Key Points
Question
Which mechanisms underlie the negative association of posttraumatic stress disorder (PTSD) with traits related to educational attainment (EA)?
Findings
In this mendelian randomization study based on large-scale genomic data sets, including data from more than 1 million individuals, EA was negatively associated with PTSD, also supporting the role of economic status as a mediator in this association.
Meaning
This study suggests that economic status may mediate the association of EA with PTSD independent of the brain mechanisms associated with EA.
This mendelian randomization study investigates the association of posttraumatic stress disorder and traits associated with educational attainment.
Abstract
Importance
There is a well-established negative association of educational attainment (EA) and other traits related to cognitive ability with posttraumatic stress disorder (PTSD), but the underlying mechanisms are poorly understood.
Objectives
To investigate the association of PTSD with traits related to EA.
Design, Setting, and Participants
Genetic correlation, polygenic risk scoring, and mendelian randomization (MR) were conducted including 23 185 individuals with PTSD and 151 309 control participants from the Psychiatric Genomics Consortium for PTSD and up to 1 131 881 individuals assessed for EA and related traits from UK Biobank, 23andMe, and the Social Science Genetic Association Consortium. Data were analyzed from July 3 through November 19, 2018.
Main Outcomes and Measures
Genetic correlation obtained from linkage disequilibrium score regression, phenotypic variance explained by polygenic risk scores, and association estimates from MR.
Results
Summary association data from multiple genome-wide association studies were available for a total of 1 180 352 participants (634 391 [53.7%] women). Posttraumatic stress disorder showed negative genetic correlations with EA (rg = −0.26; SE = 0.05; P = 4.60 × 10−8). Mendelian randomization analysis, conducting considering a random-effects inverse-variance weighted method, indicated that EA has a negative association with PTSD (β = −0.23; 95% CI, −0.07 to −0.39; P = .004). Investigating potential mediators of the EA-PTSD association, propensity for trauma exposure and risk-taking behaviors were observed as risk factors for PTSD independent of EA (trauma exposure: β = 0.37; 95% CI, 0.19 to 0.52; P = 2.57 × 10−5; risk-taking: β = 0.76; 95% CI, 0.38 to 1.13; P = 1.13 × 10−4), while income may mediate the association of EA with PSTD (MR income: β = −0.18; 95% CI, −0.29 to −0.07; P = .001; MR EA: β = −0.23; 95% CI, −0.39 to −0.07; P = .004; multivariable MR income: β = −0.32; 95% CI, −0.57 to 0.07; P = .02; multivariable MR EA: β = −0.04; 95% CI, −0.29 to 0.21; SE, 0.13; P = .79).
Conclusions and Relevance
Large-scale genomic data sets add further evidence to the negative association of EA with PTSD, also supporting the role of economic status as a mediator in the association observed.
Introduction
Posttraumatic stress disorder (PTSD) is a psychological condition that occurs in some individuals after exposure to a major traumatic event. Prospective studies have suggested that many variables previously considered outcomes of trauma are likely to be pretrauma risk factors.1 Among these complex associations, that of PTSD with cognitive ability and educational attainment (EA; ie, the number of years of schooling that individuals complete) is among the most puzzling.
While there is a robust epidemiologic literature on the negative association of PTSD with cognitive ability and EA,2,3,4,5 the underlying mechanisms remain unclear. Reverse causation (ie, when an outcome precedes and causes the exposure)6,7,8 is a key obstacle to disentangling the direction of the mechanisms that associate these 2 phenotypes.
Adequately powered genome-wide association studies (GWASs) are able to dissect the predisposition to complex traits. This requires extremely large sample sizes to detect the polygenic architecture of complex traits, overcoming their heterogeneity to have sufficient power to find risk loci of very small effect.9 By combining such small-effect loci, it is possible to build genetic instruments that can be used to investigate complex epidemiological associations, such as the underlying mechanisms connecting PTSD, traits related to EA, and the potential mediation of other pretrauma risk factors. In particular, genetic information can remove the bias of reverse causation from analysis of the association of PTSD with cognition and education. Genetic variants are allocated at conception and do not change throughout life, and they can be used to define reliable genetic instruments that can be applied in a mendelian randomization (MR) analysis.10,11,12,13,14 The basic principle in MR is that an instrumental variable based on genetic variants associated with a phenotype can be used to represent, or mirror, the disease risk associated with that phenotype without the presence of possible environmental confounders.15
Previous large-scale GWASs conducted by the Social Science Genetic Association Consortium (SSGAC)16,17,18,19 investigated traits related to EA, identifying a number of loci and biological pathways regulating brain mechanisms at the basis of human cognitive ability. A 2018 genome-wide analysis of multiple brain disorders and phenotypes20 showed pervasive shared heritability of these traits with EA and related traits. Based on the GWAS regarding PTSD newly generated by the Psychiatric Genomics Consortium (PGC),21 we applied multiple statistical methods to large-scale genomic data sets to investigate the mechanisms involved in the association of PTSD with traits related to EA and potential pretrauma risk factors.
Methods
This study was conducted using summary association data generated by previous studies. Owing to the use of previously collected, deidentified, aggregated data, this study did not require institutional review board approval. Ethical approval had been obtained in all original studies.16,21 Summary data were available for a total of 1 180 352 participants. Multiple statistical methods were applied to these data sets to investigate the association of PTSD with EA and related traits. A schematic workflow summarizing the analyses conducted is reported in eFigure 1 in the Supplement. These analyses were conducted from July 3 through November 19, 2018. The study was reported in accordance to the Strengthening the Reporting of Genetic Association Studies (STREGA) reporting guideline.22
Cohorts Investigated
Genome-wide information regarding PTSD was derived from the freeze-2 analysis conducted by the Psychiatric Genomics Consortium Posttraumatic Stress Disorder (PGC-PTSD) Working Group.21 In this analysis, lifetime and/or current PTSD status was assessed using various instruments and different versions of the Diagnostic and Statistical Manual of Mental Disorders (Third Edition Revised, Fourth Edition, and Fifth Edition). We focused on the data generated from the analysis of individuals of European descent (23 185 individuals with PTSD; 151 309 control participants) because the genome-wide analyses of traits associated with EA were conducted only on this ancestry group.
Genome-wide information regarding traits related to EA were derived from the GWAS meta-analysis by the SSGAC,16 which investigated EA as the primary phenotype in a total of 1 131 881 individuals (EA1M). Additional information about the definition of the EA phenotype is available in the eAppendix in the Supplement. In the same SSGAC study,16 3 additional phenotypes were investigated. Of these, 2 were analyzed exclusively among research participants of the personal genomics company 23andMe. Participants were asked to rate their mathematical ability (MA; n = 564 698; very poor, 0; poor, 1; about average, 2; good, 3; excellent, 4) and to list the most advanced math course they had successfully completed (MC; n = 430 445; prealgebra, 1; algebra, 2; geometry, 3; trigonometry, 4; precalculus, 5; calculus, 6; vector calculus, 7; >vector calculus, 8). The third phenotype investigated, cognitive performance, was assessed in 257 828 participants from the Cognitive Genomics Consortium study and the UK Biobank.16 For the data sets including participants from 23andMe, we had access only to summary association data of the top 10 000 variants. Accordingly, the data sets derived from 23andMe were not used for the reverse analysis (ie, estimating the association of PTSD with traits related to EA).
A summary of the data sets tested is reported in the Table. Since UK Biobank participants were included in both PGC-PTSD and SSGAC studies, some analyses were conducted on a PGC-PTSD subsample that excluded the UK Biobank cohort (PGC-PTSD freeze-1.5 data set: 12 823 individuals with PTSD; 35 648 control participants). Excluding UK Biobank, negligible overlap is present between PGC-PTSD and SSGAC cohorts (eAppendix in the Supplement).
Table. Traits Tested With Corresponding Information Regarding Sample Size, Data Available, and Cohorts Included.
| Trait | Abbreviation | Sample Size | Cohort | Summary Statistics |
|---|---|---|---|---|
| Posttraumatic stress disorder | PTSD freeze-1.5 | 12 823 with PTSD; 35 648 controls | PGC | Full |
| Posttraumatic stress disorder | PTSD freeze-2 | 23 185 with PTSD; 151 309 controls | PGC, UKB | Full |
| Cognitive performance | CP | 257 828 | SSGAC, UKB | Full |
| Educational attainment | EA | 766 345 | SSGAC, UKB | Full |
| Educational attainment | EA1M | 1 131 881 | SSGAC, UKB, 23andMe | Top 10 000 |
| Self-reported math ability | MA | 564 692 | 23andMe | Top 10 000 |
| Most advanced math course completed | MC | 430 439 | 23andMe | Top 10 000 |
| Risk-taking behaviors | RT | 348 549 | UKB | Full |
| Income | Income | 311 028 | UKB | Full |
| Physically abused by family as a child | PAC | 117 838 | UKB | Full |
Abbreviations: PGC, Psychiatric Genomics Consortium; PTSD, posttraumatic stress disorder; SSGAC, Social Science Genetic Association Consortium; UKB, UK Biobank.
We also used data from the UK Biobank to investigate the possible mediation of PTSD risk factors in their associations with traits related to EA. Self-reported risk-taking behaviors were assessed with the question, “Would you describe yourself as someone who takes risks?” (UK Biobank data field 2040). Income was assessed with, “Average total household income before tax” (UK Biobank data field 738). We also considered traumatic events assessed in the UK Biobank (eTable 1 in the Supplement). Genome-wide information regarding these traits was derived from GWAS summary association data generated from the UK Biobank.23
Genetic Correlation and Definition of the Genetic Instruments
Linkage disequilibrium (LD) score regression was used to estimate the genetic correlation among the traits investigated.24 Since sample overlap between the GWAS tested does not affect the results obtained from this method,24 we were able to calculate pairwise genetic correlations, including those between data sets that had overlapping information with the UK Biobank participants.
The polygenic risk scores (PRSs) were calculated after using P value–informed clumping with an LD cutoff of R2 = 0.001 within a 10 000-kilobase window, excluding the major histocompatibility complex region of the genome because of its complex LD structure and including only variants with a minor allele frequency less than 1%. The European samples from the 1000 Genomes Project were used as the LD reference panel.25 The PRS analysis was conducted on the basis of the GWAS summary association data using the gtx R package incorporated in PRSice software.26 For each PRS analysis, we calculated an approximate estimate of the explained variance from a multivariate regression model.27 For the traits related to EA, we considered a genome-wide significance threshold (P < 5.00 × 10−8). For other traits (ie, PTSD, income, risk-taking behaviors, and trauma exposure), owing to the limited power to detect a large number of genome-wide significant loci, we evaluated multiple P value thresholds (PT; PT = 5.00 × 10−8, 10−7, 10−6, 10−5, 10−4, .001, .05, .1, .3, .5, and PT < 1) to increase the variance explained by the genetic instruments. To account for the multiple PTs tested, we considered P < .005 as the significance threshold in the PRS analysis. Tests of statistical significance were 2-tailed. The results of the PRS analyses were used to define the genetic instruments for each pairwise comparison to be investigated further via the MR approach.
Mendelian Randomization
To assess the association among the traits tested, we used GWAS summary association data to conduct 2-sample MR analyses.28 As mentioned earlier, genetic instruments were based on the results obtained in the PRS analysis. Since different MR methods have different sensitivities to different potential issues, accommodate different scenarios, and vary in their statistical efficiency,11 we considered a range of MR methods. The primary analysis was conducted considering a random-effects inverse-variance weighted (IVW) method.29 The secondary MR methods included MR Egger,30 simple mode,31 weighted median,32 and weighted mode.31 These MR analyses were conducted using the TwoSampleMR R package.29 Additionally, owing to the fact that some traits showed a limited number of associated genome-wide significant variants, genetic instruments associated with PTSD, income, risk-taking behaviors, and trauma exposure were based on suggestive PTs similar to previous MR studies.33,34,35 We verified these IVW estimates using the MR–Robust Adjusted Profile Score (MR-RAPS) approach, which is a method designed to identify and estimate confounded associations using weak genetic instrument variables.36,37 We conducted multiple sensitivity analyses with respect to the MR tests conducted to exclude possible biases (horizontal pleiotropy, ie, the variants included in the genetic instrument having an effect on disease outside their effects on the exposure in MR38,39) under different scenarios in the MR estimates. These included the IVW heterogeneity test,29 the MR-Egger intercept,30 the MR-RAPS overdispersion test,36 and the MR–Pleiotropy Residual Sum and Outlier (MR-PRESSO) global test.40 Finally, a leave-1-out analysis was conducted to identify potential outliers among the variants included in the genetic instruments tested. The MR results without evidence of horizontal pleiotropy and heterogeneity were entered in the multivariable MR (MVMR) analysis41 conducted using the IVW approach. This method permits evaluation of the independent association of each risk factor with the outcome, similar to the simultaneous assessment of several treatments in a factorial randomized trial.41
Comparing the utility of MR with MVMR methods, MR estimates the total association of the exposure with the outcome, whereas MVMR estimates the direct association of each exposure with the outcome.42 In this scenario, MVMR is not a form of mediation analysis but instead estimates the direct association of the exposure with the outcome that does not act via the mediator.42 The MVMR analysis was conducted using the MendelianRandomization R package.43
Enrichment Analysis
We tested for functional differences between the traits of interest by means of enrichment analyses based on tissue-specific and cell type–specific gene expression reference panels.44,45,46,47,48 These analyses were conducted using the MAGMA tool49 implemented in FUMA.50
Results
Data were available for a total of 1 180 352 participants (634 391 [53.7%] women). Information regarding PTSD was available for 23 185 individuals with PTSD and 151 309 control participants (174 494 individuals; 15%) from the PGC-PSTD Working Group and regarding EA for 1 131 881 individuals (96%) from the total sample.
Genetic Correlation and Polygenic Risk Scoring
Our study investigated multiple data sets generated from different cohorts and with different data availability (Table). Using LD score regression and data sets with full GWAS summary association data, we observed a negative genetic correlation of PTSD (freeze-2) with EA (rg = −0.26; SE = 0.05; P = 4.60 × 10−8) and cognitive performance (rg = −0.16; SE = 0.05; P = 9.00 × 10−4). The PRS analyses were conducted using PTSD as the target and traits related to EA as the training data set, considering only genome-wide significant variants (P < 5.00 × 10−8; eFigure 2 in the Supplement). This analysis was conducted excluding those pairwise comparisons that would have included data sets with the UK Biobank as an overlapping cohort. The most significant PRS association was observed between MC genome-wide significant PRS with respect to PGC-PTSD freeze-2 data (R2 = 0.04%; SE = 0.0001; P = 1.13 × 10−8). The same PTSD data set showed a weaker association with MA PRS (R2 = 0.01%; SE =0.00003; P = .004). Significant associations were also observed with respect to PGC-PTSD freeze-1.5 outcome for EA (R2 = 0.03%; SE = 0.0001; P = 6.16 × 10−4), MC PRS (R2 = 0.03%; SE = 0.0001; P = 7.86 × 10−4), and EA1M PRS (R2 = 0.03%; SE = 0.0001; P = .002). The phenotypic variance explained by the significant PRS is in line with the cross-phenotype association expected between 2 complex traits with a moderate genetic correlation. No association of the PRS of cognitive performance and MA was observed with respect to PGC-PTSD freeze-1.5 data set (cognitive performance: R2 < 0.01%; P = .15; MA: R2 < 0.01%; P = .11), and accordingly, these phenotypes were not investigated further. We tested the reverse direction (ie, PTSD as base and traits related to EA as target). Owing to the limited number of genome-wide significant loci in the PGC-PTSD analysis, the PRS analysis was conducted considering multiple-association PTs to include at least 10 LD-independent variants in each PRS tested (eFigure 3 in the Supplement). We observed a nominally significant association between the PGC-PTSD freeze-1.5 PRS (PT = 10−5) and EA (R2 = 0.0004%; SE = 0.000002; P = .04) that would not survive a Bonferroni correction for the number of PTs tested.
Mendelian Randomization
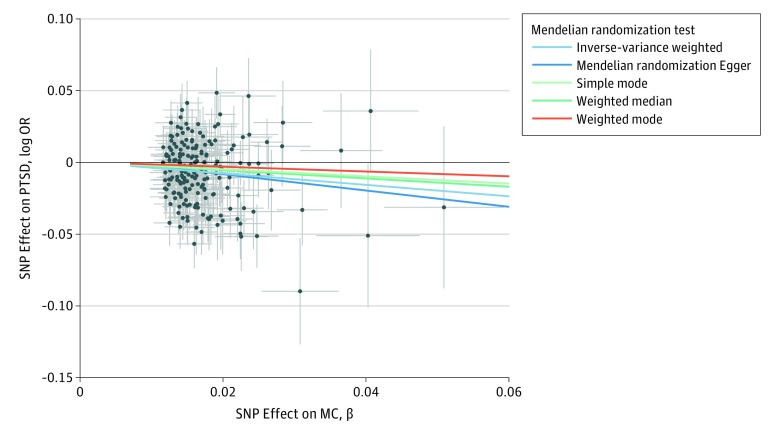
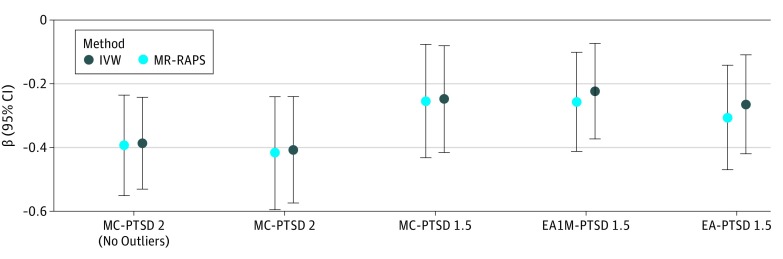
Based on the PRS results, we conducted MR tests using 3 genetic instruments based on genome-wide significant variants. These included MC, EA, and EA1M, tested with respect to PGC-PTSD data sets (freeze-2 and/or freeze-1.5, depending on UK Biobank overlap). The reverse MR test was conducted on the basis of PGC-PTSD freeze-1.5 data (PT = 5.00 × 10−5) with respect to the EA data set. It was not possible to conduct reverse analyses for the MC and EA1M data sets because we did not have access to the full GWAS summary association data for these studies. A significant association was found between MC and PGC-PTSD freeze-2 data (IVW: β = −0.41; 95% CI, −0.59 to −0.23; P = 3.46 × 10−6). Concordant results were observed when considering other MR methods (Figure 1). The significance was replicated with the MR-RAPS approach (β = −0.42; 95% CI, −0.60 to −0.24; SE = 0.09; P = 3.70 × 10−6), and no outliers were identified by the leave-1-out analysis (eFigure 4 in the Supplement). However, we observed the presence of possible bias in this result owing to heterogeneity and/or pleiotropy (IVW heterogeneity test: Q = 267.4; df = 192; P = 2.61 × 10−4; MR-RAPS overdispersion test: estimated pleiotropy variance, 0.0001; P = .007; MR-PRESSO global test: observed residual sum of squares, 281.4; P = 5.00 × 10−4). Thus, we identified the outliers on the basis of the MR-RAPS standardized residuals (−1.96 > z > 1.96) and verified their contributions on the results of the IVW heterogeneity test (eFigure 5 in the Supplement). Removing the outliers from the MC genetic instrument, we confirmed the association of MC with PTSD in the freeze-2 data set (IVW: β = −0.39; 95% CI, −0.57 to 0.21; P = 4.25 × 10−7; MR-RAPS: β = −0.39; 95% CI, −0.55 to −0.23; P = 1.06 × 10−6) and the lack of evidence of possible confounders from the sensitivity analyses (eTable 2 and eFigure 6 in the Supplement). We verified the reliability of this MR finding considering PGC-PTSD freeze-1.5 data as the outcome and MC, EA, and EA1M as exposures. We observed comparable MR results across the genetic instruments generated from different cohorts and the 2 versions of the PGC-PTSD data sets (Figure 2; eFigure 7 in the Supplement). However, consistent with the lower power of PGC-PTSD freeze-1.5 data, we observed a reduction of the significance (MC for PGC-PTSD freeze-2 data set: IVW: β = −0.41; 95% CI, −0.59 to −0.23; P = 3.46 × 10−6; MC for PGC-PTSD freeze-1.5 data set: IVW: β = −0.22; 95% CI, −0.38 to −0.06; P = .004; EA for PGC-PTSD freeze-1.5 data set: β = −0.23; 95% CI, −0.39 to −0.07; P = .004). We verified that no bias in the MR results was present owing to palindromic variants with an ambiguous allele frequency51 and to the presence of assortative mating in EA52 (eTable 3 and eAppendix in the Supplement).
Figure 1. Single-Nucleotide Polymorphism (SNP) Repeated Effects on Posttraummatic Stress Disorder (PTSD) and Most Advanced Math Course Completed (MC).
SNP exposure (MC associations, β) and SNP outcome (PTSD freeze-2 associations, log odds ratio [OR]) coefficients used in the mendelian randomization analysis. Crosses represent 95% CIs for each association.
Figure 2. Estimated Associations Considering Different Traits Associated With Educational Attainment (EA) and 2 Versions of the Posttraumatic Stress Disorder (PTSD) Data Set.
EA1M indicates sample including participants from Social Science Genetic Association Consortium, UK Biobank, and 23andMe; IVW, inverse-variance weighted; MC, most advanced math course completed; MR-RAPS, mendelian randomization–robust adjusted profile score; PTSD2, PTSD freeze 2 data set; and PTSD1.5, PTSD freeze 1.5 data set.
To support further that MC and EA are associated with the same mechanism, we conducted a MVMR analysis, which showed that these 2 associations are not independent from each other (eFigure 8 in the Supplement), and their relationship with PTSD should be shared. Accordingly, we used EA as a proxy of MC in the subsequent analyses because we had full access only to the former data set.
We tested the reverse association, considering PTSD as the risk factor (ie, exposure) and EA as the outcome of the MR analysis. We included PTSD genetic instrument variants with a PTSD GWAS P = 10−5 considering the PGC-PTSD freeze-1.5 data. No significant association was observed (IVW: β = 0.006; 95% CI, −0.002 to 0.014; P = .16; MR-RAPS: β = 0.0006; 95% CI, −0.007 to 0.008; P = .10). To further confirm the absence of reverse association, we conducted an MR analysis including all LD-independent variants in the genetic instrument and applied the MR-RAPS method only. No directional association of PTSD with EA was observed (β = −0.0006; 95% CI, −0.002 to 0.002; P = .55), but we confirmed a directional association of EA with PSTD (β = −0.27; 95% CI, −0.38 to −0.15; P = 8.06 × 10−6). These outcomes were stable across different adjustments of the MR-RAPS method (eTable 4 in the Supplement).
Multivariable MR Analysis
To further investigate the association of EA with PTSD, we tested 3 potential mediators (risk-taking behaviors, income, and trauma exposure) in an MVMR analysis. Before entering these potential mediators in the MVMR analysis, we verified the reliability of each genetic instrument by conducting a standard 2-sample MR and verifying the evidence of bias owing to heterogeneity and horizontal pleiotropy. Because of the limited number of genome-wide significant variants with respect to these traits, we conducted a PRS considering multiple PTs as described earlier, to determine the best genetic instrument for each trait with respect to the PGC-PTSD freeze-1.5 data set (eFigure 9 in the Supplement). The best results were observed for PT = 5.00 × 10−4 with risk-taking behaviors (R2 = 0.06%; SE = 0.0001; P = 1.53 × 10−5) and PT = .001 for income (R2 = 0.09%; SE = 0.0002; P = 5.67 × 10−8). The MR analysis based on these genetic instruments confirmed that PTSD is associated with the genetic instruments related to risk-taking behaviors (IVW: β = 0.76; 95% CI, 0.38 to 1.13; P = 1.13 × 10−4; MR-RAPS: β = 0.76; 95% CI, 0.32 to 1.21; P = 6.75 × 10−4), and income (IVW: β = −0.18; 95% CI, −0.29 to −0.07; P = .001; MR-RAPS: β = −0.19; 95% CI, −0.31 to −0.07; P = .003). No evidence of heterogeneity or pleiotropy was observed in either analysis (eTable 5 in the Supplement).
Multiple traumatic events were assessed in the UK Biobank (eTable 1 in the Supplement), and they showed genetic correlation with each other (eTable 6 in the Supplement). We selected 4 traumatic experiences that showed a similar pattern of genetic correlation with respect to PTSD, EA, and the other 2 potential mediators (eFigure 10 in the Supplement). The most informative PRS across the 4 traumatic experiences tested was observed at PT = .001 (eFigure 11 in the Supplement). Then, we conducted an MR analysis testing different trauma-related genetic instruments with respect to PGC-PTSD freeze-1.5 data. We observed significant associations not affected by confounders (eTable 7 in the Supplement) for 3 traumatic experiences and, conducting an MVMR analysis, identified “physically abused by family as a child” (UK Biobank data field 20488) as the most informative genetic instrument for trauma exposure (MR analysis: β = 0.36; 95% CI, 0.19 to 0.52; P = 2.57 × 10−5; MVMR analysis: β = 0.26; 95% CI, −0.51 to 0.01; SE = 0.129; P = .04) (eFigure 12 in the Supplement).
In the MVMR analysis, we observed that trauma exposure and risk-taking behaviors were independent risk factors for PTSD (ie, the results obtained from the MR IVW and MVMR IVW analyses were both significant; Figures 3A, B, and C). Conversely, the genetic instrument related to income potentially mediates the association of the EA genetic instrument with PTSD (Figure 3A and D). Educational attainment has a significant association with respect to PTSD (β = −0.23; 95% CI, −0.39 to −0.07; P = .004), but when adjusted by income, this result is null (β = −0.04; 95% CI −0.30 to 0.21; P = .79). Conversely, the association of income with PTSD was still significant when adjusted by EA (unadjusted: β = −0.18; 95% CI, −0.30 to −0.06; P = .001; adjusted: β = −0.32; 95% CI, −0.57 to −0.07; SE = 0.13; P = .02).
Figure 3. Multivariable Mendelian Randomization Analysis Considering Associations of Educational Attainment (EA), Posttraumatic Stress Disorder (PTSD), and Other Traits.
IVW indicates inverse-variance weighted.
Enrichment Analysis
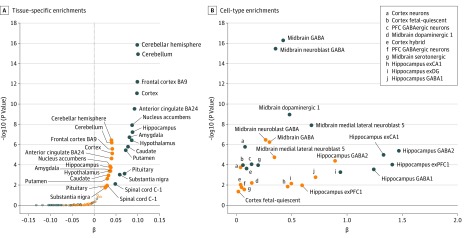
Although there is a large genetic correlation between EA and income (r = 0.81; P < 6.10 × 10−308), income is significantly more correlated with PTSD than EA (EA: r = −0.26; SE = 0.05; P = 4.60 × 10−8; income: r = −0.45; SE = 0.06; P = 9.98 × 10−16; z for EA × income = 2.65; P for EA × income = 0.008). Both traits are enriched for the transcriptomic profile of multiple brain tissues (eg, cerebellar hemisphere, EA: β = 0.01; 95% CI, 0.07-0.12; P = 1.49 × 10−16; income: β = 0.04; 95% CI, 0.02-0.06; P = 3.83 × 10−7) and neuronal cell types (eg, γ-aminobutyric acid [GABA]–ergic neurons, EA: β = 0.14; 95% CI, 0.07-0.21; P = 8.96 × 10−5; income: β = 0.05; 95% CI, 0.01-0.09; P = .02), but the enrichment signals of the EA GWAS are more significant than the ones observed in the income GWAS (Figure 4; eTable 8 in the Supplement), showing that EA data are more informative for the brain processes expected to be associated with cognition than income data.
Figure 4. Enrichment Analyses for Educational Attainment and Income .
A, Tissue-specific enrichments (brain tissues are indicated with solid circles; nonbrain tissues are indicated with open circles). B, Cell-type enrichments that were at least nominally significant (P < .05) in both traits. BA9 indicates Brodmann area 9; BA24, Brodmann area 24; exCA1, excitatory neurons in hippocampal subfield CA1; exDG, excitatory neurons in dentate gyrus; exPFC1, excitatory neurons in prefrontal cortex 1; GABA, γ-aminobutyric acid; and PFC, prefrontal cortex.
Discussion
Our analysis was based mainly on data from investigations of traits associated with EA. Previous studies have demonstrated that genetic results deriving from them are mainly informative regarding the brain mechanisms at the basis of human cognitive ability.16,17,18,19 Our analysis made use of these data to show that traits associated with cognitive ability have an association with PTSD. This result was consistent across traits in independent cohorts, even when adjusting the analysis for assortative mating present in these traits.52 In line with an association direction from cognition to PTSD, no evidence of reverse association was observed.
Although our findings are consistent with a specific direction, we had to evaluate whether other factors could be responsible for this association. Accordingly, we tested 3 phenotypes of known relevance for PTSD: propensity to trauma exposure,53 risk-taking behaviors,54 and economic status.55 Our MVMR analysis clearly showed that, while propensity to trauma exposure and risk-taking behaviors are independent PTSD risk factors, the directional association of EA with PTSD is associated with economic status, which is the driving force of the association. Educational attainment and income showed a large genetic overlap, and while PTSD showed a higher correlation with income than with EA, our investigation showed that EA is more informative for the brain mechanisms that are considered responsible for a predisposition to high cognitive ability. Since income appears to be responsible for the EA-PTSD association, we hypothesize that this mechanism is not related to cognitive ability but rather to other risk factors. This is also supported by our finding that, unlike EA and MC (traits that should be more associated with socioeconomic status), cognitive performance and MA showed a lower genetic association with PTSD. A 2018 genome-wide investigation of social stratification56 showed that cognition and socioeconomic status are correlated with a wide range of factors, including personality, psychological traits, mental health, substance use, physical health, reproductive behaviors, and anthropometric traits. Additionally, income may reflect indirect effects, such as those induced by genetic nurture on EA.57,58 Socioeconomic factors might also be associated with the outcome of PTSD, given that individuals with poorer outcomes are more likely to be included in studies of prevalent cases.
To our knowledge, this study represents the first MR analysis to investigate the underlying mechanisms linking cognitive ability to PTSD. It is based on the largest genome-wide data sets for these traits available at this time. The EA-PTSD association observed is in line with several prospective studies,1 and the association of socioeconomic status with PTSD is also a well-established risk factor reported in several observational studies.55,59 Compared with these previous investigations, the current analyses are based on a much larger population (>1 million individuals) than would ever be feasible for a traditional experimental design (which would include randomized interventions and measurements of outcomes) and without the ethical quandaries that would accompany such randomizations. Also, these results are not expected to be affected by reverse association because of the genetic information used. The findings show that income may explain the EA-PTSD association, suggesting that brain mechanisms related to cognitive ability are not directly responsible for the association observed.
Limitations
Our study has limitations. The present study is based on genetic information generated from the investigation of heterogeneous, clinically defined phenotypes such as PTSD. We tested this complex trait with respect to a series of complex, socially contextualized phenotypes, including EA, income, risk-taking behaviors, and predisposition to traumatic events. Although we used appropriate statistical methods and conducted the analyses across multiple independent cohorts, findings related to genetic data associated with these phenotypes need to be interpreted cautiously.58,60 The results of our current analysis are also limited by the statistical power of the PTSD GWAS data sets, which may have limited our ability to observe the reverse association of PTSD with EA. However, we used multiple methods that showed significant associations when applied to similarly powered GWAS data sets. Another potential limitation is owing to the pervasive presence of horizontal pleiotropy among complex traits.39 We applied multiple sensitivity analyses that accounted for different scenarios related to the potential confounding effect of horizontal pleiotropy and heterogeneity in the genetic instruments applied in our MR analyses. Although no evidence of bias was observed by the methods used, our current findings could be affected by an unaccounted confounder.
Conclusions
This study provides new evidence to elucidate the association of EA with PTSD, pointing toward risk factors associated with economic status rather than brain pathways. These findings have relevant implications with respect to our understanding of the pretrauma risk factors associated with increased vulnerability to PTSD. Additionally, MR analysis should consider testing the independence of multiple correlated risk factors with respect to the outcome of interest. This is particularly relevant when investigating the potential role of EA in human phenotypes and disorders.
eAppendix. Phenotype Definitions, Sample Overlap, Palindromic Variants, and Assortative Mating
eReferences.
eTable 1. Traumatic Experiences Assessed in the UK Biobank
eTable 2. Results of the Sensitivity Analyses Conducted With Respect to the MathClass→PTSD Test With and Without the Outlier Variants in the MathClass Genetic Instrument
eTable 3. Results of the IVW Analyses Considering Genetic Instruments With and Without Palindromic Variants With Ambiguous Allele Frequencies (PAL and noPAL, Respectively)
eTable 4. MR-RAPS Analysis Considering Various Adjustments Based on Genome-Wide Genetic Instruments
eTable 5. Results of the Sensitivity Analyses Conducted With Respect to the Income→PTSD and Risk-Tak→PTSD Tests
eTable 6. Genetic Correlation Among Trauma Experiences Assessed in the UK Biobank
eTable 7. Results (Causal Effects and Sensitivity Analyses) of the MR Test Conducted Using Trauma-Related Genetic Instruments With Respect to PTSD
eTable 8. Results of the Enrichment Analysis Based on Tissue-Specific and Cell Type–Specific Transcriptomic Data
eFigure 1. Schematic Workflow of the Analyses Conducted
eFigure 2. Genetic Correlations Estimated Between Traits Related to Cognitive Ability and 2 Versions of the Posttraumatic Stress Disorder Data Set, PGC-PTSD Freeze-2 (2) and PGC-PTSD Freeze-1.5 (1.5)
eFigure 3. Effect of the PTSD PRS on Educational Attainment (Yellow) and Cognitive Performance (Green) Considering Different Inclusion Thresholds
eFigure 4. Leave-1-Out Analysis Conducted With Respect to the MathClass→PTSD2 Result
eFigure 5. Identification of Potential Outliers (in Red) in MathClass Genetic Instrument Based on IVW Heterogeneity Test and MR-RAPS Standardized Residuals
eFigure 6. Results of the MathClass→PTSD2 Analysis After the Removal of the Potential Outliers From the Genetic Instrument
eFigure 7. Results of the Sensitivity Analyses With Respect to All MR Analyses Conducted
eFigure 8. Multivariable Mendelian Randomization Analysis Considering the Effects of EdAtt and MathClass on PTSD
eFigure 9. Effect of Income and RiskTak PRS (Green and Blue, Respectively) on PTSD Considering Different Inclusion Thresholds
eFigure 10. Genetic Correlations Between Traumatic Experiences and the Other Traits of Interest
eFigure 11. Effect of Trauma-Related PRS on PTSD Considering Different Inclusion Thresholds
eFigure 12. Multivariable Mendelian Randomization Analysis Considering the Effects of Physically Abused by Family as a Child and Belittlement by Partner or Ex-Partner as an Adult on PTSD
References
- 1.DiGangi JA, Gomez D, Mendoza L, Jason LA, Keys CB, Koenen KC. Pretrauma risk factors for posttraumatic stress disorder: a systematic review of the literature. Clin Psychol Rev. 2013;33(6):-. doi: 10.1016/j.cpr.2013.05.002 [DOI] [PubMed] [Google Scholar]
- 2.Goldstein RB, Smith SM, Chou SP, et al. The epidemiology of DSM-5 posttraumatic stress disorder in the United States: results from the National Epidemiologic Survey on Alcohol and Related Conditions-III. Soc Psychiatry Psychiatr Epidemiol. 2016;51(8):1137-1148. doi: 10.1007/s00127-016-1208-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frankenberg E, Sikoki B, Sumantri C, Suriastini W, Thomas D. Education, vulnerability, and resilience after a natural disaster. Ecol Soc. 2013;18(2):16. doi: 10.5751/ES-05377-180216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gale CR, Deary IJ, Boyle SH, Barefoot J, Mortensen LH, Batty GD. Cognitive ability in early adulthood and risk of 5 specific psychiatric disorders in middle age: the Vietnam Experience Study. Arch Gen Psychiatry. 2008;65(12):1410-1418. doi: 10.1001/archpsyc.65.12.1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kremen WS, Koenen KC, Boake C, et al. Pretrauma cognitive ability and risk for posttraumatic stress disorder: a twin study. Arch Gen Psychiatry. 2007;64(3):361-368. doi: 10.1001/archpsyc.64.3.361 [DOI] [PubMed] [Google Scholar]
- 6.Bowling A, Ebrahim S. Handbook of Health Research Methods. Maidenhead, England: McGraw-Hill, Open University Press; 2005. [Google Scholar]
- 7.Szklo M, Nieto FJ. Epidemiology: Beyond the Basics. 4th ed Sudbury, MA: Jones and Bartlett Publishers; 2006. [Google Scholar]
- 8.Rothman KJ, Greenland S, Lash TL. Modern Epidemiology. 3rd ed Philadelphia, PA: Wolters Kluwer/Lippincott Williams & Wilkins; 2008. [Google Scholar]
- 9.Dick DM. Mapping risk from genes to behavior: the enduring and evolving influence of Irving Gottesman’s endophenotype concept. Twin Res Hum Genet. 2018;21(4):306-309. doi: 10.1017/thg.2018.35 [DOI] [PubMed] [Google Scholar]
- 10.Ravera S, Carrasco N, Gelernter J, Polimanti R. Phenomic impact of genetically-determined euthyroid function and molecular differences between thyroid disorders. J Clin Med. 2018;7(10):E296. doi: 10.3390/jcm7100296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Polimanti R, Amstadter AB, Stein MB, et al. ; Psychiatric Genomics Consortium Posttraumatic Stress Disorder Workgroup . A putative causal relationship between genetically determined female body shape and posttraumatic stress disorder. Genome Med. 2017;9(1):99. doi: 10.1186/s13073-017-0491-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Polimanti R, Gelernter J, Stein DJ. Genetically determined schizophrenia is not associated with impaired glucose homeostasis. Schizophr Res. 2018;195:286-289. doi: 10.1016/j.schres.2017.10.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Polimanti R, Peterson RE, Ong JS, et al. Evidence of causal effect of major depression on alcohol dependence: findings from the Psychiatric Genomics Consortium [published online September 9, 2018]. bioRxiv. doi: 10.1101/412098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wendt FR, Carvalho C, Gelernter J, Polimanti R. DRD2 and FOXP2 are implicated in the associations between computerized device use and psychiatric disorders [published online December 17, 2018]. bioRxiv. doi: 10.1101/497420 [DOI] [Google Scholar]
- 15.Davey Smith G, Ebrahim S. Mendelian randomization: genetic variants as instruments for strengthening causal inference in observational studies In: Weinstein M, Vaupel JW, Wachter KW, eds. Biosocial Surveys. Washington, DC: The National Academies Press; 2008:428. [PubMed] [Google Scholar]
- 16.Lee JJ, Wedow R, Okbay A, et al. ; 23andMe Research Team; COGENT (Cognitive Genomics Consortium); Social Science Genetic Association Consortium . Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nat Genet. 2018;50(8):1112-1121. doi: 10.1038/s41588-018-0147-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okbay A, Beauchamp JP, Fontana MA, et al. ; LifeLines Cohort Study . Genome-wide association study identifies 74 loci associated with educational attainment. Nature. 2016;533(7604):539-542. doi: 10.1038/nature17671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Savage JE, Jansen PR, Stringer S, et al. Genome-wide association meta-analysis in 269,867 individuals identifies new genetic and functional links to intelligence. Nat Genet. 2018;50(7):912-919. doi: 10.1038/s41588-018-0152-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Selzam S, Krapohl E, von Stumm S, et al. Predicting educational achievement from DNA. Mol Psychiatry. 2017;22(2):267-272. doi: 10.1038/mp.2016.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anttila V, Bulik-Sullivan B, Finucane HK, et al. ; Brainstorm Consortium . Analysis of shared heritability in common disorders of the brain. Science. 2018;360(6395):eaap8757. doi: 10.1126/science.aap8757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nievergelt CM, Maihofer AX, Klengel T, et al. Largest genome-wide association study for PTSD identifies genetic risk loci in European and African ancestries and implicates novel biological pathways [published online November 1, 2018]. bioRxiv. doi: 10.1101/458562 [DOI] [Google Scholar]
- 22.Little J, Higgins JP, Ioannidis JP, et al. ; Strengthening the Reporting of Genetic Association Studies . Strengthening the Reporting of Genetic Association Studies (STREGA): an extension of the STROBE statement. PLoS Med. 2009;6(2):e22. doi: 10.1371/journal.pmed.1000022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neale Lab GWAS of the UK Biobank. http://www.nealelab.is/uk-biobank/. Accessed August 2, 2018.
- 24.Bulik-Sullivan B, Finucane HK, Anttila V, et al. ; ReproGen Consortium; Psychiatric Genomics Consortium; Genetic Consortium for Anorexia Nervosa of the Wellcome Trust Case Control Consortium 3 . An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47(11):1236-1241. doi: 10.1038/ng.3406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Auton A, Brooks LD, Durbin RM, et al. ; 1000 Genomes Project Consortium . A global reference for human genetic variation. Nature. 2015;526(7571):68-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Euesden J, Lewis CM, O’Reilly PF. PRSice: Polygenic Risk Score software. Bioinformatics. 2015;31(9):1466-1468. doi: 10.1093/bioinformatics/btu848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dastani Z, Hivert MF, Timpson N, et al. ; DIAGRAM+ Consortium; MAGIC Consortium; GLGC Investigators; MuTHER Consortium; DIAGRAM Consortium; GIANT Consortium; Global B Pgen Consortium; Procardis Consortium; MAGIC investigators; GLGC Consortium . Novel loci for adiponectin levels and their influence on type 2 diabetes and metabolic traits: a multi-ethnic meta-analysis of 45,891 individuals. PLoS Genet. 2012;8(3):e1002607. doi: 10.1371/journal.pgen.1002607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol. 2013;37(7):658-665. doi: 10.1002/gepi.21758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hemani G, Zheng J, Elsworth B, et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife. 2018;7:e4408. doi: 10.7554/eLife.34408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44(2):512-525. doi: 10.1093/ije/dyv080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hartwig FP, Davey Smith G, Bowden J. Robust inference in summary data mendelian randomization via the zero modal pleiotropy assumption. Int J Epidemiol. 2017;46(6):1985-1998. doi: 10.1093/ije/dyx102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent estimation in mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol. 2016;40(4):304-314. doi: 10.1002/gepi.21965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Choi KW, Chen CY, Stein MB, et al. ; Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium . Assessment of bidirectional relationships between physical activity and depression among adults: a 2-sample mendelian randomization study. JAMA Psychiatry. 2019;76(4):398-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gage SH, Jones HJ, Burgess S, et al. Assessing causality in associations between cannabis use and schizophrenia risk: a two-sample mendelian randomization study. Psychol Med. 2017;47(5):971-980. doi: 10.1017/S0033291716003172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hartwig FP, Borges MC, Horta BL, Bowden J, Davey Smith G. Inflammatory biomarkers and risk of schizophrenia: a 2-sample mendelian randomization study. JAMA Psychiatry. 2017;74(12):1226-1233. doi: 10.1001/jamapsychiatry.2017.3191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao Q, Wang J, Hemani G, Bowden J, Small DS. Statistical inference in two-sample summary-data mendelian randomization using robust adjusted profile score. arXiv. 2018. https://arxiv.org/abs/1801.09652. Accessed March 25, 2019.
- 37.Zhang Q, Yang C, Wang J, Small DS. Powerful genome-wide design and robust statistical inference in two-sample summary-data mendelian randomization. arXiv. 2018. https://arxiv.org/abs/1804.07371. Accessed March 25, 2019. [DOI] [PubMed]
- 38.Hemani G, Bowden J, Davey Smith G. Evaluating the potential role of pleiotropy in mendelian randomization studies. Hum Mol Genet. 2018;27(R2):R195-R208. doi: 10.1093/hmg/ddy163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jordan DM, Verbanck M, Do R. The landscape of pervasive horizontal pleiotropy in human genetic variation is driven by extreme polygenicity of human traits and diseases [published online April 30, 2018]. bioRxiv. doi: 10.1101/311332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Verbanck M, Chen CY, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from mendelian randomization between complex traits and diseases. Nat Genet. 2018;50(5):693-698. doi: 10.1038/s41588-018-0099-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Burgess S, Thompson SG. Multivariable mendelian randomization: the use of pleiotropic genetic variants to estimate causal effects. Am J Epidemiol. 2015;181(4):251-260. doi: 10.1093/aje/kwu283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sanderson E, Davey Smith G, Windmeijer F, Bowden J. An examination of multivariable mendelian randomization in the single-sample and two-sample summary data settings [published online December 10, 2018]. Int J Epidemiol. doi: 10.1093/ije/dyy262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yavorska OO, Burgess S. MendelianRandomization: an R package for performing mendelian randomization analyses using summarized data. Int J Epidemiol. 2017;46(6):1734-1739. doi: 10.1093/ije/dyx034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Battle A, Brown CD, Engelhardt BE, Montgomery SB; GTEx Consortium; Laboratory, Data Analysis &Coordinating Center (LDACC)—Analysis Working Group; Statistical Methods groups—Analysis Working Group; Enhancing GTEx (eGTEx) groups; NIH Common Fund; NIH/NCI; NIH/NHGRI; NIH/NIMH; NIH/NIDA; Biospecimen Collection Source Site—NDRI; Biospecimen Collection Source Site—RPCI; Biospecimen Core Resource—VARI; Brain Bank Repository—University of Miami Brain Endowment Bank; Leidos Biomedical—Project Management; ELSI Study; Genome Browser Data Integration &Visualization—EBI; Genome Browser Data Integration &Visualization—UCSC Genomics Institute, University of California Santa Cruz; Lead analysts; Laboratory, Data Analysis &Coordinating Center (LDACC); NIH program management; Biospecimen collection; Pathology; eQTL manuscript working group . Genetic effects on gene expression across human tissues. Nature. 2017;550(7675):204-213. doi: 10.1038/nature24277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Darmanis S, Sloan SA, Zhang Y, et al. A survey of human brain transcriptome diversity at the single cell level. Proc Natl Acad Sci U S A. 2015;112(23):7285-7290. doi: 10.1073/pnas.1507125112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.La Manno G, Gyllborg D, Codeluppi S, et al. Molecular diversity of midbrain development in mouse, human, and stem cells. Cell. 2016;167(2):566-580.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhong S, Zhang S, Fan X, et al. A single-cell RNA-seq survey of the developmental landscape of the human prefrontal cortex. Nature. 2018;555(7697):524-528. doi: 10.1038/nature25980 [DOI] [PubMed] [Google Scholar]
- 48.Habib N, Avraham-Davidi I, Basu A, et al. Massively parallel single-nucleus RNA-seq with DroNc-seq. Nat Methods. 2017;14(10):955-958. doi: 10.1038/nmeth.4407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol. 2015;11(4):e1004219. doi: 10.1371/journal.pcbi.1004219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun. 2017;8(1):1826. doi: 10.1038/s41467-017-01261-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hemani G, Tilling K, Davey Smith G. Orienting the causal relationship between imprecisely measured traits using GWAS summary data. PLoS Genet. 2017;13(11):e1007081. doi: 10.1371/journal.pgen.1007081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Robinson MR, Kleinman A, Graff M, et al. Genetic evidence of assortative mating in humans. Nature Human Behaviour. 2017;1(1). https://www.nature.com/articles/s41562-016-0016. Accessed April 1, 2019. [Google Scholar]
- 53.Betancourt TS, Newnham EA, Birman D, Lee R, Ellis BH, Layne CM. Comparing trauma exposure, mental health needs, and service utilization across clinical samples of refugee, immigrant, and US-origin children. J Trauma Stress. 2017;30(3):209-218. doi: 10.1002/jts.22186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.James LM, Strom TQ, Leskela J. Risk-taking behaviors and impulsivity among veterans with and without PTSD and mild TBI. Mil Med. 2014;179(4):357-363. doi: 10.7205/MILMED-D-13-00241 [DOI] [PubMed] [Google Scholar]
- 55.Lowe SR, Galea S, Uddin M, Koenen KC. Trajectories of posttraumatic stress among urban residents. Am J Community Psychol. 2014;53(1-2):159-172. doi: 10.1007/s10464-014-9634-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Abdellaoui A, Hugh-Jones D, Kemper KE, et al. Genetic consequences of social stratification in Great Britain [published online October 30, 2018]. bioRxiv. doi: 10.1101/457515 [DOI] [Google Scholar]
- 57.Kong A, Thorleifsson G, Frigge ML, et al. The nature of nurture: effects of parental genotypes. Science. 2018;359(6374):424-428. doi: 10.1126/science.aan6877 [DOI] [PubMed] [Google Scholar]
- 58.Trejo S, Domingue BW. Genetic nature or genetic nurture? quantifying bias in analyses using polygenic scores [published online January 18, 2019]. bioRxiv. doi: 10.1101/524850 [DOI] [Google Scholar]
- 59.Pietrzak RH, Feder A, Singh R, et al. Trajectories of PTSD risk and resilience in World Trade Center responders: an 8-year prospective cohort study. Psychol Med. 2014;44(1):205-219. doi: 10.1017/S0033291713000597 [DOI] [PubMed] [Google Scholar]
- 60.Martschenko D, Trejo S, Domingue BW. Genetics and education: recent developments in the context of an ugly history and an uncertain future. AERA Open. 2019;5(1). doi: 10.1177/2332858418810516 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eAppendix. Phenotype Definitions, Sample Overlap, Palindromic Variants, and Assortative Mating
eReferences.
eTable 1. Traumatic Experiences Assessed in the UK Biobank
eTable 2. Results of the Sensitivity Analyses Conducted With Respect to the MathClass→PTSD Test With and Without the Outlier Variants in the MathClass Genetic Instrument
eTable 3. Results of the IVW Analyses Considering Genetic Instruments With and Without Palindromic Variants With Ambiguous Allele Frequencies (PAL and noPAL, Respectively)
eTable 4. MR-RAPS Analysis Considering Various Adjustments Based on Genome-Wide Genetic Instruments
eTable 5. Results of the Sensitivity Analyses Conducted With Respect to the Income→PTSD and Risk-Tak→PTSD Tests
eTable 6. Genetic Correlation Among Trauma Experiences Assessed in the UK Biobank
eTable 7. Results (Causal Effects and Sensitivity Analyses) of the MR Test Conducted Using Trauma-Related Genetic Instruments With Respect to PTSD
eTable 8. Results of the Enrichment Analysis Based on Tissue-Specific and Cell Type–Specific Transcriptomic Data
eFigure 1. Schematic Workflow of the Analyses Conducted
eFigure 2. Genetic Correlations Estimated Between Traits Related to Cognitive Ability and 2 Versions of the Posttraumatic Stress Disorder Data Set, PGC-PTSD Freeze-2 (2) and PGC-PTSD Freeze-1.5 (1.5)
eFigure 3. Effect of the PTSD PRS on Educational Attainment (Yellow) and Cognitive Performance (Green) Considering Different Inclusion Thresholds
eFigure 4. Leave-1-Out Analysis Conducted With Respect to the MathClass→PTSD2 Result
eFigure 5. Identification of Potential Outliers (in Red) in MathClass Genetic Instrument Based on IVW Heterogeneity Test and MR-RAPS Standardized Residuals
eFigure 6. Results of the MathClass→PTSD2 Analysis After the Removal of the Potential Outliers From the Genetic Instrument
eFigure 7. Results of the Sensitivity Analyses With Respect to All MR Analyses Conducted
eFigure 8. Multivariable Mendelian Randomization Analysis Considering the Effects of EdAtt and MathClass on PTSD
eFigure 9. Effect of Income and RiskTak PRS (Green and Blue, Respectively) on PTSD Considering Different Inclusion Thresholds
eFigure 10. Genetic Correlations Between Traumatic Experiences and the Other Traits of Interest
eFigure 11. Effect of Trauma-Related PRS on PTSD Considering Different Inclusion Thresholds
eFigure 12. Multivariable Mendelian Randomization Analysis Considering the Effects of Physically Abused by Family as a Child and Belittlement by Partner or Ex-Partner as an Adult on PTSD