Abstract
Current asthma therapies fail to target airway remodeling that correlates with asthma severity driving disease progression that ultimately leads to loss of lung function. Macroautophagy (hereinafter “autophagy”) is a fundamental cell-recycling mechanism in all eukaryotic cells; emerging evidence suggests that it is dysregulated in asthma. We investigated the interrelationship between autophagy and airway remodeling and assessed preclinical efficacy of a known autophagy inhibitor in murine models of asthma. Human asthmatic and nonasthmatic lung tissues were histologically evaluated and were immunostained for key autophagy markers. The percentage area of positive staining was quantified in the epithelium and airway smooth muscle bundles using ImageJ software. Furthermore, the autophagy inhibitor chloroquine was tested intranasally in prophylactic (3 wk) and treatment (5 wk) models of allergic asthma in mice. Human asthmatic tissues showed greater tissue inflammation and demonstrated hallmark features of airway remodeling, displaying thickened epithelium (P < 0.001) and reticular basement membrane (P < 0.0001), greater lamina propria depth (P < 0.005), and increased airway smooth muscle bundles (P < 0.001) with higher expression of Beclin-1 (P < 0.01) and ATG5 (autophagy-related gene 5) (P < 0.05) together with reduced p62 (P < 0.05) compared with nonasthmatic control tissues. Beclin-1 expression was significantly higher in asthmatic epithelium and ciliated cells (P < 0.05), suggesting a potential role of ciliophagy in asthma. Murine asthma models demonstrated effective preclinical efficacy (reduced key features of allergic asthma: airway inflammation, airway hyperresponsiveness, and airway remodeling) of the autophagy inhibitor chloroquine. Our data demonstrate cell context–dependent and selective activation of autophagy in structural cells in asthma. Furthermore, this pathway can be effectively targeted to ameliorate airway remodeling in asthma.
Keywords: autophagy, asthma, remodeling, Beclin-1, immunohistochemistry
Asthma is a complex and heterogeneous condition characterized by spontaneous bronchoconstriction accompanied by widespread but variable airflow obstruction. Various genetic and environmental factors interplay in an array of disorders amalgamating in the classical triadic asthma phenotype of airway inflammation, remodeling, and airway hyperresponsiveness (AHR) (1). Therapeutically, only inflammation and AHR can be sufficiently attenuated. The third pathophysiological link in the triadic asthma phenotype is airway remodeling, which needs attention. Airway smooth muscle (ASM) mass, mucous gland hypertrophy, neoangiogenesis in the submucosa, subepithelial fibrosis, increased mucus secretion (by goblet cells), epithelial fragility, and epithelial–mesenchymal transition (EMT) are defining features of airway remodeling in asthma (2). ASM hyperplasia and hypertrophy in the central airways are both indicators and traits of asthma severity (3). Current therapeutics do not effectively target progressive airway fibrosis and remodeling. Elevated basal concentrations of transforming growth factor (TGF)-β in patients with asthma have been associated with sustained characteristic airway remodeling (4, 5). Alongside having a multitude of effects in homeostasis and pathophysiology of disease, TGF-β is believed to drive airway remodeling through the activation of myofibroblasts and smooth muscle cells, subsequently inducing the release of fibrogenic extracellular matrix (ECM) proteins (6). Emerging data suggest that autophagy promotes and influences this classical TGF-β–driven airway remodeling (7). Autophagy modulation may therefore be a novel effective therapy for unmanageable asthma.
Autophagy is a fundamental cellular and physiological process that occurs in all eukaryotic cells (8). The cellular process can be informally referred to as the “inner recycling mechanism” or as a process of “self-eating.” Targeted components are sequestered in double-membrane autophagosomes and ultimately degraded upon fusion with a lysosome with the cytoplasmic components recycled (9). Autophagy is a crucial regulator in the pathogenesis of human disease because it has the potential to influence innate immune responses and promote programmed cell death together with the ability to regularly remove aging proteins, large molecular complexes, and obsolete or damaged organelles (9). The Beclin-1/class III PI3K complex is integral to autophagosomal membrane nucleation (10). ATG5 (autophagy-related gene 5) is covalently conjugated with ATG12 and interacts with ATG16 to form the ATG12–ATG5–ATG16 complex. This complex is associated with autophagosome elongation and is essential for autophagosome formation (9). During autophagy, a truncated cytosolic form of LC3 (microtubule-associated proteins 1A/1B light chain 3A), LC3-I, is conjugated to phosphatidylethanolamine to form the membrane-bound LC3-II (9). Punctate LC3-II is visible with immunostaining and indicates complete formation of autophagosomes (11). SQSTM1 (sequestosome 1)/p62 is a ubiquitin-binding protein that targets and binds to other proteins for selective autophagy, and these proteins are ultimately degraded in the lysosome (12). p62 accumulates when autophagy is inhibited, and inversely, concentrations of p62 decrease when autophagy is induced (13, 14). Although autophagy routinely plays a protective role, its functions, such as cell survival, can be deleterious (15).
Autophagy is involved in the pathogenesis of various diseases, and links between autophagy and asthma are emerging (16–18). A positive correlation of ATG5 and collagen alpha-1 (V) gene expression in the airways of patients with refractory asthma supports this link between dysregulated autophagy and fibrosis in the airways (19). ECM-regulated autophagy is proposed to maintain tissue homeostasis, and thus dysfunctional autophagy in the presence of increased TGF-β may propel the progression of airway remodeling (20). In this study, we found evidence of activation of the autophagy pathway in the small and large airways of patients with asthma. The localization of autophagy proteins in the asthmatic airways is restricted to structural cells in the airway wall and is associated with features of airway remodeling in a TGF-β–dependent manner. We found that TGF-β concomitantly induced autophagy and profibrotic signaling in ASM cells. This induction was prevented by chloroquine (CQ) in vitro. Furthermore, using mouse models of allergic asthma, we demonstrated that targeting the autophagy pathway is an efficient way of providing therapeutic benefit in asthma.
Methods
Acquisition of Human Lung Tissue
Human lung tissue was obtained from surgical resections, explanted lungs, and postmortem organ donors with ethical approval from Royal Prince Alfred Hospital, Concord Repatriation General Hospital, and St. Vincent’s Hospital (HREC14-0045; Sydney).
Human Subject Classification
See the data supplement for full subject classification.
Human Lung Tissue Processing and Section Preparation
Dissected lung tissues were fixed, processed, and embedded in paraffin for analyses (21). After microtome sectioning, hematoxylin and eosin (H&E) staining and Masson’s trichrome staining were used to assess structural integrity, inflammation, and features of airway remodeling. See the data supplement for full methods.
Morphometric Analysis of Inflammation and Airway Remodeling Features
Lamina propria (LP) depth was measured perpendicularly from multiple points at the base of the reticular basement membrane (RBM) to the outer edge of ASM bundles, and the proportion of ASM in the airway wall (ASM/LP as a percentage) was calculated by measuring the total area of ASM mass per airway and dividing by the total area of the LP. Overall tissue inflammation in the lung was assessed, and immune cells were counted manually in the lung tissue as described in the data supplement.
Immunohistochemistry and Immunofluorescence Staining
Immunostaining for Beclin-1, ATG5, LC3B, p62, and ACTA2 was performed as previously described (22–24). See the data supplement for full methods.
Image Analysis
Computer-assisted image analysis was performed with a NanoZoomer-SQ Digital Slide Scanner (Hamamatsu), an Olympus BX51 upright epifluorescence microscope fitted with a DP70 charge-coupled device camera (Olympus), and ImageJ software.
Cell Culture
Human ASM cells were obtained from human lung by using a method described previously. See the data supplement for full methods.
Mouse Models of Allergic Asthma
Experiments were conducted according to the institutional guidelines and the code for the care and use of animals. The animal care committees of Thomas Jefferson University and University of Technology Sydney approved the protocol. All surgeries were performed with the animals under tribromoethanol anesthesia, and all efforts were made to minimize suffering. BALB/c mice (female) were subjected to a subchronic (prophylactic) model of allergic asthma as described. Thirty minutes before house dust mite (HDM) challenges, selected mice were administered either CQ intranasally (50 mg/kg) or saline as a vehicle. In a separate study, BALB/c mice (female) were subjected to a treatment model (chronic allergic asthma model) of asthma as described. At Week 4 and commencing for 2 weeks, 30 minutes before HDM challenges, selected mice were administered either CQ intranasally (50 mg/kg) or vehicle (saline). In both studies, 24 hours after the last HDM challenge, lung function measurements were performed (flexiVent; SCIREQ Scientific Respiratory Equipment Inc.), BAL fluid was collected, and lungs were formalin fixed or flash frozen for histopathological and biochemical analyses. See the data supplement for full methods.
Mouse BAL Immune Cell Staining and Lung H&E, Periodic Acid–Schiff, and Masson’s Trichrome Staining
BAL sample cytospins were prepared and stained with the Hema 3 Staining Kit (Fisher Scientific). The fixed lung tissues embedded in paraffin were cut and stained with H&E, periodic acid–Schiff (PAS), and Masson’s trichrome stains using a protocol described previously (25–27). See the data supplement for full methods.
Measurement of TGF-β1
The content of TGF-β1 in BAL fluid was measured by using Multiplexing LASER Bead Technology with a custom TGF-β 3-Plex Cytokine Array (Eve Technologies).
Western Blotting
Protein concentrations of collagen IA, pSMAD2/3, SMAD2/3, Beclin-1, and LC3B in ASM cell lysates or murine lung tissues were measured by immunoblotting. All immunoblotting was performed using protocols described previously (26, 28). See the data supplement for full methods.
Soluble Collagen Assay
Total soluble collagen content in the lung lysates was assessed using Sircol collagen assay (Biocolor) (29). See the data supplement for full methods.
Statistical Analysis
Data were analyzed using unpaired t tests or one-way or two-way ANOVA as appropriate and are presented as mean ± SD or mean ± SEM. All data were analyzed with Prism version 7.04 software (GraphPad Software), and P < 0.05 was considered statistically significant.
Results
Histological Evidence of Airway Remodeling and Inflammation
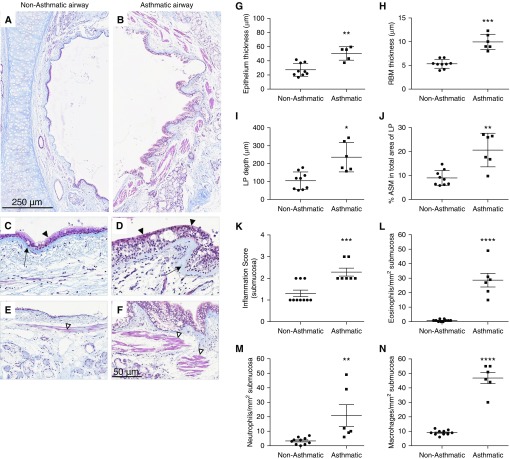
Because the link between autophagy and airway remodeling has been established, we first chose to histologically measure remodeling features of our selected patients with and without asthma. Asthmatic airways in comparison with nonasthmatic airways displayed greater features of airway remodeling. Gross remodeling changes between nonasthmatic and asthmatic large airways were observed using trichrome staining (Figures 1A–1F). The average epithelial thickness was significantly greater in patients with asthma than in those without asthma (asthma [A], 50.46 ± 9.52 μm; nonasthma [NA], 27.46 ± 9.16 μm; P < 0.001), and with aniline blue dye staining of the fibrous RBM, we observed an overall increase in thickness of the RBM in patients with asthma (A, 9.94 ± 1.63 μm; NA, 5.32 ± 0.93 μm; P < 0.0001) (Figures 1G and 1H). The average LP depth in patients with asthma was significantly thicker than in those without asthma (A, 235.71 ± 81.97 μm; NA, 105.31 ± 48.81 μm; P < 0.05), and staining of acidophilic tissue components (cytoplasm and muscle) with Biebrich scarlet-acid fuchsin allowed us to measure and observe an increase in the percentage of ASM mass in asthmatic LP (A, 20.63 ± 7.01%; NA, 8.99 ± 3.16%; P < 0.001) (Figures 1I and 1J). These measurements classify the selected patients with asthma as having undergone associated airway remodeling and justify their inclusion in the immunohistological component of this study. We further assessed tissue inflammation in the lungs by scoring overall airway wall inflammation (Figure 1K), which showed greater influx of immune cells in the asthmatic airways. The nature of immune cell influx was determined by counting eosinophils (Figure 1L), neutrophils (Figure 1M), and macrophages (Figure 1N). As shown in Figure 1, asthmatic lungs demonstrated significantly greater influx of immune cells into the lungs.
Figure 1.
Histological evidence of airway remodeling in the large airways of the selected patients with asthma. (A) Nonasthmatic and (B) asthmatic airways were stained with Masson’s trichrome stain as described in the Methods section of the text. Scale bars: 250 μm and 50 μm. (C and D) Arrows indicate the reticular basement membrane (RBM), and solid arrowheads indicate the epithelium. (E and F) Airway smooth muscle (ASM) bundles are indicated by open arrowheads. Smooth muscle bundles are stained red, and connective tissue is stained blue. Original magnification, ×400 in C–F. (G) Epithelium; (H) RBM thickness; (I) lamina propria (LP) depth; (J) proportion of ASM in the asthmatic airway wall; (K) inflammation score in submucosa; and (L) eosinophil, (M) neutrophil, and (N) macrophage counts per square millimeter were quantified. Data are expressed as mean ± SD or mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Expression Profile of Autophagy Marker Proteins in the Large Airways of Patients with Asthma
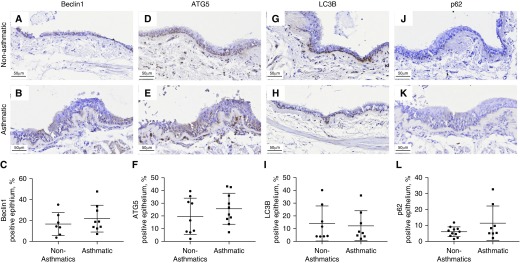
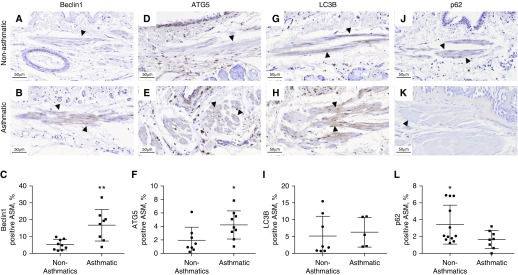
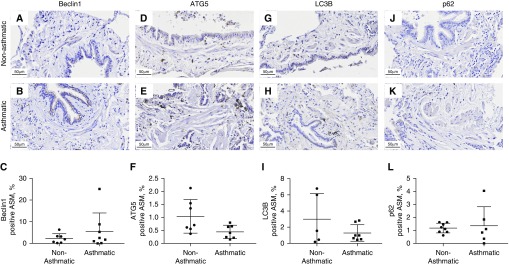
We selected four key markers of autophagy to examine their expression in the epithelial and ASM components of small and large airway walls in conjunction with normal nonasthmatic histology and remodeled asthmatic histopathology. There were no significant differences in the large airway epithelium (Figure 2), whereas in the large airway ASM bundles, we observed a marked increase in the expression (percentage area ± SD) of Beclin-1 (A, 16.78 ± 9.38%; NA, 5.16 ± 3.07%; P < 0.01) and ATG5 (A, 4.23 ± 2.09%; NA, 1.94 ± 1.94%; P < 0.05) and a decrease in p62 expression (A, 1.65 ± 1.06%; NA, 3.41 ± 2.31%; P < 0.05) in patients with asthma compared with those without asthma (Figure 3).
Figure 2.
Expression of autophagy markers in the large airway epithelium of asthmatic and nonasthmatic tissue. Asthmatic and nonasthmatic tissues were immunohistochemically stained for (A and B) Beclin-1, (D and E) ATG5 (autophagy-related gene 5), (G and H) LC3B (microtubule-associated proteins 1A/1B light chain 3B), and (J and K) sequestesome-1 (p62). The positive area for each of these markers in the epithelium was quantified (C, F, I, and L). Scale bars: 50 μm.
Figure 3.
Expression of autophagy markers in the large ASM bundles of asthmatic and nonasthmatic tissue. Asthmatic and nonasthmatic tissue were immunohistochemically stained for (A and B) Beclin-1, (D and E) ATG5, (G and H) LC3B, and (J and K) p62. Arrowheads indicate ASM bundles. The positive area for each of these markers in the ASM was quantified (C, F, I, and L). Data are expressed as mean ± SD. Scale bars: 50 μm. *P < 0.05 and **P < 0.01.
Expression Profile of Autophagy Markers in the Small Airways of Patients with Asthma
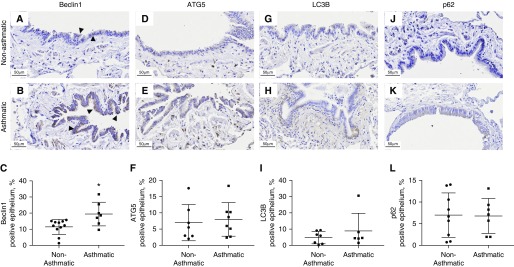
In the small airway epithelium, we observed solely a marked increase in the expression (percentage area ± SD) of Beclin-1 (A, 19.44 ± 7.31%; NA, 11.44 ± 4.63%; P < 0.05) (Figure 4), with no significant differences in small airway ASM bundles (Figure 5).
Figure 4.
Expression of autophagy markers in the small airway epithelium of asthmatic and nonasthmatic tissues. Asthmatic and nonasthmatic tissues were immunohistochemically stained for (A and B) Beclin-1, (D and E) ATG5, (G and H) LC3B, and (J and K) p62. Arrowheads indicate epithelium. The positive area for each of these markers in the epithelium was quantified (C, F, I, and L). Data are expressed as mean ± SD. Scale bars: 50 μm. *P < 0.05.
Figure 5.
Expression of autophagy markers in the small ASM bundles of asthmatic and nonasthmatic tissues. Asthmatic and nonasthmatic tissues were immunohistochemically stained for (A and B) Beclin-1, (D and E) ATG5, (G and H) LC3B, and (J and K) p62. The positive area for each of these markers in the epithelium was quantified (C, F, I, and L). Scale bars: 50 μm.
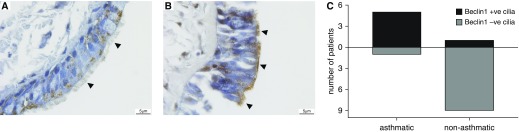
Expression of Beclin-1 in Cilia Lining the Large Airways of Patients with Asthma
In accordance with our classification, 5 of 6 patients with asthma displayed strong expression of Beclin-1 in the cilia lining the large airway epithelium, whereas only 1 of 10 patients without asthma displayed any expression of Beclin-1 in the cilia of the large airway epithelium. Overall, we demonstrated that strong expression of Beclin-1 in cilia lining large airway epithelial cells occurs mostly in patients with asthma with associated airway remodeling evident compared with patients without asthma (Figures 6A–6C). Other markers (ATG5, LC3B, p62) were not evident in the cilia lining of large airway epithelium of both patients with and without asthma.
Figure 6.
Expression of Beclin-1 in cilia lining large airway epithelium of patients with asthma. (A) Nonasthmatic large airway epithelium and (B) asthmatic large airway epithelium immunohistochemically stained for Beclin-1. Arrowheads indicate cilia. Scale bars: 5 μm. (C) Representation of the number of patients (asthma vs. nonasthma) with Beclin-1 positivity in cilia.
Concomitant Induction of Profibrotic Signaling and Autophagy Markers in Human ASM Cells
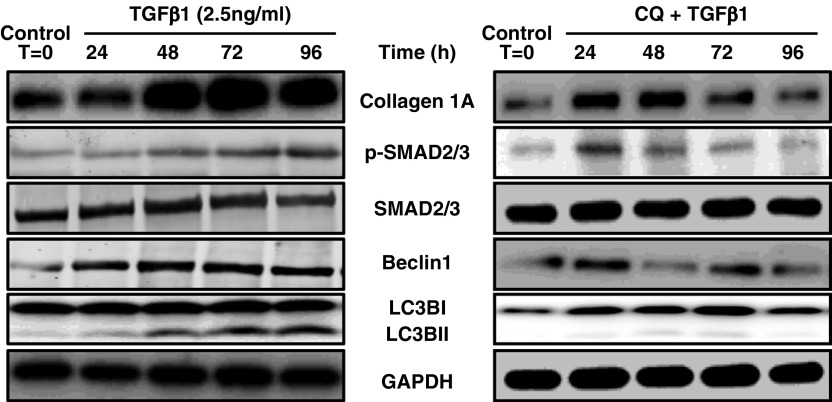
Greater tissue inflammation in the asthmatic lungs is in congruence with the published literature and supports the tenet that inflammation can partly drive airway remodeling in asthma. Therefore, we tested this hypothesis whether TGF-β1 (a pleiotropic cytokine, elevated in asthma) would concomitantly induce profibrotic signaling and autophagy in vitro. As shown in Figure 7, we found that TGF-β1, in a time-dependent manner, increased collagen I expression and SMAD2/3 phosphorylation (profibrotic signaling) and induced autophagy in ASM cells as seen with increased expression of Beclin-1 and LC3B-II (Figure 7A). This demonstrates an association between the effects of TGF-β1, the accumulation of collagen, and increased profibrotic signaling in an autophagy-dependent manner.
Figure 7.
Concomitant induction of profibrotic signaling and autophagy by transforming growth factor (TGF)-β1 in vitro. Human airway smooth muscle cells were treated with either TGF-β1 (2.5 ng/ml) or TGF-β1 (2.5 ng/ml) + chloroquine (CQ) (50 μM) for 0, 24, 48, 72, and 96 hours. Cell lysates were prepared, and immunoblotting was performed for collagen IA, phospho-SMAD2/3, total SMAD2/3, Beclin-1, and LC3B-I/II. GAPDH was used as a loading control. Data shown are representative of four independent primary human airway smooth muscle cells.
Effect of Autophagy Inhibitor on the Concomitant Induction of Profibrotic Signaling and Autophagy Markers in Human ASM Cells
Furthermore, we tested in vitro whether the treatment with autophagy inhibitor CQ would alleviate profibrotic signaling and the induction of autophagy by TGF-β1 stimulation. As shown in Figure 7, we found that treatment with CQ (50 μM) in conjunction with TGF-β1 stimulation reduced the expression of collagen IA, Beclin-1, and LCB3-II, as well as the phosphorylation of SMAD2/3, in a time-dependent manner (Figure 7). This shows that CQ reduced TGF-β1–induced airway remodeling markers in an autophagy-dependent (inhibition) manner.
Effect of Autophagy Inhibitor in a Prophylactic Model of Allergic Asthma
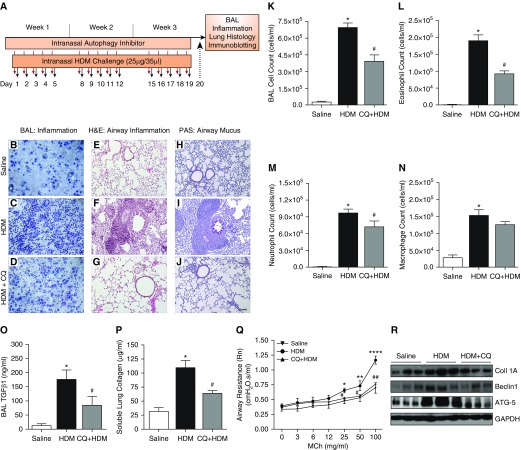
To further investigate the interplay between autophagy and asthma, we studied the effects of autophagy inhibition in allergen (HDM)-induced asthma in mice (prophylactic model) as shown in Figure 8A. CQ is a known inhibitor of autophagy, increasing the pH of lysosomes and inhibiting fusion and formation of the autophagolysosome. Increased inflammatory cell infiltration in both the airway wall and the BAL, together with increased production of mucus, was observed in the HDM-challenged mice in comparison with control mice (Figure 8). Autophagy inhibitor effectively blocked influx of immune cells into the airways (Figures 8B–8D and 8K) and significantly reduced eosinophils and neutrophils in the BAL fluid with no effect on macrophages (Figures 8L–8N). Tissue inflammation was also reduced in the mouse lungs treated with CQ (Figures 8E–8G; H&E staining), and there was a reduction in mucus production in HDM-challenged mice treated with autophagy inhibitor CQ (Figures 8H–8J; PAS staining of airway mucus).
Figure 8.
(A) Mouse model of allergic asthma. Female mice at 8 weeks old were challenged intranasally with house dust mite (HDM) allergen 5 d/wk for 3 weeks. Autophagy inhibitor CQ (50 mg/kg) was administered 30 minutes before each HDM challenge. Twenty-four hours after the last challenge, BAL and lung tissue were collected for further analysis. Inflammation in the BAL fluid was measured in (B) saline control mice, (C) HDM-challenged mice, and (D) the HDM + CQ-treated group. Hematoxylin and eosin (H&E) staining was used to measure tissue inflammation in (E) saline control mice, (F) HDM-challenged mice, and (G) the HDM + CQ-treated group. Periodic acid–Schiff (PAS) staining was performed to measure mucus production in (H) saline control mice, (I) HDM-challenged mice, and (J) the HDM + CQ-treated group. Scale bar: 100 µM. CQ treatment in the HDM-challenged group reduced influx of (K) total immune cells, (L) eosinophils, (M) and neutrophils. (N) Macrophages remained unchanged. Furthermore, inhibition of autophagy reduced (O) profibrotic cytokine TGF-β1 concentrations in BAL and also (P) the amount of soluble lung collagen in tissue lysates. One-way ANOVA: *P < 0.001 for saline versus HDM; #P < 0.05 for HDM versus HDM + CQ with Bonferroni multiple comparisons test. (Q) FlexiVent analysis revealed reduction in methacholine (MCh)-induced airway hyperresponsiveness in the CQ-treated group compared with the HDM-challenged group alone. Two-way ANOVA: *P < 0.05, **P < 0.01, and ****P < 0.0001 for saline versus HDM (at 25, 50, and 100 mg/ml); #P < 0.05 and ##P < 0.01 for HDM versus HDM + CQ (at 25, 50, and 100 mg/ml). (R) Representative protein immunoblots for collagen IA, Beclin-1, and ATG5 in lung tissue lysates from saline-, HDM-, and HDM + CQ-treated mice. Mouse data shown represent mean ± SEM from n = 6 or 7 per group.
To examine the connection of autophagy with airway remodeling, we performed assays to measure concentrations of TGF-β1 in the BAL and soluble lung collagen, and we performed immunoblot analysis of collagen I, Beclin-1, and ATG5 expression in the lung lysates. The HDM-challenged group showed a significant increase in the concentration of TGF-β1 in the BAL compared with control group (P < 0.001), whereas treatment with CQ significantly reduced TGF-β1 concentrations compared with the HDM group (P < 0.05) (Figure 8O). Furthermore, we found a significantly greater amount of soluble lung collagen in tissue lysates of the HDM-challenged group than in the control group (P < 0.001), whereas lysates from the CQ-treated group showed a significant reduction in the amount of soluble collagen in comparison with the HDM group (P < 0.05) (Figure 8P). Lung function measurements using flexiVent showed increased AHR in HDM-challenged mice in response to methacholine (MCh) when compared with control animals (increase in airway resistance at 25, 50, and 100 mg/ml), whereas CQ treatment prevented development of AHR in mice (significant reduction in airway resistance at 25, 50, and 100 mg/ml) when compared with the HDM-challenged group (Figure 8Q). Immunoblotting in lung lysates revealed higher and concomitant expression of ECM protein collagen IA and autophagy markers Beclin-1 and ATG5 in the HDM-challenged mice when compared with control animals, whereas CQ treatment prevented induction of autophagy (as shown by the reduction in Beclin-1 and ATG5) and accumulation of collagen in the lung (Figure 8R).
Effect of Autophagy Inhibitor in a Chronic Model of Allergic Asthma
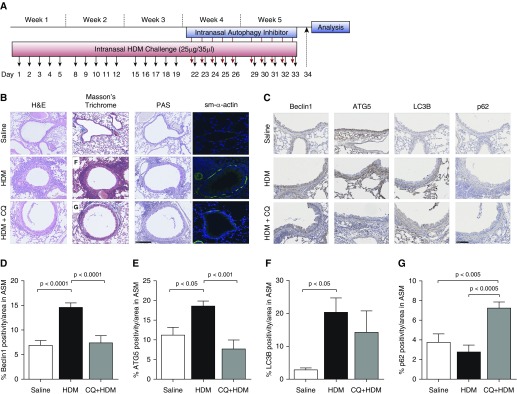
We further employed an allergen (HDM)-induced mouse model of chronic asthma (treatment model) to investigate the role of autophagy in airway remodeling in asthma (Figure 9A). Mass inflammatory cell influx was observed in the airways of mice challenged with HDM, and this was significantly attenuated in the CQ + HDM-treated mice (Figure 9B; H&E stain). Similar reduction was observed in the accumulation of fibrotic proteins in the CQ + HDM-treated group when compared with the HDM group (Figure 9B; Masson’s trichrome stain). Increased production of mucus was observed in the HDM-challenged mice in comparison with saline-treated control animals and was greatly reduced in the CQ + HDM-treated group (Figure 9B; PAS stain). We further performed immunofluorescence imaging for ASM marker ACTA2; chronic allergen challenge increased ASM bundles, which were significantly reduced by CQ treatment (Figure 9B; ACTA2 stain).
Figure 9.
(A) Mouse model of chronic allergic asthma. Female mice at 8 weeks old were challenged intranasally with HDM allergen 5 d/wk for 5 weeks. Starting at Week 4 and for the remaining 2 weeks, autophagy inhibitor CQ (50 mg/kg) was administered 30 minutes before each HDM challenge. Twenty-four hours after the last challenge, BAL and lung tissue were collected for further analysis. (B) H&E staining was used to measure tissue inflammation in the saline control, HDM-challenged, and HDM + CQ-treated mice. Masson’s trichrome staining was used to measure connective tissue deposition and smooth muscle mass in the saline control, HDM-challenged mice, and HDM + CQ-treated mice. PAS stain was used to measure mucus production in the saline control, HDM-challenged, and HDM + CQ-treated mice. Smooth muscle α-actin (sm-α-actin) was used to stain smooth muscle cells in the saline control, HDM-challenged, and the HDM + CQ-treated mice. Scale bar: 250 µm. (C) Immunohistochemical staining was used to measure the expression of autophagy proteins in the airway smooth muscle bundles of the mice. Beclin-1, ATG5, LC3B, and p62 expression were measured in the saline control, HDM-challenged, and HDM + CQ-treated mice. Scale bar: 150 µm. (D–G) Positive area for each of these markers in ASM was quantified. Data are expressed as mean ± SD or mean ± SEM. n = 4–6 mice.
Immunohistochemical staining was used to examine the expression of four key protein markers of autophagy in the airways of saline-treated control, HDM-challenged, and CQ + HDM-treated mice. In the ASM bundles of the HDM-challenged mice, we observed a marked increase in the expression (percentage area ± SD) of Beclin-1 (HDM, 14.67 ± 3.02%; control, 6.98 ± 3.08%; P < 0.0001) (Figure 9D), ATG5 (HDM, 18.7 ± 3.99%; control, 11.27 ± 6.67%; P < 0.05) (Figure 9E), and LC3B (HDM, 20.47 ± 14.67%; control, 2.97 ± 1.59%; P < 0.05) (Figure 9F) in the HDM-challenged mice compared with control mice. Treatment with CQ significantly reduced the expression of Beclin-1 (CQ + HDM, 7.5 ± 4.65%; HDM, 14.67 ± 3.02%; P < 0.0001) (Figure 9D) and ATG5 (CQ + HDM, 7.78 ± 7.18%; HDM, 18.7 ± 3.99%; P < 0.001) (Figure 9E) and increased the expression of p62 (CQ + HDM, 7.27 ± 1.97%; HDM, 2.82 ± 2.09%; P < 0.0005) (Figure 9G) in the CQ + HDM-treated mice compared with HDM-challenged mice.
Discussion
The upregulation of markers of autophagy has been linked to fibrosis and remodeling in various organs (7, 30). However, there has been limited histopathological evidence to show similar trends in the lungs of patients with asthma. Although there are early reports on increased gene expression of ATG5 with additional measurement of ATG5 protein expression in the airways of patients with refractory asthma (16, 19), our study provides a comprehensive analysis of multiple autophagy markers and their association with airway remodeling in asthma. More importantly, our data on Beclin-1 expression in both large and small airways and in the ciliated cells provide novel insight into how the autophagy pathway may regulate and control airway remodeling in asthma. Furthermore, this is also replicated in the murine models, where we found induction of autophagy in experimental asthma models with a concomitant expression of profibrotic cytokines and collagen in the lung, which was attenuated in the presence of an autophagy inhibitor.
First, we confirmed that the tissues we used displayed classical features of airway remodeling (5). A variety of factors contribute to the pathogenesis of asthma and subsequent development of airway remodeling in asthma. It is suggested that persistent insult leading to chronic inflammation with time leads to the development of structural changes in the lung that tracks clearly with significant reduction in lung function and increase in asthma severity (31). Expression of Beclin-1 and ATG5 were found to be increased in the large airway ASM of patients with asthma compared with healthy individuals without asthma, together with reduced expression of p62 in the large airway ASM of patients with asthma compared with healthy individuals without asthma. Therefore, in this patient cohort, we found concomitant expression and association of autophagy with airway remodeling and inflammation. These findings are novel in asthma because there is no evidence to date suggesting expression of autophagy being closely associated with airway remodeling. This finding is reinforced by data derived from studying our chronic HDM model of allergic asthma in mice, in which we found increased expression of Beclin-1, ATG5, and LC3B in the ASM bundles of HDM-challenged mice when compared with the control animals. This work clearly supports the published literature on the kidney, liver, and other organs, where it has been shown that autophagy regulates tissue fibrosis (32, 33). However, it was recently reported that autophagy is a necessary mechanism for changing the phenotype of lung epithelial cells to mesenchymal cells (34, 35); therefore, the present finding that shows incidence of autophagy in the epithelium of patients with asthma and HDM-challenged mice might confirm the role of EMT in the accumulation of mesenchymal cells in asthma pathogenesis.
Airway remodeling features such as thickening of the basement membrane are a key indicator of the development of asthma later in life; basement membrane thickening has been seen at an early age in children at 3–4 years of age (36). This led us to think about various signaling pathways, such as autophagy, that can be aberrant and can contribute to disease development later in life. Although this needs to be investigated in samples from children, our data from adult patients with asthma uncover the novel fundamental mechanism (autophagy) that may act as a key determinant of airway remodeling in asthma. Similarly, ASM mass is increased in asthma and correlates with poor lung function and increased airway responsiveness to a variety of contractile agonists, allergens, pollens, and so forth. Our data derived from studying human lungs clearly demonstrate a link between increased accumulation of ASM mass and increased expression of autophagy markers in the asthmatic lung, a feature totally absent in the nonasthmatic human lung. If these results are replicated in a larger cohort of patients with asthma, then we can direct our research efforts to selectively target mesenchymal cells using novel formulation and chemistry approaches, which will lead to reduced mesenchymal cell mass, reduction in release of ECM proteins, and ultimately reduction in the ability of the airways to contract in response to a variety of triggers.
An increase in the protein expression of Beclin-1 was also observed in the small airway epithelium compared with that in healthy individuals without asthma. With access to tissue from a greater number of patients, accompanying significant differences in protein expression of Beclin-1 and ATG5 in the large airway epithelium may be observed. Interestingly, asthmatic epithelium displayed strong expression of Beclin-1 in cilia, whereas nonasthmatic epithelium did not display these levels of expression. The comparative expression of Beclin-1 in cilia of asthmatic and nonasthmatic epithelium has a significant binary pattern. In chronic obstructive pulmonary disease, autophagy-dependent pathways regulate cilia length upon exposure to cigarette smoke (37). The identification of overexpression of Beclin-1 in ciliated cells of patients with asthma highlights a role for ciliophagy also in asthma and may have implications for mucociliary clearance. A question that arises is whether airway wall changes in patients with asthma contribute to increased expression of Beclin-1 in cilia or whether the increased expression of Beclin-1 in the cilia influences the cellular stimulation of airway remodeling. The effect that this upregulation of Beclin-1 in asthmatic ciliated cells has on autophagic flux in other respiratory cells of the airway wall requires further investigation. One limitation of this study is the coincidence of all patients included being male, and this coincidence is unavoidable with the limited access to asthmatic and nonasthmatic tissue we have at this time. A further limitation is our lack of addressing the different phenotypes presented in asthma diagnosis, such as eosinophilic, neutrophilic, or paucigranulocytic asthma. We believe the patient group in this study is reflective of mild-moderate asthma; that is, it represents a greater asthma population in the general public. In the future, we would like to investigate whether expression of autophagy is associated with asthma severity.
We further investigated whether an inhibitor of autophagy, CQ, has any therapeutic value in both acute (subchronic) and chronic (treatment) models of allergen (HDM)-induced asthma in mice. A few clinical studies in the early 1980s and 1990s demonstrated that hydroxychloroquine can be used as an effective treatment for severe asthma because it has a steroid-sparing effect (38). Since then, there has been a lack of big prospective clinical trials to further validate these original observations. Our preclinical data clearly demonstrate that CQ is effective in blocking the influx of immune cells both in BAL and in lung tissue. Furthermore, our data uncover novel therapeutic effects of CQ in asthma because it reduces features of airway remodeling by preventing accumulation of mucus and ECM matrix protein collagen I in the lung. The inhibition of autophagy by CQ was supported by our immunohistochemical analysis in the chronic mouse model showing decreased expression of Beclin-1 and ATG5 and increased expression of p62 in the ASM bundles of CQ + HDM-treated mice in comparison with HDM-challenged mice. We also found reduction in ASM mass, as shown by reduced staining for ACTA2 in the airways of mice treated with CQ. The mechanisms of the beneficial effect of CQ rely on blocking the release of profibrotic cytokine TGF-β1 in the BAL of HDM-challenged mice after treatment with CQ. It is well known that TGF-β1 drives fibrotic changes in the lung and that its concentrations are elevated in asthma (39), and it is emerging that the autophagy pathway may interact with the ECM and affect cytokine secretion (7, 40). We have shown in vitro that upon stimulation with TGF-β1, human ASM cells produced greater amounts of collagen IA and had increased expression of Beclin-1 and LC3B-II and greater phosphorylation of SMAD2/3 in a time-dependent manner, which supports the growing notion that TGF-β1 concomitantly induces autophagy and drives profibrotic signaling. CQ acts by inhibiting autophagy and reducing TGF-β1–dependent profibrotic signaling in asthma. This was confirmed by the cotreatment of CQ and TGF-β1 resulting in a time-dependent reduction in alpha-1 type I collagen, diminished expression of Beclin-1 and LC3B-II, and reduced phosphorylation of SMAD2/3. These beneficial effects of modulating autophagy using CQ in a disease setting (mouse models) also translated to reducing development of AHR (data not shown for chronic model) in response to MCh, which is another hallmark of allergic asthma.
Furthermore, our mouse models demonstrated clear initiation (in the 3-wk model) and establishment (in the 5-wk model) of airway remodeling, as seen by increase in mucus release, accumulation of collagen I in the lung, and activation of the autophagy pathway, reflected by an increase in autophagy markers in the lung. CQ treatment in HDM-challenged mice clearly blocked features of airway remodeling in the lung with a concomitant reduction in autophagy, as seen with reduction in Beclin-1 and ATG5 proteins. This clearly demonstrates that CQ works at multiple levels to provide therapeutic benefit in asthma: It blocked inflammation, reduced mucus secretion, inhibited TGF-β1 release in BAL, and reduced release and accumulation of ECM proteins in the lung, leading to reduction in AHR. This is achieved by modulation of the autophagy pathway because CQ inhibits overall autophagic flux by increasing the pH in the lysosome, preventing the fusion of the autophagolysosome and the subsequent degradation of lysosomal components (41). Though autophagy modulation with CQ is not highly selective, owing to the fact that it has a number of other pharmacological effects, at the concentrations used in this study, Beclin-1 and ATG5 were reduced by CQ. Therefore, we are highly confident of these findings that CQ works in an experimental asthma model by modulation of the autophagy pathway. Originally an antimalarial and antiarthritis drug (42), CQ has been shown also to target inwardly rectifying potassium channels (43), and by disrupting the blood–retina barrier, it has the potential to cause retinopathy through binding to melanin and toxic sequestration in the eye (44). Compounds with greater specificity are now being considered in the targeting of the autophagy pathway. Bafilomycins are macrolide antibiotics derived from Streptomyces griseus bacteria that inhibit vacuolar-type H+-ATPase. At high concentration, bafilomycin is capable of blocking late-phase autophagy through significant cytosolic acidification (45). 3-Methyladenine also has the ability to suppress the formation of autophagosomes specifically targeting autophagy through the inhibition of class III PI3K (46). However, 3-methyladenine is found to have a dual role in autophagy modulation. It surprisingly promotes autophagic flux with prolonged treatment under nutrient-rich conditions and also has the capability of suppressing starvation-induced autophagy (47). LY294002, derived from the flavonoid quercetin (48), inhibits PI3K activity (49) and belongs among the emerging safe therapeutics with the potential to treat airway remodeling. LY294002 treatment in mice challenged with FSTL1 (follistatin-related protein 1) (linked to the promotion of both EMT and autophagy) has shown early signs of implication in the attenuation of airway remodeling (50).
Inhibition of autophagy, as shown in the present study with CQ treatment, is entwined with an upstream TGF-β1 response and the amount of collagen production. As previously shown in hepatic cells and cardiomyocytes, TGF-β concomitantly influences features of remodeling and regulates degrees of autophagy (7, 30), and we introduce and provide evidence that a similar concomitant pathway occurs both in vitro and in vivo. This pathway, we believe, has the greatest influence in the ASM of the large airways, as seen in asthmatic lungs, and this was also replicated in vitro using primary human ASM cells, where we found TGF-β1 concomitantly induced both profibrotic signaling and autophagy. We have shown an increase (and supporting decrease for p62) in several vital autophagy-linked proteins in the large airway ASM bundles. We have shown that inhibition of autophagy in murine models can attenuate inflammation, clear mucus production, reduce concentrations of TGF-β1 in the BAL, and ultimately reduce airway remodeling and prevent bronchoconstriction. Thus, our study indicates that in asthmatic airways, autophagy is enhanced, which we believe contributes to remodeling in a TGF-β1–dependent manner, and that inhibition of autophagy is an attractive target for alleviation of airway remodeling in asthma.
Table 1.
Demographic Data
| Large airway demographics | ||
| Group (n) | Nonasthma (10) | Asthma (6) |
| Age, yr, median (range) | 55 (19–67) | 48 (15–80) |
| Males/females | 10/0 | 6/0 |
| Small airway demographics | ||
| Group (n) | Nonasthma (10) | Asthma (7) |
| Age, yr, median (range) | 52 (25–69) | 53.5 (15–80) |
| Males/females | 10/0 | 7/0 |
Data are expressed as number of subjects or median (range).
Supplementary Material
Footnotes
Supported by the Chancellor’s Fellowship Program and Rebecca Cooper Foundation (P.S.), the Clifford Craig Foundation (S.S.S.), the National Institutes of Health (National Heart, Lung, and Blood Institute grant HL137030 [D.A.D.]), and the National Health and Medical Research Council (B.G.O.).
Author Contributions: P.S.: conceived of the idea for the study; K.D.M. and P.S.: performed the experiments and data analysis and wrote the manuscript; D.A.D., S.G., and S.S.S.: helped with data analysis and reagents; and D.X., B.G.O., and M.H.: reviewed, edited, and revised the manuscript and agreed to the final content.
This article has a data supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Originally Published in Press as DOI: 10.1165/rcmb.2018-0169OC on November 1, 2018
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Royce SG, Cheng V, Samuel CS, Tang ML. The regulation of fibrosis in airway remodeling in asthma. Mol Cell Endocrinol. 2012;351:167–175. doi: 10.1016/j.mce.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 2.James AL, Elliot JG, Jones RL, Carroll ML, Mauad T, Bai TR, et al. Airway smooth muscle hypertrophy and hyperplasia in asthma. Am J Respir Crit Care Med. 2012;185:1058–1064. doi: 10.1164/rccm.201110-1849OC. [DOI] [PubMed] [Google Scholar]
- 3.Benayoun L, Druilhe A, Dombret MC, Aubier M, Pretolani M. Airway structural alterations selectively associated with severe asthma. Am J Respir Crit Care Med. 2003;167:1360–1368. doi: 10.1164/rccm.200209-1030OC. [DOI] [PubMed] [Google Scholar]
- 4.Redington AE, Madden J, Frew AJ, Djukanovic R, Roche WR, Holgate ST, et al. Transforming growth factor-β1 in asthma: measurement in bronchoalveolar lavage fluid. Am J Respir Crit Care Med. 1997;156:642–647. doi: 10.1164/ajrccm.156.2.9605065. [DOI] [PubMed] [Google Scholar]
- 5.Chakir J, Shannon J, Molet S, Fukakusa M, Elias J, Laviolette M, et al. Airway remodeling-associated mediators in moderate to severe asthma: effect of steroids on TGF-β, IL-11, IL-17, and type I and type III collagen expression. J Allergy Clin Immunol. 2003;111:1293–1298. doi: 10.1067/mai.2003.1557. [DOI] [PubMed] [Google Scholar]
- 6.Boxall C, Holgate ST, Davies DE. The contribution of transforming growth factor-β and epidermal growth factor signalling to airway remodelling in chronic asthma. Eur Respir J. 2006;27:208–229. doi: 10.1183/09031936.06.00130004. [DOI] [PubMed] [Google Scholar]
- 7.Ghavami S, Cunnington RH, Gupta S, Yeganeh B, Filomeno KL, Freed DH, et al. Autophagy is a regulator of TGF-β1-induced fibrogenesis in primary human atrial myofibroblasts. Cell Death Dis. 2015;6:e1696. doi: 10.1038/cddis.2015.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mizushima N, Levine B. Autophagy in mammalian development and differentiation. Nat Cell Biol. 2010;12:823–830. doi: 10.1038/ncb0910-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi AM, Ryter SW, Levine B. Autophagy in human health and disease. N Engl J Med. 2013;368:651–662. doi: 10.1056/NEJMra1205406. [DOI] [PubMed] [Google Scholar]
- 10.Kihara A, Kabeya Y, Ohsumi Y, Yoshimori T. Beclin-phosphatidylinositol 3-kinase complex functions at the trans-Golgi network. EMBO Rep. 2001;2:330–335. doi: 10.1093/embo-reports/kve061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stumptner C, Fuchsbichler A, Zatloukal K, Denk H. In vitro production of Mallory bodies and intracellular hyaline bodies: the central role of sequestosome 1/p62. Hepatology. 2007;46:851–860. doi: 10.1002/hep.21744. [DOI] [PubMed] [Google Scholar]
- 13.Bjørkøy G, Lamark T, Pankiv S, Øvervatn A, Brech A, Johansen T. Monitoring autophagic degradation of p62/SQSTM1. Methods Enzymol. 2009;452:181–197. doi: 10.1016/S0076-6879(08)03612-4. [DOI] [PubMed] [Google Scholar]
- 14.Shvets E, Fass E, Scherz-Shouval R, Elazar Z. The N-terminus and Phe52 residue of LC3 recruit p62/SQSTM1 into autophagosomes. J Cell Sci. 2008;121:2685–2695. doi: 10.1242/jcs.026005. [DOI] [PubMed] [Google Scholar]
- 15.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poon AH, Chouiali F, Tse SM, Litonjua AA, Hussain SN, Baglole CJ, et al. Genetic and histologic evidence for autophagy in asthma pathogenesis. J Allergy Clin Immunol. 2012;129:569–571. doi: 10.1016/j.jaci.2011.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin LJ, Gupta J, Jyothula SS, Butsch Kovacic M, Biagini Myers JM, Patterson TL, et al. Functional variant in the autophagy-related 5 gene promotor is associated with childhood asthma. PLoS One. 2012;7:e33454. doi: 10.1371/journal.pone.0033454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poon A, Eidelman D, Laprise C, Hamid Q. ATG5, autophagy and lung function in asthma. Autophagy. 2012;8:694–695. doi: 10.4161/auto.19315. [DOI] [PubMed] [Google Scholar]
- 19.Poon AH, Choy DF, Chouiali F, Ramakrishnan RK, Mahboub B, Audusseau S, et al. Increased autophagy-related 5 gene expression is associated with collagen expression in the airways of refractory asthmatics. Front Immunol. 2017;8:355. doi: 10.3389/fimmu.2017.00355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neill T, Schaefer L, Iozzo RV. Instructive roles of extracellular matrix on autophagy. Am J Pathol. 2014;184:2146–2153. doi: 10.1016/j.ajpath.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eapen MS, McAlinden K, Tan D, Weston S, Ward C, Muller HK, et al. Profiling cellular and inflammatory changes in the airway wall of mild to moderate COPD. Respirology. 2017;22:1125–1132. doi: 10.1111/resp.13021. [DOI] [PubMed] [Google Scholar]
- 22.Eapen MS, Hansbro PM, McAlinden K, Kim RY, Ward C, Hackett TL, et al. Abnormal M1/M2 macrophage phenotype profiles in the small airway wall and lumen in smokers and chronic obstructive pulmonary disease (COPD) Sci Rep. 2017;7:13392. doi: 10.1038/s41598-017-13888-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sohal SS, Reid D, Soltani A, Ward C, Weston S, Muller HK, et al. Reticular basement membrane fragmentation and potential epithelial mesenchymal transition is exaggerated in the airways of smokers with chronic obstructive pulmonary disease. Respirology. 2010;15:930–938. doi: 10.1111/j.1440-1843.2010.01808.x. [DOI] [PubMed] [Google Scholar]
- 24.Sharma P, Jha A, Stelmack GL, Detillieux K, Basu S, Klonisch T, et al. Characterization of the dystrophin-glycoprotein complex in airway smooth muscle: role of δ-sarcoglycan in airway responsiveness. Can J Physiol Pharmacol. 2015;93:195–202. doi: 10.1139/cjpp-2014-0389. [DOI] [PubMed] [Google Scholar]
- 25.Sharma P, Ryu MH, Basu S, Maltby SA, Yeganeh B, Mutawe MM, et al. Epithelium-dependent modulation of responsiveness of airways from caveolin-1 knockout mice is mediated through cyclooxygenase-2 and 5-lipoxygenase. Br J Pharmacol. 2012;167:548–560. doi: 10.1111/j.1476-5381.2012.02014.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sharma P, Ghavami S, Stelmack GL, McNeill KD, Mutawe MM, Klonisch T, et al. β-Dystroglycan binds caveolin-1 in smooth muscle: a functional role in caveolae distribution and Ca2+ release J Cell Sci 20101233061–3070. [DOI] [PubMed] [Google Scholar]
- 27.Sharma P, Basu S, Mitchell RW, Stelmack GL, Anderson JE, Halayko AJ. Role of dystrophin in airway smooth muscle phenotype, contraction and lung function. PLoS One. 2014;9:e102737. doi: 10.1371/journal.pone.0102737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deshpande DA, Wang WC, McIlmoyle EL, Robinett KS, Schillinger RM, An SS, et al. Bitter taste receptors on airway smooth muscle bronchodilate by localized calcium signaling and reverse obstruction. Nat Med. 2010;16:1299–1304. doi: 10.1038/nm.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schaafsma D, Dueck G, Ghavami S, Kroeker A, Mutawe MM, Hauff K, et al. The mevalonate cascade as a target to suppress extracellular matrix synthesis by human airway smooth muscle. Am J Respir Cell Mol Biol. 2011;44:394–403. doi: 10.1165/rcmb.2010-0052OC. [DOI] [PubMed] [Google Scholar]
- 30.Hernández-Gea V, Ghiassi-Nejad Z, Rozenfeld R, Gordon R, Fiel MI, Yue Z, et al. Autophagy releases lipid that promotes fibrogenesis by activated hepatic stellate cells in mice and in human tissues. Gastroenterology. 2012;142:938–946. doi: 10.1053/j.gastro.2011.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Busse W, Elias J, Sheppard D, Banks-Schlegel S. Airway remodeling and repair. Am J Respir Crit Care Med. 1999;160:1035–1042. doi: 10.1164/ajrccm.160.3.9902064. [DOI] [PubMed] [Google Scholar]
- 32.Aránguiz-Urroz P, Canales J, Copaja M, Troncoso R, Vicencio JM, Carrillo C, et al. β2-adrenergic receptor regulates cardiac fibroblast autophagy and collagen degradation Biochim Biophys Acta 2011181223–31. [DOI] [PubMed] [Google Scholar]
- 33.Koesters R, Kaissling B, Lehir M, Picard N, Theilig F, Gebhardt R, et al. Tubular overexpression of transforming growth factor-β1 induces autophagy and fibrosis but not mesenchymal transition of renal epithelial cells. Am J Pathol. 2010;177:632–643. doi: 10.2353/ajpath.2010.091012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alizadeh J, Glogowska A, Thliveris J, Kalantari F, Shojaei S, Hombach-Klonisch S, et al. Autophagy modulates transforming growth factor β1 induced epithelial to mesenchymal transition in non-small cell lung cancer cells. Biochim Biophys Acta Mol Cell Res. 2018;1865:749–768. doi: 10.1016/j.bbamcr.2018.02.007. [DOI] [PubMed] [Google Scholar]
- 35.Alizadeh J, Shojaei S, Sepanjnia A, Hashemi M, Eftekharpour E, Ghavami S. Simultaneous detection of autophagy and epithelial to mesenchymal transition in the non-small cell lung cancer cells. Methods Mol Biol. 2019;1854:87–103. doi: 10.1007/7651_2017_84. [DOI] [PubMed] [Google Scholar]
- 36.Payne DN, Rogers AV, Adelroth E, Bandi V, Guntupalli KK, Bush A, et al. Early thickening of the reticular basement membrane in children with difficult asthma. Am J Respir Crit Care Med. 2003;167:78–82. doi: 10.1164/rccm.200205-414OC. [DOI] [PubMed] [Google Scholar]
- 37.Cloonan SM, Lam HC, Ryter SW, Choi AM. “Ciliophagy”: the consumption of cilia components by autophagy. Autophagy. 2014;10:532–534. doi: 10.4161/auto.27641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Charous BL. Open study of hydroxychloroquine in the treatment of severe symptomatic or corticosteroid-dependent asthma. Ann Allergy. 1990;65:53–58. [PubMed] [Google Scholar]
- 39.Halwani R, Al-Muhsen S, Al-Jahdali H, Hamid Q. Role of transforming growth factor-β in airway remodeling in asthma. Am J Respir Cell Mol Biol. 2011;44:127–133. doi: 10.1165/rcmb.2010-0027TR. [DOI] [PubMed] [Google Scholar]
- 40.Lock R, Debnath J. Extracellular matrix regulation of autophagy. Curr Opin Cell Biol. 2008;20:583–588. doi: 10.1016/j.ceb.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suzuki T, Nakagawa M, Yoshikawa A, Sasagawa N, Yoshimori T, Ohsumi Y, et al. The first molecular evidence that autophagy relates rimmed vacuole formation in chloroquine myopathy. J Biochem. 2002;131:647–651. doi: 10.1093/oxfordjournals.jbchem.a003147. [DOI] [PubMed] [Google Scholar]
- 42.Homewood CA, Warhurst DC, Peters W, Baggaley VC. Lysosomes, pH and the anti-malarial action of chloroquine. Nature. 1972;235:50–52. doi: 10.1038/235050a0. [DOI] [PubMed] [Google Scholar]
- 43.Marmolejo-Murillo LG, Aréchiga-Figueroa IA, Moreno-Galindo EG, Navarro-Polanco RA, Rodríguez-Menchaca AA, Cui M, et al. Chloroquine blocks the Kir4.1 channels by an open-pore blocking mechanism. Eur J Pharmacol. 2017;800:40–47. doi: 10.1016/j.ejphar.2017.02.024. [DOI] [PubMed] [Google Scholar]
- 44.Bernstein HN, Ginsberg J. The pathology of chloroquine retinopathy. Arch Ophthalmol. 1964;71:238–245. doi: 10.1001/archopht.1964.00970010254019. [DOI] [PubMed] [Google Scholar]
- 45.Werner G, Hagenmaier H, Drautz H, Baumgartner A, Zähner H. Metabolic products of microorganisms. 224. Bafilomycins, a new group of macrolide antibiotics. Production, isolation, chemical structure and biological activity. J Antibiot (Tokyo) 1984;37:110–117. doi: 10.7164/antibiotics.37.110. [DOI] [PubMed] [Google Scholar]
- 46.Seglen PO, Gordon PB. 3-Methyladenine: specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proc Natl Acad Sci USA. 1982;79:1889–1892. doi: 10.1073/pnas.79.6.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu YT, Tan HL, Shui G, Bauvy C, Huang Q, Wenk MR, et al. Dual role of 3-methyladenine in modulation of autophagy via different temporal patterns of inhibition on class I and III phosphoinositide 3-kinase. J Biol Chem. 2010;285:10850–10861. doi: 10.1074/jbc.M109.080796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vlahos CJ, Matter WF, Hui KY, Brown RF. A specific inhibitor of phosphatidylinositol 3-kinase, 2-(4-morpholinyl)-8-phenyl-4H-1-benzopyran-4-one (LY294002) J Biol Chem. 1994;269:5241–5248. [PubMed] [Google Scholar]
- 49.Blommaart EF, Krause U, Schellens JP, Vreeling-Sindelárová H, Meijer AJ. The phosphatidylinositol 3-kinase inhibitors wortmannin and LY294002 inhibit autophagy in isolated rat hepatocytes. Eur J Biochem. 1997;243:240–246. doi: 10.1111/j.1432-1033.1997.0240a.x. [DOI] [PubMed] [Google Scholar]
- 50.Liu T, Liu Y, Miller M, Cao L, Zhao J, Wu J, et al. Autophagy plays a role in FSTL1-induced epithelial mesenchymal transition and airway remodeling in asthma. Am J Physiol Lung Cell Mol Physiol. 2017;313:L27–L40. doi: 10.1152/ajplung.00510.2016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.