Abstract
Background: Altered expression of the BCL-2 family member MCL-1 has been linked to the progression and outcome of various malignancies. Recently, MCL-1 inhibitor S63845 was reported to kill MCL-1-dependent cancer cells and has potential value in clinical application.
Purpose: Herein, we reported MCL-1 expression pattern in Chinese de novo acute myeloid leukemia (AML) and its impact on prognosis and may provide theoretical basis for AML patients using MCL-1 inhibitor in clinics. Real-time quantitative PCR was carried out to detect the transcript of MCL-1 in AML patients.
Results: MCL-1 expression was significantly up-regulated in AML compared with controls (P=0.042). We divided the patients into two groups (higher and lower expression of MCL-1) based on the median level. Among both non-acute promyelocytic leukemia (APL) and cytogenetically normal AML (CN-AML), patients with higher expression of MCL-1 correlated with lower complete remission (CR) rate (P=0.031 and 0.004, respectively) and shorter overall survival (OS) time (P=0.008 and 0.004, respectively) compared with those with lower expression of MCL-1. Meanwhile, Cox regression analyses revealed that overexpression of MCL-1 acted as an independent risk factor for OS in non-APL patients and CN-AML patients (P=0.011 and 0.045, respectively). In follow-up patients, MCL-1 expression level decreased after CR compared with newly diagnosis time (P=0.020) and increased after relapse (P=0.004).
Conclusion: Our findings suggest that higher expression of MCL-1 predicts poor prognosis and can be used for disease monitoring.
Keywords: MCL-1, expression, prognosis, recurrence, acute myeliod leukemia
Introduction
Acute myeloid leukemia (AML), a molecularly and clinically heterogeneous disease, is characterized by differentiation arrest and uncontrolled proliferation of myeloid progenitor cells, which ultimately interferes with the production of normal blood cells.1 Although recent treatment advances in hematopoietic stem cell transplantation and intensive chemotherapy, the clinical outcome for AML remained unsatisfied.2 Since treatment with all-trans retinoic acid (ATRA) has dramatically improved the outcome of acute promyelocytic leukemia (APL) by binding to retinoic acid receptor α, the development of molecular risk-adapted treatment strategies and targeted therapies may improve the clinical outcome of AML.3 Therefore, the identifying biological markers correlated with prognosis, and recurrence is required for better management of AML patients and may provide potential targets for AML therapy. B-cell leukemia/lymphoma-2 (BCL-2) protein family consists of a pro-survival group and two pro-apoptotic subgroups. The pro-survival group comprise BCL-2, myeloid cell leukemia-1 (MCL-1), BCL-2 and BCL-2-like protein 1 isoform 1 (BCL-XL), which play important roles in regulating cell apoptosis, proliferation, differentiation and tumorigenesis.4–8 Among them, MCL-1, isolated from a human myeloblastic leukemia cell line, can affect the development of tumor through the expression of its mRNA and protein in many cancers.9–11 Allen et al indicated that overexpression of MCL-1 was related to poor overall survival (OS) in non-small cell lung cancer when MYC was up-regulated simultaneously.6 In breast cancer, MCL-1 high-expressed was associated with later tumor stage and worse survival.12 Glaser et al also found that knockdown of MCL-1, but not loss or pharmacological blockade of Bcl-xL, Bcl-2 or Bcl-w, caused the death of transformed AML and could cure disease in AML-afflicted mice.13 Recently, MCL-1 protein inhibitor S63845 was reported to kill MCL-1-dependent cancer cells and has potential value in clinical application.14 Herein, we aimed to investigate the MCL-1 expression pattern as well as its clinical implication in Chinese de novo patients with AML and may provide theoretical basis for AML patients using MCL-1 protein inhibitor in clinics.
Materials and methods
Patients
This study was approved by the Institutional Ethics Committee of the Affiliated People’s Hospital of Jiangsu University. Bone marrow (BM) aspirate specimens of 151 patients newly diagnosed AML, 26 healthy donors, 52 AML patients achieving complete remission and 23 relapsed AML patients were collected after written informed consent was signed. The diagnosis and classification of the patients were based on the 2008 WHO criteria.15 All patients were treated at the Affiliated People’s Hospital of Jiangsu University from October 2005 to December 2016. Treatment protocol was described as reported previously.16 The characteristics of all patients are summarized in Table 1.
Table 1.
Comparison of clinical manifestations and laboratory features between AML patients with low and high MCL1 expression
| Patient’s parameters | High (n=75) | Low (n=76) | P-value |
|---|---|---|---|
| Sex, male/female | 44/31 | 47/29 | 0.741 |
| Median age, years (range) | 60 (10–93) | 52 (15–85) | 0.029 |
| Median WBC, ×109/L (range) | 19.5 (0.3–197.7) | 16.6 (0.9–528) | 0.248 |
| Median hemoglobin, g/L (range) | 77 (34–144) | 77 (34–123) | 0.849 |
| Median platelets, ×109/L (range) | 40 (8–399) | 34 (3–415) | 0.355 |
| BM blasts, % (range)* | 50 (1–94.5) | 43 (5.5–97.5) | 0.348 |
| Karyotype classification | 0.465 | ||
| Favorable | 18 (24%) | 25 (33%) | |
| Intermediate | 45 (60%) | 41 (54%) | |
| Poor | 10 (13%) | 8 (10%) | |
| No data | 2(3%) | 2 (3%) | |
| Karyotype | 0.042 | ||
| normal | 31 (41%) | 34 (45%) | |
| t(8;21) | 8 (11%) | 6 (8%) | |
| t(16;16) | 1 (1%) | 0 (0%) | |
| t(15;17) | 8 (11%) | 19 (25%) | |
| +8 | 2 (3%) | 3 (4%) | |
| −5/5q- | 3 (4%) | 0 (0%) | |
| −7/7q- | 0 (0%) | 1 (1%) | |
| t(9;22) | 1 (1%) | 1 (1%) | |
| others | 12 (16%) | 3 (4%) | |
| complex | 7 (9%) | 7 (9%) | |
| No data | 2 (3%) | 2 (3%) | |
| Gene mutation | |||
| CEBPA (±) | 4/60 | 8/53 | 0.234 |
| NPM1 (±) | 10/54 | 2/59 | 0.030 |
| FLT3-ITD (±) | 8/56 | 7/54 | 1.000 |
| c-KIT (±) | 2/62 | 4/57 | 0.432 |
| CR, all AML (±)** | 49/24 | 30/38 | 0.007 |
| CR, non-APL AML (±) | 48/17 | 27/24 | 0.031 |
| CR, CN-AML (±) | 24/7 | 12/18 | 0.004 |
Notes: *AML patients less than 20% BM blasts often with typical cytogenetics such as t(15;17) (q22;q12). **For CR analysis, a total of 141 patients with available follow-up data were included.
Abbreviations: AML, acute myeloid leukemia; WBC, white blood cells; BM, bone marrow; CR, complete remission; CN-AML,cytogenetically normal AML.
BMMNCs separation, RNA isolation and reverse transcription
BM mononuclear cells (BMMNCs) were separated by Lymphocyte Separation Medium (Beijing Solarbio Science & Technology Co., Ltd., Beijing, China). Total RNA was isolated using Trizol reagent (Invitrogen, Carlsbad, CA, USA). Reverse transcription was performed as reported previously.17,18
Real-time quantitative PCR
Real-time quantitative PCR (RQ-PCR) was used to detect MCL-1 expression level on a 7,300 Thermo cycler (Applied Biosystems, CA, USA). AceQ qPCR SYBR Green Master Mix Kit (Vazyme Biotech Co., Piscataway, NJ, USA) and ROX Reference Dye 1 (Invitrogen, Carlsbad, CA, USA) were applied to RQ-PCR reactions. The primers (forward 5ʹ-GATGATCCATGTTTTCAGCGAC-3ʹ and reverse 5ʹ-CTGGGATGGGTTTGTGGAG-3ʹ) were used for MCL-1 transcript detection. The housekeeping gene ABL detected by 2× SYBR Green PCR Mix (Multisciences, Hangzhou, China) was used to calculate the abundance of MCL-1 transcript. The primers of ABL expression were 5ʹ-TCCTCCAGCTGTTATCTGGAAGA-3ʹ (forward) and 5ʹ-TCCAACGAGCGGCTTCAC-3ʹ (reverse). The RQ-PCR was carried out at 95°C for 5 mins, followed by 40 cycles at 95°C for 10 s, 60°C (MCL-1) or 60°C (ABL) for 30 s, 72°C for 32 s and 80°C for 32 s to collect fluorescence, finally followed by 95°C for 15 s, 60°C for 60 s, 95°C for 15 s and 60°C for 15 s. Relative MCL-1 expression levels were calculated by 2−△△CT method.
Gene mutation detection
NPM1 and C-KIT mutations were detected by high-resolution melting analysis using the LightScanner platform (Idaho Technology Inc., Salt Lake City, Utah, USA).20 FLT3-ITD and CEBPA mutations were detected by direct DNA sequencing (BGI Tech Solutions Co., Shanghai, China).19,20 Both positive (leukemic cell lines samples, cultured in RPMI-1640 medium containing 10% fetal calf serum [ExCell Bio, Shanghai, China]) and negative controls (ddH2O) were included in each assay.
Gene expression profiling data
Gene expression profiling (GEP) data (accession number GSE12417, http://www.ncbi.nlm.nih.gov/geo/) was used to validate the prognostic value of MCL-1 expression in two independent cohorts of 162 and 78 cytogenetically normal AML (CN-AML) patients by the online web tool Genomicscape (http://genomicscape.com/microarray/survival.php).21
Statistical analysis
Statistical analyses were performed using the SPSS 20.0 software package. Mann–Whitney’s U test was carried to compare the difference of continuous variables between two groups. Pearson Chi-square analysis or Fisher exact test was employed to compare the difference of categorical variables. Kaplan–Meier analysis and Cox regression analyses were performed to analyze the impact of MCL-1 expression on OS and leukemia-free survival (LFS) in AML. Receiver operating characteristic curve (ROC) and area under the ROC curve (AUC) were conducted to assess the value of MCL-1 expression in discriminating AML patients from normal controls. For all analyses, a two-tailed P-value of 0.05 or less was determined as statistically significant.
Results
MCL-1 expression in controls and AML patients
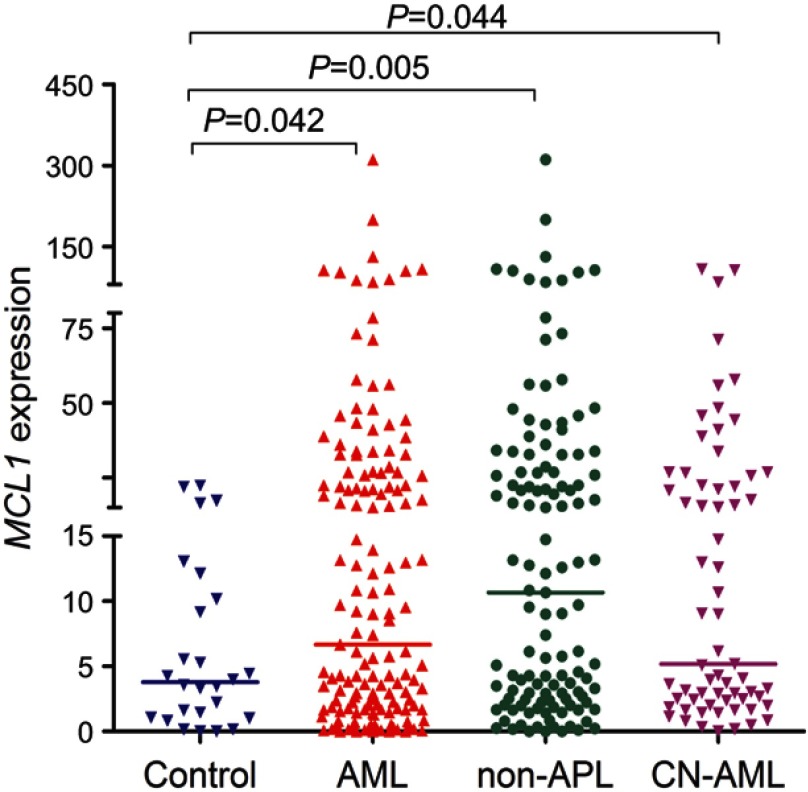
As is shown in Figure 1, MCL-1 transcript level in 151 AML patients (range 0.0000–311.0063, median 6.6821) was significantly higher as compared with 26 controls (range 0.0002–22.4120, median 3.7865), which indicated that MCL-1 expression was up-regulated in AML (P=0.042). In addition, increased MCL-1 expression was also presented in both 123 non-APL (range 0.0000–311.0063, median 10.6716) and 65 CN-AML (range 0.0023–108.2933, median 5.1723) patients (P=0.005 and 0.044, respectively, Figure 1).
Figure 1.
MCL-1 expression in AML patients. MCL-1 expression in 151 newly diagnosis (range 0.0000–311.0063, median 6.6821), 123 non-APL (range 0.0000–311.0063, median 10.6716) and 65 CN-AML (range 0.0023–108.2933, median 5.1723) patients were significantly upregulated in 26 controls (range 0.0002–22.4120, median 3.7865).
Abbreviations: AML, acute myeloid leukemia; CN-AML, cytogenetically normal AML.
Discriminative capacity of MCL-1 expression
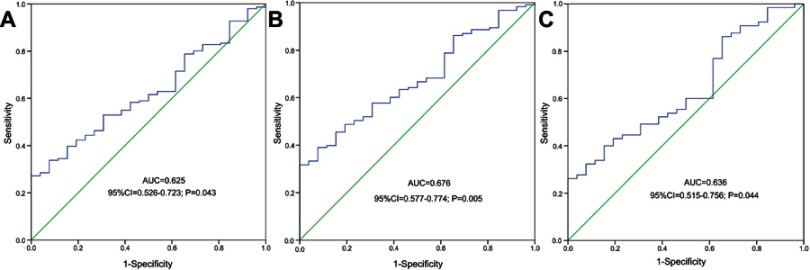
ROC was used to evaluate the discriminative capacity of MCL-1 expression. It revealed that MCL-1 expression could act as a potential biomarker for distinguishing whole-cohort AML (AUC=0.625, 95% CI: 0.526–0.723, P=0.043, Figure 2A), non-APL AML (AUC=0.676, 95% CI: 0.577–0.774, P=0.005, Figure 2B) and CN-AML (AUC=0.636, 95% CI: 0.515–0.756, P=0.044, Figure 2C) from normal controls.
Figure 2.
ROC curve analysis using MCL-1 expression for discriminating AML patients from controls. (A) all AML; (B) non-APL; (C) CN-AML.
Abbreviations: AML, acute myeloid leukemia; APL, acute promyelocytic leukemia; ROC, receiver operating characteristic curve; CN-AML, cytogenetically normal AML.
Clinical and laboratory characteristics of AML patients
In order to investigate the clinical implication of MCL-1 expression in AML, we further divided the patients into two groups (MCL-1high and MCL-1low) based on the median level of 6.6821. The comparison of clinical characteristics of two groups is presented in Table 1. MCL-1high patients showed older age as compared with MCL-1low patients (P=0.029). Moreover, there was a significant difference in the distribution of karyotypes between two groups (P=0.042). However, no significant differences were found between the two groups in the sex, white blood cell (WBC), hemoglobin, platelets, percentage of BM blasts and karyotype classification (P>0.05). MCL-1high patients had a significantly higher frequency of NPM1 mutation than MCL-1low patients (Table 1). No significant differences were observed between MCL-1high and MCL-1low groups among other gene mutations.
Effect of MCL-1 expression on chemotherapy response in AML
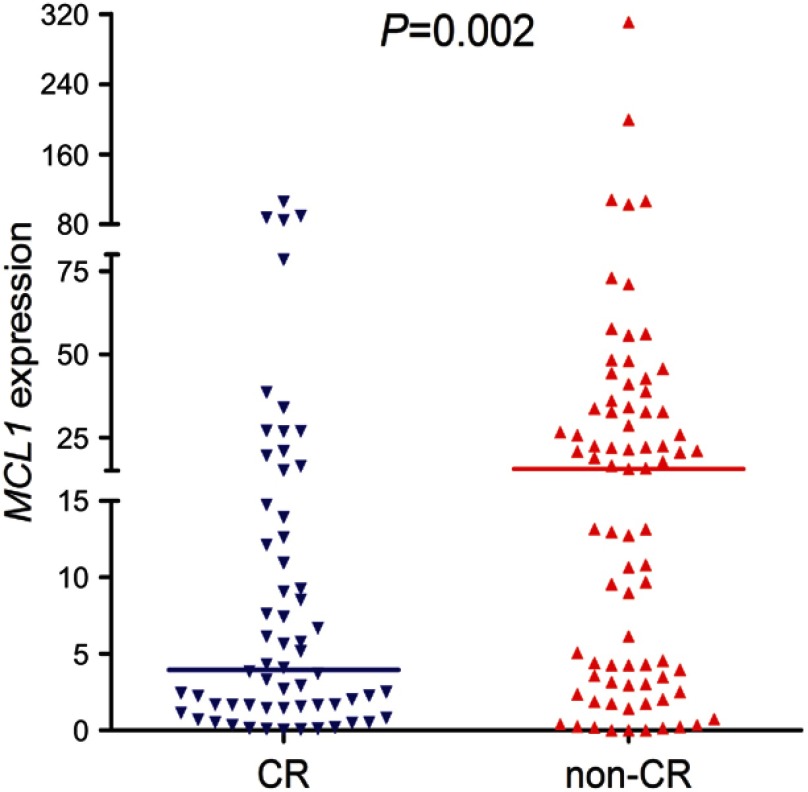
The follow-up data is available in 141 patients after induction chemotherapy. MCL-1high patients presented significantly lower CR rate than MCL-1low patients (33% vs 57%, P=0.007, Table 1). In addition, we further showed MCL-1 transcript level between the patients with and without CR (P=0.002, Figure 3). Apart from APL patients, MCL-1high cases also had markedly lower CR rate than MCL-1low cases (26% vs 47%, P=0.031, Table 1). There was also a significant difference in CR rate between the two groups in CN-AML (23% vs 60%, P=0.004, Table 1).
Figure 3.
The comparison of MCL-1 expression among AML patients with and without CR. MCL-1 expression among AML patients (n=62, range 0.0000–105.5293, median 3.9643) who could achieve CR was significantly lower than those who could not achieve CR (n=79, range 0.0000–311.0063, median 15.5237) after induction therapy.
Abbreviations: AML, acute myeloid leukemia; CR, complete remission.
Correlation between MCL-1 expression and clinical outcome
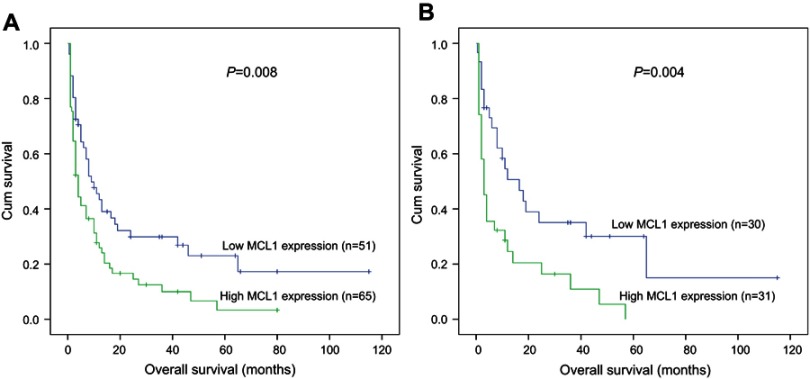
To investigate the prognostic significance of MCL-1 expression in AML, 141 AML patients with follow-up data (median 8 months, range 1–115 months) were included in survival analysis. Due to independent disease entity, APL was excluded from the analysis. Kaplan–Meier analyses revealed that there was a significant difference in OS time between MCL-1high and MCL-1low groups in non-APL patients (median 4 vs 9 months, respectively, P=0.008, Figure 4A). MCL-1high patients also had a shorter OS time than MCL-1low patients in CN-AML patients (median 3 vs 16.5 months, respectively, P=0.004, Figure 4B). In addition, multivariate analysis, including variables (age (>60/≤60y), WBC (≥30/<30*109/L), MCL-1 expression (high/low) and gene mutations) with P<0.200 in univariate analysis, demonstrated that increased expression of MCL-1 was an independent adverse prognostic factor for OS among both non-APL and CN-AML patients (Tables 2 and 3). However, there was no significant difference in LFS between two groups both in non-APL (P=0.157) and CN-AML patients (P=0.228).
Figure 4.
The impact of MCL-1 expression on prognosis of AML patietns. Overall survival between MCL-1high and MCL-1low group. (A) non-APL patients; (B) CN-AML patients.
Abbreviations: AML, acute myeloid leukemia; APL, acute promyelocytic leukemia; CN-AML, cytogenetically normal AML.
Table 2.
Univariate and multivariate analyses of prognostic factors for overall survival in non-APL patients
| Prognostic factors | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|
| Hazard ratio (95% CI) | P-value | Hazard ratio (95% CI) | P-value | |
| Age | 2.433 (1.599–3.702) | <0.001 | 2.088 (1.359–3.207) | 0.001 |
| WBC | 2.118 (1.405–3.193) | <0.001 | 1.000 (0.997–1.002) | 0.804 |
| Karyotype | 1.744 (1.404–2.165) | <0.001 | 1.751 (1.266–2.421) | 0.001 |
| MCL-1 expression | 1.710 (1.125–2.600) | 0.012 | 1.777 (1.164–2.714) | 0.008 |
| FLT3-ITD mutation | 1.403 (0.698–2.822) | 0.342 | - | - |
| NPM1 mutation | 1.484 (0.738–2.982) | 0.268 | - | - |
| CEBPA mutation | 0.818 (0.375–1.783) | 0.613 | - | - |
| c-KIT mutation | 0.613 (0.150–2.503) | 0.496 | - | - |
Notes: Multivariate analyses, including variables [age (>60/≤60 y), WBC (≥30/<30*109/L), MCL-1 expression (high/low), and gene mutations] with P<0.200 in univariate analysis.
Abbreviations: APL, acute promyelocytic leukemia; WBC, white blood cells.
Table 3.
Univariate and multivariate analyses of prognostic factors for overall survival in CN-AML patients
| Prognostic factors | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|
| Hazard ratio (95% CI) | P-value | Hazard ratio (95% CI) | P-value | |
| age | 2.437 (1.336–4.447) | 0.004 | 2.126 (1.155–3.914) | 0.015 |
| WBC | 2.348 (1.312–4.202) | 0.004 | 1.755 (0.958–3.215) | 0.069 |
| MCL1 expression | 2.235 (1.238–4.035) | 0.008 | 1.876 (1.013–3.473) | 0.045 |
| FLT3-ITD mutation | 0.941 (0.361–2,452) | 0.901 | - | - |
| NPM1 mutation | 1.311 (0.545–3.149) | 0.545 | - | - |
| CEBPA mutation | 1.055 (0.409–2.718) | 0.912 | - | - |
| c-KIT mutation | 0.631 (0.086–4.643) | 0.651 | - | - |
Notes: Multivariate analyses, including variables [age (>60/≤60 y), WBC (≥30/<30*109/L), MCL-1 expression (high/low), and gene mutations] with P<0.200 in univariate analysis.
Abbreviations: AML, acute myeloid leukemia; CN-AML: cytogenetically normal AML; WBC: white blood cells.
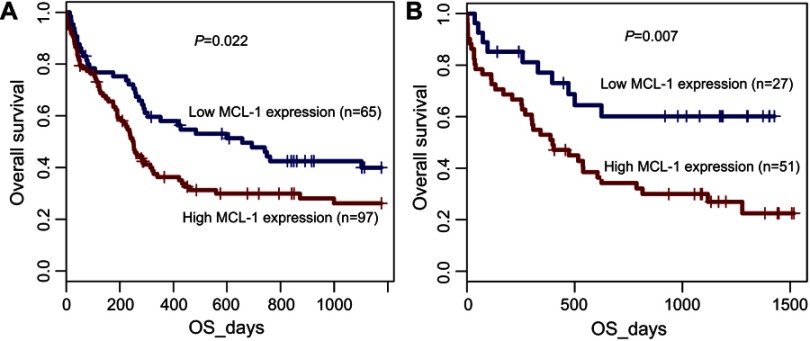
To further validate the prognostic impact of MCL-1 expression in CN-AML patients, we focused on the GEP data (accession number GSE12417) using the online web tool Genomicscape.21 MCL-1high cases showed shorter OS time than MCL-1low cases in two independent cohorts of CN-AML patients (P=0.022 and P=0.007, respectively, Figure 5).
Figure 5.
Prognostic value of MCL-1 expression validated by public database. Gene expression profiling (GEP) data (accession number GSE12417, http://www.ncbi.nlm.nih.gov/geo/) was used to validate the prognostic value of MCL-1 expression in two independent cohorts of 162 and 78 cytogenetically normal AML (CN-AML) patients by the online web tool Genomicscape (http://genomicscape.com/microarray/survival.php).
Surveillance of MCL-1 expression in follow-up AML patients
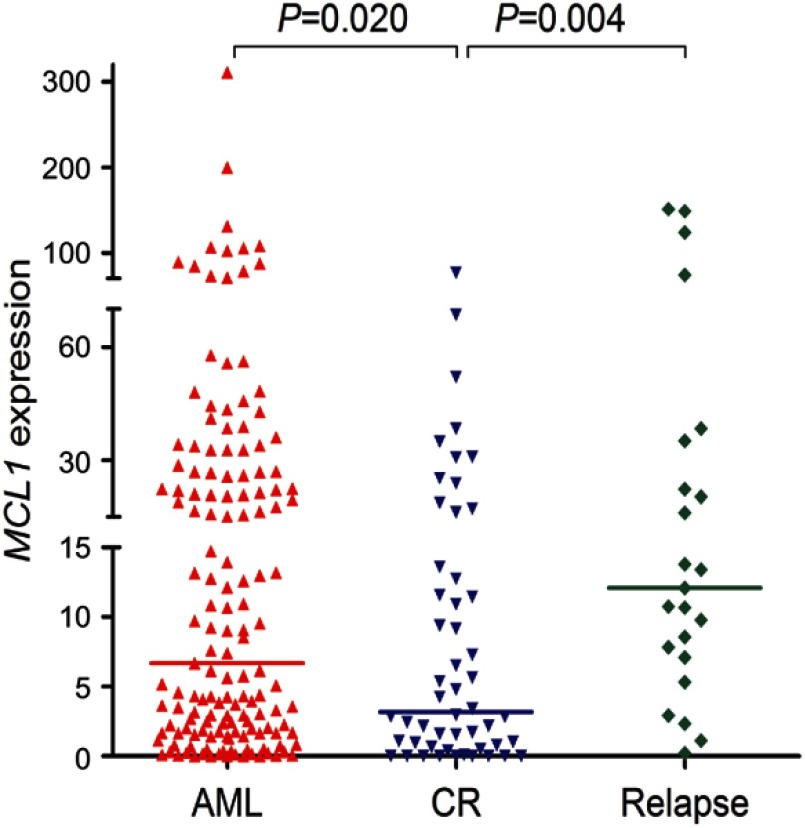
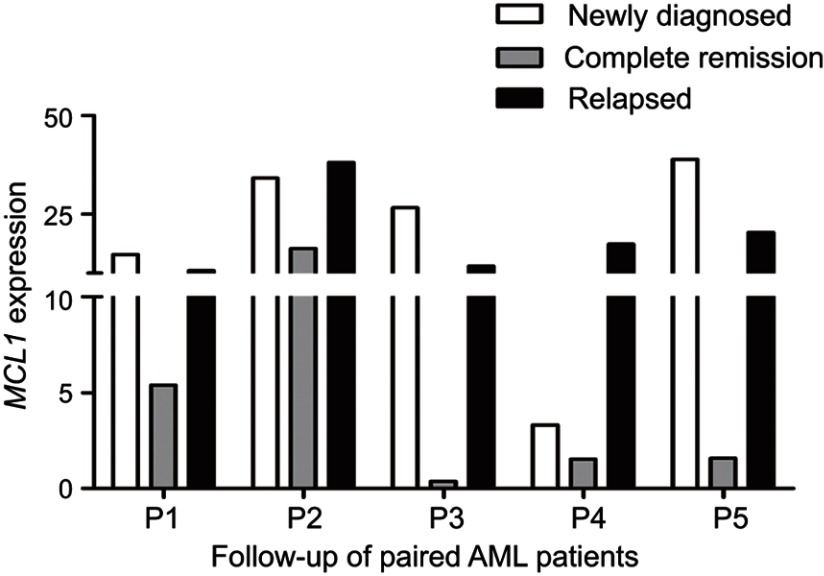
To investigate whether the expression of MCL-1 could monitor disease recurrence in AML, we assessed MCL-1 expression in 52 AML patients in CR and 23 cases in relapse. The median MCL-1 levels in patients at CR and relapse were 3.194 (range 0.000 to 76.368) and 12.076 (range 0.216–151.220), respectively. Obviously, MCL-1 expression was significantly decreased in CR time and was significantly increased in relapsed time compare with initial diagnosis time (P=0.020 and P=0.004, respectively, Figure 6). In addition, the dynamic change of MCL-1 expression in the five paired AML patients of different stages (Figure 7).
Figure 6.
MCL-1 expression in different clinical stages of AML. MCL-1 expression in 151 newly diagnosis, 52 AML patients achieving CR (range 0.000–76.368, median 3.194), and 23 relapsed (range 0.216–151.220, median 12.076) AML patients.
Abbreviations: AML, acute myeloid leukemia; CR, complete remission.
Figure 7.
Dynamic change of MCL-1 expression in the five paired AML patients of different clinical stages. White column: newly diagnosed AML patients; Gray column: AML patients who achieved CR; Black column: relapsed AML patients.
Abbreviations: AML, acute myeloid leukemia; CR, complete remission.
Discussion
Accumulating studies stated that MCL-1 expression was associated with the progression and outcome of various human malignant tumors.4–6 In multiple myeloma, MCL-1 high-expression was reported to be associated with relapse and shorter event-free survival.11 Mylin et al demonstrated that MCL-1 was up-regulated in multiple myeloma compared with monoclonal gammopathy of undetermined significance, but had no impact on OS and event-free survival.22 Moreover, the activated vascular endothelial growth factor induced the overexpression of MCL-1 and predicted a poor outcome and aggressive disease progression in non-Hodgkin’s lymphoma.23 More interestingly, Kempkensteffen et al elucidated that the antiapoptotic full-length splicing variant of MCL-1, down-regulation in clear-cell renal cancer, was related with poor tumor differentiation and predicted a higher risk for relapse; they also revealed that low MCL-1L was associated with short recurrence-free and disease-specific survival.24 Glaser et al used RNAi for knockdown of MCL-1 to determine that MCL-1 was essential for development and sustained growth of this hematological malignancy and it could be targeted for therapeutic benefit.13 In this study, we observed that up-regulation of MCL-1 expression was a frequent event in Chinese de novo AML patients. Survival analyses revealed that high MCL-1 expression was associated with shorter OS both in non-APL and in CN-AML patients. Notably, our data showed that high-expression of MCL-1 had lower CR rate. Meanwhile, expression of MCL-1 in CR and relapsed patients illustrated that MCL-1 were significantly decreased from the initial diagnosis to CR and increased after relapsed. These results together indicated that MCL-1 expression was a valuable predictor in assessing treatment outcome as well as status,and might serve as a criterion for the therapeutic evaluation in AML.
It has been well demonstrated that BCL-2 family proteins act as positive or negative regulators on cellular apoptosis and druggable targets.4–11,25 Kaufmann et al found that cells overexpressing MCL-1 were resistant to a variety of chemotherapeutic agents and raised the possibility that some chemotherapeutic regimens might select for leukemia cells with elevated levels of this particular apoptosis inhibitor.26 Recently, ABT-199, a highly potent and selective inhibitor of BCL-2, has been granted breakthrough designation by FDA for relapsed or refractory chronic lymphocytic leukemia with 17p deletion.27 Moreover, Pan et al found that ABT-199 also potently killed a diverse array of AML cell lines by establishing an aggressive mouse xenograft model of MOLM-13.27 Notably, some scholars stated that MCL-1 induced the resistance to ABT-737 (the first molecule studied extensively preclinically) and its analogs ABT-263 (the first of this series to enter the clinic) and ABT-199,28–30 while MCL-1 suppression could restore sensitivity to these compounds in leukemia cells,31,32 suggesting that drug resistance was attributed to high level of MCL-1. Kotschy et al found that the selective MCL-1 inhibitor S63845 killed MCL-1-dependent cancer cells by activating the BAX/BAK-dependent mitochondrial apoptotic pathway in diverse cancer models.14 Doi et al showed that maritoclax, a novel kind of BCL-2 family inhibitors, induced the selective degradation of MCL-1 through the proteasome to kill AML cells that express elevated levels of MCL-1.33 Recently, Wang et al revealed that inhibition of CDK9, a key component of the positive transcriptional regulator complex pTEFb which activates transcription via phosphorylation of RNA polymerase II, blocked transcription resulting in the repression of short-lived proteins such as MCL-1.34 Dey J et al found that voruciclib, a novel CDK9 inhibitor which addressed a well-characterized key-mediator of resistance to venetoclax by repressing MCL-1, and the BCL-2 specific inhibitor, venetoclax, with promise for treating high-risk subtype of diffuse large B-cell lymphoma by establishing specific models.35 Previous researches proved that higher levels of MCL-1 have been reported to result in the failure to achieve CR after fludarabine and chlorambucil treatment,36,37 while higher MCL-1/BAX ratio was related to poor response to rituximab in some patients.38 Taken together, these results suggested that MCL-1 combined with other targets have great potential in AML targeted therapies.
In addition, our study further observed that MCL-1 overexpression was associated with NPM1 mutation, which was a favorable prognostic indicator in AML and frequently occurred in patients with normal karyotype.39 In some studies, AML patients with NPM1 mutation appeared to benefit from ATRA as an adjunct to conventional chemotherapy.40,41 Moreover, it was reported that AML cells carrying NPM1 mutation were more sensitive to ATO, because of the expression of NPM1 mutant protein and its acquired C-terminus cysteine 288.42 Martelli et al also showed that ATO/ATRA induced proteasome-dependent degradation of NPM1 leukemic protein leading to cell growth inhibition and apoptosis in NPM1-mutated AML.43 Although a large number of findings suggest that the NPM1 mutation has a favorable effect on the outcome for AML.44–47 Falini et al found that NPM1 exon 12 mutations which caused cytoplasmic dislocation of NPM were related to the malignant clone and leukemogenesis.48 Moreover, a study conducted by Konoplev et al suggested that NPM1 mutations did not impact OS and event-free survival (EFS) in AML patients.49 However, Verhaak et al demonstrated that patients with intermediate cytogenetic risk AML with NPM1 mutations had a significantly better OS and EFS than those without NPM1 mutations.44 Further studies are needed to confirm and reveal the underlying association between MCL-1 expression and NPM1 mutation.
Collectively, these results demonstrated that MCL-1 is overexpressed and confers a poor prognosis in Chinese de novo AML patients and could be also used as a potential therapeutic target in the treatment of newly diagnosed and relapsed AML.
Acknowledgments
This work was supported by National Natural Science foundation of China (81270630), Medical Innovation Team of Jiangsu Province (CXTDB2017002), Six Talent Peaks Project in Jiangsu Province (2015-WSN-115), Zhenjiang Clinical Research Center of Hematology (SS2018009), Postgraduate Research & Practice Innovation Program of Jiangsu Province (KYCX17_1821, KYCX18_2281), Social Development Foundation of Zhenjiang (SH2016045, SH2017040, SH2018044), Clinical Medical Science Development Foundation of Jiangsu University (JLY20160011).
Abbreviation list
AML, acute myeloid leukemia; APL, acute promyelocytic leukemia; ATRA, all-trans retinoic acid; AUC, area under the ROC curve; BCL-2, B-cell leukemia/lymphoma-2; BCL-XL, BCL-2 and BCL-2-like protein 1 isoform 1; BM, Bone marrow; BMMNCs, BM mononuclear cells; CN-AML, cytogenetically normal AML; CR, complete remission; GEP, melting analysis; LFS, leukemia-free survival; MCL-1, myeloid cell leukemia-1; OS, overall survival; PLT, platelets; RARα, retinoic acid receptor α; ROC, Receiver operating characteristic curve; RQ-PCR, Real-time quantitative PCR; WBC, white blood cell; WHO, World Health Organization.
Ethics Statements
We confirm in the revised manuscript that a parent or legal guardian provided written informed consent for any patient under 18 years of age, and that this study was conducted in accordance with the Declaration of Helsinki.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Tenen DG. Disruption of differentiation in human cancer: AML shows the way. Nat Rev Cancer. 2003;3:89–101. doi: 10.1038/nrc989 [DOI] [PubMed] [Google Scholar]
- 2.Zhao J, Lu Q, Zhu J, Fu J, Chen Y-X. Prognostic value of miR-96 in patients with acute myeloid leukemia. Diagn Pathol. 2014;9:76. doi: 10.1186/1746-1596-9-76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grimwade D. The clinical significance of cytogenetic abnormalities in acute myeloid leukaemia. Best Pract Res Clin Haematol. 2001;14:497–529. doi: 10.1053/beha.2001.0152 [DOI] [PubMed] [Google Scholar]
- 4.Williams MM, Lee L, Hicks DJ, et al. Key survival factor, MCL-1, correlates with sensitivity to combined BCL-2/BCL-XL blockade. Mol Cancer Res. 2017;15(3):259–268. doi: 10.1158/1541-7786.MCR-16-0280-T [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goodwin CM, Rossanese OW, Olejniczak ET, Fesik SW. Myeloid cell leukemia-1 is an important apoptotic survival factor in triple-negative breast cancer. Cell Death Differ. 2015;22(12):2098–2106. doi: 10.1038/cdd.2015.73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Allen TD, Zhu CQ, Jones KD, Yanagawa N, Tsao M-S, Bishop JM. Interaction between MYC and MCL1 in the genesis and outcome of non-small-cell lung cancer. Cancer Res. 2011;71(6):2212–2221. doi: 10.1158/0008-5472.CAN-10-3590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dzhagalov I, St John A, He YW. The antiapoptotic protein MCL-1 is essential for the survival of neutrophils but not macrophages. Blood. 2007;109(4):1620–1626. doi: 10.1182/blood-2006-03-013771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peperzak V, Vikström I, Walker J, et al. Mcl-1 is essential for the survival of plasma cells. Nat Immunol. 2013;14(3):290–297. doi: 10.1038/ni.2527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pepper C, Lin TT, Pratt G, et al. Mcl-1 expression has in vitro and in vivo significance in chronic lymphocytic leukemia and is associated with other poor prognostic markers. Blood. 2008;112(9):3807–3817. doi: 10.1182/blood-2008-05-157131 [DOI] [PubMed] [Google Scholar]
- 10.Kozopas KM, Yang T, Buchan HL, Zhou P, Craig RW. MCL1, a gene expressed in programmed myeloid cell differentiation, has sequence similarity to BCL2. Proc Natl Acad Sci U S A. 1993;90(8):3516–3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wuillème-Toumi S, Robillard N, Gomez P, et al. MCL-1 is overexpressed in multiple myeloma and associated with relapse and shorter survival. Leukemia. 2005;19(7):1248–1252. doi: 10.1038/sj.leu.2403784 [DOI] [PubMed] [Google Scholar]
- 12.Mitchell C, Yacoub A, Hossein H, et al. Inhibition of MCL-1 in breast cancer cells promotes cell death in vitro and in vivo. Cancer Biol Ther. 2010;10:903–917. doi: 10.4161/cbt.10.9.13273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glaser SP, Lee EF, Trounson E, et al. Anti-apoptotic MCL-1 is essential for the development and sustained growth of acute myeloid leukemia. Genes Dev. 2012;26(2):120–125. doi: 10.1101/gad.182980.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kotschy A, Szlavik Z, Murray J, et al. The MCL1 inhibitor S63845 is tolerable and effective in diverse cancer models. Nature. 2016;538(7626):477–482. doi: 10.1038/nature19830 [DOI] [PubMed] [Google Scholar]
- 15.Swerdlow SH, Campo E, Harris NL, et al. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Lyon: IARC Press; 2008. [Google Scholar]
- 16.Zhou JD, Zhang TJ, Li XX, et al. Epigenetic dysregulation of ID4 predicts disease progression and treatment outcome in myeloid malignancies. J Cell Mol Med. 2017;21(8):1468–1481. doi: 10.1111/jcmm.13073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou JD, Wang YX, Zhang TJ, et al. Identification and validation of SRY-box containing gene family member SOX30 methylation as a prognostic and predictive biomarker in myeloid malignancies. Clin Epigenetics. 2018;10:92. doi: 10.1186/s13148-018-0523-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang TJ, Zhou JD, Zhang W, et al. H19 overexpression promotes leukemogenesis and predicts unfavorable prognosis in acute myeloid leukemia. Clin Epigenetics. 2018;10:47. doi: 10.1186/s13148-018-0486-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin J, Yang J, Wen XM, et al. Detection of SRSF2-P95 mutation by high-resolution melting curve analysis and its effect on prognosis in myelodysplastic syndrome. PLoS One. 2014;9:e115693. doi: 10.1371/journal.pone.0115693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wen XM, Hu JB, Yang J, et al. CEBPA methylation and mutation in myelodysplastic syndrome. Med Oncol. 2015;32:192. doi: 10.1007/s12032-015-0605-z [DOI] [PubMed] [Google Scholar]
- 21.Kassambara A, Rème T, Jourdan M, et al. GenomicScape: an easy-to-use web tool for gene expression data analysis. Application to investigate the molecular events in the differentiation of B cells into plasma cells. PLoS Comput Biol. 2015;11(1):e1004077. doi: 10.1371/journal.pcbi.1004077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mylin AK, Rasmussen T, Lodahl M, Dahl IM, Knudsen LM. Upregulated MCL1 mRNA expression in multiple myeloma lacks association with survival. Br J Haematol. 2009;144(6):961–963. doi: 10.1111/j.1365-2141.2008.07521.x [DOI] [PubMed] [Google Scholar]
- 23.Kuramoto K, Sakai A, Shigemasa K, et al. High expression of MCL1 gene related to vascular endothelial growth factor is associated with poor outcome in non-Hodgkin‘s lymphoma. Br J Haematol. 2002;116(1):158–161. [DOI] [PubMed] [Google Scholar]
- 24.Kempkensteffen C, Hinz S, Johannsen M, et al. Expression of MCL-1 splicing variants in clear-cell renal cancer and their correlation with histopathological parameters and prognosis. Tumour Biol. 2009;30(2):73–79. doi: 10.1159/000215826 [DOI] [PubMed] [Google Scholar]
- 25.Anderson MA, Huang D, Roberts A. Targeting BCL2 for the treatment of lymphoid malignancies. Semin Hematol. 2014;51:219–227. doi: 10.1053/j.seminhematol.2014.05.008 [DOI] [PubMed] [Google Scholar]
- 26.Kaufmann SH, Karp JE, Svingen PA, et al. Elevated expression of the apoptotic regulator MCL-1 at the time of leukemic relapse. Blood. 1998;91(3):991–1000. [PubMed] [Google Scholar]
- 27.Pan R, Hogdal LJ, Benito JM, et al. Selective BCL-2 inhibition by ABT-199 causes on-target cell death in acute myeloid leukemia. Cancer Discov. 2014;4:362–375. doi: 10.1158/2159-8290.CD-13-0609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oltersdorf T, Elmore SW, Shoemaker AR, et al. An inhibitor of BCL-2 family proteins induces regression of solid tumours. Nature. 2005;435:677–681. doi: 10.1038/nature03579 [DOI] [PubMed] [Google Scholar]
- 29.Gandhi L, Camidge DR, Ribeiro de Oliveira M, et al. Phase I study of Navitoclax (ABT-263), a novel BCL-2 family inhibitor, in patients with small-cell lung cancer and other solid tumors. J Clin Oncol. 2011;29:909–916. doi: 10.1200/JCO.2010.31.6208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Souers AJ, Leverson JD, Boghaert ER, et al. ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nat Med. 2013;19:202–208. doi: 10.1038/nm.3048 [DOI] [PubMed] [Google Scholar]
- 31.Yecies D, Carlson NE, Deng J, Letai A. Acquired resistance to ABT-737 in lymphoma cells that up-regulate MCL-1 and BFL-1. Blood. 2010;115:3304–3313. doi: 10.1182/blood-2009-07-233304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mazumder S, Choudhary GS, Al-Harbi S, Almasan A. MCL-1 phosphorylation defines ABT-737 resistance that can be overcome by increased NOXA expression in leukemic B cells. Cancer Research. 2012;72:3069–3079. doi: 10.1158/0008-5472.CAN-11-4106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Doi K, Liu Q, Gowda K, et al. Maritoclax induces apoptosis in acute myeloid leukemia cells with elevated MCL-1 expression. Cancer Biol Ther. 2014;15(8):1077–1086. doi: 10.4161/cbt.29186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang S, Fischer PM. Cyclin-dependent kinase 9: a key transcriptional regulator and potential drug target in oncology,virology and cardiology. Trends Pharmacol Sci. 2008;29(6):302–313. doi: 10.1016/j.tips.2008.03.003 [DOI] [PubMed] [Google Scholar]
- 35.Dey J, Deckwerth TL, Kerwin WS, et al. Voruciclib, a clinical stage oral CDK9 inhibitor, represses MCL-1 and sensitizes high-risk diffuse large B-cell lymphoma to BCL2 inhibition. Sci Rep. 2017;7(1):18007. doi: 10.1038/s41598-017-18368-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kitada S, Andersen J, Akar S, et al. Expression of apoptosis-regulating proteins in chronic lymphocytic leukemia: correlations with in vitro and in vivo chemoresponses. Blood. 1998;91:3379–3389. [PubMed] [Google Scholar]
- 37.Saxena A, Viswanathan S, Moshynska O, Tandon P, Sankaran K, Sheridan DP. MCL-1 and BCL-2/BAX ratio are associated with treatment response but not with Rai stage in B-cell chronic lymphocytic leukemia. Am J Hematol. 2004;75:22–33. doi: 10.1002/ajh.10453 [DOI] [PubMed] [Google Scholar]
- 38.Bannerji R, Kitada S, Flinn IW, et al. Apoptoticregulatory and complement-protecting protein expression in chronic lymphocytic leukemia: relationship to in vivo rituximab resistance. J Clin Oncol. 2003;21:1466–1471. doi: 10.1200/JCO.2003.06.012 [DOI] [PubMed] [Google Scholar]
- 39.Suzuki T, Kiyoi H, Ozeki K, et al. Clinical characteristics and prognostic implications of NPM1 mutations in acute myeloid leukemia. Blood. 2005;106:2854–2861. doi: 10.1182/blood-2005-04-1733 [DOI] [PubMed] [Google Scholar]
- 40.Schlenk RF, Döhner K, Kneba M, et al; German-Austrian AML Study Group (AMLSG). Gene mutations and response to treatment with all-trans retinoic acid in elderly patients with acute myeloid leukemia. Results from the AMLSG Trial AML HD98B. Haematologica. 2009;94(1):54–60. doi: 10.1182/blood-2006-03-013771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schlenk RF, Döhner K. Impact of new prognostic markers in treatment decisions in acute myeloid leukemia. Curr Opin Hematol. 2009;16(2):98–104. doi: 10.1097/MOH.0b013e3283257adb [DOI] [PubMed] [Google Scholar]
- 42.Huang M, Thomas D, Li MX, et al. Role of cysteine 288 in nucleophosmin cytoplasmic mutations: sensitization to toxicity induced by arsenic trioxide and bortezomib. Leukemia. 2013;27(10):1970–1980. doi: 10.1038/leu.2013.222 [DOI] [PubMed] [Google Scholar]
- 43.Martelli MP, Gionfriddo I, Mezzasoma F, et al. Arsenic trioxide and all-trans retinoic acid target NPM1 mutant oncoprotein levels and induce apoptosis in NPM1-mutated AML cells. Blood. 2015;125(22):3455–3465. doi: 10.1182/blood-2014-11-611459 [DOI] [PubMed] [Google Scholar]
- 44.Verhaak RG, Goudswaard CS, van Putten W, et al. Mutations in nucleophosmin (NPM1) in acute myeloid leukemia (AML): association with other gene abnormalities and previously established gene expression signatures and their favorable prognostic significance. Blood. 2005;106(12):3747–3754. doi: 10.1182/blood-2005-05-2168 [DOI] [PubMed] [Google Scholar]
- 45.Kiyoi H, Naoe T, Nakano Y, et al. Prognostic implication of FLT3 and N-RAS gene mutations in acute myeloid leukemia. Blood. 1999;93(9):3074–3080. [PubMed] [Google Scholar]
- 46.Thiede C, Steudel C, Mohr B, et al. Analysis of FLT3-activating mutations in 979 patients with acute myelogenous leukemia: association with FAB subtypes and identification of subgroups with poor prognosis. Blood. 2002;99(12):4326–4335. [DOI] [PubMed] [Google Scholar]
- 47.Schnittger S, Schoch C, Dugas M, et al. Analysis of FLT3 length mutations in 1003 patients with acute myeloid leukemia: correlation to cytogenetics, FAB subtype, and prognosis in the AMLCG study and usefulness as a marker for the detection of minimal residual disease. Blood. 2002;100(1):59–66. [DOI] [PubMed] [Google Scholar]
- 48.Falini B, Mecucci C, Tiacci E, et al. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N Engl J Med. 2005;352(3):254–266. doi: 10.1056/NEJMoa041974 [DOI] [PubMed] [Google Scholar]
- 49.Konoplev S, Huang X, Drabkin HA, et al. Cytoplasmic localization of nucleophosmin in bone marrow blasts of acute myeloid leukemia patients is not completely concordant with NPM1 mutation and is not predictive of prognosis. Cancer. 2009;115:4737–4744. doi: 10.1002/cncr.24543 [DOI] [PMC free article] [PubMed] [Google Scholar]