window was removed and a standard window was cemented over cortex before recovering the animal.
Structural two-photon imaging and data analysis
Imaging was done with the animal anesthetized under 1.25% isoflurane in 100% O2. Eyes were protected with Puralube ointment and body temperature maintained at 37° C. A week after virus injection, fields of view (FOVs) were chosen some distance away from the area of brightest tdTomato labeling, since relative sparseness was useful for counting cells. Two well-separated areas were chosen in each mouse, one near the edge of V1, the other either on the opposite edge of V1 or in a higher visual area (LM or PM; except in one mouse with a single V1 FOV). No systematic differences were observed in cell survival rates between V1 and higher visual areas, so all FOVs were treated as equal in the analyses.
Two-photon structural imaging was performed on a custom-built microscope with non-resonant galvanometric scanning mirrors (512×512 pixels, 1 Hz frame rate) coupled to a mode-locked Ti:sapphire laser (Chameleon Ultra II) emitting at 1020 nm. Image acquisition was controlled with a modified version of ScanImage70. Laser power exiting the 16x water-immersion objective (Nikon, NA 0.8) varied between 30–70 mW, depending on depth. Two volumes were imaged in each area at each time point: one at low magnification (600 × 600 μm per frame, from pial surface down to 300–600 μm, 1 μm spacing between frames), used to record labeling efficiency, for alignment checks, and for the volume renders (e.g., Fig. 2c); and a second, smaller volume (100–200 μm in z) taken at higher magnification (300 × 300 μm per frame, 1 μm spacing). The latter was used for cell counts. The main criterion for deciding how many frames to include in the high-magnification stacks was reducing somatic overlap when a maximum intensity z-projection was taken of the stack, so that individual cells could be unambiguously identified in the projection. The max projection for different time points of an individual imaged area were aligned using TurboReg71 in ImageJ. An affine transformation was generally appropriate, since slight cortical deformations did occur over 16 weeks. Each alignment was checked with attention paid to dendritic processes and other unique identifiers. After alignment across time points, cell bodies were manually counted in Vaa3D72. Vaa3D was also used for full volume renders of the low-magnification stacks. Images were pseudocolored for presentation in ImageJ.
15 cell-counting FOVs from eight animals were followed over time. Five RVΔGL-Cre FOVs from three animals; four RVΔG-Cre FOVs from two animals; and six RVΔG-tdTomato FOVs from three animals, giving us a total starting population of 1025 cells. All FOVs and time points are shown in Fig. 2c. No injected animals that survived beyond four weeks were excluded, so statistical analyses of cell counts (paired t-tests; Fig. 2b) were performed for all FOVs at weeks one and four. Imaging was only terminated before 16 weeks for either animal health reasons or death, or in the case of RVΔG-tdTomato because very few cells remained.
Functional two-photon imaging and data analysis
A week after virus injection, the center of the brightest GCaMP6 label in left-hemisphere V1 (assessed under conventional epifluorescence) was chosen as the imaging area. Two-photon functional imaging was performed using a custom-built, resonant-scanning microscope (30 Hz frame rate, 512 lines per frame) with the same objective and laser (at 940 nm) as in the structural imaging experiments. Laser power at the objective ranged from 50 to 150 mW. Calcium imaging data were acquired from three equispaced planes in supragranular layers (100 to 200 μm deep). Surface vasculature provided coarse fiducial markers for finding the same FOVs in subsequent weeks. Radial vasculature and cell body images from averaged data taken in the first session provided a template for fine alignment. The animal was awake, head-fixed and free to run on a circular disk. No behavioral training or reward was given.
Visual stimuli were generated in python with custom software based on Psychopy73 and shown on the same LCD screen as in the widefield mapping experiments. Each condition consisted of 2 seconds of a full-field sine wave grating drifting in one direction, presented at 80% contrast with spatial frequency of 0.04 cycles/deg, followed by 1 second of uniform mean luminance (gray). All permutations of 8 directions (45 deg steps) and 5 temporal frequencies (1, 2, 4, 8, 15 Hz) were shown, in randomized order. The complete set was repeated 15 times, for a total stimulation period of 30 min per FOV per session. Whole-frame registration based on cross-correlation was used to reduce motion artifacts, and data were downsampled to 4 Hz. Cells were then manually segmented, and single-cell fluorescence traces were extracted by averaging the fluorescence of all pixels masking the soma. The mean ΔF/F over the full 2 seconds of each stimulus condition was used to calculate orientation tuning curves, with background fluorescence (F) in ΔF/F taken as the value of the trace immediately preceding a condition, averaged over all conditions. The raw calcium traces from cells within individual FOVs (not across FOVs, given different imaging conditions across animals and time points) were sorted by mean fluorescence, and the quantification of bright-inactive cells was performed only on cells in the upper half of sorted brightness for each FOV, in order to distinguish abnormal, filled-in cells from cells that may have been healthy but quiescent.
To estimate time constants, the functional data were reprocessed using a constrained non-negative matrix factorization (CNMF) algorithm74 with Python code provided by the authors. In the CNMF framework, the matrix encoding the complete spatiotemporal activity of a calcium imaging session is expressed as the product of a spatial matrix encoding the location of each neuron in the FOV and a temporal matrix encoding the time-varying fluorescence signal of each neuron. Importantly, the framework imposes a model of calcium indicator dynamics that can be approximated as an autoregressive process of order p (AR(p)). Since GCaMP6f has a small rise time constant39 relative to the length of our imaging time bin Δt, a first-order model AR(1) was appropriate for this dataset74, in which the calcium impulse response following a spike is modeled by an instantaneous rise followed by a slowly decaying exponential function with a single decay time constant τ. The discrete AR(1) model of the calcium concentration dynamics c(t) at timesteps t=1,…T, can be written as:
where s(t) is the number of spikes estimated at timestep t and γ is an estimate of the discrete time constant. The decay time constant τ can then be calculated from γ using the relation:
Decay time constants were estimated for cells segmented with the CNMF package using data from the first 4 minutes of each imaging session. Only cells that had a peak inferred ΔF/F > 0.25 with no plateaus of elevated fluorescence (e.g., Fig. 5e, bottom trace), with some activity (no bright-inactive cells), and that were clearly imaged at their cell bodies and not apical dendrites (which generally exhibited different time constants than somatic signals), were included in the population analysis.
7 animals were used in these functional imaging experiments (21 total FOVs). Three RVΔGL-Cre cases, each imaged out to 16 weeks postinjection; two RVΔG-Cre cases, each imaged out to at least eight weeks postinjection (with one imaged at 16 weeks); and two RVΔG-GCaMP6f cases, imaged out to only four weeks because of very few surviving cells (see Supplementary Fig. S3). Since week two had brighter GCaMP6f labeling than week one in the two Cre conditions, population analyses over time (Fig. 5d,f,g) were done between weeks two and eight. Imaging termination criteria were the same as in the structural imaging experiments.
Whole-cell patch-clamp recordings
For brain slice electrophysiology recordings 6 adult male Ai14 transgenic mice (aged 8 or 14 weeks) were used. Retrobeads (Lumafluor Inc.) or the first- or second-generation rabies viral vector encoding Cre recombinase was injected into the nucleus accumbens (NAc) at the stereotaxic coordinates from bregma: +1.30 mm AP, +0.80 mm ML and −4.70 mm DV.
After 8 weeks mice were anesthetized with 90 mg/kg sodium pentobarbital and perfused transcardially with 20 mL of artificial cerebrospinal fluid (ACSF, at ~4°C) containing (in mM): 75 sucrose, 87 NaCl, 2.5 KCl, 1.3 NaH2PO4, 7 MgCl2, 0.5 CaCl2, 25 NaHCO3 and 5 ascorbic acid. The brain was extracted and glued (Roti coll 1; Carh Roth GmbH, Karlsruhe, Germany) to the platform of a semiautomatic vibrating blade microtome (VT1200; Leica, Buffalo Grove, IL) which was placed in a slicing chamber containing modified ACSF at 4°C. Coronal sections of 300 μm containing the NAc and BLA were collected and placed in a holding chamber filled with ACSF saturated with 95% O2 and 5% CO2, containing (in mM): 126 NaCl, 2.5 KCl, 1.25 NaH2PO4, 1.0 MgCl2, 2.4 CaCl2, 26.0 NaHCO3, and 10 glucose. Recordings were commenced at least 1 hr following slicing, and the temperature was maintained at approximately 31°C both in the holding chamber and recording chamber. All NAc injection sites were verified and imaged with a camera (Hamatsu Photonics K.K., Japan) attached to a microscope (BX51; Olympus, Center Valley, PA).
Recordings were made from visually identified neurons expressing tdTomato or containing retrobeads. Voltage and current-clamp recordings of BLA neurons projecting to the NAc were conducted using glass microelectrodes (4–7 MΩ) shaped with a horizontal puller (P-1000, Sutter, CA) and filled with a solution containing (in mM): 125 potassium gluconate, 20 HEPES, 10 NaCl, 3 MgATP, 8 biocytin and 2 Alexa Fluor 350 (pH 7.33; 287 mOsm). Recorded signals were amplified using a Multiclamp 700B amplifier (Molecular Devices, Sunnyvale, CA). Analog signals were digitized at 10 kHz using a Digidata 1440 and recorded using the pClamp10 software (Molecular Devices, Sunnyvale, CA). Oxygenated ACSF was perfused onto the slice via a peristaltic pump (Minipuls3; Gilson, Middleton, WI) at ~3 mL/min. The membrane properties of each cell were evaluated in voltage-clamp and current-clamp modes. The access resistance (Rs), membrane capacitance (Cm), membrane resistance (Rm) and time constant (Tau) were calculated from a membrane seal test conducted in voltage-clamp mode, in which 100 ms, 5 mV voltage steps were delivered at a frequency of 5 Hz. We also measured the holding current at a holding potential of −70 mV and calculated the current/voltage relationship in voltage clamp using a series of 200 ms voltage steps, deviating from the −70 mV holding potential by −30, −25, −20, −15, −10, −5, 0, 5, 10, 15, 20, 25, 30, 35, and 40 mV. In order to evaluate the action potential threshold and rheobase, a 1 s, 300–750 pA current ramp was delivered in current-clamp mode. Finally, to evaluate the relationship between injected current and firing frequency, we delivered a series of 2 s current pulses in current clamp, in which the first pulse was 50 pA above the holding current required to keep the cell at −70 mV, and each subsequent pulse increased by 25 pA to a maximum of 525 pA. Any significant changes in Rs or holding current were interpreted as signs of poor cell health and recordings were terminated. Off-line analysis was performed using Clampfit 10.6.1.1 (Molecular Devices, Sunnyvale, CA) or MATLAB R2013b (seal test analysis). Statistical significance was assessed with GraphPad Prism 6 (GraphPad Software, Inc.) using a one-way analysis of variance (ANOVA), with Neuman-Keuls post-hoc tests where appropriate.
Comparative viral vector injections, imaging, and quantification
CAV2-Cre with a titer of 1.0X1012 genome copies/ml was obtained from the Viral Vector Production Unit at the Universitat Autonoma de Barcelona and was produced from a vector enabling expression of Cre under control of the CMV promoter75. rAAV2-retro-Cre was generated at the Allen Institute by cotransfecting 293 cells with pAAV-EF1a-Cre-WPRE-hGHpolyA, pHelper and pAAV2-retro42, followed by iodixanol gradient purification. The Cre coding sequence with the kozak consensus sequence of GCCGCCACC was inserted into the XbaI and EcoRI restriction sites to generate the pAAV-EF1a-Cre-WPRE-hGHpolyA construct. The titer of rAAV-retro-EF1a-Cre was 5.0×1012 genome copies/ml as measured by qPCR. The titer of the undiluted RVΔGL-Cre was 4.26×1010 infectious units/ml. rAAV2/1-CAG-FLEX-EGFP-WPRE-bGH stock number V5749 with a titer of 1.34X1013 genome copies/ml (AAV-FLEX-EGFP) was obtained from the Penn Vector Core.
Viruses were injected into the left hemisphere of adult (P55-P134) male and female Ai75 (RCL-nT; Jackson Laboratory #025106) mice using stereotaxic coordinates or intrinsic signal imaging to locate the target region. A total of seven mice were injected in ACA, and eight in AM, with representative data shown from each animal between Fig. 6 and Supplementary Fig. S5. Stereotaxic coordinates were used for all three tracers in ACA (mm from bregma: AP +0.5, ML −0.25, and DV 0.9 from brain surface), and for RVΔGL-Cre and AAV-retro-EF1a-Cre injections into area AM (AP −2.18, ML −1.6, DV 0.55). CAV2-Cre injections into AM were guided by sign maps derived from intrinsic signal imaging results for each animal (injection depths 0.3 and 0.6 mm from brain surface with a 20° pipette angle76; see also methods at http://help.brain-map.org/download/attachments/2818171/Connectivity_Overview.pdf].
For paired intersectional injections in mice, AAV-FLEX-EGFP was injected into ACA at (mm from bregma: AP +1.1, ML −0.25, and DV 0.9&1.3 from brain surface) using iontophoresis (5 uA, 5 min at each depth) and RVΔGL-Cre was injected into RSP at (mm from bregma: AP −2.06, ML −0.17, and DV 0.8 from brain surface) using pressure injection. The two injections were performed during a single surgical procedure. All retrograde viruses were injected via pressure using a Nanoject II system (CAV2-Cre; 100–200 nl; RVΔGL-Cre 250 nl; rAAV2-retro-EF1a-Cre, 50 nl). Twenty-one days after virus injection, mice were perfused with 4% paraformaldehyde (PFA), then brains were dissected and post-fixed in 4% PFA at room temperature for 3–6 hours, followed by overnight at 4°C. CAV2-Cre injected brains were stored in PBS with 0.1% sodium azide before proceeding to serial two photon (STP) imaging using a TissueCyte 1000 system (TissueVision, Inc.) as described77. Briefly, coronal images were acquired every 100 μm through the entire rostral-caudal extent of the brain with xy resolution of 0.35 μm/pixel. RVΔGL-Cre and rAAV2-retro-EF1a-Cre injected brains were stored in 10% sucrose in PBS before sectioning at 50 μm intervals with a microtome (Leica SM2000 R). Sections were mounted on gelatin-coated slides and stained for 10 minutes with 5 μM 4’,6-diamidino-2-phenylindole (DAPI) in PBS, rinsed for 10 minutes in PBS, and coverslipped using Fluoromount-G (SouthernBiotech). Whole slide scanning was performed with an Olympus VS110 microscope at 4x magnification. Images were cropped, downsampled, and dynamic range-adjusted using Adobe PhotoShop CS6.
The number of retrogradely labeled cells per layer was determined by counting the number of tdTomato-positive nuclei in each figure panel, except for row iii and iv (ORBvl and RSP) in Fig. 6, where a 1mm x 1mm ROI was drawn on the image and counting was performed inside the ROI. Layer boundaries were determined by expert visualization of DAPI labeled nuclei where available and pia-white matter distance. To estimate the uptake volume of viral injections, we manually drew polygons around the borders of the densely labeled cells on the images for all tissue sections covering the rostral-caudal extent of the injection site. The area of the polygons was measured using ImageJ, and these areas were converted from pixels to microns and then multiplied by the section thickness (100 μm) to estimate the total volume of the injection site. The volume of injection spread did not differ between viruses (mean ± SD = CAV2-Cre: 1.4 ± 0.5 mm3, RV-ΔGL-Cre: 1.1 ± 0.6 mm3, AAV-retro-EF1a-Cre: 0.6 ± 0.4 mm3, p=0.22) but was dependent on injection site (1.5 ± 0.4 mm3 for ACA, 0.7 ± 0.4 mm3 for VISam, p=0.02, two-way ANOVA). All injections spanned the entire thickness of the cortex.
Statistics and reproducibility
Please see the Life Sciences Reporting Summary for further details.
Supplementary Material
Acknowledgements
SC thanks Derric Williams and Saskia de Vries for software, Adrian Cheng and Natalia Orlova for hardware construction, and Raina D’Aleo and Naveen Ouellette for experimental assistance. JAH thanks Karla E. Hirokawa and Philip Bohn for technical assistance. IRW thanks Wendy Salmon for assistance with confocal imaging. Research reported in this publication was supported primarily by the National Institute of Mental Health under award number U01MH106018 (BRAIN Initiative) to IRW and in part by the National Institute on Aging under award number R01AG047589 to JAH and the National Eye Institute under award numbers R01EY010115 and R01EY018742 to RCR. KMT is a New York Stem Cell Foundation - Robertson Investigator and McKnight Scholar. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. GGC is a JFDP Fellow. The authors wish to thank the founders of the Allen Institute for Brain Science, Paul G. Allen and Jody Allen, for their vision, encouragement and support.
Footnotes
Accession codes
Addgene accession codes for the novel plasmids described here are as follows: 98034 (pRVdG-4Cre), 98035 (pRVdG-4GCaMP6f), 98036 (pRVdGL-1EGFP), 98037 (pRVdGL-4EGFP), 98038 (pRVdGL-1Cre), 98039 (pRVdGL-4Cre), and 98040 (pRVdGL-4Flpo).
Data availability
All novel plasmids and their complete sequences have been deposited with Addgene (see Accession Codes section). The data that support the findings of this study are available from the corresponding author upon reasonable request.
Code availability
Code is available from the authors upon request. The CNMF code for modeling in vivo fluorescence time courses can be found at: https://github.com/flatironinstitute/CaImAn.
Competing financial interests statement
The authors have no competing financial interests.
References
- 1.Wickersham IR, Finke S, Conzelmann KK & Callaway EM Retrograde neuronal tracing with a deletion-mutant rabies virus. Nature Methods 4, 47–9 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wickersham IR et al. Monosynaptic restriction of transsynaptic tracing from single, genetically targeted neurons. Neuron 53, 639–47 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wall NR, Wickersham IR, Cetin A, De La Parra M & Callaway EM Monosynaptic circuit tracing in vivo through Cre-dependent targeting and complementation of modified rabies virus. Proceedings of the National Academy of Sciences of the United States of America (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marshel JH, Mori T, Nielsen KJ & Callaway EM Targeting single neuronal networks for gene expression and cell labeling in vivo. Neuron 67, 562–74 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rancz EA et al. Transfection via whole-cell recording in vivo: bridging single-cell physiology, genetics and connectomics. Nature neuroscience (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Watabe-Uchida M, Zhu L, Ogawa SK, Vamanrao A & Uchida N Whole-brain mapping of direct inputs to midbrain dopamine neurons. Neuron 74, 858–73 (2012). [DOI] [PubMed] [Google Scholar]
- 7.Wertz A et al. PRESYNAPTIC NETWORKS. Single-cell-initiated monosynaptic tracing reveals layer-specific cortical network modules. Science 349, 70–4 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Tian J et al. Distributed and Mixed Information in Monosynaptic Inputs to Dopamine Neurons. Neuron 91, 1374–1389 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wallace ML et al. Genetically Distinct Parallel Pathways in the Entopeduncular Nucleus for Limbic and Sensorimotor Output of the Basal Ganglia. Neuron 94, 138–152 e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rompani SB et al. Different Modes of Visual Integration in the Lateral Geniculate Nucleus Revealed by Single-Cell-Initiated Transsynaptic Tracing. Neuron 93, 1519 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu K et al. Lhx6-positive GABA-releasing neurons of the zona incerta promote sleep. Nature 548, 582–587 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chung S et al. Identification of preoptic sleep neurons using retrograde labelling and gene profiling. Nature 545, 477–481 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dolen G, Darvishzadeh A, Huang KW & Malenka RC Social reward requires coordinated activity of nucleus accumbens oxytocin and serotonin. Nature 501, 179–84 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kiritani T, Wickersham IR, Seung HS & Shepherd GM Hierarchical connectivity and connection-specific dynamics in the corticospinal-corticostriatal microcircuit in mouse motor cortex. J Neurosci 32, 4992–5001 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kress GJ et al. Convergent cortical innervation of striatal projection neurons. Nat Neurosci 16, 665–7 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Namburi P et al. A circuit mechanism for differentiating positive and negative associations. Nature 520, 675–8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rajasethupathy P et al. Projections from neocortex mediate top-down control of memory retrieval. Nature 526, 653–9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamawaki N, Suter BA, Wickersham IR & Shepherd GM Combining Optogenetics and Electrophysiology to Analyze Projection Neuron Circuits. Cold Spring Harb Protoc 2016, pdb prot090084 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shin Yim Y et al. Reversing behavioural abnormalities in mice exposed to maternal inflammation. Nature 549, 482–487 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Osakada F et al. New rabies virus variants for monitoring and manipulating activity and gene expression in defined neural circuits. Neuron 71, 617–31 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gong Y et al. High-speed recording of neural spikes in awake mice and flies with a fluorescent voltage sensor. Science 350, 1361–6 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roland B et al. Massive normalization of olfactory bulb output in mice with a ‘monoclonal nose’. Elife 5(2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wickersham IR, Sullivan HA & Seung HS Axonal and subcellular labelling using modified rabies viral vectors. Nat Commun 4, 2332 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reardon TR et al. Rabies Virus CVS-N2c(DeltaG) Strain Enhances Retrograde Synaptic Transfer and Neuronal Viability. Neuron 89, 711–24 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ciabatti E, Gonzalez-Rueda A, Mariotti L, Morgese F & Tripodi M Life-Long Genetic and Functional Access to Neural Circuits Using Self-Inactivating Rabies Virus. Cell 170, 382–392 e14 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Albertini AA, Ruigrok RW & Blondel D Rabies virus transcription and replication. Advances in virus research 79, 1–22 (2011). [DOI] [PubMed] [Google Scholar]
- 27.Madore HP & England JM Rabies virus protein synthesis in infected BHK-21 cells. J Virol 22, 102–12 (1977). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schnell MJ, McGettigan JP, Wirblich C & Papaneri A The cell biology of rabies virus: using stealth to reach the brain. Nature reviews. Microbiology 8, 51–61 (2010). [DOI] [PubMed] [Google Scholar]
- 29.Wickersham IR, Sullivan HA & Seung HS Axonal and subcellular labelling using modified rabies viral vectors. Nature communications 4, 2332 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cormack BP, Valdivia RH & Falkow S FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173, 33–8 (1996). [DOI] [PubMed] [Google Scholar]
- 31.Sauer B & Henderson N Site-specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1. Proceedings of the National Academy of Sciences of the United States of America 85, 5166–70 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sadowski PD The Flp recombinase of the 2-microns plasmid of Saccharomyces cerevisiae. Prog Nucleic Acid Res Mol Biol 51, 53–91 (1995). [PubMed] [Google Scholar]
- 33.Raymond CS & Soriano P High-efficiency FLP and PhiC31 site-specific recombination in mammalian cells. PLoS One 2, e162 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shaner NC et al. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol 22, 1567–72 (2004). [DOI] [PubMed] [Google Scholar]
- 35.Fenno LE et al. Targeting cells with single vectors using multiple-feature Boolean logic. Nat Methods 11, 763–72 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schwarz LA et al. Viral-genetic tracing of the input-output organization of a central noradrenaline circuit. Nature (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gomme EA, Wirblich C, Addya S, Rall GF & Schnell MJ Immune clearance of attenuated rabies virus results in neuronal survival with altered gene expression. PLoS pathogens 8, e1002971 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ohki K, Chung S, Ch’ng YH, Kara P & Reid RC Functional imaging with cellular resolution reveals precise micro-architecture in visual cortex. Nature 433, 597–603 (2005). [DOI] [PubMed] [Google Scholar]
- 39.Chen TW et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ohki K & Reid RC In vivo two-photon calcium imaging in the visual system. Cold Spring Harb Protoc 2014, 402–16 (2014). [DOI] [PubMed] [Google Scholar]
- 41.Soudais C, Laplace-Builhe C, Kissa K & Kremer EJ Preferential transduction of neurons by canine adenovirus vectors and their efficient retrograde transport in vivo. FASEB J 15, 2283–5 (2001). [DOI] [PubMed] [Google Scholar]
- 42.Tervo DG et al. A Designer AAV Variant Permits Efficient Retrograde Access to Projection Neurons. Neuron 92, 372–382 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Senn V et al. Long-range connectivity defines behavioral specificity of amygdala neurons. Neuron 81, 428–37 (2014). [DOI] [PubMed] [Google Scholar]
- 44.Kohara K et al. Cell type-specific genetic and optogenetic tools reveal hippocampal CA2 circuits. Nat Neurosci 17, 269–79 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu C et al. Distinct Hippocampal Pathways Mediate Dissociable Roles of Context in Memory Retrieval. Cell 167, 961–972 e16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schwarz LA & Luo L Organization of the locus coeruleus-norepinephrine system. Curr Biol 25, R1051–R1056 (2015). [DOI] [PubMed] [Google Scholar]
- 47.Wu Z, Yang H & Colosi P Effect of genome size on AAV vector packaging. Mol Ther 18, 80–6 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dong B, Nakai H & Xiao W Characterization of genome integrity for oversized recombinant AAV vector. Mol Ther 18, 87–92 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Finke S, Cox JH & Conzelmann KK Differential transcription attenuation of rabies virus genes by intergenic regions: generation of recombinant viruses overexpressing the polymerase gene. Journal of virology 74, 7261–9 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tian L et al. Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nature Methods 6, 875–81 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Finke S & Conzelmann KK Virus promoters determine interference by defective RNAs: selective amplification of mini-RNA vectors and rescue from cDNA by a 3’ copy-back ambisense rabies virus. J Virol 73, 3818–25 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Finke S, Mueller-Waldeck R & Conzelmann KK Rabies virus matrix protein regulates the balance of virus transcription and replication. J Gen Virol 84, 1613–21 (2003). [DOI] [PubMed] [Google Scholar]
- 53.Paterson RG, Russell CJ & Lamb RA Fusion protein of the paramyxovirus SV5: destabilizing and stabilizing mutants of fusion activation. Virology 270, 17–30 (2000). [DOI] [PubMed] [Google Scholar]
- 54.Matsuda T & Cepko CL Electroporation and RNA interference in the rodent retina in vivo and in vitro. Proceedings of the National Academy of Sciences of the United States of America 101, 16–22 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Marr RA et al. Neprilysin regulates amyloid Beta peptide levels. Journal of molecular neuroscience: MN 22, 5–11 (2004). [DOI] [PubMed] [Google Scholar]
- 56.Niwa H, Yamamura K & Miyazaki J Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108, 193–9 (1991). [DOI] [PubMed] [Google Scholar]
- 57.Atasoy D, Aponte Y, Su HH & Sternson SM A FLEX switch targets Channelrhodopsin-2 to multiple cell types for imaging and long-range circuit mapping. The Journal of neuroscience : the official journal of the Society for Neuroscience 28, 7025–30 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shaner NC et al. Improving the photostability of bright monomeric orange and red fluorescent proteins. Nat Methods 5, 545–51 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shaner NC et al. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nature biotechnology 22, 1567–72 (2004). [DOI] [PubMed] [Google Scholar]
- 60.Koresawa Y et al. Synthesis of a new Cre recombinase gene based on optimal codon usage for mammalian systems. Journal of biochemistry 127, 367–72 (2000). [DOI] [PubMed] [Google Scholar]
- 61.Wickersham IR, Sullivan HA & Seung HS Production of glycoprotein-deleted rabies viruses for monosynaptic tracing and high-level gene expression in neurons. Nature protocols 5, 595–606 (2010). [DOI] [PubMed] [Google Scholar]
- 62.Wickersham IR & Sullivan HA Rabies viral vectors for monosynaptic tracing and targeted transgene expression in neurons. Cold Spring Harb Protoc 2015, 375–85 (2015). [DOI] [PubMed] [Google Scholar]
- 63.Wickersham IR et al. Lentiviral vectors for retrograde delivery of recombinases and transactivators. Cold Spring Harb Protoc 2015, 368–74 (2015). [DOI] [PubMed] [Google Scholar]
- 64.Madisen L et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nature neuroscience 13, 133–40 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Madisen L et al. Transgenic mice for intersectional targeting of neural sensors and effectors with high specificity and performance. Neuron 85, 942–58 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tallquist MD & Soriano P Epiblast-restricted Cre expression in MORE mice: a tool to distinguish embryonic vs. extra-embryonic gene function. Genesis 26, 113–5 (2000). [DOI] [PubMed] [Google Scholar]
- 67.Mayford M et al. Control of memory formation through regulated expression of a CaMKII transgene. Science 274, 1678–83 (1996). [DOI] [PubMed] [Google Scholar]
- 68.Husson TR, Mallik AK, Zhang JX & Issa NP Functional imaging of primary visual cortex using flavoprotein autofluorescence. J Neurosci 27, 8665–75 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Andermann ML, Kerlin AM, Roumis DK, Glickfeld LL & Reid RC Functional specialization of mouse higher visual cortical areas. Neuron 72, 1025–39 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pologruto TA, Sabatini BL & Svoboda K ScanImage: flexible software for operating laser scanning microscopes. Biomed Eng Online 2, 13 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Thevenaz P, Ruttimann UE & Unser M A pyramid approach to subpixel registration based on intensity. IEEE Trans Image Process 7, 27–41 (1998). [DOI] [PubMed] [Google Scholar]
- 72.Peng H, Bria A, Zhou Z, Iannello G & Long F Extensible visualization and analysis for multidimensional images using Vaa3D. Nat Protoc 9, 193–208 (2014). [DOI] [PubMed] [Google Scholar]
- 73.Peirce JW PsychoPy--Psychophysics software in Python. J Neurosci Methods 162, 8–13 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pnevmatikakis EA et al. Simultaneous Denoising, Deconvolution, and Demixing of Calcium Imaging Data. Neuron 89, 285–99 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hnasko TS et al. Cre recombinase-mediated restoration of nigrostriatal dopamine in dopamine-deficient mice reverses hypophagia and bradykinesia. Proc Natl Acad Sci U S A 103, 8858–63 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Garrett ME, Nauhaus I, Marshel JH & Callaway EM Topography and areal organization of mouse visual cortex. J Neurosci 34, 12587–600 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Oh SW et al. A mesoscale connectome of the mouse brain. Nature 508, 207–14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Klapoetke NC et al. Independent optical excitation of distinct neural populations. Nat Methods 11, 338–46 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.