Abstract
Tetrahydrofolate coenzymes involved in one-carbon (C1) metabolism are polyglutamylated. In organisms that synthesize tetrahydrofolate de novo, dihydrofolate synthetase (DHFS) and folylpolyglutamate synthetase (FPGS) catalyze the attachment of glutamate residues to the folate molecule. In this study we isolated cDNAs coding a DHFS and three isoforms of FPGS from Arabidopsis thaliana. The function of each enzyme was demonstrated by complementation of yeast mutants deficient in DHFS or FPGS activity, and by measuring in vitro glutamate incorporation into dihydrofolate or tetrahydrofolate. DHFS is present exclusively in the mitochondria, making this compartment the sole site of synthesis of dihydrofolate in the plant cell. In contrast, FPGS is present as distinct isoforms in the mitochondria, the cytosol, and the chloroplast. Each isoform is encoded by a separate gene, a situation that is unique among eukaryotes. The compartmentation of FPGS isoforms is in agreement with the predominance of γ-glutamyl-conjugated tetrahydrofolate derivatives and the presence of serine hydroxymethyltransferase and C1-tetrahydrofolate interconverting enzymes in the cytosol, the mitochondria, and the plastids. Thus, the combination of FPGS with these folate-mediated reactions can supply each compartment with the polyglutamylated folate coenzymes required for the reactions of C1 metabolism. Also, the multicompartmentation of FPGS in the plant cell suggests that the transported forms of folate are unconjugated.
In all living cells, tetrahydrofolate (vitamin B9) is involved in one-carbon (C1) transfer reactions. It is an absolute requirement for the synthesis of purine, thymidylate, formylmethionyl-tRNA, pantothenate, and methionine, and for the interconversion of glycine and serine. The tetrahydrofolate molecule [5,6,7,8-tetrahydropteroylpoly-γ-glutamate (H4F-Glun)] capable of carrying and donating C1 units consists chemically of a pteridine ring attached to a para aminobenzoic acid (p-ABA) moiety, which in turn is attached to glutamate residues. Because the affinity of folate-dependent enzymes increases markedly with the number of glutamate residues, the folylpolyglutamates are the preferred coenzymes of C1 metabolism (reviewed in refs. 1–3). The conjugated forms of the vitamin are also involved in the retention of folate within the cell and its subcellular compartments (4). Plant folates vary in their levels of glutamyl conjugation, the pentaglutamate species being the most abundant in pea cotyledons and leaves (5). H4F-Glun is present in various compartments of the plant cell. The cytosol contains the largest pool of folates, predominantly in the form of polyglutamylated methyl derivatives. In pea leaves, mitochondria and chloroplasts also contain principally (>90%) polyglutamylated forms (5, 6). The distribution of folates among the organelles is unequal because the estimated concentration of folates in mitochondria is 100–150-fold higher than in chloroplasts (7, 8).
In the complex pathway leading to H4F-Glun synthesis, two reactions mediate the attachment of glutamate. The first reaction, catalyzed by dihydrofolate synthetase (DHFS, EC 6.3.2.12), attaches the first glutamate residue to the carboxyl moiety of para aminobenzoic acid (p-ABA) to form dihydrofolate. The enzyme is only present in organisms having the ability to synthesize H4F de novo from the 6-hydroxymethyldihydropterin, p-ABA, and glutamate precursors (e.g., plants, most bacteria, fungi, and protozoa). In all organisms, including those requiring exogenous folate for growth, the polyglutamate tail of H4F-Glun is formed by the sequential addition of γ-linked glutamate residues to H4F-Glu1, a reaction catalyzed by folylpolyglutamate synthetase (FPGS, EC 6.3.2.17). The crucial role of the glutamylation steps in H4F-Glun synthesis has been demonstrated by the characterization of mutants affected in the DHFS or FPGS activities. Thus, disruption of the FOL3 gene coding DHFS in Saccharomyces cerevisiae leads to spores requiring a stable H4F derivative for growth (9). Mutations affecting FPGS result in folylpolyglutamate deficiencies and auxotrophies for the products of C1 metabolism. Such mutants from S. cerevisiae and Neurospora crassa display methionine auxotrophy (2, 9, 10), whereas the Chinese hamster ovary AUXB1 cell line lacking FPGS activity is auxotroph for purine, thymidine, and glycine (11).
In bacteria that synthesize H4F-Glun de novo (Escherichia coli, Corynebacterium), the product of the folC gene possesses both DHFS and FPGS activities (12). Yet, the only example of an eukaryote gene encoding both DHFS and FPGS activities is from the human malaria parasite Plasmodium falciparum (13). In other eukaryotes synthesizing folate de novo (plants, fungi), distinct proteins catalyze the addition of glutamate residues to dihydropteroate or H4F. In fungi, it has been suggested that DHFS is a cytosolic enzyme and that FPGS isoforms are present in the cytosol and the mitochondria (10, 14). Analysis of the subcellular distribution of folate-synthesizing enzymes in pea leaves established that mitochondria play a major role in H4F-Glun synthesis (7). Other studies supported the mitochondrial localization of DHFS (15) and suggested the presence of FPGS in the cytosol (16). Organisms that lack DHFS activity and require exogenous folate for growth (animals and bacteria such as Lactobacillus) possess a monofunctional FPGS. In mammalian tissues, the enzyme is present in the cytosol and the mitochondria (4). Both forms of FPGS derive from the same gene by the use of two different translation initiation codons, the first AUG generating the mitochondrial enzyme (17).
During the last decade, our knowledge concerning H4F biosynthesis in plants has improved considerably (for reviews, see refs. 2 and 3). However, the existence of FPGS isoforms, their subcellular distribution, and their relative contribution to H4F biosynthesis have not been yet elucidated. In this study we describe the cloning and functional characterization of cDNAs coding the enzymes responsible for the glutamylation of dihydropteroate and H4F in Arabidopsis thaliana. DHFS is a mitochondrial protein, whereas the three FPGS isoforms characterized are respectively located in the cytosol, the mitochondria, and the chloroplasts.
Materials and Methods
Plant Materials.
A. thaliana (ecotype Wassilewskija) seeds were grown on soil with a light intensity of 100 μE m−2 s−1, a day length of 14 h, and a day/night temperature of 25/20°C. A. thaliana (ecotype Columbia) cell suspension cultures were grown under continuous white light (40 μE m−2 s−1) at 23°C with rotary agitation at 125 rpm in Gamborg's B5 medium supplemented with 1 μM 2-naphtalene acetic acid and 1.5% (wt/vol) sucrose.
cDNA Cloning.
Poly(A)+ mRNA was isolated from above-ground parts of 3-week-old A. thaliana plants and used to construct a Marathon cDNA amplification library according to the manufacturer's instructions (CLONTECH). Rapid amplification of cDNA ends (RACE) PCR was conducted with adapter-specific primers and sets of antisense (5′-RACE) and sense (3′-RACE) gene-specific primers designed to generate fragments overlapping by ≈300 bp. Amplification was done with the Advantage cDNA polymerase mix according to the manufacturer's instructions (CLONTECH). PCR products were subcloned into the pGEM T-Easy vector (Promega) and sequenced by using the ABI Prism dye terminator system (GenomeExpress, Meylan, France). Sequencing confirmed that the isolated sequences encode DHFS-FPGS homologs in Arabidopsis. Full-length cDNAs were then generated by PCR, cloned in the pGEM vector, and sequenced in both strands. The cloned sequences were designated AtDFA, -B, -C, and -D for A. thaliana DHFS-FPGS homologs.
Functional Complementation in Yeast.
The S. cerevisiae strains used in this study were CC788–2B (MATα, his3, leu2, ura3, and trp1), CD180 (MATa/α, his3/his3, leu2/leu2, ura3/ura3, trp1/trp1, and MET7/met7∷TRP1), CD180–5C (MATα, his3, leu2, ura3, trp1, and met7∷TRP1), and CD208–2B (MATα, his3, leu2, ura3, trp1, and fol3∷HIS3) (9). Growth media and genetic manipulations were as described by Cherest et al. (9). Transformation of yeast strains disrupted for the genes coding FPGS (MET7) or DHFS (FOL3) was performed by using the shuttle vector pRS316-proADH1 that contains the URA3 gene for selection and the ADH1 promoter to drive expression of the Arabidopsis cDNAs (18). For expression in yeast, the coding sequences of AtDFA, AtDFB, and AtDFC devoid of the predicted transit peptide presequences were amplified by PCR and cloned in pRS316-proADH1. Thus, the AtDFA and -B proteins started at Met-20 and Met-59, respectively. To express the recombinant AtDFC protein, a methionine codon was placed before Leu-51. The entire coding sequence of AtDFD was amplified by PCR and cloned into pRS316-proADH1. The sequence of each construct was verified before transformation.
Assays of DHFS and FPGS Activities.
Yeast strains were grown in minimal medium complemented to meet their auxotrophic requirements, harvested by centrifugation (5,000 × g, for 5 min) during exponential growth phase, and washed in 50 mM Tris⋅HCl, pH 8.0/10 mM 2-mercaptoethanol (2-ME). Cells were suspended in 50 mM Tris⋅HCl (pH 8.0), 10 mM 2-ME, 5% (vol/vol) glycerol, 1 mM PMSF, and a mixture of protease inhibitors (no. 1873580, Roche Molecular Biochemicals), and broken by passage through a French press at 10,000 psi (1 psi = 6.89 kPa). Cell lysates were centrifuged at 16,000 × g for 15 min at 4°C, and the supernatants were used for enzymatic assays. Proteins were measured by the method of Bradford (19) with BSA as standard.
DHFS and FPGS activities were measured by the incorporation of [3H]glutamate into dihydrofolate and H4F-Glun, respectively. The DHFS assay medium (100–200 μl) contained 0.1 M Tris⋅HCl (pH 8.0), 50 mM 2-mercaptoethanol, 20 mM KCl, 5 mM ATP, 10 mM MgCl2, 3 mM [3H]glutamate (2 μCi, Amersham Pharmacia), and various amounts of protein extracts. The reaction was started by the addition of 100 μM dihydropteroate and proceeded for 20–60 min at 37°C. The [3H]dihydrofolate formed was separated from unreacted [3H]glutamate by ion exchange chromatography and counted (7). The FPGS activity was determined by using a similar procedure except that the reaction was initiated with 100 μM H4F-Glu1 instead of dihydropteroate. According to Schirch (20), 10-CHO-H4F was synthesized from 5-CHO-H4F, and 5,10-CH2-H4F was generated in situ by incubating 0.1 mM H4F and 1 mM formaldehyde for 15 min at 37°C before the addition of other reaction components. The standard assay conditions for FPGS (incubation time and saturating H4F-Glu1 concentration) favored the formation of folyldiglutamates (5).
Preparation of a Cytosolic-Enriched Fraction.
Protoplasts were prepared by enzymatic digestion of 6-day-old cell suspension cultures, using the procedure described in ref. 21. Protoplasts were gently ruptured by passing through a 20-μm nylon mesh and subsequently through a 10-μm nylon mesh. The protoplast lysate was centrifuged successively at 100 × g for 5 min, 900 × g for 5 min, and 13,000 × g for 20 min. The 13,000 × g supernatant fraction was centrifuged further at 100,000 × g for 1 h to remove membranes. Pellets were pooled and used, together with the 13,000 × g supernatant fraction, to measure marker enzyme activities (7). Most of the chloroplast (83 ± 5%, mean of three independent experiments ± SE) and mitochondria (92 ± 2%) marker activities were recovered in the pooled pellets, whereas the 13,000 × g supernatant fraction contained 67 ± 3% of the cytosolic marker activity. This cytosolic-enriched fraction contained a small proportion of mitochondrial (8 ± 2%) and chloroplastic (17 ± 5%) marker enzymes.
Purification of Chloroplasts.
Chloroplasts were purified from 3-week-old A. thaliana leaves as described in ref. 22. Intact chloroplasts were purified on Percoll gradients and lysed in 10 mM Mops (pH 7.6), 4 mM MgCl2, 1 mM PMSF, and 1 mM aminocaproic acid. Chloroplast subfractions were separated on a step gradient of 0.93–0.6 M sucrose in 10 mM Mops (pH 7.6) by centrifugation at 70,000 × g for 1 h. The soluble fraction (stroma) was collected at the top of the 0.6 M sucrose layer.
Purification of Mitochondria.
Mitochondria were prepared from 6-day-old Arabidopsis cell suspension cultures as described by Douce et al. (23). Crude mitochondria were layered on discontinuous gradients consisting of 18, 23, and 40% Percoll layers in 0.3 M sucrose, 10 mM potassium phosphate (pH 7.2), 1 mM EDTA, and 0.1% BSA. Gradients were centrifuged at 39,000 × g for 40 min, and mitochondria were collected at the 23/40% Percoll interface. To obtain soluble proteins (matrix), purified mitochondria were lysed by a 10-fold dilution in 10 mM Mops (pH 7.2)/1 mM DTT and submitted to 3 freeze-thaw cycles. Membranes were removed by centrifugation at 100,000 × g for 1 h.
Ab Production and Immunoblot Analysis.
To prepare specific Abs, the AtDF proteins were produced in E. coli by using pET15b or pET20b(+) vectors (Novagen). For the AtDFA and AtDFD proteins, the complete ORF was cloned in the expression vectors whereas AtDFB and AtDFC were truncated at their N-termini (58 and 86 residues, respectively) for cloning convenience. Constructs were introduced into BL21(DE3)pLysS cells (Novagen) and protein expression was conducted for 3 h at 37°C in Luria–Bertani medium containing 0.4 or 1 mM isopropyl β-d-thiogalactoside for pET20b(+) or pET15b constructs, respectively. Most of the expressed proteins were recovered in the inclusion body fractions. Recombinant proteins were purified further by SDS/PAGE (24) and injected into New Zealand rabbits to raise Abs (Centre Valbex, Institut Universitaire de Technologie de Biologie, Villeurbanne, France). Proteins from Arabidopsis cell subfractions were resolved by SDS/PAGE and electroblotted to nitrocellulose membrane. The blots were probed by using the affinity-purified Abs, horseradish peroxidase-conjugated goat anti-rabbit IgGs, and detection was achieved by chemiluminescence.
Transient Expression of Green Fluorescent Protein (GFP) Fusion Proteins in Arabidopsis Protoplasts.
A pUC18 plasmid expressing the modified version mGFP4 of the GFP under the control of the cauliflower mosaic virus 35S promoter was used for transient expression experiments in Arabidopsis protoplasts (25). The N-terminal regions of the AtDFA, -B, -C, and D proteins fused upstream and in-frame with mGFP4 contained 82, 100, 119, and 46 residues, respectively. The transit peptide sequences of the small subunit of Rubisco from Arabidopsis (55 residues, ats1A gene, GenBank accession no. X13611) and dihydropterin pyrophosphokinase-dihydropteroate synthase (HPPK-DHPS) from pea (28 residues; ref. 26) were used as controls for the targeting of GFP to plastids and mitochondria, respectively. Transient transformation of Arabidopsis protoplasts prepared from a 6-day-old cell suspension culture was achieved with the procedure described by Abel and Theologis (27). Just before analysis, protoplasts were incubated for 5 min at room temperature with the dye MitoTracker Orange (CM-H2TMRos, Molecular Probes). Samples were analyzed by confocal laser scanning microscopy, using a Leica TCS-SP2 operating system. GFP, MitoTracker, and chlorophyll fluorescence were excited and collected sequentially (400 Hz line by line) by using 488 nm for GFP, 543 nm for MitoTracker, and 633 nm for chlorophyll excitation. Fluorescence emission was collected from 500 to 535 nm for GFP, from 553 to 600 nm for MitoTracker, and from 643 to 720 nm for chlorophyll.
Results
Isolation of DHFS-FPGS Homologs in Arabidopsis.
In all organisms studied so far, enzymes displaying DHFS and/or FPGS activities share highly conserved motives designated FPGS signatures (PROSITE PS01011 and PS01012). By using blast analysis, we searched the Arabidopsis genome database for sequences similar to these signatures. Four DHFS-FPGS-related sequences were retrieved. Sets of primers were designed to amplify the corresponding cDNAs by 5′- and 3′-rapid amplification of cDNA ends, using poly(A)+ mRNA isolated from above-ground organs of 3-week-old Arabidopsis plants. The isolated clones displayed significant homology with known DHFS-FPGS sequences and were designated AtDFA, -B, -C, and -D for A. thaliana DHFS-FPGS homologs. The main properties of the cDNAs are summarized in Table 2, which is published as supporting information on the PNAS web site, www.pnas.org.
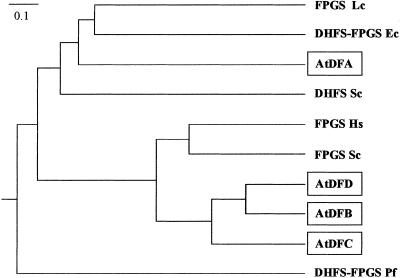
Comparison of the entire amino acid sequences deduced from the isolated cDNAs allowed us to classify the AtDF proteins into two classes (Fig. 1). AtDFB, -C, and -D share 41–47% identity (58–69% similarity) and form the first subclass. The AtDFA protein seemed to be distinct from this group because identical residues accounted for only 22% (42% similarities). Furthermore, alignment of the Arabidopsis proteins (Fig. 6, which is published as supporting information on the PNAS web site) led us to distinguish three domains. The first one located at the N terminus contained 30–70 residues for AtDFA, -B, and -C but was absent in AtDFD. Features typical for mitochondrial or chloroplastic transit peptides were found in the N-terminal extensions of AtDFA, -B, and -C (Table 2), thus suggesting that these proteins are targeted to the organelles. A similar analysis performed with AtDFD revealed no signal sequence, thus indicating that the protein is most probably cytosolic. The second region started after the N-terminal domain and extended over ≈220 residues. It is characterized by the highest similarity score among the four sequences (28% identical plus 14% similar residues) and contained a number of residues that are highly conserved in known FPGS sequences (FPGS signatures; Fig. 6). In particular, the GTKGKGS motif (P loop) and subsequent blocks are involved in the binding of ATP and Mg2+ (29). Except for the last 40 residues, the C-terminal part of the AtDF proteins was less conserved. However, several blocks of highly conserved residues are evident, in particular when comparing AtDFB, -C, and -D. Structural analysis of FPGS from Lactobacillus casei indicated that the C-terminal domain of the protein is the site of H4F binding (29). The low percentage of identity observed in this region between AtDFA on one hand and AtDFB, -C, and -D on the other hand may therefore reflect from the utilization of different substrates, namely dihydropteroate for DHFS vs. H4F for FPGS, by these proteins.
Figure 1.
Phylogenetic analysis of the Arabidopsis DHFS-FPGS proteins. The phenogram was produced from a CLUSTAL W alignment (28) by using the KITSCH program of the Phylip package available at http://www.infobiogen.fr (bootstrap analysis with 415 trees examined). Bifunctional DHFS-FPGS from E. coli (Ec, GenBank accession no. M32445), P. falciparum (Pf, AF161264), monofunctional FPGS from S. cerevisiae (Sc, NC001147), Homo sapiens (Hs, M98045), L. casei (Lc, J05221), and monofunctional DHFS from S. cerevisiae (Sc, NC001145) were included in the alignment.
A phylogenetic analysis indicated that AtDFB, -C, and -D are closely related to monofunctional FPGS from eukaryotes (Fig. 1). AtDFA belonged to another division of the tree that includes FPGS from L. casei, DHFS-FPGS from E. coli, and DHFS from S. cerevisiae. Thus, AtDFB, -C, and -D are probably FPGS, but the situation is less obvious for AtDFA. It was not possible from phylogenetic analysis to decipher whether this protein fit in the class of (i) eukaryotic DHFS or (ii) FPGS and DHFS-FPGS from prokaryotes.
Functional Analysis of Arabidopsis DHFS-FPGS Proteins in Yeast.
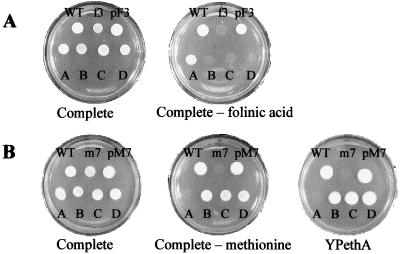
To test the function of the Arabidopsis DHFS-FPGS proteins, the yeast strains CD180 and CD208–2B disrupted for the MET7 and FOL3 genes, respectively, were used (9). Yeasts were selected as host cells for functional complementation because they possess, as do plants, monofunctional DHFS (Fol3 protein) and FPGS (Met7 protein) enzymes. The haploid strain CD208–2B bearing the fol3Δ allele grows if the medium is supplemented with 5-CHO-H4F, a stable H4F derivative. Transformation of CD208–2B (fol3Δ) with the shuttle vector containing AtDFA allowed yeast growth on medium lacking 5-CHO-H4F (Fig. 2A), providing the first evidence that the AtDFA cDNA encodes a functional DHFS protein. Expression of AtDFB, -C, or -D did not correct the 5-CHO-H4F requirement (Fig. 2A), thus indicating that these proteins have no DHFS activity.
Figure 2.
Complementation of the fol3Δ and met7Δ yeast strains by the Arabidopsis DHFS-FPGS homologs. (A) Growth of strains CC788–2B (WT), CD208–2B (fol3Δ; f3), and CD208–2B transformed with pF3 (harboring the wild type FOL3 gene) or with the pRS-AtDF vectors (A–D) on complete synthetic medium and complete synthetic medium lacking 5-CHO-H4F. (B) Growth of strains CC788–2B (WT), CD180–5C (met7Δ; m7) and the Ura+/Trp+ segregants resulting from the sporulation of CD180 (met7/MET7) diploid cells transformed with pM7 (harboring the wild type MET7 gene) or with the pRS-AtDF vectors (A–D) on complete synthetic medium, complete synthetic medium lacking methionine, and on ethanol-containing YPethA medium (9).
In yeast, FPGS is essential for methionine biosynthesis and for maintenance of mitochondrial DNA (9). Indeed, cells bearing the met7Δ mutation were methionine prototrophs and “petites,” i.e., the cells were respiration-deficient and unable to use ethanol as a carbon source. The diploid CD180 (met7Δ/MET7) strain was transformed with the pRS-AtDF plasmids, and the progeny was analyzed on a medium lacking methionine (glucose as a carbon source) and on medium containing ethanol as a carbon source. Spores disrupted for the MET7 gene (Trp+, met7∷TRP1) and expressing the AtDFB, -C, or -D cDNAs (Ura+, presence of the pRS vector) did not require methionine for growth (Fig. 2B). These cells were also able to grow on a medium containing ethanol as a carbon source, thus indicating that they are respiration-competent (Fig. 2B). These results are similar to that obtained when a plasmid harboring the wild-type MET7 gene is used to transform the diploid CD180 (met7Δ/MET7) strain (Fig. 2B; ref. 9), thus indicating that the AtDFB, -C, and -D cDNAs encode three isoforms of functional FPGS. On the other hand, the expression of the AtDFA protein did not correct either the methionine requirement or the respiration-deficient phenotype (Fig. 2B).
To validate the results described above, DHFS and FPGS activities were determined in vitro by using protein extracts prepared from the different strains. The DHFS activity detected in CD208–2B (fol3Δ) transformed with pRS-AtDFA (0.26 nmol dihydrofolate formed per h/mg of protein) is similar to that measured in the parental FOL3 strain (0.22 nmol/h/mg of protein). Because no activity could be detected for CD208–2B (fol3Δ), this result confirmed that the AtDFA cDNA encodes a functional DHFS enzyme. When FPGS was assayed by using H4F-Glu1 as a substrate, no activity could be detected in CD180–5C (met7Δ), as expected for a null mutation of the MET7 gene (9). The activities measured in the met7Δ cells expressing AtDFB, -C, or -D ranged from 0.08 to 0.30 nmol glutamate incorporated per h/mg of protein (Table 1), thus confirming that these proteins are functional FPGS enzymes. The three FPGS isoforms from Arabidopsis exhibited broad substrate specificity because they were able to add glutamates to H4F-Glu1 carrying various C1 units (Table 1). This property is characteristic for the eukaryotic FPGS so far studied (1, 2).
Table 1.
Substrate specificity of FPGS in the different yeast strains
| Yeast strain | H4F | CH2-H4F | CH3-H4F | 5-CHO-H4F | 10-CHO-H4F |
|---|---|---|---|---|---|
| CC788-2B (MET7) | 0.78 | 0.86 | 0.34 | 0.69 | 1.10 |
| CD180-5C + pRS-AtDFB | 0.11 | 0.16 | 0.17 | 0.17 | 0.20 |
| CD180-5C + pRS-AtDFC | 0.08 | 0.11 | 0.14 | 0.11 | 0.15 |
| CD180-5C + pRS-AtDFD | 0.30 | 0.36 | 0.19 | 0.27 | 0.39 |
Activities were determined at 37°C as described under Materials and Methods by using 100 μM H4F-Glu1 and its derivatives. Specific activities are expressed in nmol/h/mg of protein. Data are representative of 2–5 independent measurements. No activity was detected in the original CD180-5C (met7Δ) strain by using H4F-Glu1 as a substrate.
Subcellular Localization of DHFS and FPGS Isoforms.
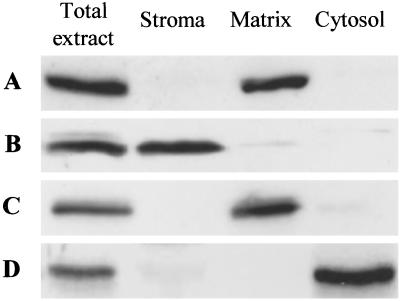
To analyze the localization of DHFS and the three FPGS isoforms in the plant cell, we prepared specific Abs against each protein. Intact chloroplasts and mitochondria were purified on Percoll density gradients, thus providing organelles devoid of contamination from the other compartments. Also, a cytosolic-enriched fraction was prepared. Preliminary experiments indicated that DHFS and FPGS isoforms are soluble proteins in Arabidopsis (data not shown). Soluble proteins from purified chloroplasts (stroma), mitochondria (matrix), and the cytosolic-enriched fractions were then analyzed by Western blotting. Abs raised against the AtDFA protein identified a 53-kDa polypeptide in the total cell protein extract and the mitochondrial matrix fraction (Fig. 3A). A similar pattern was obtained for the isoform C of FPGS, which had an estimated molecular mass of 66 kDa (Fig. 3C). These results strongly suggest that in Arabidopsis DHFS and FPGS C are located in the matrix of mitochondria. This localization is consistent with a previous study performed in pea leaves (7), and the estimated Mr of the mature proteins fits well with the calculated molecular mass of the corresponding ORF minus the predicted mitochondrial transit peptide presequence. Fig. 3B shows that the 60-kDa polypeptide recognized by the AtDFB Abs in the total cell protein extract was also detected in the stromal fraction, thus indicating that the isoform B of FPGS is present in the chloroplast. The estimated molecular mass was consistent with a transit peptide of ≈25 residues. The immunoblots probed with the Abs raised against FPGS D indicated that the protein is present in the cytosol (Fig. 3D). Indeed, a 55-kDa polypeptide that corresponds to the calculated molecular mass for FPGS D is detected only in the total cell protein extract and the cytosolic-enriched fraction.
Figure 3.
Immunolocalization of DHFS and FPGS isoforms in Arabidopsis. Proteins (35 μg for total extract, 17 μg for chloroplast stroma, 5 μg for mitochondrial matrix, and 17 μg for the cytosolic-enriched fraction) were separated by SDS/PAGE using 10% acrylamide gels, transferred to nitrocellulose membranes, and probed with the Abs specific to each protein. A–D correspond to membranes probed with the Abs to AtDFA, -B, -C, and -D, respectively.
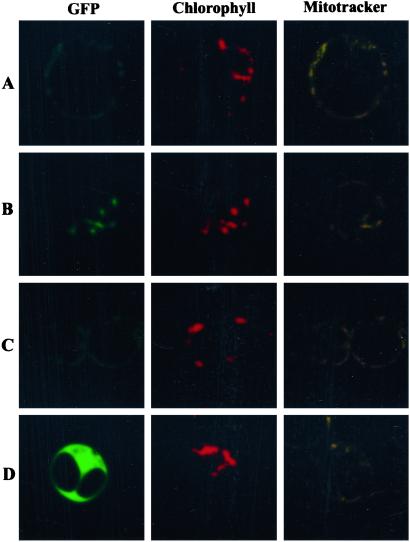
As a complementary approach, the subcellular localization of DHFS and FPGS isoforms was analyzed in vivo by using fusions between the N terminus of the individual Arabidopsis proteins and a GFP marker protein. As shown in Fig. 4D, the expression of AtDFD-GFP in Arabidopsis protoplasts resulted in green fluorescence throughout the cytoplasm and the nucleus, a pattern similar to the one observed with GFP alone. The AtDFB-GFP fusion protein colocalized with the red autofluorescence of chlorophyll (Fig. 4B), a situation that is identical to that obtained with GFP fused to the transit peptide of the Rubisco small subunit. This result demonstrates that FPGS B contains a functional plastid-targeting presequence. The staining pattern obtained for GFP fused with either AtDFA or -C was consistent with a mitochondrial localization of these proteins (Fig. 4 A and C). Indeed, similar patterns were observed with a fusion between GFP and the mitochondrial transit peptide of HPPK-DHPS, and the green fluorescence of the AtDFA and -C fusion proteins colocalized with the mitochondrial probe MitoTracker Orange. This result confirmed that DHFS and the isoform C of FPGS are mitochondrial proteins in Arabidopsis.
Figure 4.
Expression of DHFS and FPGS isoforms fused to GFP in Arabidopsis protoplasts. Constructs encoding fusion proteins between mGFP4 and the N terminus of DHFS and FPGS isoforms were introduced into Arabidopsis protoplasts as described under Materials and Methods. GFP (green pseudocolor), chlorophyll (red pseudocolor), and MitoTracker (yellow pseudocolor) fluorescence was observed by confocal laser scanning microscopy. A–D correspond to GFP fused to AtDFA, -B, -C, and -D, respectively. In D only the central cell is transformed. Optical sections are 0.6 μm thick.
Discussion
Plants have the ability to synthesize H4F-Glun de novo from the dihydropterin, para aminobenzoic acid, and glutamate precursors through the sequential operation of five enzymes, including DHFS and FPGS (Fig. 5). In pea leaf, Neuburger et al. (7) measured the five activities only in the mitochondria, suggesting that these organelles play a major role in H4F-Glun synthesis. Other biochemical and genetic studies supported the mitochondrial localization of HPPK-DHPS (26), DHFS (15), and dihydrofolate reductase-thymidylate synthase (DHFR-TS; ref. 7). Also, the presence of DHFR-TS in the plastids (31) and FPGS in the cytosol (16) have been reported.
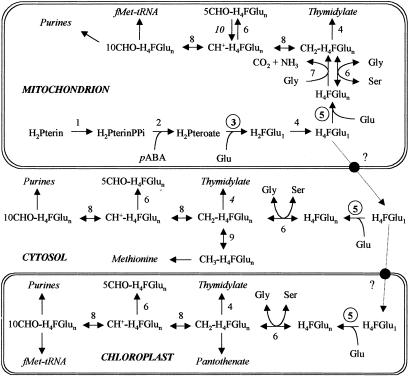
Figure 5.
Proposed model for the compartmentation of tetrahydrofolate synthesis and C1-tetrahydrofolate interconversion in the plant cell. Tetrahydrofolate-synthesizing enzymes are 1, dihydropterin pyrophosphokinase; 2, dihydropteroate synthase; 3, DHFS; 4, dihydrofolate reductase–thymidylate synthase; and 5, FPGS. Enzymes involved in the interconversion of H4F coenzymes are 6, serine hydroxymethyltransferase; 7, glycine decarboxylase; 8, methylene H4F dehydrogenase/methenyl H4F cyclohydrolase; 9, methylene H4F reductase; and 10, 5-formyl H4F cycloligase. Enzymes 1, 2, 3, 7, and 10 (S. Roje and A. D. Hanson, unpublished data) are exclusively mitochondrial, whereas enzyme 9 is found only in the cytosol. The occurrence of reaction 4 in the cytosol is predicted from DNA sequence data. The compartmentation of the enzymes involved in the synthesis of C1-metabolism end products is discussed in ref. 30. H2Pterin(PPi), dihydropterin(pyrophosphate); H2Pteroate, dihydropteroate; H2F-Glu1, dihydrofolate monoglutamate.
In this study, a genomic-based approach allowed us to clone a cDNA coding DHFS in Arabidopsis. The subcellular localization of DHFS in the mitochondria and the estimated molecular mass of the mature protein (53 kDa) corroborate previous data concerning the plant enzyme (7, 15). Also, this result definitely demonstrates that the first three reactions of H4F-Glun synthesis catalyzed by the bifunctional HPPK-DHPS (26) and DHFS are present only in the mitochondria (Fig. 5). Three cDNAs coding FPGS isoforms were also characterized, thus providing new insights into the subcellular compartmentation of folate biosynthesis in the plant cell. The plant proteins resemble FPGS from other organisms in having an N-terminal domain involved in ATP and Mg2+ binding (catalytic site), and a C-terminal region dedicated to the binding of the folate substrate (29). However, there is a marked difference among plants, mammals, and fungi regarding the organization of the genes coding FPGS. In mammalian cells, a single gene encodes both the cytosolic and mitochondrial isoforms of the enzyme by the use of alternate transcription and translation starts (17). A similar situation has been proposed for the met6 gene encoding FPGS in N. crassa (32). In S. cerevisiae, the existence of a mitochondrial isoform of FPGS resulting from an alternate translation start is still controversial (9, 10). In Arabidopsis, FPGS isoforms are encoded by separate genes (Table 2), thus suggesting that the expression of each protein may be controlled separately through the interaction of transcriptional regulatory elements to the corresponding promoter regions.
The most important finding from our studies of plant FPGSs concerns their cellular distribution. Western blot analyses and expression of GFP-fusion proteins in vivo have demonstrated for the first time that each compartment of the plant cell containing γ-glutamyl-conjugated H4F derivatives possesses one isoform of the enzyme responsible for the extension of the glutamate side chain of folate coenzymes. First, the localization of one FPGS isoform in the mitochondria fits well with the biochemical data reported by Neuburger et al. (7), confirming that the mitochondrial compartment is the only one that contains the complete set of enzymes required for de novo synthesis of H4F-Glun (Fig. 5). On the other hand, the multicellular compartmentation of FPGS in the plant cell is in agreement with the presence of folate-dependent reactions in the cytosol, the mitochondria, and the chloroplasts (Fig. 5). Serine hydroxymethyltransferase, the enzyme catalyzing the conversion of serine to glycine with the concomitant formation of 5,10-CH2-H4F from unsubstituted H4F, is found in these three compartments (33) and exhibits a marked preference for polyglutamate coenzymes (6). Also, the mitochondria, the cytosol, and the chloroplast each contain a parallel set of enzymes catalyzing the interconversion of 5,10-CH2-, 5,10-CH+-, and 10-CHO-substituted folate coenzymes (7, 34). Thus, the occurrence of FPGS isoforms in the three main compartments of the plant cell matches with the predominance of glutamyl-conjugated H4F derivatives and the presence of serine hydroxymethyltransferase and C1-interconverting enzymes in the cytosol, the mitochondria, and the plastids. Because serine is the principle source of C1 units in the plant cell (35), the combination of these folate-mediated reactions can supply each compartment with the polyglutamylated C1-substituted H4F coenzymes required for their nucleotides and formylmethionyl-tRNA synthesis (Fig. 5).
The complex compartmentation of folate-mediated reactions in the plant cell implies the transport of H4F species between the organelles via the cytosol. In mammalian cells, the traffic of folates first involves the uptake of the monoglutamate species found in the blood. Once internalized, folates are either converted to polyglutamate forms to promote their retention in the cytosol or transported to the mitochondria where they are polyglutamylated to minimize their diffusion out of the organelle. The transport systems involved in the cellular uptake (36) and the translocation of folates from the cytosol to the mitochondria (37) have been characterized in mammals. Nothing is known concerning the traffic of folate coenzymes in the plant cell. The presence of FPGS isoforms in the cytosol and the chloroplast strongly suggests that monoglutamate folates, which are initially synthesized in the mitochondria, are the transported forms. Indeed, the presence of a glutamate tail markedly increases the anionic nature of folate coenzymes, thus impairing their diffusion through hydrophobic barriers (4). The reduction status of the transported form(s) of folate has to be established. Mitochondria from rat liver have the ability to import oxidized (folate, dihydrofolate) or reduced (5-CHO- and 5-CH3-H4F) species from the cytosol (38, 39). In plants, the traffic of oxidized folates would imply the presence of dihydrofolate reductase-thymidylate synthase (DHFR-TS) isoenzymes in the cytosol and the chloroplasts to reduce dihydro- to tetrahydrofolate before the addition of glutamate residues by FPGS. The presence of a DHFR-TS in the plastids has been suggested by immunological techniques (31), whereas the existence of a cytosolic isoform is supported only by DNA sequence data. The intracellular transport of reduced folates is also questionable because of their sensitivity to oxidative degradation (40).
Supplementary Material
Acknowledgments
We thank Drs. M. Block and J. Bourguignon for their expertise in the purification of organelles.
Abbreviations
- H4F-Glun
5,6,7,8-tetrahydrofolate with n glutamate residues
- DHFS
dihydrofolate synthetase
- FPGS
folylpolyglutamate synthetase
- AtDFA
-B, -C, and -D, isolated cDNAs displaying homology with known DHFS-FPGS
- GFP
green fluorescent protein
- HPPK-DHPS
dihydropterin pyrophosphokinase-dihydropteroate synthase
Footnotes
References
- 1.Shane B. Vitam Horm (San Francisco) 1989;45:263–335. doi: 10.1016/s0083-6729(08)60397-0. [DOI] [PubMed] [Google Scholar]
- 2.Cossins E A, Chen L. Phytochemistry. 1997;45:437–452. doi: 10.1016/s0031-9422(96)00833-3. [DOI] [PubMed] [Google Scholar]
- 3.Scott J, Rébeillé F, Fletcher J. J Sci Food Agric. 2000;80:795–824. [Google Scholar]
- 4.Appling D R. FASEB J. 1991;5:2645–2651. doi: 10.1096/fasebj.5.12.1916088. [DOI] [PubMed] [Google Scholar]
- 5.Imeson H C, Zheng L, Cossins E A. Plant Cell Physiol. 1990;31:223–231. [Google Scholar]
- 6.Besson V, Rébeillé F, Neuburger M, Douce R, Cossins E A. Biochem J. 1993;292:425–430. doi: 10.1042/bj2920425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Neuburger M, Rébeillé F, Jourdain A, Nakamura S, Douce R. J Biol Chem. 1996;271:9466–9472. doi: 10.1074/jbc.271.16.9466. [DOI] [PubMed] [Google Scholar]
- 8.Gambonnet B, Jabrin S, Ravanel S, Karan M, Douce R, Rébeillé F. J Sci Food Agric. 2001;81:835–841. [Google Scholar]
- 9.Cherest H, Thomas D, Surdin-Kerjan Y. J Biol Chem. 2000;275:14056–14063. doi: 10.1074/jbc.275.19.14056. [DOI] [PubMed] [Google Scholar]
- 10.DeSouza L, Shen Y, Bognar A L. Arch Biochem Biophys. 2000;376:299–312. doi: 10.1006/abbi.2000.1741. [DOI] [PubMed] [Google Scholar]
- 11.McBurney M W, Whitmore G F. Cell. 1974;2:173–182. doi: 10.1016/0092-8674(74)90091-9. [DOI] [PubMed] [Google Scholar]
- 12.Bognar A L, Osborne C, Shane B, Singer S C, Ferone R. J Biol Chem. 1985;260:5625–5630. [PubMed] [Google Scholar]
- 13.Salcedo E, Cortese J F, Plowe C V, Sims P F, Hyde J E A. Mol Biochem Parasitol. 2001;112:239–252. doi: 10.1016/s0166-6851(00)00370-4. [DOI] [PubMed] [Google Scholar]
- 14.McDonald D, Atkinson I J, Cossins E A, Shane B. Phytochemistry. 1995;38:327–333. doi: 10.1016/0031-9422(94)00615-z. [DOI] [PubMed] [Google Scholar]
- 15.Iwai K, Ikeda M, Kobashi M. Methods Enzymol. 1980;66:581–585. doi: 10.1016/0076-6879(80)66511-2. [DOI] [PubMed] [Google Scholar]
- 16.Chan P Y, Coffin J W, Cossins E A. Plant Cell Physiol. 1986;27:431–441. [Google Scholar]
- 17.Freemantle S J, Taylor S M, Krystal G, Moran R G. J Biol Chem. 1995;270:9579–9584. doi: 10.1074/jbc.270.16.9579. [DOI] [PubMed] [Google Scholar]
- 18.Sikorski R S, Hieter P. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bradford M M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 20.Schirch V. Methods Enzymol. 1997;281:81–87. doi: 10.1016/s0076-6879(97)81012-9. [DOI] [PubMed] [Google Scholar]
- 21.Garcia I, Rodgers M, Lenne C, Rolland A, Sailland A, Matringe M. Biochem J. 1997;325:761–769. doi: 10.1042/bj3250761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Awai K, Maréchal E, Block M A, Brun D, Masuda T, Shimada H, Takamiya K, Ohta H, Joyard J. Proc Natl Acad Sci USA. 2001;98:10960–10965. doi: 10.1073/pnas.181331498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Douce R, Bourguignon J, Brouquisse J, Neuburger M. Methods Enzymol. 1987;148:403–415. [Google Scholar]
- 24.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 25.Haseloff J, Siemering K R, Prasher D C, Hodge S. Proc Natl Acad Sci USA. 1997;94:2122–2127. doi: 10.1073/pnas.94.6.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rébeillé F, Macherel D, Mouillon J M, Garin J, Douce R. EMBO J. 1997;16:947–957. doi: 10.1093/emboj/16.5.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abel S, Theologis A. Plant J. 1994;5:421–427. doi: 10.1111/j.1365-313x.1994.00421.x. [DOI] [PubMed] [Google Scholar]
- 28.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun X, Bognar A L, Baker E N, Smith C A. Proc Natl Acad Sci USA. 1998;95:6647–6652. doi: 10.1073/pnas.95.12.6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hanson A D, Roje S. Annu Rev Plant Physiol Plant Mol Biol. 2001;52:119–137. doi: 10.1146/annurev.arplant.52.1.119. [DOI] [PubMed] [Google Scholar]
- 31.Luo M, Orsi R, Patrucco E, Pancaldi S, Cella R. Plant Mol Biol. 1997;33:709–722. doi: 10.1023/a:1005798207693. [DOI] [PubMed] [Google Scholar]
- 32.Atkinson I J, Nargang F E, Cossins E A. Phytochemistry. 1998;49:2221–2232. doi: 10.1016/s0031-9422(98)00317-3. [DOI] [PubMed] [Google Scholar]
- 33.Besson V, Neuburger M, Rébeillé F, Douce R. Plant Physiol Biochem. 1995;33:665–673. [Google Scholar]
- 34.Kirk C D, Chen L, Imeson H C, Cossins E A. Phytochemistry. 1995;39:1309–1317. [Google Scholar]
- 35.Mouillon J M, Aubert S, Bourguignon J, Gout E, Douce R, Rébeillé F. Plant J. 1999;20:197–205. doi: 10.1046/j.1365-313x.1999.00591.x. [DOI] [PubMed] [Google Scholar]
- 36.Antony A C. Annu Rev Nutr. 1996;16:501–521. doi: 10.1146/annurev.nu.16.070196.002441. [DOI] [PubMed] [Google Scholar]
- 37.Titus S A, Moran R G. J Biol Chem. 2000;275:36811–36817. doi: 10.1074/jbc.M005163200. [DOI] [PubMed] [Google Scholar]
- 38.Cybulski R L, Fisher R R. Biochim Biophys Acta. 1981;646:329–333. doi: 10.1016/0005-2736(81)90339-4. [DOI] [PubMed] [Google Scholar]
- 39.Horne D W, Holloway R S, Said H M. J Nutr. 1992;122:2204–2209. doi: 10.1093/jn/122.11.2204. [DOI] [PubMed] [Google Scholar]
- 40.Rébeillé F, Neuburger M, Douce R. Biochem J. 1994;302:223–228. doi: 10.1042/bj3020223. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.