Abstract
Long non-coding RNAs (lncRNAs) have been shown to play important roles in regulating host immune and inflammatory responses to bacterial infection. Infection with Clostridium perfringens (C. perfringens), a food-borne zoonotic pathogen, can lead to a series of inflammatory diseases in human and piglet, greatly challenging the healthy development of global pig industry. However, the roles of lncRNAs involved in piglet immune response against C. perfringens type C infection remain unknown. In this study, the regulatory functions of ileum lncRNAs and mRNAs were investigated in piglet immune response to C. perfringens type C infection among resistance (IR), susceptibility (IS) and sham-inoculation (control, IC) groups. A total of 480 lncRNAs and 3,669 mRNAs were significantly differentially expressed, the differentially expressed lncRNAs and mRNAs in the IR and IS groups were enriched in various pathways of ABC transporters, olfactory transduction, PPAR signaling pathway, chemokine signaling pathway and Toll-like receptor signaling pathway, involving in regulating piglet immune responses and resistance during infection. There were 212 lncRNAs and 505 target mRNAs found to have important association with C. perfringens infectious diseases, furthermore, 25 dysregulated lncRNAs corresponding to 13 immune-related target mRNAs were identified to play potential roles in defense against bacterial infection. In conclusion, the results improve our understanding on the characteristics of lncRNAs and mRNAs on regulating host immune response against C. perfringens type C infection, which will provide a reference for future research into exploring C. perfringens-related diseases in human.
Keywords: piglet, lncRNA, mRNA, Clostridium perfringens type C, ileum, immune response
Introduction
Clostridium perfringens (C. perfringens) is a Gram-positive anaerobic rod and ranks as the second most common bacteria that causes fulminant, fatal infectious and immune diseases (Scharff, 2012; Grass et al., 2013). These diseases are characterized by fever, pain, gas production, local edema, and severe tissue destruction, they usually further develop into systemic toxemia, necrotic enteritis, shock, sepsis, or even death in many animal species, including human and piglet (Songer et al., 2010), which are estimated one million illnesses reported every year (Low et al., 2018). Considering the four major lethal toxins produced by C. perfringens (alpha, beta, epsilon, and iota), it has been subdivided into five types (A to E) (Hassan et al., 2015). C. perfringens infection in pigs is mainly caused by the type C strain, the infection in younger piglets (<1 week of age) usually results in enteric inflammatory diseases, which have a high mortality rate; in older pigs, it results in asymptomatic intestinal inflammation with persistent shedding of the organisms in feces. The economic loss, in general, has severely crippled development of the global pig industries (Chan et al., 2012). Therefore, reducing the incidence and severity of C. perfringens infectious diseases (CPID) is an urgent problem that needs to be resolved.
Pigs are the important transmitter and reservoirs of C. perfringens type C, the colonization and shedding of this bacterium occurs within infected or asymptomatic pigs, posing a huge risk to herd and public health (Kich et al., 2014). Controlling the transmission of C. perfringens type C from pigs to humans, through the environment and pork products, and simultaneously decreasing the prevalence of C. perfringens type C in pig litters and herds may effectively reduce CPID in humans, especially in large pork-producing and consuming countries. According to the bacteria's pathogenicity, the host's immune activities and the cross-action between them, hosts generally manifest different severities of responses to bacterial infection (Zanella et al., 2011), which resulted in the dynamic profiles of host resistance/susceptibility to specific pathogen. Enhancing the disease resistance of host may serve as a useful alternative approach to control the spread of C. perfringens type C.
Long non-coding RNA (lncRNA) is a type of nucleotide transcripts, longer than 200 nucleotides in length, without coding potential. The lncRNAs can participate in regulation of mRNA expression, through their abilities to activate or restrain protein coding genes (Engreitz et al., 2016). Researches have clearly demonstrated that the various lncRNAs have participated in the development of immune system and in regulation of host inflammatory response against pathogenic bacteria (Ding et al., 2016). For example, Salmonella infection has been shown to cause changes in expression of certain sensitive lncRNAs in the early stage of infection in HeLa cells (Westermann et al., 2016). The long intergenic non-coding RNA (lincRNA)-cyclooxygenase 2 (Cox2) has been shown to regulate transcription of inflammation-related genes in the innate immune response through the NF-κB signaling pathway, and Listeria monocytogenes infection has been shown to cause the up-regulation of lincRNA-Cox2 in macrophages (Carpenter et al., 2013). The lncRNA p50-assocated Cox-2 extragenic RNA (known as PACER) has been shown to be up-regulated in LPS-infected human macrophages, subsequently regulating expression of prostaglandin endoperoxide synthase 2 (PTGS2) via the NF-κB signaling pathway (Krawczyk and Emerson, 2014). LncRNA HOX transcript antisense RNA (HOTAIR), a positive regulator of inflammation, has been shown to induce production of TNF-α by NF-κB activation, to regulate infection response in lipopolysaccharide (LPS)-induced sepsis (Wu et al., 2016). The up-regulation of lncRNA TCONS00183659 in resistant piglets has been shown to regulate the expressions of inflammatory factors MX1, MX2, and IFIT2, through combining with histone H4, to improve the resistance of weaning piglet defense against Escherichia coli (E. coli) F18 infection (Wu, 2018). In addition, in the E. coli F18-infected mice diarrhea model, the up-regulation of lncRNA ENSMUST00000122226 has been shown to improve the expression of adhesion molecule CD28 on T cell surface to facilitate the secretion of various inflammatory cytokines, including IFN and IL, finally leading intestinal inflammatory response and diarrhea disease (Zhao, 2016). While these collected studies have highlighted the array of dynamic changes and roles of lncRNAs in regulating host immune and inflammatory responses during bacterial and viral infections, the molecular interaction and regulatory roles of lncRNAs involved in host immune responses to C. perfringens type C infection have not yet been defined.
The intestine serves as the important physical barrier to prevent systemic invasion of pathogen. Invasion of pathogen in the intestine first triggers a series of antibacterial responses of immune cells residing in the tissue, the interactions between the two manifest the resistance capacities of each (the invading pathogen and the responding host). To evaluate functions of lncRNAs and mRNAs in regulating piglet immune responses to C. perfringens type C, we performed a comprehensive analysis of the expression profiles of lncRNAs and mRNAs in ileum of C. perfringens type C-infected piglets using RNA sequencing, the results could provide support for the merit of exploring molecular mechanisms of lncRNAs and mRNAs underlying a mammalian immune response to the C. perfringens infection.
Materials and Methods
Preparation of C. perfringens Type C Strain
The C. perfringens type C strain (CVCC 2032) was obtained from the China Veterinary Culture Collection Center (Beijing, China). The bacteria were cultured in bouillon culture-medium (HopeBio, Qingdao, China) under anaerobic conditions with 5% (v/v) H2: 5% (v/v) CO2 and 90% (v/v) N2 mixtures, at 37°C for 16 h with shaking before infection. The numbers of colony-forming units (CFUs) of C. perfringens type C were determined by plate colony counting method, and an expected concentration of 1 × 109 CFU/mL C. perfringens type C medium was used to inoculate piglets.
Sample Collection
Experimental piglets were from a C. perfringens seronegative Landrace × Yorkshire healthy nucleus herd in Dingxi, Gansu, China. Blood samples were collected from every piglet via anterior vena cava and into sterile coagulation-promoting tubes. Serum samples were obtained after centrifugation at 2,000 × g for 10 min at 4°C, and then confirmed antibody-negative for detection of porcine E. coli, Salmonella, and C. perfringens by the commercial enzyme-linked immunosorbent assay kits (Jiancheng Bioengineering Institute, Nanjing, China). Finally, thirty 7-day-old piglets (15 males and 15 females) were selected and randomly assigned into two groups (inoculated group: n = 25; control group (IC): n = 5). Piglets in the inoculated group were orally inoculated with 1 mL 1 × 109 CFU/mL C. perfringens type C medium for 5 consecutive days, while piglets in the IC group received orally sham-inoculation with sterile culture media. All piglets were separately housed and maintained in a climate-controlled and fully isolated environment, with water and diets provided ad libitum. After infection, piglets were monitored twice per day for general health, behavior, appetite, body condition, hair coat and dehydration. Fecal consistency was evaluated 4–5 times daily, and scored based on the visual symptom traits (Yang et al., 2013; Huang et al., 2016): 0 = normal, solid feces; 1 = slight diarrhea, soft and loose feces; 2 = moderate diarrhea, semi-liquid feces; 3 = severe diarrhea, watery feces.
Grouping were made as follows: firstly, recording fecal consistency of every defecation of each piglet, then calculating and ranking the total fecal scores of every piglet, and lastly, defining the top five piglets with highest and lowest fecal scores as the susceptibility (IS) and resistance (IR) groups, respectively. A total of 15 piglets from IR, IS and IC groups were humanely euthanized at 6 days post-challenge (dpc). Ileum tissues were collected and flushed clean with sterile PBS buffer (pH 7.4), and then quickly frozen in liquid nitrogen and stored at −80°C until RNA extraction.
Effect of C. perfringens Type C Infection on Number of Bacteria in Feces
The fecal samples were collected by sterile rectal swab every day and serially diluted in sterile PBS. From each dilution, 100 μL sample was plated in triplicate on 100 mL tryptose sulfite cycloserine (TSC) agar base supplemented with 10 mL D-cycloserine and 10 mL egg yolk emulsion (HopeBio, Qingdao, China). Plates were anaerobically incubated at 37°C for 24h. The fecal C. perfringens type C CFUs in feces of piglets from the IR, IS, and IC groups were compared using the Student's t-test in SPSS 22.0 software (IBM Corp., Armonk, NY, USA). The P < 0.05 was considered statistically significant.
Total RNA Isolation
Total RNA was isolated from each individual sample using Trizol™ reagent (Invitrogen, Carlsbad, CA, USA). RNA concentration was measured using Qubit® RNA Assay Kit in Qubit® 2.0 Flurometer (Life Technologies, Frederick, MD, USA). Purity and integrity of total RNA were assessed using the NanoPhotometer® spectrophotometer (IMPLEN, Westlake Village, CA, USA) and RNA Nano6000 Assay Kit of the Agilent Bionalyzer 2100 system (Agilent Technologies, Palo Alto, CA, USA). Sample with the RNA integrity number (RIN) > 7.0 were used for library preparation.
Library Preparation and RNA-Seq Data Acquisition
Approximately 3 μg total RNA per sample was used for preparing RNA sequencing libraries. Ribosomal RNA (rRNA) was removed from total RNA by epicenter Ribo-zero™ rRNA Removal Kit (epicenter, USA), and rRNA free residue was cleaned up by ethanol precipitation. Subsequently, rRNA-depleted RNA (Ribo-Zero RNA) was used to generate strand-specific RNA sequencing libraries by NEBNext® Ultra™ Directional RNA Library Prep Kit for Illumina® (NEB, Ipswich, MA, UK), which can capture all transcripts with and without poly A. To select cDNA fragments of preferentially 150–200 bp in length, the library fragments were purified with AMPure XP system (Beckman Coulter, Beverly, USA). Then 3 μL USER Enzyme (NEB, USA) was used with size-selected, adaptor-ligated cDNA at 37°C for 15 min followed by 5 min at 95°C before PCR, then PCR reaction was performed using Phusion High-Fidelity DNA polymerase, Universal PCR primers and Index (X) Primer. At last, products were purified (AMPure XP system) and library quality were assessed on the Agilent Bioanalyzer 2100 system.
The clustering of the index-coded samples was performed on a cBot Cluster Generation System using the TruSeq PE Cluster Kit v3-cBot-HS (Illumina®). After cluster generation, RNA libraries were sequenced on an Illumina Hiseq 4000 platform (Illumina, San Diego, CA, USA) to generate 150bp paired-end (PE150) reads at the Novogene Bioinformatics Institute (Beijing, China).
Transcriptome Assembly
Firstly, clean reads were obtained by discarding reads that contained adapter, ploy-N and low quality (>50% of bases with Phred scores <5) reads from raw data (fastq format) processed through in-house perl scripts. Simultaneously, the Phred score (Q20, Q30), and GC content of the clean data were calculated. Secondly, the paired-end clean reads were mapped to the pig reference genome sequence (Sus scrofa 10.2) by Tophat v2.0.9 (Kim et al., 2013). Lastly, the mapped reads of every sample were assembled by Scripture (Guttman et al., 2010) and Cufflinks (Trapnell et al., 2010) in a reference-based approach.
Analyses of Coding Potential and Conservation
We used four tools to distinguish mRNA from lncRNA, namely Coding-Non-Coding-Index (CNCI) (Sun et al., 2013), Coding Potential Calculator (CPC) (Kong et al., 2007), Pfam-scan v1.3 (E-value < 0.001) (Punta et al., 2012), and phylogenetic codon substitution frequency (phyloCSF) (Lin et al., 2011), respectively. Transcripts with coding potential predicted by any one of these four tools were filtered out, and those without coding potential were filtered as the candidate set of lncRNA.
The Phast software (version 1.3) was generally used for phylogenetic analysis (Siepel et al., 2005) and PhastCons is a conservation scoring and identifying program of conserving elements. We used phyloFit to compute phylogenetic models for conserved and non-conserved regions and then set the model and HMM transition parameters for phyloP to calculate the conservation scores of lncRNA and coding genes.
Prediction of lncRNA Target Genes
Cis and trans analyses were used to predict the target genes of differentially expressed lncRNAs. The cis role of lncRNAs indicated their actions on neighboring target genes, the coding genes close to 10 k upstream and downstream regions of lncRNA were considered as the cis role target genes (Guil and Esteller, 2012). The target genes of lncRNA in trans role were identified by expression levels, according to Pearson's correlation coefficient (|r| > 0.95).
Identification of Differentially Expressed lncRNA and mRNA
The FPKMs (fragments per kilo-base of exon per million fragments mapped) of lncRNA and mRNA were calculated by Cuffdiff software (Trapnell et al., 2010). The expression levels of gene were computed by summing the FPKMs of transcripts in each group. Differential expression levels were determined by Cuffdiff using a model based on the negative binomial distribution model. Transcripts with a corrected P-value < 0.05 were considered significantly differentially expressed.
GO and KEGG Enrichment Analyses
Analyses of Gene Ontology (GO) enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) signaling pathway (www.kegg.jp/kegg/kegg1.html) were performed to investigate the roles of differentially expressed lncRNA and mRNAs by the GOseq R package and KOBAS v2.0 software (Xie et al., 2011), respectively. Corrected P-value < 0.05 was considered significantly enriched.
Association Analysis Between lncRNA and CPID
To deepen our understanding of the relationship between lncRNAs, these target genes and CPID, three criteria were performed to screen lncRNA and the target mRNA pairs. Firstly, the predicted lncRNAs and their target genes were both significantly differentially expressed in IR vs. IC and/or IS vs. IC groups through cis and trans roles. Secondly, the CPID-related functional mRNA set was searched and downloaded from the GeneCard database: Genes Associated with Diseases (http://www.genecards.org/cgi-bin/listdiseasecards.pl), the CPID-associated keywords were used to screen gene pairs, such as C. perfringens, diarrhea, inflammation, immune, infection, and inflammatory bowel disease (IBD). Lastly, compared with functional mRNA set, only the matched gene pairs were selected and considered as potential CPID-related gene pairs. The screening process was outlined in Figure S1. Function analyses were also performed to investigate the functions of these lncRNAs and target genes in regulating CPID. Furthermore, these gene pairs were processed to predict the potential regulatory relationship between lncRNAs and immune-related genes, which were utilized to further explore the immune function of lncRNAs in regulating CPID.
Quantitative PCR Validation
To validate RNA-seq data, the expression levels of 12 genes from multiple groups were quantified by quantitative PCR (qPCR) using 2−ΔΔCt value methods. Ileum total RNA used for RNA-seq was performed to synthesize cDNA using reverse transcriptase Kit (TaKaRa, Dalian, China). Specific primers of these genes were designed using NCBI database, porcine GAPDH gene was designed as an endogenous control (Table S1). The qPCR reactions were performed in 20 μL system involved in 9.5 μL 2 × SYBR Green Realtime PCR Master Mix (TaKaRa, Dalian), 1 μL of forward and reverse primers, 1 μL cDNA and 7.5 μL RNase free ddH2O using LightCycler 480 II Real-Time PCR System. The cycling conditions included an initial denaturation (95°C for 3 min), followed by 30 cycles (95°C for 15 s; 60 ± 1°C for 15 s; 72°C for 20 s). Three independent biological replicates were performed in triplicate.
Results
Effect of C. perfringens Type C Infection on Fecal Bacterial Shedding
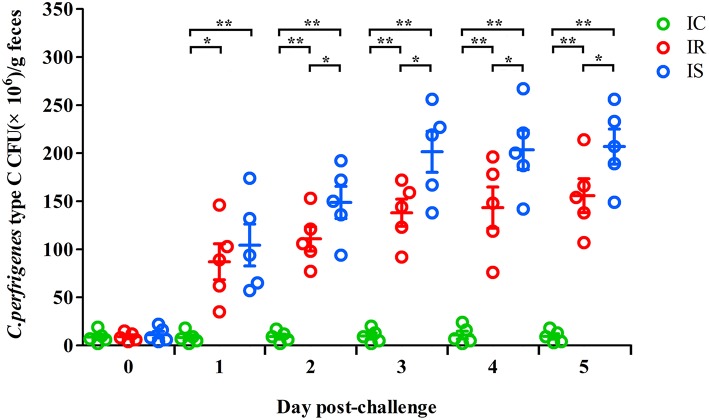
To explore the effect of C. perfringens type C infection on the fecal bacterial shedding, the numbers of fecal bacterial shedding of piglets from the IR, IS and IC groups were measured on 1–5dpc. The fecal C. perfringens type C shedding in the IR and IS groups were increasing over time. The mean values of C. perfringens type C CFUs in the IR group were significantly lower for the IR group than those in the IS group, which were both significantly higher than those in the IC group (P < 0.01) (Figure 1). The results suggested that C. perfringens type C infection could increase the numbers of fecal bacterial shedding, the hosts with more severe diarrhea shed more bacteria in their feces.
Figure 1.
The fecal shedding levels of piglets in IR, IS, and IC groups from 0 to 5 days post challenge (dpc). Fecal shedding CFUs were determined by plate count method. The horizontal line represents the mean. Green circle represents results of IC; Red circle represents results of IR; Blue circle represents results of IS. An asterisk denotes a significant difference (*P < 0.05; **P < 0.01).
Summarized RNA Sequencing Data
To study the expression profiles of lncRNA and mRNA of piglets, RNA sequencing was performed on ileum tissues of 15 piglets from the IR, IS, and IC groups. Through sequencing, each tissue sample generated a total of 90 to 130 million raw reads, after discarding reads with poly-N >10%, adapters, low quality reads, and/or any other contaminants, ~83–125 million clear reads were obtained. The percentage of clean reads in each library ranged from 88.95 to 96.36%, and an average of 92.97% clean reads passed initial quality thresholds. Upon aligning to the pig reference genome (Sus scrofa 10.2), ~59–89 million clean reads (average 65.89% clean reads) were mapped (for details of sequencing results see Table S2).
Identification of lncRNA and mRNA
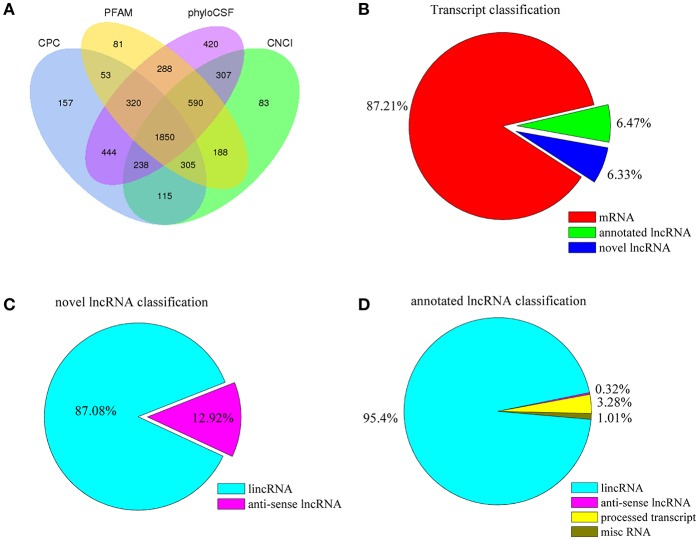
To identify the high-confidence lncRNAs, we first reconstructed and assembled 163,927 non-redundant transcripts by two assemblers: Cufflinks and Scripture. Then, a series of highly stringent bioinformatics filtering pipelines was applied to screen putative lncRNAs (Figure S2). Since transcripts involved immature mRNA fragments, four bioinformatic tools, namely CPC, PFAM, phyloCSF, and CNCI, were used to assess the coding potential of transcripts, and only transcripts that were simultaneously shared by none of four tools were designated as putative lncRNA. This strategy yielded a “high-confidence” set of 1,850 putative novel lncRNAs (Figure 2A, Figure S3), corresponding to 1,393 lncRNA genes and representing 1,611 lincRNAs (87.08%) and 239 anti-sense lncRNAs (12.92%) (Figure 2C). In addition, 1,890 annotated lncRNAs were identified, corresponding to 1,650 lncRNA genes and representing 1,803 lincRNAs (95.4%), 62 processed transcripts (3.28%), 19 miscRNAs (1%), and 6 anti-sense lncRNAs (0.32%) (Figure 2D and Table S3-1). In addition, a total of 25,491 mRNAs were identified (Figure 2B and Table S3-2). We henceforth referred these sets as the “pig intestinal transcriptome,” and all subsequent analyses were based on these transcripts.
Figure 2.
(A) Identification of 1,850 putative lncRNAs without protein-coding potential evaluated by CPC, PFAM, phyloCSF, and CNCI. (B) Classification of lncRNAs, (C) novel lncRNAs, (D) annotated lncRNAs, and mRNAs identified in piglet ileums by RNA-seq.
Genomic Feature Analyses of lncRNA and mRNA
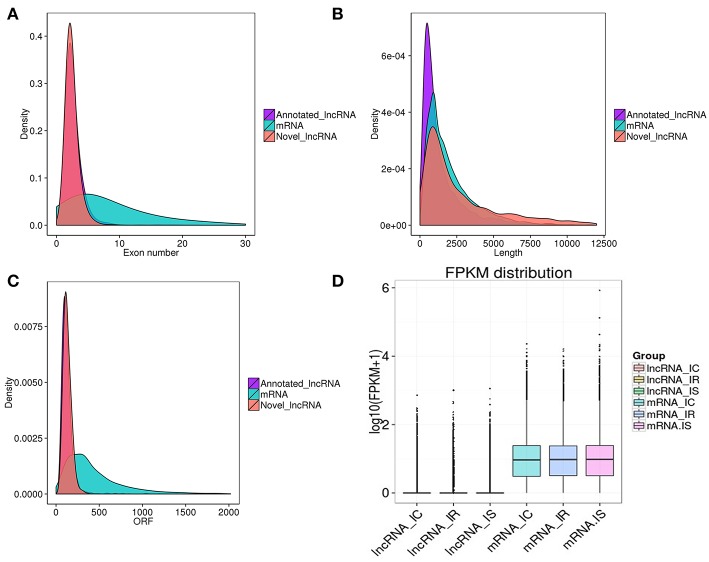
To investigate the genomic features of these predicted transcripts, we characterized the exon number, transcript length, open reading frame (ORF) length and sequence conservation of lncRNAs and mRNAs. In agreement with other lncRNAs, the predicted lncRNAs in this study were shorter in length and had less exons, ORF and lower conservation than mRNAs (Figures 3A–C). Specifically, the 1,890 annotated lncRNAs had an average of 1,546bp lengths, 2.77 exons and 129bp ORF, being shorter than the 1,850 novel lncRNAs, which had an average of 3,496bp lengths, 2.44 exons and 131bp ORF (Table S4). Interestingly, we also found that lncRNAs in piglet ileums were not only longer in length than those in pig endometrium (1,454bp on average) (Wang et al., 2016), skeletal muscle (1,043bp on average) (Zhao et al., 2015), testis (1,240bp on average) (Ran et al., 2016) and thyroid gland (2,337bp on average) (Shen et al., 2016), but also longer than those in human (1kb on average), mouse (550bp on average), and zebrafish (1,113bp on average). In addition, the predicted lncRNAs of pig contained the fewer numbers of exon than those of human (2.9 exon on average), mouse (3.7 exon on average) and zebrafish (2.8 exon on average) (Siepel et al., 2005; Cabili et al., 2011; Pauli et al., 2012).
Figure 3.
Genomic characteristic of lncRNAs and mRNAs in piglet ileums. (A) Exon number distribution of lncRNAs and mRNAs; (B) Length distribution of 1,850 annotated lncRNAs (purple), 25,491 mRNAs (green), and 1,890 putative novel lncRNAs (red); (C) ORF length distribution of lncRNAs and mRNAs; (D) Box plots showing the expression levels (log 10 FPKM) of lncRNAs and mRNAs in IR, IS, and IC groups.
Differential Expression Analyses of lncRNA and mRNA
To identify the expression levels of lncRNAs and mRNAs, Cuffdiff software was first applied to screen differentially expressed lncRNAs and mRNAs by FPKM. Consistent with other species, the expression levels of lncRNAs were much lower than those of mRNAs (Figure 3D). With the false discovery rate (FDR) set at 4% and corrected P-value of < 0.05, a total of 480 lncRNAs and 3,669 mRNAs were significantly differentially expressed among the pairwise comparisons (IR vs. IC, IS vs. IC, and IR vs. IS) (Table S5). The 480 dysregulated lncRNAs represented 237 novel lncRNAs and 243 annotated lncRNAs, corresponding to 478 lncRNA genes (Table S5-1), a total of 359, 16 and 419 lncRNAs were specifically expressed in IR vs. IC, IS vs. IC, and IR vs. IS groups, respectively (Table 1). Meanwhile, the 3,669 dysregulated mRNAs corresponded to 3,663 mRNA genes (Table S5-2), and 2,588, 126 and 3,283 mRNAs were found to be differentially expressed in IR vs. IC, IS vs. IC, and IR vs. IS groups (Table 1), respectively.
Table 1.
Number of differentially expressed lncRNAs and mRNAs in each comparison.
| Type | Groups | IR vs. IC | IR vs. IS | IS vs. IC | Total genes |
|---|---|---|---|---|---|
| lncRNA | Up-regulated | 84 | 6 | 108 | 480 |
| Down-regulated | 275 | 10 | 311 | ||
| Sum | 359 | 16 | 419 | ||
| mRNA | Up-regulated | 1,419 | 41 | 1,825 | 3,669 |
| Down-regulated | 1,169 | 85 | 1,458 | ||
| Sum | 2,588 | 126 | 3,283 |
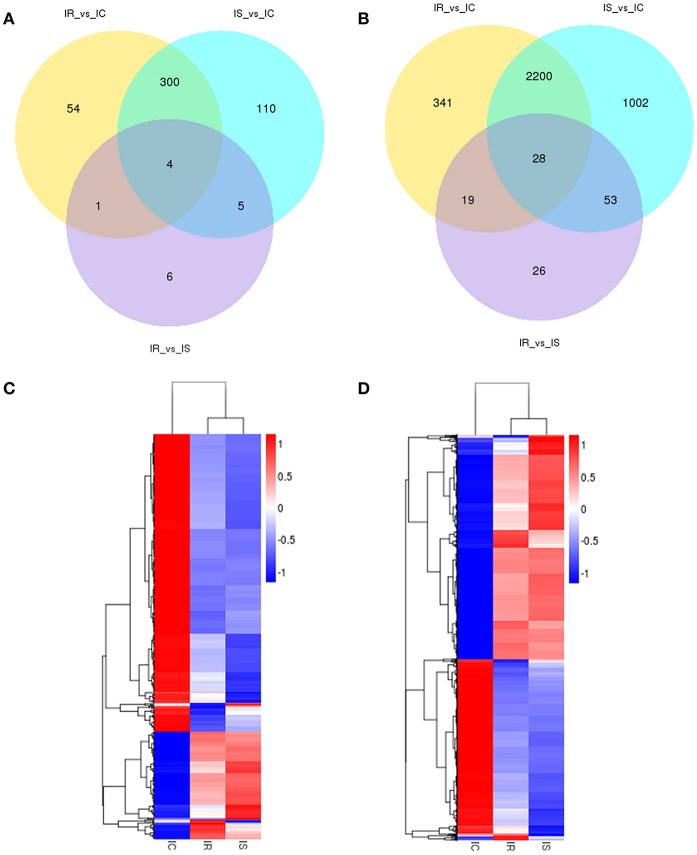
Venn diagraming was performed to describe overlaps among the differentially expressed lncRNAs and mRNAs from pairwise comparisons. In total, 54, 110 and 6 stage-specific lncRNAs were differentially expressed in IR vs. IC, IS vs. IC, and IR vs. IS groups, respectively. Four dysregulated lncRNAs (ENSSSCT00000032859, ENSSSCT00000018610, LNC_001066 and LNC_001186) were shared among the IR, IS, and IC groups (Figure 4A). Moreover, a set of 341, 1,002, and 26 dysregulated mRNAs were found to be specifically expressed in the IR vs. IC, IS vs. IC, and IR vs. IS groups, respectively, and 28 dysregulated mRNAs were identified as the transcripts shared among IR, IS, and IC groups (Figure 4B). Taken together, the set of lncRNAs and mRNAs suggested the existence of a global coordination for regulatory responses in piglet intestine immune response to C. perfringens type C infection. In addition, systematic cluster analyses of differentially expressed lncRNA and mRNAs among IR, IS, and IC groups were revealed by heat map. The lncRNAs (Figure 4C) and mRNAs (Figure 4D) of IR and IS showed similar expression patterns being clustered together.
Figure 4.
Gene expression number and profiling analyses of differentially expressed lncRNAs and mRNAs among IR, IS, and IC groups after C. perfringens type C infection. Venn diagram of differentially expressed lncRNAs (A) and mRNAs (B) in three comparison groups. Cluster analyses of differentially expressed lncRNAs (C) and mRNAs (D) of 15 piglets infected by C. perfringens type C by hierarchical heat map. Data are expressed as FPKM. Red, relatively higher expression level; Blue, relatively lower expression level.
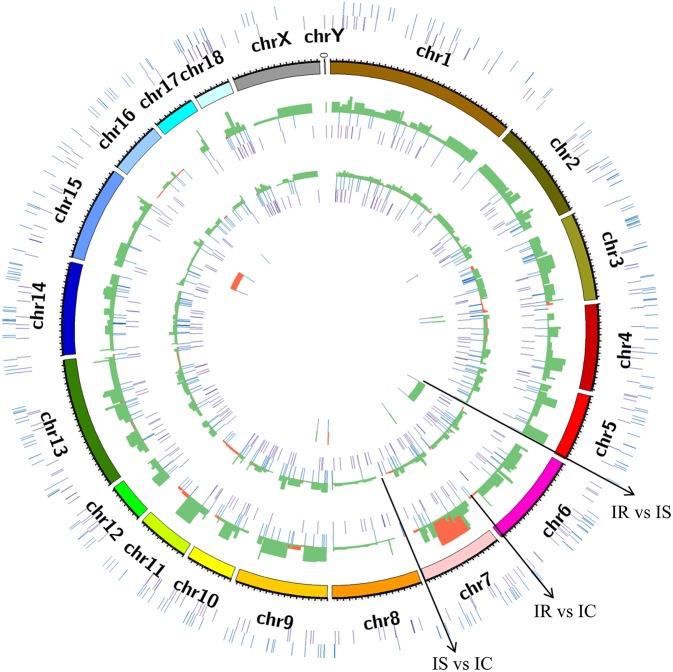
To explore the genomic distribution of differentially expressed lncRNAs, a circular chromosomal distribution was generated to display the differential expression patterns of novel and annotated lncRNAs in IR vs. IC, IS vs. IC, and IR vs. IS groups (Figure 5). The distribution and expression patterns of dysregulated lncRNAs were similarly between the IR vs. IC and IS vs. IC comparisons, and the dysregulated lncRNAs mainly represented the down-regulated transcripts, evenly distributed among nearly all chromosomes. In contrast, the majority of up-regulated lncRNAs were distributed among chromosomes 7, 9, and 11 for the IR vs. IC comparison, and chromosomes 3, 4, 7, and 11 for the IS vs. IC comparison. For the IR vs. IS comparative expressions, the up- and down-regulated lncRNAs were mainly distributed in chromosomes 15 and 6, respectively.
Figure 5.
Circular chromosome distribution of lncRNA identified in piglet ileums across the reference genome and different comparison groups. The outermost circle displays the sum of novel and annotated lncRNAs identified in three comparison groups. The second circle displays pig chromosomes scale. The remaining three circles display the novel and annotated lncRNAs in IR vs. IC, IS vs. IC, and IR vs. IS, respectively. Purple: novel lncRNA; Blue: annotated lncRNA; Red: the up-regulated lncRNA; Green: the down-regulated lncRNA, the red and green were plotted by the log2 (fold change) among IR vs. IC, IS vs. IC, and IR vs. IS groups.
Function Enrichment Analysis of Differentially Expressed lncRNAs
To gain insight into the potential function of the infection-related differentially expressed lncRNAs, GO and KEGG enrichment analyses of lncRNAs in cis and trans regulatory roles were performed. We first predicted the potential target genes located in the 10-kb (upstream and downstream) regions of lncRNAs. A total of 1,728 lncRNAs were found to transcribe close to 2,104 mRNAs, corresponding to 2,672 lncRNA: coding gene pairs (Table S6-1). Through no significantly enriched GO terms were identified in the IR vs. IC and IS vs. IC groups (corrected P-value > 0.05, Tables S7, S8), target genes of these lncRNAs were also found to enrich in some immune-related signaling pathways, such as the B cell receptor signaling pathway, chemokine signaling pathway, NF-κB signaling pathway, NOD-like receptor signaling pathway and T cell receptor signaling pathway. It was notably that up-regulated lncRNAs in the IS vs. IC group were significantly enriched in the ABC transporters pathway (corrected P-value = 0.0178). GO survey indicated that the cis lncRNA target genes in 13 GO terms were significantly enriched in IR vs. IS group (corrected P-value < 0.05, Table S7-1), these enriched terms were mostly associated with the receptor activity and detection of stimulus, such as olfactory receptor activity, G protein-coupled receptor activity, transmembrane signaling receptor activity, detection of chemical stimulus involved in sensory perception and smell. Notably, in IR vs. IS group, the target genes of up-regulated lncRNAs were mainly enriched in two pathways, olfactory transduction (corrected P-value = 0.0071) and nitrogen metabolism (corrected P-value = 0.0362), while the target genes of down-regulated lncRNAs were enriched in one pathway (protein processing in endoplasmic reticulum; corrected P-value = 0.0071), these enriched GO classes and KEGG pathways mainly revealed two genes, CPS1 and OR6A2, hinting the key roles of these two genes. Importantly, these pathways enriched in the IR vs. IS group might be significantly associated with the resistance and susceptibility of host response to C. perfringens infection (Table S8-1).
We further predicted potential target genes of lncRNAs in trans regulation based on Pearson's correlation coefficients (|r| > 0.95). A total of 17,895 interaction relationships (17,654 positive and 241 negative correlations) were identified, representing 781 dysregulated lncRNAs and 2,949 target genes (Table S6-2). Furthermore, the trans target genes of lncRNAs were significantly enriched in 198, 3 and 187 GO terms for IR vs. IC, IR vs. IS, and IS vs. IC comparisons, respectively (corrected P-value < 0.05, Table S7-2). Remarkably, the 3 enriched GO terms in the IR vs. IS group included cytokine production, regulation of immune response and positive regulation of immune system process, and four immune-related target genes, IRAK3, LCP2, TLR8, and CD84, were annotated in them (corrected P-value < 0.05, Table S7-2). Similar to lncRNAs in cis role, the major categories of target genes of lncRNAs in trans roles from IR vs. IC, IS vs. IC, and IR vs. IS groups mainly included olfactory transduction, transcriptional misregulation in cancer (IL-8, CD86, and MMP-3), osteoclast differentiation (TGFBR1, IL-1A, and TNFRSF11A), immune- and inflammation-related pathways (inflammation signaling-JUK and NLRP3; cytokine-CCL5 and IL-1; immune receptor- NOD1, TLR3, and IRAK3) (Table S8-2).
Function Enrichment Analyses of Differentially Expressed mRNAs
Gene set enrichment analyses of differentially expressed mRNAs of piglets revealed that a total of 145, 23, and 257 highly enriched GO terms were derived from IR vs. IC, IR vs. IS, and IS vs. IC groups, respectively (corrected P-value < 0.05, Table S7-3). The most enriched GO terms in three comparison groups included the broad functional clustering, especially various inflammatory-related functions (cytokine, leukocyte activation, chemotaxis, and lymphocyte differentiation) and cell metabolic process (catalytic activity, hydrolase, hemopoiesis). In the KEGG analysis, the most significant pathways of mRNAs included cGMP-PKG signaling pathway (INOS), ABC transporters (ABCB1) and PPAR signaling pathway (PPAR-α). It's worth noting that the dysregulated mRNAs in the IR vs. IS group significantly activated some immune-related GO functions, these terms included mucosal immune response, defense response, antioxidant activity, sulfiredoxin activity (corrected P-value < 0.05, Table S7-3). Furthermore, the enrichment pathways of up-regulated mRNAs in the IR vs. IS group were significantly enriched in only two immune-related pathways: chemokine signaling pathway (corrected P-value = 0.0161) and Toll-like receptor signaling pathway (corrected P-value = 0.0218), in which, 6 immune-related mRNAs (CXCL9, CXCL10, CCL17, CCR5, and TLR8) were found to annotate in these pathways. Other pathways of dysregulated mRNAs included glycosphingolipid biosynthesis and protein processing in endoplasmic reticulum (corrected P-value = 0.0357) (Table S8-3). In general, the significant functions of mRNAs that changed during infection might be linked to the induction of this gene in the regulation process of host immune response to C. perfringens type C infection.
Association Analysis Between lncRNA, Target Genes and CPID
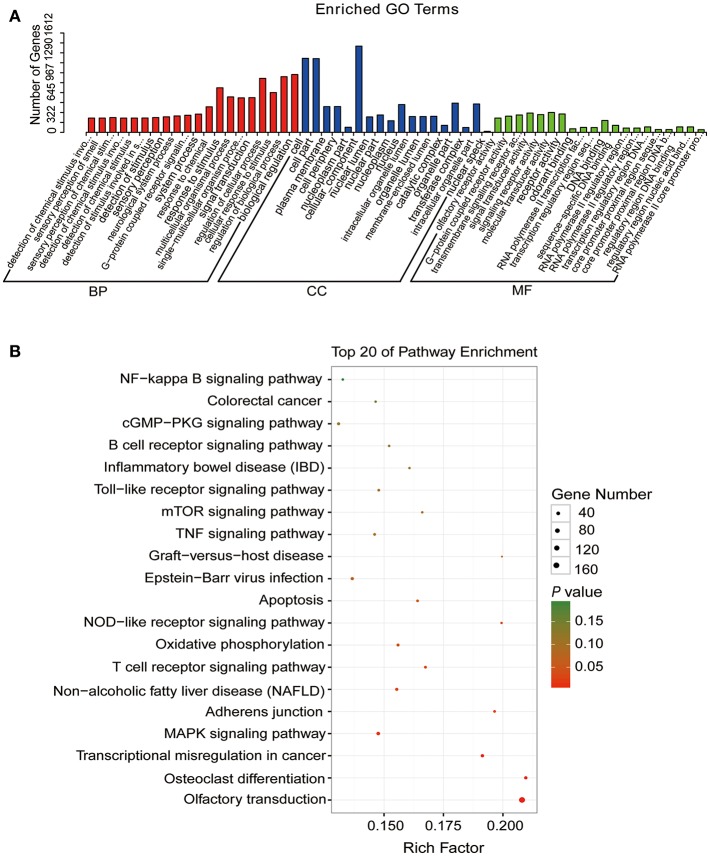
To study the association between lncRNAs and CPID, we had implemented three criteria to screen for the most likely lncRNAs involved in the regulation of CPID. A total of 212 lncRNAs, corresponding to 505 target genes were filtered (Table S9). GO analysis of these candidate gene pairs showed that these lncRNA target genes were mainly involved in cellular response to stimulus, regulation of cellular and biological processes, signal transduction and activity, cellular component, and G protein-coupled receptor activity and so on (Figure 6A). The significantly enriched KEGG pathways involved MAPK signaling pathway, T cell receptor signaling pathway, NOD-like receptor signaling pathway, apoptosis, adhesion junction, Toll-like receptor signaling pathway and NF-κB signaling pathway (Figure 6B). The potential candidate gene pairs could regulate the process of C. perfringens type C infection through these functions and pathways.
Figure 6.
The functional enrichment analyses of target genes of the disease-related lncRNAs. (A) Gene ontology (GO) function annotation of target genes of the disease-related lncRNAs. The X-axis indicates the gene numbers and the Y-axis indicates the detailed terms. (B) Kyoto encyclopedia of genes and genomes (KEGG) pathways of target genes of the disease-related lncRNAs. The X-axis indicates the gene ratio and the Y-axis indicates the name of KEGG pathway. The size of dots indicates the numbers of target genes, and the color of dots indicates P-value (Fisher's Exact Test).
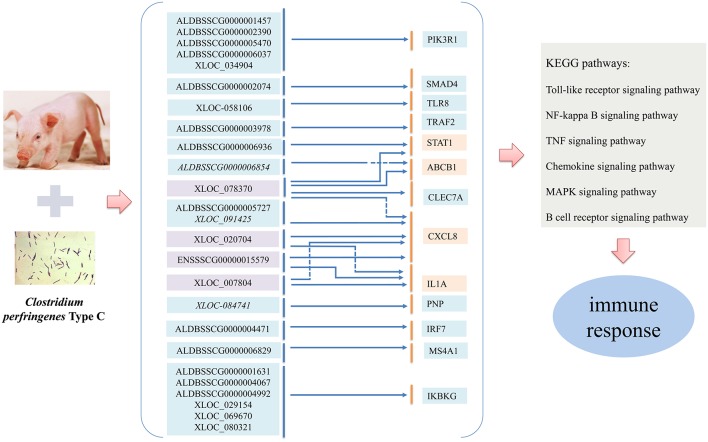
To study the immune regulatory functions of lncRNAs, the potential relationships of differentially expressed lncRNAs and immune-related genes were predicted. Finally, a total of 25 dysregulated lncRNAs targeting 13 immune-related genes passed the filter (Figure 7) and were used to perform the enrichment analyses. Several of the lncRNAs had multiple immune-related target genes and a given immune target gene could be regulated by several lncRNAs. In particular, ABCB1, a significantly dysregulated immune-related gene, was found to be regulated by XLOC_078370 in cis and by ALDBSSCG0000006854 in trans, respectively. Simultaneously, XLOC_078370, together with ALDBSSCG0000005727, ENSSSCG00000015579, XLOC_007804, and XLOC_020704, could co-regulate target gene CXCL8 in trans; CXCL8 was also regulated by XLOC_091425 in cis. In addition, XLOC_078370 could also target STAT1 and CLEC7A genes, both of which were significantly differentially expressed in IR, IS and IC groups.
Figure 7.
Overview of the regulatory relationship between the differentially expressed lncRNAs and their immune-related target genes involved in CPID. Italics and non-italics fonts of lncRNA indicate the cis and trans regulatory functions of lncRNAs, respectively.
Validation of Expression Levels of Genes Detected in RNA-Seq
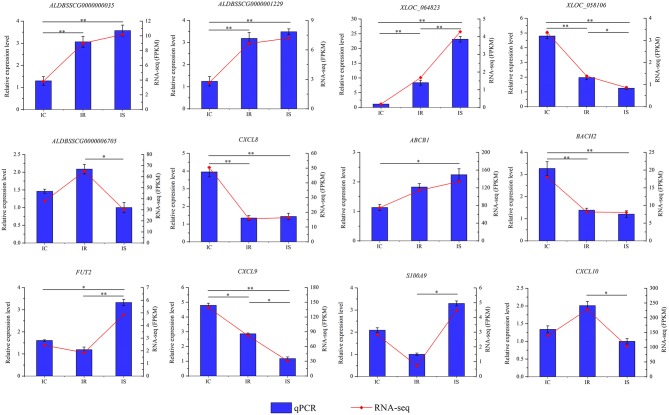
To validate the RNA-seq result and detect expression levels of differentially expressed genes in three comparison groups, a total of 12 dysregulated genes were subjected to qPCR detection, including 5 lncRNAs (XLOC_064823, XLOC_058106, ALDBSSCG0000000035, ALDBSSCG0000001229, and ALDBSSCG0000006705) and 7 mRNAs (CXCL8, CXCL9, ABCB1, BACH2, FUT2, CXCL10, and S100A9). The qPCR results of all 12 genes were perfectly consistent with the RNA-seq data (Figure 8), suggesting the high reliability and accuracy of RNA-seq.
Figure 8.
Validation of RNA-seq results by quantitative reverse transcription polymerase chain reaction (qPCR). Porcine GAPDH gene is used as the endogenous control. Expression was quantified using the comparative cycle threshold (2−ΔΔCt) value method. The data were expressed as the mean ± SEM). An asterisk denotes a significant difference (*P < 0.05; **P < 0.01).
Discussion
C. perfringens has been described for decades years, it is generally recognized as one of the most widespread potential bacterial pathogens in nature as well as in the gastrointestinal tracts of most animal species. However, due to a paucity of studies on mechanisms of host response to C. perfringens type C infection, no adequate treatment or cure is yet available for the manifested diseases. In general, the complexity of mechanisms underlying human and animal infections by pathogenic bacteria is appreciated, and the related research has begun to focus on the coordinated posttranscriptional regulatory functions of host genes, such as microRNAs seem to have a relationship with chicken necrotic enteritis disease caused by C. perfringens infection (Hong et al., 2014), lncRNAs have been found to be implicated in regulating the bacterial infection diseases in piglets and mice (Zhao, 2016; Wu, 2018). However, no studies to date had yet performed a comprehensive assessment of lncRNA in regulation of piglet defense response against C. perfringens type C infection, therefore, in our study, the primary goal was to identify C. perfringens type C infection-related lncRNAs and mRNAs as well as the potential functions in the 7-day-old piglets from IR, IS and IC groups. According to the results, these lncRNAs and mRNAs may constitute potential candidates for future application of preventing and treating CPID.
The degree of bacterial shedding in feces is an important parameter that affects herd disease spread, as well as a measure of host's immunologic control of bacteria replication. In this study, the numbers of fecal bacterial shedding of piglets were significantly higher in IS than in IR and IC, and that in IR was significantly higher than that in IC. These findings were in accordance with the feces' consistency of three groups, suggesting that C. perfringens infection could induce different bacterial shedding levels of piglets that might be associated with the different immune abilities of piglets. Therefore, screening pigs with reduced C. perfringens numbers, may decrease environmental contamination and lower pathogen transmission to other animals and humans. Certainly, the piglets with low shedding level of feces need to be further investigated.
Recently, although a total of 12,107 pig lncRNAs have been submitted to the animal lncRNA database (ALDB Release v1.0), it is still far below the total listing of 15,778 human lncRNAs (version 27) in the GENCODE database. In the present study, the identified 3,740 lncRNAs (1,850 novel lncRNAs and 1,890 annotated lncRNAs) and 25,491 mRNAs will greatly enrich the pig transcript genomic database. Non-coding and protein-coding genes are distinguished by their coding potential. In line with other organism studies (Jia et al., 2010; Esteve-Codina et al., 2011; Ni et al., 2011; Ran et al., 2016), the putative lncRNAs of our study had the fewer exons, shorter lengths, lower expression levels and less conservation than mRNAs (Figure 3), these common characteristics of lncRNAs might be associated with the pivotal functions of regulation, control and guidance. Herein, the 480 differentially expressed lncRNAs and 3,669 differentially expressed mRNAs were used to subsequently investigated into potential regulatory functions of the host in response to C. perfringens type C infection, though these genes still needing to be validated by experiments.
A dominant feature of differentially expressed lncRNAs and mRNAs activated in our work was that they were primarily associated with olfactory transduction and inflammasome activation, including chemokine signaling pathway and NF-κB signaling pathway. Indeed, in our study, C. perfringens type C infection leaded to the significantly dysregulated expressions of proinflammatory factors (TNFAIP8, IL-1A, IL-1B, IL-4, IL-6, and IL-7), interferon (IFNE and IFNG) and toll-like receptor (TLR6, TLR8, TLR9, and TLR10) of infected piglets, suggesting inflammatory response had occurred in the injury intestine portions during infection. Inflammatory response stimulated by microbial agents or necrotic cells could activate NF-κB signaling, which was required for C. perfringens induced reactive oxygen species (ROS) production and cytotoxicity (Li et al., 2001), thus NF-κB-regulated proinflammatory response, TLRs and interferon (IFN) regulatory responses were activated in the process of piglet immune defense response to C. perfringens type C infection (Kamada et al., 2013). In addition, we also found olfactory receptor family 51 subfamily E member 1 (OR51E1) gene was significantly decreased in the IR and IS groups, the decreased OR51E1 were found to suppress the transcription of intestinal epithelial cell surface receptors. Similar to other study, Chen had reported the complex relationship between transduction of olfactory signaling and inflammatory response, and thought that inflammatory cytokines could contribute to olfactory neural regeneration in immune process that fighting infection or injury through the NF-κB/JNK pathway (Chen et al., 2017). Therefore, we hypothesized olfactory transduction and receptor activity were triggered by inflammatory responses and involved in the response process of piglet immune response to C. perfringens type C infection, which may contribute to the survive and regeneration of injured tissue.
Remarkably, the activation process of differentially expressed mRNAs mainly corresponded to several functions of antioxidant activity and sulfiredoxin activity, and pathways of the cGMP-PKG signaling pathway (NOS2), ABC transporters (ABCB1), PPAR signaling pathway (PPARA, ANGPTL4), and amino acid biosynthesis metabolism. The activated PPAR signaling pathway is found to limit oxygen availability by driving the energy metabolism of epithelial cells and prevent the expansion of potentially pathogenic bacteria, while it leads to an increase oxygen concentration in injured intestines during function dysfunction of host (Laura et al., 2014; Mariana et al., 2017). NOS2, the gene encoding inducible nitric oxide synthase (iNOS), is elevated in the absence of PPAR signaling pathway, and then catalyze the synthesis of NO in tissues or cells stimulated by cytokines or microbes through cGMP-PKG, PPAR, and NF-κB signaling pathways in the process of regulating cell proliferation, apoptosis and angiogenesis, while excess NO will lead to cytotoxicity, tissue damage and necrosis, and further promote the occurrence and development of inflammatory diseases (Kim et al., 2006; Vacca, 2017). In this study, the PPARA gene was up-expressed both in the IR and IS groups after infection, the increased expression of PPARA might contribute to provide a microaerophilic and anaerobic condition that benefited to growth of anaerobic pathogen C. perfringens type C in intestine tissues, aggravating the inflammatory and diarrhea disease of piglets. Furthermore, affected by up-expression of PPARA, the expressions of NOS2 were down-regulated in the IR and IS groups, indicating that C. perfringens type C infection had stimulated and induced the activations of PPAR and cGMP-PKG signaling pathways, which further suppressed NOS2 expression in infected piglets, the reduced synthesis of NO might decrease the secretion of inflammatory factors and prevent the deterioration of inflammatory disease. These results suggested that piglets might regulate the expressions of PPARA and NOS2 genes to mediate immune defense responses against C. perfringens type C infection by cGMP-PKG and PPAR signaling pathways.
The total 212 significantly dysregulated lncRNAs corresponded to 505 unique target genes, suggested the potential relationship between the dysregulated lncRNAs, target genes and CPID. Function enrichment analyses showed that these lncRNAs and target genes primarily corresponded to several key immune-related pathways participated in regulating piglet immune responses to C. perfringens infection, such as MAPK signaling pathway, T cell receptor signaling pathway, NOD-like receptor signaling pathway, apoptosis, Toll-like receptor signaling pathway and NF-κB signaling pathway (Lu et al., 2009; Nagahama et al., 2013; Low et al., 2018). Specially, the 25 lncRNAs responded to 13 immune-related target genes were found to be highly associated with CPID, further exploring the regulatory relationships of them can improve the understanding of characteristics of lncRNAs and mRNAs in regulating host immunomodulation responses to C. perfringens infection. Due to the highly complex and diverse roles of lncRNAs and the incomplete functional prediction method, the regulatory roles of lncRNAs in host immune response to infection may be explored through mediating expressions and functions of mRNAs.
Remarkably, among the 25 lncRNAs and 13 target genes, the significantly dysregulated immune-related mRNAs (ABCB1, STAT1, CXCL8, and IL1A) are considered as the highly immune-active genes. The ATP-binding cassette subfamily B member 1 (ABCB1), known as multidrug resistance 1 (MDR1 gene), was found to be targeted by ALDBSSCG0000006854 and XLOC_078370. ABCB1 encodes the P-glycoprotein (P-gp) to protect cells from xenobiotics damage by affecting the combination between them (Dudarewicz et al., 2013). ABCB1 is recognized as a potential target gene of high risk of IBD, colorectal cancer (CRC), Crohn's disease and ulcerative colitis (Onnie et al., 2010; Senhaji et al., 2015), ABCB1 may regulate functions of transporter activity, ATPase activity and transmembrane movement of substances in the immune response through ABC transporters signaling pathway during infection. Thus, the overexpression of ABCB1 gene in the IR and IS groups may be the danger signal for inflammatory intestinal disease caused by C. perfringens type C infection.
The signal transducer and activator of transcription factors STAT1 was regulated by lncRNAs ALDBSSCG0000006936 and XLOC_078370. It has also been reported that activated STAT1 mediates the expressions of proinflammatory cytokines (CXCL10) and interleukin production (IFN-1, IL-8, IL-10) in human IBD (Tao et al., 2013; Giles et al., 2016), down-regulated STAT1 has been proved to reduce tumorigenesis, inflammation and gastrointestinal diseases (Ernst et al., 2008; Lamarthée et al., 2015). Therefore, low expression of STAT1 in the IR and IS groups might reduce the cellular damage and antibacterial state by regulating cellular response to interferon and cytokine through JAK-STAT signaling pathway, which was very important for cell viability in response to different stimuli and pathogen.
CXCL8 is an important inflammatory mediator regulated by NF-κB transcription factor and serves to modify and enhance human innate immune responses in defense against bacterial infections (Krupa et al., 2015). The expression of CXCL8 is significantly increased in lactobacilli-treated pig intestinal epithelial cells infected by viruses, indicating that up-regulated CXCL8 can increase protection against intestinal pathogen infections (Albarracin et al., 2017). CXCL8 has been considered to be associated with resistance of CRC (Xiao et al., 2015), with overexpression of CXCL8 can induce the proliferation and migration of intestinal epithelial cell (Shen et al., 2017). CXCL8 also serves to attract T cells, dampening tissue inflammation in response to bacterial infection, such as C. perfringens, through NF-κB signaling pathway, chemokine signaling pathway, NOD-like receptor signaling pathway and Toll-like receptor signaling pathway. The IL1A gene, encoding a member of the interleukin 1 cytokine family, is a pleiotropic cytokine involving in various virus infections, immune responses and inflammatory processes (Anders, 2016). Produced by activated macrophages, IL1A is processed by proteolytic enzymes and releases in response to cell injury, subsequently inducing apoptosis through MAPK signaling pathway. The up-regulation of IL1A gene has been suggested to play an important role in preventing colon tumor in patients with IBD (Yoshikawa et al., 2017). In our study, the expressions of CXCL8 and IL1A genes were significantly decreased both in the IR and IS groups, the low expressions might exert a negative impact on the piglet defense against C. perfringens type C infections. In general, these results strongly suggest a linkage between expressions of lncRNAs, immune-related genes and C. perfringens type C infection. Future studies should seek to detail mechanisms by which these lncRNAs function to regulate the target genes in pig and to confirm the findings in humans.
In summary, this study provides an overview of the expression patterns and enrichment functions of lncRNAs and mRNAs involved in the piglet immune response to C. perfringens type C infection. These insights into the characteristics of lncRNAs and mRNAs underlying host (piglet) immunomodulation responses to counteract the C. perfringens type C infection and related diseases, particularly of resistance, and provide a foundation for future studies of this pathogen infection in humans.
Ethics Statement
This study was carried out in accordance with the recommendations of Institutional Animal Care and Use Committee (IACUC) of Gansu Research Center for Swine Production Engineering and Technology. The protocol was approved by the College of Animal Science and Technology, Gansu Agricultural University.
Author Contributions
XH conceived and designed the study, analyzed the data, and wrote the manuscript. SG contributed to critical revising of the manuscript and final approval of the version to be published. HS and WS did the laboratory work in the expression and statistical analysis. QY, PW, and SL contributed to data analysis and interpretation. LL, SZ, and ZY participated in the analysis and interpretation of data. All the authors read and approved the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Glossary
Abbreviations
- CPID
C. perfringens infectious diseases.
Footnotes
Funding. This work was supported by Science and Technology Innovation Funds of Gansu Agricultural University, Scientific Research start-up Funds for openly-recruited Doctors (GAU-KYQD-2018-27) and the National Natural Science Foundation of China (31660646).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2019.00130/full#supplementary-material
Screening principles of association analysis between dysregulated lncRNAs and mRNAs.
Identification pipeline for lncRNAs. Each step is described in detail in the methods section.
One thousand eight hundred and fifty non-coding lncRNAs were selected by Cufflinks and Scripture.
Primers and location of lncRNAs and mRNAs used in qPCR validation.
Summary of RNA-seq data and reads mapped to the Sus scrofa reference genome.
Catalog of lncRNAs and mRNAs identified in the 15 cDNA libraries of piglets infected by C. perfringens type C.
Characteristics of lncRNAs in piglet ileums.
List of differentially expressed lncRNAs and mRNAs detected in piglet ileums using the RNA-seq.
Target genes prediction of significantly differentially expressed lncRNAs in cis and trans.
GO enrichment analysis of the significantly dysregulated lncRNAs target genes and mRNAs among IR, IS, and IC groups.
KEGG enrichment analysis of the significantly dysregulated lncRNAs target genes and mRNAs among IR, IS, and IC groups.
Relationship between dysregulated lncRNAs and mRNAs in cis and trans involved in CPID.
Refereces
- Albarracin L., Kobayashi H., Iida H., Sato N., Nochi T., Aso H., et al. (2017). Transcriptomic analysis of the innate antiviral immune response in porcine intestinal epithelial cells: influence of immunobiotic lactobacilli. Front. Immunol. 8:57. 10.3389/fimmu.2017.0005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders H. J. (2016). Of inflammasomes and alarmins: IL-1β and IL-1α in Kidney Disease. J. Am. Soc. Nephrol. 27, 2564–2575. 10.1681/ASN.2016020177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabili M. N., Trapnell C., Goff L., Koziol M., Tazon-Vega B., Regev A., et al. (2011). Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 25, 1915–1927. 10.1101/gad.17446611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter S., Atianand M., Aiello D., Ricci E., Gandhi P., Hall L. L., et al. (2013). A long noncoding RNA induced by TLRs mediates both activation and repression of immune response genes. Science 341, 789–792. 10.1126/science.1240925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan G., Farzan A., Soltes G., Nicholson V. M., Pei Y. L., Friendship R., et al. (2012). The epidemiology of Clostridium perfringens type A on Ontario swine farms, with special reference to cpb2-positive isolates. BMC Vet. Res. 8:156. 10.1186/1746-6148-8-156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Reed R. R., Lane A. P. (2017). Acute inflammation regulates neuroregeneration through the NF-κB pathway in olfactory epithelium. Proc. Natl. Acad. Sci. U.S.A. 114, 8089–8094. 10.1073/pnas.1620664114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y. Z., Zhang Z. W., Liu Y. L., Shi C. X., Zhang J., Zhang Y. G. (2016). Relationship of long noncoding RNA and viruses. Genomics 107, 150–154. 10.1016/j.ygeno.2016.01.007 [DOI] [PubMed] [Google Scholar]
- Dudarewicz M., Baranska M., Rychlik-Sych M., Trzcinski R., Dziki A., Skretkowicz J. (2013). The importance of C1236T polymorphism in the ABCB1/MDR1 gene in assessment of susceptibility to inflammatory bowel diseases in the Polish population. Prz. Gastroen. Terol. 8, 38–43. 10.5114/pg.2013.34181 [DOI] [Google Scholar]
- Engreitz J. M., Haines J. E., Perez E. M., Munson G., Chen J., Kane M., et al. (2016). Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature 539, 452–455. 10.1038/nature20149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst M., Najdovska M., Grail D., Lundgrenmay T., Buchert M., Tye H., et al. (2008). STAT3 and STAT1 mediate IL-11–dependent and inflammation-associated gastric tumorigenesis in gp130 receptor mutant mice. J. Clin. Invest. 118, 1727–1738. 10.1172/JCI34944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteve-Codina A., Kofler R., Palmieri N., Bussotti G., Notredame C., Pérez-Enciso M. (2011). Exploring the gonad transcriptome of two extreme male pigs with RNA-seq. BMC Genomics 12:552. 10.1186/1471-2164-12-552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giles E. M., Sanders T. J., Mccarthy N. E., Lung J., Pathak M., Macdonald T. T., et al. (2016). Regulation of human intestinal T-cell responses by type 1 interferon-STAT1 signaling is disrupted in inflammatory bowel disease. Mucosal Immunol. 10, 184–193. 10.1038/mi.2016.44 [DOI] [PubMed] [Google Scholar]
- Grass J. E., Gould L. H., Mahon B. E. (2013). Epidemiology of foodborne disease outbreaks caused by Clostridium perfringens, United States, 1998–2010. Foodborne Pathog. Dis. 10, 131–136. 10.1089/fpd.2012.1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guil S., Esteller M. (2012). Cis-acting noncoding RNAs: friends and foes. Nat. Struct. Mol. Biol. 19, 1068–1075. 10.1038/nsmb.2428 [DOI] [PubMed] [Google Scholar]
- Guttman M., Garber M., Levin J. Z., Donaghey J., Robinson J., Adiconis X., et al. (2010). Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat. Biotechnol. 28, 503–510. 10.1038/nbt.1633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan K. A., Elbourne L. D., Tetu S. G., Melville S. B., Rood J. I., Paulsen I. T. (2015). Genomic analyses of Clostridium perfringens isolates from five toxinotypes. Res Microbiol. 166, 255–263. 10.1016/j.resmic.2014.10.003 [DOI] [PubMed] [Google Scholar]
- Hong Y. H., Dinh H., Lillehoj H. S., Song K. D., Oh J. D. (2014). Differential regulation of microRNA transcriptome in chicken lines resistant and susceptible to necrotic enteritis disease. Poult. Sci. 93, 1383–1395. 10.3382/ps.2013-03666 [DOI] [PubMed] [Google Scholar]
- Huang X. Y., Yang Q. L., Yuan J. H., Liu L. X., Sun W. Y., Jiang Y. D., et al. (2016). Effect of genetic diversity in swine leukocyte antigen-DRA gene on piglet diarrhea. Genes 7:36. 10.3390/genes7070036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia H., Osak M., Bogu G. K., Stanton L. W., Johnson R., Lipovich L. (2010). Genome-wide computational identification and manual annotation of human long noncoding RNA genes. RNA 16, 1478–1487. 10.1261/rna.1951310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada N., Seo S. U., Chen G. Y., Núñez G. (2013). Role of the gut microbiota in immunity and inflammatory disease. Nat. Rev. Immunol. 13, 321–335. 10.1038/nri3430 [DOI] [PubMed] [Google Scholar]
- Kich J. D., Uthe J. J., Benavides M. V., Cantão M. E., Zanella R., Tuggle C. K., et al. (2014). TLR4 single nucleotide polymorphisms (SNPs) associated with Salmonella shedding in pigs. J. Appl. Genet. 55, 267–271. 10.1007/s13353-014-0199-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B. J., Lee J. H., Jun J. Y., Chang I. Y., So I., Kim K. W., et al. (2006). Vasoactive intestinal polypeptide inhibits pacemaker activity via the nitric oxide-cGMP-protein kinase G pathway in the interstitial cells of Cajal of the murine small intestine. Mol. Cells 21, 337–342. [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S. L. (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14:R36. 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L., Zhang Y., Ye Z. Q., Liu X. Q., Zhao S. Q., Wei L., et al. (2007). CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 35, W345–W349. 10.1093/nar/gkm391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawczyk M., Emerson B. M. (2014). p50-associated COX-2 extragenic RNA (PACER) activates COX-2 gene expression by occluding repressive NF-κB complexes. Elife 3:e01776. 10.7554/eLife.01776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupa A., Fol M., Dziadek B. R., Kepka E., Wojciechowska D., Brzostek A., et al. (2015). Binding of CXCL8/IL-8 to Mycobacterium tuberculosis modulates the innate immune response. Mediators Inflamm. 2015:124762. 10.1155/2015/124762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamarthée B., Malard F., Gamonet C., Bossard C., Couturier M., Renauld J. C., et al. (2015). Donor interleukin-22 and host type I interferon signaling pathway participate in intestinal graft-versus-host disease via STAT1 activation and CXCL10. Mucosal Immunol. 9, 309–321. 10.1038/mi.2015.61 [DOI] [PubMed] [Google Scholar]
- Laura M. G., Marietta F. D., Maria J. P., Ana C. C., Alberto A. G. (2014). Clostridium perfringens phospholipase C induced ROS production and cytotoxicity require PKC, MEK1 and NF kappa B activation. PLoS ONE 9:e86475 10.1371/journal.pone.0086475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Carpio D. F., Zheng Y., Bruzzo P., Singh V., Ouaaz F., et al. (2001). An essential role of the NF-kappa B/Toll-like receptor pathway in induction of inflammatory and tissue-repair gene expression by necrotic cells. J. Immunol. 166, 7128–7135. 10.4049/jimmunol.166.12.7128 [DOI] [PubMed] [Google Scholar]
- Lin M. F., Jungrei I., Kellis M. (2011). PhyloCSF:a comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics 27, i275–i282. 10.1093/bioinformatics/btr209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low L. Y., Harrison P. F., Gould J., Powell D. R., Choo J. M., Forster S. C., et al. (2018). Concurrent host-pathogen transcriptional responses in a Clostridium perfringens murine myonecrosis infection. mBio 9:e00473–e00418. 10.1128/mBio.00473-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y., Sarson A. J., Gong J., Zhou H. J., Zhu W. Y., Kang Z. M., et al. (2009). Expression profiles of genes in Toll-like receptor-mediated signaling of broilers infected with Clostridium perfringens. Clin. Vaccine Immunol. 16, 1639–1647. 10.1128/CVI.00254-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariana X. B., Erin E. O., Fabian R. C., Connor R. T., Stephanie A. C., Kristen L. L., et al. (2017). Microbiota-activated PPAR-γ-signaling inhibits dysbiotic Enterobacteriaceae expansion. Science 357, 570–575. 10.1126/science.aam9949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagahama M., Shibutani M., Seike S., Yonezaki M., Takagishi T., Masataka O., et al. (2013). The p38 MAPK and JNK pathways protect host cells against Clostridium perfringens beta-toxin. Infect. Immun. 81, 3703–3708. 10.1128/IAI.00579-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni M. J., Hu Z. H., Liu Q., Liu M. F., Lu M. H., Zhang J. S., et al. (2011). Identification and characterization of a novel non-coding RNA involved in sperm maturation. PLoS ONE 6:e26053. 10.1371/journal.pone.0026053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onnie C. M., Fisher S. A., Pattni R., Sanderson J., Forbes A., Lewis C. M., et al. (2010). Associations of allelic variants of the multidrug resistance gene (ABCB1 or MDR1) and inflammatory bowel disease and their effects on disease behavior: a case-control and meta-analysis study. Inflamm. Bowel Dis. 12, 263–271. 10.1097/01.MIB.0000209791.98866.ba [DOI] [PubMed] [Google Scholar]
- Pauli A., Valen E., Lin M. F., Garber M., Vastenhouw N. L., Levin J. Z., et al. (2012). Systematic identification of long noncoding RNAs expressed during zebrafish embryogenesis. Genome Res. 22, 577–591. 10.1101/gr.133009.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punta M., Coggill P. C., Eberhardt R. Y., Mistry J., Tate J., Boursnell C., et al. (2012). The Pfam protein families database. Nucleic Acids Res. 40, D290–D301. 10.1093/nar/gkr1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran M. L., Chen B., Li Z., Wu M. S., Liu X. C., He C. Q., et al. (2016). Systematic identification of long noncoding RNAs in immature and mature porcine testes. Biol. Reprod. 94:77. 10.1095/biolreprod.115.136911 [DOI] [PubMed] [Google Scholar]
- Scharff R. L. (2012). Economic burden from health losses due to foodborne illness in the United States. J. Food Protect. 75, 123–131. 10.4315/0362-028X.JFP-11-058 [DOI] [PubMed] [Google Scholar]
- Senhaji N., Kassogue Y., Fahimi M., Serbati N., Badre W., Nadifi S. (2015). Genetic polymorphisms of multidrug resistance gene-1 (MDR1/ABCB1) and glutathione S-transferase gene and the risk of inflammatory bowel disease among moroccan patients. Mediators Inflamm. 2015:248060. 10.1155/2015/248060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen T., Yang Z. B., Cheng X. S., Xiao Y. C., Yu K., Cai X. Y., et al. (2017). CXCL8 induces epithelial-mesenchymal transition in colon cancer cells via the PI3K/Akt/NF-κB signaling pathway. Oncol. Rep. 37, 2095–2100. 10.3892/or.2017.5453 [DOI] [PubMed] [Google Scholar]
- Shen Y. F., Mao H. G., Huang M. J., Chen L. X., Chen J. C., Cai Z. W., et al. (2016). Long noncoding RNA and mRNA expression profiles in the thyroid gland of two phenotypically extreme pig breeds using Ribo-zero RNA sequencing. Genes 7:34. 10.3390/genes7070034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A., Bejerano G., Pedersen J. S., Hinrichs A. S., Hou M., Rosenbloom K., et al. (2005). Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 15, 1034–1050. 10.1101/gr.3715005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Songer J. G., Pfeffer M., Truyen U., Gaastra W. (2010). Clostridia as agents of zoonotic disease. Vet. Microbiol. 140, 399–404. 10.1016/j.vetmic.2009.07.003 [DOI] [PubMed] [Google Scholar]
- Sun L., Luo H., Bu D., Zhao G., Yu K., Zhang C., et al. (2013). Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res. 41:e166. 10.1093/nar/gkt646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao F. F., Qian C., Guo W. J., Luo Q., Xu Q., Sun Y. (2013). Inhibition of Th1/Th17 responses via suppression of STAT1 and STAT3 activation contributes to the amelioration of murine experimental colitis by a natural flavonoid glucoside icariin. Biochem. Pharmacol. 85, 798–807. 10.1016/j.bcp.2012.12.002 [DOI] [PubMed] [Google Scholar]
- Trapnell C., Williams B. A., Pertea G., Mortazavi A., Kwan G., Marijke J., et al. (2010). Transcript assembly and abundance estimation from RNA-Seq reveals thousands of new transcripts and switching among isoforms. Nat. Biotechnol. 28, 511–515. 10.1038/nbt.1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacca I. (2017). The microbiota maintains oxygen balance in the gut. Nat. Rev. Microbiol. 15, 574–575. 10.1038/nrmicro.2017.112 [DOI] [PubMed] [Google Scholar]
- Wang Y., Xue S., Liu X., Liu H., Hu T., Qiu X., et al. (2016). Analyses of long non-coding RNA and mRNA profiling using RNA sequencing during the pre-implantation phases in pig endometrium. Sci. Rep. 6:20238. 10.1038/srep20238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann A. J., Forstner K. U., Amman F., Barquist L., Chao Y. J., Schulte L. N., et al. (2016). Dual RNA-seq unveils noncoding RNA functions in host-pathogen interactions. Nature 529:496. 10.1038/nature16547 [DOI] [PubMed] [Google Scholar]
- Wu H., Liu J., Li W., Liu G., Li Z. (2016). LncRNA-HOTAIR promotes TNF-α production in cardiomyocytes of LPS-induced sepsis mice by activating NF-κB pathway. Biochem. Biophys. Res. Commun. 471, 240–246. 10.1016/j.bbrc.2016.01.117 [DOI] [PubMed] [Google Scholar]
- Wu Z. C. (2018). Screening and Regulatory Mechanism Analysis of Genes and Long Noncoding RNA (lncRNA) Related to E. coli F18 Resistance in Weaned Piglets. Dissertation/Doctor's thesis. Yangzhou: Yangzhou University. [Google Scholar]
- Xiao Y. C., Yang Z. B., Cheng X. S., Fang X. B., Shen T., Xia C. F., et al. (2015). CXCL8, overexpressed in colorectal cancer, enhances the resistance of colorectal cancer cells to anoikis. Cancer Lett. 361, 22–32. 10.1016/j.canlet.2015.02.021 [DOI] [PubMed] [Google Scholar]
- Xie C., Mao X., Huang J., Ding Y., Wu J., Dong S., et al. (2011). KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 39, W316–W322. 10.1093/nar/gkr483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q. L., Kong J. J., Wang D. W., Zhao S. G., Gun S. B. (2013). Swine leukocyte antigen-DQA gene variation and its association with piglet diarrhea in large white, landrace and duroc. Asian-Australas J. Anim. Sci. 26, 1065–1071. 10.5713/ajas.2013.13067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa T., Wu J., Otsuka M., Kishikawa T., Suzuki N., Takata A., et al. (2017). Repression of microRNA function mediates inflammation-associated colon tumorigenesis. Gastroenterology 152, 631–643. 10.1053/j.gastro.2016.10.043 [DOI] [PubMed] [Google Scholar]
- Zanella R., Settles M. L., Mckay S. D., Schnabel R., Taylor J., Whitlock R. H., et al. (2011). Identification of loci associated with tolerance to Johne's disease in holstein cattle. Anim. Genet. 42, 28–38. 10.1111/j.1365-2052.2010.02076.x [DOI] [PubMed] [Google Scholar]
- Zhao N. (2016). LncRNA Expression Signatures in Response to Enterotoxigenic Escherichia coli Infection in Mice. Dissertation/Master's thesis. Yinchuan: University of Ningxia. [Google Scholar]
- Zhao W., Mu Y., Ma L., Wang C., Tang Z., Yang S., et al. (2015). Systematic identification and characterization of long intergenic non-coding RNAs in fetal porcine skeletal muscle development. Sci. Rep. 5:8957. 10.1038/srep08957 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Screening principles of association analysis between dysregulated lncRNAs and mRNAs.
Identification pipeline for lncRNAs. Each step is described in detail in the methods section.
One thousand eight hundred and fifty non-coding lncRNAs were selected by Cufflinks and Scripture.
Primers and location of lncRNAs and mRNAs used in qPCR validation.
Summary of RNA-seq data and reads mapped to the Sus scrofa reference genome.
Catalog of lncRNAs and mRNAs identified in the 15 cDNA libraries of piglets infected by C. perfringens type C.
Characteristics of lncRNAs in piglet ileums.
List of differentially expressed lncRNAs and mRNAs detected in piglet ileums using the RNA-seq.
Target genes prediction of significantly differentially expressed lncRNAs in cis and trans.
GO enrichment analysis of the significantly dysregulated lncRNAs target genes and mRNAs among IR, IS, and IC groups.
KEGG enrichment analysis of the significantly dysregulated lncRNAs target genes and mRNAs among IR, IS, and IC groups.
Relationship between dysregulated lncRNAs and mRNAs in cis and trans involved in CPID.