Abstract
Background
Noninvasive genotyping using plasma cell-free DNA (cfDNA) has the potential to obviate the need for some invasive biopsies in cancer patients while also elucidating disease heterogeneity. We sought to develop an ultra-deep plasma next-generation sequencing (NGS) assay for patients with non-small-cell lung cancers (NSCLC) that could detect targetable oncogenic drivers and resistance mutations in patients where tissue biopsy failed to identify an actionable alteration.
Patients and methods
Plasma was prospectively collected from patients with advanced, progressive NSCLC. We carried out ultra-deep NGS using cfDNA extracted from plasma and matched white blood cells using a hybrid capture panel covering 37 lung cancer-related genes sequenced to 50 000× raw target coverage filtering somatic mutations attributable to clonal hematopoiesis. Clinical sensitivity and specificity for plasma detection of known oncogenic drivers were calculated and compared with tissue genotyping results. Orthogonal ddPCR validation was carried out in a subset of cases.
Results
In 127 assessable patients, plasma NGS detected driver mutations with variant allele fractions ranging from 0.14% to 52%. Plasma ddPCR for EGFR or KRAS mutations revealed findings nearly identical to those of plasma NGS in 21 of 22 patients, with high concordance of variant allele fraction (r = 0.98). Blinded to tissue genotype, plasma NGS sensitivity for de novo plasma detection of known oncogenic drivers was 75% (68/91). Specificity of plasma NGS in those who were driver-negative by tissue NGS was 100% (19/19). In 17 patients with tumor tissue deemed insufficient for genotyping, plasma NGS identified four KRAS mutations. In 23 EGFR mutant cases with acquired resistance to targeted therapy, plasma NGS detected potential resistance mechanisms, including EGFR T790M and C797S mutations and ERBB2 amplification.
Conclusions
Ultra-deep plasma NGS with clonal hematopoiesis filtering resulted in de novo detection of targetable oncogenic drivers and resistance mechanisms in patients with NSCLC, including when tissue biopsy was inadequate for genotyping.
Keywords: plasma cell-free DNA, next-generation sequencing, lung cancer, oncogenic drivers
Key Message
Ultra-deep plasma NGS can play a complementary role to tissue biopsy in detecting a wide range of oncogenic drivers and drug resistance mechanisms in NSCLC, including in settings where tissue biopsy is inadequate for genomic analysis. These findings support the potential clinical use of plasma NGS and the development of cfDNA-guided intervention studies in lung cancers.
Introduction
Therapeutic advances in genome-driven precision oncology rely upon the prospective molecular identification of oncogenic alterations and resistance mechanisms to guide precise treatments [1]. Recent technological advances in genetic sequencing of plasma cell-free DNA (cfDNA) have enabled ‘liquid biopsies’ that have the potential to identify oncogenic drivers in tumor derived DNA present in blood and capture intra-tumoral heterogeneity not addressed by biopsy of a single site, potentially obviating the need for invasive tissue biopsies in some circumstances [2]. As failure rates from tissue biopsy-based next-generation sequencing (NGS) are approximately 14% at large academic cancer centers, cfDNA analysis may be useful in guiding treatment selection in patients for whom tissue-based NGS is not an option [3].
Previous studies of cfDNA genotyping in lung cancers have largely focused on assay validation and concordance between plasma and tissue genotyping of specific mutant alleles, typically using digital PCR for detecting EGFR mutations [4–6]. More recent studies employing plasma cfDNA NGS have shown promise in detecting a broader variety of genetic alterations with similar sensitivity to that of digital PCR, with potential to change clinical practice [7–10]. A recent joint review by the American Society of Clinical Oncology and College of American Pathologists, however, concluded that there is currently insufficient evidence to recommend routine clinical use of cfDNA analysis in most clinical settings and called for more clinical validity and utility studies [11]. One goal of the Actionable Genome Consortium, formed in 2014 by academic cancer centers in collaboration with Illumina, Inc., was to develop an ultra-deep cfDNA NGS assay that could, with high sensitivity and specificity, detect key driver oncogenes and resistance mechanisms in patients with advanced non-small-cell lung cancers (NSCLCs).
To demonstrate the clinical utility of cfDNA in guiding the care of NSCLC patients, we systematically assessed a novel cfDNA assay in three groups of patients: those whose cancers were known to be oncogenic driver-positive (i.e. harboring a known driver genetic alteration), driver-negative (i.e. lacking a genetic alteration affecting pre-specified driver oncogenes after tissue-based NGS), and driver-unknown (patients in whom tissue biopsy was unavailable or insufficient to detect an oncogenic driver). We also assessed the sensitivity of detecting somatic genetic alterations in cfDNA de novo (i.e. without a priori knowledge of tissue genotyping results) to compare performance in an unbiased manner and to extrapolate the utility of this approach for patients in whom adequate tissue was unavailable for tumor genomic profiling.
Methods
Patients and study design
Patients were accrued across three academic cancer centers as part of the Illumina-sponsored Actionable Genome Consortium: Memorial Sloan Kettering Cancer Center, MD Anderson Cancer Center, and Dana-Farber Cancer Center. Patients with newly diagnosed, progressing stage IV, or recurrent metastatic NSCLC consented to collection of blood and clinical data under plasma collection protocols approved by each local institutional review board. The study collected blood prospectively from three pre-defined groups of patients. Group 1 included patients with known lung cancer oncogenic drivers based on tissue genotyping, defined by current National Comprehensive Cancer Network (NCCN) Guidelines, consisting of known activating alterations in the EGFR, ALK, ROS1, BRAF, KRAS, MET, RET, and HER2 oncogenes, identified by either tissue NGS or other methods [12]. Group 2 included patients who tested driver-negative via tissue NGS. Group 3 included patients who were driver-unknown due to insufficient or inadequate tissue for NGS, or in whom biopsy was not deemed feasible.
The primary objective was to evaluate the clinical sensitivity and specificity of de novo oncogenic driver detection in cfDNA in relation to tissue genotyping; this allowed performance estimation of de novo driver detection in patients unable to undergo tissue NGS. Secondary objectives included detection of drug resistance mechanisms; calculation of concordance between tissue and plasma NGS; orthogonal validation of plasma NGS with another plasma assay, and evaluation of factors associated with driver detection in plasma, including total cfDNA input, time lapse between tissue biopsy and plasma collection, and treatment status.
Tumor genotyping
Tumor genotyping was carried out per the standard-of-care at each participating cancer center (supplementary Table S1, available at Annals of Oncology online). Most patients (90 of 127) underwent NGS-based tumor genotyping. NGS methods included MSK-IMPACTTM (Memorial Sloan Kettering Cancer Center, New York, NY), FoundationOne CDxTM (Foundation Medicine, Cambridge, MA), OncoMap (Sequenom, San Diego, CA), and Ion Torrent OncomineTM (Thermo Fisher Scientific, Waltham, MA), all of which were designed to detect oncogenic alterations in the lung cancer driver oncogenes listed above. All tumor genomic testing was conducted in CLIA laboratories. The heterogeneity in approaches to tumor genomic profiling reflects tumor diagnosis and genotyping differences in real-world clinical practices.
Sample collection, DNA isolation, sequencing, and analysis
Whole-blood sample collection, plasma cfDNA, white blood cell (WBC) genomic DNA (gDNA) isolation, and library construction are described in the supplementary Materials, available at Annals of Oncology online. Plasma cfDNA and genomic DNA from WBCs were subjected to targeted NGS (Figure 1A). Sequencing data were analyzed using a variant-calling pipeline designed to detect single nucleotide variants and small insertions/deletions (indels) at allele frequencies (AFs) below typical sequencing error rates that was developed and characterized using a separate cohort of cancer and non-cancer samples (Jung, Aravanis et al. unpublished data). This variant calling and error correction methodology consisted of stitching of read pairs, read-collapsing for error correction, and de novo assembly of localized fragments. A novel machine learning-based cfDNA somatic mutation detection approach was employed. Candidate variants were scored against a hierarchical Bayesian noise model to assign calibrated quality scores. Only cfDNA variants with an error probability of <1 in 100 000 and a variant allele fraction (VAF) of ≥3x that of the matching WBC gDNA VAF were called in order to filter likely clonal hematopoiesis somatic variants from the cfDNA signal. Detection of fusions utilized the Manta structural variant caller whereas detection of somatic copy number alterations (CNAs) was based on normalized coverage in target genes.
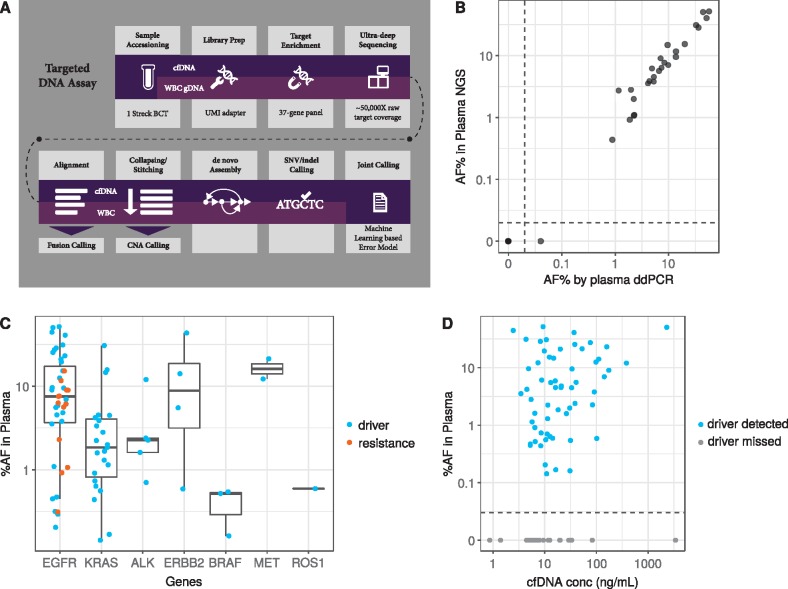
Figure 1.
Plasma next-generation sequencing (NGS) assay workflow, comparison of variant allelic fraction with orthogonal plasma ddPCR, and variant detection in cfDNA. (A) Workflow describing the targeted DNA plasma cell-free DNA (cfDNA) assay. Additional details are provided in the ‘Patients and Methods’ section. (B) Allele frequency (AF) in plasma as determined by NGS were compared those calculated by ddPCR in a subset of group 1 and 3 samples with NGS-identified EGFR or KRAS mutations. Plasma AF as measured by NGS (y-axis, log scale) was plotted against plasma AF as measured by ddPCR (x-axis, log scale). Correlation was calculated using Pearson’s correlation coefficient (0.98, P < 0.001). Dotted line indicates a break in the axis to allow representation of variants which were not detected by either assay. (C) Box plot of plasma AF (y-axis) by gene (x-axis) according to mutation type (driver, blue; resistance, orange) depicting median as well as first and third quartiles. Data falling outside the Q1–Q3 range were plotted as outliers (outside of the vertical lines). (D) Correlation of plasma variant AF (y-axis, log scale) and total cfDNA concentration (x-axis, log scale) (Spearman’s correlation = 0.3 P-value = 0.006) for samples in which the driver was detected (blue dots) or missed (gray dots). Dotted line indicates a break in the vertical axis to allow representation of samples in which the driver identified by tissue-based testing was not detected by cfDNA analysis.
Concordance analysis of cfDNA variants with tissue variants focused on known driver, resistance, and actionable mutations in NSCLC. Orthogonal validation for the presence of EGFR or KRAS mutations was conducted using paired plasma ddPCR in a subset of group 1 (n = 18) and group 3 (n = 4) patients [6].
Statistical considerations
Clinical sensitivity for plasma driver detection was defined as the proportion of patients with known oncogenic drivers (group 1) detected in tissue (considered ‘gold-standard’) who were found to harbor the identical oncogenic driver on plasma NGS. Clinical specificity for plasma driver detection was defined as the proportion of patients known to be oncogenic driver negative based on tissue NGS analysis (group 2) who were found to be driver negative on plasma NGS. The unweighted Cohen’s kappa coefficient was calculated for concordance agreement between tissue and plasma NGS results. Spearman’s correlation coefficient was calculated for the relationship between plasma variant AF and total cfDNA concentration. Descriptive statistics were used for correlating detection with the time between tissue and plasma.
Results
A total of 127 of 136 enrolled patients were included in the primary analysis (all enrolled from January to December 2015); two patients were excluded for sample contamination and seven were excluded for lack of matching WBC samples. In the remaining 127 patients (supplementary Table S2, available at Annals of Oncology online), plasma NGS using consensus-based error correction enabled the confident detection of driver mutations with VAFs ranging from 0.14% to 52%. Orthogonal validation with plasma ddPCR for EGFR or KRAS mutations showed concordant findings to plasma NGS in 21 of 22 patients, and the ddPCR-derived VAFs correlated well with those obtained with NGS (Pearson’s correlation = 0.98, P < 0.001; Figure 1B).
Overall, 91 patients had driver mutations identified by tissue genotyping (group 1), including 64 by NGS and 27 by other methods (supplementary Table S1, available at Annals of Oncology online). Of those, 68 had identical driver variants detectable in cfDNA, resulting in a clinical sensitivity of 75% [95% confidence interval (CI): 65% to 83%]. Mutations, fusions, and amplifications across eight driver oncogenes were identified in plasma, as depicted in Tables 1 and supplementary Table S3, available at Annals of Oncology online. EGFR and KRAS variants were the most common in tumor and cfDNA. There was no obvious bias towards detection of gene-specific mutations in plasma versus tissue.
Table 1.
Number of patients with driver or resistance mutations detected in tumor and plasma, as well as sensitivity and specificity. Sensitivity estimates are based on Group 1, in which tumor variants were detected in the plasma. Specificity estimates are based on Group 2, in which tumor samples tested negative for driver mutations by NGS
| Group 1 (n = 91) Driver positive on tissue genotyping |
Group 2 (n = 19) Driver negative on tissue NGS | Group 3 (n=17) Driver unknown insufficient tissue | |||
|---|---|---|---|---|---|
| Detected in tumor | Tumor variant detected in plasma, N (sensitivity) | Detected in plasma, N (specificity) | Detected in plasma | ||
| Driver | Total Subjects | 91 | 68 (75%) | 0 (100%) | 4 |
| EGFR | 37 | 29 (78%) | 0 | 0 | |
| KRAS | 29 | 23 (79%) | 0 | 4 | |
| ALK | 8 | 5 (62%) | 0 | 0 | |
| MET | 6 | 3 (50%) | 0 | 0 | |
| ERBB2 | 4 | 4 (100%) | 0 | 0 | |
| BRAF | 3 | 3 (100%) | 0 | 0 | |
| ROS1 | 3 | 1 (33%) | 0 | 0 | |
| RET | 1 | 0 (0%) | 0 | 0 | |
| Resistance | EGFR | 13 | 11 (85%) | NA | NA |
| MET | 2 | 0 (0%) | NA | NA | |
NGS, next-generation sequencing.
Group 2 was used to evaluate clinical specificity, as this group tested negative for driver mutations by tissue NGS. Overall, specificity was 100% (19/19, 95% CI: 82% to 100%); there were no driver mutations identified in cfDNA that were not identified in tissue. Given that the tissue NGS assays across the three academic centers differed in their breadth of coverage for non-driver alterations, we did not assess concordance of genetic alterations other than the pre-defined oncogenic drivers.
Concordance analyses, calculated for driver alterations detected at least once in tissue or plasma among 83 group 1 or group 2 patients who underwent tissue genotyping by NGS (supplementary Table S4, available at Annals of Oncology online), revealed kappa scores of 0.83 (95% CI: 0.69–0.97) for EGFR, 0.86 (95% CI: 0.75–0.98) for KRAS and 0.69 (95% CI: 0.52–0.87) for other drivers including ALK, ROS1, and RET fusions, and BRAF, ERBB2, and MET alterations.
Among the 17 group 3 patients for whom tumor tissue was insufficient for NGS at enrollment, KRAS G12 driver mutations were detected by cfDNA analysis in four patients. Of these four patients, one patient had a subsequent repeat biopsy at which time the KRAS G12 mutation detected in cfDNA was identified on tissue genomic analysis. The remaining three patients with KRAS G12-mutant cfDNA did not have subsequent biopsies. Of the remaining 13 patients without cfDNA-detected KRAS G12 mutations, two had a subsequent repeat biopsy; a KRAS G12 mutation was identified in both tissue samples. The remaining 11 patients did not have subsequent biopsy.
The relationship between detection of genomic aberrations in cfDNA and cfDNA concentration, a potential indicator of tumor shedding, was examined to identify effects on concordance (supplementary Figure S1A, available at Annals of Oncology online). Overall, the percent of subjects with oncogenic drivers detected in cfDNA increased with increasing cfDNA concentration (P = 0.012), likely due to higher tumor-derived cfDNA levels increasing the overall cfDNA concentration and/or increased probability of sampling the relevant mutant DNA sequence with increased input.
We next investigated associations between time between blood draw and tissue biopsy to assess for potential confounding effects of tumor evolution or intervening treatment (supplementary Figure S1B, available at Annals of Oncology online). Samples were grouped according to time between blood draw and biopsy. Overall, a trend of increased concordance with decreased time interval between biopsy and blood draw was observed but was not statistically significant (P = 0.1). While a numerically higher proportion of treatment-naive patients than patients on treatment had drivers detected in cfDNA (supplementary Table S2, available at Annals of Oncology online), this difference was also not statistically significant (85% versus 72%, P = 0.36), possibly due to small sample size.
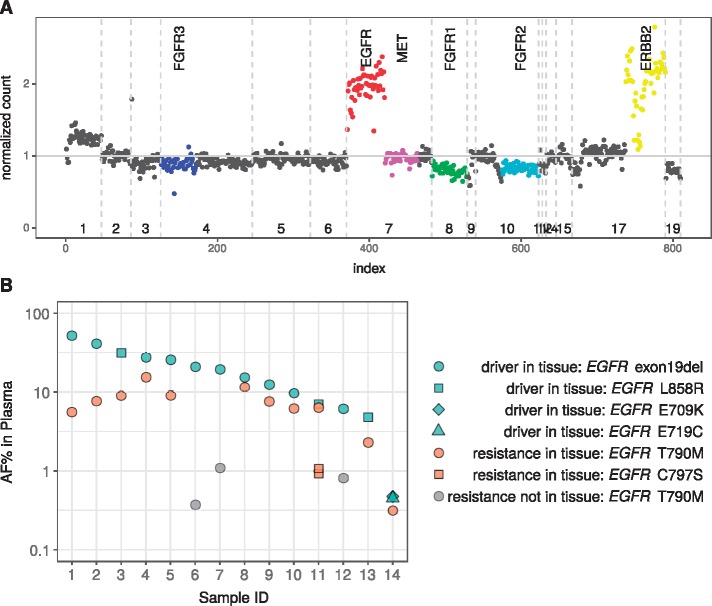
In 23 EGFR mutant cases with acquired resistance to targeted therapy, plasma genotyping detected potential resistance mechanisms, including 13 EGFR T790M and 1 EGFR C797S mutations, and 2 ERBB2 amplifications. Tissue biopsy and plasma detection of EGFR T790M were concordant for 19 of 23 cases (83% concordance; 95% CI: 61% to 95%). Of the remaining four patients, two had neither the primary driver mutation (EGFR exon 19 deletion) nor the EGFR T790M resistance mutation detected in cfDNA, potentially due to low tumor DNA shedding; for two patients, EGFR T790M was detected in plasma at low AF (0.4% and 0.8%) despite not being detected in tissue biopsy. Figure 2A provides an example of EGFR and ERBB2 amplification detected in cfDNA as resistance mechanisms to erlotinib.
Figure 2.
Capturing drug resistance mutations in plasma cfDNA. (A) Plot depicting one patient with EGFR and ERBB2 amplifications in plasma after treatment with erlotinib. Normalized sequencing read counts in comparison to healthy samples (y-axis) were plotted against chromosome number (x-axis); each dot represents an exon or intron target region covered by the 37-gene panel. Vertical dotted lines indicate chromosome boundaries. Specific genes are indicated by color: FGFR3, blue; EGFR, red; MET, pink; FGFR1, green; FGFR2, aqua; ERBB2, yellow. (B) Relationship between allelic fraction of driver and resistance mutations detected in plasma. The allele frequency (AF) (percent) of plasma cell-free DNA (cfDNA) variants detected in plasma (y-axis) was plotted for individual samples (x-axis). Driver and resistance mutations detected in tissue and plasma cfDNA are indicated by green and orange dots, respectively; resistance mutations identified in plasma cfDNA but not detected in tissue are indicated in gray. Shapes indicate specific mutations.
There was a significant but weak positive correlation between VAF (Figure 1C) and total cfDNA concentration (Spearman’s correlation = 0.3, P = 0.006; Figure 1D). Resistance mutations typically had lower VAF than driver mutations in the same patient, consistent with the notion that these resistance alterations were likely subclonal and arose later in tumor evolution (Figure 2B).
Discussion
In this multi-center study, we prospectively collected plasma for a blinded analysis of clinical sensitivity and specificity of ultra-deep NGS of plasma cfDNA for de novo detection of oncogenic drivers in lung cancers. Using a novel approach that incorporated WBC sequencing to address potential contamination of cfDNA results by somatic mutations attributable to clonal hematopoiesis, plasma NGS achieved overall high concordance with tissue testing across a variety of actionable oncogenic drivers specifically recognized by NCCN practice guidelines [12]. This study also demonstrated that targetable oncogenic drivers can be identified prospectively by plasma NGS in NSCLC patients when tissue biopsy failed or could not be carried out due to patient safety concerns. These results reveal the potential utility of NGS assays that use cfDNA as input for detecting actionable driver alterations and both de novo and emergent resistance mechanisms in the clinical setting [13]. Whilst clinical utility was not assessed here, the results illustrate that NGS-based cfDNA analysis has the potential to guide patient care and treatment selection.
The 75% sensitivity for detection of oncogenic drivers in lung cancers using the ultra-deep NGS assay described here compares favorably with non-ultra-deep NGS-based approaches [14, 15], as well as with ultra-deep ddPCR methods reported for the detection of lung cancer driver mutations [4, 6]. Additionally, orthogonal validation with a validated ddPCR assay for EGFR or KRAS mutations demonstrated similar performance; the only driver mutation not detected by the NGS assay had a VAF of 0.04% by ddPCR. This suggests that ultra-deep NGS assays can overcome several historical limitations of NGS-based profiling including shallow sequencing depth to achieve analytic sensitivity similar to that of ddPCR while allowing for the concurrent interrogation of many more driver alterations in cancer genes [16].
Of the 23 cases in which oncogenic drivers were detected in tumor tissue biopsy but not detected in plasma, the median time difference between tissue biopsy and plasma collection was 207 days (range: 0–1758 days). A trend toward decreasing plasma driver detection with increasing time interval and intervening treatment between tissue and plasma collection was also observed in a previous study suggesting that tumor shedding of cfDNA may vary over time [9]. Further, effective targeted therapies or immune response may allow clearance of cfDNA fragments from plasma, and tumors may evolve under selective therapeutic pressure against the driver as alterations and pathways other than those mediated by the original driver oncogene may emerge as dominant clones. These hypotheses, however, cannot be addressed in this study, which focused on a limited number of driver oncogenes, and will need to be explored using broader assays that are currently in development and designed to detect a wider spectrum of potential oncogenic alterations [17].
Several tissue genotyping assays were used as the reference standard in this study reflecting the variability of clinical practice across the participating institutions. As each of these tissue profiling assays have variable performance characteristics, we could not fully account for the possibility of false-positives on tissue genotyping, thus potentially underestimating the true sensitivity of plasma NGS. We also found that plasma VAF varied widely from patient to patient and may have been influenced by a variety of biologic and technical factors, including total cfDNA concentration which may not be tumor specific [18]. The 100% specificity for the plasma detection of drivers was consistent with its high concordance with ddPCR findings. As oncogenic drivers in lung cancers focused upon in this study are typically truncal mutations, high specificity is needed for the development of plasma NGS technology to guide treatment independent of tissue sampling [19]. This high specificity also validates our novel approach for the incorporation of WBC sequencing into cfDNA analysis which helped to filter potential contamination from somatic mutations attributable to clonal hematopoiesis [20].
The finding of resistance mechanisms in plasma, including EGFR T790M, EGFR C797S, and ERBB2 amplification, is of important clinical value, as they are now actionable based upon the development of novel targeted therapies and combination approaches (Figure 2). Serial tissue biopsies collected in patients receiving targeted therapies and after acquisition of drug resistance are invasive and not always feasible in clinical practice, highlighting the need for highly specific and sensitive non-invasive methods to identify potentially actionable resistance mechanisms in the clinical setting [21]. Early detection of subclonal resistance mechanisms may be useful to guide patient management and future drug development.
Finally, although our study only accrued a small number of group 3 patients, the detection of four drivers in 17 patients from plasma serves as proof-of-concept that plasma NGS may be clinically useful when tissue fails to yield interpretable results. These data thus support a complementary role for plasma cfDNA NGS in the diagnostic approach to lung cancer patients, particularly when tissue is not readily available for testing. Due to its high specificity, non-invasive nature, and fast turnaround time [14], we suggest that plasma NGS may be carried out at diagnosis or progression, even preceding tissue genotyping and be used to guide therapy. It should be noted that cfDNA analysis is not a replacement for histologic confirmation and immunohistochemistry from a diagnostic biopsy. Owing to its modest sensitivity, failure to detect a known oncogenic driver or resistance mechanism in plasma should then prompt tissue-based molecular profiling. Indeed, two recently published studies have separately demonstrated the potential clinical utility of plasma cfDNA guided treatments independent of tissue genotyping [14, 15].
This study had several limitations. The heterogeneous tissue genotyping assays used as a reference standard were without central confirmation. Although NGS is now recommended by NCCN guidelines for treatment of lung cancers [1, 12], this is not always possible in clinical practice, even at specialized cancer centers, as was shown in this study. Furthermore, clinical utility was not assessed in this study, as the plasma NGS assay studied was not carried out in real-time in a CLIA laboratory and the plasma results were thus not reported back to physicians or patients. Nevertheless, in this clinical validity study, the tissue NGS assays used incorporated all lung cancer drivers currently including in NCCN guidelines and were carried out in a CLIA environment, and plasma was prospectively collected in pre-defined patient subgroups for blinded analysis.
In conclusion, this prospective multi-center study demonstrated that ultra-deep NGS of plasma cfDNA with clonal hematopoiesis filtering accurately detected a wide variety of oncogenic drivers and resistance mechanisms in patients with advanced lung cancers. The sensitivity of detection by NGS was comparable to that of established ddPCR methods. Its high concordance with tissue genotyping and the detection of drivers in settings where tissue biopsy had failed or was not feasible lend credence to the potential clinical use of plasma cfDNA NGS and the development of cfDNA-guided intervention studies.
Supplementary Material
Acknowledgements
We thank Curtis Tom, Ruth Mauntz, Ting-Chun Liu, Youngsoon Park, Suchitra Ramani, Jessica Nguyen, KC Shashidhar, Eric Scott, and John Hyunsung Kim for technical assistance.
Funding
This work was supported by, and sample collection and sequencing were carried out at, Illumina, Inc. (San Francisco, CA). Authors from Memorial Sloan Kettering Cancer Center were supported by the Comprehensive Cancer Center Core Grant (P30 CA008748) from the National Institutes of Health, USA. Authors from the University of Texas MD Anderson Cancer Center were supported by the Comprehensive Cancer Center Core Grant (P30 CA016672) from the National Institutes of Health, USA.
Disclosure
BTL received consulting/advisory board fees from Genentech, Thermo-Fisher Scientific, and Guardant Health outside of this work. PR received consulting/advisory board fees from Novartis outside of this work. BJ, CH, NE, SG, RVS, HX, AJS, and AA are or were employees of Illumina, Inc. (San Francisco, CA) and GRAIL, Inc. (Menlo Park, CA) with stock or options to hold stock. AWB, NH, and MPH are or were employees of GRAIL, Inc. with stock or options to hold stock. AWB is an employee of Foresite Capital Management, which holds equity in GRAIL, Inc. RVS holds options to hold stock in Guardant Health, outside of this work. BTL, PR, JSR-F, RS, JMI, DB, ML, CMR, DBS, and GJR received institutional project funding from Illumina, Inc., and GRAIL, Inc. GRO received consulting/advisory board fees from AstraZeneca; DropWorks; GRAIL, Inc.; Inivata; and Sysmex. GBM received consulting/advisory board fees from AstraZeneca, Catena Pharmaceuticals, Critical Outcome Technologies, ImmunoMet, Ionic, Medimmune, Nuevolution, Pfizer, Precision Medicine, Signalchem Lifesciences, Symphogen, Takeda/Millenium Pharmaceuticals, and Tarveda; holds stock or options to hold stock in Catena Pharmaceuticals, ImmunoMet, SignalChem, Spindletop Ventures, and Tarveda; has licensed technology to Myriad Genetics; and has received research funding from Abbvie, Adelson Medical Research Foundation, AstraZeneca, the Breast Cancer Research Foundation, Critical Outcome Technologies, Horizon Diagnostics, Illumina, Immunomet, Ionis, Karus Therapeutics, the Komen Research Foundation, Nanostring, the Ovarian Cancer Research Foundation, Pfizer, the Prospect Creek Foundation, Takeda/Millennium Pharmaceuticals, and Tesaro; all were outside of this work. CP received consultant/advisory board fees from DropWorks, honoraria fees from Bio-Rad and AstraZeneca, and received individual research grants from the National Institutes of Health (NIH R41_CA228854-01) and Department of Defense (DOD W81XWH-17-1-0689); all were outside of this work. FJ received consultant/advisory board fees from Trovagene; GRAIL, Inc.; Illumina; and Guardant Health; and received an institutional research grant from Illumina and Trovagene; and has an ownership interest in Trovagene; FJ also received consultant/advisory board fees from Deciphera, IFM Therapeutics, Immunomet, Novartis, PureTech Health, Sotio, Synlogic, and Valeant; and received institutional research grants from Novartis, FujiFilm, Plexxikon, Deciphera, Symphogen, Biomevalley Discoveries, Asana, Genentech, Bristol-Myers Squibb, Agios, Upsher-Smith Laboratories, and Bayer, outside of this work. ML has received advisory board compensation from Boehringer Ingelheim, AstraZeneca, Bristol-Myers Squibb, Takeda, and Bayer, and research support from LOXO Oncology and Helsinn Healthcare; all were outside of this work. PAJ has received honoraria payments/fees from Biodesix, and consultant payments/fees as well as research grants from AstraZeneca, both were outside of this work. CMR has received consultant fees from Abbvie, AstraZeneca, Bristol-Myers Squibb, Celgene, Genentech/Roche, Harpoon, and Seattle Genetics, as well as a research grant from Daiichi Sankyo, all outside of this work. All remaining authors have declared no conflicts of interest.
References
- 1. Sholl LM, Aisner DL, Varella-Garcia M. et al. Multi-institutional oncogenic driver mutation analysis in lung adenocarcinoma: the Lung Cancer Mutation Consortium Experience. J Thorac Oncol 2015; 10(5): 768–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bettegowda C, Sausen M, Leary RJ. et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 2014; 6: 224ra224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zehir A, Benayed R, Shah RH. et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med 2017; 23(6): 703–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Oxnard GR, Paweletz CP, Kuang Y. et al. Noninvasive detection of response and resistance in EGFR-mutant lung cancer using quantitative next-generation genotyping of cell-free plasma DNA. Clin Cancer Res 2014; 20(6): 1698–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Oxnard GR, Thress KS, Alden RS. et al. Association between plasma genotyping and outcomes of treatment with osimertinib (AZD9291) in advanced non-small-cell lung cancer. J Clin Oncol 2016; 34(28): 3375–3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sacher AG, Paweletz C, Dahlberg SE. et al. Prospective validation of rapid plasma genotyping for the detection of EGFR and KRAS mutations in advanced lung cancer. JAMA Oncol 2016; 2(8): 1014–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Newman AM, Bratman SV, To J. et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med 2014; 20: 548–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Guibert N, Hu Y, Feeney N. et al. Amplicon-based next-generation sequencing of plasma cell-free DNA for detection of driver and resistance mutations in advanced non-small cell lung cancer. Ann Oncol 2018; 29(4): 1049–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Thompson JC, Yee SS, Troxel AB. et al. Detection of therapeutically targetable driver and resistance mutations in lung cancer patients by next-generation sequencing of cell-free circulating tumor DNA. Clin Cancer Res 2016; 22(23): 5772–5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Janku F, Zhang S, Waters J. et al. Development and validation of an ultradeep next-generation sequencing assay for testing of plasma cell-free DNA from patients with advanced cancer. Clin Cancer Res 2017; 23(18): 5648–5656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Merker JD, Oxnard GR, Compton C. et al. Circulating tumor DNA analysis in patients with cancer: American Society of Clinical Oncology and College of American Pathologists Joint Review. J Clin Oncol 2018; 36(16): 1631–1641. [DOI] [PubMed] [Google Scholar]
- 12. Ettinger DS, Wood DE, Aisner DL. et al. Non-small cell lung cancer, Version 5.2017, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw 2017; 15(4): 504–535. [DOI] [PubMed] [Google Scholar]
- 13. Teutsch SM, Bradley LA, Palomaki GE. et al. The Evaluation of Genomic Applications in Practice and Prevention (EGAPP) initiative: methods of the EGAPP Working Group. Genet Med 2009; 11(1): 3–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sabari JK, Offin M, Stephens D. et al. A prospective study of circulating tumor DNA to guide matched targeted therapy in lung cancers. J Natl Cancer Inst 2019; 111(6). doi: 10.1093/jnci/djy156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Aggarwal C, Thompson JC, Black TA. et al. Clinical implications of plasma-based genotyping with the delivery of personalized therapy in metastatic non-small cell lung cancer. JAMA Oncol 2019; 5(2): 173–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Thierry AR, Mouliere F, El Messaoudi S. et al. Clinical validation of the detection of KRAS and BRAF mutations from circulating tumor DNA. Nat Med 2014; 20(4): 430–435. [DOI] [PubMed] [Google Scholar]
- 17. Razavi P, Li BT, Abida W. et al. Performance of a high-intensity 508-gene circulating-tumor DNA (ctDNA) assay in patients with metastatic breast, lung, and prostate cancer. JCO 2017; 35(Suppl 15): LBA11516 [Google Scholar]
- 18. Li BT, Drilon A, Johnson ML. et al. A prospective study of total plasma cell-free DNA as a predictive biomarker for response to systemic therapy in patients with advanced non-small-cell lung cancers. Ann Oncol 2016; 27(1): 154–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang Z, Cheng Y, An T. et al. Detection of EGFR mutations in plasma circulating tumour DNA as a selection criterion for first-line gefitinib treatment in patients with advanced lung adenocarcinoma (BENEFIT): a phase 2, single-arm, multicentre clinical trial. Lancet Respir Med 2018; 6(9): 681–690. [DOI] [PubMed] [Google Scholar]
- 20. Hu Y, Ulrich B, Supplee J. et al. False positive plasma genotyping due to clonal hematopoiesis. Clin Cancer Res 2018; 24(18): 4437–4443. [DOI] [PubMed] [Google Scholar]
- 21. Adalsteinsson VA, Ha G, Freeman SS. et al. Scalable whole-exome sequencing of cell-free DNA reveals high concordance with metastatic tumors. Nat Commun 2017; 8(1): 1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.