Abstract
Biocatalytic transformations employed by chemists are often restricted to simple functional group interconversions. In contrast, Nature has developed complexity-generating biocatalytic reactions within natural product pathways. These sophisticated catalysts are rarely employed by chemists as the substrate scope, selectivity and robustness of these catalysts are unknown. Our strategy to bridge the gap between the biosynthesis and synthetic chemistry communities leverages the diversity of catalysts available within natural product pathways. Starting from a suite of biosynthetic enzymes, catalysts with complementary substrate scope as well as selectivity can be identified. This strategy has been applied to the oxidative dearomatization of phenols, a chemical transformation that rapidly builds molecular complexity from simple starting materials and cannot be accomplished with high site- and enantioselectivity using existing catalytic methods. Using enzymes from biosynthetic pathways, we have successfully developed a method to produce ortho-quinol products with controlled site- and stereoselectivity. Further, we have capitalized on the scalability and robustness of this method in gram-scale reactions as well as multi-enzyme and chemoenzymatic cascades.
Graphical Abstract:

Oxidative dearomatization of phenolic compounds is a powerful transformation for the synthesis of complex molecules, providing an avenue for simultaneously introducing stereochemical information and generating products that are primed for further reaction.1 For example, chemical methods exist for the conversion of simple phenols to dearomatized products with concomitant formation of new C–C, C–N, C–halogen and C–O bonds.2 A number of reagents for dearomatization to afford ortho-quinol products (see 2, Fig. 1A),3–5 including IIII, IV, PbIV and CuI, have been developed and leveraged for the chemical synthesis of a range of bioactive natural products (Fig. 1B).1,4,6–8 However, in addition to the requirement of stoichiometric amounts of these reagents, two major challenges associated with these chemical methods are site-selectivity and product stability. For example, highly substituted resorcinol substrates such as 3 are chemically oxidized to afford mixtures of isomers such as 4, 5 and 6, which can fragment to afford quinone products (see 11, Fig. 1C).9 Furthermore, the desired quinol products are often difficult to use productively, as side reactions such as dimerization10 (7), rearomatization11,12 (8, 9 and 10) and rearrangement13 are facile under the requisite reaction conditions. Moreover, the development of asymmetric catalytic versions of these oxidative dearomatizations has proven challenging, particularly for cases involving concomitant C–O bond formation (1 to 2, Fig. 1A).14 While high enantioselectivities have been achieved with stoichiometric amounts of chiral hypervalent iodine reagents, superstoichiometric chiral metal complexes4 and in cases where an intramolecular cyclization is possible,15 a highly enantioselective catalytic method has yet to be reported.
Figure 1 ∣. Strategies for oxidative dearomatization of phenolic compounds and application in complex molecule synthesis.
a, Oxidative dearomatization of phenolic substrates to afford ortho-quinol products and small molecule reagents employed for this transformation. b, Natural products accessible from ortho-quinol intermediates. c, Potential products afforded by conditions for resorcinol oxidative dearomatization including potential product isomers, dimers, and undesired products, d, biocatalytic oxidative dearomatization leveraging the diversity of a suite of flavin-dependent monooxygenases from natural product biosynthetic pathways, protein structure shown is a homology model of TropB based on the structure of PDB ID 2DKH using the Phyre2 server.
The limitations of traditional methods to achieve site- and enantioselective oxidative dearomatization led us to consider utilizing the tools employed by Nature to effect this transformation. Oxidative dearomatization has been proposed as the chirality-generating step in a number of natural product biosynthetic pathways. While enzymes capable of the dearomatization of arenes through dihydroxylation or oxidative ring cleavage have been known for decades, the proteins responsible for oxidative dearomatization of phenols to generate ortho-quinol products, such as 2, have remained more elusive.16 Only recently have in vivo and in vitro studies identified enzymes responsible for mediating oxidative dearomatization of phenolic compounds to form ortho-quinol products in a handful of natural product pathways.17 We reasoned that these FAD-dependent monooxygenases represent ideal catalysts for oxidative dearomatization reactions, particularly since they require only molecular oxygen and a nicotinamide cofactor to enable catalysis under mild reaction conditions, with high site- and enantioselectivity.18
In order to harness the advantages offered by FAD-dependent monooxygenases, the challenges classically associated with biocatalysts, including limited substrate scope and the ability to achieve the desired selectivity, must be overcome.19 A common strategy in biocatalyst development involves: (1) first selecting a single enzyme; (2) evaluating its substrate scope, site- or enantioselectivity; and then (3) engineering the protein to operate on a different suite of compounds or with altered selectivity.20 Given the diversity that Nature has evolved in the context of natural product biosynthetic pathways, we hypothesized that greater synthetic utility could be accessed by profiling the substrate promiscuity and selectivity of a number of different FAD-dependent monooxygenases capable of catalyzing the oxidative dearomatization of phenol and resorcinol substrates.
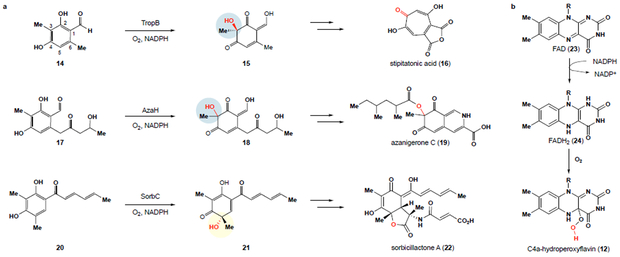
Our initial studies focused on a set of enzymes with orthogonal site- and stereoselectivities. We were guided by the work of the Cox,17,21 Tang22 and Watanabe23 groups, which have each biochemically characterized FAD-dependent monooxygenases that mediate the oxidative dearomatization of resorcinol substrates to ortho-quinol products that are further elaborated into various classes of natural products by downstream biosynthetic enzymes. Cox and coworkers first demonstrated this transformation through in vitro characterization of a FAD-dependent monooxygenase, TropB, included in the gene cluster that encodes for the fungal tropolone natural product stipitatonic acid (16). TropB catalyzes the site- and stereoselective oxidative dearomatization of resorcinol 14 to alcohol 15 (Fig. 2). Also in 2012, Tang identified a similar FAD-dependent monooxygenase from a silent Aspergillus niger ATCC 1015 gene cluster and demonstrated the role of this enzyme, AzaH, in the site-selective oxidative dearomatization of resorcinol substrate 17 to afford 18 with unknown configuration at the newly formed stereocenter. While TropB and AzaH operate with the same site-selectivity, a third enzyme in this class, SorbC, has evolved orthogonal site- and facial selectivity within the sorbicillactone A (22) pathway, oxidizing sorbicillin (20) to the C5-hydroxylation product 21.24
Figure 2 ∣. Nature’s tools for oxidative dearomatization of resorcinol compounds.
a, Secondary metabolite pathways containing FAD-dependent monooxygenases that mediate the oxidative dearomatization of resorcinol substrates with orthogonal selectivities including the biosynthetic pathways to stipitatonic acid (16), azanigerone C (19) and sorbicillactone A (22). b, Generation of C4a-hydroperoxyflavin (12) from FAD (23) through NADPH reduction of FAD (23) to FADH2 (24) and subsequent oxidation to 12.
We began by synthesizing the native substrates for TropB, AzaH and SorbC (14, 17, and 20, respectively), which were subsequently used as positive controls for activity of each FAD-dependent monooxygenase (see supporting information for details). In parallel, the three proteins were heterologously expressed in E. coli and purified using standard Ni-affinity chromatography. Analytical-scale in vitro reactions with each enzyme and the corresponding native substrate confirmed productive catalysis. In the case of TropB, we observed complete consumption of the starting material in one hour, which corresponds to 1,000 turnovers of the catalyst (Table 1, entry 1). Partial conversions were seen in the analogous assays for AzaH and SorbC, with total turnover numbers (TTNs) of 725 and 816, respectively (entries 17 and 29).
Table 1 ∣.
Promiscuity profile substrate scope of FAD-dependent monooxygenases TropB, AzaH and SorbC.
 |
Values given are total turnover numbers (TTNs) of each enzyme/substrate pair. Complete conversion of substrate to dearomatized product: TTN = 1000 (dark green). No observed conversion of starting material is represented by 0 (light grey). Reaction conditions: 2.5 mM substrate, 2.5 μM FAD-dependent monooxygenase, 1 mM NADP+, 5 mM glucose-6-phosphate (G6P), 1 U/mL glucose-6-phosphate dehydrogenase (G6PDH), 50 mM potassium phosphate buffer, pH 8.0, 30 °C, 1 h. TTN was assessed by quantifying the remaining substrate after 1 h by UPLC, where the area of the substrate peak was divided by the area of the internal standard and compared to a substrate standard curve prepared with the internal standard.
With activity on the natural substrates benchmarked, we next assessed the reactivity of each monooxygenase on a panel of compounds designed to probe the steric and electronic requirements of a substrate for productive catalysis. As shown in Table 1, each enzyme demonstrated a unique footprint of substrate scope. For example, TropB can tolerate a n-butyl ketone in place of the C1 aldehyde present in the native substrate (entries 2 and 8–10), but a benzoyl substituent was not compatible (see Supplementary Fig. 55). In contrast, SorbC exhibited the highest TTNs on ketone substrates with butyl or pentyl substituents that mirror the native SorbC substrate (20) (entries 8, 10, 26, 28–33). AzaH demonstrated the most flexibility in the carbonyl substituent, operating with TTNs exceeding 250 for a range of aldehyde and ketone substrates (entries 1, 4, 5, 11–13, 17–21, 27–28 and 30–33). Variation of the substitution pattern on the aryl ring was also tolerated. In the case of TropB, substrates lacking substituents at the C5 and C6 positions (entries 3 and 7) were converted to dearomatized products with TTNs approaching those observed with the native substrate, but AzaH and SorbC both required at least one substituent at either C5 or C6. The more sterically demanding biaryl and 6,7-bicyclic substrates were converted to dearomatized products by both TropB and AzaH (entries 11 and 6).
Since these transformations are anticipated to proceed through nucleophilic attack of the electron-rich phenol substrate onto the hydroperoxyflavin cofactor (12), we were interested in evaluating more electron-poor substrates with the three FAD-dependent monooxygenases. Interestingly, brominated substrates (entries 4 and 5) were dearomatized by TropB and AzaH. However, even the bromine-containing substrate that meets the steric preferences of SorbC showed no detectable conversion to oxidized product with SorbC, but was hydroxylated by AzaH (entry 21). AzaH was the only monooxygenase, of the three tested, with the ability to perform an oxidative dearomatization on even less electron-rich substrates such as a nitroresorcinol compound (entry 19) and monophenols (entries 20 and 22). These results prompted us to examine additional monophenol substrates (entries 23–25). AzaH catalyzed the oxidative dearomatization of brominated monophenols with low turn over numbers. Although the origin of this reactivity difference between AzaH and the other two monooxygenases is unclear, these results highlight the advantage of our approach of concurrently examining similar enzymes, which has allowed for the rapid identification of a promising general biocatalyst for the asymmetric transformation of phenols into ortho-quinols.
As a first step toward evaluating the selectivity and scalability of this oxidative dearomatization, substrates with the highest TTNs were selected for milligram-scale reactions. Based on the robust expression of these monooxygenases (>100 mg protein/1 L E. coli culture), purified protein was used in these reactions. An NADPH recycling system was employed to reduce cofactor cost.25 As shown in Table 2, these conditions led to efficient transformation of substrates to dearomatized products with conversions mirroring those observed for initial analytical-scale reactions. In some cases with excellent conversions of starting material to product, the isolated yields do not mirror these conversions. In these instances, the mass balance can be accounted for as the product dimer arising through [4+2] cycloaddition, which occurs upon concentration of the isolated product. Characterization of products from these reactions provided information on the site- and stereoselectivity of each enzyme. Operating on their natural substrates, both TropB and AzaH selectively hydroxylate the C3 position (see 15 and 36, Table 2). This site-selectivity is conserved across a panel of unnatural substrates (Tables 2A and B). Even in cases where the substrate bears a substituent at C5, the preference for C3 hydroxylation is maintained (see 28 and 35). In contrast, SorbC demonstrates orthogonal site-selectivity, exclusively hydroxylating at C5 across the library of phenols (Table 2C).
Table 2 ∣.
Isolation and characterization of ortho-quinol products from reactions with (A) TropB, (B) AzaH and (C) SorbC.
 |
Reaction conditions: 2.5 mM substrate, 2.5 μM FAD-dependent monooxygenase, 1 mM NADP+, 5 mM glucose-6-phosphate (G6P), 1 U/mL glucose-6-phosphate dehydrogenase (G6PDH), 50 mM potassium phosphate buffer, pH 8.0, 30 °C, 1 h.
With their native substrates, both TropB and SorbC deliver the expected product in >99% ee (Table 2). The configuration of the two stereocenters present in the native AzaH product 36 was previously unknown. Therefore, racemic 17 was synthesized and presented to AzaH. Both enantiomers of 17 were converted to azaphilone product 36 to afford a 1:1 mixture of diastereomers each formed in >99% ee. Without knowledge of the absolute configuration at C3 of azaphilone products from the aza gene cluster, we compared the products of TropB and AzaH with a common substrate 14. Both enzymes gave rise to the same stereoisomer, providing evidence that AzaH installs a hydroxyl group to generate (R)-C3 products. Analysis of TropB and AzaH reactions with a range of unnatural substrates indicates that a high-level of stereoselectivity is maintained with product ee’s typically >99%. These results can be compared favorably to the state-of-the-art copper-oxo-sparteine oxidant developed by Porco and coworkers, which, when used superstoichiometrically, delivers o-quinols from resorcinol precursors in up to 98% ee.4 SorbC displays altered facial selectivity producing (S)-C5 products (Table 2). Excellent stereoselectivity is observed in all cases examined, except when the C3 methyl group is replaced with an ethyl group or the carbonyl substituent is shortened to a butyl chain, eroding the stereoselectivity to deliver 40 and 43 in 94% and 95% ee, respectively. Notably, racemic 38 has been employed as a key intermediate in a number of natural product syntheses, however, to date no enantioselective method for accessing this class of o-quinols has been reported. The stereochemical outcome of the biotransformation is likely controlled by the pose the substrate adopts in the enzyme active site, wherein a specific face of the substrate is presented to the hydroperoxyflavin cofactor (12).
In contrast, isolation of racemic standards of these dearomatized products from traditional chemical methods employing IBX26 or Pb(OAc)427 proved challenging, often affording low yields of the desired product (listed in grey, Table 2), with the mass balance accounted for as isomers, quinol dimers arising through [4+2] cycloaddition, overoxidation and product decomposition pathways. Notably, a subset of the products generated enzymatically were not accessible using standard chemical methods, including nitroquinol 35 and 45, thus precluding the determination of the enantioselectivity of the enzymatic transformation by comparison to racemic material. Additionally, products 31-33 were observed as a mixture of tautomers (see 33 and 34) in a variety of solvents, which prohibited successful chiral separation and measurement of the enantiopurity of these compounds.
While enzymatic reactions are routinely carried out on sub-milligram quantities to characterize their biochemical function, the challenges associated with preparative-scale biocatalytic reactions can serve as a barrier to enzymes being embraced as tools for synthesis.19 Complications with enzyme supply, labor-intensive protein purification, substrate solubility in aqueous buffer, and cofactor expense can all be hurdles to preparative-scale biocatalysis. To move from milligram to gram-scale reactions and beyond, we envisioned transitioning from in vitro reactions, conducted with purified protein, to a scalable platform that would be readily accessible to synthetic chemists. Toward this goal, a series of experiments utilizing whole E. coli cells containing heterologously expressed protein in place of purified protein was undertaken. Whole cell reactions were sufficient for complete conversion of aldehyde 44 in 2 hours (Table 3, entry 1). Additionally, gram-scale reactions on a given substrate could be carried with whole cells to give comparable conversions as observed on analytical scale. To enhance the accessibility of this method to traditional synthetic labs, wet whole cells were freeze-dried and stored as a lyophilized powder, which could be weighed on the benchtop and directly used in reactions without suffering a significant loss in activity compared to experiments with wet whole cells (entries 3–6). A number of excipients commonly employed to aid in the long-term stabilization of proteins were screened.28 When lyophilized in the presence of 10 wt% sucrose or PEG 4 × 103, full activity of the enzyme was retained and the freeze-dried catalyst could be stored in the freezer at −80 °C for at least six months without any impact on reactivity.
Table 3 ∣.
Whole cell TropB reactions with wet cells versus freeze-dried cells.
 | |||
| entry | cell preparation | additive | % conversion |
|---|---|---|---|
| 1 | wet | none | >99% |
| 2 | wet | none | >99%a |
| 3 | lyophilized | none | 98% |
| 4 | lyophilized | 10 wt. % skim milk | 83% |
| 5 | lyophilized | 10 wt. % sucrose | >99% |
| 6 | lyophilized | PEG 4 × 103 | >99% |
Reaction conditions: 2.5 mM substrate, 10 mg wet cell mass/mL, 1 mM NADP+, 5mM glucose-6-phosphate (G6P), 1 U/mL glucose-6-phosphate dehydrogenase (G6PDH), 50 mM potassium phosphate buffer, pH 8.0, 30 °C, 2 h. aReaction on 1 g 44.
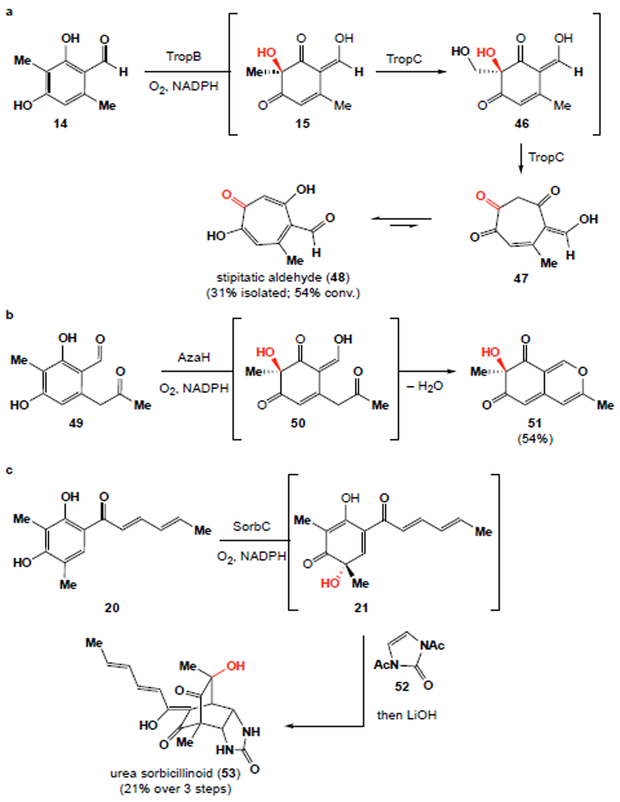
To rapidly build molecular complexity from enzymatically generated ortho-quinols, we sought to leverage the reactivity of these compounds in cascade reactions by performing subsequent in situ enzymatic or chemical transformations. We anticipated that various natural product scaffolds could be accessed in one-pot using biocatalytic dearomatization-initiated cascades. Toward this goal, we explored the synthesis of the tropolone (see 48), azaphilone (see 51) and sorbicillinoid (see 53)29 cores (Fig. 3). To access the tropolone natural product stipitatic aldehyde (48), TropB dearomatization of aldehyde 14 produced ortho-quinol 15. Next, 15 was further hydroxylated by an alpha-ketoglutarate dependent non-heme iron enzyme, TropC, to afford 1,2-diol 46. Diol 46 readily underwent ring expansion in the presence of TropC to afford seven-membered ring 47 which tautomerized to afford the tropolone, stipitatic aldehyde (48). Additionally, the first asymmetric synthesis of azaphilone natural product 5130 was achieved from methyl ketone 49. Initial AzaH-mediated dearomatization delivered enol 50 which spontaneously cyclized to the natural azaphilone 51, which could be isolated in 54% yield and >99% ee. Finally, urea sorbicillinoid 53 was generated directly from ketone 20 in a cascade that commenced with SorbC oxidation to deliver sorbicilinol (21). The addition of bisacylated urea 52 led to facile [4+2] cycloaddition. Addition of LiOH to the reaction completed the first synthesis of urea sorbicillinoid natural product 53 in 21% yield over three steps.
Figure 3 ∣. One-pot cascades featuring biocatalytic oxidative dearomatization to access natural products.
a, Stipitatic aldehyde (48) synthesis from aldehyde 14 through a two-enzyme cascade. b, Synthesis of natural azaphilone 51 from methyl ketone 49, c, and the first synthesis of the natural product, urea sorbicillinoid 53 through a chemoenzymatic sequence initiated by the SorbC-catalyzed oxidative dearomatization of 20.
In sum, the results disclosed herein demonstrate the potential that FAD-dependent monooxgenases offer for site- and stereoselective oxidative dearomatization. The structural diversity available from various natural product pathways has provided a robust platform for developing a suite of biocatalysts with orthogonal selectivity and complementary substrate scope. The catalytic efficiency and exquisite stereoselectivity of these catalysts paired with the mild reaction conditions provide an excellent opportunity to apply these biocatalysts in complexity generating chemoenzymatic and multi-enzyme reaction cascades. This work provides an initial data set that will fuel the engineering of these catalysts and exploration of alternative reaction pathways.
Methods
Analytical-scale reactions:
Analytical-scale reactions were performed on 50 μL scale. Each reaction contained 25 μL 100 mM potassium phosphate buffer, pH 8.0, 2.5 mM substrate (2.5 μL of a 50 mM stock solution in DMSO), 2.5 μM FAD-dependent monooxygenase, 5 mM G6P (0.5 μL, 500 mM), 1 mM NADP+ (0.5 μL, 100 mM), 1 U/mL G6P-DH (0.5 μL, 100 U/mL), and Milli-Q water to a final volume of 50 μL. The reaction was carried out at 30 °C for 1 h and quenched by addition of 75 µL acetonitrile with 25 mM pentamethylbenzene as an internal standard. Precipitated biomolecules were pelleted by centrifugation (16,000 x g, 12 min). The supernatant was analyzed by UPLC-DAD and conversion obtained by comparison to calibration curves of the substrate.
Whole cell preparative-scale reactions:
Whole-cell enzymatic reactions were conducted on 1 g of substrate under the following conditions: 20 weight equivalents of wet cell pellet, 5 mM substrate, 10% (v/v) toluene, 0.1 mM NADP+, 0.1 U/mL G6PDH, and 10 mM G6P for NADPH regeneration in reaction buffer (50 mM potassium phosphate buffer, pH 8.0). The reaction mixture was added to a 1 L Erlenmeyer flask and incubated at 30 °C with shaking at 100 rpm. After 2 h, the reaction mixture was filtered through Celite, acidified to pH 2.0 and extracted with EtOAc (3 × 500 mL). The combined organic layers were dried over sodium sulfate, filtered and concentrated under reduced pressure. The resulting mixture was purified on silica gel (MeOH/AcOH/DCM, 1:1:10) to afford the o-quinol product.
Data availability.
Data supporting the findings of this study are available in the corresponding Supplementary Information file, or from the corresponding author upon request. The Supplementary Information file contains full details on the synthesis and characterization of compounds as well as the expression and purification of proteins employed in this work.
Supplementary Material
Acknowledgements:
This work was supported by funds from the University of Michigan Life Sciences Institute and Department of Chemistry. We are grateful to Prof. Yi Tang from the University of California Los Angles for providing a plasmid containing azaH. S.A.B.D. thanks the National Institutes of Health Chemistry Biology Interface Training Grant (T32 GM008597). A.L.L. acknowledges Graduate Assistance of Areas in National Need (GAANN P200A150164) for funding. We thank Carolyn Suh and Jianxin Liu for assistance in the synthesis of substrates.
Footnotes
Competing financial interests: The authors declare no competing financial interests.
References
- 1.Roche SP & Porco JA Dearomatization Strategies in the Synthesis of Complex Natural Products. Angew. Chem.-Int. Edit 50, 4068–4093, doi: 10.1002/anie.201006017 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu WT, Zhang LM & You SL Catalytic asymmetric dearomatization (CADA) reactions of phenol and aniline derivatives. Chem. Soc. Rev 45, 1570–1580, doi: 10.1039/c5cs00356c (2016). [DOI] [PubMed] [Google Scholar]
- 3.Volp KA & Harned AM Chiral aryl iodide catalysts for the enantioselective synthesis of para-quinols. Chem. Commun 49, 3001–3003, doi: 10.1039/c3cc00013c (2013). [DOI] [PubMed] [Google Scholar]
- 4.Zhu JL, Grigoriadis NP, Lee JP & Porco JA Synthesis of the azaphilones using copper-mediated enantioselective oxidative dearomatization. Journal of the American Chemical Society 127, 9342–9343, doi: 10.1021/ja052049g (2005). [DOI] [PubMed] [Google Scholar]
- 5.Bosset C et al. Asymmetric Hydroxylative Phenol Dearomatization Promoted by Chiral Binaphthylic and Biphenylic Iodanes. Angew. Chem.-Int. Edit 53, 9860–9864, doi: 10.1002/anie.201403571 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Wang WX et al. Antibacterial Azaphilones from an Endophytic Fungus, Colletotrichum sp BS4. J. Nat. Prod 79, 704–710, doi: 10.1021/acs.jnatprod.5b00436 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Yang QL et al. Evolution of an oxidative dearomatization enabled total synthesis of vinigrol. Org. Biomol. Chem 12, 330–344, doi: 10.1039/c3ob42191k (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shiao HY, Hsieh HP & Liao CC First total syntheses of (+/−)-annuionone B and (+/−)-tanarifuranonol. Org. Lett 10, 449–452, doi: 10.1021/ol7028178 (2008). [DOI] [PubMed] [Google Scholar]
- 9.Nicolaou KC et al. Biomimetic total synthesis of bisorbicillinol, bisorbibutenolide, trichodimerol, and designed analogues of the bisorbicillinoids. J. Am. Chem. Soc 122, 3071–3079, doi: 10.1021/ja9942843 (2000). [DOI] [PubMed] [Google Scholar]
- 10.Pettus LH, Van de Water RW & Pettus TRR Synthesis of (+/−)-epoxysorbicillinol using a novel cyclohexa-2,5-dienone with synthetic applications to other sorbicillin derivatives. Org. Lett 3, 905–908, doi: 10.1021/ol0155438 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morrow GW & Schwind B SYNTHESES OF PARA-TERPHENYL VIA REDUCTIVE DEOXYGENATION OF QUINOL DERIVATIVES. Synth. Commun 25, 269–276, doi: 10.1080/00397919508010816 (1995). [DOI] [Google Scholar]
- 12.Schultz AG & Antoulinakis EG Photochemical and acid-catalyzed rearrangements of 4-carbomethoxy-4-methyl-3-(trimethylsilyl)-2,5-cyclohexadien-1-one. J. Org. Chem 61, 4555–4559, doi: 10.1021/jo952240g (1996). [DOI] [PubMed] [Google Scholar]
- 13.Dong SW, Zhu JL & Porco JA Enantioselective synthesis of bicyclo 2.2.2 octenones using a copper-mediated oxidative dearomatization/ 4+2 dimerization cascade. J. Am. Chem. Soc 130, 2738–2739, doi: 10.1021/ja711018z (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun WS, Li GF, Hong L & Wang R Asymmetric dearomatization of phenols. Org. Biomol. Chem 14, 2164–2176, doi: 10.1039/c5ob02526e (2016). [DOI] [PubMed] [Google Scholar]
- 15.Uyanik M, Yasui T & Ishihara K Hydrogen Bonding and Alcohol Effects in Asymmetric Hypervalent Iodine Catalysis: Enantioselective Oxidative Dearomatization of Phenols. Angew. Chem.-Int. Edit 52, 9215–9218, doi: 10.1002/anie.201303559 (2013). [DOI] [PubMed] [Google Scholar]
- 16.Ullrich R & Hofrichter M Enzymatic hydroxylation of aromatic compounds. Cell. Mol. Life Sci 64, 271–293, doi: 10.1007/s00018-007-6362-1 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davison J et al. Genetic, molecular, and biochemical basis of fungal tropolone biosynthesis. Proc. Natl. Acad. Sci. U. S. A 109, 7642–7647, doi: 10.1073/pnas.1201469109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Berkel WJH, Kamerbeek NM & Fraaije MW Flavoprotein monooxygenases, a diverse class of oxidative biocatalysts. J. Biotechnol 124, 670–689, doi: 10.1016/j.jbiotec.2006.03.044 (2006). [DOI] [PubMed] [Google Scholar]
- 19.Khalil AS & Collins JJ Synthetic biology: applications come of age. Nat. Rev. Genet 11, 367–379, doi: 10.1038/nrg2775 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Turner NJ Directed evolution drives the next generation of biocatalysts. Nat. Chem. Biol 5, 568–574, doi: 10.1038/nchembio.203 (2009). [DOI] [PubMed] [Google Scholar]
- 21.Abood A et al. Kinetic characterisation of the FAD dependent monooxygenase TropB and investigation of its biotransformation potential. RSC Adv. 5, 49987–49995, doi: 10.1039/c5ra06693j (2015). [DOI] [Google Scholar]
- 22.Zabala AO, Xu W, Chooi YH & Tang Y Characterization of a Silent Azaphilone Gene Cluster from Aspergillus niger ATCC 1015 Reveals a Hydroxylation-Mediated Pyran-Ring Formation. Chem. Biol 19, 1049–1059, doi: 10.1016/j.chembiol.2012.07.004 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sato M et al. Combinatorial Generation of Chemical Diversity by Redox Enzymes in Chaetoviridin Biosynthesis. Org. Lett 18, 1446–1449, doi: 10.1021/acs.orglett.6b00380 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.al Fahad A et al. Oxidative dearomatisation: the key step of sorbicillinoid biosynthesis. Chem. Sci 5, 523–527, doi: 10.1039/c3sc52911h (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chenault HK & Whitesides GM REGENERATION OF NICOTINAMIDE COFACTORS FOR USE IN ORGANIC-SYNTHESIS. Appl. Biochem. Biotechnol 14, 147–197, doi: 10.1007/bf02798431 (1987). [DOI] [PubMed] [Google Scholar]
- 26.Zhu JL, Germain AR & Porco JA Synthesis of azaphilones and related molecules by employing cycloisomerization of o-alkynylbenzaldehydes. Angew. Chem.-Int. Edit 43, 1239–1243, doi: 10.1002/anie.200353037 (2004). [DOI] [PubMed] [Google Scholar]
- 27.Barnes-Seeman D & Corey EJ A two-step total synthesis of the natural pentacycle trichodimerol, a novel inhibitor of TNF-alpha production. Org. Lett 1, 1503–1504, doi: 10.1021/ol991070h (1999). [DOI] [PubMed] [Google Scholar]
- 28.Wessman P, Hakansson S, Leifer K & Rubino S Formulations for Freeze-drying of Bacteria and Their Influence on Cell Survival. J. Vis. Exp, 5, doi: 10.3791/4058 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cabrera GM et al. A sorbicillinoid urea from an intertidal Paecilomyces marquandii. J. Nat. Prod 69, 1806–1808, doi: 10.1021/np060315d (2006). [DOI] [PubMed] [Google Scholar]
- 30.Zhang SP et al. Antiviral anthraquinones and azaphilones produced by an endophytic fungus Nigrospora sp from Aconitum carmichaeli. Fitoterapia 112, 85–89, doi: 10.1016/j.fitote.2016.05.013 (2016). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data supporting the findings of this study are available in the corresponding Supplementary Information file, or from the corresponding author upon request. The Supplementary Information file contains full details on the synthesis and characterization of compounds as well as the expression and purification of proteins employed in this work.