Among the hallmark features of asthma (airway inflammation, airway hyperresponsiveness, and remodeling), airway remodeling has been the most intractable. There are several reasons why an effective therapeutic approach for airway remodeling has yet to emerge (1). One of the biggest reasons is undoubtedly the complexity of the phenomenon, coupled with the difficulty of characterizing it under relevant conditions. The lack of noninvasive approaches for visualizing airway remodeling; the chronic, slowly developing nature of the phenomenon; and the infeasibility of clinical trials for assessing antiremodeling drugs have hampered both research into and the development of new drugs that address airway remodeling.
The poor progress in understanding and addressing airway remodeling suggests the need for a fundamentally new approach inspired by novel insight into its pathogenesis. As has been the case with the development of many successful disease-management approaches, success in managing airway remodeling may rely on understanding a fundamental biological process and how it functions (or malfunctions) in the disease context.
In this issue of the Journal, McAlinden and colleagues (pp. 541–553) significantly advance a growing body of literature suggesting that autophagy plays a critical role in the airway remodeling that is common in asthma (2). Autophagy (“self-eating”) is a highly conserved process whereby misfolded proteins, damaged organelles, or invading bacteria are trafficked to lysosomes, where they are degraded and the constituents are recycled by the cell. Although autophagy can serve as a protective mechanism by which the cell can ward off death and maintain cellular building blocks and energy substrates, it can also serve other important functions depending on the cell type in which it occurs (3). For example, peptides generated by autophagy provide a source of antigens for CD4+ T cells, one of several ways in which autophagy plays a role in the regulation of cellular immunity (4, 5). Autophagy is also believed to play a role in various forms of programmed cell death in multiple cell types, and thereby serves either a pathogenic or a protective role in various diseases (6).
Macroautophagy, the best-characterized form of autophagy, involves a reorganization of subcellular membranes that form an “autophagosome” that engulfs cellular proteins and organelles. Multiple autophagy-related (ATG) genes are involved in the process and serve as autophagy markers. Autophagy is initiated by protein kinase ULK1, which acts as an effector of mTOR and AMPK, which are responsible for sensing the energy status of the cell. The mTOR pathway is activated by growth factors and inhibits autophagy, whereas AMPK is activated during cell starvation and activates autophagy. ULK1 forms a complex with Atg1, a serine/threonine kinase, to phosphorylate and activate Beclin-1 to form the class III phosphoinositide 3-kinase (PI3K) (7). The PI3K complex is responsible for PIP2 to PIP3 conversion, which helps in sequestration of microtubule-associated protein 1A/1B light chain 3A (LC3). LC3 is synthesized into the inactive proform, which is cleaved by Atg4, a cysteine protease, into LC3B I (the free form). LC3B I is then activated by Atg7/Atg3 in an ATP-dependent manner. Finally, LC3B I is conjugated to phosphatidylethanolamine to be recognized as LC3B II, and this phenomenon is known as lipidation. Atg7 is also involved in Atg5/Atg12 complex formation, which is indispensable for recruitment of LC3 II to the autophagosome. Maturation of the autophagosome involves the ubiquitin sequestration domain-containing protein p62, which helps in selective autophagy (8).
In previous studies, autophagy has been implicated in asthma pathogenesis and airway remodeling (3). Variations in multiple autophagy genes are associated with familial asthma (9), and variation in the ATG5 gene is associated with childhood asthma (10). Beyond its fundamental role in immune cell function and adaptive immunity, autophagy regulates important processes in resident airway cells that influence the asthma phenotype and airway remodeling. Autophagy appears to be important in regulating mucus and cytokine secretion in airway epithelium (9, 11). A handful of studies have examined autophagy in airway smooth muscle (ASM). The antimitogenic effect of azithromycin on cultured rabbit ASM cells was associated with the induction of autophagy markers (12), and simvastatin-induced caspase activation and cell death were augmented by either chemical or molecular inhibition of autophagy in both human ASM and fibroblasts (13). Silencing Atg5 and Atg7 inhibited transforming growth factor β1 (TGF-β1)–induced collagen A1 and fibronectin in human ASM cells, as reported in abstract form (14). In a murine model of allergic lung inflammation induced by ovalbumin inhalation, increased levels of Beclin-1, LC3A/B, and α-smooth muscle actin were observed in the airway subepithelium, effects that were reversed by oral treatment with the antiinflammatory agent astragalin (15).
McAlinden and colleagues provide new insight into this issue by demonstrating the prevalence of autophagy markers in sections of asthmatic airways possessing features of airway remodeling. Tissue sections of airways derived from subjects with asthma post mortem were shown to stain for multiple markers of autophagy, including increased expression (relative to that in sections of nonasthmatic airways) of markers of autophagy such as Beclin-1 and ATG5, and decreased expression of p62, while demonstrating increased airway inflammation and thickness of the epithelial cell layer, ASM, basement membrane, and lamina propria. Increased expression of the autophagy markers Beclin-1 and ATG5, and reduced p62 expression occurred primarily in the large smooth muscle bundles, and increased Beclin-1 expression was also observed in the cilia of airway epithelial cells.
To further explore the potential role of autophagy in mediating airway remodeling through its role in regulating profibrotic cellular functions, cultures of human ASM cells were treated with TGF-β1 in the presence or absence of chloroquine, a known inhibitor of autophagy. In a time-dependent manner, TGF-β1 induced increases in collagen-1 expression and SMAD2/3 phosphorylation (profibrotic signaling), and concomitantly caused the induction of the autophagy markers Beclin-1 and LC3BII; each of these features was inhibited by chloroquine pretreatment. Consistent with these findings, treatment of mice with house dust mite (HDM) allergen over the course of 3 weeks elicited significant airway inflammation, airway hyperresponsiveness, and increased soluble lung collagen and TGF-β1 expression, and the autophagy markers Beclin-5 and ATG-5 in lung tissue lysates. All of these effects were reversed by pretreatment of the mice with chloroquine.
Lastly, the authors used a more chronic model of murine asthma to explore the mechanistic role of autophagy in the development of airway remodeling in asthma. Five weeks of HDM exposure induced robust inflammation and airway remodeling, as evidenced by increases in ASM bundle size and α-smooth muscle actin abundance. Moreover, immunostaining of ASM bundles demonstrated increases in Beclin-1, ATG-5, and LC3B staining. Chloroquine treatment introduced during the last 2 weeks of the HDM challenge reversed airway inflammation, as well as the increases in ASM bundle size, α-smooth muscle actin, Beclin-1, and ATG-5.
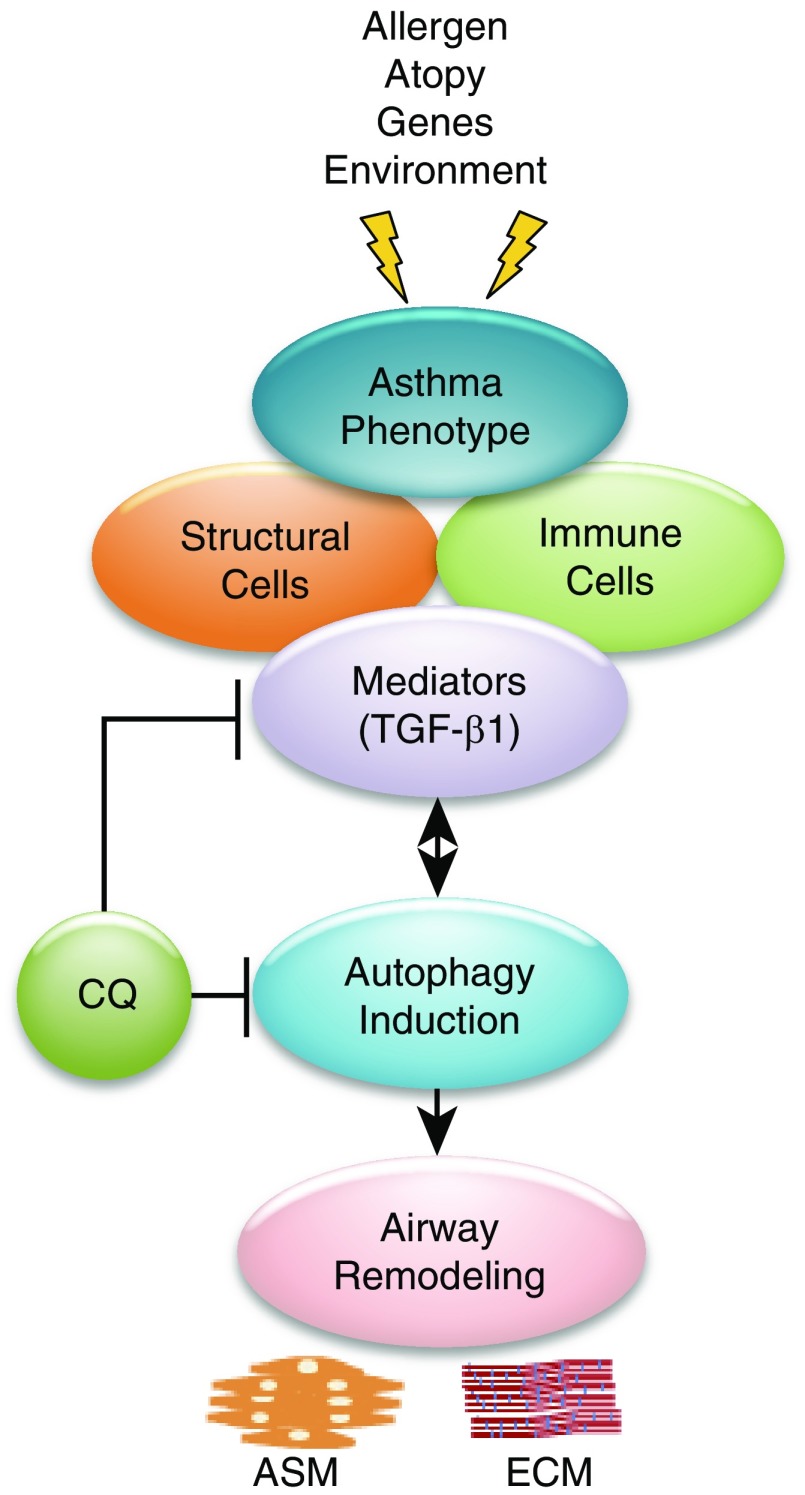
Collectively, these data from human tissue samples, cultured ASM cells, and murine models of asthma strongly suggest that increased autophagy plays a permissive or causal role in the development of airway remodeling in asthma (Figure 1). Increased autophagy in resident human airway cells is coincident with the cardinal features of airway remodeling observed in human asthmatic airways. The profibrotic response of cultured human ASM cells to TGF-β1 is associated with increased autophagy and is reversed by chemical inhibition of autophagy, and features of fibrosis and airway remodeling observed in murine models of allergic lung inflammation/asthma are similarly inhibited by either prophylaxis or treatment with a chemical inhibitor of autophagy.
Figure 1.
Factors that promote autophagy-mediated airway remodeling in asthma. Asthma is a complex disease whose pathology is influenced by multiple factors, including genetics, allergen exposure, atopy, environmental triggers, and respiratory infections. The recruitment of immune cells into the lung leads to chronic allergic inflammation, which directly affects structural cells, and it is this complex interaction and cross-talk between a variety of immune cells and structural cells that leads to the secretion/release of many pro- and antiinflammatory cytokines in the lung. Transforming growth factor β1 (TGF-β1) is an important cytokine that drives chronic structural changes in the airway in asthma through its direct effect on inducing the autophagy pathway in mesenchymal cells, leading to increased profibrotic signaling and accumulation of extracellular matrix (ECM) proteins such as collagen in the airways. The beneficial effect of chloroquine (CQ) in asthmatic airway remodeling is a result of its actions in both blocking TGF-β1 release and reducing autophagy signaling in mesenchymal cells. ASM = airway smooth muscle.
Although these findings significantly advance our understanding of the role of autophagy in asthma and airway remodeling pathogenesis, the question of whether targeting autophagy for the management of airway remodeling in asthma will be effective is a long way from being answered. Although investigators have demonstrated their ability to reverse airway remodeling features in vitro and in murine in vivo models (and have cured murine asthma in dozens of ways), relevant interventional studies in humans are lacking. Moreover, the disparate functions of autophagy among the many invading and resident airway cells in the asthmatic lung suggest that the effects of drugs inhibiting autophagy in the lung are likely to produce both pro- and antiinflammation/asthma/remodeling effects in different cell types, with an unclear integrative effect. Indeed, the role of autophagy in regulating profibrotic functions in lung fibroblasts is far from settled and appears to be highly dependent on the disease context (16). Autophagy, after all, appears to be a homeostatic and protective mechanism for most cell types; perhaps, as is often the case in disease, such adaptive and protective mechanisms can turn maladaptive and pathogenic.
Also, it is important to point out the limitations of the findings of McAlinden and colleagues. The critical human data used to assess the relationship between autophagy and airway remodeling remain associative. The critical mechanistic evidence is limited by the relevance of the species they assessed (mouse) and the promiscuous nature of the autophagy inhibitor (chloroquine) they used. Although chloroquine, Baf-A1, and 3-MA are arguably the current drugs of choice for implicating autophagy, they are far from specific, and a collective investigation into any role for autophagy demands a robust approach that includes multiple inhibitory strategies—both pharmacologic and molecular—under a range of experimental conditions. For example, the effects of chloroquine in vivo reported in the current study cannot necessarily be attributed to direct effects on ASM, fibroblasts, or epithelia, and likely involve inhibition of inflammation that is only partly dependent on the inhibition of autophagy in immune cells. Despite these limitations, the findings of McAlinden and colleagues provide a strong foundation for future studies that (ideally) will 1) clarify the role of autophagy in the fate and function of the individual cell types that contribute to asthma and the development of airway remodeling; 2) use tissue and in vivo models in creative ways to understand the integrated effects of targeting autophagy in the multicellular lung; and 3) test the most promising drugs (developed on the back of the aforementioned strong basic-science, preclinical studies) when feasible and use informative clinical research designs to investigate airway remodeling drugs in human subjects.
Supplementary Material
Footnotes
Supported by National Institutes of Health grants R01 HL58506, R01 AI110007, R01 HL136209, and P01 HL114471.
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Prakash YS, Halayko AJ, Gosens R, Panettieri RA, Jr, Camoretti-Mercado B, Penn RB ATS Assembly on Respiratory Structure and Function. An official American Thoracic Society research statement: current challenges facing research and therapeutic advances in airway remodeling. Am J Respir Crit Care Med. 2017;195:e4–e19. doi: 10.1164/rccm.201611-2248ST. [DOI] [PubMed] [Google Scholar]
- 2.McAlinden KD, Deshpande DA, Ghavami S, Xenaki D, Sohal SS, Oliver BG, et al. Autophagy activation in asthma airways remodeling. Am J Respir Cell Mol Biol. 2019;60:541–553. doi: 10.1165/rcmb.2018-0169OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zeki AA, Yeganeh B, Kenyon NJ, Post M, Ghavami S. Autophagy in airway diseases: a new frontier in human asthma? Allergy. 2016;71:5–14. doi: 10.1111/all.12761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chaturvedi A, Pierce SK. Autophagy in immune cell regulation and dysregulation. Curr Allergy Asthma Rep. 2009;9:341–346. doi: 10.1007/s11882-009-0050-1. [DOI] [PubMed] [Google Scholar]
- 5.Valečka J, Almeida CR, Su B, Pierre P, Gatti E. Autophagy and MHC-restricted antigen presentation. Mol Immunol. 2018;99:163–170. doi: 10.1016/j.molimm.2018.05.009. [DOI] [PubMed] [Google Scholar]
- 6.Doherty J, Baehrecke EH. Life, death and autophagy. Nat Cell Biol. 2018;20:1110–1117. doi: 10.1038/s41556-018-0201-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kihara A, Kabeya Y, Ohsumi Y, Yoshimori T. Beclin-phosphatidylinositol 3-kinase complex functions at the trans-Golgi network. EMBO Rep. 2001;2:330–335. doi: 10.1093/embo-reports/kve061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stumptner C, Fuchsbichler A, Zatloukal K, Denk H. In vitro production of Mallory bodies and intracellular hyaline bodies: the central role of sequestosome 1/p62. Hepatology. 2007;46:851–860. doi: 10.1002/hep.21744. [DOI] [PubMed] [Google Scholar]
- 9.Poon AH, Chouiali F, Tse SM, Litonjua AA, Hussain SN, Baglole CJ, et al. Genetic and histologic evidence for autophagy in asthma pathogenesis. J Allergy Clin Immunol. 2012;129:569–571. doi: 10.1016/j.jaci.2011.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martin LJ, Gupta J, Jyothula SS, Butsch Kovacic M, Biagini Myers JM, Patterson TL, et al. Functional variant in the autophagy-related 5 gene promotor is associated with childhood asthma. PLoS One. 2012;7:e33454. doi: 10.1371/journal.pone.0033454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dickinson JD, Alevy Y, Malvin NP, Patel KK, Gunsten SP, Holtzman MJ, et al. IL13 activates autophagy to regulate secretion in airway epithelial cells. Autophagy. 2016;12:397–409. doi: 10.1080/15548627.2015.1056967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stamatiou R, Paraskeva E, Boukas K, Gourgoulianis KI, Molyvdas PA, Hatziefthimiou AA. Azithromycin has an antiproliferative and autophagic effect on airway smooth muscle cells. Eur Respir J. 2009;34:721–730. doi: 10.1183/09031936.00089407. [DOI] [PubMed] [Google Scholar]
- 13.Ghavami S, Mutawe MM, Sharma P, Yeganeh B, McNeill KD, Klonisch T, et al. Mevalonate cascade regulation of airway mesenchymal cell autophagy and apoptosis: a dual role for p53. PLoS One. 2011;6:e16523. doi: 10.1371/journal.pone.0016523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghavami S, Yeganeh B, Serebrin A, Mutawe MM, Sharma P, McNeill KD, et al. Autophagy regulates TGF-beta1 induced fibrosis in human airway smooth muscle cells. Am J Respir Crit Care Med. 2011;183:A2110. [Google Scholar]
- 15.Cho IH, Choi YJ, Gong JH, Shin D, Kang MK, Kang YH. Astragalin inhibits autophagy-associated airway epithelial fibrosis. Respir Res. 2015;16:51. doi: 10.1186/s12931-015-0211-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghavami S, Yeganeh B, Zeki AA, Shojaei S, Kenyon NJ, Ott S, et al. Autophagy and the unfolded protein response promote profibrotic effects of TGF-β1 in human lung fibroblasts. Am J Physiol Lung Cell Mol Physiol. 2018;314:L493–L504. doi: 10.1152/ajplung.00372.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.