Abstract
Intermittent fasting (IF) increases lifespan and decreases metabolic disease phenotypes and cancer risk in model organisms, but the health benefits of IF in humans are less clear. Human plasma derived from clinical trials is one of the most difficult sample sets to analyze using mass spectrometry-based proteomics due to the extensive sample preparation required and the need to process many samples to achieve statistical significance. Here, we describe an optimized and accessible device (Spin96) to accommodate up to 96 StageTips, a widely used sample preparation medium enabling efficient and consistent processing of samples prior to LC–MS/MS. We have applied this device to the analysis of human plasma from a clinical trial of IF. In this longitudinal study employing 8-weeks IF, we identified significant abundance differences induced by the IF intervention, including increased apolipoprotein A4 (APOA4) and decreased apolipoprotein C2 (APOC2) and C3 (APOC3). These changes correlated with a significant decrease in plasma triglycerides after the IF intervention. Given that these proteins have a role in regulating apolipoprotein particle metabolism, we propose that IF had a positive effect on lipid metabolism through modulation of HDL particle size and function. In addition, we applied a novel human protein variant database to detect common protein variants across the participants. We show that consistent detection of clinically relevant peptides derived from both alleles of many proteins is possible, including some that are associated with human metabolic phenotypes. Together, these findings illustrate the power of accessible workflows for proteomics analysis of clinical samples to yield significant biological insight.
Keywords: human, plasma, intermittent fasting, 96-well, sample cleanup, solid-phase extraction (SPE), liquid chromatography, mass spectrometry (MS), 3D-printing
Introduction
Intermittent fasting (IF) is a dietary modification that generally consists of alternating periods where food is freely available (ad libitum), followed by fasting periods of up to 24 h, for 1–4 days/week. The fasting period that is most often tested in animal models is 24 h. IF initiated in young adult and middle-aged chow-fed mice increases maximal lifespan.1−3 Anson and colleagues4 re-stimulated interest in this model when they observed that alternating periods of 24 h fasting and ad libitum feeding reduced fasting glucose and insulin levels as effectively as 60% daily calorie restriction, even though there was a minimal reduction in either caloric intake, or body weight, versus pair fed controls. This work suggests that intermittent energy deprivation is sufficient to improve metabolic health.5 Intermittent fasting in model animals generally produces little or no weight loss, but clearly reduces fasting glucose and insulin,6,7 reduces blood pressure,7,8 improves cardiac remodeling, increases left ventricle ejection fraction response to myocardial infarction,8 reduces cell proliferation,9 increases cancer survival,10 increases adiponectin and lipid oxidation in muscle and liver and reduces visceral fat.11 However, far fewer studies are reported for IF interventions in humans.
Mass spectrometry (MS)-based proteomics has become a mainstay technology for elucidating the biological functions of proteins on a large scale.12 In preparing samples for peptide-level proteomic analysis (bottom-up proteomics), it is often necessary to concentrate peptides and remove contaminants.13 One of the most commonly used methods in MS-based proteomics sample preparation is offline reversed-phase solid phase extraction (SPE),14−16 in which peptides are bound to a stationary phase, typically either C18, or a mixed-mode resin, enabling simultaneous desalting, contaminant removal, and concentration. Single-use StageTips are a widely used implementation of SPE, as their small void volumes enable processing of microgram amounts of proteins.14,17 StageTips can also accommodate a variety of stationary phases, expanding their application to peptide fractionation, or to increase flexibility in washing steps. For example, a mixed mode strong cation exchange (SCX) and reversed-phase material may be used, which enables either the removal of contaminants such as lipids from plasma-derived samples, or the fractionation of peptides.16−19 Additionally, the StageTip format is often incorporated into other proteomics workflows, such as in the “in-StageTip” protocol in which samples are lysed and digested directly within StageTips,17 or in the case of phosphoproteomics workflows to capture and retain beads used for peptide enrichment.20−23
Here we describe a device (Spin96) that can be rapidly produced using widely available 3D-printers designed to accommodate up to 96 StageTips simultaneously. We have applied our Spin96 device to the analysis of human plasma from a longitudinal clinical trial of IF. Our intervention employed 8-weeks of IF with plasma samples and physiological data collected before and after this period. Using established StageTip methods and our device, we have identified significant differences in plasma protein abundance induced by the IF intervention, particularly in apolipoproteins. In addition, we have applied a protein variant database to detect clinically relevant protein variants across the participants. These data are the first unbiased analysis of plasma proteome changes induced during IF. The complete design files for the Spin96 are made freely available to the scientific community, enabling local production of the device.
Experimental Procedures
Chemicals and Reagents
Acetonitrile (Optima grade), acetone, water (Optima grade), ammonia, formic acid, and isopropanol (Optima grade) were from Thermo Fisher Scientific (Massachusetts, USA). Ethyl acetate LC–MS grade was from Millipore (Massachusetts, USA). CBQCA reagent was from Applied Bioprobes (Maryland, USA). Proteomics-grade trypsin (Catalogue number T6567) and all other reagents were from Sigma-Aldrich (Missouri, USA).
Intermittent Fasting without Weight Loss Clinical Trial
Between 1 March 2013 and 4 September 2015, 119 women were screened following advertisement in local newspapers and media to participate in this single-center, randomized controlled trial in Adelaide, South Australia. The Royal Adelaide Hospital Research Ethics Committee approved the study protocol, and all participants provided written, informed consent prior to their inclusion. The PREFER study24 was registered with Clinicaltrials.gov (NCT01769976). A total of 88 women were enrolled in the study, of which 25 were assigned to the intermittent fasting with weight maintenance group (IF100) analyzed in this manuscript; 3 participants withdrew during the diet period (2 due to time, 1 no longer wished to participate). The resulting 44 paired plasma samples from 22 subjects in the IF100 group were the subject of this proteomic analysis. Inclusion criteria were: aged 35–70 years; BMI 25–42 kg/m2; weight-stable (within 5% of their screening weight) for >6 months prior to study entry; no diagnosis of type 1 or type 2 diabetes; nonsmoker; sedentary or lightly active (i.e., <2 moderate to high-intensity exercise sessions per week); consumed <140 g alcohol/week; no personal history of cardiovascular disease, no diagnosis of eating disorders or major psychiatric disorders (including those taking antidepressants); not pregnant or breastfeeding; and not taking medication that may affect study outcomes (e.g., phentermine, orlistat, metformin, excluding antihypertensive/lipid lowering medication). The primary outcome of this study was insulin sensitivity, assessed by hyperinsulinemic-euglycemic clamp. Secondary outcomes were plasma markers of blood glucose control, body fat and anthropometric measures, adherence, hunger and appetite, weight, plasma markers of cardiovascular risk, and hepatic biomarkers.
The active trial period was 10 weeks, comprised of a 2-week lead-in period, and 8 weeks of dietary intervention. During the lead-in period, participants consumed their normal diet and maintained their weight. Following this, participants were placed on an intermittent fasting diet at 100% of calculated baseline energy requirements per week (i.e., weight maintenance). Energy requirements were calculated using an average of published equations, both of which use age, gender, height, and weight variables.25,26 Due to the nature of the intervention, blinding was not possible.
Diet
On fed days, participants were provided with food equal to ∼145% of energy requirements. On fasting days, participants consumed breakfast before 8 am (∼37% of energy requirements were given at breakfast on fasting days) and were then instructed to “fast” for 24 h, until 8 am the following day. Participants were advised to fast on 3 nonconsecutive weekdays per week. During the fasting period participants were allowed to consume water and limited amounts of energy-free foods (e.g., “diet” drinks, chewing gum, mints), black coffee and/or tea, and were provided with 250 mL of a very low energy broth (86 kJ/250 mL, 2.0 g protein, 0.1 g fat, 3.0 g carbohydrate) for either lunch, or dinner. Participants were free-living, and foods were provided by fortnightly delivery to their home, except for fresh fruits and vegetables. Portions of fruits and vegetables were standardized and participants allowed to self-select according to the number of servings specified in their individual menus (∼10% overall energy intake).
Adherence
All participants completed daily diet checklists to monitor dietary adherence. Energy intake in weeks 1, 4, and 7 was calculated from 7-day food diaries using FoodWorks (version 8, Xyris Software). Participants also attended weekly check-ins at the clinic, where they returned the 7-day diet checklists from the previous week, were weighed and received individual counselling to maintain compliance.
Procedures for Metabolic Testing
To minimize the influence of the menstrual cycle on outcomes, premenopausal women were studied in the follicular phase. Participants were provided with a standardized diet (100% of calculated energy requirements, 35% fat, 15% protein, 50% carbohydrate) for 3 days prior to the first metabolic testing visit (“baseline”). Participants were instructed to avoid exercise, alcohol, and caffeine for 24 h and fast for 12 h overnight, prior to arrival at both the baseline “Before IF” visit and the “After IF” visit (i.e., after the 8 week IF intervention). Testing days began at 0730 h, participants were weighed in a hospital gown after voiding, waist and hip measurements taken, and blood pressure measured with the participant in a seated position after 10 min rest. Intravenous cannulae were placed, baseline samples collected and a primed 120 min hyperinsulinemic-euglycemic (60 mU/m2/min) clamp commenced according to previously described methods.27 Peripheral insulin sensitivity was calculated as the mean exogenous glucose infusion rate during steady-state (last 30 min). We adjusted for the estimated size of the metabolizing fat free mass (FFM; FFM + 17.7) as described by others (μmol/min/mU insulin/kg FFM + 17.7).28 Due to scheduling conflicts or a technical issue arising on the day of the clamp, 3 clamps were not conducted. Homeostatic model assessment of insulin resistance (HOMA-IR) was calculated as: (fasting serum insulin (mU/L) × fasting plasma glucose (mmol/L) × 22.5). Total body composition was assessed by DXA (Lunar Prodigy; GE Healthcare, NSW, Australia).
Blood Collection and Analysis
Blood samples were collected directly into purple K2-EDTA vacutainers (Becton Dickinson) and placed on ice immediately after collection. Samples were centrifuged within 10 min of collection at 4 °C and the plasma frozen at −80 °C in cryotubes. Blood lipids and fasting blood glucose were examined by photometric assays in the laboratory of SA Pathology (Adelaide, South Australia, Australia). Serum insulin was measured by radioimmunoassay (HI-14K, Millipore, MA, USA). Serum nonesterified fatty acids (NEFA) were measured by enzymatic colorimetric assay (NEFA-HR (2), Wako Diagnostics, CA, USA). Plasma β-hydroxybutyrate (RANBUT D-3 Hydroxybutyrate kit; Randox, Antrim, UK) and high-sensitivity c-reactive protein (HS-CRP) were measured using commercially available enzymatic kits (Beckman Coulter Inc., CA, USA) on a Beckman AU480 clinical analyzer (Beckman Coulter Inc.). Samples from each participant were analyzed within the same run to reduce instrument variation, and these measures were used as some of the variables for the unbiased correlation analysis.
Spin96 Design and 3D-Printing
The Spin96 components (Figure S1) were designed in SolidWorks (version 2017) and exported as STL files for slicing in either Z-suite, or Simplify3D v4.0.1 with a layer height of 0.14 mm. Sliced models were printed using Z-HIPS Natural White filament on either a Zortrax M200, or an Intamsys FUNMAT HT printer. Printing on the Zortrax M200 was performed according to the high-quality HIPS profile with either maximum infill (used for the holders, wash-bottom and bottom components), or low infill (used for the top component). Printing of the Intamsys FUNMAT HT used the following settings (extruder temperature 255 °C, build-plate temperature 80 °C, chamber temperature 40 °C, fan speed 20%, layer height 0.14 mm, print speed 3600 mm/min, retraction distance 0.5 mm, retraction speed 1800 mm/min, infill percentage 95%, use support with maximum overhang angle of 45° and use raft with 5 mm offset). Exported STL models and the original SolidWorks SLDPRT files are available as Supporting Information (File S1). Two different designs of the bottom component were generated to accommodate either trimmed unskirted 96-well PCR plates (File S1), or PCR tubes (File S1).
Plasma Sample Preparation and Cleanup Using SDB-RPS StageTips
Plasma sample preparation was performed according the protocol in the Supporting Methods, which was adapted from 3 previous studies.17−19 All steps in this protocol were completed without the aid of any robotics. Briefly, 1 μL of plasma (60–70 μg protein) was added to 24 μL of SDC buffer (1% sodium deoxycholate, 10 mM TCEP, 40 mM chloroacetamide, and 100 mM Tris-HCl pH 8.5) in either a 500 μL 96-well Protein Lo-bind plate, or Protein Lo-bind 1.5 mL tubes (Eppendorf). After sealing the tube or plate (with a silicone mat, Eppendorf), samples were heated to 95 °C for 10 min at 1,000 rpm in an Eppendorf Thermomixer-C with a ThermoTop (heated lid) to denature, reduce, and alkylate proteins. Once cooled to RT and diluted 10-fold with water, LysC and Trypsin were added (from 1 mg/mL stock solutions in either water, or 50 mM acetic acid, respectively) to digest proteins at 1:100 ratio (each protease:protein, μg/μg) and digested at 37 °C for 16 h at 1000 rpm in an Eppendorf Thermomixer-C with a ThermoTop (heated lid). An equal volume (250 μL) of 99% ethyl acetate/1% TFA was added to the digested peptides for a final concentration of 49.5% ethyl acetate and 0.5% TFA.
SDB-RPS StageTips were generated by punching double-stacked SDB-RPS discs (Sigma, Cat # 66886-U) with an 18-gauge needle and mounted in 200 μL tips (Eppendorf), as shown in Supporting Methods. For StageTip SPE processing using the Spin96, StageTips were inserted into a holder and placed in the top, which was then stacked onto the wash-bottom containing a polypropylene 96-well microtiter plate to collect the sample flow-through and washes (Figure S1a). Each tip was wetted with 100 μL of 100% acetonitrile and centrifuged at 1000g for 1 min. Following wetting, each StageTip was equilibrated with 100 μL of 0.1% TFA in H2O and 30% methanol/1% TFA with centrifugation for each at 1000g for 3 min. Each StageTip was then loaded with the equivalent of ∼10 μg peptide in 49.5% ethyl acetate and 0.5% TFA (equal volumes of each phase used). The peptides were washed twice with 100 μL of 99% ethyl acetate/1% TFA, which was followed by one wash with 100 μL of 0.2% TFA in water. For elution of peptides, the wash-bottom was exchanged with a bottom containing a holder supporting an unskirted PCR plate that has been trimmed to fit (Figure S1b). To elute, 100 μL of 5% ammonium hydroxide/80% acetonitrile was added to each tip and centrifuged as above for 5 min. Samples in the PCR plate were dried using a GeneVac EZ-2 using the ammonia setting at 40 °C for 1 h. Dried peptides were resuspended in 30 μL of 5% formic acid and a CBQCA assay (see below) was performed to ensure even sample loading. Samples were then stored at 4 °C until analyzed by LC–MS.
CBQCA Peptide Assay
The concentration of peptide in each sample after resuspension in 5% formic acid was determined by CBQCA assay (Life Technologies, Cat # C6667). The assay was performed as per manufacturer’s instructions with the following modifications. Standards consisting of trypsin digested BSA peptides were prepared in borate buffer (0.1 M H3BO3–NaOH pH 9.3) from 655,360 ng/mL to 40 ng/mL in a 4-fold dilution series. Samples in 5% formic acid were diluted 25-fold in borate buffer, borate buffer blanks and standards were added to separate wells of a black flat-bottom 96-well microtiter plate with a final volume of 25 μL per well. To each well, 95 μL of 1 mM KCN in borate buffer was added, followed by 30 μL of 0.66 mM CBQCA reagent in borate buffer. The plate was incubated in the dark at RT for 1 h and the fluorescence detected by excitation at 465 nm and emission at 550 nm.
High pH Reversed Phase Chromatography
Pooled trypsin digested plasma peptides (100 μg total) in 5% formic acid were subjected to high pH reversed phase chromatography on a Thermo Scientific Dionex Ultimate 3000 BioRS system with a fractionation autosampler, using a Waters XBridge Peptide BEH C18 column (130 Å, 3.5 μm, 4.6 mm × 250 mm, Cat No. 186003570). The column was incubated at 30 °C with a constant flow rate of 1 mL/min, with buffer A containing 2% acetonitrile (ACN) and 10 mM ammonium formate (pH 9.0) and buffer B containing 80% ACN and 10 mM ammonium formate (pH 9.0). Fractions were collected every 8.75 s from a retention time of 2 to 16 min (96 pseudo fractions, concatenated into 16 fractions total). Peptides were separated by a linear gradient from 10% to 40% buffer B for the first 11 min and 100% buffer B for the remaining time. The fractions were collected in a 2 mL protein Lo-bind 96-well deepwell plate (Eppendorf) across 16 wells in a concatenated pattern using tube wrapping.
LC–MS/MS and Analysis of Spectra
Using a Thermo Easy-nLC 1200 nanoUHPLC, peptides in 5% (v/v) formic acid (injection volume 3 μL) were directly injected onto a 45 cm × 75 μm C18AQ (Dr. Maisch, Ammerbuch, Germany, 1.9 μm) fused silica analytical column with a ∼10 μm pulled tip, coupled online to a nanospray ESI source. Peptides were resolved over gradient from 5% acetonitrile to 40% acetonitrile over 45 min with a flow rate of 300 nL min–1 at 60 °C. Peptides were ionized by electrospray ionization at 2.3 kV. Tandem mass spectrometry analysis was carried out on a Q-Exactive HF mass spectrometer (ThermoFisher) using HCD fragmentation in positive mode. The data-dependent acquisition method used acquired MS/MS spectra of the top 10 most abundant ions at any one point during the gradient. MS1 scans were acquired from 350–1400 m/z (60 000 resolution, 3 × 106 AGC target, 50 ms maximum injection time) and MS2 scans having a fixed first m/z of 140 (15 000 resolution, 1 × 105 AGC target, 25 ms maximum injection time, 27 NCE, 1.4 m/z isolation width). RAW MS data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the data set identifier PXD009324. RAW data were analyzed using the quantitative proteomics software MaxQuant29 (http://www.maxquant.org, version 1.5.7.0), and the MaxQuant output has also been uploaded to the ProteomeXchange Consortium under the same identifier. This version of MaxQuant includes an integrated search engine, Andromeda.30 Peptide and protein level identification were both set to a false discovery rate of 1% using a target-decoy based strategy, and proteins were filtered such that they must have >2 razor and unique peptides. The database supplied to the search engine for peptide identifications contained both the human UniProt database downloaded on the 30th September 2017, containing 42 170 protein sequence entries and the MaxQuant contaminants database. For the variant protein analysis, a customized human plasma protein variant database was generated from each Uniprot entry using Perl (File S2) and combined with the entire human Uniprot database used above. Mass tolerance was set to 4.5 ppm for precursor ions and MS/MS mass tolerance was 20 ppm. Enzyme specificity was set to trypsin (cleavage C-terminal to Lys and Arg) with a maximum of 2 missed cleavages permitted. Deamidation of Asn and Gln, oxidation of Met, pyro-Glu (with peptide N-term Gln) and protein N-terminal acetylation were set as variable modifications. Either N-ethylmaleimide (Spin96 Optimization), carbamidomethyl (All plasma analysis) on Cys was searched as a fixed modification. We used the MaxLFQ algorithm for label-free quantitation, integrated into the MaxQuant environment.29,31 MaxQuant output was processed and statistical tests performed using the R software package (version 3.4.3). Processed data was plotted using Tableau (version 10.0.2).
Experimental Design and Statistical Rationale
PREFER Trial
The number of participants was established from past studies,32−34 which suggested n = 22 per group would allow detection of a mean difference in glucose infusion rate (GIR) of 15 μmol/kg FFM + 17.7 between groups, with β = 0.8 and α = 0.05. We allowed for a 10% drop out rate, and thus recruited a total of n = 25 per group. In this manuscript we analyzed the intermittent fasting with weight maintenance group (IF100), with plasma samples collected before and after intermittent fasting. To these data we applied a Wilcox robust test to allow for proteins whose distribution for the difference between treatment groups across participants was not normally distributed. Specifically, we used Yuen’s test on trimmed means for dependent (paired) samples without correcting for multiple testing, which was restricted to those proteins with at least 2 razor and unique peptides identified in each sample. Fold changes comparing plasma protein abundance before and after intermittent fasting were calculated using the median. For plotting the variation in individual subject protein intensity measurements and clinical measures in response to IF, we calculated adjusted values that force all participants to have the same mean value for each measure, thus allowing us to focus on the effect of the IF treatment by eliminating the variation in protein abundance between individuals.35 The Pearson correlation was calculated between all clinical measures and all measured proteins, and the associated p-values were adjusted using the Benjamini–Hochberg correction to control for multiple testing. For comparison of allele frequencies between the detected variant peptides and the genotypes derived from the European background population a Fisher’s exact test was used as it is robust to small frequencies. For all data sets statistical analyses were performed using R (version 3.4.3), and processed data was plotted using Tableau (version 10.0.2). Data are shown as median ± 95% confidence interval, unless otherwise stated. Significance was set at p < 0.05.
Results
Spin96 Design and Optimization for StageTip Processing
We aimed to design a 96-well device for use in deep-well centrifuges to increase throughput and consistency when processing StageTips, while also minimizing the likelihood of cross-contamination. Our device (Spin96) addresses these issues and was designed with two bottom configurations and several components (Figure S1a,b) to allow for a quick changeover between washing and elution steps. The characterization of the Spin96 device is detailed in the Supporting Methods. Several different plastics for the 3D-printing of Spin96 were tested for durability and usability, with high impact polystyrene (HIPS) chosen for its durability and ease of printing (Figure S1c). To optimize the Spin96 protocol, an ideal force of 1000g was determined as the minimum force required to efficiently pass liquid through StageTips with the Spin96 (Figure S1d). The protocol for processing SDB-RPS StageTips was also optimized to minimize centrifugation time without compromising the method (Figure S1e). This protocol was then tested against more conventional methods of StageTip processing (Tube-based centrifugation) and found to perform at an equal level in terms of peptides identified while being much faster (Figure S2). The final optimized protocol for the use of the Spin96 is included in the Supporting Methods.
Analysis of Human Plasma Samples from an Intermittent Fasting Clinical Trial
To demonstrate the capabilities of the Spin96 we utilized the optimized protocol to analyze plasma samples from a clinical trial in which 22 overweight participants underwent 8-weeks of IF.24 The primary aim of the PREFER trial was to examine the effects of IF on insulin sensitivity (hyperinsulinemic-euglycemic clamp) in these patients (see Experimental Procedures). The study was longitudinal with plasma collected both before and after the IF intervention. In parallel, many other clinical phenotypes were measured including body composition by DXA, and plasma lipids (see Figure S3 and Table S3). Analysis of these clinical parameters demonstrated that there was a significant decrease in plasma triglycerides (23%) due to the IF intervention. There was no change in the glucose infusion rate (insulin and fat-mass normalized), but there was a significant increase in the insulin-resistance as measured by HOMA-IR scoring. These data are consistent with previous studies in lean humans and in mice36,37 where periods of fasting have been shown to increase insulin resistance.
To investigate the IF-mediated perturbation on the plasma proteome, we used the Spin96 device to process all 44 plasma samples using trypsin digestion and mixed mode SDB-RPS StageTip SPE prior to LC–MS/MS analysis (see Supporting Methods). The plasma samples were randomized before sample preparation and StageTip processing to control technical variation and each sample was analyzed using a 45 min gradient separation during LC–MS/MS. In addition, a small portion of peptides from each participant were mixed to generate a pooled peptide sample for offline high pH reversed phase fractionation into 16 concatenated fractions. Each of these fractions was analyzed by LC–MS/MS with the same method as the individual plasma samples. The data derived from each plasma sample and the peptide fractions were processed together in MaxQuant using match-between-runs to increase peptide/protein identifications. This yielded the identification of 172 protein groups (Table S4) that were quantified from more than 2 razor and unique peptides across all 22 participants (no missing values were allowed) before and after the IF intervention, from a total of >7500 peptides (Table S5).
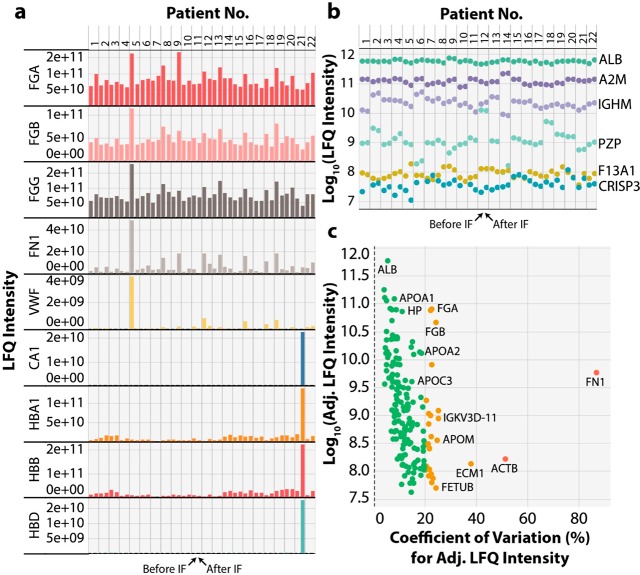
Initially, we performed some QC analysis on the samples from our proteomic data set to determine if either coagulation, or red blood cell lysis, occurred in any of the plasma samples (Figure 1a). This showed that no participant displayed a >10-fold loss in fibrinogen proteins (FGA, FGB, FGG), indicating that the anticoagulant (K+-EDTA) had functioned effectively. A plasma sample from one participant before the IF intervention showed 4-fold higher than the PREFER population average for von Willebrand factor (VWF) and fibronectin (FN1), which may potentially indicate either arterial disease or recent injury. One participant sample showed significant increases in hemoglobins (HBA, HBB, HBD) and carbonic anhydrase (CA1), indicating that this blood sample underwent some red blood cell lysis during processing. We also examined the variance in protein abundance (LFQ intensity) both between and within participants before and after the IF intervention using six marker plasma proteins covering 5 orders of magnitude in protein abundance (Figure 1b). Among these proteins, albumin (ALB) the most abundant protein in plasma showed little variation both between and within individuals with a CV across all samples of 10.6%. In contrast, other marker proteins showed large variability in relative abundance between individuals, with lower variability within an individual’s samples. The protease inhibitor alpha2-macroglobin (A2M) had an overall CV of 24.9% and had marked abundance differences in several participants. However, both immunoglobulin class mu heavy chain (IGHM) and pregnancy zone protein (PZP) showed 10-fold or larger abundance differences between individuals with CVs of 40.3% and 144.9%, respectively. The large variability in PZP between subjects is not surprising given its low abundance in nonpregnant women and abundance variance by 3–4 fold between individuals.38 The lower abundance plasma proteins Cysteine-rich secretory protein 3 (CRISP3) and Coagulation factor XIIIa (F13A1) also showed strong participant-specific protein abundance with CVs of 42.1% and 47%, respectively. These data agree with previous studies demonstrating participant-specific protein abundance profiles and demonstrate the value of longitudinal analysis rather than cross-sectional studies for plasma proteome characterization.19 To examine variation in the proteome in response to IF and minimize variance due to the differences in protein abundances between participants, we calculated an adjusted LFQ intensity, which forces all participants to have the same mean intensity for each protein. From these values we calculated the % CV for each protein during the IF response across all participants except 2 participants who were removed due to the sample quality issues above (Figure 1c). These proteins have been classified into 3 groups, those with % CV < 20 (green ∼65% of proteins), % CV of 20–40 (orange ∼30% of proteins), and % CV > 40 (red ∼5% of proteins). These data show that higher abundance proteins generally have lower % CV values, which is to be expected given the high signal-to-noise ratio for quantitation of the abundant proteins. These data also showed that many IgG-related proteins displayed % CV values >40, as did cadherin-5 (CDH5), beta actin (ACTB), VWF, and FN1.
Figure 1.
PREFER clinical trial plasma quality control. Plasma from participants before and after IF were analyzed using our Spin96 proteomics workflow. Label-free quantitation (LFQ) intensity was plotted for (a) proteins indicative of blood clotting and hemolysis for each participant, before and after fasting, and (b) LFQ intensity plot for example proteins of very similar abundance before and after the IF intervention, which showed participant-specific abundance patterns. Each color represents a different protein. (c) Plot for each plasma protein of the adjusted LFQ (Adj. LFQ) intensity versus the Adj. LFQ percentage coefficient of variation (% CV). Green circles indicate <20% CV, orange circles indicate 20–40% CV and red circles indicate >40% CV.
Intermittent Fasting Regulated Plasma Proteins
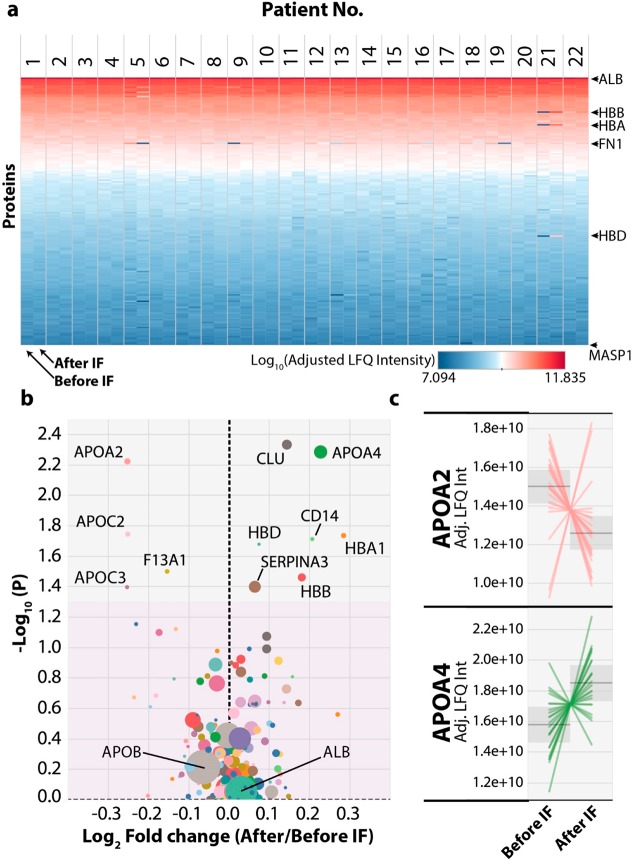
To provide an overview of the changes induced by the IF intervention across all participants we plotted a heat map of the adjusted LFQ intensity for all 172 proteins ranked by protein abundance (Figure 2a). This confirmed that very few proteins drastically changed abundance due to the IF intervention and only a few changes were immediately obvious by eye. These include the increased hemoglobin isoforms from the participant whose plasma sample taken after the IF intervention displayed hemolysis, which therefore makes these proteins appear highly differential between the two time points. In addition, fibronectin (FN1) appears to be significantly perturbed across several participants. To identify statistically significant differences due to the IF intervention, we generated a volcano plot showing fold-changes versus a paired test statistic (Figure 2b). Only 11 of the 172 proteins had a p-value less than 0.05, with modest median fold-changes between 5 and 30% in either direction. Of these proteins apolipoprotein A4 (APOA4) and clusterin (CLU) were the most significantly up-regulated, while apolipoprotein C2 (APOC2) and apolipoprotein A2 (APOA2) were the most significantly down-regulated proteins. All five of these proteins are functionally related in the regulation of HDL particle metabolism. To examine more closely the response of each participant to the IF intervention we plotted the adjusted LFQ intensity for the proteins showing significant changes due to the IF intervention and for a negative control albumin (ALB), with each participant as a separate line on the plot (Figure 2c and Figure S4). For each of these proteins, many participants displayed much larger fold changes of up to 2-fold after the IF intervention, compared to the fold change calculated from the median values used in the volcano plot (Figure 2b). For each of these proteins there were also participants who displayed the opposite trend in abundance change; however, these participants were not the same for each of these significant proteins.
Figure 2.
Intermittent fasting induces changes in protein abundance in human plasma. (a) Heat map showing each protein ranked by its total log10 adjusted Label-Free Quantitation (Adj. LFQ) intensity (y-axis) versus the data for each participant from both before and after the IF intervention. Blue colors represent low abundance and red colors high abundance. Only complete cases were used; any protein with a missing value in any sample was excluded. (b) Volcano plot of plasma protein abundance changes after IF were plotted with the y-axis showing the −log10 (p-value, paired) and the x-axis showing the log2 fold-change of protein abundance (after IF/before IF) calculated from the LFQ intensity values. The gray area denotes significant (p < 0.05) changes with IF and the pink area denotes nonsignificant (p > 0.05) changes. The size of each circle represents the minimum number of razor and unique peptides across all participants, larger size indicates more razor and unique peptides. (c) Line graphs for the two most significant up and down regulated proteins after the IF intervention were plotted with the y-axis showing the Adj. LFQ intensity and each line representing one participant’s response.
Global Correlation Analysis
Given the number of proteins for which we have collected quantitative data and the large number of clinical parameters measured, we wanted to examine if our data set could yield novel correlations between any of these factors. To this end we have calculated the Pearson correlation between all measurements including clinical (e.g., weight, plasma HDL, plasma triglycerides) and protein (LFQ Intensities). These analysis used data derived from all participants including the before and after IF measurements (Figure 3). Initially, we wanted to confirm that we had observed well-known correlations. This include the positive correlations between LDL (Clin. LDL) with apolipoprotein B (APOB: the main LDL protein constituent), PZP with sex hormone binding globulin (SHBG), and HOMAIR (Clin. HOMAIR) with either complement C3 (C3), proteoglycan 4 (PRG4), Alpha-1-acid glycoprotein 1 (ORM1), or Serum amyloid A-4 protein (SAA4).19,39,40 All these positive controls were seen in our data set with particularly significant correlation between APOB and LDL levels (Figure 3 and Figure S5). We also observed a strong positive association between von Willebrand factor (VWF) and either fibronectin (FN1) (Figure 3 and Figure S5), extracellular matrix protein 1 (ECM1), fibrinogen proteins (FGA, FGB, FGG), or coagulation factor XIII (F13A1 and F13B). In addition, we observed significant negative correlations between alpha1-antitrypsin (SERPINA1) and either the Framingham Risk Score (Clin. FRS), or plasma LDL, which is a parameter used in the calculation of the FRS. As would be expected, the glucose infusion rate (Clin. GIR, normalized to insulin levels and fat mass) was positively correlated with HDL levels (Clin. HDL), and negative correlated with the inflammatory protein alpha-1-acid glycoprotein 2 (ORM2). To identify any associations between the proteins significantly changing due to the IF intervention and the remaining clinical or protein parameters, these proteins were specifically examined. Interestingly, APOA4 showed a significant positive correlation with plasma gelsolin (GSN) a protein whose function in plasma is not well understood, and a negative correlation with plasma total triglycerides (Clin. TG) (Figure S5). Conversely, APOC2 was positively correlated with total plasma triglycerides, LDL levels, the Framingham Risk Score, PRG4, and fasting insulin levels.
Figure 3.
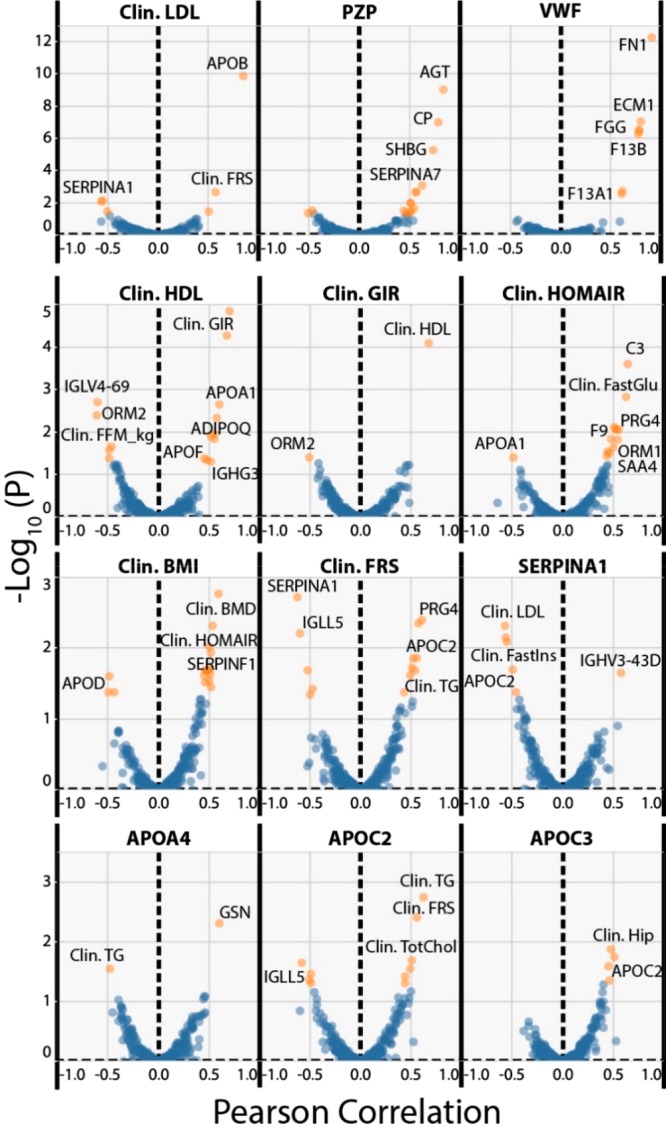
Correlation analysis of plasma protein abundance versus clinical parameters monitored in the PREFER trial. Data derived from all detected proteins and all clinical parameters were correlated against each other across all 44 participant samples, disregarding any grouping information. Correlations were calculated using the Pearson’s correlation coefficient and the resulting p-values were subjected to Benjamini–Hochberg correction. The y-axis of each plot shows the −log10 (corrected p-value) versus the Pearson correlation. Values in blue are insignificant (FDR > 5%) and values in orange are significant (FDR < 5%). Each measure starting with Clin. (Clinical) represents an individual clinical measurement. Proteins are indicated by their corresponding gene name.
Unbiased Identification of Human Plasma Protein Variants
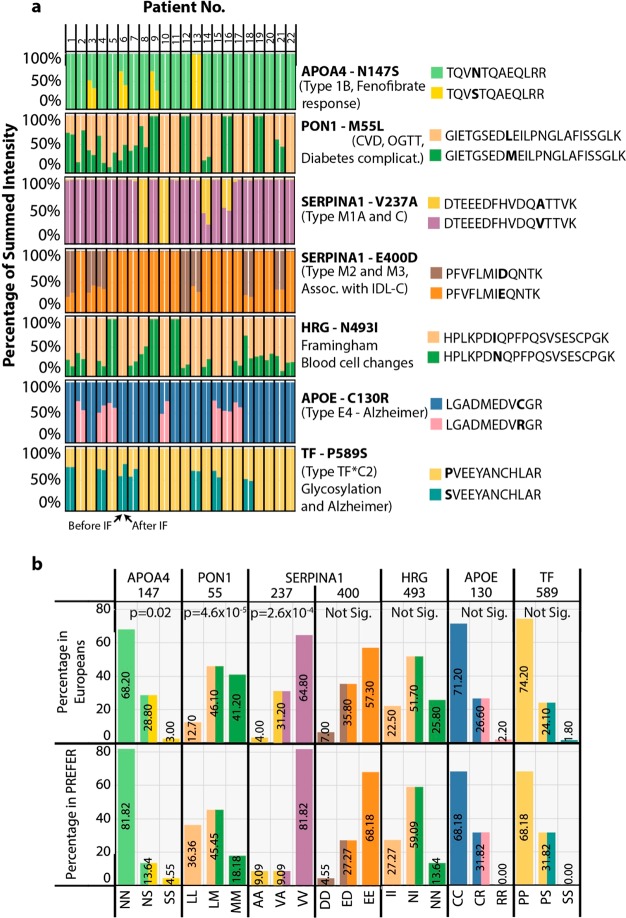
Given the depth of proteome coverage we have achieved, we wanted to identify peptides characteristic of known human protein coding variants (derived from SNPs), some of which are known to be associated with metabolic phenotypes. To achieve this, we generated a database of the 172 plasma proteins consistently identified in our samples and for each protein used the Uniprot Natural Variant annotations to generate additional database entries (full protein sequence) containing the associated amino acid change (File S2). We then used this variant database in a combined search with a full human database from SwissProt in MaxQuant to identify and quantify the peptides present in our plasma samples (Table S6). To our surprise we were able to identify >20 protein coding variants, where peptides specific to natural alleles could be observed across the participants. To narrow our list of high-quality variants, we ignored those that had either any missing values, where peptides from both alleles could not be observed, or have been shown previously to cause benign changes to the protein’s function. This yielded 7 variants in APOA4, paraoxonase (PON1), alpha1-antitrypsin (SERPINA1), histidine-rich glycoprotein (HRG), apolipoprotein E (APOE) and transferrin (TF) (see Figure S6 for annotated spectra). Using the intensity values for the peptides specific to each allele we generated a bar plot for each of the 44 participant samples where the summed intensity of both peptides was set to 100% (Figure 4). Most of these peptides had conservative amino acid changes and only moderate intensity differences between the allele-specific peptides was seen, likely due to similar peptide ionization efficiencies. Thus, participants that had peptides detected from both alleles have colored bars with each showing ∼50% of the summed intensity, while participants that only had one type of allele detected have bars of a single color (Figure 4). In six of these protein variants, clinically relevant phenotypes have been associated including drug response (APOA4 N147S),41 metabolic dysfunction (PON1 L55M and SERPINA1 E400D),42,43 blood cell type changes (HRG N493I)44 and Alzheimer’s disease (APOE C130R and TF P589S).45,46 Using a Fisher’s exact test, we compared the frequency of our observed peptide alleles to the known genomic allele frequency in the Caucasian European population,47 which is representative of the PREFER participant ethnicity. This would allow us to determine if the PREFER population was either enriched, or depleted, for particular genotypes compared to their ethnic background. This analysis showed APOA4 N147S, PON1 L55M and SERPINA1 V237A displayed peptide observation frequencies significantly different (p < 0.05) from the known genotype frequencies. The most significant difference was observed for the PON1 L55M variant with the homozygous LL (associated with CVD and lower oral glucose tolerance) and MM genotype frequencies in Europeans having higher and lower observed frequency, respectively, at the peptide level in the PREFER population.
Figure 4.
Detection of clinically relevant natural protein variants in the PREFER plasma proteomes. Mass spectrometry-based proteomics data was reanalyzed using a modified plasma-specific protein database containing all known forms of naturally occurring clinically-relevant variants. (a) Bar plot for each protein variant where the y-axis shows the summed intensity of peptides derived from each allele as a percentage of total, and the x-axis shows the 44 participant samples with one bar per sample. The gene name for each variant and its position in the protein are indicated in bold to the right of each plot. Also shown are the associated human phenotypes. Peptides from each allele are shown in different colors on the same plot and correspond to each legend shown on the right. (b) Bar plot showing either the frequency of the genotype in the European population sampled by the 1000Genomes study, or the frequency of the corresponding peptide variants in the PREFER plasma proteome samples. The y-axis shows the frequency in percentage of total, and the x-axis shows the corresponding genomic allele and peptide variant combinations. A comparison of the frequencies for each allele was made using Fisher’s Exact Test, which is shown at the top of the plot.
Discussion
Here we present an unbiased analysis of human plasma following a clinical trial of intermittent fasting (IF). These samples were analyzed using a 3D-printed device, the Spin96, to increase efficiency and consistency of the sample preparation with a low-cost and readily accessible device. From this analysis, we identified ∼200 plasma proteins that we correlated with a swath of clinical phenotypes to examine the molecular adaptations that occur with IF. In addition, we were able to detect a number of clinically relevant protein variants at the peptide level using a unique protein variant database. Together, this study provides a novel device and protocol for processing of large numbers of plasma samples and novel insights into the potential mechanisms behind IF beneficial effects in humans.
Plasma is an easily accessible sample from human clinical trials, which when analyzed with current proteomics workflows, can provide insight into possible mechanisms in the development of clinical phenotypes.19,48−51 Particularly useful are intervention-based longitudinal trials where the same participant is monitored over an extended period to identify treatment-related protein changes.19,52−54 These studies provide useful statistical power in reasonably small number of samples (≤96), compatible with the current nanoLC–MS/MS based proteomics throughput of ∼1 sample per hour. To date, proteomics studies of plasma have focused on the analysis of the top 200–1000 most abundant proteins, using either no depletion strategy, or with employment of abundant protein depletion with affinity columns. As suggested previously, depletion columns likely have their own caveats regarding lot-to-lot reproducibility55 and excessive cost. Our device offers a faster approach to StageTip sample preparation, and a similar number of proteins identified compared to previous nondepleted plasma analysis studies.17 We observed far fewer proteins when compared to studies using either more in-depth fractionation,55 or recently improved MS acquisition strategies.56 However, these strategies still do not provide easy access to very low abundance plasma proteins such as insulin, GLP-1, or chemokines. The field would be greatly accelerated by the implementation of improved plasma analysis workflows to access these clinically relevant low abundance proteins/peptides.
Our analysis detected the up-regulation of the apolipoprotein A IV (APOA4) protein in human plasma after 8-weeks of intermittent fasting. This is interesting given that higher levels of APOA4 have been associated with many beneficial phenotypes such as promotion of reverse cholesterol transport, increased satiety, and decreased LDL particle oxidation.57 APOA4 has been quantified as strongly expressed in the small intestine by the luminal epithelial cells and to a much lower extent by cells in the liver and other tissues of both humans and mice.58,59 APOA4 is present in plasma in either a lipid-free form, associated with chylomicrons, or associated with HDL particles, which makes sense given its known functional associations. In previous studies, plasma abundance of the APOA4 protein has been shown to increase after either a high fat diet60 or bariatric surgery, where it was positively correlated with increased HDL cholesterol.61−63 In contrast, APOA4 protein abundance after weight loss has been reported to either decrease19 or slightly increase,64 indicating that weight loss alone does not strongly influence the abundance of this potentially beneficial protein. Several human protein-coding SNPs of APOA4 have been associated with clinical phenotypes, which includes the N147S variant detected in this study that is associated with the response to fenofibrate treatment.41 Another phenotype associated with several APOA4 variants is altered plasma total triacylglycerol abundance,65,66 which is in agreement with our correlation analysis.
In addition to APOA4, we also observed significant changes in CLU, APOC2, APOC3, and APOA2, which are all related to either lipid, chylomicron, or HDL-particle metabolism (Figure 5). Of particular interest is the decrease in protein abundance of APOC3 in plasma with IF (Figure 2b), as it has been previously demonstrated to decrease triglyceride lipolysis through direction inhibition of lipoprotein lipase (LPL)67 and to also decrease the uptake of triglycerides into tissues such as the liver.68 Together, this suggests a combined regulatory effect of intermittent fasting to increase triglyceride lipolysis in chylomicrons and increase the production of mature spherical HDL-particles. This hypothesis fits with our clinical observations that plasma total triglycerides were significantly decreased after intermittent fasting (Table S3); however, total HDL abundance was not significantly changed after IF. Future analysis of participant plasma samples using size-exclusion chromatography (SEC) to detect HDL particle size distributions before and after the intervention would help address these aspects of APOA4 function. Validation of these findings in different cohorts with larger numbers of participants will be important to confirm their functional significance.
Figure 5.
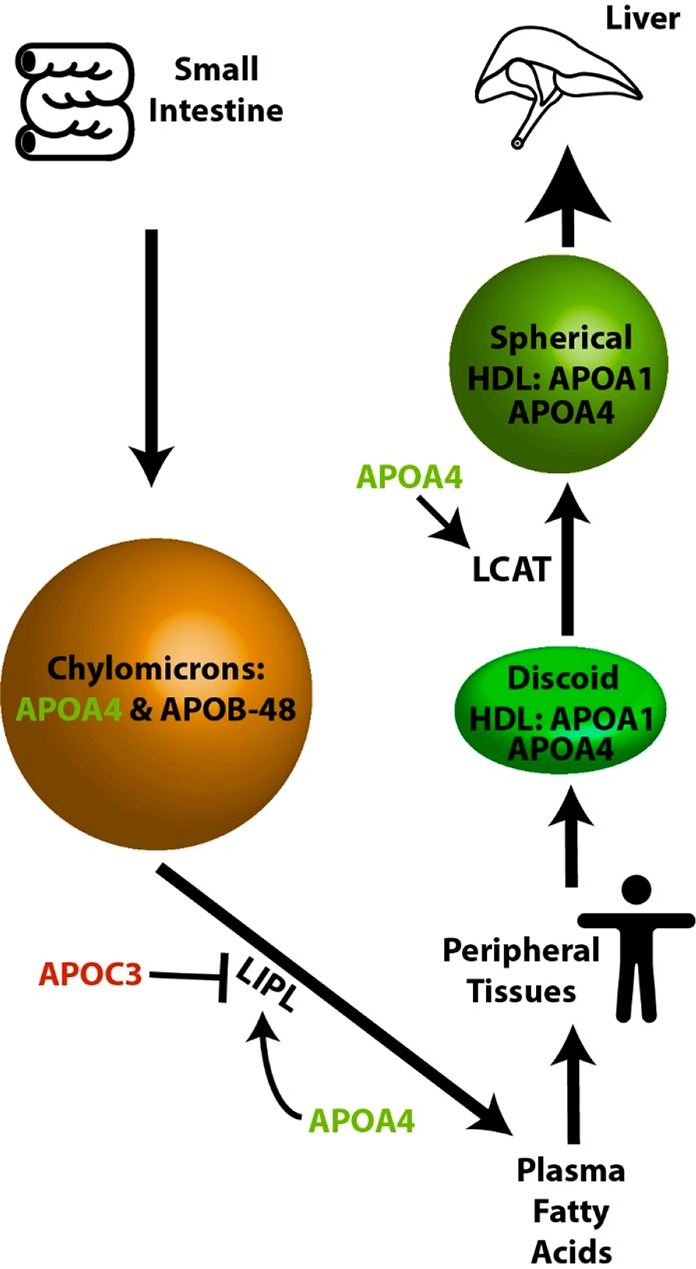
Model of intermittent fasting modulation of apolipoprotein plasma abundance and metabolism. Proteins in green are up-regulated and proteins in red down-regulated by IF in human plasma. Proteins shown in black do not change significantly.
Global correlation analysis was performed on all the protein-level plasma data and the clinical measures for each subject. From this analysis, the highly abundant plasma protein, SERPINA1, was negatively correlated with the Framingham Risk Score (FRS). To calculate FRS for an individual, several lifestyle factors including smoker status, age, and gender are considered alongside biological measures including LDL, blood pressure, and heart rate.69 Given the short time scale of our IF intervention, and that most patients maintained a constant BMI, it is most likely that a change in LDL is the reason for the positive FRS change. This agrees with the observed negative correlation between plasma SERPINA1 abundance and LDL levels (Figure 3). Indeed, previous studies have demonstrated a clear interaction between SERPINA1 and the LDL-associated protein, lipoprotein A (LPA).70 LPA is a key component of LDL particles where it is directly bound to the major LDL component, apolipoprotein B-100 (APOB100).71 Our data support the hypothesis that increasing plasma SERPINA1 levels will lead to a decrease in blood LDL levels, through its association with the LDL-associated protein LPA.
The analysis of protein coding SNPs offers a direct link between genetic mutation and clinical outcomes as it relates to human disease. We have adapted a human protein variant database for the detection and quantitation of protein variant-derived peptides. Most of these amino acid variations were conservative changes and only moderate intensity differences between the allele-specific peptides were observed. However, SNPs that introduce more divergent amino acids could pose a detection/quantification problem. For example, we observed some peptide variants such as HRG (N493I), where intensity differed by 3-fold between alleles in heterozygous individuals, which may reflect different ionization efficiencies. Given that these data are protein based, we cannot discriminate between the true genotype of these individuals and any allele-specific expression that may occur for each gene analyzed. However, the fact that we observed the expected allele frequencies in 4 of the 7 clinically relevant protein variants suggests that we are observing peptide abundance in concordance with the genomic alleles. For the 3 protein variants that showed a significant deviation from the European population allele distribution there are two main reasons for the observed deviation. First, those participants selected for the PREFER trial (including BMI > 25) may be enriched/depleted for those specific genotypes, which may provide a genetic predisposition to becoming overweight, or how the subject would respond to IF. Second, the proteins we have examined may display allele-specific expression. Unfortunately, matching tissue samples for genotyping were not available in the PREFER trial. To confidently associate specific SNPs to observed phenotypes future studies need to apply this method across larger cohorts of participants to provide greater statistical power. Preferably, these measurements can then be matched to tissue samples to correlate these data with the DNA-derived genotype of each participant.
The Spin96 device offers fast and reproducible proteomics analysis, further facilitating the use of proteomics over conventional protein analysis methods such as either ELISA assays, or Western blotting. Proteomics-based approaches offer an unbiased quantitative analysis at a reduced cost per detected analyte at an increased speed and with higher specificity. While more conventional approaches are still useful for their great precision, they are not easily compatible with the analysis of large numbers of samples and the simultaneous detection of many proteins.
Conclusions
In summary, we developed a 3D printed 96 well device for StageTip-based SPE of peptide samples derived from human plasma. The application of this device and optimized protocol enabled the detection of significant changes in the plasma proteome after IF, which may mediate some of the observed beneficial phenotypes derived from IF. Furthermore, we have demonstrated the ability to identify several novel correlations between clinical measures and the identified proteins from this clinical trial. Lastly, we have detected many peptides derived from nonsynonymous protein-coding variants of plasma proteins including several that are clinically significant. Together, these findings illustrate novel changes in the human plasma proteome in response to intermittent fasting using a new sample preparation device.
Acknowledgments
M.L. is a Cancer Institute New South Wales Future Research Leader Fellow (15/FRL/1-06A). This work was supported by grants from the NHMRC (APP1120475). We thank SydneyMS for providing the instrumentation used in this study. The clinical research study was funded by a National Health and Medical Research Council Project Grant APP1023401. L.K.H. was supported by an Australian Research Council Future Fellowship FT120100027.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jproteome.9b00090.
Author Contributions
∥ D.J.H. and A.H. contributed equally to this work. L.K.H. and M.L. conceived and supervised the project, and are co-senior authors. A.H., L.K.H., and S.H. collected the human plasma samples and associated clinical data. D.J.H., L.H., and M.L. manufactured the Spin96 and performed all proteomics experiments and associated analysis. D.J.H., A.H., L.H., S.H., S.J.H., D.E.J., L.K.H., and M.L. wrote the manuscript.
The authors declare no competing financial interest.
Notes
RAW MS data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the data set identifier PXD009324. RAW data were analyzed using MaxQuant, and the output has also been uploaded to the ProteomeXchange Consortium under the same identifier.
Supplementary Material
References
- Goodrick C. L.; Ingram D. K.; Reynolds M. A.; Freeman J. R.; Cider N. Effects of intermittent feeding upon body weight and lifespan in inbred mice: interaction of genotype and age. Mech. Ageing Dev. 1990, 55 (1), 69–87. 10.1016/0047-6374(90)90107-Q. [DOI] [PubMed] [Google Scholar]
- Wan R.; Camandola S.; Mattson M. P. Intermittent food deprivation improves cardiovascular and neuroendocrine responses to stress in rats. J. Nutr. 2003, 133 (6), 1921–9. 10.1093/jn/133.6.1921. [DOI] [PubMed] [Google Scholar]
- Xie K.; Neff F.; Markert A.; Rozman J.; Aguilar-Pimentel J. A.; Amarie O. V.; Becker L.; Brommage R.; Garrett L.; Henzel K. S.; Holter S. M.; Janik D.; Lehmann I.; Moreth K.; Pearson B. L.; Racz I.; Rathkolb B.; Ryan D. P.; Schroder S.; Treise I.; Bekeredjian R.; Busch D. H.; Graw J.; Ehninger G.; Klingenspor M.; Klopstock T.; Ollert M.; Sandholzer M.; Schmidt-Weber C.; Weiergraber M.; Wolf E.; Wurst W.; Zimmer A.; Gailus-Durner V.; Fuchs H.; Hrabe de Angelis M.; Ehninger D. Every-other-day feeding extends lifespan but fails to delay many symptoms of aging in mice. Nat. Commun. 2017, 8 (1), 155. 10.1038/s41467-017-00178-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anson R. M.; Guo Z.; de Cabo R.; Iyun T.; Rios M.; Hagepanos A.; Ingram D. K.; Lane M. A.; Mattson M. P. Intermittent fasting dissociates beneficial effects of dietary restriction on glucose metabolism and neuronal resistance to injury from calorie intake. Proc. Natl. Acad. Sci. U. S. A. 2003, 100 (10), 6216–20. 10.1073/pnas.1035720100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo V. D.; Mattson M. P. Fasting: molecular mechanisms and clinical applications. Cell Metab. 2014, 19 (2), 181–92. 10.1016/j.cmet.2013.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mager D. E.; Wan R.; Brown M.; Cheng A.; Wareski P.; Abernethy D. R.; Mattson M. P. Caloric restriction and intermittent fasting alter spectral measures of heart rate and blood pressure variability in rats. FASEB J. 2006, 20 (6), 631–7. 10.1096/fj.05-5263com. [DOI] [PubMed] [Google Scholar]
- Wan R.; Ahmet I.; Brown M.; Cheng A.; Kamimura N.; Talan M.; Mattson M. P. Cardioprotective effect of intermittent fasting is associated with an elevation of adiponectin levels in rats. J. Nutr. Biochem. 2010, 21 (5), 413–7. 10.1016/j.jnutbio.2009.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmet I.; Wan R.; Mattson M. P.; Lakatta E. G.; Talan M. Cardioprotection by intermittent fasting in rats. Circulation 2005, 112 (20), 3115–21. 10.1161/CIRCULATIONAHA.105.563817. [DOI] [PubMed] [Google Scholar]
- Varady K. A.; Roohk D. J.; McEvoy-Hein B. K.; Gaylinn B. D.; Thorner M. O.; Hellerstein M. K. Modified alternate-day fasting regimens reduce cell proliferation rates to a similar extent as daily calorie restriction in mice. FASEB J. 2008, 22 (6), 2090–6. 10.1096/fj.07-098178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel I.; Liu T. L.; Nepomuceno N.; Gleicher N. Effects of short-term dietary restriction on survival of mammary ascites tumor-bearing rats. Cancer Invest. 1988, 6 (6), 677–80. 10.3109/07357908809078034. [DOI] [PubMed] [Google Scholar]
- Varady K. A.; Allister C. A.; Roohk D. J.; Hellerstein M. K. Improvements in body fat distribution and circulating adiponectin by alternate-day fasting versus calorie restriction. J. Nutr. Biochem. 2010, 21 (3), 188–95. 10.1016/j.jnutbio.2008.11.001. [DOI] [PubMed] [Google Scholar]
- Larance M.; Lamond A. I. Multidimensional proteomics for cell biology. Nat. Rev. Mol. Cell Biol. 2015, 16 (5), 269–80. 10.1038/nrm3970. [DOI] [PubMed] [Google Scholar]
- Elias J. E.; Haas W.; Faherty B. K.; Gygi S. P. Comparative evaluation of mass spectrometry platforms used in large-scale proteomics investigations. Nat. Methods 2005, 2 (9), 667–75. 10.1038/nmeth785. [DOI] [PubMed] [Google Scholar]
- Ishihama Y.; Rappsilber J.; Mann M. Modular stop and go extraction tips with stacked disks for parallel and multidimensional Peptide fractionation in proteomics. J. Proteome Res. 2006, 5 (4), 988–94. 10.1021/pr050385q. [DOI] [PubMed] [Google Scholar]
- Rappsilber J.; Ishihama Y.; Mann M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 2003, 75 (3), 663–70. 10.1021/ac026117i. [DOI] [PubMed] [Google Scholar]
- Rappsilber J.; Mann M.; Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2007, 2 (8), 1896–906. 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- Kulak N. A.; Pichler G.; Paron I.; Nagaraj N.; Mann M. Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat. Methods 2014, 11 (3), 319–24. 10.1038/nmeth.2834. [DOI] [PubMed] [Google Scholar]
- Geyer P. E.; Kulak N. A.; Pichler G.; Holdt L. M.; Teupser D.; Mann M. Plasma Proteome Profiling to Assess Human Health and Disease. Cell Systems 2016, 2 (3), 185–95. 10.1016/j.cels.2016.02.015. [DOI] [PubMed] [Google Scholar]
- Geyer P. E.; Wewer Albrechtsen N. J.; Tyanova S.; Grassl N.; Iepsen E. W.; Lundgren J.; Madsbad S.; Holst J. J.; Torekov S. S.; Mann M. Proteomics reveals the effects of sustained weight loss on the human plasma proteome. Mol. Syst. Biol. 2016, 12 (12), 901. 10.15252/msb.20167357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Graaf E. L.; Giansanti P.; Altelaar A. F.; Heck A. J. Single-step enrichment by Ti4+-IMAC and label-free quantitation enables in-depth monitoring of phosphorylation dynamics with high reproducibility and temporal resolution. Mol. Cell. Proteomics 2014, 13 (9), 2426–34. 10.1074/mcp.O113.036608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey S. J.; Azimifar S. B.; Mann M. High-throughput phosphoproteomics reveals in vivo insulin signaling dynamics. Nat. Biotechnol. 2015, 33 (9), 990–5. 10.1038/nbt.3327. [DOI] [PubMed] [Google Scholar]
- Kanshin E.; Bergeron-Sandoval L. P.; Isik S. S.; Thibault P.; Michnick S. W. A cell-signaling network temporally resolves specific versus promiscuous phosphorylation. Cell Rep. 2015, 10 (7), 1202–14. 10.1016/j.celrep.2015.01.052. [DOI] [PubMed] [Google Scholar]
- Olsen J. V.; Blagoev B.; Gnad F.; Macek B.; Kumar C.; Mortensen P.; Mann M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 2006, 127 (3), 635–48. 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- Hutchison A. T.; Liu B.; Wood R. E.; Vincent A. D.; Thompson C. H.; O’Callaghan N. J.; Wittert G. A.; Heilbronn L. K. Effects of Intermittent Versus Continuous Energy Intakes on Insulin Sensitivity and Metabolic Risk in Women with Overweight. Obesity 2019, 27 (1), 50–58. 10.1002/oby.22345. [DOI] [PubMed] [Google Scholar]
- Redman L. M.; Heilbronn L. K.; Martin C. K.; de Jonge L.; Williamson D. A.; Delany J. P.; Ravussin E. Metabolic and behavioral compensations in response to caloric restriction: implications for the maintenance of weight loss. PLoS One 2009, 4 (2), e4377 10.1371/journal.pone.0004377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trumbo P.; Schlicker S.; Yates A. A.; Poos M. Dietary reference intakes for energy, carbohydrate, fiber, fat, fatty acids, cholesterol, protein and amino acids. J. Am. Diet. Assoc. 2002, 102 (11), 1621–30. 10.1016/S0002-8223(02)90346-9. [DOI] [PubMed] [Google Scholar]
- Samocha-Bonet D.; Campbell L. V.; Viardot A.; Freund J.; Tam C. S.; Greenfield J. R.; Heilbronn L. K. A family history of type 2 diabetes increases risk factors associated with overfeeding. Diabetologia 2010, 53 (8), 1700–8. 10.1007/s00125-010-1768-y. [DOI] [PubMed] [Google Scholar]
- DeFronzo R. A.; Tobin J. D.; Andres R. Glucose clamp technique: a method for quantifying insulin secretion and resistance. Am. J. Physiol. 1979, 237 (3), E214–23. 10.1152/ajpendo.1979.237.3.E214. [DOI] [PubMed] [Google Scholar]
- Cox J.; Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26 (12), 1367–1372. 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- Cox J. r.; Neuhauser N.; Michalski A.; Scheltema R. A.; Olsen J. V.; Mann M. Andromeda: A Peptide Search Engine Integrated into the MaxQuant Environment. J. Proteome Res. 2011, 10 (4), 1794–1805. 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- Cox J.; Hein M. Y.; Luber C. A.; Paron I.; Nagaraj N.; Mann M. Accurate Proteome-wide Label-free Quantification by Delayed Normalization and Maximal Peptide Ratio Extraction, Termed MaxLFQ. Mol. Cell. Proteomics 2014, 13 (9), 2513–2526. 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albu J. B.; Heilbronn L. K.; Kelley D. E.; Smith S. R.; Azuma K.; Berk E. S.; Pi-Sunyer F. X.; Ravussin E.; Look A. A. R. G. Metabolic changes following a 1-year diet and exercise intervention in patients with type 2 diabetes. Diabetes 2010, 59 (3), 627–33. 10.2337/db09-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilbronn L. K.; Rood J.; Janderova L.; Albu J. B.; Kelley D. E.; Ravussin E.; Smith S. R. Relationship between serum resistin concentrations and insulin resistance in nonobese, obese, and obese diabetic subjects. J. Clin. Endocrinol. Metab. 2004, 89 (4), 1844–1848. 10.1210/jc.2003-031410. [DOI] [PubMed] [Google Scholar]
- Samocha-Bonet D.; Campbell L. V.; Mori T. A.; Croft K. D.; Greenfield J. R.; Turner N.; Heilbronn L. K. Overfeeding reduces insulin sensitivity and increases oxidative stress, without altering markers of mitochondrial content and function in humans. PLoS One 2012, 7 (5), e36320 10.1371/journal.pone.0036320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loftus G. R.; Masson M. E. J. Using Confidence-Intervals in within-Subject Designs. Psychon. B. Rev. 1994, 1 (4), 476–490. 10.3758/BF03210951. [DOI] [PubMed] [Google Scholar]
- Park S.; Yoo K. M.; Hyun J. S.; Kang S. Intermittent fasting reduces body fat but exacerbates hepatic insulin resistance in young rats regardless of high protein and fat diets. J. Nutr. Biochem. 2017, 40, 14–22. 10.1016/j.jnutbio.2016.10.003. [DOI] [PubMed] [Google Scholar]
- Soeters M. R.; Sauerwein H. P.; Dubbelhuis P. F.; Groener J. E.; Ackermans M. T.; Fliers E.; Aerts J. M.; Serlie M. J. Muscle adaptation to short-term fasting in healthy lean humans. J. Clin. Endocrinol. Metab. 2008, 93 (7), 2900–3. 10.1210/jc.2008-0250. [DOI] [PubMed] [Google Scholar]
- Sottrup-Jensen L.; Folkersen J.; Kristensen T.; Tack B. F. Partial primary structure of human pregnancy zone protein: extensive sequence homology with human alpha 2-macroglobulin. Proc. Natl. Acad. Sci. U. S. A. 1984, 81 (23), 7353–7. 10.1073/pnas.81.23.7353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azrad M.; Gower B. A.; Hunter G. R.; Nagy T. R. Intra-abdominal adipose tissue is independently associated with sex-hormone binding globulin in premenopausal women. Obesity 2012, 20 (5), 1012–5. 10.1038/oby.2011.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominiczak M. H.; Caslake M. J. Apolipoproteins: metabolic role and clinical biochemistry applications. Ann. Clin. Biochem. 2011, 48 (6), 498–515. 10.1258/acb.2011.011111. [DOI] [PubMed] [Google Scholar]
- Liu Y.; Ordovas J. M.; Gao G.; Province M.; Straka R. J.; Tsai M. Y.; Lai C. Q.; Zhang K.; Borecki I.; Hixson J. E.; Allison D. B.; Arnett D. K. Pharmacogenetic association of the APOA1/C3/A4/A5 gene cluster and lipid responses to fenofibrate: the genetics of lipid-lowering drugs and diet network study. Pharmacogenet. Genomics 2009, 19 (2), 161–9. 10.1097/FPC.0b013e32831e030e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garin M. C.; James R. W.; Dussoix P.; Blanche H.; Passa P.; Froguel P.; Ruiz J. Paraoxonase polymorphism Met-Leu54 is associated with modified serum concentrations of the enzyme. A possible link between the paraoxonase gene and increased risk of cardiovascular disease in diabetes. J. Clin. Invest. 1997, 99 (1), 62–6. 10.1172/JCI119134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye M.; Ripatti S.; Kettunen J.; Lyytikainen L. P.; Oksala N.; Laurila P. P.; Kangas A. J.; Soininen P.; Savolainen M. J.; Viikari J.; Kahonen M.; Perola M.; Salomaa V.; Raitakari O.; Lehtimaki T.; Taskinen M. R.; Jarvelin M. R.; Ala-Korpela M.; Palotie A.; de Bakker P. I. Novel Loci for metabolic networks and multi-tissue expression studies reveal genes for atherosclerosis. PLoS Genet. 2012, 8 (8), e1002907 10.1371/journal.pgen.1002907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox C. S.; Heard-Costa N.; Cupples L. A.; Dupuis J.; Vasan R. S.; Atwood L. D. Genome-wide association to body mass index and waist circumference: the Framingham Heart Study 100K project. BMC Med. Genet. 2007, 8 (Suppl 1), S18. 10.1186/1471-2350-8-S1-S18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauwe J. S.; Bertelsen S.; Mayo K.; Cruchaga C.; Abraham R.; Hollingworth P.; Harold D.; Owen M. J.; Williams J.; Lovestone S.; Morris J. C.; Goate A. M. Suggestive synergy between genetic variants in TF and HFE as risk factors for Alzheimer’s disease. Am. J. Med. Genet., Part B 2010, 153b (4), 955–9. 10.1002/ajmg.b.31053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corder E. H.; Saunders A. M.; Strittmatter W. J.; Schmechel D. E.; Gaskell P. C.; Small G. W.; Roses A. D.; Haines J. L.; Pericak-Vance M. A. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science 1993, 261 (5123), 921–3. 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- Auton A.; Brooks L. D.; Durbin R. M.; Garrison E. P.; Kang H. M.; Korbel J. O.; Marchini J. L.; McCarthy S.; McVean G. A.; Abecasis G. R. A global reference for human genetic variation. Nature 2015, 526 (7571), 68–74. 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candia J.; Cheung F.; Kotliarov Y.; Fantoni G.; Sellers B.; Griesman T.; Huang J.; Stuccio S.; Zingone A.; Ryan B. M.; Tsang J. S.; Biancotto A. Assessment of Variability in the SOMAscan Assay. Sci. Rep. 2017, 7 (1), 14248. 10.1038/s41598-017-14755-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold L.; Ayers D.; Bertino J.; Bock C.; Bock A.; Brody E. N.; Carter J.; Dalby A. B.; Eaton B. E.; Fitzwater T.; Flather D.; Forbes A.; Foreman T.; Fowler C.; Gawande B.; Goss M.; Gunn M.; Gupta S.; Halladay D.; Heil J.; Heilig J.; Hicke B.; Husar G.; Janjic N.; Jarvis T.; Jennings S.; Katilius E.; Keeney T. R.; Kim N.; Koch T. H.; Kraemer S.; Kroiss L.; Le N.; Levine D.; Lindsey W.; Lollo B.; Mayfield W.; Mehan M.; Mehler R.; Nelson S. K.; Nelson M.; Nieuwlandt D.; Nikrad M.; Ochsner U.; Ostroff R. M.; Otis M.; Parker T.; Pietrasiewicz S.; Resnicow D. I.; Rohloff J.; Sanders G.; Sattin S.; Schneider D.; Singer B.; Stanton M.; Sterkel A.; Stewart A.; Stratford S.; Vaught J. D.; Vrkljan M.; Walker J. J.; Watrobka M.; Waugh S.; Weiss A.; Wilcox S. K.; Wolfson A.; Wolk S. K.; Zhang C.; Zichi D. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS One 2010, 5 (12), e15004 10.1371/journal.pone.0015004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberger G.; Liu Y.; Rost H. L.; Ludwig C.; Buil A.; Bensimon A.; Soste M.; Spector T. D.; Dermitzakis E. T.; Collins B. C.; Malmstrom L.; Aebersold R. Inference and quantification of peptidoforms in large sample cohorts by SWATH-MS. Nat. Biotechnol. 2017, 35 (8), 781–788. 10.1038/nbt.3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun B. B.; Maranville J. C.; Peters J. E.; Stacey D.; Staley J. R.; Blackshaw J.; Burgess S.; Jiang T.; Paige E.; Surendran P.; Oliver-Williams C.; Kamat M. A.; Prins B. P.; Wilcox S. K.; Zimmerman E. S.; Chi A.; Bansal N.; Spain S. L.; Wood A. M.; Morrell N. W.; Bradley J. R.; Janjic N.; Roberts D. J.; Ouwehand W. H.; Todd J. A.; Soranzo N.; Suhre K.; Paul D. S.; Fox C. S.; Plenge R. M.; Danesh J.; Runz H.; Butterworth A. S. Genomic atlas of the human plasma proteome. Nature 2018, 558 (7708), 73–79. 10.1038/s41586-018-0175-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cominetti O.; Nunez Galindo A.; Corthesy J.; Oller Moreno S.; Irincheeva I.; Valsesia A.; Astrup A.; Saris W. H.; Hager J.; Kussmann M.; Dayon L. Proteomic Biomarker Discovery in 1000 Human Plasma Samples with Mass Spectrometry. J. Proteome Res. 2016, 15 (2), 389–99. 10.1021/acs.jproteome.5b00901. [DOI] [PubMed] [Google Scholar]
- Liu Y.; Buil A.; Collins B. C.; Gillet L. C.; Blum L. C.; Cheng L. Y.; Vitek O.; Mouritsen J.; Lachance G.; Spector T. D.; Dermitzakis E. T.; Aebersold R. Quantitative variability of 342 plasma proteins in a human twin population. Mol. Syst. Biol. 2015, 11 (1), 786. 10.15252/msb.20145728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmstrom E.; Kilsgard O.; Hauri S.; Smeds E.; Herwald H.; Malmstrom L.; Malmstrom J. Large-scale inference of protein tissue origin in gram-positive sepsis plasma using quantitative targeted proteomics. Nat. Commun. 2016, 7, 10261. 10.1038/ncomms10261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshishian H.; Burgess M. W.; Specht H.; Wallace L.; Clauser K. R.; Gillette M. A.; Carr S. A. Quantitative, multiplexed workflow for deep analysis of human blood plasma and biomarker discovery by mass spectrometry. Nat. Protoc. 2017, 12 (8), 1683–1701. 10.1038/nprot.2017.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier F.; Geyer P. E.; Virreira Winter S.; Cox J.; Mann M. BoxCar acquisition method enables single-shot proteomics at a depth of 10,000 proteins in 100 minutes. Nat. Methods 2018, 15 (6), 440–448. 10.1038/s41592-018-0003-5. [DOI] [PubMed] [Google Scholar]
- Wang F.; Kohan A. B.; Lo C. M.; Liu M.; Howles P.; Tso P. Apolipoprotein A-IV: a protein intimately involved in metabolism. J. Lipid Res. 2015, 56 (8), 1403–18. 10.1194/jlr.R052753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger T.; Wehner A.; Schaab C.; Cox J.; Mann M. Comparative Proteomic Analysis of Eleven Common Cell Lines Reveals Ubiquitous but Varying Expression of Most Proteins. Mol. Cell. Proteomics 2012, 11 (3), M111.014050. 10.1074/mcp.M111.014050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlen M.; Fagerberg L.; Hallstrom B. M.; Lindskog C.; Oksvold P.; Mardinoglu A.; Sivertsson A.; Kampf C.; Sjostedt E.; Asplund A.; Olsson I.; Edlund K.; Lundberg E.; Navani S.; Szigyarto C. A.; Odeberg J.; Djureinovic D.; Takanen J. O.; Hober S.; Alm T.; Edqvist P. H.; Berling H.; Tegel H.; Mulder J.; Rockberg J.; Nilsson P.; Schwenk J. M.; Hamsten M.; von Feilitzen K.; Forsberg M.; Persson L.; Johansson F.; Zwahlen M.; von Heijne G.; Nielsen J.; Ponten F. Proteomics. Tissue-based map of the human proteome. Science 2015, 347 (6220), 1260419. 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- Weinberg R. B.; Dantzker C.; Patton C. S. Sensitivity of serum apolipoprotein A-IV levels to changes in dietary fat content. Gastroenterology 1990, 98 (1), 17–24. 10.1016/0016-5085(90)91285-E. [DOI] [PubMed] [Google Scholar]
- Bochman M. L.; Schwacha A. The Mcm Complex: Unwinding the Mechanism of a Replicative Helicase. Microbiol. Mol. Biol. Rev. 2009, 73 (4), 652–683. 10.1128/MMBR.00019-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culnan D. M.; Cooney R. N.; Stanley B.; Lynch C. J. Apolipoprotein A-IV, a putative satiety/antiatherogenic factor, rises after gastric bypass. Obesity 2009, 17 (1), 46–52. 10.1038/oby.2008.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffaelli M.; Guidone C.; Callari C.; Iaconelli A.; Bellantone R.; Mingrone G. Effect of gastric bypass versus diet on cardiovascular risk factors. Ann. Surg. 2014, 259 (4), 694–9. 10.1097/SLA.0b013e31829d6989. [DOI] [PubMed] [Google Scholar]
- Oller Moreno S.; Cominetti O.; Nunez Galindo A.; Irincheeva I.; Corthesy J.; Astrup A.; Saris W. H. M.; Hager J.; Kussmann M.; Dayon L. The differential plasma proteome of obese and overweight individuals undergoing a nutritional weight loss and maintenance intervention. Proteomics: Clin. Appl. 2018, 12 (1), 1600150. 10.1002/prca.201600150. [DOI] [PubMed] [Google Scholar]
- Hockey K. J.; Anderson R. A.; Cook V. R.; Hantgan R. R.; Weinberg R. B. Effect of the apolipoprotein A-IV Q360H polymorphism on postprandial plasma triglyceride clearance. J. Lipid Res. 2001, 42 (2), 211–7. [PubMed] [Google Scholar]
- Mack S.; Coassin S.; Vaucher J.; Kronenberg F.; Lamina C. Evaluating the Causal Relation of ApoA-IV with Disease-Related Traits - A Bidirectional Two-sample Mendelian Randomization Study. Sci. Rep. 2017, 7 (1), 8734. 10.1038/s41598-017-07213-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan D. C.; Chen M. M.; Ooi E. M.; Watts G. F. An ABC of apolipoprotein C-III: a clinically useful new cardiovascular risk factor?. Int. J. Clin Pract 2008, 62 (5), 799–809. 10.1111/j.1742-1241.2007.01678.x. [DOI] [PubMed] [Google Scholar]
- Yao Z.; Wang Y. Apolipoprotein C-III and hepatic triglyceride-rich lipoprotein production. Curr. Opin. Lipidol. 2012, 23 (3), 206–12. 10.1097/MOL.0b013e328352dc70. [DOI] [PubMed] [Google Scholar]
- Wilson P. W.; D’Agostino R. B.; Levy D.; Belanger A. M.; Silbershatz H.; Kannel W. B. Prediction of coronary heart disease using risk factor categories. Circulation 1998, 97 (18), 1837–47. 10.1161/01.CIR.97.18.1837. [DOI] [PubMed] [Google Scholar]
- von Zychlinski A.; Kleffmann T.; Williams M. J.; McCormick S. P. Proteomics of Lipoprotein(a) identifies a protein complement associated with response to wounding. J. Proteomics 2011, 74 (12), 2881–91. 10.1016/j.jprot.2011.07.008. [DOI] [PubMed] [Google Scholar]
- Utermann G. The mysteries of lipoprotein(a). Science 1989, 246 (4932), 904–10. 10.1126/science.2530631. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.