Abstract
Aim:
The chromosomal aberrations induced by radiation appear about nonrandomly distributed across the whole genome. Previous studies have shown that chromosomes with high DNA content are less frequently involved in the formation of symmetrical translocations and dicentric chromosomes than expected, whereas smaller chromosomes are more frequently involved. We hypothesized that these translocation regions are linked to radiation sensitivity.
Materials and methods:
We investigated the frequencies of chromosome translocations induced by radiation exposure and adjusted the results according to chromosome length. We specifically analyzed whole blood samples from 3 participants. The samples were irradiated using 60Co at doses of 0.5, 1, 2.5, and 5 Gy. Traditional Giemsa-trypsin-Wright band staining was performed to identify the translocations in the chromosomes, and results were compared with microarray data generated in our previous study.
Results:
Our analysis indicated that chromosomes 9q were the most sensitive to translocations after various doses of radiation, and such translocations occurred in the euchromatin region. Chromosomes 1, 9, 15, and 17 were more sensitive to radiation doses of 0.5 Gy. This observation could be useful when selecting sensitive reference chromosomes in the low-dose region. The results of expression profiling analysis for radiation-sensitive regions were similar to those of chromosome translocation analysis.
Conclusion:
This study shows that some chromosomes or genomic regions are more sensitive to alteration by radiation exposure.
Keywords: radiation, chromosome aberrations, gene expression
Introduction
The effects of radiation depend on the dose and quality of the radiation as well as the sensitivity of the organism to radiation which can lead to a variety of different chromosome aberrations. Radiation exposure can affect cellular structural integrity, immune functions, cell cycle control, and apoptosis.1,2 Radiation-induced alterations in the expression of genes responsible for the formation and maintenance of cellular structure and cell cycle control may lead to chromosome instability and carcinogenesis.2-5 In human cells, metabolic activities, environmental factors, or radiation can induce DNA damage, resulting in as many as 1 million individual molecular lesions per cell per day.6 When the repair system fails or is not entirely successful, and cellular apoptosis does not occur, irreparable DNA damage may occur, including double-strand breaks and DNA cross linkages (interstrand crosslinks).7,8 This damage may be stable, allowing the cells to pass through mitosis.
The dicentric chromosome assay (DCA) is a sensitive and specific method for assessing biodosimetry. At present, the DCA is the gold standard for radiation dose assessment according to ISO19238 and the International Atomic Energy Agency. It can be used to assess the effects of radiation doses from 0.2 to 5.0 Gy, independent of age and gender.9 ,10 Fluorescence in situ hybridization is also used to measure the frequency of translocations and evaluate the effects of radiation dose.11 Although the methods mentioned earlier identify the chromosomal rearrangements and translocation frequencies, they are not optimal for indicating the regions on chromosomes that are sensitive to aberrations. In this investigation, we observed the large-scale changes in chromosomes for obtaining the significant and obvious chromosome aberrance, especially in low-dose radiation. Furthermore, the adjustment for chromosome length was not performed in previous studies. More detailed observation and analysis will be used in the further study.
Radiation-induced DNA damage can be detected as large-scale rearrangements of the genome, and chromosomal aberrations induced by radiation appear to be approximately random across the whole genome.12-15 A previous study, however, has shown that chromosomes with high DNA content were less frequently involved in the formation of balanced translocations and dicentric chromosomes than would be expected, whereas smaller chromosomes were more frequently involved in the formation of these aberrations.11
In this study, we attempted to identify chromosome regions that were susceptible to radiation exposure. We performed traditional Giemsa-trypsin-Wright (GTW)-banding staining to determine the specific translocation regions on the chromosomes and compared this with gene expression array data. We hypothesized that these translocation regions are linked to radiation sensitivity.
Materials and Methods
Sample Preparation
Whole blood samples (30 mL) from each of 3 apparently healthy participants were collected into vacutainer tubes containing sodium heparin. The groups enrolled in our study were having similar age (25∼30 years old), health status (no illness and no medicine), and 2 genders (included female and male). The samples were irradiated using 60Co at a dose rate of 0.546 Gy/min (0.0091 Gy/ sec) at the Institute of Nuclear Energy Research, in Taoyuan. The radiation doses used in these experiments were 0.5 (exposed to 55 seconds), 1 (exposed to 110 seconds), 2.5 (exposed to 275 seconds), and 5 Gy (exposed to 549 seconds). The control samples were not exposed to any radiation. Informed consent was obtained from all participants, and all procedures were approved by the Institutional Review Board at Tzu Chi General Hospital, Hualien.
Cytogenetic Analysis
After irradiation, the samples were incubated at 37°C for 48 hours before being harvested.9 Metaphase preparation was performed using a standard protocol for lymphocyte cultures based on the general guidance provided by the International Atomic Energy Agency and ISO 19238.9,10 We used the l-form phytohemagglutinin (PHA), which is the mitogen that induces the T cell into cell division. We decided to analyze the T cells first. The GTW-banded chromosome was done using a standard protocol.16 The breakpoints of chromosomal translocation were investigated on the metaphase chromosomes.
Scoring Chromosome Aberrations
The samples were scored for radiation-induced dicentrics and translocations according to previously published references.17,18 For each dose, 50 metaphase cells were analyzed per sample as stipulated by the criteria for performing triage quality dose assessments.19,20 The accuracy of dose assessments after scoring only 50 cells is considered sufficient in a preliminary triage during a mass-casualty event.17-20 The slides were scanned at low magnification (100×) to avoid personal bias and then analyzed at high magnification (1000×). Only spread metaphase cells with 46 centromeres were scored.
Chromosome Aberration Analysis
The identity and number of chromosome translocation sites were mapped on each chromosome. The chromosomal aberration sites were considered for aberrations that were detected at least twice or with more than 2 radiation doses. The hit frequencies of translocations in each chromosome at different radiation doses were estimated by using the translocation hit number adjusted according to the length (Mbp) of each chromosome arm. We used ISCN 2016 guideline to describe the chromosome aberration.
Microarray Data Acquisition and Analysis
Microarray data generated in our previous study21 were reanalyzed in this study. The alterations in genes (including increased and decreased gene expression after different doses of radiation exposure) selected from microarray analysis according to a fold-change >3 or <−3 and a P value <.0001 were mapped on each chromosome according to the location of the genes. The number of altered genes in each chromosome arm at various radiation doses was estimated by adjusting the number of altered genes according to the total number of genes in each arm of the chromosome.
Chromosome aberrant regions annotation
We try to find radiation sensitive chromosome regions, and these aberrant regions should link to different dose of ionizing radiation. Furthermore, we investigate the aberrant chromosome regions may harbor some disease susceptibility genes. We search potential related genes involved in the break chromosome regions from these websites: the Atlas of Genetics and Cytogenetics in Oncology and Hematology website (http://www.atlasgeneticsoncology.org/), Cancer Genetics Web (http://www.cancerindex.org/geneweb/), and NCBI/OMIM (https://www.ncbi.nlm.nih.gov/omim).
Results
Examples of chromosome aberrations induced by different doses of radiation exposure are shown in Figure 1. The chromosome aberrations include different structural abnormalities such as ring chromosomes in Figure 1, acentric chromosomes, deletions, and double minutes, translocation, and dicentric chromosome. Figure 2 shows 2 sample karyotypes for the cells all exposed to 5 Gy of radiation with more detail.
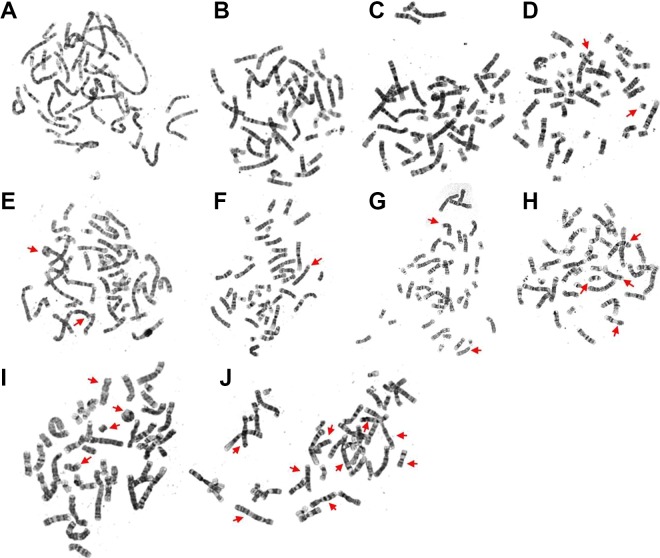
Figure 1.
Chromosomal variations induced by different radiation doses. The arrows indicate the translocation and chromosome aberrations, which were observed at various radiation dose as follows: (A and B): 0 Gy; (C and D): 0.5 Gy; (E and F): 1 Gy; (G and H): 2.5 Gy; (I and J): 5 Gy. The number (count) of chromosome aberrations was as follows: 0 for (A and B), 2 for (C and D), 3 for (E and F), 6 for (G and H), and 12 for (I and J).
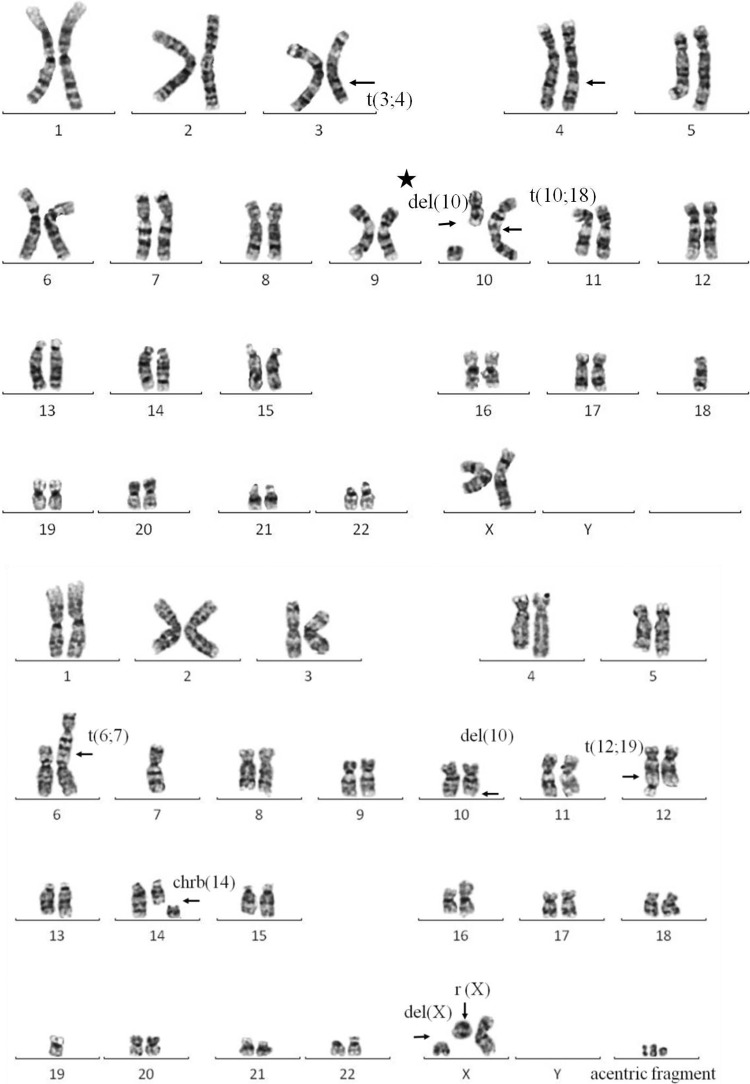
Figure 2.
Karyotypes demonstrated in cells exposed to 5 Gy of radiation. Arrows indicate deletions (del), translocations (t), chromosome breaks (chrb), ring chromosomes (r), and acentric fragments; t(3;4): breakage and reunion have occurred at chromosome 3q and 4q. The segments distal to these q arms have been exchanged; del (10) (with a star): interstitial deletion without reunion; chrb(14): chromosome 14 break which is synonymous with isochromatid break; r(X) and del(X): t derivative deleted chromosome X, one part reunion became a ring chromosome, another part remained as a chromosome X fragment, and other parts of this chromosome X was missing.
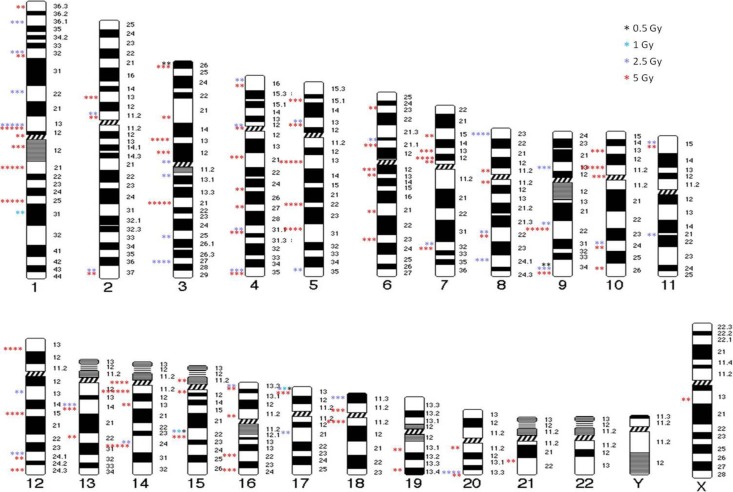
Figure 3 shows the chromosome aberrations mapped by radiation exposure. This map indicates the chromosome aberrations that were induced by radiation exposure, which appeared in locations in a nonrandom manner, that is, more than once or induced by more than 1 dose of radiation. Some specific chromosome regions were highly radiosensitive, and the translocations occurred mainly in light G-bands (ie, the euchromatic region).
Figure 3.
Chromosome aberration map after different doses of 60Co radiation exposure. Karyotypes illustrate the positions of the translocations. Stars show the number of translocations from the 150 cells analyzed.
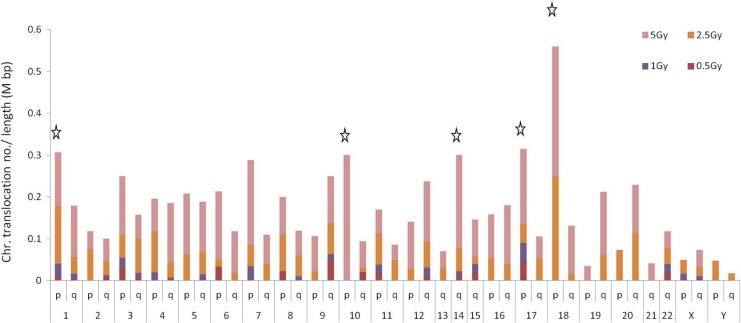
Figure 4 shows the random count of translocation for each of the chromosome arms. The estimated frequency for chromosome translocations is adjusted according to the chromosome length (obtained from the NCBI). Chromosomes 18, 17, and 1 were the chromosomes with the highest frequencies of translocations per length of chromosome. However, chromosomes 3 and 9 showed the highest sensitivity to 0.5 Gy of radiation. The data revealed that chromosome 18 had the highest frequency of translocations with high doses of radiation. This chromosome was not sensitive to radiation of 0.5 Gy compared to other chromosomes. Supplementary Table S1 shows the all translocation positions in each chromosome.
Figure 4.
Hit frequencies of translocations in each chromosome at different radiation doses. The stars indicate the top 5 chromosomes. The frequencies were estimated for sensitive chromosomal regions using the translocation hit number adjusted according to the length of the chromosome arms (Mbp).
Table 1 shows the translocations that occurred on chromosomes 9q, 15q, and 17q more than 5 times and for at least 3 doses. In these regions, most of the genes were related to leukemia, particularly in chromosomes 9q and 15q. Translocations that appeared more than 5 times at 2 different radiation doses were associated with multiple types of cancer-related and tumorigenesis-related genes, such as NFkB2 (Table 2). Table 3 shows the translocations that appeared more than 4 times in the same chromosomal region after exposure to 5 Gy of radiation. XRCC4 gene locates to chromosome 5q and acts in DNA double-strand break repair. The TCRD gene is involved in Ataxia telangiectasia mutated (ATM)-deficient thymic lymphoma. Translocations that appeared more than 4 times in the same chromosomal region following exposure to 1 or 2.5 Gy of radiation are shown in Table 4. Two examples, BCL6 and PTPRC genes, are associated with B-cell lymphoma and acute myeloid leukemia, respectively. The results showed in Tables 1 and 2 were searched from the Atlas of Genetics and Cytogenetics in Oncology and Hematology website, Cancer Genetics Web, and NCBI/OMIM.
Table 1.
Chromosome Regions With More Than 5 Translocations Following Radiation Exposure at Doses of 0.5, 1, 2.5, and 5 Gy (the Number of Translocations Appeared With at least 3 Different Radiation Doses).
| Location | Genea | Diseaseb | Common Cytogenetic Aberrationsb |
|---|---|---|---|
| 9q34 | SET | Myeloid leukemia-associated Wilms’ tumor |
t(6;9)(p23;q34) t(7;9)(q34;q34.3) t(9;22) |
| 9q34.1 | ABL1 | Chronic myelogenous leukemia | |
| NUP214 | Acute myeloid leukemia | ||
| 9q34.3 | NOTCH1 | Neoplastic transformation | |
| 15q21-q22 | B2M | Multiple myeloma | t(15;17) |
| 15q22 | PML | Promyelocytic leukemia | |
| 15q22-q24 | CYP1A1 | Childhood acute lymphoblastic leukemia | |
| 17p13.1 | TP53 (P53) | Li-Fraumeni syndrome Neuroectodermal tumors Medulloblastoma JPAs in children |
Loss of 17p |
aCancer-related genes within the chromosome translocation sites.
bData obtained from the Atlas of Genetics and Cytogenetics in Oncology and Hematology website, Cancer Genetics Web, and NCBI/OMIM.
Table 2.
Chromosome Regions With More Than 5 Translocations Following Radiation Exposure at 0.5, 1, 2.5, or 5 Gy (the Number of Translocations Appeared With at least 2 Different Radiation Doses).
| Location | Gene | Diseases | Common Cytogenetic Aberrations |
|---|---|---|---|
| 1p13.3 | RAP1A | Human tumors | |
| 1p32 | TAL1 (SCL, TCL5) | T-cell acute lymphocytic leukemia | |
| EPS15 (MLLT5, AF-1P) | |||
| CDKN2C (p18) | Inhibits CDK4 | ||
| 1p32-p31 | JUN | V-jun avian sarcoma virus 17 oncogene homolog | |
| 3p26-p25 | VHL | Von Hippel-Lindau syndrome | |
| 4p11-q22 | KIT | Gastrointestinal stromal tumors | |
| 7q32-q36 | EPHA1 | Hepatoma | |
| 9q22 | NR4A3 (CHN, CSMF) | Extra-skeletal myxoid chondrosarcoma | t(9;22)(q22-31;q11-12) |
| SYK | Breast cancer B-cell lymphomas Colorectal cancer |
||
| 9q22.3 | PTCH | Nevoid basal cell carcinoma syndrome Gorlin syndrome |
|
| FANCC | Fanconi anemia | ||
| 10q24 | NFKB2 (p49/p100) | Abnormal cell
proliferation Tumorigenesis Hepatocarcinogenesis |
t(10;14)(q24;q11) t(1;10) |
| HOX11 (TCL3) | T-cell lymphoma | ||
| PLAU | Multiple types of cancer | ||
| 10q24.1 | TNFRSF6 | Tumor formation and defense Leukemia |
|
| 10q24.1-q24.3 | MGEA5 | Hyaluronidase in meningioma Bladder cancer |
|
| 11p15 | NUP98 | Myelodysplastic syndromes Adult acute myeloid leukemia |
t(7;11)(p15;p15) t7;11) t(11;14)(p15;q11) |
| LMO1 | T-cell acute lymphoblastic leukemia cell line activates multiple transcripts | ||
| 11p15.2-p15.1 | TSG101 | Breast cancer Neoplasm |
|
| 11p15.5 | WT2 (MTACR1) | Second Wilms’ tumor Familial Wiedemann-Beckwith syndrome |
|
| 11pter-p13 | CD44 | Malignancies lymphomas Neuroblastoma |
|
| 11p15.5 | IGF2 | Wide range of cancers Wilms’ tumor |
|
| IGF2AS | Wilms’ tumor | ||
| 12q24.13 | BCL7A | Directly involved with Myc and IgH | t(8;14;12)(q24.1;q32.3;q24.1) |
| 13q14.1-q14.3 | LCP1 (PLS2) | B-cell non-Hodgkin lymphoma Retinoblastoma |
t(3;13)(q27;q14) 13q deletion |
| 14q24.3 | FOS | Ovarian cancer Bladder cancer Inflammation-mediated skin tumorigenesis |
|
| 14q24.3-q31 | CHES1 | Areca nut extract-induced oral cancer | |
| 16p13.13-p13.12 | MYH11 | Acute myelomonocytic leukemia Acute myelomonoblastic leukemia Acute myeloid leukemia |
|
| 20q13.3 | GNAS | Pseudohypoparathyroidism type 1a Pseudohypoparathyroidism type 1b Albright hereditary osteodystrophy, pseudopseudohypoparathyroidism McCune-Albright syndrome Progressive osseous heteroplasia Polyostotic fibrous dysplasia of bone Pituitary tumors |
Table 3.
Number of Translocations That Appeared More Than 4 Times in the Same Chromosomal Region After Exposure to 5-Gy Radiation, Along With the Related Genes and Their Functions.
| Location | Gene | Diseases | Common Cytogenetic Aberrations |
|---|---|---|---|
| 1q21 | MCL1 | Preneoplastic and neoplastic disease Pancreatic cancers |
t(1;14)(q21;q32) t(X;1)(p11;q21) t(1;12)(q25;p13) t(1;11)(q21;q23) |
| AF1Q (MLLT11, RP11-316M1.10) | Leukemia Acute myelomonocytic leukemia |
||
| 1q21-q22 | NTRK1 | ||
| 1q21.3-q22 | CD48 | Lymphoid leukemia Arthritis |
|
| 1q24-q25 | HPC1 | Hereditary prostate cancer | |
| ABL2 | Leukemia | ||
| 3q21-q27 | MME | Acute lymphoblastic leukemia | |
| 3q21-q28 | TERC | Leukemia | |
| 5q13-q14 | XRCC4 | ||
| 10p11.2 | COT | Human cancer | t(10;11) |
| 14q11.2 | TCRD | ATM-deficient thymic lymphoma Acute lymphoblastic leukemia |
t(11;14)(p15;q11) |
| 18p11.2 | MC2R | Adrenocortical neoplasms | t(X;18)(p11.2;q11.2) |
Table 4.
Translocations That Appeared More Than 4 Times in Chromosomal Regions After Exposure to Radiation at 1 and 2.5 Gy, Along With Related Genes and Their Functions.
| Location | Gene | Diseases | Common Cytogenetic Aberrations |
|---|---|---|---|
| 1 Gy | |||
| 1q31-q32 | PTPRC (CD45, LCA) | Acute myeloid leukemia | |
| 2.5 Gy | |||
| 3q27 | BCL6 | B-cell lymphoid neoplasms B-cell non-Hodgkin’s lymphoma Diffuse large B-cell lymphomas |
t(3;11)(q27;q23.1) t(3;14)(q27;q32) |
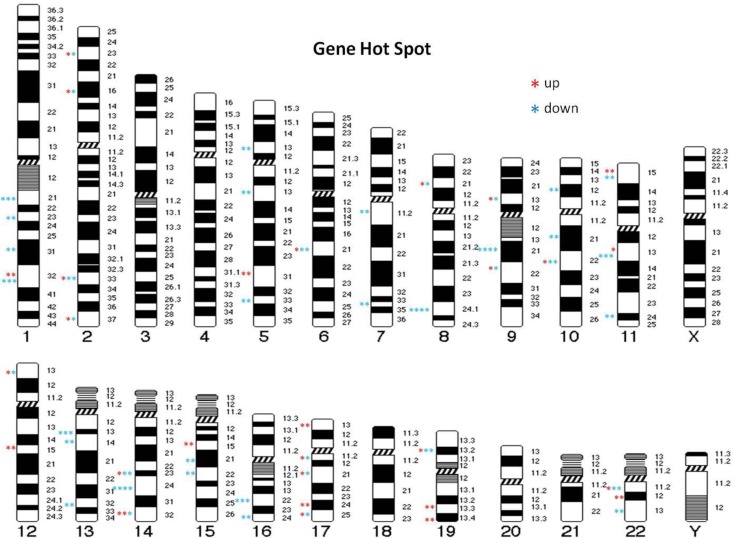
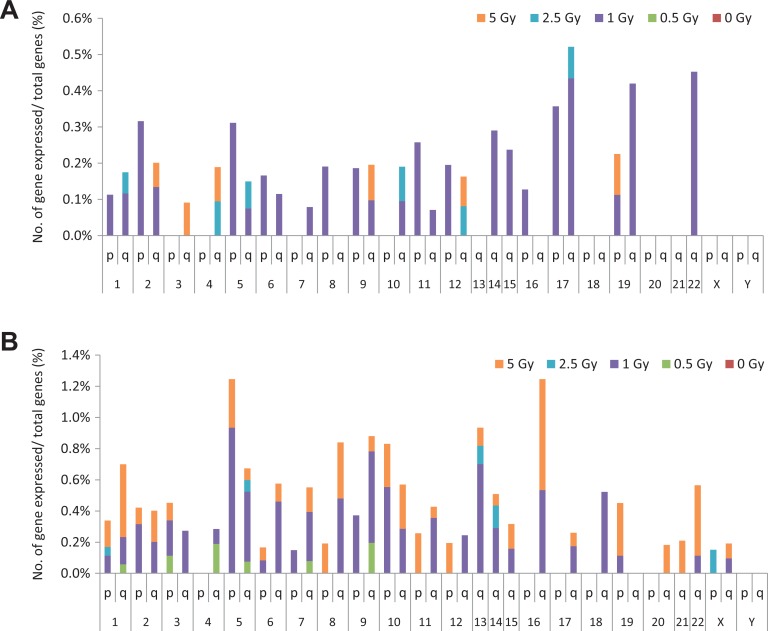
The microarray analysis demonstrated that chromosomes 1, 5, 9, 11, 13, 14, 17, 19, and 22 exhibited suppression and enhancement of gene expression induced by radiation exposure, and the incidences of change were higher than that of other chromosomes (Figure 5). We analyzed the percentage of altered genes among all genes in each chromosome arm and found that chromosomes 5, 9, 13, 16, 17, 19, and 22 had the highest percentages. In particular, the maximum percentage of upregulated genes was observed in chromosomes 17p/q, 19q, and 22q, whereas the majority of suppressed genes were observed in chromosomes 5p, 9q, 13q, and 16q (Figure 6A and B).
Figure 5.
Gene expression map after different doses of 60Co radiation exposure. Karyotypes illustrate the position of up- or downregulated genes. Stars show the number of genes expressed (fold-change >3 or <−3; P < .0001).
Figure 6.
Number of up- and downregulated genes. (A) A high percentage of upregulated genes was found in chromosomes 17p/q, 19q, and 22q. Upregulated genes did not appear with low-radiation doses of 0.5 Gy, and 5-Gy doses also appeared to have fewer effects on genes. (B) A high percentage of downregulated genes was found in chromosomes 5p, 9q, 13q, and 16q (fold-change >3 or <−3; P < .0001).
Discussion
In this study, we used G-banding to investigate the location of chromosome aberrations induced by various radiation doses and at the banding level of at least 400 bands. We found that some chromosome regions were more sensitive to radiation-induced, large-scale genomic rearrangement. Moreover, the greatest numbers of translocation occurred in the euchromatic regions. In addition, we compared our results to microarray data and found that chromosomes such as 9 and 17 were more sensitive to radiation.
The GC-rich regions are more frequently involved in transcription. Dark G-Bands are heterochromatic, AT-rich regions are less frequently involved with transcription. In heterochromatic regions, most genes are inactivated tissue-specific genes,22 which are rarely involved in transcription. Heterochromatin protects the genes while they are not in use. Euchromatin participates in the active transcription of regulatory proteins and the binding of RNA polymerase complexes to DNA sequences for the initiation of the transcription process.23 The results of the present study suggest the interaction of irradiation with more sensitive regions of the chromosome can induced more damage than when interacts with other regions of the chromosome as previously supposed. Indeed, our data indicate that the euchromatic region is more sensitive to radiation exposure. These results are similar to those of previous studies.24
In chromosomes 1q, 9q, 15q, and 17, translocations appeared more than 5 times with different radiation doses; in 15q22 and 17p13, translocations appeared with 4 different doses. A comparison to the previous study showed that the aberration in chromosome 17p13 was found only in the bone marrow sample.25 The TP53 and HIC1 genes were involved in the 17p13 region, whereas B2M, PML, and CYP1A1 genes were involved in the 15q22 region. PML is a tumor suppressor protein from the dynamic macromolecular nuclear structure, the PML-nuclear body (PML-NB). The PML gene generates the oncogenic fusion protein PML-retinoic acid receptor-alpha. Disruption of PML-NBs is implicated in the pathogenesis of acute promyelocytic leukemia.25,26
We found that 1p13 and 1q25, 3q21, 5q13, 10p11.2, and 14q11.2/q24 were more sensitive to high radiation dose after 5-Gy 60Co exposure. These results were inconsistent with a previous study,24 which showed that chromosome regions 1p36, 2q33, 3p21, 4q31.1, 7q22, 8q22, and 12q24.1 of the lymphocyte sample were sensitive to 4-Gy 60Co exposure. Chromosome regions 3p21, 1q42, 7q32, 9p11/q11, 10q24, 14q24, and 17p13 of bone marrow samples were also sensitive to 4-Gy 60Co exposure.24 This inconsistency between the 2 studies could be due to counting the regions with translocations and not counting the breakpoint distribution in the chromosomes in our investigation. The previous study also showed the breakpoint distribution within relatively fragile sites, either in the bone marrow sample or the lymphocyte sample. The breakpoints observed at 14q24 and 17p13 within these sites were only significantly found in bone marrow, and another breakpoint was observed at 2q25 in lymphocytes only.
However, we found that the chromosome 18p region had a high frequency of translocations when subjected to a high dose (2.5 Gy and 5 Gy) of radiation after adjusting for the length of the chromosome arms. This region reportedly possesses a significant loss of heterozygosity (LOH; 63%), with 56% occurring early in ductal carcinoma in situ in 96 microdissected breast cancer samples. Compared to LOH data, 18p LOH was found in conjunction with allelic deletions on 3p, 9p, 17p, and 17q.27 The putative tumor suppressor gene DAL-1 has recently been mapped to chromosome band 18p11.3.28 High doses of radiation may influence these regional events, which are likely to be more varied depending on the stage and/or type of tumor foci analyzed. After adjusting for the length of the chromosome arms (Mbp), we found that 1p and 10p, 14q, 17p, and 18p had high frequencies of translocations, especially in 17p, where translocations were observed at all radiation doses (0.5 Gy, 1 Gy, 2.5 Gy, and 5 Gy).
In our previous study, we showed that gene expression was altered in some genes following exposure to 1 Gy of radiation. With 0.5 Gy of radiation dose, gene regulation remained unchanged; similarly, 5 Gy of radiation appeared not to affect some genes.21 We analyzed the gene expression array data with the criteria of P < .0001 and change >3 or <−3. We found that the number of downregulated genes increased in chromosomes 5p, 9q, 13q, and 16q, whereas the number of upregulated genes increased in chromosomes 17p/q, 19q, and 22q. By comparison with our chromosome data, it appears that chromosomes 9q and 17q are those most sensitive to radiation. In contrast to a previous study,24 our findings demonstrated that some chromosome regions were more susceptible to radiation than others and that chromosome alteration occurred mainly in the light G-bands of the euchromatic region. In the previous review discussing the intrachromosomal and interarm distribution of radiation-induced breakpoints in the human genome, the data demonstrated that chromosomes 1, 9, 13, 14, 15, 21, and 22 had a significant probability of radiation-induced breakpoints per Mbp of DNA.29 In comparison to our data, we used the expression array and adjusted it by the total number of genes in the chromosomes and arms. Chromosomes 1, 9, 15, and 17 were found to be more sensitive to radiation at 0.5 Gy.
Radiation exposure can cause the alteration in messenger RNA expression through direct or indirect effects. The direct effects may have been induced by nonobserved mutations caused by radiation, such as microdeletions/insertions and point mutations. The indirect effects are caused by their regulatory gene(s) being affected by radiation. The limitation of our comparison of chromosome aberrations and expression profiling is that the direct effects are not confirmed, and further investigation is needed. However, we still found some distinguished patterns from our preliminary data. Next-generation sequencing should be performed to investigate nonobserved chromosome/genomic aberrations.
In summary, our data, along with that of previous studies, showed that chromosomes 1, 3, 7, 10, 14, and 17 had aberration distributions similar to lymphocyte samples in previous studies. Chromosomes 5, 14, and 17 showed results similar to a bone marrow sample from a previous study.24,29 This finding provided information for the selection of sensitive reference chromosomes in relationship with low-radiation doses (under 0.5 Gy). In particular, focusing on these 4 chromosomes should be helpful in the determination of chromosome variation of dose less than 1 Gy.
Supplemental Material
Supplementary_Table for The Potential Effect of Different Doses of Ionizing Radiation on Genes and Disease by Cheng-Chia Lin, Lawrence Shih-Hsin Wu and Kuei-Fang Lee in Dose-Response
Footnotes
Authors’ Note: Lawrence Shih-Hsin Wu and Kuei-Fang Lee contributed equally to this article.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
ORCID iD: Kuei-Fang Lee  https://orcid.org/0000-0002-3407-429X
https://orcid.org/0000-0002-3407-429X
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Horn S, Barnard S, Rothkamm K. Gamma-H2AX-based dose estimation for whole and partial body radiation exposure. PLoS One. 2011;6(9):e25113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Paul S, Smilenov LB, Amundson SA. Widespread decreased expression of immune function genes in human peripheral blood following radiation exposure. Radiat Res. 2013;180(6):575–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Turtoi A, Sharan RN, Srivastava A, Schneeweiss FH. Proteomic and genomic modulations induced by γ-irradiation of human blood lymphocytes. Int J Radiat Biol. 2010;86(10):888–904. [DOI] [PubMed] [Google Scholar]
- 4. Kabacik S, Mackay A, Tamber N, et al. Gene expression following ionising radiation: identification of biomarkers for dose estimation and prediction of individual response. Int J Radiat Biol. 2011;87(2):115–129. [DOI] [PubMed] [Google Scholar]
- 5. Amundson SA, Do KT, Shahab S, et al. Identification of potential mRNA biomarkers in peripheral blood lymphocytes for human exposure to ionizing radiation. Radiat Res. 2000;154(3):342–346. [DOI] [PubMed] [Google Scholar]
- 6. Lodish H, Berk A, Matsudaira P, et al. Molecular Biology of the Cell. 5th ed New York, NY: WH Freeman; 2004:963. [Google Scholar]
- 7. Acharya PV. The isolation and partial characterization of age-correlated oligo-deoxyribo-ribonucleotides with covalently linked aspartyl-glutamyl polypeptides. Johns Hopkins Med J. 1971;(1):254–260. [PubMed] [Google Scholar]
- 8. Bjorksten J, Acharya PV, Ashman S, Wetlaufer DB. Gerogenic fractions in the tritiated rat. J Am Geriatr Soc. 1971;19(7):561–574. [DOI] [PubMed] [Google Scholar]
- 9. International Organization for Standardization (ISO). Radiation Protection–Performance Criteria for Service Laboratories Performing Biological Dosimetry by Cytogenetics, ISO 19238. Geneva, Switzerland: ISO; 2004. [Google Scholar]
- 10. International Atomic Energy Agency (IAEA). Cytogenetic analysis for radiation dose assessment: a manual. Technical Report 405 Vienna, Austria: IAEA; 2001. [Google Scholar]
- 11. Knehr S, Zitzelsberger H, Braselmann H, Nahrstedt U, Bauchinger M. Chromosome analysis by fluorescence in situ hybridization: further indications for a non-DNA proportional involvement of single chromosomes in radiation-induced structural aberrations. Int J Radiat Biol. 1996;70(4):385–392. [DOI] [PubMed] [Google Scholar]
- 12. Nakano M, Kodama Y, Ohtaki K, et al. Detection of stable chromosome aberrations by FISH in A-bomb survivors: comparison with previous solid Giemsa staining data on the same 230 individuals. Int J Radiat Biol. 2001;77(9):971–977. [DOI] [PubMed] [Google Scholar]
- 13. Coco Martin JM, Mooren E, Ottenheim C, et al. Potential of radiation-induced chromosome aberrations to predict radiosensitivity in human tumor cells. Int J Radiat Biol. 1999;75(9):1161–1168. [DOI] [PubMed] [Google Scholar]
- 14. Barber JBP, Burrill W, Spreadborough AR, et al. Relationship between in vitro chromosomal radiosensitivity of peripheral blood lymphocytes and the expression of normal tissue damage following radiotherapy for breast cancer. Radiother Oncol. 2000;55(2):179–186. [DOI] [PubMed] [Google Scholar]
- 15. Richardson C, Jasin M. Frequent chromosomal translocations induced by DNA double-strand breaks. Nature. 2000;405(6787):697–700. [DOI] [PubMed] [Google Scholar]
- 16. Barch MJ, Knutsen T, Spurbeck JL, et al. The ACT Cytogenetics Laboratory Manual. 3rd ed Philadelphia: Raven Press; 1991. [Google Scholar]
- 17. Ainsbury EA, Barquinero JF. Biodosimetric tools for a fast triage of people accidentally exposed to ionising radiation. Statistical and computational aspects. Ann Ist Super Sanità. 2009;45(3):307–312. [PubMed] [Google Scholar]
- 18. Pereira de Lemos Pinto MM, Santos NF, Amaral A. Current status of biodosimetry based on standard cytogenetic methods. Radiat Environ Biophys. 2010;49(4):567–581. [DOI] [PubMed] [Google Scholar]
- 19. Lloyd DC, Edwards AA, Moquet JE, Guerrero-Carbajal YC. The role of cytogenetics in early triage of radiation casualties. Appl Radiat Isot. 2000;52(5):1107–1112. [DOI] [PubMed] [Google Scholar]
- 20. Doloy MT, Malarbet JL, Guedeney G, et al. Use of unstable chromosome aberrations for biological dosimetry after the first post-irradiation mitosis. Radiat Res. 1991;125(2):141–151. [PubMed] [Google Scholar]
- 21. Lee KF, Weng JTY, Hsu PWC, et al. Gene expression profiling of biological pathway alterations by radiation exposure. Biomed Res Int. 2014;2014:834087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Di Tomaso MV, Liddle P, Lafon-Hughes L, Reyes-Ábalos AL, Folle G. Chromatin damage patterns shift according to eu/heterochromatin replication In: Stuart D, ed. The mechanisms of DNA replication. Rijeka, Croatia: InTech; 2013. ISBN: 978-953-51-0991-4 doi: 10.5772/51847. [Google Scholar]
- 23. International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431(7011):931–945. [DOI] [PubMed] [Google Scholar]
- 24. Tanaka K, Kamada N. Distribution of breakpoints on chromatid-type aberration induced by three different radiations, in relation to fragile sites. Indian J Sci Technol. 2009;2(9):1–9. [Google Scholar]
- 25. Bernardi R, Pandolfi PP. Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat Rev Mol Cell Biol. 2007;8(12):1006–1016. [DOI] [PubMed] [Google Scholar]
- 26. Salomoni P, Ferguson BJ, Wyllie AH, Rich T. New insights into the role of PML in tumour suppression. Cell Res. 2008;18(6):622–640. [DOI] [PubMed] [Google Scholar]
- 27. Kittiniyom K, Gorse KM, Dalbegue F, Lichy JH, Taubenberger JK, Newsham IF. Allelic loss on chromosome band 18p11.3 occurs early and reveals heterogeneity in breast cancer progression. Breast Cancer Res. 2001;3(3):192–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yi C, McCarty JH, Troutman SA, Eckman MS, Bronson RT, Kissil JL. Loss of the putative tumor suppressor band 4.1B/Dal1 gene is dispensable for normal development and does not predispose to cancer. Mol Cell Biol. 2005;25(22):10052–10059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Johnson KL, Brenner DJ, Nath J, Tucker JD, Geard CR. Radiation-induced breakpoint misrejoining in human chromosomes: random or non-random? Int J Radiat Biol. 1999;75(2):131–141. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary_Table for The Potential Effect of Different Doses of Ionizing Radiation on Genes and Disease by Cheng-Chia Lin, Lawrence Shih-Hsin Wu and Kuei-Fang Lee in Dose-Response