Abstract
Ischemic stroke (IS) is the leading cause of disability. Researchers have demonstrated that IS is more a multifactorial disorder than a single-factor disease. At present, no consistent conclusions have been reached on susceptibility loci for IS on chromosome 9p21. We conducted this meta-analysis to verify whether genetic loci on chromosome 9p21 reported domestically and abroad could be responsible for IS in Chinese populations. We analyzed data from eight studies that covered a total of 9756 individuals with Chinese ancestry comprising 4254 cases and 5502 controls. Each of the four reported susceptibility loci (rs2383206, rs2383207, rs10757274, and rs10757278) was analyzed separately. The odds ratios (ORs) of rs2383206 and rs10757274 were 1.09 (95% confidence interval (CI): 1.02–1.06, P = 0.01) and 1.09 (95% CI: 1.01–1.17, P = 0.03), respectively. For rs2383207, OR value was 0.91 (95% CI: 0.84–0.98, P = 0.01). No statistical association was identified for rs10757278. We have verified previous associations for IS in Chinese populations on chromosome 9p21. Loci rs2383206 and rs10757274 may increase susceptibility to IS. Mutation at locus rs2383207 may be beneficial. However, we are unable to identify any association between rs10757278 and IS.
Keywords: Chinese, chromosome 9p21, ischemic stroke, rs2383206, rs2383207, rs10757274, rs10757278
Introduction
Stroke is one of the most common diseases and leads to a high rate of disability and mortality. In the United States, stroke is the fifth leading cause of death, and a person dies from stroke every 4 min on average.1 Apart from the influence of traditional risk factors, the evidence suggests that genetic variants could also affect the occurrence and progression of stroke.
Given that stroke has a complex genetic background, genome-wide association studies (GWAS) were used to explore genetic variations that could be responsible for stroke. As the most common subtype of stroke,1 ischemic stroke (IS) has received the most attention in research studies. Kubo et al.2 first described that the PRKCH gene plays a role in the pathogenesis of cerebral infarction in a Japanese population in 2007. Soon afterwards, several loci on chromosome 9p21, the HDAC9 gene on chromosome 7p21, and rs556621 on chromosome 6p21 were found to correlate with large artery atherosclerotic stroke (LAS).
In the last 10 years, there were controversies about the actual relationship between chromosome 9p21 and LAS, whether in Caucasians or Chinese. Wahlstrand et al.3 first found that the allele G on rs2383207 and rs10757278 could increase risk of stroke in 2009. These two loci have been studied intensively, but no conclusion has been reached. In addition, the study of loci such as rs10757274, rs1537378, and rs1333040 has become very popular in Caucasian populations. In China, rs1333049, rs10757274, rs10757278, rs2383207, and rs2383206 have attracted a fair amount of attention.
Traylor et al.,4 Lancet Neurology 2012, performed a large-scale study that analyzed data from a 14-year IS cohort and conducted a replication with 13,347 cases and 29,083 controls. It verified associations between the PITX2 gene, the ZFHX3 gene, and cardio embolic stroke (CES) and between chromosome 9p21, the HDAC9 gene, and LAS, but not between the PRKCH gene and stroke.4 The frequencies of locus rs2230500 on the PRKCH gene are different between East Asian and European populations. The frequency of the allele G on rs2230500 is nearly 100% in European but 73% in East Asian people (data from 1000 Genomes Project Phase 3). Therefore, it might be that studies including people of different races came to different conclusions on the PRKCH gene at home and abroad.
NINDS Stroke Genetics Network (SiGN) and International Stroke Genetics Consortium (ISGC),5 Lancet Neurology 2016, identified risks of variants of the PITX2, ZFHX3, and HDAC9 genes via a two-stage GWAS with 37,893 cases and 397,209 controls. For chromosome 6p21 and 9p21, evidence was weak. In their analysis, only one study included East Asian participants.
Frequencies of A-allele and G-allele are different in different races. Therefore, similar to the PRKCH gene, the role of chromosome 9p21 in IS may be different in Chinese and Caucasian people. Considering inconsistent results and differences in population frequencies of different races, we performed this meta-analysis to obtain a more precise estimation on relevant IS risk loci in Chinese populations.
Materials and methods
Literature search strategy
Relevant studies published before the end of April 2017 focused on IS were identified through a search of MEDLINE (via PubMed), EMBASE (via Ovid), Cochrane, Chinese National Knowledge Infrastructure (CNKI), and WANFANG MED ONLINE. Searches were performed independently by two of the authors (Man Li and Jing Liu) without language restrictions. Search keywords were exemplary terms of IS (e.g. “cerebral infarction,” “cerebral ischemia,” “ischemic stroke,” “ischemic infarction,” “ischaemic stroke,” “ischaemic infarction,” “cerebral ischaemia,” and the Mesh word “cerebral infarction”) in combination with words relating to locus (e.g. “chromosome 9,” “9p21,” “rs10757278,” “rs10757274,” “rs2383207,” “rs2383206,” and “CDKN2B-AS1”). The terms IS and loci were connected via the Boole logical operator “or” separately. Then, an “and” was used to link the two keywords to get the most comprehensive search results. Each locus was searched separately. Titles and abstracts of search results were screened to determine their relevance. Studies that overlapped with other published studies were compared to pick the most comprehensive ones. Full texts of potential articles were read to determine the usefulness of the information they contained.
Selection criteria
Concerning the distributional difference, loci whose minimum allele frequencies were higher than 5% in the Chinese Han population were taken into consideration. We searched each controversial locus on 9p21 reported in papers for each allele frequency. Four loci (rs2383206, rs2383207, rs10757274, and rs10757278) were ultimately included. Meta-analyses on associations between rs2383206, rs2383207, rs10757274, and IS have not been performed. Two meta-analyses have been performed on associations between rs10757278 and IS. rs1537378 was another locus that was studied in Caucasian populations; however, only two studies have concentrated on it. We did not obtain the complete data from one of the studies because the authors only displayed the comparison between patients without carotid plaques and controls.
Case-control and nested case-control designs were eligible to be used in all participating studies. The Newcastle–Ottawa Scale (NOS) was used to grade those potential articles. NOS was designed to assess the quality of non-randomized studies and was recommended for meta-analyses (http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp). The selection, comparability of the study groups, and ascertainment of the outcome of interest are the three factors that were used for judging. An NOS score of 5 or higher for each study was feasible. In addition, independent data must be contained in original papers, so we excluded the studies without full text for which we failed to correspond with the authors. In addition, case-only studies, reviews, and meta-analyses were excluded.
Data extraction
According to the criteria listed above, we (Man Li and Jing Liu) extracted information from the eligible publications independently. Lei Zhao resolved discrepancies in data extraction and obtained a consensus. Self-reported identifications on race and cases were collected from each paper. IS cases were confirmed through imaging data and clinical manifestations that met the World Health Organization (WHO)/International Classification of Diseases (9th revision; ICD-9) criteria. Data were grouped according to different races self-reported in each paper.
Statistical methods
OR and corresponding 95% confidence interval (95% CI) were used to assess the strength of the associations between single-nucleotide polymorphisms (SNPs) and IS risk. A chi-square test was used to examine the deviation from Hardy–Weinberg equilibrium (HWE). I2 (percentage of effect size attributable to heterogeneity) was used to estimate heterogeneity through the Cochran Q test. A fixed-effects model was adopted when I2 was lower than 50%. Otherwise, a random-effects model was used. Publication bias was presented by P value of the Harbord test (threshold for significance was 0.05). P < 0.05 was indicative of an absence of publication bias. Sensitivity analyses were performed if controls of studies were unable to meet HWE or I2 was higher than 50%. Since there were no multiple comparisons of rate in the comparisons of genetic models (additive, homozygous, heterozygous, dominant, and recessive models), the P value threshold for significance was 0.05.
Results
Selections of studies
Although some loci are in high linkage disequilibrium with each other, they are not in complete linkage disequilibrium, and conclusions of correlative studies are thus incompatible. Therefore, we searched the database for each locus separately. The workflow used in this meta-analysis is shown in Figure 1. For rs2383206, five studies were ultimately included, with 3499 cases and 4026 controls.6–10 For rs2383207, there were three studies, with 2584 cases and 3695 controls.8,10,11 Four studies with 2512 cases and 2981 controls7–10 met the criteria for inclusion for rs10757274, and four studies with 3074 cases and 3517 controls were included for rs10757278.6,8,10,12 Table 2 scored all included studies according to NOS.
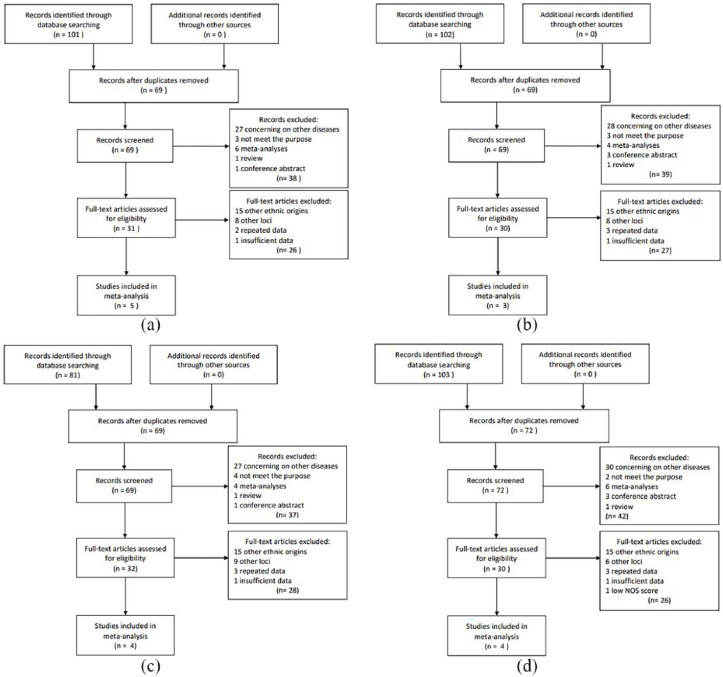
Figure 1.
A search of MEDLINE (via PubMed), EMBASE (via Ovid), and Cochrane was performed for English articles, while Chinese National Knowledge Infrastructure (CNKI) and WANFANG MED ONLINE were searched for Chinese articles. After study collection, we first removed duplicates. Then, we expurgated other types of articles or studies concerning on other diseases by screening abstracts. Full-text articles were browsed to eliminate repeated, ineligible, or insufficient data (we had to exclude insufficient data because of failure of making contact with the authors). Finally, articles that had a Newcastle–Ottawa score lower than 5 were removed. We finally included five studies for rs2383206, three for rs2383207, four for rs10757274, and four for rs10757278: (a) selection process of rs2383206, (b) selection process of rs2383207, (c) selection process of rs10757274, and (d) selection process of rs10757278.
Table 2.
Summary of ORs in this meta-analysis.
| Locus | No. of studies | Homozygous model |
Heterozygous model |
Dominant model |
Recessive model |
||||
|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | ||
| rs2383206 | 6a | GG/AA (fixed-effect model, I2 = 10%, P = 0.35) | AG/AA (fixed-effect model, I2 = 24%, P = 0.25) | AG + GG/AA (fixed-effect model, I2 = 3%, P = 0.40) | GG/AG + AA (fixed-effect model, I2 = 43%, P = 0.12) | ||||
| 1.18 (1.03, 1.34) | 0.01 | 1.06 (0.95, 1.18) | 0.28 | 1.10 (0.99, 1.21) | 0.07 | 1.14 (1.02, 1.27) | 0.02 | ||
| rs2383207 | 3 | AA/GG (fixed-effect model, I2 = 0, P = 0.43) | AG/GG (fixed-effect model, I2 = 0, P = 0.61) | AG + AA/GG (fixed-effect model, I2 = 0, P = 0.49) | AA/AG + GG (fixed-effect model, I2 = 0, P = 0.51) | ||||
| 0.83 (0.71, 0.99) | 0.03 | 0.89 (0.79, 0.99) | 0.03 | 0.87 (0.79, 0.97) | 0.01 | 0.89 (0.76, 1.04) | 0.14 | ||
| rs10757274 | 4 | GG/AA (fixed-effect model, I2 = 0, P = 0.51) | AG/AA (fixed-effect model, I2 = 43%, P = 0.15) | AG + GG/AA (fixed-effect model, I2 = 5%, P = 0.37) | GG/AG + AA (fixed-effect model, I2 = 44%, P = 0.15) | ||||
| 1.18 (1.01, 1.37) | 0.03 | 1.05 (0.93, 1.19) | 0.43 | 1.09 (0.97, 1.23) | 0.15 | 1.15 (1.01, 1.31) | 0.04 | ||
| rs10757278 | 5a | GG/AA (random-effect model, I2 = 55%, P = 0.06) | AG/AA (fixed-effect model, I2 = 39%, P = 0.16) | AG + GG/AA (random-effect model, I2 = 57%, P = 0.06) | GG/AG + AA (random-effect model, I2 = 59%, P = 0.04) | ||||
| 1.18 (0.94, 1.49) | 0.16 | 0.98 (0.87, 1.10) | 0.70 | 1.05 (0.88, 1.25) | 0.60 | 1.17 (0.95, 1.45) | 0.14 | ||
CI: confidence interval; OR: odds ratio.
For rs2383206, rs10757274, and rs10757278, allele A was the major allele. Therefore, the homozygous model was GG versus AA, the heterozygous model was AG versus AA, the dominant model was the sum of AG and GG versus AA, and the recessive model was GG versus the summation of AG and AA. For rs2383207, since allele G accounts for the majority, the homozygous model was AA versus GG, the heterozygous model was AG versus GG, the dominant model was the sum of AG and AA versus GG, and the recessive model was AA versus the summation of AG and GG.
The values given in boldface mean that the ORs were of statistical significance.
Ding et al.6 was a two-stage research, so we divided it into two studies.
Table 1 displays detailed characteristics of studies. The diagnostic bases of cases, allele designations and frequencies, matching criteria for controls, genotyping methods, and numbers of cases and controls were extracted, and the HWE values of the controls were calculated.
Table 1.
Characteristics of the included studies.
| Study | Allele designations and frequencies | Case confirmation | Match criteria for controls | No. of cases/controls (values of H-W in controls) | Quality assessments | Reference |
|---|---|---|---|---|---|---|
| Ding et al.6 | rs2383206 First stage: (A→G, P = 0.164) Second stage: (A→G, P = 0.051) rs10757278 (A→G, P = 0.249) Second stage: (A→G, P = 0.030) |
NE, CT, or MRI according to ICD-9 criteria | Ethnicity and resident area | First stage: 551/556 (0.252) Second stage: 440/498 (0.122) First stage: 558/554 (0.204) Second stage: 441/501 (0.102) |
6 | 6 |
| Hu et al.7 | rs2383206 (A→G, P = 0.062) rs10757274 (A→G, P = 0.181) |
CT and/or MRI | Sex and ethnicity | 352/423 (0.169) 353/430 (0.579) |
5 | 7 |
| Zhang et al.10 | rs2383206 (A→G, P = 0.010) rs2383207 (G→A, P = 0.180) rs10757274 (A→G, P = 0.001) rs10757278 (A→G, P = 0.002) |
NE, CT or MRI according to WHO criteria | Age, sex, ethnicity, and resident area | 1190/1664 (0.529) 1190/1664 (0.266) 1190/1664 (0.376) 1190/1664 (0.999) |
9 | 10 |
| Yue et al.8 | rs2383206 (A→G, P = 0.500, adjusted P = 0.610) rs2383207 (G→A, P = 0.040, adjusted P = 0.030) rs10757274 (A→G, P = 0.880, adjusted P = 0.950) rs10757278 (A→G, P = 0.870, adjusted P = 0.950) |
CT or MRI | Age, sex, and ethnicity | 766/680 (0.429) 767/682 (0.504) 769/682 (0.909) 769/680 (0.831) |
7 | 8 |
| Xiong et al.9 | rs2383206 (A→G, P = 0.636) rs10757274 (A→G, P = 0.449) |
According to WHO criteria | Age, sex, and ethnicity | 200/205 (0.579) 200/205 (0.403) |
7 | 9 |
| Lin et al.11 | rs2383207 (G→A, P = 0.057, adjusted P = 0.097) |
CT or MRI according to WHO criteria | Ethnicity | 627/1349 (0.660) | 6 | 11 |
| Bi et al.12 | rs10757278 (A→G, P = 0.003) |
NE, CT, or MRI according to ICD-9 criteria | Age, sex, and ethnicity | 116/118 (0.307) | 7 | 12 |
NE: neurological examination; CT: computed tomography; MRI: magnetic resonance imaging; ICD-9 criteria: International Classification of Diseases (9th revision); H-W: Hardy–Weinberg equilibrium; WHO: World Health Organization.
Quality assessment was done according to Newcastle–Ottawa Scale; 5/9 was thought to be an eligible score for quality assessments.
Association between loci polymorphisms and IS susceptibility
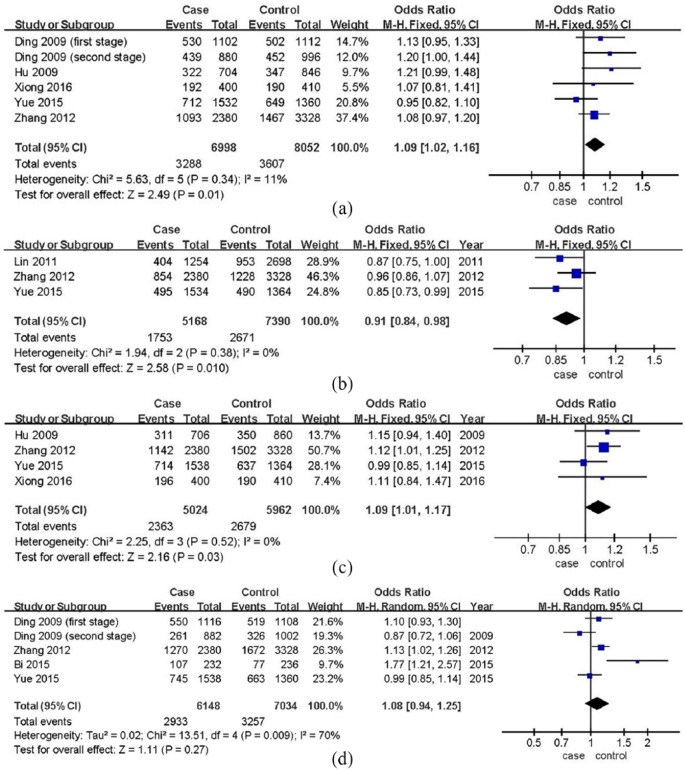
For rs2383206, allele G was the minor allele (the variant allele); the additive model was G versus A. A fixed-effect model (I2 = 11%, P = 0.34) was used in comparison of alleles (Figure 2(a)). rs2383206 polymorphism was significantly associated with IS risk, and allele G increased risk (OR: 1.09, 95% CI: 1.02–1.06, P = 0.01, Figure 2(a)). No significant heterogeneity was identified, so fixed-effects models were applied in other genetic models. The homozygous (OR: 1.18, 95% CI: 1.03–1.34, P = 0.01, Table 2) and recessive models (OR: 1.14, 95% CI: 1.02–1.27, P = 0.02, Table 2) appeared to be statistically significant. No significant association was found in the heterozygous (P = 0.28, Table 2) and dominant models (P = 0.07, Table 2).
Figure 2.
displayed the additive models (comparisons of alleles). Since Ding et al.6 was a two-stage study, we divided it into two studies in comparisons of rs2383206 and rs10757278. No significant heterogeneity was identified in allele comparisons of rs2383206, rs2383207, and rs10757274 (I2 < 50%), so fixed-effects models were applied. For rs10757278, a random-effects model was used (I2 > 50%). Polymorphisms of rs2383206 and rs10757274 were significantly associated with IS risk, and allele G increased the risks (allele G was mutant type). In addition, a mutant from allele G to A of rs2383207 could protect subjects from IS. No statistical association was identified in allele comparison of rs10757278: (a) allele comparison of rs2383206 (G versus A), (b) allele comparison of rs2383207 (A versus G), (c) allele comparison of rs10757274 (G versus A), and (d) allele comparison of rs10757278 (G versus A).
Allele A was the variant allele of rs2383207; the additive model was A versus G. Allele A could protect subjects from IS in a fixed-effect model in allele comparison (I2 = 0%, P = 0.38), with per-allele OR 0.91 (95% CI: 0.84–0.98, P = 0.01, Figure 2(b)). Fixed-effects models were used in other genetic models due to the absence of significant heterogeneity. Statistically significant differences were not found in the recessive model (P = 0.14, Table 2) but were found in the homozygous model (OR: 0.83, 95% CI: 0.71–0.99, P = 0.03, Table 2), heterozygous (OR: 0.89, 95% CI: 0.79–0.99, P = 0.03, Table 2), and dominant models (OR: 0.87, 95% CI: 0.79–0.97, P = 0.01, Table 2).
A combination of four studies indicated an elevated risk of IS with allele G of rs10757274 (OR: 1.09, 95% CI: 1.01–1.17, P = 0.03, Figure 2(c)) in a fixed-effect model (I2 = 0%, P = 0.52). The additive model was G versus A. Similarly, fixed-effects models were applied in all genetic models. The homozygous (OR: 1.18, 95% CI: 1.01–1.37, P = 0.03, Table 2) and recessive models (OR: 1.15, 95% CI: 1.01–1.31, P = 0.04, Table 2) were statistically significant; no difference was found when comparing the heterozygous (P = 0.43, Table 2) and dominant models (P = 0.15, Table 2).
For rs10757278, the additive model was G versus A. No statistical association (P = 0.27, Figure 2(d)) was identified in the allele comparison (random-effects model, I2 = 70%, P = 0.009, Figure 2(d)) or any genetic model (Table 2).
Publication bias
A Harbord test was used to evaluate publication bias. No publication bias was found in studies of rs2383206 (P = 0.516), rs10757274 (P = 0.998), or rs10757278 (P = 0.687). For rs2383207, there might be publication bias (P = 0.046).
Sensitivity analysis
A fixed-effects model was used when I2 was below 50%; otherwise, a random-effects model was applied. All models of rs2383206, rs2383207, and rs10757274 had low heterogeneities, and fixed-effects models were used. For comparisons of rs10757278, excluding comparison of the heterozygous model, I2 was high. Therefore, we removed each study one at a time to examine the influence of each individual study. I2 was higher than 50% throughout the analysis in the allele comparison.
Discussion
Chromosome 9p21 was initially found to be associated with CAD. The etiology and pathogenesis of coronary artery disease (CAD) and stroke share some similarities, and 9p21 is a shared susceptibility locus.13 The relationship between chromosome 9p21 and stroke has been studied intensively. However, no consistent conclusion has been reached. In our meta-analysis, significant associations between polymorphisms of rs2383206, rs2383207, and rs10757274 and IS in the Chinese population were demonstrated.
In comparisons of each genetic model of rs2383206, both the homozygous and recessive models were statistically significant, while the other two models were not. rs2383206 was tested only in the Chinese population. We have confirmed that mutation from the A-allele to the G-allele might increase susceptibility to stroke, and the GG genotype is associated with a high risk of stroke. I2 remained lower than 30% in sensitivity analyses of the allele comparison, so the above conclusion was stable.
Statistical significance existed in all comparisons of rs2383207 except for the recessive model. With the significant outcome in allele comparison, we can come to the conclusion that carrying the mutated allele A on rs2383207 protects against IS.
For comparisons of rs10757274, statistical outcomes were the same as those for comparisons of rs2383206, which means that GG of rs10757274 may be the only pathogenic genotype.
rs10757278 has been extensively studied in both Caucasian and Chinese populations. Many studies have included large numbers of participants, but there was no consistent conclusion. In sensitivity analysis of rs10757278, I2 remained high throughout the allele comparison analysis, which indicates that rs10757278 allele polymorphism is not related to IS; this appears to be relatively stable.
IS comprises five subtypes and each has different causes, so some studies have divided cases into different subgroups according to the TOAST Classification. However, several studies6,8 include no subgroups. Hence, there were no subgroup analysis in this meta-analysis, which represents a limitation of our conclusion in that the mechanisms of subtype susceptibility to IS cannot be investigated.
In summary, this meta-analysis arrives at the conclusion that 9p21 is a susceptibility region for IS in the Chinese population. This result is quite different from those of previous meta-analyses. Allowing for the limitation that subgroup analyses could not be conducted in our analysis, strict selection and classification of patients and well-matched controls should be included in future studies.
Footnotes
Author contributions: Man Li, Jing Liu, and Ji-Xiang Chen conceived and designed the experiments. Man Li, Jing Liu, and Lei Zhao performed the experiments. Man Li, Jing Liu, and Lei Zhao analyzed the data. Feng Hu contributed analysis tools. Feng Hu, Man Li, and Jing Liu wrote the paper.
Declaration of conflicting interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the National Natural Science Foundation of China (81371840, 81400970, and 81502735) and Research Project of Health and Family Planning Commission of Hubei Province of China (JX6B40).
ORCID iDs: Jing Liu  https://orcid.org/0000-0002-7888-2619
https://orcid.org/0000-0002-7888-2619
References
- 1. Mozaffarian D, Benjamin EJ, Go AS, et al. (2016) Heart disease and stroke statistics-2016 update: A report from the American Heart Association. Circulation 133(4): e38–e360. [DOI] [PubMed] [Google Scholar]
- 2. Kubo M, Hata J, Ninomiya T, et al. (2007) A nonsynonymous SNP in PRKCH (protein kinase C eta) increases the risk of cerebral infarction. Nature Genetics 39(2): 212–217. [DOI] [PubMed] [Google Scholar]
- 3. Wahlstrand B, Orho-Melander M, Delling L, et al. (2009) The myocardial infarction associated CDKN2A/CDKN2B locus on chromosome 9p21 is associated with stroke independently of coronary events in patients with hypertension. Journal of Hypertension 27(4): 769–773. [DOI] [PubMed] [Google Scholar]
- 4. Traylor M, Farrall M, Holliday EG, et al. (2012) Genetic risk factors for ischaemic stroke and its subtypes (the METASTROKE Collaboration): A meta-analysis of genome-wide association studies. The Lancet Neurology 11(11): 951–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. NINDS Stroke Genetics Network (SiGN) and International Stroke Genetics Consortium (ISGC) (2016) Loci associated with ischaemic stroke and its subtypes (SiGN): A genome-wide association study. The Lancet Neurology 15(2): 174–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ding KH, Xu Y, Wang X, et al. (2009) 9p21 is a shared susceptibility locus strongly for coronary artery disease and weakly for ischemic stroke in Chinese Han population. Circulation: Cardiovascular Genetics 2(4): 338–346. [DOI] [PubMed] [Google Scholar]
- 7. Hu WL, Li SJ, Liu DT, et al. (2009) Genetic variants on chromosome 9p21 and ischemic stroke in Chinese. Brain Research Bulletin 79(6): 431–435. [DOI] [PubMed] [Google Scholar]
- 8. Yue X, Tian L, Fan X, et al. (2015) Chromosome 9p21.3 variants are associated with cerebral infarction in Chinese population. Journal of Molecular Neuroscience 56(3): 546–552. [DOI] [PubMed] [Google Scholar]
- 9. Xiong L, Zhang B, Liu H, et al. (2016) ANRIL genetic variation is associated with atherothrombotic stroke in Chinese Han population in Northeast Sichuan. Medical Journal of West China 28: 1354–1359. [Google Scholar]
- 10. Zhang W, Chen Y, Liu P, et al. (2012) Variants on chromosome 9p21.3 correlated with ANRIL expression contribute to stroke risk and recurrence in a large prospective stroke population. Stroke 43(1): 14–21. [DOI] [PubMed] [Google Scholar]
- 11. Lin HF, Tsai PC, Liao YC, et al. (2011) Chromosome 9p21 genetic variants are associated with myocardial infarction but not with ischemic stroke in a Taiwanese population. Journal of Investigative Medicine 59: 926–930. [DOI] [PubMed] [Google Scholar]
- 12. Bi J, Yang L, Liu D, et al. (2015) Sequence variants on chromosome 9p21 are associated with ischemic stroke and the lipids level in Chinese Han population. Journal of Stroke and Cerebrovascular Diseases 24(4): 894–900. [DOI] [PubMed] [Google Scholar]
- 13. Matarin M, Brown WM, Singleton A, et al. (2008) Whole genome analyses suggest ischemic stroke and heart disease share an association with polymorphisms on chromosome 9p21. Stroke 39(5): 1586–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]