Abstract
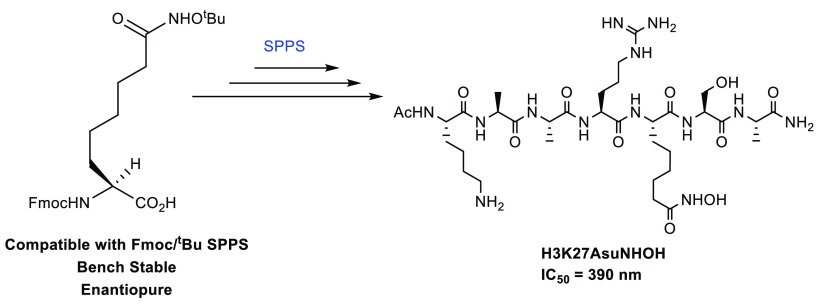
Syntheses of Fmoc amino acids having zinc-binding groups were prepared and incorporated into substrate inhibitor H3K27 peptides using Fmoc/tBu solid-phase peptide synthesis (SPPS). Peptide 11, prepared using Fmoc-Asu(NHOtBu)-OH, is a potent inhibitor (IC50 = 390 nM) of the core NuRD corepressor complex (HDAC1–MTA1–RBBP4). The Fmoc amino acids have the potential to facilitate the rapid preparation of substrate peptidomimetic inhibitor (SPI) libraries in the search for selective HDAC inhibitors.
Epigenetic regulation of gene expression is a key process in the cell that facilitates changes in heritable phenotype without a change in DNA sequence. Gene expression is, in part, regulated by the acetylation state of lysine residues in histone protein tails that coordinate the packaging of DNA into chromatin. Two different classes of lysine-modifying enzymes, the histone deacetylases (HDACs) and the histone acetyltransferases (HATs), are responsible for applying and removing this post-translational modification.1,2 HDAC enzymes have been found to deacetylate other nonhistone proteins and in this context can also be referred to as KDACs.3 Modifications in HDAC function have been linked to numerous diseases including neurological disorders, muscular dystrophy, cardiac hypertrophy, cancer, and HIV infection.4
There are 18 human HDAC enzymes, which have been subdivided into four different classes, based on their sequence homology with yeast proteins.5 The majority of cellular protein acetylation (70–80%) is performed by the class I (1, 2, 3, and 8) HDACs, so an understanding of their specific role in cell biochemistry is important and will provide insight into the epigenetic working of the cell.6,7 Most class I HDAC enzymes (1, 2, and 3) must be recruited to larger corepressor complexes to be fully active. There are at least five corepressor complexes (NuRD, CoREST, MiDAC, SIN3, SMRT/NCoR), and all are believed to have distinct roles in gene expression.8 Understanding the regulation of HDAC activity in the context of corepressor complexes presents a promising strategy to understand the specific role of each complex on gene expression.
Four HDAC inhibitors have been FDA approved for clinical application including Vorinostat (SAHA), Romidepsin (FK228, a prodrug), Belinostat (PXD-101), and Panobinostat (LBH-589, Farydak), with at least 20 others in clinical trials.9 Unfortunately, these inhibitors are nonspecific across various HDACs, and lack of specificity contributes to the cytotoxicity observed in clinical trials.10 Therefore, there is a persistent need to develop isoform-specific and complex-selective HDAC inhibitors.
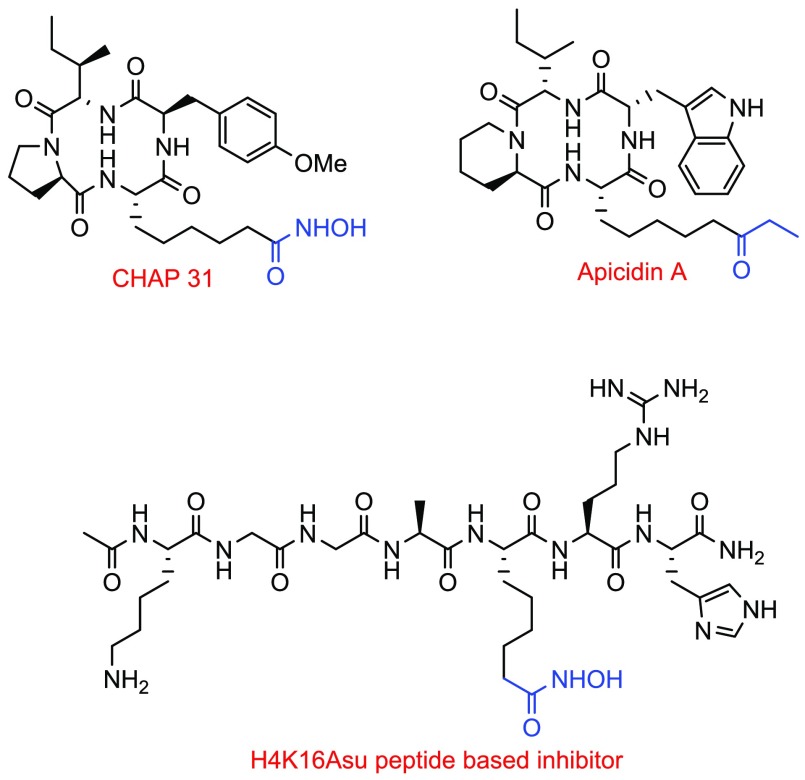
Many cyclic peptide HDAC inhibitors exist in nature such as CHAP31 and Apicidin A, which show class I selective HDAC inhibition in the nanomolar range (Figure 1).11 These inhibitors function by coordinating with the HDAC active site zinc through a side chain that has zinc binding properties such as a hydroxamic acid or ethyl ketone. Recently, we reported a linear substrate peptidomimetic inhibitor (SPI) based on the histone H4 sequence (H4K16Asu) which inhibits the HDAC1–MTA1 complex with IC50 366 nM (Figure 1).12
Figure 1.
Examples of HDAC inhibitors having zinc binding groups.
Despite their attractiveness as tool compounds and as potential therapeutic lead compounds, the synthesis of this type of peptide still poses a significant challenge. Numerous synthetic strategies have been reported to prepare these peptides; however, they all involve complex multistep syntheses. For example, the multistep solution-phase syntheses of CHAP31 and Apicidin A have been reported but require the purification of intermediates.13−15 Furumai et al. reported the synthesis of CHAP31 starting from Z-d-proline and requires purification after each peptide coupling. The unnatural Asu amino acid was initially introduced as the benzyl-protected ester. Following peptide N- to C-terminal macrocyclization the Asu side-chain benzyl-protecting group was removed, and the activated carboxylic acid reacted with hydroxylamine to provide the desired hydroxamic acid (AsuNHOH). This synthetic strategy is only appropriate for peptides such as CHAP31 and Apicidin A with aliphatic amino acid residues. However, histone peptides that incorporate reactive amino acid side chains would require a complex orthogonal protection strategy.
Recently, our group reported a linear inhibitor peptidomimetic (H4K16Asu) using solid-phase peptide synthesis (SPPS) using hydroxylamine resin.12 The reported method required a multistep procedure starting from the Fmoc–oxooctanoic acid–OAll (prepared via Grubbs cross metathesis). The desired peptide was synthesized with a poor overall yield of 1%. The foremost limitation in moving forward to generate this type of peptide-based inhibitor is the lack of availability of the nonproteogenic zinc binding Fmoc-protected amino acids that would provide straightforward synthetic access to this class of molecules via standard Fmoc/tBu SPPS.
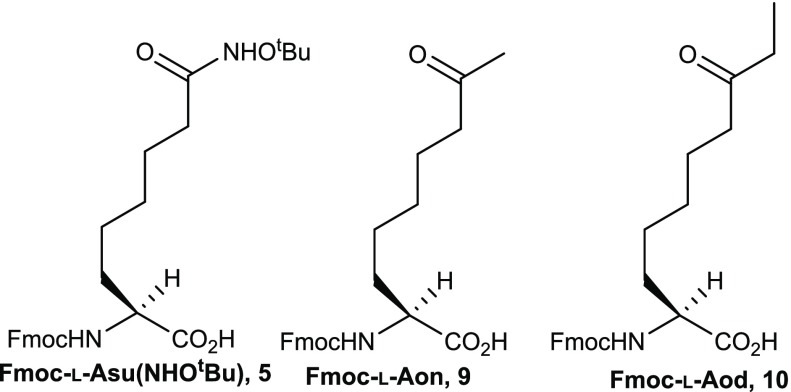
As part of our ongoing research program to develop SPIs, we developed the synthesis of Fmoc amino acids having side chains with functional groups suitable for zinc binding. Here we describe the development of an asymmetric synthesis of these Fmoc-AAs (Figure 2), which are compatible building blocks with standard Fmoc/tBu SPPS.
Figure 2.
Structure of amino acids for SPPS.
Different types of ZBG have been selected to fine-tune the binding affinity with the active site zinc atom, in the search for selective and less toxic HDAC inhibitors. We selected two ZBGs, a ketone with relatively low predicted binding affinity and hydroxamic acid with strong binding affinity.16 The length of the aliphatic side chain in these amino acids is based on the distance between the α-carbon and the side-chain carbonyl on the acetyl lysine residue.17 The crystal structure of the H4K16Asu peptide bound to HDAC1–MTA1 validates this as the correct length and side chains of longer and shorter length have previously been demonstrated to have limited potency.12
Several methods have been reported for the synthesis of nonproteinogenic α-amino acids: Strecker synthesis,18 Grubbs metathesis,19 alkylation of glycine derivatives,20 and oxazolones.21 Initially we focused our attention on the synthesis of the Fmoc amino acids using Grubbs cross metathesis as the key step to prepare the required side-chain functionality. Due to extremely poor yields (5–7% for cross metathesis due to homocoupling) we turned our attention to alkylation of glycine derivatives first reported by Belokon and co-workers.22 This method provided an efficient and highly stereoselective way to prepare the amino acids required for this work and results in the desired Fmoc-protected amino acid analogues required for solid-phase peptide synthesis.
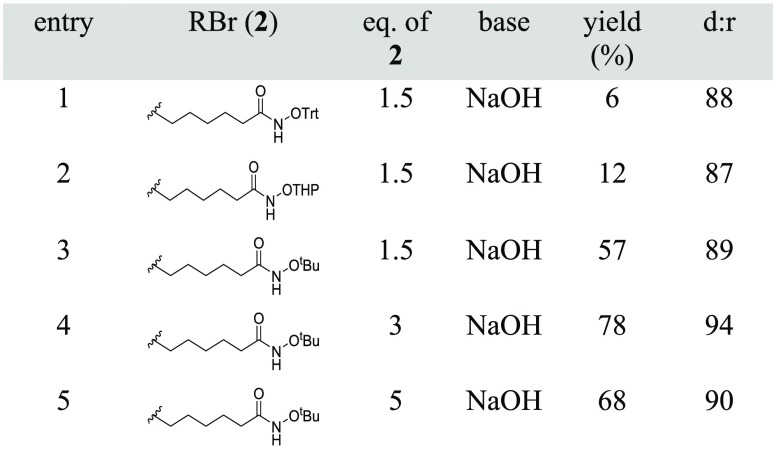
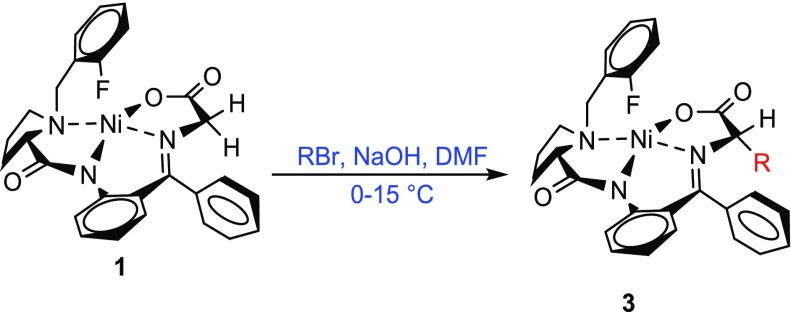
The chiral NiII Schiff base complex (S)-Ni-Gly-2FBPB 1 was prepared by a modified version of our previously reported route (Supporting Information Scheme S1).23,24 Alkylation of (S)-Ni-Gly-2FBPB 1 required orthogonally protected electrophiles compatible with basic conditions for the amino acid synthesis and also Fmoc/tBu SPPS (Figure 2).25 The alkylation reaction was initially attempted using electrophiles 6-bromo-N-(trityl)hexanamide (2a) (Table 1, entry 1) and 6-bromo-N-(THP)hexanamide (2b) (Table 1, entry 2) and resulted in degradation of the product under the reaction conditions and low yields. Stereoselective alkylation was achieved using 6-bromo-N-(tert-butoxy)hexanamide (2c) to provide the desired product 3 in 57% yield (Table 1, entry 3). The reaction conditions were optimized by increasing the equivalents of base (3 equiv) to provide complex 3 diastereoselectively (94:6 d.r.) (Table 1, entry 4). At reduced temperature no bisalkylation was observed. The NiII Schiff base complex 3 was isolated in excellent yield and in diastereomerically pure form (as confirmed by 1H and 19F NMR and LCMS) as a red crystalline solid after automated flash column chromatography.
Table 1. Screening of Protecting Groups for Alkylation of 1 to Generate Fmoc-l-Asu(NHOtBu)-OH.
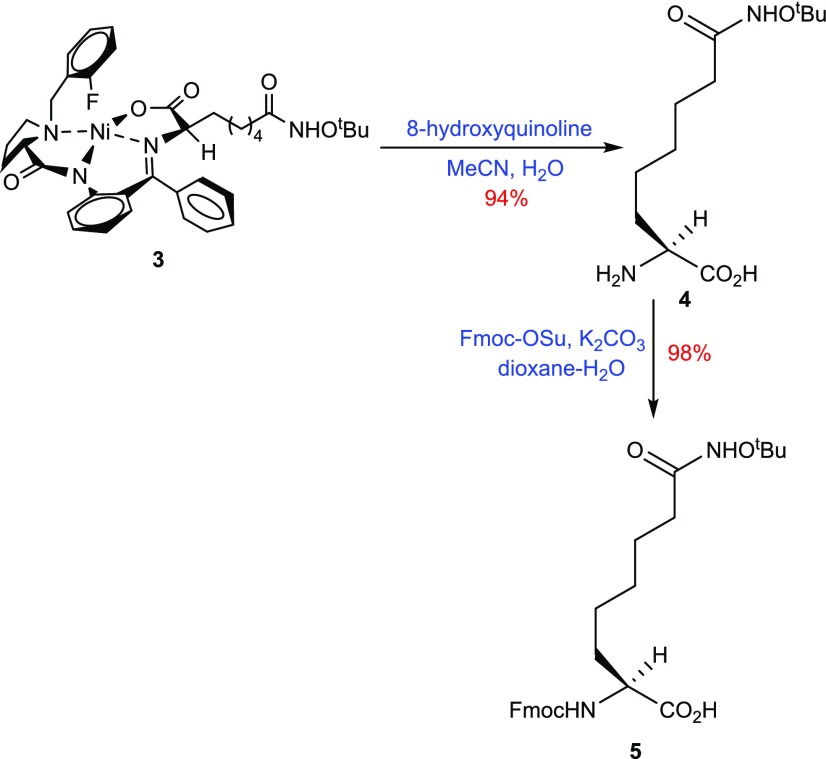
Decomplexation of 3 was achieved under basic conditions using 8-hydroxyquinoline to give the amino acid l-Asu(NHOtBu) 4 and chiral auxiliary (2S)-FBPB, which can be recycled.26 Finally, l-Asu(NHOtBu) 4 was N-protected using N-(9-fluoromethoxycarbonyloxy)succinimide to provide the desired Fmoc-l-Asu(NHOtBu)-OH 5 in excellent 98% yield (Scheme 1).27
Scheme 1. Synthesis of Fmoc-l-Asu(NHOtBu)-OH (5).
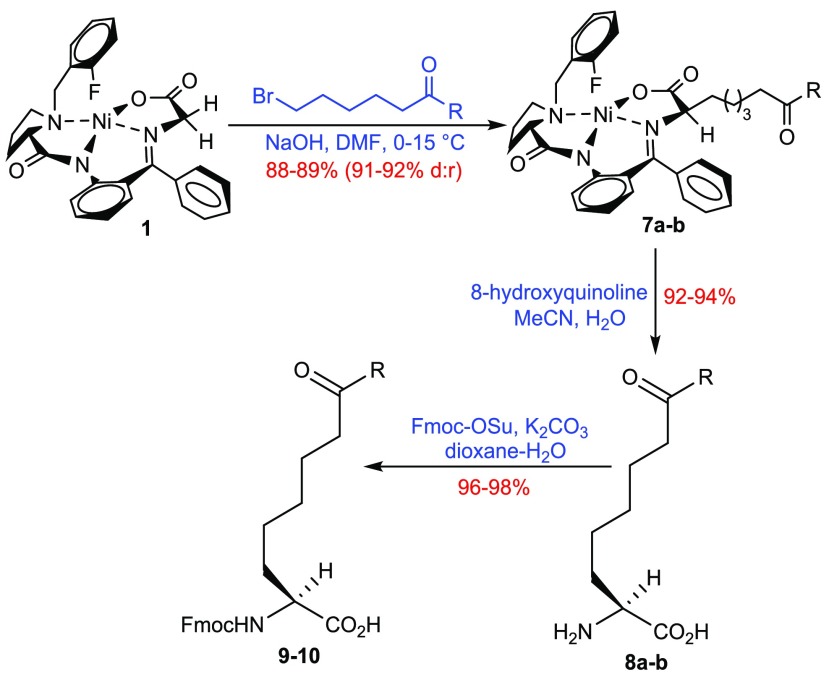
Synthesis of amino acids Fmoc-l-Aon-OH 9 and Fmoc-l-Aod-OH 10 was achieved using the same route and with electrophiles 7-bromoheptan-2-one (6a) and 8-bromooctan-3-one (6b), which were prepared from commercially available 6-bromohexanoic acid (Supporting Information Schemes 3 and 4) via Weinreb amides.28 Alkylation of NiII Schiff base complex 1 with 6a or 6b followed by decomplexation and Fmoc protection gave the desired amino acids [9 (R = −CH3), 10 (R = −C2H5)] with excellent yield at each step (Scheme 2).
Scheme 2. Synthesis of Fmoc-l-Aon 9 (R = −CH3) and Fmoc-l-Aod 10 (R = −C2H5).
To access the chiral integrity of 5, the Fmoc-d-Asu(NHOtBu)-OH 5a isomer was produced for comparison. Chiral integrity was then analyzed by chiral HPLC. As evident from the chromatograms of l- and d-Asu(NHOtBu), we conclude that chirality is retained, and the protocol described herein did not result in any racemization (e.e. > 99%).
The synthesis of these amino acids 5, 9, and 10 was achieved on scale to provide gram quantities of the bench-stable Fmoc-protected amino acid building blocks. The foremost advantage of these amino acids (AAs) is that they can be incorporated at any position in a peptide sequence using standard Fmoc/tBu SPPS conditions. These Fmoc amino acid building blocks therefore facilitate the rapid generation of peptide inhibitors.
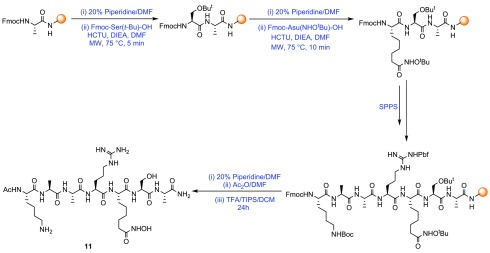
The Fmoc-AA building blocks 5, 9, and 10 were then used to synthesize histone tail derived SPIs using SPPS. Heptapeptide SPI 11 based on residues 23–29 of the unstructured tail region of histone H3 and incorporating the non-native amino acid l-Asu(NHOtBu)-OH in place of K27Ac was prepared by microwave-assisted Fmoc/tBu SPPS on Rink Amide resin (Scheme 3). The histone H3K27Ac (23–29) peptide 14 was also produced as a negative control. Finally, the peptides were cleaved from resin and underwent global deprotection under acidic conditions.
Scheme 3. Synthesis of Peptide (H3K27) Incorporating Fmoc-l-Asu(NHOtBu)-OH.
An impurity corresponding to the hydroxymate t-butyl-protected peptide byproduct was identified in the crude sample following cleavage of peptide 11 from the solid support. Extended deprotection reaction times (up to 48 h) or increased temperature (up to 40 °C) results in hydrolysis of the hydroxamic acid. Hydroxamic acid peptides 11 were obtained when cleavage was performed with [TFA/TIPS/DCM (98:1:1)] for 24 h, providing SPIs 11 in good 29% yield following purification by reverse-phase high-performance liquid chromatography (RP-HPLC).
During acid-mediated cleavage of peptide 12 in the presence of TIPS scavenger, silicon hydride reduction of the ketone functionality occurs, followed by esterification to provide the trifluoroacetate ester (Supporting Information Scheme S5) as detected by LCMS. Water-mediated hydrolysis of the ester then takes place and results in formation of the corresponding alcohol.29 Therefore, TIPS was excluded from the cleavage cocktail for peptides 12 and 13, containing ketone functionality, and successful cleavage and deprotection were achieved using TFA/DCM (95:5, v/v) for 3 h.
SPIs 11–14 were assessed in a fluorescence-based inhibition assay to determine potency as HDAC inhibitors (Table 2). SPI 11 incorporating the hydroxamic acid amino acid has an IC50 = 390 nM against the HDAC1–MTA1–RBBP4 corepressor complex, which is comparable to our previously reported SPI based on the H4 peptide sequence.12 SPI 12 incorporating the ethyl ketone amino acid side chain has an IC50 = 17 μM. Unexpectedly, SPI 13 with methyl ketone side chain was inactive at the tested concentration. The lower activity of SPIs 12–13 can be attributed to the lower zinc binding affinity of ketones. In the future these amino acids will be incorporated in various histone tails to generate a library to identify important binding interactions through additional crystal structure studies. The cell permeability of these peptides will also be investigated to determine their usefulness as cellular probes.
Table 2. Synthesis of H3K27 Peptidomimetics via SPPS.
| sequence | yield (%) | purity (%) | IC50 |
|---|---|---|---|
| Ac-KAARAsuSA-NH211 | 29 | 99 | 390 nM |
| Ac-KAARAodSA-NH212 | 32 | 99 | 17 μM |
| Ac-KAARAonSA-NH213 | 28 | 99 | ND |
| Ac-KAARK(Ac)SA-NH214 | 34 | 99 | ND |
In summary, we have developed the synthesis of three novel Fmoc amino acids incorporating zinc binding group side chains. The foremost advantage of these amino acid building blocks is that they can be incorporated at any position in a peptide sequence using standard Fmoc/tBu SPPS. As proof of principle a series of SPIs based on the structure of the H3K27Ac tail were synthesized by SPPS. One of these analogues 11 was active (IC50 = 390 nM) against the core NuRD corepressor complex (HDAC1–MTA1–RBBP4). This synthetic strategy has the potential to facilitate the rapid generation of peptide-based HDAC inhibitor libraries.
Acknowledgments
We thank the University of Glasgow and the EPSRC (Research Project Grant EP/N034295/1) for financial support of this research. L.A. thanks the EPSRC for a studentship (EP/M506539/1 and EP/N509668/1). JWRS is supported by a Senior Investigator Award (WT100237) from the Wellcome Trust. JWRS is a Royal Society Wolfson Research Merit Award holder. The authors also thank Andrew Monaghan (high-resolution mass spectrometry, University of Glasgow) for technical assistance.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.orglett.9b00885.
Experimental procedures and characterization data for all compounds, copies of 1H,13C, COSY, HSQC, and 19F NMR spectra for building blocks, and analytical HPLC traces for peptides (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- a Allfrey V. G.; Faulkner R.; Mirsky A. E. Proc. Natl. Acad. Sci. U. S. A. 1964, 51, 786–794. 10.1073/pnas.51.5.786. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Struhl K. Genes Dev. 1998, 12, 599–606. 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]; c Biel M.; Wascholowski V.; Giannis A. Angew. Chem., Int. Ed. 2005, 44, 3186–3216. 10.1002/anie.200461346. [DOI] [PubMed] [Google Scholar]; d Narita T.; Weinert B. T.; Choudhary C. Nat. Rev. Mol. Cell Biol. 2019, 20, 156–174. 10.1038/s41580-018-0081-3. [DOI] [PubMed] [Google Scholar]
- a Perri F.; Longo F.; Giuliano M.; Sabbatino F.; Favia G.; Ionna F.; Addeo R.; Scarpati G. D. V.; Di Lorenzo G.; Pisconti S. Crit. Rev. Oncol. Hematol. 2017, 111, 166–172. 10.1016/j.critrevonc.2017.01.020. [DOI] [PubMed] [Google Scholar]; b Conley B. A.; Wright J. J.; Kummar S. Cancer 2006, 107, 832–840. 10.1002/cncr.22064. [DOI] [PubMed] [Google Scholar]; c Bernstein B. E.; Meissner A.; Lander E. S. Cell 2007, 128, 669–681. 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- Choudhary C.; Kumar C.; Gnad F.; Nielsen M. L.; Rehman M.; Walther T. C.; Olsen J. V.; Mann M. Science 2009, 325, 834–840. 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- a Fraga M. F.; Ballestar E.; Villar-Garea A.; Boix-Chornet M.; Espada J.; Schotta G.; Bonaldi T.; Haydon C.; Ropero S.; Petrie K.; Iyer N. G.; et al. Nat. Genet. 2005, 37, 391–400. 10.1038/ng1531. [DOI] [PubMed] [Google Scholar]; b Esteller M. N. Engl. J. Med. 2008, 358, 1148–1159. 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]; c Falkenberg K. J.; Johnstone R. W. Nat. Rev. Drug Discovery 2014, 13, 673–691. 10.1038/nrd4360. [DOI] [PubMed] [Google Scholar]
- a Gregoretti I.; Lee Y.-M.; Goodson H. V. J. Mol. Biol. 2004, 338, 17–31. 10.1016/j.jmb.2004.02.006. [DOI] [PubMed] [Google Scholar]; b Witt O.; Deubzer H. E.; Milde T.; Oehme I. Cancer Lett. 2009, 277, 8–21. 10.1016/j.canlet.2008.08.016. [DOI] [PubMed] [Google Scholar]; c New M.; Olzscha H.; La Thangue N. B. Mol. Oncol. 2012, 6, 637–656. 10.1016/j.molonc.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Haberland M.; Montgomery R. L.; Olson E. N. Nat. Rev. Genet. 2009, 10, 32–42. 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Yang X. J.; Seto E. Nat. Rev. Mol. Cell Biol. 2008, 9, 206–218. 10.1038/nrm2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Micelli C.; Rastelli G. Drug Discovery Today 2015, 20, 718–735. 10.1016/j.drudis.2015.01.007. [DOI] [PubMed] [Google Scholar]; b Kelly R.; Chandru A.; Watson P.; Song Y.; Blades M.; Robertson N.; Jamieson A. G.; Schwabe J. W. R.; Cowley S. M. Sci. Rep. 2018, 8, 14690. 10.1038/s41598-018-32927-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Kalin J. H.; Wu M.; Gomez A. V.; Song Y.; Das J.; Hayward D.; Adejola N.; Wu M.; Panova I.; Chung H. J.; Kim E.; Roberts H. J.; Roberts J. M.; Prusevich P.; Jeliazkov J. R.; Roy Burman S. S.; Fairall L.; Milano C.; Eroglu A.; Proby C. M.; Dinkova-Kostova A. T.; Hancock W. W.; Gray J. J.; Bradner J. E.; Valente S.; Mai A.; Anders N. M.; Rudek M. A.; Hu Y.; Ryu B.; Schwabe J. W. R.; Mattevi A.; Alani R. M.; Cole P. A. Nat. Commun. 2018, 9, 53. 10.1038/s41467-017-02242-4. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Watson P. J.; Fairall L.; Schwabe J. W. R. Mol. Cell. Endocrinol. 2012, 348, 440–449. 10.1016/j.mce.2011.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Millard C. J.; Watson P. J.; Fairall L.; Schwabe J. W. R. Trends Pharmacol. Sci. 2017, 38, 363–377. 10.1016/j.tips.2016.12.006. [DOI] [PubMed] [Google Scholar]
- a Duvic M.; Talpur R.; Ni X.; Zhang C.; Hazarika P.; Kelly C.; Chiao J. H.; Reilly J. F.; Ricker J. L.; Richon V. M.; Frankel S. R. Blood 2007, 109, 31–39. 10.1182/blood-2006-06-025999. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Butler L. M.; Zhou X.; Xu W. S.; Scher H. I.; Rifkind R. A.; Marks P. A.; Richon V. M. Proc. Natl. Acad. Sci. U. S. A. 2002, 99, 11700–11705. 10.1073/pnas.182372299. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Taori K.; Paul V. J.; Luesch H. J. Am. Chem. Soc. 2008, 130, 1806–1807. 10.1021/ja7110064. [DOI] [PubMed] [Google Scholar]; d Sawas A.; Radeski D.; O’Connor O. A. Ther. Adv. Hematol. 2015, 6, 202–208. 10.1177/2040620715592567. [DOI] [PMC free article] [PubMed] [Google Scholar]; e Atadja P. Cancer Lett. 2009, 280, 233–241. 10.1016/j.canlet.2009.02.019. [DOI] [PubMed] [Google Scholar]
- a Kang S. P.; Ramirez J.; House L.; Zhang W.; Mirkov S.; Liu W.; Haverfield E.; Ratain M. J. A. Pharmacogenet. Genomics 2010, 20, 638–641. 10.1097/FPC.0b013e32833e1b37. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Garcia-Manero G.; Minden M.; Estrov Z.; Verstovsek S.; Newsome W. M.; Reid G.; Besterman J.; Li Z.; Pearce L.; Martell R. J. Clin. Oncol. 2006, 24, 337. 10.1200/JCO.2005.01.5784. [DOI] [PubMed] [Google Scholar]; c Mottamal M.; Zheng S.; Huang T. L.; Wang G. Molecules 2015, 20, 3898–3941. 10.3390/molecules20033898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Han J. W.; Ahn S. H.; Park S. H.; Wang S. Y.; Bae G. U.; Seo D. W.; Kwon H. K.; Hong S.; Lee H. Y.; Lee Y. W.; Lee H. W. Cancer Res. 2000, 60, 6068–6074. [PubMed] [Google Scholar]; b Yoshida M.; Furumai R.; Nishiyama M.; Komatsu Y.; Nishino N.; Horinouchi S. Cancer Chemother. Pharmacol. 2001, 48, S20–S26. 10.1007/s002800100300. [DOI] [PubMed] [Google Scholar]; c Newkirk T. L.; Bowers A. A.; Williams R. M. Nat. Prod. Rep. 2009, 26, 1293–1320. 10.1039/b817886k. [DOI] [PubMed] [Google Scholar]; d Mwakwari S.; Patil V.; Guerrant W.; K Oyelere A. Curr. Top. Med. Chem. 2010, 10, 1423–1440. 10.2174/156802610792232079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson P. J.; Millard C. J.; Riley A. M.; Robertson N. S.; Wright L. C.; Godage H. Y.; Cowley S. M.; Jamieson A. G.; Potter B. V.; Schwabe J. W. R. Nat. Commun. 2016, 7, 11262. 10.1038/ncomms11262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Furumai R.; Komatsu Y.; Nishino N.; Khochbin S.; Yoshida M.; Horinouchi S. Proc. Natl. Acad. Sci. U. S. A. 2001, 98, 87–92. 10.1073/pnas.98.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Komatsu Y.; Tomizaki K. Y.; Tsukamoto M.; Kato T.; Nishino N.; Sato S.; Yamori T.; Tsuruo T.; Furumai R.; Yoshida M.; Horinouchi S. Cancer Res. 2001, 61, 4459–4466. [PubMed] [Google Scholar]
- a Mou L.; Singh G. Tetrahedron Lett. 2001, 42, 6603–6606. 10.1016/S0040-4039(01)01331-4. [DOI] [Google Scholar]; b Berst F.; Ladlow M.; Holmes A. B. Chem. Commun. 2002, 5, 508–509. 10.1039/b110068h. [DOI] [PubMed] [Google Scholar]
- a Dose A.; Sindlinger J.; Bierlmeier J.; Bakirbas A.; Schulze-Osthoff K.; Einsele-Scholz S.; Hartl M.; Essmann F.; Finkemeier I.; Schwarzer D. Angew. Chem., Int. Ed. 2016, 55, 1192–1195. 10.1002/anie.201508174. [DOI] [PubMed] [Google Scholar]; b Wu M.; Hayward D.; Kalin J. H.; Song Y.; Schwabe J. W. R.; Cole P. A. eLife 2018, 7, 37231. 10.7554/eLife.37231. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Maolanon A. R.; Kristensen H. M.; Leman L. J.; Ghadiri M. R.; Olsen C. A. ChemBioChem 2017, 18, 5–49. 10.1002/cbic.201600519. [DOI] [PubMed] [Google Scholar]
- a Chen K.; Xu L.; Wiest O. J. Org. Chem. 2013, 78, 5051–5055. 10.1021/jo400406g. [DOI] [PMC free article] [PubMed] [Google Scholar]; b McCarren P.; Hall M. L.; Whitehead L. Chem. Biol. Drug Des. 2012, 80, 203–214. 10.1111/j.1747-0285.2012.01382.x. [DOI] [PubMed] [Google Scholar]
- Finnin M. S.; Donigian J. R.; Cohen A.; Richon V. M.; Rifkind R. A.; Marks P. A.; Breslow R.; Pavletich N. P. Nature 1999, 401, 188. 10.1038/43710. [DOI] [PubMed] [Google Scholar]
- a Harada K. Nature 1963, 200, 1201. 10.1038/2001201a0. [DOI] [PubMed] [Google Scholar]; b Yet L. Angew. Chem., Int. Ed. 2001, 40, 875–877. . [DOI] [PubMed] [Google Scholar]; c Wang J.; Liu X.; Feng X. Chem. Rev. 2011, 111, 6947–6983. 10.1021/cr200057t. [DOI] [PubMed] [Google Scholar]
- a Miller S. J.; Blackwell H. E.; Grubbs R. H. J. Am. Chem. Soc. 1996, 118, 9606–9614. 10.1021/ja961626l. [DOI] [Google Scholar]; b Chatterjee A. K.; Choi T.-L.; Sanders D. P.; Grubbs R. H. J. Am. Chem. Soc. 2003, 125, 11360–11370. 10.1021/ja0214882. [DOI] [PubMed] [Google Scholar]; c Wang Z. J.; Spiccia N. D.; Jackson W. R.; Robinson A. J. J. Pept. Sci. 2013, 19, 470–476. 10.1002/psc.2522. [DOI] [PubMed] [Google Scholar]
- a O’Donnell M. J.; Bennett W. D.; Wu S. J. Am. Chem. Soc. 1989, 111, 2353–2355. 10.1021/ja00188a089. [DOI] [Google Scholar]; b Belokon Y. N.; Bespalova N. B.; Churkina T. D.; Císařová I.; Ezernitskaya M. G.; Harutyunyan S. R.; Hrdina R.; Kagan H. B.; Kočovský P.; Kochetkov K. A.; Larionov O. V.; et al. J. Am. Chem. Soc. 2003, 125, 12860–12871. 10.1021/ja035465e. [DOI] [PubMed] [Google Scholar]; c Sorochinsky A. E.; Aceña J. L.; Moriwaki H.; Sato T.; Soloshonok V. A. Amino Acids 2013, 45, 691–718. 10.1007/s00726-013-1539-4. [DOI] [PubMed] [Google Scholar]
- a Wang T.; Yu Z.; Hoon D. L.; Phee C. Y.; Lan Y.; Lu Y. J. Am. Chem. Soc. 2016, 138, 265–271. 10.1021/jacs.5b10524. [DOI] [PubMed] [Google Scholar]; b Cabrera S.; Reyes E.; Aleman J.; Milelli A.; Kobbelgaard S.; Jørgensen K. A. J. Am. Chem. Soc. 2008, 130, 12031–12037. 10.1021/ja804567h. [DOI] [PubMed] [Google Scholar]; c Fisk J. S.; Mosey R. A.; Tepe J. J. Chem. Soc. Rev. 2007, 36, 1432–1440. 10.1039/b511113g. [DOI] [PubMed] [Google Scholar]
- Belokon Y. N.; Bakhmutov V. I.; Chernoglazova N. I.; Kochetkov K. A.; Vitt S. V.; Garbalinskaya N. S.; Belikov V. M. J. Chem. Soc., Perkin Trans. 1 1988, 2, 305–312. 10.1039/P19880000305. [DOI] [Google Scholar]
- Aillard B.; Robertson N. S.; Baldwin A. R.; Robins S.; Jamieson A. G. Org. Biomol. Chem. 2014, 12, 8775–8782. 10.1039/C4OB01832J. [DOI] [PubMed] [Google Scholar]
- Li B.; Zhang J.; Xu Y.; Yang X.; Li L. Tetrahedron Lett. 2017, 58, 2374–2377. 10.1016/j.tetlet.2017.05.007. [DOI] [Google Scholar]
- Wuts P. G.; Greene T. W. In Greene’s protective groups in organic synthesis; John Wiley & Sons, 2006. [Google Scholar]
- Yuan G.; Lingfang K.; Yingchun L.. Method for preparing chiral alpha-unnatural amino acid by transition metal complex, Patent China, 2013.
- Obkircher M.; Stähelin C.; Dick F. J. Pept. Sci. 2008, 14, 763–766. 10.1002/psc.1001. [DOI] [PubMed] [Google Scholar]
- a Jones P.; Altamura S.; De Francesco R.; Paz O. G.; Kinzel O.; Mesiti G.; Monteagudo E.; Pescatore G.; Rowley M.; Verdirame M.; Steinkühler C. J. Med. Chem. 2008, 51, 2350–2353. 10.1021/jm800079s. [DOI] [PubMed] [Google Scholar]; b Nahm S.; Weinreb S. M. Tetrahedron Lett. 1981, 22, 3815–3818. 10.1016/S0040-4039(01)91316-4. [DOI] [Google Scholar]; c Qu B.; Collum D. B. J. Org. Chem. 2006, 71, 7117–7119. 10.1021/jo061223w. [DOI] [PubMed] [Google Scholar]
- a Doyle M. P.; DeBruyn D. J.; Donnelly S. J.; Kooistra D. A.; Odubela A. A.; West C. T.; Zonnebelt S. M. J. Org. Chem. 1974, 39, 2740–2747. 10.1021/jo00932a015. [DOI] [Google Scholar]; b Kursanov D. N.; Parnes Z. N.; Bassova G. I.; Loim N. M.; Zdanovich V. I. Tetrahedron 1967, 23, 2235–2242. 10.1016/0040-4020(67)80059-0. [DOI] [Google Scholar]; c Doyle M. P.; West C. T. J. Org. Chem. 1975, 40, 3835–3838. 10.1021/jo00914a004. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.